Abstract
Newly synthesized proteins co-translationally inserted into the endoplasmic reticulum (ER) lumen may be recruited into anterograde transport vesicles by their association with specific cargo receptors. We recently identified a role for the cargo receptor SURF4 in facilitating the secretion of PCSK9 in cultured cells. To examine the function of SURF4 in vivo, we used CRISPR/Cas9-mediated gene editing to generate mice with germline loss-of-function mutations in Surf4. Heterozygous Surf4+/- mice exhibit grossly normal appearance, behavior, body weight, fecundity, and organ development, with no significant alterations in circulating plasma levels of PCSK9, apolipoprotein B, or total cholesterol, and a detectable accumulation of intrahepatic apoliprotein B. Homozygous Surf4-/- mice exhibit embryonic lethality, with complete loss of all Surf4-/- offspring between embryonic days 3.5 and 9.5. In contrast to the milder murine phenotypes associated with deficiency of known SURF4 cargoes, the embryonic lethality of Surf4-/- mice implies the existence of additional SURF4 cargoes or functions that are essential for murine early embryonic development.
Introduction
The coatomer protein complex II (COPII) coat assembles on the cytoplasmic surface of endoplasmic reticulum (ER) exit sites to drive the formation of membrane-bound transport vesicles. Efficient recruitment of proteins and lipids into these vesicles occurs via physical interaction with the COPII coat[1]. For cargoes accessible on the cytoplasmic surface of the ER membrane, this interaction may be direct. For soluble cargoes in the ER lumen, however, transmembrane cargo receptors serve as intermediaries for this interaction[2]. Although thousands of human proteins traffic through the secretory pathway, a corresponding cargo receptor has been identified for only a few, and the size and identity of the cargo repertoire for each individual cargo receptor remains largely unknown.
Through unbiased genome-scale CRISPR screening, we recently discovered a role for the ER cargo receptor Surfeit locus protein 4 (SURF4) in the secretion of Proprotein convertase subtilisin/kexin type 9 (PCSK9)[3], a protein that modulates mammalian cholesterol homeostasis through its negative regulation of the Low-density lipoprotein receptor (LDLR)[4]. Consistent with a role as a PCSK9 cargo receptor, SURF4 was found to localize to the ER and the ER-Golgi intermediate compartment, to physically associate with PCSK9, and to promote the ER exit and extracellular secretion of PCSK9. These experiments relied on heterologous expression of PCSK9 in HEK293T cells, however, and the physiologic relevance of this interaction in vivo remains uncertain. Additionally, although SURF4 deletion did not affect the secretion of a control protein, alpha-1 antitrypsin, a broader role for SURF4 in protein secretion remains possible and is supported by the recent identification of other potential cargoes including apolipoprotein B, growth hormone, dentin sialophosphoprotein, and amelogenin[5, 6].
To investigate the physiologic functions of SURF4, we generated mice with targeted disruption of the Surf4 gene. We found that partial loss of SURF4 in heterozygous mice led to a modest accumulation of intrahepatic apolipoprotein B, with no effect on steady state plasma levels. However, complete genetic deletion of Surf4 resulted in early embryonic lethality.
Results
Generation of mice with germline deletion of Surf4
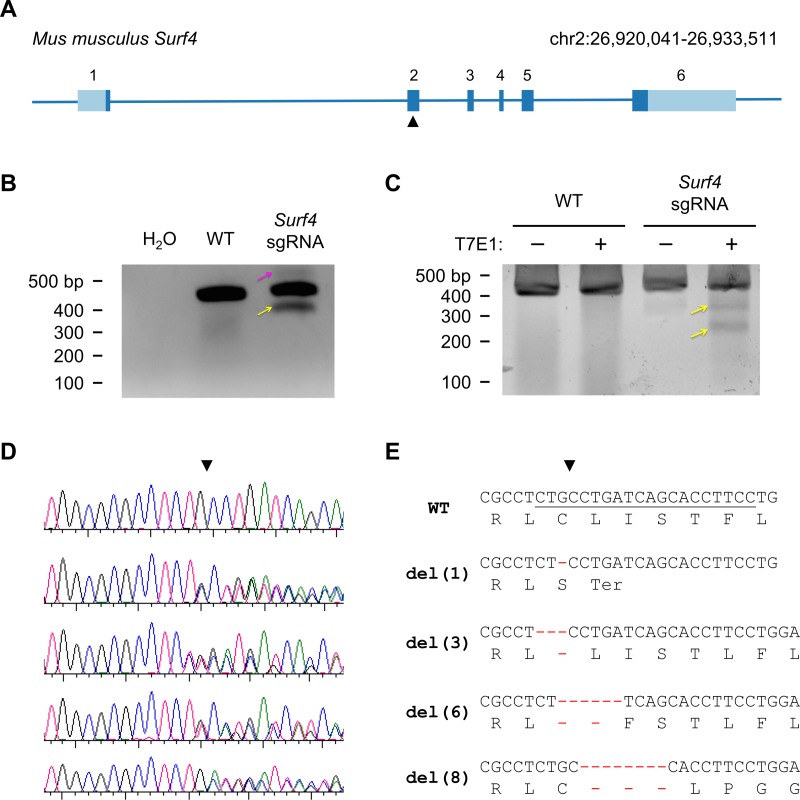
The Mus musculus Surf4 gene is composed of 6 exons and 5 introns spanning approximately 14 kb in the tightly clustered surfeit gene locus on chromosome 2[7, 8]. We targeted exon 2 of Surf4 for CRISPR/Cas-mediated mutagenesis (Fig 1A), verified sgRNA efficiency in embryonic stem (ES) cells (Fig 1B and 1C), and generated mice from microinjected zygotes. Sanger sequencing identified 4 of 57 mice with disruption of the target site (Fig 1D). These mice were then mated to C57BL/6J wild-type mice and their progeny genotyped, confirming germline transmission for each of the 4 alleles (Fig 1E). Two alleles introduced frameshift deletions both leading to early termination codons, with the other alleles containing in-frame deletions of 3 and 6 DNA base pairs, respectively.
Fig 1. Generation of Surf4 mutant alleles.
(A) Surf4 gene structure. Exons are shaded light blue for untranslated regions or dark blue for coding sequence. The target site for the sgRNA used for oocyte editing is indicated by the black triangle. (B) Mouse ES cells were either untreated or electroporated with plasmids for CRISPR/Cas9 disruption of the Surf4 target site. PCR amplification of genomic DNA or water control across the Surf4 target site revealed higher and lower molecular weight DNA fragments suggestive of nonhomologous endjoining repair of Surf4 indels. (C) The major PCR product was gel purified and subjected to T7 endonuclease I digestion. T7E1 digestion produced novel DNA fragments (arrows) indicating the presence of insertions/deletions in Surf4 exon 2. Wild type DNA was resistant to T7E1 digestion. (D) Sanger sequencing chromatograms of Surf4 target site amplicons of progeny from matings between Surf4-targeted founder mice and wild-type C57BL6/J mice. (E) DNA and predicted protein sequences for the 4 individual allele generated by CRISPR/Cas9 gene-editing of Surf4.
Effect of SURF4 haploinsufficiency on cholesterol regulation
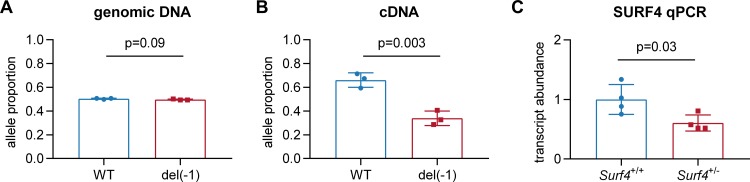
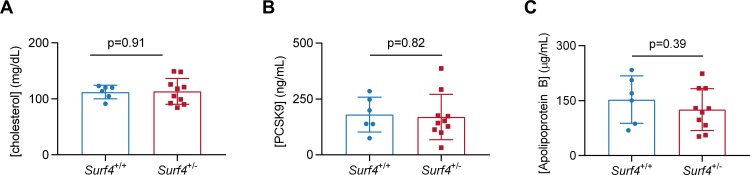
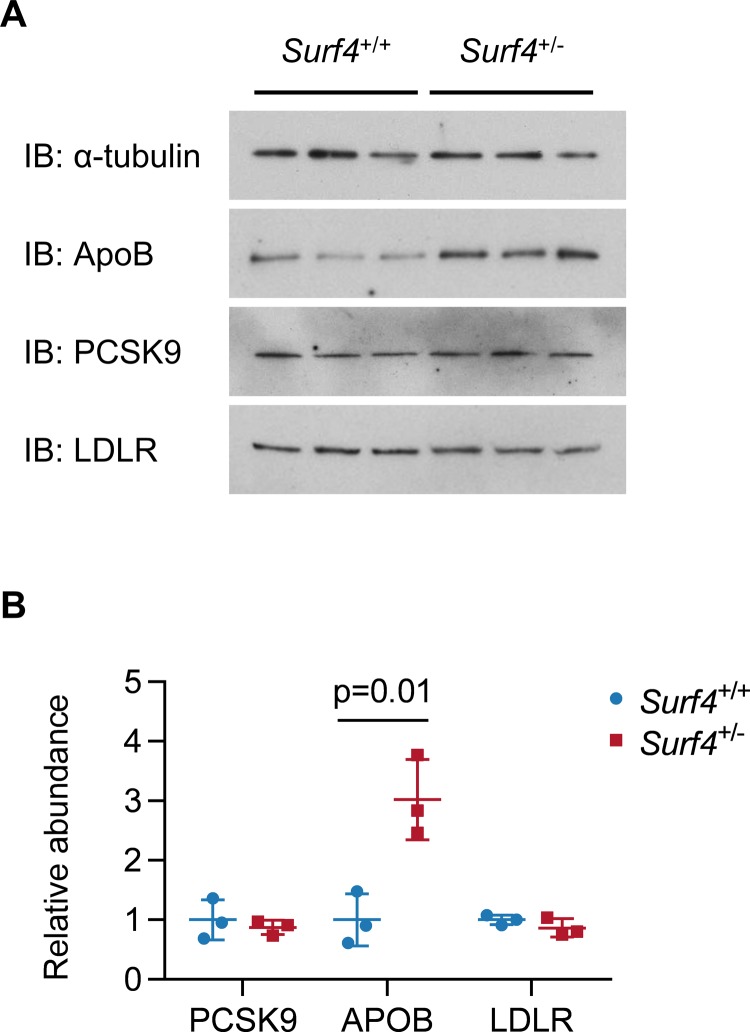
Surf4+/- mice were observed at expected Mendelian ratios at weaning (Table 1) and exhibited grossly normal appearance, behavior, and organ development by necropsy. Analysis of mRNA from Surf4+/- mouse liver tissue confirmed a reduction in total Surf4 transcripts with a relative decrease in the mutant allele, consistent with nonsense-mediated decay (Fig 2). Surf4+/- mice heterozygous for the del(1) allele also showed no significant differences in plasma PCSK9, cholesterol, and apolipoprotein B levels compared to Surf4+/+ litter-mate controls (Fig 3). Similarly, no differences were seen in the intrahepatic accumulation of the putative SURF4 cargo, PCSK9, or its downstream target, LDL receptor. In contrast, despite its normal steady state levels in plasma, an approximately 3-fold increase in intrahepatic apolipoprotein B was observed in Surf4+/- mice (Fig 4), consistent with selective retention of this putative SURF4 cargo in the setting of Surf4 haploinsufficiency.
Table 1. Mice heterozygous for each of 3 independent Surf4 targeted alleles are observed at expected Mendelian ratios at weaning.
Mice heterozygous for the indicated Surf4 allele were crossed with wild-type C57BL/6J mice and the resulting litters genotyped for the corresponding Surf4 alleles at approximately 2 weeks of age. The proportion of mice with the heterozygous mutant genotype was compared to expected Mendelian ratios by the chi-square test.
| Surf4+/- x Surf4+/+ | |||
|---|---|---|---|
| Allele | Progeny | p (Χ2) | |
| +/+ | +/- | ||
| del (3) | 36 | 39 | 0.81 |
| del (6) | 22 | 15 | 0.41 |
| del (1) | 19 | 18 | 0.91 |
Fig 2. Surf4+/- mice exhibit partial reduction of Surf4 transcripts with preferential loss of the mutant allele.
Sanger sequencing of the Surf4 target site was performed on PCR amplicons derived from genomic DNA (A) or reverse-transcribed cDNA (B) prepared from liver tissue of 3 Surf4+/- mice. Decomposition of chromatograms was performed to quantify the relative proportion of each allele in each sample. (C) Total Surf4 transcript levels in liver tissue from 4 Surf4+/+ and 4 Surf4+/- mice were quantified and normalized to a panel of housekeeping genes by qRT-PCR.
Fig 3. Surf4 haploinsufficiency does not affect baseline plasma levels for PCSK9, ApoB, or cholesterol levels.
Plasma samples collected from 10 Surf4+/- mice (heterozygous for the del(1) allele) and 6 wild-type littermate controls were assayed for plasma levels of total cholesterol (A), PCSK9 (B), and ApoB (C). Values were measured and averaged for each of two independent phlebotomies from each mouse. Both male and female mice were tested for each genotype. Significance testing was calculated by Student’s t-test between genotype groups.
Fig 4. Surf4 haploinsufficiency causes hepatic accumulation of apolipoprotein B but not PCSK9.
Liver lysates from 3 male Surf4+/- mice harboring the del(1) allele and 3 male Surf4+/+ littermate controls were immunoblotted for PCSK9, LDL receptor, apolipoprotein B, and alpha-tubulin. Densitometry values for PCSK9, LDLR, and apolipoprotein B were normalized to alpha-tubulin. Significance testing was performed by Student’s t-test between genotype groups.
Surf4 function is required for embryonic development
Intercrosses were performed for Surf4+/- mice carrying each of the 3 independent Surf4 deletion alleles described above. Genotyping at the time of weaning demonstrated the expected number of heterozygous progeny, with complete absence of homozygous Surf4-/- pups (Table 2). Timed matings of mice heterozygous for the del(1) allele were performed, with no Surf4-/- embryos identified at E9.5 or later (Table 3). However, analysis of E3.5 blastocysts generated by in vitro fertilization revealed the expected proportion of Surf4-/- genotypes with no gross morphologic abnormalities on visual assessment by an experienced expert in murine embryology Thus, complete genetic deficiency of Surf4 results in embryonic lethality occurring sometime between E3.5 and E9.5.
Table 2. Germline deletion of Surf4 causes embryonic lethality.
Mice heterozygous for the indicated Surf4 alleles were intercrossed and progeny genotyped for Surf4 at weaning. The proportion of mice with the homozygous null genotype was compared to expected Mendelian ratios by the chi-square test.
| Surf4+/- x Surf4+/- | |||||
|---|---|---|---|---|---|
| Allele | Stage | Progeny | p (Χ2) | ||
| +/+ | +/- | -/- | |||
| del (3) | weaning | 9 | 22 | 0 | <0.01 |
| del (6) | weaning | 12 | 32 | 0 | <0.01 |
| del (1) | weaning | 40 | 72 | 0 | <0.01 |
Table 3. Germline deletion of Surf4 results in lethality between embryonic day 3.5 and 9.5.
Timed matings were performed between Surf4+/- mice carrying the del(1) allele and embryos harvested at E9.5, E12.5, E.14.5 or at the time of weaning. For analysis at E3.5, blastocysts were collected following in vitro fertilization of oocytes from Surf4+/- females with sperm from Surf4+/- males. The proportion of mice with the homozygous null genotype was compared to expected Mendelian ratios by the chi-square test.
| Surf4+/- x Surf4+/- | |||||
|---|---|---|---|---|---|
| Allele | Stage | Progeny | p (Χ2) | ||
| +/+ | +/- | -/- | |||
| del (1) | weaning | 40 | 72 | 0 | <0.01 |
| E14.5 | 2 | 3 | 0 | >0.99 | |
| E12.5 | 3 | 12 | 0 | 0.100 | |
| E9.5 | 5 | 15 | 0 | 0.047 | |
| E3.5 | 1 | 7 | 4 | >0.99 | |
Discussion
Identification of the molecular machinery underlying eukaryotic protein secretion has been elucidated by elegant work in model systems including yeast and cultured mammalian cells. Recent characterizations of mice with genetic deficiency of COPII components have extended these findings to mammalian physiology, revealing a variety of complex phenotypes[9–18]. Comparatively little is known about the physiologic role of mammalian cargo receptors in vivo. In humans, genetic deletion of either subunit of a cargo receptor complex, LMAN1/MCFD2, results in a rare bleeding disorder due to the impaired secretion of coagulation factors V and VIII[19, 20], with a similar phenotype in Lman1-/- and MCFD2-/- mice[21, 22].
We set out to investigate the physiologic function of murine SURF4 in vivo, with a particular focus on its putative function in the secretion of PCSK9[3] and apolipoprotein B[6], both of which play central roles in mammalian cholesterol regulation. We generated multiple independent gene targeted Surf4 alleles, with heterozygous Surf4+/- mice exhibiting no gross developmental abnormalities and normal circulating levels of cholesterol, PCSK9, and apolipoprotein B. Consistent with our observations that SURF4 haploinsufficiency is well-tolerated in mice, a number of loss-of-function variants have been observed in human SURF4, including a p.Gln185Ter nonsense variant with an allele frequency of 0.1%[23]. Of note, previous human genome-wide association studies for lipid traits have not detected a significant signal near the SURF4 gene[24].
To assess the impact of Surf4 haploinsufficiency on the physiologic secretion of putative cargoes, we measured the levels of PCSK9 and apolipoprotein B in circulation and in the liver.
We found that plasma and intrahepatic levels of PCSK9 were unaffected by partial SURF4 reduction in Surf4+/- mice. The Surf4 exon targeted by our gene editing approach is expressed in all currently annotated mouse Surf4 splice variants (Ensembl release 98[25]). Analysis of liver mRNA from Surf4+/- mice confirmed a reduction in total Surf4 transcript levels, with reduced levels of the mutant allele relative to the wild-type allele consistent with nonsense-mediated decay (Fig 2). This observation is similar to the normal plasma levels of LMAN1 cargo proteins reported in heterozygous Lman1+/- mice[21]. In contrast, we found that apolipoprotein B accumulated approximately 3-fold in liver cell lysates prepared from Surf4+/- mice compared to controls, suggesting greater sensitivity of apolipoprotein B than PCSK9 to partial SURF4 depletion. Nonetheless, plasma levels of apolipoprotein B and total cholesterol were unaffected by haploinsufficiency for Surf4, possibly due to downstream effects on complex cholesterol regulatory pathways which could alter the rate of clearance and/or expression of ApoB or other related components of this network. Together, these observations indicate a complexity in the degree of dependence of different cargoes on the partial or complete reduction of their corresponding cargo receptor. The mechanistic basis for this variability remains unknown but may be related to different stoichiometries or cargo receptor binding affinities.
In cultured cells, secretion defects of PCSK9[3] and apolipoprotein B[6] are observed upon complete deletion of SURF4. Our attempts to generate adult mice with complete loss of Surf4 were precluded by the embryonic lethality caused by Surf4 deletion. A loss of Surf4-/- mice at weaning was unlikely to have been caused by a linked spontaneous or off-target CRISPR-generated passenger mutation[26] as this phenotype was observed for each of 3 independent alleles. Timed matings revealed that loss of Surf4-/- embryos occurs between E3.5 and E9.5. The mechanism for this observation is unclear. Deficiency of the SURF4 homologue SFT-4 is similarly associated with embryonic lethality in C. elegans[6], but SURF4 is not essential for cellular viability in cultured HEK293T cells[3, 5] and its homologue, Erv29p, is dispensable in yeast[27, 28]. Deficiencies of PCSK9 or apoliporotein B alone cannot account for this developmental phenotype, given that PCSK9-/- mice are viable[29] and that ApoB-/- mice survive past E9.5[30]. Likewise, mice with genetic deletion of 3 other putative SURF4 cargoes, growth hormone[31], amelogenin[32], and dental sialophosphoprotein[33], are viable. The embryonic lethality of SURF4 deficiency may therefore result from additive effects of disrupted secretion of known cargoes, or the presence of additional unknown SURF4 cargoes or functions that are essential for early embryonic development. A role for SURF4 in global protein secretion is unlikely, as previous studies demonstrated no effect of SURF4 deletion on the secretion of a number of other proteins[3, 5]. A broader role for SURF4 in the secretion of additional unknown cargoes however is suggested by the observation that SURF4 has been evolutionarily conserved in yeast and other organisms lacking homologues of PCSK9. An N-terminal tripeptide “ER-ESCAPE motif”, present on a large number of potential cargoes, has recently been proposed to mediate cargo recruitment by SURF4[5]. A comprehensive identification of SURF4 cargoes and the nature of their interaction with SURF4 should clarify the function of SURF4 in cholesterol regulation and in mammalian development.
Materials and methods
Generation of Surf4 mutant mice
All animal protocols used in this study were approved by the University of Michigan Committee on the Use and Care of Animals. We used CRISPR/Cas9 technology[34, 35] to generate a new genetically modified mouse strain with a Surf4 gene knockout. The presence of a premature termination codon in exon 2 is predicted to result in loss of protein expression due to nonsense mediated decay of mRNA[36]. A single guide RNA (sgRNA) target and protospacer adjacent motif was identified in exon 2 (ENSMUSE00000232711.1) with CRISPOR[37]. The sgRNA is 5’ CTGCCTGATCAGCACCTTCC TGG 3’ on the non-coding strand (chromosome 2; coordinates 26926892–26926911) with a predicted cut site 47 bp downstream of the first exon 2 codon. The sgRNA target was cloned into plasmid pX330 (Addgene.org plasmid #42230, a kind gift of Feng Zhang) as described[38]. The sgRNA was validated in mouse JM8.A3 ES cells[39] prior to use for mouse zygote microinjection. The sgRNA plasmid (15 μg) was electroporated into 8 X 10E6 ES cells. To the electroporation, 5 μg of a PGK1-puromycin resistance plasmid[40] was added for transient puromycin selection (2 μg/ml puromycin applied 48–72 hours after electroporation). ES cell culture and electroporation was carried out as described[41]. After selection, DNA was extracted from surviving cells, PCR was used to amplify the sequences across the sgRNA cut site, and T7 endonuclease 1 assays were used to detect small deletions/insertions at the predicted Cas9 DNA cut site[42]. The circular sgRNA plasmid was resuspended in microinjection buffer as described[43]. The plasmid mixture was used for pronuclear microinjection of zygotes obtained from the mating of superovulated C57BL/6J female mice (The Jackson Laboratory Stock No. 0006640) and C57BL/6J male mice as described[44]. A total of 305 zygotes were microinjected and 285 zygotes were transferred to pseudopregnant B6D2F1 female mice (The Jackson Laboratory Stock No. 100006). 18 mouse pups were born and four of them transmitted gene edited Surf4 alleles.
Mouse genotyping
Surf4 genotyping was performed by PCR of genomic DNA with primers mSurf4-ex2-for [TGCTGAGGGCCTCTCTGTCT] and mSurf4-ex2-rev [CAGGTAGCCACAGCTCCAGG]. Sanger sequencing was performed with the same genotyping primers and chromatograms were inspected both manually and by automated deconvolution[45] to determine the presence or absence of target site indels.
Analysis of Surf4+/- mice
Mice were housed and monitored in accordance with University of Michigan Unit of Laboratory Animal Medicine (ULAM) guidelines. Blood was collected at 6–12 weeks of age by retro-orbital bleeding into heparin-coated collection tubes from mice anesthetized with isoflurane. Plasma was prepared by centrifugation at 2,000 g for 10 min at 4°C. A second blood collection was performed 1 week following the initial collection. Plasma samples were analyzed by total cholesterol colorimetric assay (SB-1010-225, Fisher Scientific, Hampton NH) and ELISAs for PCSK9 (MPC900, R&D Systems, Minneapolis MN) and apolipoprotein B (ab230932, Abcam, Cambridge UK). Liver tissue was perfused with PBS and harvested from mice at the time of sacrifice. Liver mRNA was prepared with RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and oligo(dT)-primed first strand cDNA generated with Superscript III reverse transcriptase (Invitrogen, Carlsbad CA) according to manufacturer’s instructions. Quantitative PCR was performed using 20 ng of cDNA per reaction with PowerSYBR Green PCR Master Mix (Applied Biosystems, Foster City CA). Normalization of qPCR data was performed using a panel of 4 selected housekeeping genes[46]. The relative proportion of each Surf4 allele was quantified by decomposition of Sanger sequencing chromatograms with TIDE indel analysis[45]. The primer sequences for qRT-PCR were: Surf4-forward [CTGTTGGCCTCATCCTTCGT], Surf4-reverse [GGCAATTGTCTGCAGTGCG], Actb-forward [CCACTGCCGCATCCTCTTCC], Actb-reverse [CTCGTTGCCAATAGTGATGACCTG], B2m-forward [CATGGCTCGCTCGGTGACC], B2m-reverse [AATGTGAGGCGGGTGGAACTG], Tbp-forward [CCCCACAACTCTTCCATTCT], Tbp-reverse [GCAGGAGTGATAGGGGTCAT], Ppia-forward [CAAATGCTGGACCAAACACAAACG], Ppia-reverse [GTTCATGCCTTCTTTCACCTTCCC].
Protein lysates were prepared from liver tissue by mechanical homogenization, resuspension in RIPA lysis buffer (Pierce Manufacturing, Appleton WI), and collection of supernatant after centrifugation for 15 minutes at 21,000xg. Protein concentrations of lysates were measured by DC Protein Assay (5000111, Bio-Rad Laboratories, Hercules CA). Equal amounts of lysate were analyzed by immunoblotting with antibodies against Apolipoprotein B (70R-15771, 1:1000, Fitzgerald Industries International, Acton MA), PCSK9 (ab31762, 1:1000, Abcam), LDLR (ab52818, 1:1000, Abcam), and alpha-tubulin (ab176560, 1:2000, Abcam) and densitometry analysis performed with ImageJ[47].
Timed matings and in vitro fertilization
For analysis of embryonic day 9.5 and later, Surf4+/- male and female mice were co-housed overnight and females with copulatory vaginal plugs the following morning were assigned embryonic day 0.5. Pregnant females were then sacrificed at indicated time points and genomic DNA prepared from isolated embryos. For analysis of embryonic day 3.5, Surf4+/- females were superovulated with anti-inhibin serum as described[48]. The collected oocytes were fertilized with sperm from Surf4+/- males as described[49]. Resulting fertilized eggs were maintained in cell culture in KSOM medium (Zenith Biotech) until visual inspection, harvesting, and genomic DNA preparation from blastocysts at embryonic day 3.5.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by NIH grants R35-HL135793T (DG), T32-HL007853, KL2-TR002241 and K08-HL148552 (BTE). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14(1):20–8. Epub 2011/12/24. 10.1038/ncb2390 . [DOI] [PubMed] [Google Scholar]
- 2.Barlowe C, Helenius A. Cargo Capture and Bulk Flow in the Early Secretory Pathway. Annu Rev Cell Dev Biol. 2016;32:197–222. 10.1146/annurev-cellbio-111315-125016 . [DOI] [PubMed] [Google Scholar]
- 3.Emmer BT, Hesketh GG, Kotnik E, Tang VT, Lascuna PJ, Xiang J, et al. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife. 2018;7 Epub 2018/09/27. 10.7554/eLife.38839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci. 2007;32(2):71–7. Epub 2007/01/12. 10.1016/j.tibs.2006.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin Y, Garcia MR, Novak AJ, Saunders AM, Ank RS, Nam AS, et al. Surf4 (Erv29p) binds amino-terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol. 2018;16(8):e2005140 Epub 2018/08/08. 10.1371/journal.pbio.2005140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saegusa K, Sato M, Morooka N, Hara T, Sato K. SFT-4/Surf4 control ER export of soluble cargo proteins and participate in ER exit site organization. J Cell Biol. 2018;217(6):2073–85. Epub 2018/04/13. 10.1083/jcb.201708115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams T, Yon J, Huxley C, Fried M. The mouse surfeit locus contains a very tight cluster of four "housekeeping" genes that is conserved through evolution. Proc Natl Acad Sci U S A. 1988;85(10):3527–30. Epub 1988/05/01. 10.1073/pnas.85.10.3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huxley C, Fried M. The mouse surfeit locus contains a cluster of six genes associated with four CpG-rich islands in 32 kilobases of genomic DNA. Mol Cell Biol. 1990;10(2):605–14. Epub 1990/02/01. 10.1128/mcb.10.2.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen XW, Wang H, Bajaj K, Zhang P, Meng ZX, Ma D, et al. SEC24A deficiency lowers plasma cholesterol through reduced PCSK9 secretion. Elife. 2013;2:e00444 Epub 2013/04/13. 10.7554/eLife.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merte J, Jensen D, Wright K, Sarsfield S, Wang Y, Schekman R, et al. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nat Cell Biol. 2010;12(1):41–6; sup pp 1–8. Epub 2009/12/08. 10.1038/ncb2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams EJ, Chen XW, O'Shea KS, Ginsburg D. Mammalian COPII coat component SEC24C is required for embryonic development in mice. J Biol Chem. 2014;289(30):20858–70. Epub 2014/05/31. 10.1074/jbc.M114.566687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baines AC, Adams EJ, Zhang B, Ginsburg D. Disruption of the Sec24d gene results in early embryonic lethality in the mouse. PLoS One. 2013;8(4):e61114 Epub 2013/04/19. 10.1371/journal.pone.0061114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao J, Zhu M, Wang H, Afelik S, Vasievich MP, Chen XW, et al. SEC23B is required for the maintenance of murine professional secretory tissues. Proc Natl Acad Sci U S A. 2012;109(29):E2001–9. Epub 2012/06/30. 10.1073/pnas.1209207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu M, Tao J, Vasievich MP, Wei W, Zhu G, Khoriaty RN, et al. Neural tube opening and abnormal extraembryonic membrane development in SEC23A deficient mice. Sci Rep. 2015;5:15471 Epub 2015/10/27. 10.1038/srep15471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khoriaty R, Everett L, Chase J, Zhu G, Hoenerhoff M, McKnight B, et al. Pancreatic SEC23B deficiency is sufficient to explain the perinatal lethality of germline SEC23B deficiency in mice. Sci Rep. 2016;6:27802 Epub 2016/06/15. 10.1038/srep27802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoriaty R, Hesketh GG, Bernard A, Weyand AC, Mellacheruvu D, Zhu G, et al. Functions of the COPII gene paralogs SEC23A and SEC23B are interchangeable in vivo. Proc Natl Acad Sci U S A. 2018;115(33):E7748–E57. Epub 2018/08/02. 10.1073/pnas.1805784115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khoriaty R, Vasievich MP, Jones M, Everett L, Chase J, Tao J, et al. Absence of a red blood cell phenotype in mice with hematopoietic deficiency of SEC23B. Mol Cell Biol. 2014;34(19):3721–34. Epub 2014/07/30. 10.1128/MCB.00287-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khoriaty R, Vogel N, Hoenerhoff MJ, Sans MD, Zhu G, Everett L, et al. SEC23B is required for pancreatic acinar cell function in adult mice. Mol Biol Cell. 2017;28(15):2146–54. Epub 2017/05/26. 10.1091/mbc.E17-01-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA, et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93(1):61–70. Epub 1998/04/18. 10.1016/s0092-8674(00)81146-0 . [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Cunningham MA, Nichols WC, Bernat JA, Seligsohn U, Pipe SW, et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat Genet. 2003;34(2):220–5. Epub 2003/04/30. 10.1038/ng1153 . [DOI] [PubMed] [Google Scholar]
- 21.Zhang B, Zheng C, Zhu M, Tao J, Vasievich MP, Baines A, et al. Mice deficient in LMAN1 exhibit FV and FVIII deficiencies and liver accumulation of alpha1-antitrypsin. Blood. 2011;118(12):3384–91. Epub 2011/07/29. 10.1182/blood-2011-05-352815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu M, Zheng C, Wei W, Everett L, Ginsburg D, Zhang B. Analysis of MCFD2- and LMAN1-deficient mice demonstrates distinct functions in vivo. Blood Adv. 2018;2(9):1014–21. Epub 2018/05/08. 10.1182/bloodadvances.2018018317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. Epub 2016/08/19. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–83. Epub 2013/10/08. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, et al. Ensembl variation resources. Database (Oxford). 2018;2018 Epub 2018/12/24. 10.1093/database/bay119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westrick RJ, Mohlke KL, Korepta LM, Yang AY, Zhu G, Manning SL, et al. Spontaneous Irs1 passenger mutation linked to a gene-targeted SerpinB2 allele. Proc Natl Acad Sci U S A. 2010;107(39):16904–9. Epub 2010/09/15. 10.1073/pnas.1012050107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otte S, Belden WJ, Heidtman M, Liu J, Jensen ON, Barlowe C. Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J Cell Biol. 2001;152(3):503–18. Epub 2001/02/07. 10.1083/jcb.152.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276(26):23296–303. Epub 2001/04/24. 10.1074/jbc.M102962200 . [DOI] [PubMed] [Google Scholar]
- 29.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, et al. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci U S A. 2005;102(15):5374–9. 10.1073/pnas.0501652102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farese RV Jr., Ruland SL, Flynn LM, Stokowski RP, Young SG. Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes. Proc Natl Acad Sci U S A. 1995;92(5):1774–8. Epub 1995/02/28. 10.1073/pnas.92.5.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508–14. Epub 2016/09/15. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276(34):31871–5. Epub 2001/06/15. 10.1074/jbc.M104624200 . [DOI] [PubMed] [Google Scholar]
- 33.Sreenath T, Thyagarajan T, Hall B, Longenecker G, D'Souza R, Hong S, et al. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;278(27):24874–80. Epub 2003/05/02. 10.1074/jbc.M303908200 . [DOI] [PubMed] [Google Scholar]
- 34.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. Epub 2013/01/05. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. Epub 2013/01/05. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp MW, Maquat LE. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell. 2016;165(6):1319–22. Epub 2016/06/04. 10.1016/j.cell.2016.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17(1):148 Epub 2016/07/07. 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. Epub 2013/10/26. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6(7):493–5. Epub 2009/06/16. 10.1038/nmeth.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBurney MW, Fournier S, Jardine K, Sutherland L. Intragenic regions of the murine Pgk-1 locus enhance integration of transfected DNAs into genomes of embryonal carcinoma cells. Somat Cell Mol Genet. 1994;20(6):515–28. Epub 1994/11/01. 10.1007/bf02255842 . [DOI] [PubMed] [Google Scholar]
- 41.Hughes ED, Saunders TL. In: Pease S, Saunders TL, editors. Advanced Protocols for Animal Transgenesis: An ISTT Manual: Springer-Verlag, Berlin: p. 291–325. [Google Scholar]
- 42.Sakurai T, Watanabe S, Kamiyoshi A, Sato M, Shindo T. A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol. 2014;14:69 Epub 2014/07/22. 10.1186/1472-6750-14-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355 Epub 2013/11/29. 10.1038/srep03355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker K, Jerchow B. Advanced Protocols for Animal Transgenesis: An ISTT Manual. In: Pease S, Saunders TL, editors. Berlin: Springer-Verlag; 2011. p. 99–116. [Google Scholar]
- 45.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168 Epub 2014/10/11. 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling D, Salvaterra PM. Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One. 2011;6(3):e17762 Epub 2011/03/23. 10.1371/journal.pone.0017762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. 10.1038/nmeth.2089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeo T, Nakagata N. Immunotherapy using inhibin antiserum enhanced the efficacy of equine chorionic gonadotropin on superovulation in major inbred and outbred mice strains. Theriogenology. 2016;86(5):1341–6. Epub 2016/06/01. 10.1016/j.theriogenology.2016.04.076 . [DOI] [PubMed] [Google Scholar]
- 49.Ostermeier GC, Wiles MV, Farley JS, Taft RA. Conserving, distributing and managing genetically modified mouse lines by sperm cryopreservation. PLoS One. 2008;3(7):e2792 Epub 2008/07/31. 10.1371/journal.pone.0002792 [DOI] [PMC free article] [PubMed] [Google Scholar]