Abstract
BACKGROUND
Asymmetric arginine dimethylation of histone H4R3 to H4R3me2a by protein arginine methyltransferase 1 (PRMT1) has been implicated to play a key role in gene activation throughout vertebrates. PRMT1 knockout in mouse leads to embryonic lethality. This and the uterus-enclosed nature of the mouse embryo make it difficult to determine the development role of PRMT1 in mammals.
METHODS
We took advantage of the external development of the diploid anuran Xenopus tropicalis and adapted the TALEN genome editing technology to knock out PRMT1 in order to investigate how PRMT1 participates in vertebrate development.
RESULTS
We observed that PRMT1 knockout had no apparent effect on embryogenesis because normally feeding tadpoles were formed, despite the reduced asymmetric H4R3 di-methylation (H4R3me2a) due to the knockout. However, PRMT1 knockout tadpoles had severely reduced growth even with normal growth hormone gene expression. These tadpoles were also stalled in development shortly after feeding began at stages 44/45 and died within 2 weeks, well before the onset of metamorphosis. In situ analyses revealed broad cessation or drastic reduction in cell proliferation in diverse organs including the eye, brain, spinal cord, liver, and intestine.
CONCLUSIONS
Our findings suggest that PRMT1 is not required for embryogenesis but is a key regulator for normal progression of vertebrate development and growth.
GENERAL SIGNIFICANCE
The similarities and differences between PRMT1 knockout Xenopus tropicalis and mouse suggest that two distinct phases of vertebrate development: early embryogenesis and subsequent growth/organ maturation, have different but evolutionally conserved requirement for epigenetic modifications.
Keywords: Epigenetics, histone modification, activation mark, thyroid hormone receptor, organogenesis, Xenopus tropicalis
1. INTRODUCTION
The core histones, consisting of H2A, H2B, H3 and H4, are subjected to a large number of post-translational modifications, including acetylation, methylation, phosphorylation, and ubiquitylation, which can influence many cellular processes via gene activation or silencing [1–4]. Among them, arginine methylation of histone tails can promote or prevent the docking of key transcription factors, and thus contribute to transcriptional activation or repression depends on the methylated residues and positions [5]. There are three main forms of methylated arginine: monomethylarginines (MMA); asymmetric dimethylarginines (ADMA); and symmetric dimethylarginines (SDMA), which are products of protein arginine methyltransferases (PRMTs). There are nine known vertebrate PRMT members (PRMT1–9) and are classified as type I, type II, or type III enzymes. Type I and type II enzymes catalyze the formation of MMA, which can be used by type I PRMTs (PRMT1, 2, 3, 4, 6 and 8) to further catalyze the production of ADMA, or type II PRMTs (PRMT5 and 7) to further catalyze the formation of SDMA [5, 6]. PRMT7 is referred as type III enzyme because it has only monomethylation activity [5, 7]. PRMT1 is the most abundant type I methyltransferase in mammals and methylation by PRMT1 has been implicated in diverse cellular processes such as interferon signaling pathway and transcriptional regulation through inhibition of FOXO1 phosphorylation by AKT [5, 8, 9]. PRMT1 also acts as a coactivator for different nuclear hormone receptors including thyroid hormone (TH) receptor (TR) [10–13].
While the molecular properties and biological functions of PRMT1 have been studied extensively in vitro and in cultured cells, much less is known about its involvement in vertebrate development. Gene knockout studies in mouse have shown that no homozygous PRMT1 knockout embryos can be detected by E8.5, indicating that PRMT1 is essential for mouse embryogenesis [8]. However, the mechanism underlying the epigenetic regulation during early vertebrate development has been difficult to study due to the uterus-enclosed nature of mammalian embryos.
The development of anuran amphibians such as Xenopus tropicalis offer a unique opportunity to study epigenetic regulation of vertebrate development, especially the so-called postembryonic development, a period around birth that is regulated by TH and when many organs/tissues mature into the adult form [14, 15]. We have previously shown that PRMT1 is upregulated during Xenopus laevis metamorphosis [16], a process resembling postembryonic development in mammals [14, 15]. We have further demonstrated that PRMT1 functions as a TR coactivator and transgenic overexpression of PRMT1 in Xenopus laevis accelerates frog metamorphosis and increases adult epithelial stem cells proliferation during intestinal remodeling [16, 17]. Here, we have investigated the role of endogenous PRMT1 gene in Xenopus tropicalis, a diploid anuran species that is highly related to Xenopus laevis [18], by using TALEN genome editing technology to knock out PRMT1. We showed that knockout of PRMT1 had no observable effect on embryogenesis as homozygous PRMT1 knockout animals developed normally into feeding stage tadpoles (stage 44/45 or 4 days old). On the other hand, all homozygous PRMT1 knockout animals died within 2 weeks after fertilization and were stalled developmentally at stage 46, well before the onset of metamorphosis at stage 54. The homozygous knockout animals also had severe growth inhibition despite normal levels of growth hormone gene expression. Furthermore, the knockout animals had reduced levels of H4R3me2a methylation and drastically reduced cell proliferation in broad organs/tissues. Our data suggest that histone H4R3me2a methylation by PRMT1 is essential for tadpole growth and development via regulating cell proliferation in diverse organs but is not required for embryogenesis.
2. MATERIALS AND METHODS
2.1. Animal rearing and staging
All animals were cared for and treated as approved by the Animal Use and Care Committee of Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), U.S. National Institutes of Health (NIH). Adult X. tropicalis were purchased from NASCO (Fort Atkinson, WI, USA). Embryos were staged according to the description for X. laevis [19]. Embryos were reared in 0.1M Marc’s modified Ringers (MMR) in agar coated petri dish for 4 d at 25°C, and then transferred to a large volume (9-L) container as described [20, 21]. Different groups of tadpoles were reared at a similar density. For tadpole survival studies, tadpole density was maintained the same across treatment groups by removing tadpoles as necessary. To account for these changes, the tadpole survival rate was calculated by dividing the number of live tadpoles at a given day by the number of live tadpoles on the day earlier and then multiplying the resulting value by the survive rate for the day earlier.
2.2. TALEN assembly and TALEN mRNA preparation
A TALEN pair targeting X. tropicalis PRMT1 were assembled as described (16, 33, 34). The PRMT1 TALEN left (TALEN-L) arm recognizes the sequence GAATGGATGGGATATTGTCT and the TALEN right (TALEN-R) arm recognizes the sequence CGATAGATAACAGTGTTCAG in the PRMT1 coding region. To generate the TALEN mRNA in vitro, the individual TALEN plasmid was linearized with NotI. Capped RNA was produced by using the linearized DNA and the Ambion (Grand Island, NY, USA) in vitro transcription kit. After removing the DNA template by DNaseI digestion, capped RNA was purified with RNAeasy kit (Qiagen, Valencia, CA, USA).
2.3. Generation of PRMT1 knockout Xenopus tropicalis animals by using TALEN genome editing technology and genotyping
Mature adult X. tropicalis frogs, a few females and a male, were primed 1 day before the experiment with 20 U of human chorionic gonadotropin (hCG; Novarel; Ferring Pharmaceuticals Inc. Parsippany, NJ, USA). The injected frogs were boosted with 200 U of hCG on the second day. Just before the females started to lay eggs, the male was sacrificed to obtain testes. A sperm suspension was prepared in 300 μl 1×MMR by using one testes. For in vitro fertilization, freshly squeezed eggs from an hCG-injected female were mixed with the sperm suspension for about 2 min. The sperms in the mixture were then activated by diluting the mixture to 0.1×MMR. The fertilized eggs were dejellied in 3% cysteine solution at pH 8.0. After washing with 0.1×MMR several times, the fertilized eggs were placed on an agar-coated plate. For TALEN mRNA injection, equal amounts of the TALEN-L and -R arm mRNAs were mixed and injected into the fertilized egg at 400 pg/egg for each mRNA.
The TALEN-injected embryos were reared to adults (F0-generation frogs). A sexually mature F0 frog was mated with a wild type frog, and their offspring were screened to identify PRMT1 heterozygous mutant tadpoles [PRMT1(+/−)]. After PRMT1(+/−) mutants were sexually mature (F1 frogs), female and male mutant frogs were primed with 20 U of hCG (Novarel), one day before egg laying. They were then boosted with another injection of 200 U of hCG on the second day for natural mating to obtain PRMT1 total knockout [PRMT1(+/−)] animals (F2 generation). The resulting fertilized eggs/embryos were collected and reared for 3–4 days at 25°C to reach the onset feeding stage (stage 45). The tadpoles were then transferred to a 4-L container and fed.
Tadpoles were anesthetized with MS222 for photography, tail clipping, and body length measurement. For genotyping, the tadpole tail tip (about 5 mm or less) was clipped and lysed in 20 μL QuickExtract DNA extraction solution (Epicentre) at 65°C for 20 minutes. After incubating at 95°C for 5 minutes, 1 μl of the DNA extraction solution was immediately used for genotyping by PCR. For the F1 animals, genotyping was performed with PCR amplification of the PRMT1 targeted regions (Supplemental Table 1). After the sequence analysis, an 8 base out-of-frame deletion PRMT1 line (Fig. 1) was chosen for further studies. For F2 generation, the genotyping of PRMT1 gene was done by PCR with two primer sets to detect wild type and mutant allele respectively: forward primer f1, 5′- GGGATATTGTCTGTTCTATGAATC −3′ and reverse primer R, 5′- GTGTAGCACGATCTGGAAATATTAACCCA-3′, forward primer F, 5′- GAGTGCTCAAGTATATCTGACTATGCCA −3′ and reverse primer r1, 5′- GATAACAGTGTTCAGCATAGAAC-3′, for 33 cycles of 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 20 seconds. The PCR products were analyzed by gel electrophoresis
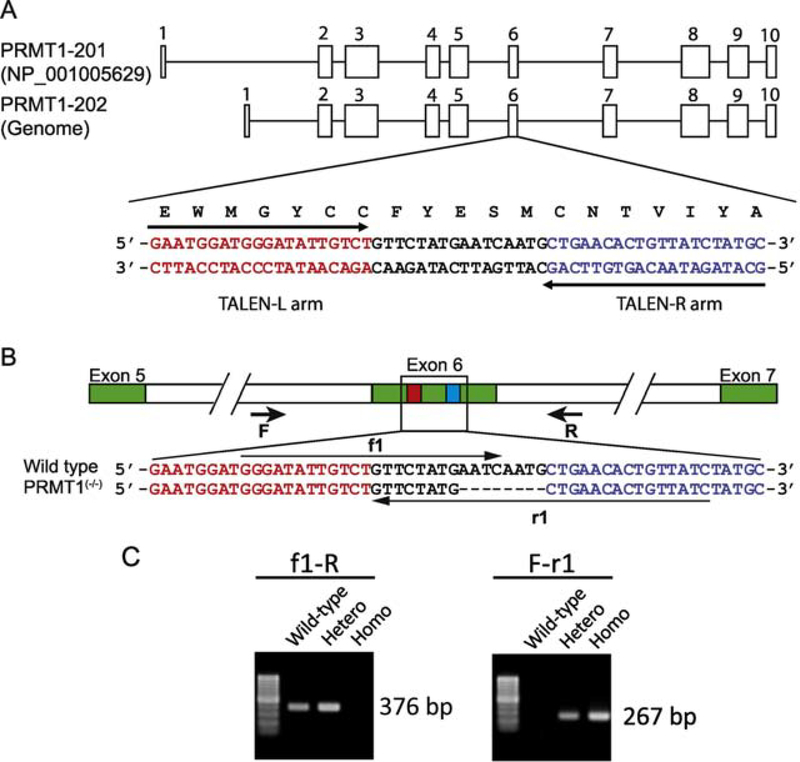
Figure 1. Generation of PRMT1 knockout Xenopus tropicalis.
(A) PRMT1 genomic structure and design of TALEN against X. tropicalis PRMT1 locus. There are two PRMT1 transcripts encoded by 10 exons each. PRMT1 specific TALEN-arms (marked with arrows) were designed to target E153 in the functional domain for di-methylation activity, in the exon 6.
(B) Schematic diagram depicting sequences of the TALEN targeted region in the wild type and a PRMT1 mutant (8 bases deletion) line. Red box and blue box in exon 6 indicate left and right TALEN arms, respectively. Arrows represent primers used for genotyping: the forward primer F and f1, and the reverse primer R and r1. Green boxes are exons.
(C) Representative examples of genotyping by PCR. Genotyping was carried out on genomic DNA by using two different primer set, primer f1 and primer R for detecting wild type allele and primer F and primer r1 for detecting the mutant allele, respectively. The presence of only wild type PCR product (376 bp) or mutant PCR product (267 bp) in the PCR reaction indicates the animals as wild type or a homozygous mutant, respectively, while the presence of both PCR products indicates a heterozygous mutant.
2.4. RNA extraction and quantitative RT-PCR
Total RNA of the intestine, tail, or limb was extracted with RNeasy® Mini Kit 250 (Qiagen). The homogenates of individual tissues from at least five animals, or five whole animals, of each genotype were combined together for RNA extraction. The RNA concentration was measured by using a NanoDrop (Thermo Scientific). The same amount of RNA from each of the three genotypes (PRMT1: wild-type, heterozygous and homozygous knockout) was reverse-transcribed with the QuantiTect reverse transcription kit (Qiagen). The cDNA was analyzed by qPCR by using the SYBR Green method. The PCR primers for the internal control genes odc (ornithine decarboxylase) and rpl8 (ribosomal protein L8) were described previously [22] (note that odc and rpl8 mRNA expression levels were similar in all genotypes; data not shown). All gene expression data were normalized against that of the internal control gene odc or rpl8. The expression analyses were performed at least twice, with similar results. The primer sequences are listed on Supplemental Tables 1–2.
2.5. Protein extraction and Western blot
Xenopus tropicalis embryos at indicated ages/stages were pooled together and homogenized into 10 μl per embryo of M-PER solution with proteinase inhibitor cocktail (Roche, Basel, Switzerland) and 50 μM GSK-LSD1 (Sigma-Aldrich). The lysates were centrifuged at 4°C, 12,000 g for 20 min. The supernatants were mixed with equal volume of 2× loading buffer and boiled for 5 min. Western blot was carried out as previously described by using antibodies against beta-actin (diluted 1:1000; Millipore-Sigma), histone H4 (diluted 1:1000, Sigma-Aldrich), H4R3me2a (diluted 1:200; Millipore-Sigma) or PRMT1 (diluted 1:250; Sigma-Aldrich).
2.6. 5-Ethynyl-2’-deoxyuridine (EdU) labeling for cell proliferation
EdU staining was performed as described (35) with a few modifications. Briefly, genotyped, 9-day-old wild type and PRMT1 homozygous knockout tadpoles were kept in the 10 ml water with 0.25 μg/mL EdU in 6-well cell culture plate. After 16 hours of treatment at 25°C, the tadpoles were sacrificed, and the whole body was fixed in 4% PFA/PBS and processed for paraffin-sectioning. Tissue sections cut at 5 μm were subjected to EdU staining by using the Click-iT Plus EdU Alexa Fluor 594 Imaging kit (Thermo Fisher Scientific). EdU positive areas were measured by using ImageJ software (National Institutes of Health).
2.7. Statistical analysis
Data are presented as mean ±SE. The significance of differences between groups was evaluated by one-way ANOVA followed by Bonferroni multiple comparison test or Student’s t test by using Prism 5 (GraphPad Software).
3. RESULTS
3.1. Generation of Xenopus tropicalis PRMT1 total knockout animals by using TALEN genome editing technology
To investigate the role of endogenous PRMT1 during vertebrate development, we adapted transcriptional activator–like effector nuclease (TALEN) technology to generate mosaic PRMT1 knockdown frogs (F0). We designed a TALEN nuclease made of a pair of left and right arms targeting E153 in exon 6 of X. tropicalis PRMT1 that is critical for PRMT1 methylase activity (Fig. 1A) [23]. We microinjected mRNAs encoding the 2 TALEN arms into one cell stage embryos to generate the F0 animals with mosaic mutations in PRMT1. Analysis of the mutation in the target region of the F0 animals showed 20–30% mutation efficiency (data not shown). We raised some of these F0 animals to sexual maturity and then crossed them with wild type animals to generate F1 PRMT1 heterozygous animals. After genotyping the F1 animals, we chose one mutant line with an 8 base deletion, which is an out of frame mutation and thus inactivates the gene, for further analysis (Fig. 1B). We produced PRMT1 homozygous knockout animals in the F2 generation by intercrossing F1 frogs and identified them by genotyping PCR (Fig. 1C).
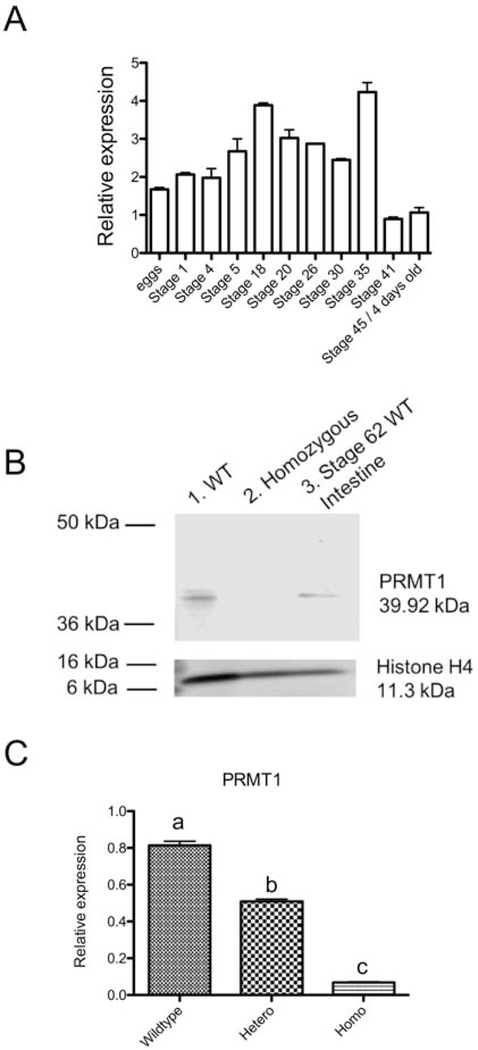
We confirmed whether we indeed had complete PRMT1 knockout by using western blot analysis and quantitative RT-PCR. First, we determined PRMT1 mRNA level during embryogenesis by RT-PCR analysis of total animal RNA (Fig. 2A). PRMT1 mRNA was present maternally in the egg and was upregulated slightly during embryogenesis. After hatching at stage 35, the PRMT1 mRNA levels were reduced significantly by feeding stage (stage 45) (Fig. 2A). Second, we analyzed the protein level in tadpoles. Total protein was isolated from 10-day old tadpoles or the intestine of tadpoles at stage 62, the climax of metamorphosis when PRMT1 is highly expressed in the intestine [16, 17]. Western blot result showed that PRMT1 was expressed expectedly in the wild type tadpoles or the intestine at the metamorphic climax (Fig. 2B). However, PRMT1 protein was not detected in the homozygous mutant animals (Fig. 2B), confirming the complete knockout of PRMT1 in the homozygous mutant animals. Interestingly, when we analyzed PRMT1 mRNA level on total tadpole RNA in wild-type, heterozygous and homozygous PRMT1 mutant animals with a primer pair located in the 5’-untranslated region [16], we observed that PRMT1 mRNA level was very low in the homozygous mutant animals (Fig. 2C) and was significantly reduced in the heterozygous animals, suggesting that most of the mutant PRMT1 mRNA underwent nonsense-mediated RNA decay due to the out of frame deletion.
Figure 2. TALEN-mediated mutation leads to reduction in PRMT1 mRNA level and the loss of PRMT1 protein.
(A) RT-PCR analysis of PRMT1 mRNA level during Xenopus tropicalis embryogenesis. PRMT1 mRNA levels were normalized against that of the control gene ornithine decarboxylase. PRMT1 mRNA levels rose slightly until end of embryogenesis at stage 35, the tailbud stage when hatching begins. It then dropped to lower levels by stage 45 (about 4 day old), when tadpole feeding begins.
(B) PRMT1 protein is absent in PRMT1 homozygous tadpoles. Proteins were extracted from whole body of 12-day-old wild type and PRMT1 homozygous tadpoles. Western blot analysis was performed to determine the protein levels of PRMT1 and histone H4 (loading control). Lane 3 shows the protein sample from wild type intestine at stage 62 as a positive control.
(C) TALEN-induced PRMT1 mutation results in reduced PRMT1 mRNA level in tadpoles. Total RNA was isolated from the intestine of wild type, and PRMT1 heterozygous and homozygous knockout tadpoles at 10 days of age and used for real-time RT-PCR analysis of PRMT1 expression. The mRNA levels were normalized against that of rpl8. The groups included 4 wild, 5 PRMT1 heterozygous, and 5 PRMT1 homozygous animals. These results were plotted with the mean, marked as a line, and SE. Different lower-case letters denote statistically significant differences (P < 0.01) as determined by Bonfferoni comparison test.
3.2. Xenopus tropicalis PRMT1 is essential for tadpole development and growth during the postembryonic development
The homozygous PRMT1 knockout tadpoles had normal morphology at stage 45, 4 days after fertilization and the onset of tadpole feeding (Fig. 3A), suggesting that PRMT1 knockout did not affect Xenopus embryogenesis. To determine if PRMT1 knockout affected tadpole development, we genotyped 30 randomly selected tadpoles generated from mating of two PRMT1 heterozygous knockout frogs every two days starting 4 days after fertilization (feeding stage). The results in Fig. 3B showed that PRMT1 homozygous knockout tadpoles were present at 12 days of age but not at 14 days of age, whereas the wild type and the heterozygous tadpoles were present at 14 days of age with the expected Mendelian ratio. Furthermore, of the 180 tadpoles randomly selected for genotyping during the two week period, a Mendelian distribution was observed for the three genotypes (48:95:37, or a ratio of 1.1:2.1:0.82). Thus, PRMT1 is not required for embryogenesis but is essential for tadpole survival up to two weeks of age.
Figure 3. PRMT1 knockout leads to tadpole lethality.
(A) Representative photos of 4-day-old tadpoles. Ventral view of a wild-type (upper panel) and PRMT1 homozygous knockout (lower panel) tadpole. Bars: 1 mm.
(B) Genotype distribution of tadpoles at the age of 4, 6, 8, 10, 12 and 14 days. 30 tadpoles were randomly selected and genotyped by PCR (Figure 1C) at indicated age (days). Note that no homozygous knockout animals were found at day 14.
(C) PRMT1 knockout leads to lethality within two weeks after fertilization. Tadpoles were genotyped at 4 days of age and the survival rate of wild-type (N=35), heterozygous (PRMT1(+/−), N=83) and homozygous (PRMT1(+/−), N=32) was determined from the age of day 4 to day 14. Note that PRMT1(+/−) tadpoles started to die around day 12 and completely died by day 14.
To determine when the homozygous knockout tadpoles die, 150 randomly selected F2 siblings were kept in a 9-L container together and the dead ones were picked up each day for 3 weeks. The dead ones and the remaining live ones were all genotyped by PCR at the end of the 3 week period. We found that overall, 35/150 (23.3%) were the wild type, 83/150 (55.3%) were heterozygous and 32/150 (21.3%) were homozygous, again following the expected Mendelian ratio. However, the survival rates of the different genotypes were very different. The homozygous knockout tadpoles began to die around 12 days after fertilization and completely died after 14 days (Fig. 3B, C). On the other hand, 91.4 % (32/35) of the wild type and 79.5% (66/83) of the heterozygous tadpoles survived till the end of the 3 week period. Thus, PRMT1 is essential for tadpole development while removing a single copy of the PRMT1 gene has little effect, consistent with our ability to obtain adult heterozygous PRMT1 mutant frogs.
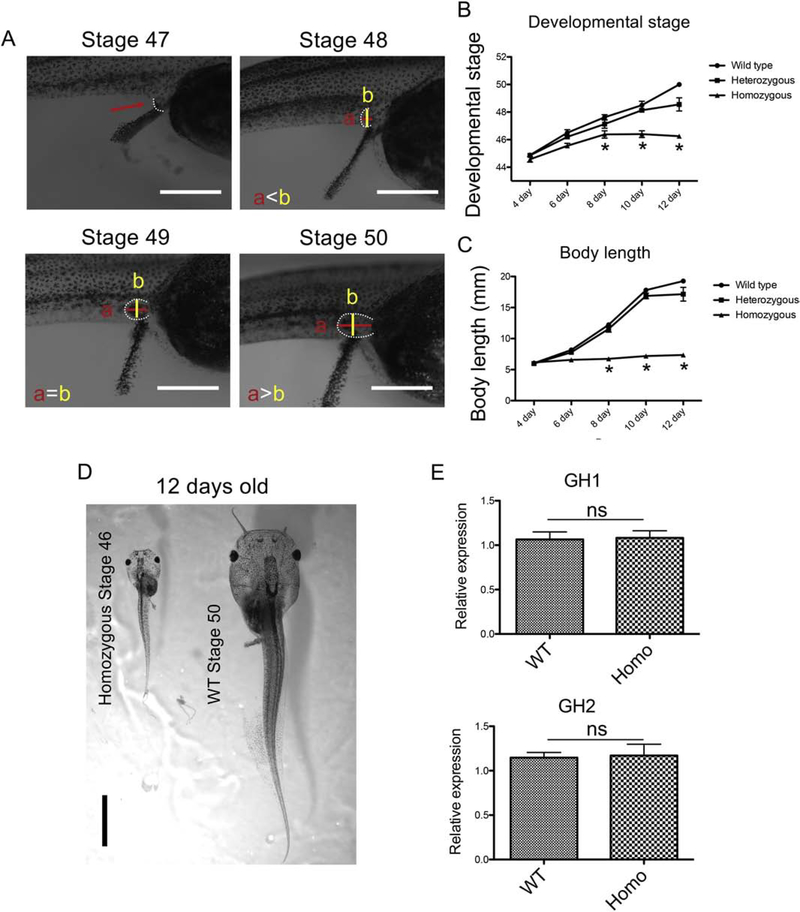
Next, we analyzed the phenotype of PRMT1 knockout during tadpole growth and development up to 14 days old. All tadpoles were kept in the same plastic container (9-L tank) and 30 tadpoles were randomly picked up for genotyping, measuring the body length and judging the developmental stages on each day. According to [19], we determined the developmental stages of the animals based on intestinal morphology until stage 46 and limb morphology after stage 46 (Fig. 4A). The morphological analysis revealed that there was no significant difference between the wild type (Stage: 45±0, Length: 6.2±0.1 mm) and the homozygous (Stage: 44±1.7, Length: 6.2±0.2 mm) tadpoles at 4 days after fertilization (stages 44/45, onset of tadpole feeding) (Fig. 4B, C). Subsequently, the homozygous tadpoles had delayed or stalled development and growth up to 12 days of age, with the wild type reaching stage 50±0.1 and 19.3±0.4 mm in length compared to the homozygous at stage 46±0 and 7.4±0.1 mm in length at 12 days old (Fig. 4B–D). Thus, homozygous PRMT1 knockout animals could develop only to stage 46 and had little growth although they began feeding around stage 45 as reflected by the presence of the food in the intestine at 4 days old. Interestingly, the expression of both growth hormone (GH) genes was the same in 10-day old PRMT1 knockout tadpoles compare with wild type siblings (Fig. 4E), suggesting that the stalled growth was not due to the lack of GH in the knockout tadpoles.
Figure 4. PRMT1 knockout tadpoles are stalled in development after feeding begins.
(A) Representative photos of tadpoles at stages 47–50, depicting the criteria used to determine the stages. Arrowhead indicates limb bud at stage 47. At stage 48, the vertical length (b) of limb bud is longer than the horizontal length (a) (a<b). At stage 49, both lengths are the same (a=b). At stage 50, the horizontal length is longer than the vertical length (a>b). Bars: 1 mm.
(B) PRMT1 homozygous knockout tadpoles stop development shortly after tadpole feeding begins around stage 44/45 (day 4). The tadpoles as genotyped in Figure 3B were allowed to develop for up to 2 weeks and their developmental stages were determined at indicated ages based on intestinal morphology (stage 44-stage 46) or limb morphology (stage 47-stage 50, see A). The average developmental stage for each of the three genotypes, wild type, PRMT1 heterozygous, and PRMT1 homozygous, was determined and plotted. Note that PRMT1 knockout tadpoles were stalled at stage 46 and eventually died by day 14 (see Fig. 3) The asterisks indicate a significant difference among the 3 genotypes (P < 0.01).
(C) PRMT1 homozygous tadpoles have a severe growth inhibition. The tadpoles as genotyped in Figure 3B were allowed to develop for up to 2 weeks and their total body lengths were measured at indicated ages. The average body length for each of the three genotypes was determined and plotted. The asterisks indicate a significant difference among the 3 genotypes (P < 0.01).
(D) Representative photos of 12-day-old tadpoles. The wild-type tadpole (right) is much bigger than the PRMT1 homozygous knockout tadpole (left) (dorsal view). Bar: 5 mm.
(E) No significant difference in the expression of two growth hormone genes between 10-day-old wild type and PRMT1(+/−) tadpoles. Total RNA was isolated from the intestine of wild type and PRMT1 homozygous knockout tadpoles at 10 days of age and used for real-time RT-PCR analysis of the expression of growth hormone genes: gh1 and gh2. The mRNA levels were normalized against that of rpl8. The groups included 4 wild and 5 PRMT1 homozygous animals. These results were plotted with the mean, marked as a line, and SE. ns: No significant difference between knockout and wild-type animals as determined by student-t test. The experiments were done with two or more batches of tadpoles with similar results.
3.3. Knocking out PRMT1 reduces Histone H4R3me2a methylation
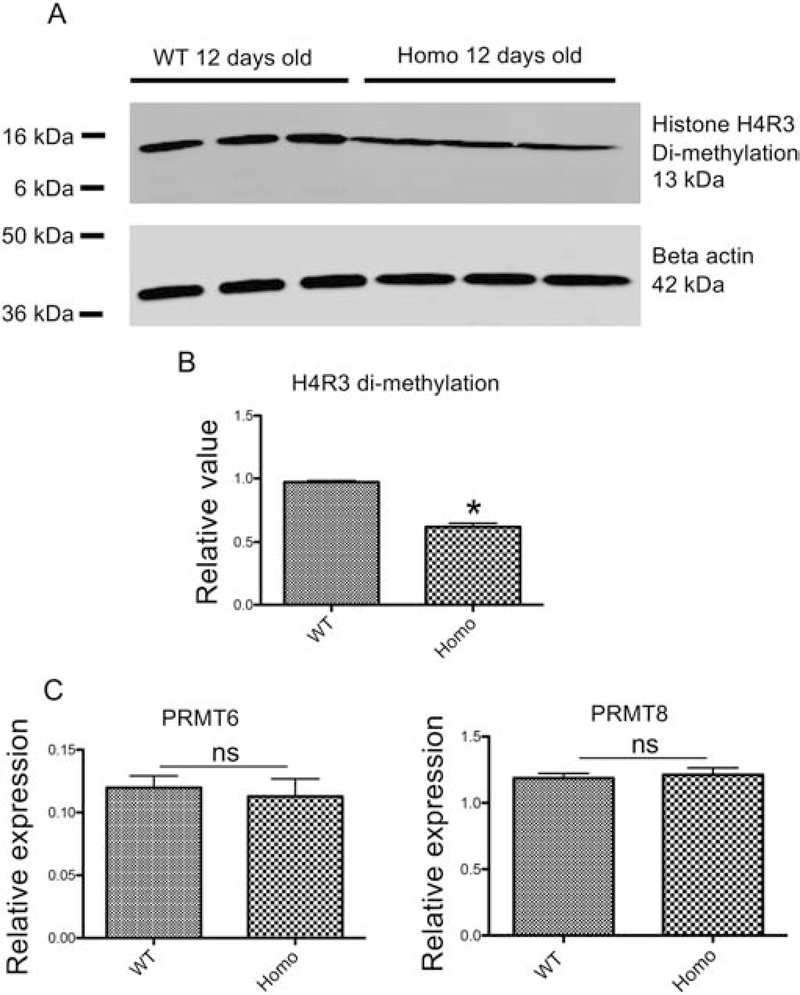
To investigate how PRMT1 knockout affected tadpole development, we first investigated if histone methylation was altered. As indicated above, PRMT1 catalyzes histone H4R3 asymmetric di-methylation (H4R3me2a) [5]. We determined H4R3me2a level by using protein extracted from whole tadpoles. Western blot analysis showed that H4R3me2a level was drastically reduced in the homozygous PRMT1 knockout tadpoles compared to that in the wild type siblings (Fig. 5A and B), suggesting that PRMT1 is the main methylase for H4R3me2a and that normal Xenopus development requires asymmetric di-methylation of H4R3. Earlier studies reported that PRMT6 and PRMT8 could also contribute asymmetric di-methylation of H4R3 [24–28]. We thus investigated the mRNA expression of PRMT6 and PRMT8 by RT-PCR and found no significant difference between wild type and PRMT1 homozygous knockout tadpoles (Fig. 5C), suggesting that PRMT1 knockout did not alter the expression of other PRMTs to compensate for the change in histone H4R3me2a methylation activity.
Figure 5. Histone H4R3me2a level is reduced in PRMT1 homozygous knockout tadpoles.
(A) Representative photos of western blot analysis. Proteins were isolated from three different pooled wild-type (lanes 1–3) and PRMT1 homozygous knockout (lanes 4–6) tadpoles at 12 days of age and subjected to Western blot analyses with antibodies against methylated histone H4R3 (H4R3me2a) (Upper) and beta-actin (lower) as the loading control, respectively.
(B) Quantification of histone H4R3me2a levels in wild-type and PRMT1 homozygous tadpoles. The expression levels as shown in (A) were normalized against that of beta-actin. The groups included 3 wild-type tadpoles and 4 PRMT1 homozygous tadpoles at 12 days of age, respectively. Asterisks (*) indicate a significant difference between knockout and wild-type animals (P<0.001).
(C) The expression of PRMT6 and PRMT8 is not altered in PRMT1 homozygous knockout tadpoles. Total RNA was isolated from the intestine of 10-day old wild type and PRMT1 homozygous tadpoles and used for real-time RT-PCR analysis of the expression of two PRMTs known to have H4R3me2a methylation activity: PRMT6 and PRMT8. The mRNA levels were normalized against that of rpl8. The groups included 4 wild and 5 PRMT1 homozygous knockout animals. These results were plotted with the mean, marked as a line, and SE. ns: No significant difference between knockout and wild type animals as determined by student-t test.
3.4. PRMT1 knockout tadpoles have drastically reduced cell proliferation in broad tissues/organs
Given the severe growth inhibition and animal lethality of the homozygous PRMT1 knockout tadpoles, we next investigated if normal cell proliferation required PRMT1 by in vivo labeling with 5-Ethynyl-2’-deoxyuridine (EdU) in 10-day old tadpoles. The 10 randomly selected tadpoles generated from mating of heterozygous PRMT1 knockout frogs were treated with or without 0.25 μg/ml EdU in 10 ml rearing water for 16 hours in a 6-well cell growth plate. The animals were subsequently sacrificed, genotyped, and sectioned for EdU staining to detect EdU positive cells, i.e., proliferating cells, in the brain and the spinal cord (Fig. 6), the cornea of eye, the intestine, and the liver (Fig. 7). EdU staining clearly detected active cell proliferation in all these tissues/organs in the wild type tadpoles. However, few EdU positive cells were detected in the PRMT1 knockout tadpoles in any of these tissues/organs (Figs. 6, 7). Thus, PRMT1 knockout prevented or drastically inhibited cell proliferation in broad organs/tissues, which likely underlay the observed cessation in development after stage 46 and severe growth reduction.
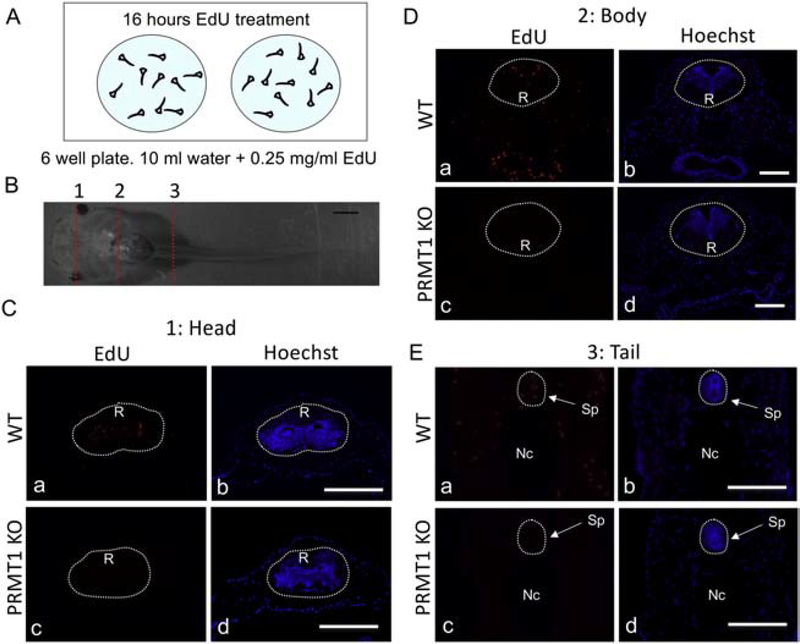
Figure 6. Reduced proliferating cells in brain and spinal cord of PRMT1 knockout tadpoles.
(A) Schematic diagram for EdU treatment. The 10-day old tadpoles were treated with or without 0.25 μg/ml EdU in 10 ml water for 16 hours in the 6-wells plate, each well contained 10 randomly selected tadpoles. They were then genotyped and sectioned for cell proliferation analyses (EdU staining).
(B) Representative photo of a 10-day-old tadpole. The 10-day old of wild-type tadpole from dorsal view showing three regions as marked with dotted lines: 1 or the upper head regions, 2 or lower head/upper body region, and 3 or tail region, for tissue sections. Bar: 1 mm.
(C) PRMT1 homozygous knockout tadpoles (PRMT1 KO) have drastically reduced cell proliferation in the upper head region. Sections from region 1 (see B) of wild type ( a, b) and PRMT1 knockout (c, d) tadpoles were stained for EdU to detect proliferating cells. The dotted lines depict the brain region, drawn based on morphological differences in the pictures of the stained tissues, under enhanced contrast and/or brightness by using Photoshop, if needed. EdU, red-color (a, c) and Hoechst, blue-color (b, d). R: rhombencephalon.
(D) PRMT1 KO tadpoles have drastically reduced cell proliferation in the lower head/upper body region. Sections from region 2 (see B) of wild type ( a, b) and PRMT1 knockout (c, d) tadpoles were stained for EdU to detect proliferating cells. The dotted lines depict the brain region, drawn based on morphological differences in the pictures of the stained tissues, under enhanced contrast and/or brightness by using Photoshop, if needed. EdU, red-color (a, c) and Hoechst, blue-color (b, d). R: rhombencephalon.
(E) PRMT1 KO tadpoles have drastically reduced cell proliferation in spinal cord of the tail region. Sections from region 3 (see B) of wild type ( a, b) and PRMT1 knockout (c, d) tadpoles were stained for EdU to detect proliferating cells. The dotted lines depict the spinal cord region, drawn based on morphological differences in the pictures of the stained tissues, under enhanced contrast and/or brightness by using Photoshop, if needed. EdU, red-color (a, c) and Hoechst, blue-color (b, d). Nc: notochord, Sp: spinal cord. At least 3 tadpoles were analyzed for all genotypes. Bars: 100 μm. At least three different animals and three sections from each animal were analyzed. The similar results were obtained from at least two different batches of tadpoles.
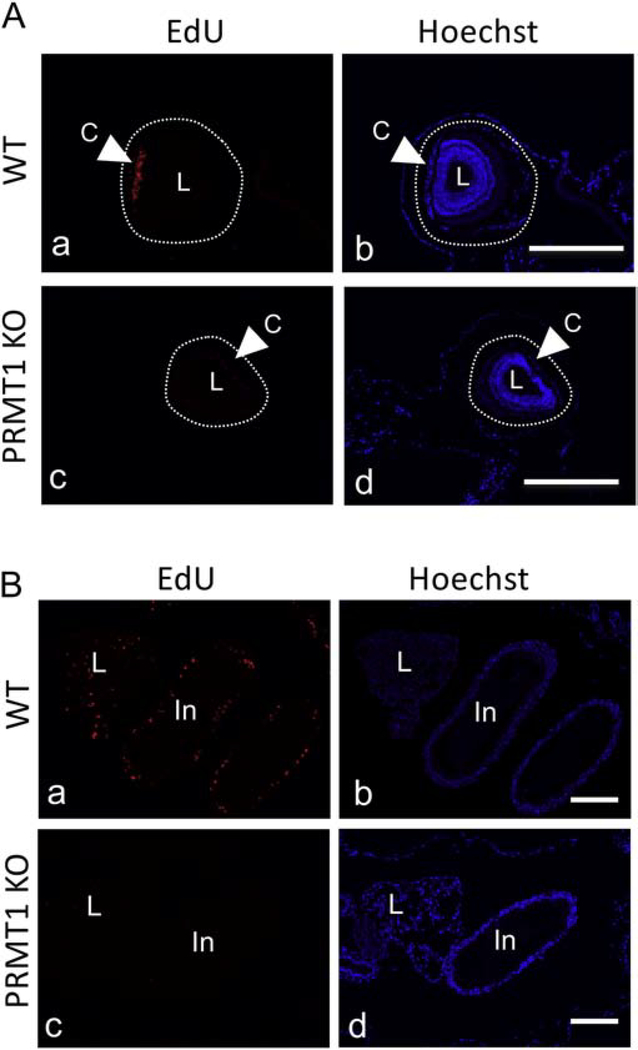
Figure 7. Reduced proliferating cells in eye, liver and intestine in PRMT1 knockout tadpoles as revealed EdU staining.
(A) PRMT1 KO tadpoles have drastically reduced cell proliferation on the cornea. Sections from region 1 (see Fig. 6B) of wild type ( a, b) and PRMT1 knockout (c, d) tadpoles were stained for EdU to detect proliferating cells. The dotted lines depict the eye region, drawn based on morphological differences in the pictures of the stained tissues, under enhanced contrast and/or brightness by using Photoshop, if needed. EdU, red-color (a, c) and Hoechst, blue-color (b, d). C: cornea, L: lens.
(B) PRMT1 KO tadpoles have drastically reduced cell proliferation in the liver and intestine. Sections from region 2 (see Fig. 6B) of wild type ( a, b) and PRMT1 knockout (c, d) tadpoles were stained for EdU to detect proliferating cells. EdU, red-color (a, c) and Hoechst, blue-color (b, d). L: liver, In: intestine. At least 3 tadpoles were analyzed for all genotypes. Bars: 100 μm. At least three different animals and three sections from each animal were analyzed. The similar results were obtained from at least two different batches of tadpoles.
4. DISCUSSION
Posttranslational histone modifications including acetylation, methylation, and ubiquitination have been reported as activation or repression marks of genes expression in yeast and cultured cells [1, 29, 30]. Importantly, epigenetic inheritance of abnormal histone lysine methylation (H3K4 or H3K27) is associated with aberrant development in mouse and Xenopus [22, 31]. PRMT1, capable of histone H4R3 methylation, is known to be responsible for most of the total protein arginine methylation in mammalian cells [5, 32]. Additionally, we have shown previously that transgenic overexpression of PRMT1 causes the acceleration of Xenopus laevis metamorphosis and knocking down the endogenous PRMT1 results the reduction in stem cell proliferation during Xenopus laevis intestinal metamorphosis [16, 17], implicating an important role of PRMT1 during metamorphosis, a period resembling mammalian postembryonic development around birth [14, 15]. Here, by using TALEN-mediated gene editing technology, we have knocked out PRMT1 in Xenopus tropicalis and demonstrated that PRMT1 is not required for embryogenesis but is essential for tadpole growth and development prior to metamorphosis, likely via histone H4R3 methylation.
PRMT1 mRNA is present in unfertilized eggs and its level rises slightly after zygotic transcription starts during Xenopus tropicalis embryogenesis before dropping to lower levels after hatching at stage 35. Despite this early expression, PRMT1 knockout embryos could completed the embryogenesis to become normal feeding stage tadpoles around 4 days of age (Fig. 3), which seems to differ from the embryonic lethal phenotype of the PRMT1-null mice [8]. Thus, maternal or even early zygotic expression of PRMT1 is not required for Xenopus embryogenesis. This may suggest that either H4R3me2a methylation activity is not important for embryogenesis or other PRMTs can compensate for the loss of PRMT1 during embryogenesis. Potential contribution from other PRMTs is supported by the reduction of only about ½ of the H4R3me2a methylation in homozygous PRMT1 knockout tadpoles.
In contrast to embryogenesis, the homozygous PRMT1 knockout tadpoles had severe inhibition in both development and growth, accompanied by reduced histone H4R3me2a methylation (Figs. 4 and 5). These animals developed only to stage 46, just after the onset of feeding at stage 45, and then were stalled in development until eventually died around 12–14 days of age. In addition, the homozygous knockout animals had a nearly complete cessation of growth after the onset of feeding stage (stage 45, 4 days old). Consistently, cell proliferation activity was significantly reduced or abolished in diverse organs such as the brain, the cornea, the liver and the intestine (Figs. 6 and 7).
It is interesting that there is an apparent difference between the embryonic lethal phenotype of the homozygous PRMT1 knockout mice and the normal formation of the feeding stage homozygous PRMT1 knockout tadpoles. On the other hand, a careful examination suggests a similar role of PRMT1 in vertebrate development. Homozygous PRMT1 knockout mouse embryos can be detected on E7.5 but not on E8.5 [8]. E7.5 or TS17 (Theiler Stage 11) is the neural plate/presomite stage when head folds are formed and continue to enlarge and foregut pocket begins to form (http://php.med.unsw.edu.au/embryology/index.php?title=Mouse_Stages). This may temporally fall in the early feeding stages in Xenopus. Like other anuran, Xenopus undergoes a biphasic development. After fertilization, the embryo undergoes embryogenesis to produces a feeding tadpole, which after a growth period, transforms into a frog. Earlier studies have demonstrated that the tadpole-to-frog transformation, or metamorphosis, is controlled by TH in a process that mimics the TH-dependent mouse neonatal period [14, 15, 33, 34]. Thus, the period between onset feeding and metamorphosis in Xenopus temporally correlates with the middle of mouse embryogenesis when PRMT1 knockout leads to the lethal phenotype. The findings here therefore reveal interesting similarities and differences between Xenopus and mouse development and supports a conserved role of PRMT1 for late development in vertebrates, likely via H4R3 methylation. It is interesting that a similar observation was made from knockout studies on another histone methyltransferase, Dot1L [22], which, like PRMT1, is upregulated during Xenopus metamorphosis and functions as a coactivator for TR [35, 36]. It is likely that the compression of vertebrate development from a biphasic process (embryogenesis and subsequent metamorphosis) in anurans into a single phase in mammals masks these two separate phases of vertebrate development with distinct requirements for epigenetic modifications.
How arginine methylation of histone and/or other proteins by PRMT1 regulates vertebrate development and how PRMT1 knockout leads to developmental lethality (during embryogenesis for mouse and premetamorphic tadpole growth for Xenopus) remain to be investigated. PRMT1 is known as a transcriptional coactivator for different transcription factors such as TR. It forms a large complex that also contains acetyltransferases SRC1, 2, or 3, and p300/CBP, which has been shown to be important for the progression of metamorphosis as well as adult stem cell development in the intestine [16, 17]. PRMT1 thus may affect cell fate and behavior by functioning both as a transcriptional coactivator and to methylate proteins that act in cellular processes other than transcription. Clearly, identifying the transcriptional targets or methylation substrates of PRMT1 during early development is essential for understanding how PRMT1 knockout causes growth reduction and animal lethality. Our PRMT1 knockout model combined with external development of anurans will thus offer a unique opportunity for future mechanistic studies on this late embryonic developmental period.
Supplementary Material
Highlights.
PRMT1 is a major enzyme for H4R3me2a during Xenopus development
PRMT1 knockout stalls tadpole development around feeding stages
PRMT1 knockout inhibits tadpole growth after tadpole feeding begins
PRMT1 is essential for tadpole survival
5.
Funding:
This work was supported by the intramural Research Program of NICHD, NIH. Y. Shibata was supported in part by Japan Society for the Promotion of Science (NIH) Fellowship. The funding sources had no roles in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Abbreviations
- PRMT1
protein arginine methyltransferase 1
- TALEN
transcription activator-like effector nuclease
- MMA
monomethylarginines
- ADMA
asymmetric dimethylarginines
- SDMA
symmetric dimethylarginines
- H4R3me2a
asymmetrically dimethylated of histone H4R3
- TH
thyroid hormone
- TR
TH receptor
Footnotes
COMPETING INTERESTS
None.
7. REFERENCES
- [1].Li B, Carey M, Workman JL, The role of chromatin during transcription., Cell, 128 (2007) 707–719. [DOI] [PubMed] [Google Scholar]
- [2].Kouzarides T, Chromatin modifications and their function., Cell, 128 (2007) 693–705. [DOI] [PubMed] [Google Scholar]
- [3].Barth TK, Imhof A, Fast signals and slow marks: the dynamics of histone modifications., Trends Biochem Sci., 35 (2010) 618–626. [DOI] [PubMed] [Google Scholar]
- [4].Blanc RS, Richard S, Arginine Methylation: The Coming of Age, Mol Cell, 65 (2017) 8–24. [DOI] [PubMed] [Google Scholar]
- [5].Di Lorenzo A, Bedford MT, Histone arginine methylation, FEBS Lett, 585 (2011) 2024–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bedford MT, Richard S, Arginine methylation an emerging regulator of protein function., Mol Cell, 18 (2005) 263–272. [DOI] [PubMed] [Google Scholar]
- [7].Miranda TB, Miranda M, Frankel A, Clarke S, PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity, J Biol Chem, 279 (2004) 22902–22907. [DOI] [PubMed] [Google Scholar]
- [8].Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE, Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable., Mol. Cell. Biol, 20 (2000) 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A, Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt, Mol Cell, 32 (2008) 221–231. [DOI] [PubMed] [Google Scholar]
- [10].Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD, Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1., Curr Biol, 11 (2001) 996–1000. [DOI] [PubMed] [Google Scholar]
- [11].Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR, Regulation of transcription by a protein methyltransferase., Science, 284 (1999) 2174–2177. [DOI] [PubMed] [Google Scholar]
- [12].Koh SS, Chen DG, Lee YH, Stallcup MR, Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities., J. Biol. Chem, 276 (2001) 1089–1098. [DOI] [PubMed] [Google Scholar]
- [13].Bartosch C, Lopes JM, Jeronimo C, Epigenetics in endometrial carcinogenesis - part 2: histone modifications, chromatin remodeling and noncoding RNAs, Epigenomics, 9 (2017) 873–892. [DOI] [PubMed] [Google Scholar]
- [14].Shi Y-B, Amphibian Metamorphosis: From morphology to molecular biology, John Wiley & Sons, Inc, Place Published, 1999. [Google Scholar]
- [15].Tata JR, Gene expression during metamorphosis: an ideal model for post-embryonic development., Bioessays, 15 (1993) 239–248. [DOI] [PubMed] [Google Scholar]
- [16].Matsuda H, Paul BD, Choi CY, Hasebe T, Shi Y-B, Novel functions of protein arginine methyltransferase 1 in thyroid hormone receptor-mediated transcription and in the regulation of metamorphic rate in Xenopus laevis., Mol. Cell. Biol, 29 (2009) 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Matsuda H, Shi YB, An essential and evolutionarily conserved role of protein arginine methyltransferase 1 for adult intestinal stem cells during postembryonic development, Stem Cells, 28 (2010) 2073–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang X, Matsuda H, Shi Y-B, Developmental regulation and function of thyroid hormone receptors and 9-cis retinoic acid receptors during Xenopus tropicalis metamorphosis., Endocrinology, 149 (2008) 5610–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nieuwkoop PD, Faber J, Normal table of Xenopus laevis., North Holland Publishing, Amsterdam, (1965). [Google Scholar]
- [20].Shibata Y, Bao L, Fu L, Shi B, Shi YB, Functional Studies of Transcriptional Cofactors via Microinjection-Mediated Gene Editing in Xenopus, Methods Mol Biol, 1874 (2019) 507–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wen L, Shibata Y, Su D, Fu L, Luu N, Shi Y-B, Thyroid hormone receptor α controls developmental timing and regulates the rate and coordination of tissue specific metamorphosis in Xenopus tropicalis., Endocrinology, 158 (2017) 1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wen L, Fu L, Guo X, Chen Y, Shi YB, Histone methyltransferase Dot1L plays a role in postembryonic development in Xenopus tropicalis, FASEB J, 29 (2015) 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang X, Cheng X, Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides., Structure, 11 (2003) 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG, Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain, J Biol Chem, 282 (2007) 36444–36453. [DOI] [PubMed] [Google Scholar]
- [25].Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT, PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family, J Biol Chem, 280 (2005) 32890–32896. [DOI] [PubMed] [Google Scholar]
- [26].Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT, Arginine methylation of the histone H3 tail impedes effector binding, J Biol Chem, 283 (2008) 3006–3010. [DOI] [PubMed] [Google Scholar]
- [27].Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM, PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation, Genes Dev, 21 (2007) 3369–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dillon MB, Rust HL, Thompson PR, Mowen KA, Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity, J Biol Chem, 288 (2013) 27872–27880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K, High-resolution profiling of histone methylations in the human genome, Cell, 129 (2007) 823–837. [DOI] [PubMed] [Google Scholar]
- [30].Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K, Combinatorial patterns of histone acetylations and methylations in the human genome, Nat Genet, 40 (2008) 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters AH, Kimmins S, Disruption of histone methylation in developing sperm impairs offspring health transgenerationally, Science, 350 (2015) aab2006. [DOI] [PubMed] [Google Scholar]
- [32].Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR, PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells, J Biol Chem, 275 (2000) 7723–7730. [DOI] [PubMed] [Google Scholar]
- [33].Hasebe T, Fu L, Miller TC, Zhang Y, Shi YB, Ishizuya-Oka A, Thyroid hormone-induced cell-cell interactions are required for the development of adult intestinal stem cells, Cell Biosci, 3 (2013) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ishizuya-Oka A, Shi YB, Evolutionary insights into postembryonic development of adult intestinal stem cells, Cell Biosci, 1 (2011) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wen L, Fu L, Shi YB, Histone methyltransferase Dot1L is a coactivator for thyroid hormone receptor during Xenopus development, FASEB J, 31 (2017) 4821–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Matsuura K, Fujimoto K, Das B, Fu L, Lu CD, Shi YB, Histone H3K79 methyltransferase Dot1L is directly activated by thyroid hormone receptor during Xenopus metamorphosis, Cell Biosci, 2 (2012) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.