Abstract
Metastases are responsible for the majority of breast cancer-associated deaths. The contribution of epithelial-to-mesenchymal transition (EMT) in the establishment of metastases is still controversial. To obtain in vivo evidence of EMT in metastasis, we established an EMT lineage tracing model (Tri-PyMT), in which tumor cells undergoing EMT would irreversibly switch their fluorescent marker from RFP+ to GFP+ due to mesenchymal-specific Cre expression. Surprisingly, we found that lung metastases were predominantly derived from the epithelial compartment of breast tumors. However, concerns were raised on the fidelity and sensitivity of RFP-to-GFP switch of this model in reporting EMT of metastatic tumor cells. Here, we evaluated Tri-PyMT cells at the single-cell level using single-cell RNA-sequencing, and found that the Tri-PyMT cells exhibited a spectrum of EMT phenotypes, with EMT-related genes concomitantly expressed with the activation of GFP. The fluorescent color switch in these cells precisely marked an unequivocal change in EMT status, defining the pre-EMT and post-EMT compartments within the tumor. Consistently, the pre-EMT cells played dominant roles in metastasis, while the post-EMT cells were supportive in promoting tumor invasion and angiogenesis. Importantly, the post-EMT (GFP+) cells in the Tri-PyMT model were not permanently committed to the mesenchymal phenotype; they were still capable of reverting to the epithelial phenotype and giving rise to secondary tumors, suggesting their persistent EMT plasticity. Our study addressed major concerns with the Tri-PyMT EMT lineage tracing model, which provides us with a powerful tool to investigate the dynamic EMT process in tumor biology.
Keywords: Epithelial to mesenchymal transition (EMT), breast cancer, metastasis, invasion, angiogenesis, metastatic seeding, colonization
Introduction
Metastasis represents the primary cause of mortality in cancer patients. Therefore, exploring the mechanisms of metastasis stands as a major task in cancer research. Epithelial-to-mesenchymal transition (EMT), originally characterized in embryo development as a process of cellular phenotypic transdifferentiation from stationary epithelial cells to motile mesenchymal cells, is also hijacked by tumor cells(1,2). Through EMT, epithelial tumor cells lose their polarity and tight connection with neighboring cells, gain the ability to migrate and invade, exhibit resistance to apoptosis, and retrieve stemness properties(3–5). EMT has been enthusiastically proposed as an essential step for metastasis, in that EMT-associated features adequately meet the requirements for metastasis formation.
However, the mesenchymal phenotype was rarely observed in secondary tumors. Metastatic lesions usually resemble the epithelial phenotype of primary tumors(6–8). This observation has been tentatively explained by the dynamic nature of EMT. When the mesenchymal tumor cells had seeded at metastatic sites, they regained the epithelial features by undergoing a mesenchymal-to-epithelial transition (MET). To obtain in vivo evidence of the reversible EMT in metastasis, we established an EMT lineage tracing model in a multiple-transgenic mouse (MMTV-PyMT/Fsp1-Cre/Rosa26mT/mG, Tri-PyMT)(9). In this model, breast tumor cells that underwent EMT would irreversibly switch their fluorescent marker from RFP+ to GFP+ due to the mesenchymal specific Cre expression. Surprisingly, we found that metastatic lesions did not convert their fluorescent marker(9). Lung metastases were predominantly composed of RFP+ cells exhibiting epithelial phenotypes. Although EMT tumor cells (GFP+) were detected in the primary tumor, circulation, and metastatic lungs, they were significantly outnumbered by their epithelial precursors (RFP+). Similar observations were obtained by using MMTV-Her2-driven breast tumors, or vimentin-driven Cre mediated EMT lineage tracing models(9). These findings challenge the concept that EMT is required for metastasis, and have aroused vigorous discussion about its true contributions to metastasis(6,10–12).
One of the major concerns with the Tri-PyMT model is that the expression of GFP only indicates the complete EMT. The partial-EMT, a concept describing tumor cells exhibiting both epithelial and mesenchymal features(10,12,13), could possibly fail to launch the Fsp1-Cre-mediated fluorescence switch. Here, we reevaluated the efficiency and fidelity of the Tri-PyMT model by performing single-cell RNA-sequencing (scRNA-seq). We aimed to clarify the advantages and disadvantages of the RFP+ and GFP+ Tri-PyMT cells in the metastatic cascade.
Materials and Methods
Animals
CB-17 SCID mice (Charles River, MA) were used in the orthotopic and metastatic Tri-PyMT models. Animal works were approved and conducted following the guidance of the IACUC at WCM.
Tri-PyMT cells
Tri-PyMT cells were derived from primary Tri-PyMT tumors (9). Cell authentication was performed by RT-PCR of PyMT antigen expression (9).Cells were cultured in DMEM with 10% FBS, 2mM glutamine, and antibiotics. Experiments were performed with cells of 5th −10th passages from the primary culture. The mycoplasma free culture condition was confirmed by MycoAlert PLUS kit (Lonza).
Flow cytometry
Single-cell suspensions were prepared by digesting lung or tumor tissues with Collagenase A and DNase I (Roche Applied Science) in HBSS at 37°C for 30 min. GFP+ and RFP+ cells were detected via LSRII flow cytometer (BD Biosciences).
RNA-sequencing analysis
Total RNA extracted from RFP+ and GFP+ Tri-PyMT cells with the RNeasy Kit (Qiagen). RNA-Seq libraries were constructed and sequenced following standard protocols (Illumina). RNA-seq data were analyzed with Cufflinks and Cuffdiff2 packages. GSEA was performed following descriptions at http://www.broad.mit.edu/gsea.
Single-cell RNA-sequencing analysis
Single-cell suspension was prepared following a standard protocol of Drop-seq sample preparation at the Genomics and Epigenetics Core Facility at WCM. The Drop-seq libraries were prepared and sequenced on HiSeq 2500 (Illumina). For in vivo samples, RFP+ and GFP+ cells were FACS-sorted from Tri-PyMT primary tumors and remixed at 1:1. The sequencing library was prepared following 10X Genomics protocol and sequenced on HiSeq 4000 (Illumina).
The Drop-seq data analyses were performed with the Seurat R package (14). The data quality was controlled by the total number of genes (200 – 5000 genes), UMIs (> 200) and the percentage of mitochondria gene (< 2%). The mapping of RFP and GFP sequences were used to identify RFP+ and GFP+ cells, respectively. After filtering, 871 RFP cells and 3,357 GFP cells were preserved for further analyses. The top 10 principal components (PC) were selected for tSNE visualization. The Wilcoxon rank sum test in the Seurat package was employed for differential expression analysis.
EMT score calculation of single cells
Using the identified signature genes in RFP+ and GFP+ cells, we normalized their expression matrix, and calculated the sum of expression values across all signature genes (unweighted) for each cell, and added 1,000 to ensure positive values. The ratio of the sum of mesenchymal to that of epithelial genes was defined as the EMT score, which was further employed to build a binary classifier and ROC curve.
RT-PCR analysis
Total RNA was extracted using the RNeasy Kit (Qiagen), and converted to cDNA using qScriptTM_cDNA_SuperMix (Quanta Biosciences). PCR was performed with primers and iQTM SYBER Green master mix on a CFX96 System (Bio-Rad).
Gapdh: GGTCCTCAGTGTAGCCCAAG, AATGTGTCCGTCGTGGATCT
E-cad: ACACCGATGGTGAGGGTACACAGG, GCCGCCACACACAGCATAGTCTC
Vimentin: TGACCTCTCTGAGGCTGCCAACC, TTCCATCTCACGCATCTGGCGCTC
Snai1: ACTGGTGAGAAGCCATTCTCCT, CTGGCACTGGTATCTCTTCACA
Fn1: CGAAGAGCCCTTACAGTTCCA, ATCTGTAGGCTGGTTCAGGC
Col18a: GCAGTGCCATTCCAAGTTCTC, AACATTCTCTGGGAAGTCTGGT
Mmp14: TTGTCTTCAAGGAGCGATGGT, AGGGAGGCTTCGTCAAACAC
Tgfb: ACGTCACTGGAGTTGTACGG, GGGGCTGATCCCGTTGATT
Ccl2: CACTCACCTGCTGCTACTCA, GCTTGGTGACAAAAACTACAGC
Cxcl12: CTTCAGATTGTTGCACGGCTG, CTCGGGGGTCTACTGGAAAG
Il1b: TGCCACCTTTTGACAGTGATG, ATGTGCTGCTGCGAGATTTG
Il6: AGACAAAGCCAGAGTCCTTCAG, TTAGGAGAGCATTGGAAATTGG
Vegfc: CTTGTCTCTGGCGTGTTCCC, TTCAAAAGCCTTGACCTCGCC
Vegfd: GCCTGGGACAGAAGACCACT, GCAGCAGCTCTCCAGACTTT
Fgf2: GGCTGCTGGCTTCTAAGTGT, TCTGTCCAGGTCCCGTTTTG
Angpt1: TTCCAGAACACGACGGGAAC, TAATTCTCAAGTTTTTGCAGCCAC
Pdgfa: GGAGGAGACAGATGTGAGGTG, GGAGGAGAACAAAGACCGCA
Endothelial cell proliferation assay
Mouse endothelial cells (2H11, ATCC) were seeded in 96-well plates (2×103 cells/well) in 2% FBS medium overnight, and then stimulated with supernatant collected from RFP+ or GFP+ Tri-PyMT cells for 3 days. Cell proliferation was measured with the CellTiter-Glo® Luminescent Kit (Promega).
Orthotopic breast tumor model
RFP+ Tri-PyMT cells were FACS-sorted and injected (5×105 cells/mouse) into the mammary fat pad (#4, right) of 8-week-old female SCID mice. Primary tumors were removed when tumor sizes reach ~1.5cm in diameter. Lung metastasis was analyzed at 2–4 weeks after primary tumor removal.
Tissue processing, Immunofluorescence, and Microscopy
The tumor and lung tissues were fixed in 4% paraformaldehyde overnight, followed by desiccation in 30% sucrose for 2 days. Serial sections (10–20μm) were prepared from O.C.T. embedded blocks. H&E and immunofluorescent staining were performed following standard protocols. Primary antibodies include E-cadherin (DECMA-1, BioLegend), vimentin (sc-7557, Santa Cruz), CD31 (MEC13.3, Biolegend). Fluorescent images were obtained using a Zeiss fluorescent microscope (Axiovert 200M), fitted with an apotome and an HRM camera.
Statistical Analysis
Experiment results were expressed as mean ± SD. Data distribution in groups and significance between groups was analyzed by using the Mann-Whitney T-test in GraphPad Prism software. P values < 0.05 were considered significant.
Results
The fluorescence switch in Tri-PyMT cells precisely reports a specific EMT program on the single-cell level
Tri-PyMT cells were derived from primary tumors of an MMTV-PyMT/Fsp1-Cre/Rosa26mT/mG transgenic mouse. The cells switch their fluorescence from RFP+ to GFP+ in culture with 10% FBS (Fig. 1A), concomitant with changing in morphologies, altering the expression of key EMT markers (including Ecad, Occl, Fn1, Vim, Snail, and Zeb1/2), and gaining mobility and resistance to apoptosis(9). Bulk RNA-sequencing of purified RFP+ and GFP+ cells revealed significant upregulation of “Hallmark Epithelial_Mesenchymal_Transition”, “Angiogenesis” and “Hypoxia” pathways in the GFP+ cells with the gene set enrichment analysis (GSEA) (Supplementary Fig. 1).
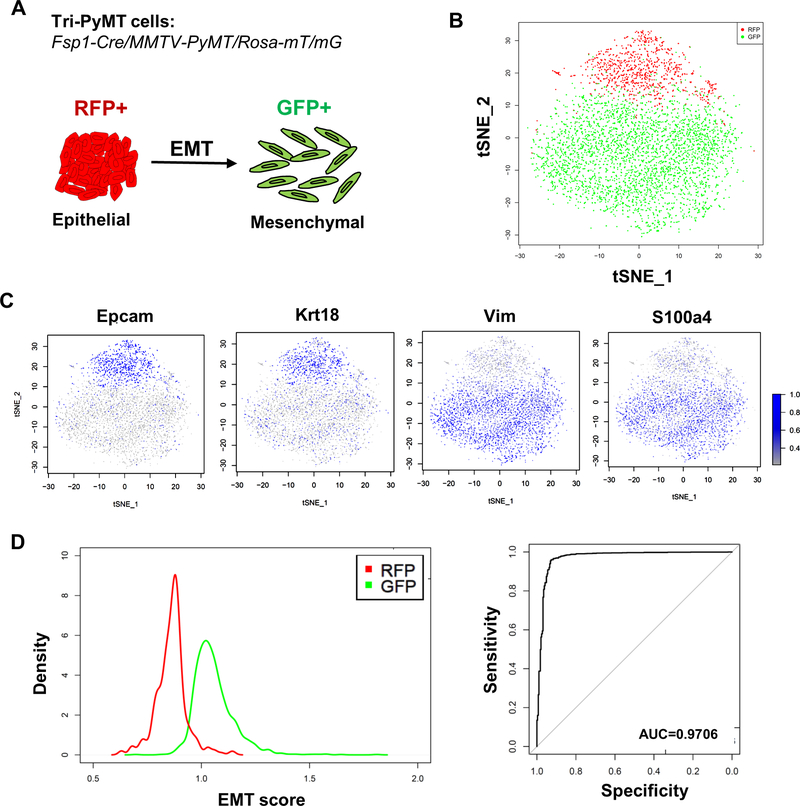
Figure 1. The Tri-PyMT model precisely reports the EMT status of Tri-PyMT cells on the single cell level.
A, A Scheme of the Tri-PyMT EMT lineage tracing model. Epithelial tumor cells will switch their expression of fluorescent marker from RFP+ to GFP+ due to the activation of Fsp1 promoter during EMT. B, The tSNE visualization of RFP+ (in red) and GFP+ (in green) Tri-PyMT cells with the top 10 principal components (PCs) of their scRNA-seq data. C, The relative expression of epithelial markers (EpCam and Krt18) and mesenchymal markers (Vim and S100a4) are projected to individual cells. D, Plots show the distribution of individual RFP+ (in red) and GFP+ (in green) cells according to their EMT scores (left), and the quantification of the sensitivity and specificity (GFP+ versus RFP+) with ROC curve (right). AUC = 0.9706.
To evaluate the single-cell EMT statuses, we performed scRNA-seq with Tri-PyMT cells. Interestingly, RFP+ and GFP+ cells were preferentially clustered on opposite sides in the dimensionality reduction tSNE plot (Fig. 1B). The adjacent and continuous localization of the two clusters suggested a gradient differentiation from RFP+ to GFP+ according to their overall transcriptome. Projecting the expression of EMT markers on the tSNE plot revealed that the epithelial (i.e. Epcam and Krt18) and mesenchymal markers (i.e. Vim and S100a4) were largely confined to RFP+ and GFP+ subpopulations, respectively (Fig. 1C).
We next performed differential expression analyses of RFP and GFP cells. With criteria of log fold change ≥0.25 and p-value <0.05, 241 genes in RFP+ cells and 324 genes in GFP+ cells were differentially expressed (Supplementary Table 1). Many well-characterized epithelial genes (i.e. Epcam, Cdh1, Krt14, Cldn7, and Jup), and mesenchymal genes (i.e. Vim, Fn1, S100a4, Spp1, Col8a1, Cxcl12, Prrx1and Zeb1/2) were included in the signature. When compared with published EMT hallmark gene sets, the EMT signature in Tri-PyMT cells overlapped with 61 out of 200 genes in the “Hallmark_EMT” of MSigDB database, and with 50 out of 359 genes of the dbEMT gene sets(15). Of note, only 39 overlapping genes exist between these two databases. These results suggested that a specific gene set was activated during the RFP-to-GFP transition in Tri-PyMT cells.
To assess the efficiency of the Tri-PyMT model, we quantified the EMT status of individual cells. Basically, we calculated the sum expression level of identified mesenchymal and epithelial genes of each cell. The ratio of mesenchymal and epithelial sums was defined as the EMT score. Accordingly, epithelial-like cells were expected with low EMT scores, while mesenchymal-like cells would display high scores. Density plots of RFP+ and GFP+ cells showed a well-defined separation with minor overlaps (Fig. 1D, left). To evaluate the performance of the fluorescence switch in predicting the EMT status of single cells, we used their EMT scores to build a binary classifier. ROC curve revealed a high specificity and sensitivity, with an accuracy of up to 97% (Fig. 1D, right).
Together, the scRNA-seq analyses of Tri-PyMT cells revealed a continuous EMT spectrum and a specific EMT signature of this model. The fluorescence switch reported the EMT statuses of single cells with high sensitivity and specificity. Therefore, in the Tri-PyMT model, RFP+ and GFP+ cells would well represent the “pre-EMT” and “post-EMT” subpopulations, respectively.
The differential contributions of pre-EMT and post-EMT cells in tumor progression
To further evaluate the roles of pre-EMT and post-EMT tumor cells in metastasis, we established orthotopic model by implanting sorted RFP+ Tri-PyMT cells in the mammary fat pad of mice. Histological (Fig. 2A) and flow cytometry (Fig. 2B) analyses revealed that the primary tumors comprised mostly RFP+ cells at 4 weeks after implantation. Interestingly, a relatively higher percentage of GFP+ was observed in the early stage tumors (~15% at 1 week, Supplementary Fig.2A, 2B). This percentage decreased to 1–2% at 4 weeks, suggesting that post-EMT tumor cells may play a more important role in the early stage of tumor progression. Of note, post-EMT tumor cells were also detected in the blood of tumor-bearing mouse, which comprised approximately 10% of total circulating tumor cells (Supplementary Fig 2C).
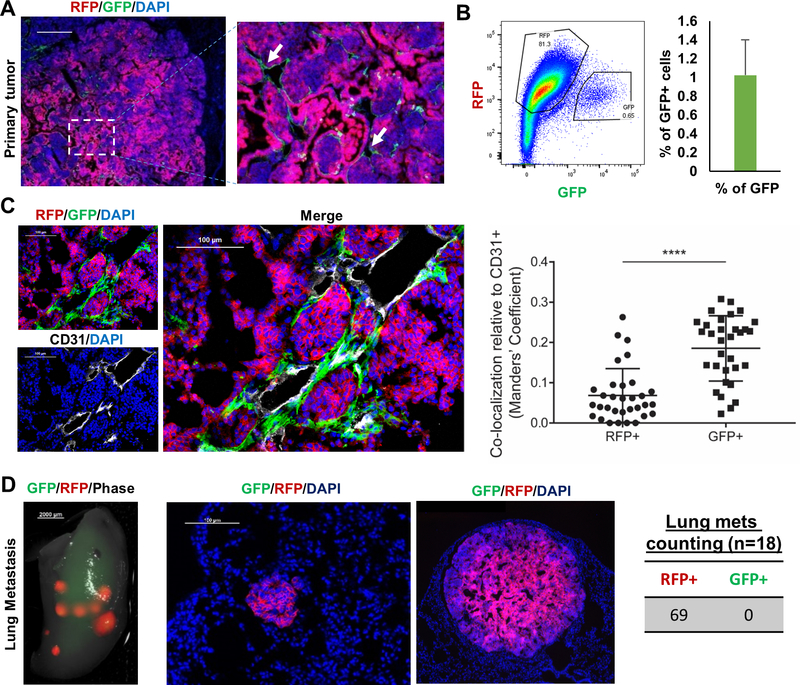
Figure 2. The differential contributions of pre-EMT (RFP+) and post-EMT (GFP+) cells in the Tri-PyMT tumor progression.
A, Fluorescent images of Tri-PyMT primary tumor. Post-EMT cells (GFP+) are indicated by arrows. B, Flow cytometry plot and quantification of GFP+ tumor cells (n=5). C, Immunefluorescent image and quantification plot showing the co-localization of EMT (GFP+) tumor cells and tumor vasculature (CD31+). D, Fluorescent images showing the predominant role of RFP+ cells in lung metastases (n=18).
To further evaluate the fidelity of Tir-PyMT model in vivo, we performed scRNA-seq with sorted RFP+ and GFP+ cells from the orthotopic primary tumors. Two major clusters representing epithelial and mesenchymal phenotypes were detected with dimensionality reduction tSNE analysis (Supplementary Fig. 3). Consistently, RFP and GFP expression were closely associated with the epithelial (Ecad, Epcam, and Krt18) and mesenchymal (Vim, Fn1, s100a4, Snail, and Zeb1) markers, respectively. These results strongly suggested that the Tri-PyMT model reported the transition of epithelial-to-mesenchymal phenotype with high fidelity.
In the primary tumor, post-EMT (GFP+) cells were detected in close proximity to the necrotic region (Supplementary Fig.4) and at the edge of pre-EMT (RFP+) cell clusters, directly connecting with tumor stroma (Fig. 2C). Immunofluorescent staining of CD31 (endothelial cell marker) revealed that the post-EMT (GFP+) cells were more preferentially adjacent to tumor vasculatures than pre-EMT (RFP+) cells (Fig. 2C). Consistent with the upregulated “Angiogenesis” pathway in GSEA (Supplementary Fig.1), RT-PCR analyses of proangiogenic factors confirmed that the expression of Vegfd, Pdgfa, Angpt1, IL6, Tgfb, Cxcl12, and Fn1 were upregulated in GFP+ cells when compared with that in RFP+ cells (Supplementary Fig.5A). Moreover, supernatant collected from GFP+ cells significantly enhance the proliferation of mouse endothelial (2H11) cells (Supplementary Fig.5B). These results suggested that post-EMT cells account for a minority of primary tumor cells, however, they may play supportive roles in tumor angiogenesis.
Consistent with previous studies, lung metastases were mostly derived from the pre-EMT, but not the post-EMT compartment (Fig. 2D). RFP-dominated metastases were observed in all 69 lung nodules from 18 animals bearing Tri-PyMT tumors, suggesting a robust advantage of the pre-EMT in lung metastasis formation.
The important role of the pre-EMT tumor cells in metastasis formation was also confirmed in MMTV-PyMT/Vim-CreERT/Rosa26mT/mG model(9), in which the fluorescent marker switch was driven by Vimentin promoter. Both primary tumors and lung metastases were mainly composed of RFP+ cells (Supplementary Fig. 6), indicating the dominant role of pre-EMT cells in metastasis.
Pre-EMT tumor cells possess advantages in lung colonization
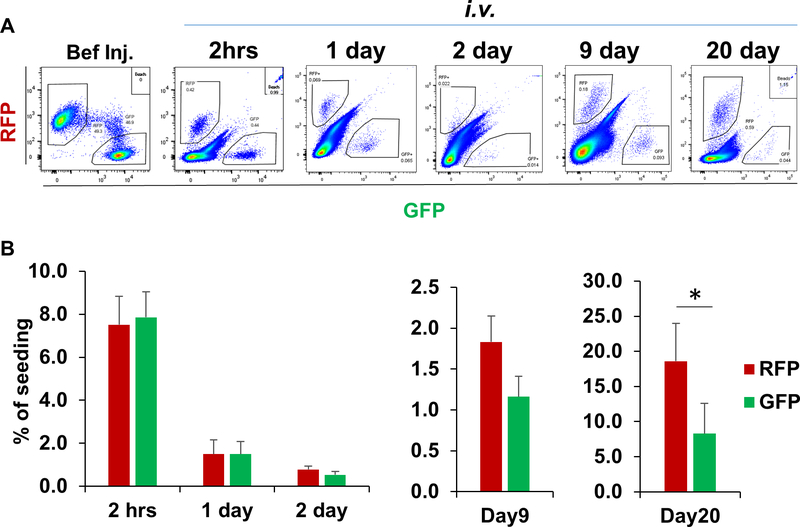
The predominant ratio of pre-EMT to post-EMT cells in the primary tumor may not be sufficient to explain the outgrowth of RFP+ metastases in the lung. We further asked whether there were differences between pre-EMT and post-EMT cells in seeding and colonizing at the metastatic site. A comparative metastasis assay was performed with sorted RFP+ and GFP+ Tri-PyMT cells (2.5×105 cells of each) from culture via tail vein injection. Lung-seeding tumor cells were quantified via flow cytometry at various time points (Fig. 3A). We found that both RFP+ and GFP+ tumor cells seeded the lung with similar low efficiencies on 2nd day (0.78 ± 0.15% and 0.53 ± 0.15%, respectively, Fig 3B). However, a significantly higher number of RFP+ than GFP+ cells were detected in the lung on 20th day after injection (Fig. 3B). These results suggest that both pre-EMT and post-EMT cells possess similar seeding capacities, whereas the pre-EMT cells have advantages in the metastasis outgrowth.
Figure 3. Characterizing the seeding and outgrowth of pre-EMT (RFP+) and post-EMT (GFP+) tumor cells in the lung.
RFP+ and GFP+ Tri-PyMT cells were sorted from culture, remixed at 1:1 and injected into mice through tail vein. Flow cytometry plots (A) and quantification (B) show the recovery of RFP+ and GFP+ cells at different time (2 hrs, 1, 2, 9 and 20 days) after tail vein injection (* P < 0.01, n = 3–6 mice).
Post-EMT tumor cells formed secondary tumors through MET
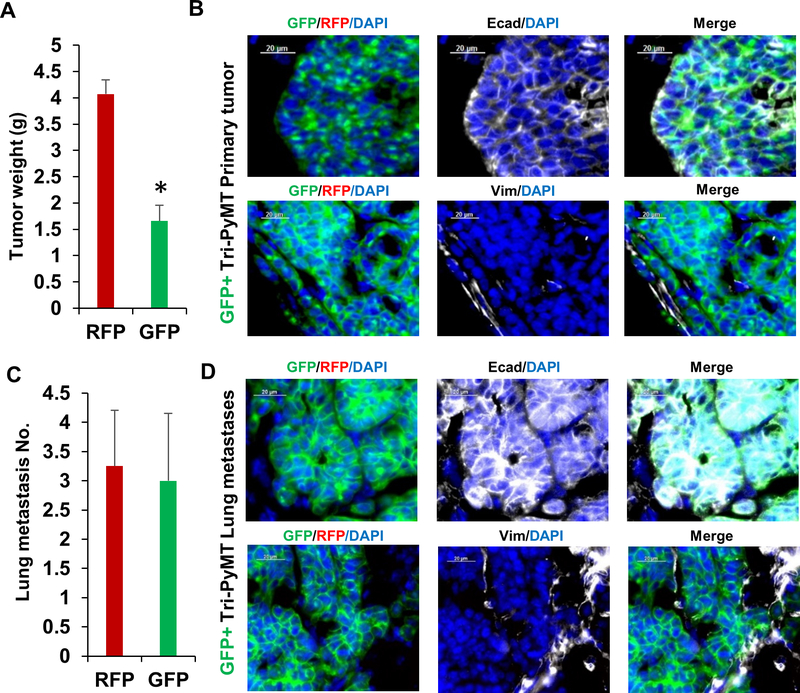
Next, we asked whether the post-EMT cells were capable to form secondary tumors. GFP+ Tri-PyMT cells were sorted from culture and injected into the mammary fat pad of animals. We found that they were capable, however, formed secondary tumors at a slower growth rate as compared with their RFP+ counterparts (Fig. 4A). At the 4th week after inoculation, the GFP+ tumors were approximately 50% smaller than the RFP+ ones. Immune staining showed that the GFP+ cells regained epithelial phenotypes (Ecad+/Vim-, Fig. 4B). Given the mesenchymal features of GFP+ cells in culture, these results suggested that the post-EMT tumor cells underwent “MET” to form tumors. Importantly, similar numbers of lung metastases were detected in animals bearing either GFP+ or RFP+ tumors (Fig. 4C). The GFP+ metastatic lesions also exhibited epithelial phenotypes (Ecad+/Vim-, Fig. 4D). These results indicated that the post-EMT cells were not permanently committed to the mesenchymal lineage. Instead, they still possessed EMT plasticity to regain epithelial phenotype for initiating secondary tumors and metastases.
Figure 4. Post-EMT Tri-PyMT cells form secondary primary tumors and metastases with epithelial phenotype.
A, Weight of GFP+ and RFP+ primary tumors at 4 weeks post-injection, n=5, *p<0.05. B, E-cadherin and vimentin staining (white pseudocolor) of GFP+ Tri-PyMT primary tumors. C, Quantification of lung metastases in mice bearing either RFP+ or GFP+ primary tumor, n=5. D, Fluorescent images of Ecad and Vim staining of the GFP+ Tri-PyMT lung metastases.
Discussion
The unexpected observations that EMT is not required for tumor metastasis with Tri-PyMT model urged us to carefully evaluate the fidelity of this EMT lineage tracing model. We performed the scRNA-seq of Tri-PyMT cells from both culture and primary tumors. A specific EMT signature was identified as comparing the RFP+ with GFP+ cells on the single-cell level. This signature contains many well-characterized EMT markers such as Vim, Fn1, S100a4, Prrx1 and Zeb1/2. However, its overlap with the published EMT gene sets (such as the Hallmark_EMT in MSigDB and dbEMT) was limited, suggesting the diversity of EMT program in different tumors. We preferred to describe the fluorescence switch of Tri-PyMT cells as a specific EMT program rather than a partial or hybrid EMT status, due to the lack of standardized criteria of these statuses(13). Indeed, the limited overlaps of the published EMT gene sets also suggest that different EMT programs are adopted by different tumors.
In the Tri-PyMT model, GFP+ cells were derived from RFP+ cells, sharing the same origin with their precursors. We assessed the sensitivity and specificity of Tri-PyMT model in reporting the overall EMT status at the single-cell level. An evident separation of RFP+ and GFP+ subpopulations was observed via an EMT-scoring assay, which suggested the fluorescent marker switch reported a gain of mesenchymal features in Tri-PyMT cells with high accuracy.
Conflicting conclusions of EMT in metastasis are, at least in part, due to the lack of standardized EMT scoring system. Efforts have been made to describe the variety of EMT states existing in different tumor types. However, the quantified EMT statuses did not necessarily correlate with overall survival of cancer patients(15,16). The extremely mesenchymal phenotype did not favor the metastasis development in breast cancer patients(16). We also evaluated the EMT statuses of Tri-PyMT cells with the EMT scoring metric(16). Both RFP+ and GFP+ cells were categorized as E/M hybrid phenotype (Supplementary Fig. 7). Of note, such E/M hybrid statuses were referred to NCI60 cell lines, which include a wide range of fully differentiated epithelial and mesenchymal cells. This provided another clue that the EMT program could be tumor type-dependent. Importantly, the Tri-PyMT model enabled us to compare tumor cells with different EMT statuses within the same tumor and assess their relative contributions to metastasis. The GFP+ cells were clearly more askew to the mesenchymal phenotypes with higher Vim/Cdh1 ratio and lower Cldn7 expression when compared to RFP+ cells (Supplementary Fig.7). The lack of GFP+ metastases in Tri-PyMT model suggested that further activation of EMT programming did not grant advantages in metastasis.
With the Tri-PyMT model, we confirmed that post-EMT tumor cells were not the metastasis-initiating cells. These results do not decline the biological contributions of EMT in tumor progression. Instead, EMT is believed to endow tumor cells with many metastasis-related features including migration, invasion and apoptosis resistance(10,12,17,18). Cooperations between EMT and non-EMT tumor cells were also demonstrated(19,20). While the EMT tumor cells invade and degrade extracellular matrix, the non-EMT tumor would follow and metastasize to the secondary organ. The EMT tumor cells could also assist non-EMT tumor cells to metastasize through a non-cell autonomous activation of the GLI signal(20). In the Tri-PyMT tumors, post-EMT cells co-localize with tumor vasculature and secrete more proangiogenic factors, suggesting their supportive role in tumor angiogenesis. Nevertheless, our results do not support the hypothesis that the post-EMT tumor cells are the metastasis-initiating cells. The biological involvement of post-EMT tumor cells in metastasis still need further elucidation.
Supplementary Material
Acknowledgments
This work was supported by NIH (5R01CA205418), the Neuberger Berman Foundation Lung Cancer Research Center, and by generous donations to the Division of Thoracic Surgery to N.K.A. JTS and STCW support is from NIH U01CA-188388, John S Dunn Research Foundation, TT & WF Chao Foundation, and Johnston Estate Endowment.
Footnotes
The authors declare no potential conflicts of interest
References
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. United States 2009. p 871–90. [DOI] [PubMed] [Google Scholar]
- 2.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. England 2012. p 3471–86. [DOI] [PubMed] [Google Scholar]
- 3.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525:256–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166:21–45 [DOI] [PubMed] [Google Scholar]
- 7.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev 2016;35:645–54 [DOI] [PubMed] [Google Scholar]
- 8.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 2013;27:2192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015;527:472–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, et al. Upholding a role for EMT in breast cancer metastasis. Nature 2017;547:E1–e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer KR, Altorki NK, Mittal V, Fischer Gao D., et al. reply. Nature 2017;547:E5–e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol 2017;11:28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigore AD, Jolly MK, Jia D, Farach-Carson MC, Levine H. Tumor Budding: The Name is EMT. Partial EMT. J Clin Med 2016;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, et al. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med 2014;6:1279–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George JT, Jolly MK, Xu S, Somarelli JA, Levine H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res 2017;77:6415–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, et al. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev Cell 2018;45:681–95.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Lee DK, Feng Z, Bu W, Li Y, Liao L, et al. Breast tumor cell-specific knockout of Twist1 inhibits cancer cell plasticity, dissemination, and lung metastasis in mice. Proc Natl Acad Sci U S A 2017;114:11494–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, et al. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res 2008;68:10377–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun 2017;8:15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.