Abstract
Background
Obesity is a major risk factor for many chronic diseases, including reduced lung function. The role of polymorphisms of the adiponectin gene, though linked with cardiometabolic consequences of obesity, has not been studied in relation to lung function.
Objectives
The aim of this study is to examine polymorphisms in the ADIPOQ, ADIPOR1, and ADIPOR2 genes in relation to adiponectin serum levels, BMI, and adiposity in 18-year old Cypriot males, as well as determine whether BMI, adipokines levels and polymorphisms in adipokine related genes are associated with lung function levels.
Results
From the participants, 8% were classified as obese, 22% as overweight, and the remaining 71% as normal. We found that rs266729 and rs1501299 in ADIPOQ and rs10920531 in ADIPOR1 were significantly associated with serum adiponectin levels, after adjusting for ever smoking. In addition, there was an overall significant increase in FEV1% predicted with increasing BMI (β = 0.53, 95% CI: 0.27, 0.78) and in FVC % predicted (β = 1.02, 95% CI: 0.73, 1.30). There was also a decrease in FEV1/FVC with increasing BMI (β = -0.53, 95% CI: -0.71, -0.35). Finally, rs1501299 was associated with lung function measures.
Discussion
Functional variants in the ADIPOQ gene were linked with lung function in young males. Further studies should concentrate on the role of adipokines on lung function which may direct novel therapeutic approaches.
Introduction
Obesity is a major public health concern and worldwide 150 million adults and 15 million children are obese [1]. Although in the past obesity was considered a problem only for high income countries, overweight and obesity are nowadays increasing dramatically in low- and middle-income countries as well, particularly in urban settings [2]. In Cyprus, a small European island in the Eastern Mediterranean region, it has reached epidemic proportions with data from a cross-sectional study suggesting that 36% and 28% of the adult population are overweight and obese, respectively [3].
Obesity is a disorder which is a major risk factor for many other chronic diseases, including type 2 diabetes, cardiovascular disease, and cancer, as well as for lung diseases and reduced lung function [4–7]. Different studies have shown associations between increased Body Mass Index (BMI) and asthma [8], and evidence suggests that obese individuals aged less than 18 years old show a reduction in lung volume and capacity as compared to healthy individuals [9]. Obesity causes deleterious effects on lung volume and capacity in children and adolescents, mainly by reducing functional residual capacity, expiratory reserve volume, and residual volume [10]. However, initial increases in weight increase lung function among the non-obese. These increases reflect an increase in muscle force. Afterwards, further increases in body mass represent obesity, and as a consequence lung function, decreases [11]. Additionally, among young males’ the lungs grow up to the age of 21, justifying the fact that an increase in BMI and thus lung function is usually associated with the natural growth of lungs rather than obesity.
Adipose tissue produces adipokines which are proteins that regulate inflammation and metabolism in an autocrine, paracrine, and systemic manner [12,13]. Adiponectin in particular is an adipokine, which is associated with systemic anti-inflammatory effects, and serum adiponectin concentrations are known to be reduced in obese and asthmatic individuals [14]. Adiponectin and its receptors are expressed on multiple cell types in the lung and a recent study suggested that lower serum adiponectin concentrations may be associated with lower lung function in young adults, independent of obesity [15].
Adiponectin cellular signaling is mediated by two adiponectin receptors, namely ADIPOR1 (located on chromosome 1q32) and ADIPOR2 (located on chromosome 12p13.33) [16]. Adiponectin levels are in part determined by genetic influences, with a recent study reporting heritability estimates of 51% [17], and they are encoded by the gene ADIPOQ, located on chromosome 3q27. As adiponectin is inversely associated with BMI and other measures of adiposity [13] the adiponectin related genes are considered important candidate genes for being related with obesity phenotypes, and thus could help in unraveling the genetic underpinnings of obesity. Although in various studies the polymorphisms of the ligand adiponectin gene, ADIPOQ, have been linked with cardiometabolic consequences of obesity, their role in lung function has not been studied yet.
The primary goal of this study was to examine the polymorphisms in ADIPOQ, ADIPOR1, and ADIPOR2 in relation to adiponectin serum levels and adiposity in an 18-year old male population of Cyprus. Our secondary aim was to examine whether lung function levels in young males vary in association with BMI, adipokines’ levels, and polymorphisms in adipokine related genes.
Methods
Study participants
The Cyprus Metabolism Study included 1,056 healthy 18-year old male individuals, recruited on a voluntary basis during the July 2006 and July 2007 enrolment cycles of the Cyprus Army. This sample included participants from all Cypriot regions and socioeconomic levels, recruited while performing their two-year mandatory service in the Cyprus Army, and it is considered representative of the 18-year old male population of Cyprus. Exclusion criteria included health reasons for which a person was found unable to participate in the military service, including most chronic diseases. The study had approval by the Cyprus Bioethics Committee and the Institutional Review Board (IRB) of the Harvard T.H. Chan School of Public Health; all participants were informed about the study before enrolling and signed an informed consent.
Clinical and other measurements
Participants underwent detailed clinical investigations. In brief, anthropometric parameters, including height, weight, waist circumference, hip circumference, skinfolds, and body fat were measured in all subjects according to standard protocols by trained personnel. BMI was calculated as weight in kilograms divided by the square of height in meters. Waist-to-height ratio was calculated as waist circumference in centimeters divided by height in centimeters. Information on ever smoking was collected using standard questionnaires together with demographic information, medical history, physical activity, and other. Fasting blood samples were drawn to assay the levels of serum adiponectin (μg/ml) using a standard immunoassay protocol. Spirometry was performed in 416 participants only and lung capacity was evaluated using a hand-held Micro-Spirometer Vitalograph 2120 in accordance to ATS/NIOSH guidelines. Participants were first instructed by trained personnel on how to use the spirometer and then three attempts were made keeping the best one.
Selection and genotyping of SNPs
Twenty SNPs including 5 ADIPOQ SNPs (rs266729, rs822395, rs822396, rs1501299, and rs2241766), 5 ADIPOR1 SNPs (rs2232853, rs12733285, rs1342387, rs7539542 and rs10920531), and 10 ADIPOR2 SNPs (rs1029629, rs7975600, rs11612383, rs1058322, rs11061973, rs2108642, rs767870, rs12342, rs1044471, and rs7294540) were selected a priori. These SNPs were chosen based on the following criteria: (1) they capture variations in the major blocks in each gene, (2) they are functionally relevant and have been studied by others in relation to adiponectin levels or diseases, such as diabetes, metabolic and insulin resistance syndromes [16–20], and (3) they have a minor allele frequency greater than 10% in European individuals in the HapMap database. SNPs were genotyped with the Taqman/MALDI-TOF SNP allelic discrimination using a standard protocol. A total of 9.4% of samples were genotyped in duplicate and showed 98.8% concordance. The overall genotyping success rate was 99.0%.
Statistical analysis
Continuous characteristics are presented as mean ± SD and compared among groups using the one-way analysis of variance technique, whereas categorical variables are presented as frequency (%) and compared among groups using the chi-square test of independence. BMI was used to classify individuals into obese (BMI ≥ 30 kg/m2), overweight (25 kg/m2 ≤ BMI < 30 kg/m2) and normal/underweight (reference weight) (BMI<25 kg/m2) categories. Linear regression models were used to assess the effect of genetic variation in the adiponectin-related genes on adiponectin levels and BMI after adjusting for smoking as well as with lung function. Results are reported as beta coefficients with the corresponding 95% confidence interval (CI). Similarly, logistic regression models were utilized to estimate the impact of genetic variations in the adiponectin-related genes on the probability of being overweight or obese, after adjusting for smoking, and results are reported as OR (95% CI). Dominant models were used which grouped the heterozygous with the homozygous for the minor allele. The referent genotype was selected to be the most common homozygous genotype. Haploview was used to determine the linkage disequilibrium (LD) between SNPs and departure from Hardy-Weinberg equilibrium was assessed using an exact test. In addition, linear regression models were used to assess the association of BMI and adiponectin serum levels with lung function after adjusting for smoking. Statistical analysis was performed using SAS software Version 9.3 (SAS Inc., Cary, NC, USA) and all tests performed were two-sided with P<0.05 indicating statistical significance. Bonferroni correction method was not used because in this hypothesis driven study selection of included SNPs of candidate genes was based on known functionality and previous findings of associations of SNPs with adiponectin levels or metabolic diseases.
Results
Out of the 1,056 participants in the study 1,043 had available BMI information. Of them, 7.8% (n = 81) were classified as obese, 21.7% (n = 226) as overweight, and the remaining 70.6% (n = 736) as normal, which was the reference group (Table 1). The mean age of the participants was 18.4 ± 0.6 years and 40.8% of them were smokers. Obese participants had worse anthropometric measures than overweight and reference weight individuals and they were more likely to be smokers (p = 0.02). Furthermore, obese participants had similar adiponectin levels as overweight individuals but differed significantly from the reference weight individuals (p <0.01).
Table 1. Characteristics of participants by obesity group.
| All | Obese | Overweight | Normal | P-value | |
|---|---|---|---|---|---|
| N | 1043 | 81 | 226 | 736 | |
| Age (years) | 18.4 ± 0.6 | 18.4 ± 0.4 | 18.4 ± 0.6 | 18.4 ± 0.6 | 0.78 |
| BMI (Kg/m2) | 23.5 ± 4.1 | 33.2 ± 2.7 | 26.9 ± 1.4 | 21.4 ± 2.1 | <0.01 |
| Waist circumference (cm) | 81.7 ± 10.7 | 105.4 ± 7.4 | 90.1 ± 6.1 | 76.6 ± 5.9 | <0.01 |
| Waist-to-height ratio | 0.47 ± 0.06 | 0.60 ± 0.04 | 0.51 ± 0.03 | 0.44 ± 0.03 | <0.01 |
| Sums of skinfolds (cm) | 50.9 ± 24.2 | 105.0 ± 21.6 | 67.7 ± 18.6 | 40.0 ± 12.6 | <0.01 |
| Fat percentage | 12.9 ± 6.7 | 27.1 ± 4.9 | 18.0 ± 4.0 | 9.7 ± 3.8 | <0.01 |
| Smokers—n (%) | 419 (40.8) | 45 (55.6) | 90 (40.7) | 284 (39.2) | 0.02 |
| Adiponectin (μg/mL) | 7.9 ± 3.8 | 6.8 ± 2.8 | 6.9 ± 3.6 | 8.3 ± 3.9 | <0.01 |
The data for continuous variables is mean ± SD. The data for categorical variable is shown as n (%).
The location, genotypic frequencies, and the major and minor alleles of the SNPs selected for the three adiponectin-related genes are described in S1 Table while the results for the linkage disequilibrium (LD) of SNPs are shown in S1 Fig. The genotypes of all the SNPs were in Hardy-Weinberg equilibrium in each group (p>0.05). In addition, the minor allele frequencies of the SNPs were virtually identical to those reported in the HapMap database for Central European (CEU) population.
Associations with BMI and adiposity
SNPs rs266729 and rs1501299 in ADIPOQ and rs10920531 in ADIPOR1 were significantly associated with serum adiponectin levels, after adjusting for smoking (β and (95% CI): -0.58 (-1.09, -0.07), 0.60 (0.10, 1.10), and 0.57 (0.05, 1.09), with p-values 0.03, 0.02, and 0.03, respectively (Table 2)). These associations were not altered when BMI was added as a covariate to the corresponding models (S2 Table). In addition, there were no significant associations of SNP with BMI (Table 2) or with the probability of being overweight or obese (Table 3), though rs266729 in ADIPOQ and rs7975600 in ADIPOR2 had a borderline significant association with the probability of being overweight (OR = 1.31, 95% CI: 0.95, 1.79, p = 0.10 and OR = 0.74, 95% CI: 0.51, 1.06, p = 0.10, respectively) and rs2232853 with the probability of being obese (OR = 1.61, 95% CI: 0.99, 2.61, p = 0.05). We further investigated whether there was an association between the SNPs and other anthropometric indices, such as waist circumference, waist-to-height ratio, fat percentage, and sum of skinfolds. Our results showed that none of the SNPs included was significantly associated with any of these anthropometric indices (S3 Table).
Table 2. Associations of genetic variants with adiponectin levels and Body Mass Index.
| Adiponectin (μg/mL) | BMI (kg/m2) | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Chr. | Gene | M/m | β (95% CI) | P-value | β (95% CI) | P-value |
| rs266729 | 3 | ADIPOQ | C/G | -0.58 (-1.09, -0.07) | 0.03 | 0.27 (-0.26, 0.81) | 0.32 |
| rs822395 | 3 | ADIPOQ | A/C | 0.26 (-0.24, 0.77) | 0.31 | 0.25 (-0.27, 0.78) | 0.34 |
| rs822396 | 3 | ADIPOQ | A/G | -0.12 (-0.68, 0.43) | 0.66 | 0.30 (-0.28, 0.88) | 0.31 |
| rs2241766 | 3 | ADIPOQ | T/G | -0.05 (-0.59, 0.48) | 0.84 | 0.06 (-0.50, 0.62) | 0.84 |
| rs1501299 | 3 | ADIPOQ | G/T | 0.60 (0.10, 1.10) | 0.02 | -0.20 (-0.72, 0.33) | 0.46 |
| rs2232853 | 1 | ADIPOR1 | G/A | 0.16 (-0.36, 0.67) | 0.55 | 0.40 (-0.13, 0.94) | 0.14 |
| rs12733285 | 1 | ADIPOR1 | C/T | -0.49 (-1.03, 0.05) | 0.08 | -0.14 (-0.70, 0.43) | 0.63 |
| rs1342387 | 1 | ADIPOR1 | T/C | 0.15 (-0.39, 0.69) | 0.59 | 0.27 (-0.30, 0.84) | 0.36 |
| rs7539542 | 1 | ADIPOR1 | C/G | 0.28 (-0.23, 0.78) | 0.29 | -0.04 (-0.58, 0.49) | 0.87 |
| rs10920531 | 1 | ADIPOR1 | C/A | 0.57 (0.05, 1.09) | 0.03 | 0.01 (-0.54, 0.56) | 0.97 |
| rs1029629 | 12 | ADIPOR2 | T/G | 0.01 (-0.50, 0.52) | 0.98 | -0.20 (-0.73, 0.34) | 0.47 |
| rs7975600 | 12 | ADIPOR2 | A/T | -0.12 (-0.68, 0.44) | 0.68 | -0.22 (-0.81, 0.36) | 0.45 |
| rs11612383 | 12 | ADIPOR2 | G/A | -0.06 (-0.56, 0.44) | 0.81 | 0.09 (-0.43, 0.61) | 0.74 |
| rs1058322 | 12 | ADIPOR2 | C/T | -0.05 (-0.55, 0.45) | 0.83 | -0.20 (-0.72, 0.33) | 0.46 |
| rs11061973 | 12 | ADIPOR2 | G/A | 0.27 (-0.26, 0.81) | 0.31 | 0.18 (-0.38, 0.74) | 0.53 |
| rs2108642 | 12 | ADIPOR2 | C/A | -0.22 (-0.79, 0.35) | 0.45 | -0.33 (-0.93, 0.27) | 0.28 |
| rs767870 | 12 | ADIPOR2 | A/G | -0.19 (-0.73, 0.36) | 0.50 | -0.14 (-0.71, 0.43) | 0.63 |
| rs12342 | 12 | ADIPOR2 | C/T | 0.01 (-0.50, 0.52) | 0.97 | -0.03 (-0.56, 0.50) | 0.90 |
| rs1044471 | 12 | ADIPOR2 | C/T | 0.17 (-0.40, 0.74) | 0.56 | 0.16 (-0.44, 0.75) | 0.60 |
| rs7294540 | 12 | ADIPOR2 | C/A | 0.14 (-0.38, 0.67) | 0.59 | -0.21 (-0.76, 0.34) | 0.46 |
Chr., chromosome. SNP: single nucleotide polymorphisms. M/m indicates major/minor allele.
All models are adjusted for smoking.
Table 3. Associations of genetic variants with probability of being overweight or obese.
| Overweight | Obese | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chr. | Position | Gene | M/m | OR (95% CI) | P-value | OR (95% CI) | P-value |
| rs266729 | 3 | 186559474 | ADIPOQ | C/G | 1.31 (0.95, 1.79) | 0.10 | 1.15 (0.71, 1.88) | 0.57 |
| rs822395 | 3 | 186566807 | ADIPOQ | A/C | 0.97 (0.71, 1.33) | 0.87 | 0.98 (0.60, 1.59) | 0.93 |
| rs822396 | 3 | 186566877 | ADIPOQ | A/G | 1.17 (0.83, 1.65) | 0.36 | 0.97 (0.57, 1.67) | 0.93 |
| rs2241766 | 3 | 186570892 | ADIPOQ | T/G | 1.22 (0.88, 1.69) | 0.23 | 1.08 (0.66, 1.83) | 0.72 |
| rs1501299 | 3 | 186571123 | ADIPOQ | G/T | 0.78 (0.57, 1.07) | 0.13 | 0.84 (0.52, 1.38) | 0.50 |
| rs2232853 | 1 | 202931958 | ADIPOR1 | G/A | 1.24 (0.90, 1.70) | 0.19 | 1.61 (0.99, 2.61) | 0.05 |
| rs12733285 | 1 | 202922040 | ADIPOR1 | C/T | 0.80 (0.58, 1.11) | 0.19 | 0.91 (0.54, 1.53) | 0.73 |
| rs1342387 | 1 | 202914356 | ADIPOR1 | T/C | 1.32 (0.93, 1.88) | 0.12 | 1.03 (0.61, 1.74) | 0.90 |
| rs7539542 | 1 | 202909974 | ADIPOR1 | C/G | 0.87 (0.64, 1.20) | 0.40 | 1.14 (0.69, 1.87) | 0.61 |
| rs10920531 | 1 | 202908836 | ADIPOR1 | C/A | 0.89 (0.64, 1.23) | 0.48 | 1.11 (0.67, 1.86) | 0.68 |
| rs1029629 | 12 | 1799267 | ADIPOR2 | T/G | 0.84 (0.61, 1.15) | 0.28 | 0.86 (0.53, 1.41) | 0.56 |
| rs7975600 | 12 | 1815252 | ADIPOR2 | A/T | 0.74 (0.51, 1.06) | 0.10 | 0.90 (0.52, 1.55) | 0.70 |
| rs11612383 | 12 | 1831355 | ADIPOR2 | G/A | 1.01 (0.74, 1.38) | 0.95 | 1.10 (0.68, 1.78) | 0.70 |
| rs1058322 | 12 | 1836979 | ADIPOR2 | C/T | 0.88 (0.64, 1.20) | 0.41 | 0.78 (0.48, 1.26) | 0.31 |
| rs11061973 | 12 | 1865936 | ADIPOR2 | G/A | 1.20 (0.86, 1.67) | 0.28 | 1.21 (0.73, 2.01) | 0.46 |
| rs2108642 | 12 | 1866799 | ADIPOR2 | C/A | 0.79 (0.56, 1.12) | 0.18 | 0.82 (0.48, 1.40) | 0.46 |
| rs767870 | 12 | 1889823 | ADIPOR2 | A/G | 0.79 (0.56, 1.12) | 0.18 | 0.77 (0.45, 1.33) | 0.35 |
| rs12342 | 12 | 1896880 | ADIPOR2 | C/T | 0.92 (0.67, 1.26) | 0.60 | 1.10 (0.67, 1.80) | 0.70 |
| rs1044471 | 12 | 1896956 | ADIPOR2 | C/T | 1.33 (0.92, 1.93) | 0.13 | 1.00 (0.58, 1.73) | 0.99 |
| rs7294540 | 12 | 1899714 | ADIPOR2 | C/A | 0.84 (0.60, 1.16) | 0.28 | 0.81 (0.49, 1.33) | 0.40 |
Models are adjusted for smoking status.
Chr., chromosome. Position SNP: single nucleotide polymorphisms. M/m indicates major/minor allele.
Associations with lung function level
When considering lung function (Table 4) we observed an overall significant increase in FEV1% predicted with increasing BMI (β = 0.53, 95% CI: 0.27, 0.78), in FVC % predicted (β = 1.02, 95% CI: 0.73, 1.30), and a decrease in FEV1/FVC with increasing BMI (β = -0.53, 95% CI: -0.71, -0.35). The serum levels of adiponectin were not associated with FEV1% predicted (β = 0.07, 95% CI: -0.20, 0.34), nor with FVC % predicted (β = -0.15, 95% CI: -0.46, 0.16), though adiponectin levels were significantly associated with FEV1/FVC (β = 0.21, 95% CI: 0.02, 0.40).
Table 4. Association of BMI and adiponectin with lung function.
| FEV1% predicted | FVC % predicted | FEV1/FVC | ||||
|---|---|---|---|---|---|---|
| β-coefficient | 95% CI | β-coefficient | 95% CI | β-coefficient | 95% CI | |
| BMI (kg/m2)* | 0.53 | 0.27, 0.78 | 1.02 | 0.73, 1.30 | -0.53 | -0.71, -0.35 |
| Adiponectin(ng/ml)** | 0.07 | -0.20, 0.34 | -0.15 | -0.46, 0.16 | 0.21 | 0.02, 0.40 |
BMI: Body Mass Index, FEV1: Forced expiratory volume at 1s, FVC: Forced expiratory volume
*adjusted for smoking
**adjusted for BMI and smoking
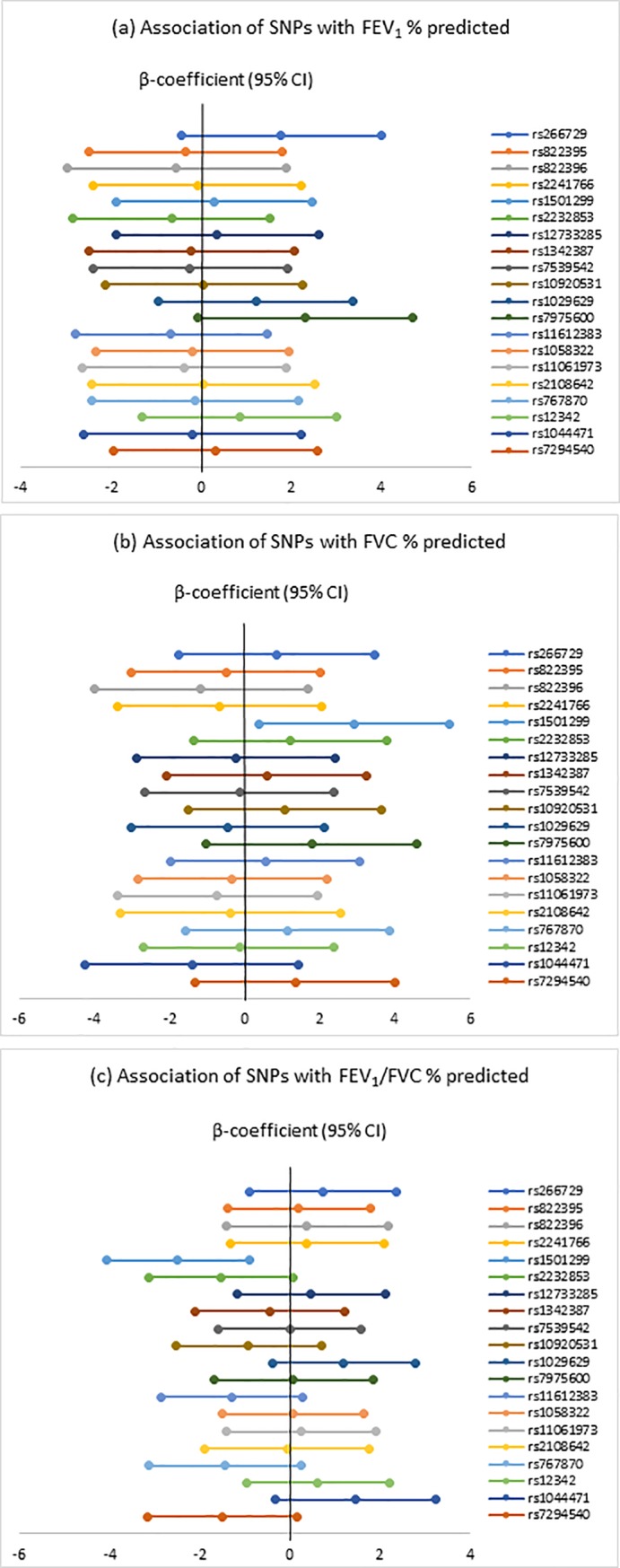
Fig 1 and S4 Table summarize the associations of SNPs in ADIPOQ, ADIPOR1, ADIPOR2 with FEV1 and FVC % predicted, and FEV1/FVC. There was a borderline statistically significant association of rs7975600 in ADIPOR2 with FEV1% predicted (β = 2.31, 95% CI: -0.07, 4.70, p = 0.06) and of rs2232853 in ADIPOR1 with FEV1/FVC (β = -1.52, 95% CI: -3.12, 0.08, p = 0.06). In addition, rs1501299 in ADIPOQ was associated with FVC % predicted (β = 2.91, 95% CI: 0.38, 5.44, p = 0.02) and FEV1/FVC (β = -2.48, 95% CI: -4.05, -0.90, p<0.01), but not with FEV1% predicted (β = 0.30, 95% CI: -1.87, 2.46, p = 0.79).
Fig 1. Association of genetic variants with FEV1% predicted, FVC % predicted and FEV1/FVC % predicted.
Discussion
The study examined the variation in three adiponectin-related genes in relation to adiponectin levels, BMI, lung function, and the risk of overweight and obesity in a sample of 18-year-old Cypriot men. We found a significant association between adiponectin levels and three SNPs in this population. The upstream ADIPOQ rs266729 was significantly associated with lower adiponectin while rs1501299 was associated with higher levels of adiponectin. Both the direction and associations of these two SNPs recapitulate that of previous candidate gene studies that evaluated these associations with adiponectin concentrations [21,22].
The SNPs considered to be functional in the present study were not significantly associated with BMI, however, rs266729 in ADIPOQ had a borderline association with the probability of being overweight, but not with the probability of being obese. This is in agreement with the findings from other studies, including the Framingham Offspring Study [23], the Heritage Family Study [24], the Oulu Diabetic Study [25], the Quebec Family Study of French-Canadians[26] and the more recent study by Cohen and colleagues [27]. Cohen et al. investigated 19 SNPs in ADIPOR1 (including rs12733285, rs1342387, rs7539542, and rs10920531 genotyped in our study) and 27 SNPs in ADIPOR2 (including rs12342, rs1044471, and rs7294540 also genotyped in our study) in relation to adiponectin levels and BMI and found no statistically significant associations [27]; we also observed no association of these SNPs with BMI but we obtained an association with adiponectin levels for rs10920531 in ADIPOR1. We noted that in the Cohen et al. study the relation between adiponectin levels and rs10920531 approached significance among black women but not among white women. Similarly the GEMS Study and the Diabetes Prevention Program Study found no genome-wide significant associations between adiponectin levels and SNPs in ADIPOR1 in a GWAS of Europeans [28] and that of mixed race/ethnic groups of White, African American, Hispanic, Asian/Pacific Islanders, and American Indian ancestry [29], respectively. We speculate that one plausible reason for the discrepancy seen in the case of this SNP could be due to differences between race and gender. Interestingly, recent genome-wide studies have identified variants in a third adiponectin receptor, T-cadherin encoding CHD [13] that also affect circulating adiponectin levels, but unfortunately this information was not available in our study [28,30]. Similarly, while some studies have reported positive associations between BMI and some ADIPOQ polymorphisms [31–34] these were on other SNPs that were not available in our study.
We also had the opportunity to examine the association of BMI, adiponectin levels, and genetic variation with lung function measurements. The findings suggest that FEV1 and FVC % predicted in this age group increase with increasing BMI, while FEV1/FVC tends to decrease as BMI increases. Even though findings in adult populations suggest that lung function decreases with increasing BMI [8], our results are supported by studies in children aged 8 to 17 years old, showing that larger BMI and larger waist circumference are associated with higher FEV1 and FVC in both genders in children aged 8 to 17 [35] as their lungs are still growing. It is likely that the larger effect of BMI on FVC % predicted compared to FEV1% predicted results in lower FEV1/FVC % predicted in these children, a finding which is in line with previous studies [36,37].
Notably, rs266729 which was found to be associated with decreased adiponectin levels in our study, increases FEV1% predicted, while rs1501299 which is linked to increased adiponectin it is also associated with airflow obstruction (decreased FEV1/FVC). These findings are consistent with Thyagarajan et al. positively linking adiponectin levels with lung function[38]. However, the null association with BMI suggests that ADIPOQ and adiponectin levels may play a role in lung biologic and lung function level in young male individuals, independent of obesity.
Our study has several limitations as well as strengths. First, we analyzed data only from a group of 18-year old men and as gender is an important determinant of adiponectin levels the results from this study cannot be easily generalized to women or other ethnic or age groups. Furthermore, it was difficult to find a similar population (young 18 years old, males only, with genetic data, lung function and serum adipokines available) to run a replication study. In addition, the study analyzed total serum adiponectin levels measured at a single point in time which may not fully characterize long-term adiponectin exposures and effects; however, studies have shown adiponectin levels to be very reproducible, with adiponectin concentrations when measured repeatedly, on average one year apart, being highly correlated [39]. Another limitation is the possibility of false positive findings arising from the multiple comparisons made; however, the SNPs analyzed were those earlier known to be functionally relevant from a pool of SNPs tagging the two major haplotype blocks within each gene among Europeans. The few SNPs that were significantly associated with adiponectin levels in this study recapitulated results of earlier studies, thereby validating this approach. Moreover, this study was of limited power to detect modest differences between genotypes. The wide CIs in this study were due largely to the small sample size especially in the pairwise comparisons of BMI groups where the small number of people particularly in the obesity group was a limiting factor. Finally, the SNPs analyzed in the present study were not powered to detect rare variants; it could be that other genes exist, working in concert with adiponectin-related genes, which would need to be included to observe an effect and such would require a much larger sample size. A system genetics analysis of the molecular mechanisms underlying variations in lung function has revealed that SNPs associated with lung function were enriched for lung expression quantitative trait loci (eQTLs) [40]. In such study, a large number of SNPs that determine the variation in lung function measures were also enriched in developmental and inflammatory pathways. Interestingly, our study has identified SNPs within the ADIPOQ gene underlying variation in lung function independent of BMI in young Cypriot males, representing testable hypothesis for future GWAS and more sophisticated post-GWAS analyses including pathway and gene set enrichment analysis and ultimately, causality investigation using Mendelian randomization to estimate the influence of blood adiponectin on lung function and lung function risk factors in the Cypriot population.
One could suggest that the lack of multiple testing correction is responsible for the current results. We decided not to apply a sequential (classical) Bonferroni correction for a number of reasons. First, our choice for the current study was explicitly driven by previous observations suggesting adiponectin levels are associated with lung function. Secondly, the independent variables in our analyses (e.g. BMI and lung function) are related, indicating that a rigid statistical procedure, such as Bonferroni correction for multiple testing, would not do justice to their biologically linked nature. Finally, although adjustment for multiple testing will decrease the chance of a type I error, it will also increase the chance of a type II error, so that a true association is not found. This is especially possible in a relatively small study such as ours. Thus, we followed the advice given by Perneger [41]: “Simply describing what was done and why, and discussing the possible interpretations of each result, should enable the reader to reach a reasonable conclusion without the help of Bonferroni adjustments”.
In conclusion, the present findings confirmed and further strengthened the current knowledge regarding the role of adiponectin on lung function. This is the first study to link functional variants in ADIPOQ gene with lung function in young males and confirmed that adiponectin levels influence lung function independent of obesity. Further studies should concentrate on the role of adipokines on lung function which may direct novel therapeutic approaches.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(SAS7BDAT)
Data Availability
We have already requested permission from the Cyprus Bioethics Committee in order to release the de-identified data; we do not expect this to be a problem and we will forward the data as soon as we have the official approval.
Funding Statement
The study was funded by the Cyprus Research Promotion Foundation and the Harvard Cyprus Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Home. May 2018. http://www.euro.who.int/en/home. Accessed May 8, 2018.
- 2.Seidell JC, Halberstadt J. The global burden of obesity and the challenges of prevention. Ann Nutr Metab. 2015;66 Suppl 2(Suppl. 2):7–12. 10.1159/000375143 [DOI] [PubMed] [Google Scholar]
- 3.Andreou E, Hajigeorgiou P, Kyriakou K, et al. Risk factors of obesity in a cohort of 1001 Cypriot adults: An epidemiological study. Hippokratia. 2012;16(3):256–260. http://www.ncbi.nlm.nih.gov/pubmed/23935294. Accessed November 29, 2016. [PMC free article] [PubMed] [Google Scholar]
- 4.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444(7121):875–880. 10.1038/nature05487 [DOI] [PubMed] [Google Scholar]
- 5.McCarthy MI. Genomics, Type 2 Diabetes, and Obesity. Feero WG, Guttmacher AE, eds. N Engl J Med. 2010;363(24):2339–2350. 10.1056/NEJMra0906948 [DOI] [PubMed] [Google Scholar]
- 6.Adams KF, Schatzkin A, Harris TB, et al. Overweight, Obesity, and Mortality in a Large Prospective Cohort of Persons 50 to 71 Years Old. N Engl J Med. 2006;355(8):763–778. 10.1056/NEJMoa055643 [DOI] [PubMed] [Google Scholar]
- 7.Rizvi AA. Hypertension, Obesity, and Inflammation: The Complex Designs of a Deadly Trio. Metab Syndr Relat Disord. 2010;8(4):287–294. 10.1089/met.2009.0116 [DOI] [PubMed] [Google Scholar]
- 8.Liu P-C, Kieckhefer GM, Gau B-S. A systematic review of the association between obesity and asthma in children. J Adv Nurs. 2013;69(7):1446–1465. 10.1111/jan.12129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melo LC, Silva MAM da, Calles AC do N. Obesity and lung function: a systematic review. Einstein (Sao Paulo). 12(1):120–125. http://www.ncbi.nlm.nih.gov/pubmed/24728258. Accessed January 29, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winck AD, Heinzmann‐Filho JP, Soares RB, da Silva JS, Woszezenki CT, Zanatta LB. Efeitos da obesidade sobre os volumes e as capacidades pulmonares em crianças e adolescentes: uma revisão sistemática. Rev Paul Pediatr. 2016;34(4):510–517. 10.1016/j.rpped.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Horne SL, Dosman JA. Body weight and weight gain related to pulmonary function decline in adults: A six year follow up study. Thorax. 1993. 10.1136/thx.48.4.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jartti T, Saarikoski L, Jartti L, et al. Obesity, adipokines and asthma. Allergy. 2009;64(5):770–777. 10.1111/j.1398-9995.2008.01872.x [DOI] [PubMed] [Google Scholar]
- 13.Balsan GA, Vieira JL da C, Oliveira AM de, et al. Relationship between adiponectin, obesity and insulin resistance. Rev Assoc Med Bras. 2015;61(1):72–80. 10.1590/1806-9282.61.01.072 [DOI] [PubMed] [Google Scholar]
- 14.Salah A, Ragab M, Mansour W, Taher M. Leptin and adiponectin are valuable serum markers explaining obesity/bronchial asthma interrelationship. Egypt J Chest Dis Tuberc. 2015;64(3):529–533. 10.1016/J.EJCDT.2015.02.012 [DOI] [Google Scholar]
- 15.Thyagarajan B, Jacobs DR, Smith LJ, et al. Serum adiponectin is positively associated with lung function in young adults, independent of obesity: the CARDIA study. Respir Res. 2010;11(1):176 10.1186/1465-9921-11-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423(6941):762–769. 10.1038/nature01705 [DOI] [PubMed] [Google Scholar]
- 17.Henneman P, Aulchenko YS, Frants RR, et al. Genetic architecture of plasma adiponectin overlaps with the genetics of metabolic syndrome-related traits. Diabetes Care. 2010;33(4):908–913. 10.2337/dc09-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniades C, Antonopoulos AS, Tousoulis D, Stefanadis C. Adiponectin: from obesity to cardiovascular disease. Obes Rev. 2009;10(3):269–279. 10.1111/j.1467-789X.2009.00571.x [DOI] [PubMed] [Google Scholar]
- 19.Li S, Shin HJ, Ding EL, van Dam RM. Adiponectin Levels and Risk of Type 2 Diabetes. JAMA. 2009;302(2):179 10.1001/jama.2009.976 [DOI] [PubMed] [Google Scholar]
- 20.Heid IM, Henneman P, Hicks A, et al. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: Results of genome-wide association analyses including 4659 European individuals. Atherosclerosis. 2010;208(2):412–420. 10.1016/j.atherosclerosis.2009.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menzaghi C, Trischitta V, Doria A. Genetic Influences of Adiponectin on Insulin Resistance, Type 2 Diabetes, and Cardiovascular Disease. Diabetes. 2007;56(5):1198–1209. 10.2337/db06-0506 [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 23.Cupples LA, Arruda HT, Benjamin EJ, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(Suppl 1):S1 10.1186/1471-2350-8-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hivert M-F, Manning AK, McAteer JB, et al. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes. 2008;57(12):3353–3359. 10.2337/db08-0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ukkola O, Santaniemi M, Rankinen T, et al. Adiponectin polymorphisms, adiposity and insulin metabolism: HERITAGE family study and Oulu diabetic study. Ann Med. 2005;37(2):141–150. http://www.ncbi.nlm.nih.gov/pubmed/16028335. Accessed December 4, 2016. 10.1080/07853890510007241 [DOI] [PubMed] [Google Scholar]
- 26.Loos RJF, Ruchat S, Rankinen T, Tremblay A, Pérusse L, Bouchard C. Adiponectin and adiponectin receptor gene variants in relation to resting metabolic rate, respiratory quotient, and adiposity-related phenotypes in the Quebec Family Study. Am J Clin Nutr. 2007;85(1):26–34. http://www.ncbi.nlm.nih.gov/pubmed/17209173. Accessed December 4, 2016. 10.1093/ajcn/85.1.26 [DOI] [PubMed] [Google Scholar]
- 27.Cohen SS, Gammon MD, North KE, et al. ADIPOQ, ADIPOR1, and ADIPOR2 polymorphisms in relation to serum adiponectin levels and BMI in black and white women. Obesity (Silver Spring). 2011;19(10):2053–2062. 10.1038/oby.2010.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling H, Waterworth DM, Stirnadel HA, et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring). 2009;17(4):737–744. 10.1038/oby.2008.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mather KJ, Christophi CA, Jablonski KA, et al. Common variants in genes encoding adiponectin (ADIPOQ) and its receptors (ADIPOR1/2), adiponectin concentrations, and diabetes incidence in the Diabetes Prevention Program. Diabet Med. 2012;29(12):1579–1588. 10.1111/j.1464-5491.2012.03662.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jee SH, Sull JW, Lee J-E, et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet. 2010;87(4):545–552. 10.1016/j.ajhg.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stumvoll M, Tschritter O, Fritsche A, et al. Association of the T-G polymorphism in adiponectin (exon 2) with obesity and insulin sensitivity: interaction with family history of type 2 diabetes. Diabetes. 2002;51(1):37–41. http://www.ncbi.nlm.nih.gov/pubmed/11756320. Accessed May 8, 2018. 10.2337/diabetes.51.1.37 [DOI] [PubMed] [Google Scholar]
- 32.Yang W-S, Tsou P-L, Lee W-J, et al. Allele-specific differential expression of a common adiponectin gene polymorphism related to obesity. J Mol Med. 2003;81(7):428–434. 10.1007/s00109-002-0409-4 [DOI] [PubMed] [Google Scholar]
- 33.Sutton BS, Weinert S, Langefeld CD, et al. Genetic analysis of adiponectin and obesity in Hispanic families: the IRAS Family Study. Hum Genet. 2005;117(2–3):107–118. 10.1007/s00439-005-1260-9 [DOI] [PubMed] [Google Scholar]
- 34.Warodomwichit D, Shen J, Arnett DK, et al. ADIPOQ Polymorphisms, Monounsaturated Fatty Acids, and Obesity Risk: The GOLDN Study. Obesity. 2009;17(3):510–517. 10.1038/oby.2008.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekkers MB, Wijga AH, Gehring U, et al. BMI, waist circumference at 8 and 12 years of age and FVC and FEV1 at 12 years of age; the PIAMA birth cohort study. BMC Pulm Med. 2015;15(1):39 10.1186/s12890-015-0032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cibella F, Bruno A, Cuttitta G, et al. An Elevated Body Mass Index Increases Lung Volume but Reduces Airflow in Italian Schoolchildren. Cormier SA, ed. PLoS One. 2015;10(5):e0127154 10.1371/journal.pone.0127154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henrique Tenório LS, da Cruz Santos A, Sarmento de Oliveira A, Myrna de Lima AJ, do Socorro Brasileiro-Santos M. Obesity and pulmonary function tests in children and adolescents: a systematic review. Rev Paul Pediatr. 2012;30(3):423–430. http://www.scielo.br/pdf/rpp/v30n3/en_18.pdf. Accessed June 28, 2018. [Google Scholar]
- 38.Thyagarajan B, Jacobs DR Jr, Smith LJ, Kalhan R, Gross MD, Sood A. Serum adiponectin is positively associated with lung function in young adults, independent of obesity: The CARDIA study. Respir Res. 2010;11:176 10.1186/1465-9921-11-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guenther M, James R, Marks J, Zhao S, Szabo A, Kidambi S. Adiposity distribution influences circulating adiponectin levels. Transl Res. 2014;164(4):270–277. 10.1016/j.trsl.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obeidad M, Hao K, Bosse Y, et al. Molecular mechanisms underlying variations in lung function: a systems genetics analysis. Lancet Respir Med. 2015;3:782–795. 10.1016/S2213-2600(15)00380-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perneger TV. What’s wrong with Bonferroni adjustments. BMJ. 1998;316:1236–1238. 10.1136/bmj.316.7139.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(SAS7BDAT)
Data Availability Statement
We have already requested permission from the Cyprus Bioethics Committee in order to release the de-identified data; we do not expect this to be a problem and we will forward the data as soon as we have the official approval.


