Abstract
Probiotics are routinely administered to hospitalized patients for a myriad of potential indications1 but have been associated with adverse effects that may outweigh their potential benefits2–7. Particularly alarming is the possibility that probiotic strains can cause bacteremia8,9, yet direct evidence for an ancestral link between blood isolates and administered probiotics is lacking. Here, focusing on intensive care unit (ICU) patients, we find markedly increased risk of Lactobacillus bacteremia among patients treated with probiotics, and we provide genomics data that support direct clonal transmission of probiotics to the bloodstream. Whole-genome based phylogeny showed that Lactobacilli isolated from the blood of these treated patients were phylogenetically inseparable from Lactobacilli isolated from the associated probiotic product. Indeed, the minute genetic diversity among the blood isolates mostly mirrored preexisting genetic heterogeneity found in the probiotic product. Some blood isolates also had de novo mutations, including a non-synonymous SNP conferring antibiotic resistance in one patient. Our findings support that probiotic strains can directly cause bacteremia and adaptively evolve within ICU patients.
Probiotics are increasingly used in hospitalized patients1. These supplementary products have shown benefit in acute infectious diarrhea, antibiotic-associated diarrhea, and ulcerative colitis2,10,11. In the intensive care unit (ICU), additional indications are being explored, including prevention of ventilator-associated pneumonia, pancreatitis, and sepsis12–14. However, studies on the efficacy and adverse effects of probiotics in the ICU are conflicting, and their use remains controversial4–7,15. Adverse outcomes, including bacteremia, have been reported and may preclude their use in specific populations such as those with immune compromise or disintegrity of the gastrointestinal tract8,9,16,17.
Bacteremia appearing during the course of probiotic treatment can involve Lactobacillus species similar to those in probiotics, yet as these species are also common in the human gastrointestinal microbiome, pinpointing the source of these infections has been challenging18. Studies using Pulsed-Field Gel Electrophoresis (PFGE) have previously revealed strain-level similarity between blood and probiotic isolates19, but higher genomic resolution is required to establish direct clonal ancestry and the possibility of direct transmission of probiotic bacteria to the blood.
In the context of bacterial pathogens, whole-genome methods have been powerful in identifying transmission links and within-host adaptation20–23. Constructing a SNP-level phylogeny of isolates from patients can unravel ancestral links between lineages and likely paths of transmission. Whole-genome comparison of isolates can also reveal adaptive mutations important for the survival of the pathogen within the host20,22. Yet, despite their established power, the use of these whole-genome approaches for tracing of probiotic strains has so far been limited. Here, we applied whole-genome analysis and phenotyping to blood isolates and probiotic strains administered to ICU patients.
Analyzing cases of Lactobacillus bacteremia at Boston Children’s Hospital, we found that ICU patients receiving Lactobacillus rhamnosus GG (LGG) probiotics had a markedly high risk of developing Lactobacillus bacteremia. Over a period of 5.5 years, a total of 22,174 patients were treated in an ICU, with 522 of these patients receiving LGG-containing probiotic, typically through a feeding tube, as part of their treatment. Analyzing recorded Lactobacillus bacteremia among these patients, we found a significantly greater risk for patients receiving the LGG-containing probiotic; 6 of these 522 patients had Lactobacillus bacteremia (1.1%, Patients R1–R6, Supplementary Table 1a) compared to only 2 out of the 21,652 patients not receiving the LGG probiotic (0.009%, Patients N1–N2, Supplementary Table 1a; P=4.8×10−9, Fisher exact test). Furthermore, all 6 of the ICU blood isolates from patients receiving the LGG probiotic were identified by MALDI-TOF as Lactobacillus rhamnosus, while the 2 isolates from patients not receiving the LGG probiotic were identified as other Lactobacillus species (Supplementary Table 1a). The ICU patients receiving probiotics containing LGG are therefore at markedly higher risk of developing Lactobacillus rhamnosus bacteremia (6 out of 522 compared to 0 out of 21,652; P=1.8×10−10, Fisher exact test). Further, the LGG probiotic bacteremia rate of 1.1% that we observed is also much higher than the annual rate of LGG probiotic bacteremia (0.00007%) reported in the general population19. Yet, L. rhamnosus bacteremia can occasionally appear also in patients not receiving these probiotics: during the study period, there were an additional 10 cases of Lactobacillus bacteremia among approximately 93,000 non-ICU patients (Patients N3–N12, Supplementary Table 1b), and 4 of these 10 isolates were identified by MALDI-TOF as Lactobacillus rhamnosus (Patients N5, N9–N11, Supplementary Table 1b). None of the 10 non-ICU patients were receiving a probiotic at the time of the bacteremia. Taken together, these results suggest that ICU patients receiving probiotics containing LGG are at much higher risk of developing Lactobacillus bacteremia, but it is difficult to derive direct links due to the occasional L. rhamnosus bacteremia appearing also in patients not receiving these probiotics.
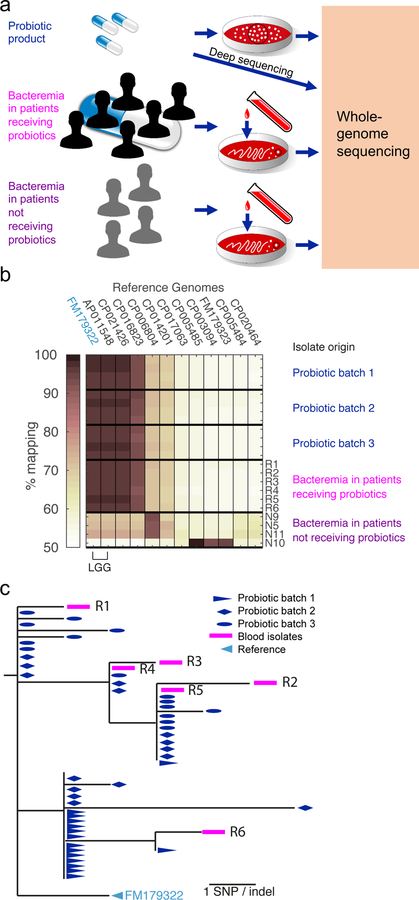
To allow better ancestral resolution, we next used whole-genome sequencing to determine strain-level similarity among the blood and probiotic isolates. We performed whole-genome sequencing of all 10 L. rhamnosus blood isolates (6 from patients receiving probiotic and 4 from patients not receiving probiotic), as well as 16 isolates from each of 3 probiotic capsules of different lots (probiotic batches 1–3, Supplementary Table 2; Methods; Figure 1a). To quantify strain-level relatedness among these isolates, we started by measuring their distance to all available L. rhamnosus genomes (GenBank October 2017, Supplementary Table 3). Illumina reads of each isolate were aligned to each of these genomes, and the fraction of aligned reads, affected both by gene content similarity and SNP density, was quantified as a measure of similarity. We found that all 6 blood isolates and all probiotic product isolates shared the same closest reference genome - an LGG genome (GenBank chromosome ID: FM179322) - suggesting high relatedness between these two sets of isolates (Figure 1b). In contrast, all 4 L. rhamnosus blood isolates from patients not receiving probiotics were more similar to other strains, indicating that they were not derived from the probiotic product (Figure 1b).
Fig. 1. Genomic evidence for Lactobacillus rhamnosus transmission from probiotic capsule to the blood of patients.
(a) Schematic for whole-genome sequencing of Lactobacillus rhamnosus probiotic isolates, blood isolates from ICU patients (n=6) receiving probiotics, and blood isolates from non-ICU patients (n=4) who were not receiving probiotics. Black circles represent sequencing multiple individual colonies for each probiotic batch but a single colony for each blood isolate. (b) Similarity between L. rhamnosus isolates and available reference genomes shown as the fraction of reads aligned to each reference. Isolates are identified by their source: four representative isolates from each of three probiotic product batches, the six blood isolates from patients receiving probiotics, and the four blood L. rhamnosus isolates from patients not receiving probiotics. (c) Phylogenetic analysis of all 54 sequenced L. rhamnosus GG (LGG) isolates: 16 isolates from each of 3 separate probiotic batches (blue), and the 6 blood isolates from Patients R1 to R6 (magenta).
To further increase genomic resolution, we next compared the genomes of the blood and LGG probiotic isolates by alignment to the reference genome. Analyzing gene content of the isolates, only a single deletion was identified: one of the probiotic isolates of batch 2 had a large deletion of a region spanning 82 genes of the reference genome FM179322 (genes 384–465, Figure 2). Strains were also almost identical at the single-nucleotide level; analyzing SNP level variations, we identified a total of only 23 SNPs among all isolates (Methods; for the list of SNPs, see Supplementary Table 4). Indeed, the highest SNP distance between any isolate and the last common ancestor was not more than 6 SNPs. Two SNPs were shared by all isolates, separating them from the reference genome, indicating that the blood and probiotic isolates share a more recent last common ancestor than the LGG clone deposited in GenBank (Figure 1c). Moreover, the blood and probiotic isolates were phylogenetically inseparable - there was no mutation that strictly separated these two groups (Figure 1c, Supplementary Table 4).
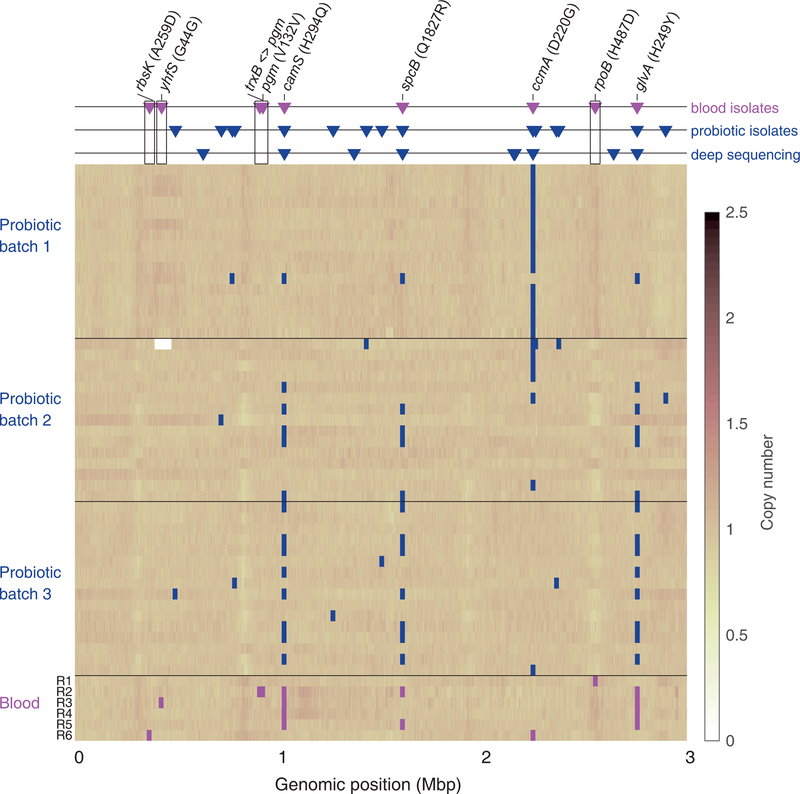
Fig. 2. Coverage of the LGG reference genome for the probiotic and blood Lactobacillus rhamnosus isolates.
For each isolate (row in matrix) SNPs are marked as squares (magenta for blood isolates, blue for product isolates). Triangles (top panel) indicate all mutations identified in blood isolates (magenta triangles) and probiotic product (blue triangles) compared to the LGG reference genome FM179322. For the probiotic product, these are either high-quality SNPs in whole-genome sequencing (middle row) or diversity identified by deep sequencing of the product (bottom row, see Methods). Annotation is included for all SNPs identified in blood isolates. SNPs identified only in blood isolates are indicated with black frame.
Much of the genetic diversity among blood isolates mirrored pre-existing genetic diversity within the probiotic capsules. We identified 11 genomic positions that were polymorphic across blood isolates (Figure 2). Three of these mutations, all of which were non-synonymous, were recurring mutations, observed in more than a single blood isolate (H294Q in CamS, H248Y in GlvA, and Q1827R in SpcB; Figure 2, Supplementary Table 4). These repeatedly occurring blood isolate mutations were all identified as pre-existing in the probiotic product (these same loci were diverse within each of the three batches of the probiotic product, Figure 2). Furthermore, the camS and glvA SNPs were genotypically linked in both the blood and probiotic isolates. One other polymorphic locus identified in a single blood isolate was also found pre-existing in the probiotic product (a D220G mutation in the ABC transporter CcmA). Overall, correspondence between blood isolate mutations and pre-existing diversity within the product further supports the likelihood of transmission of bacteria from probiotic to blood.
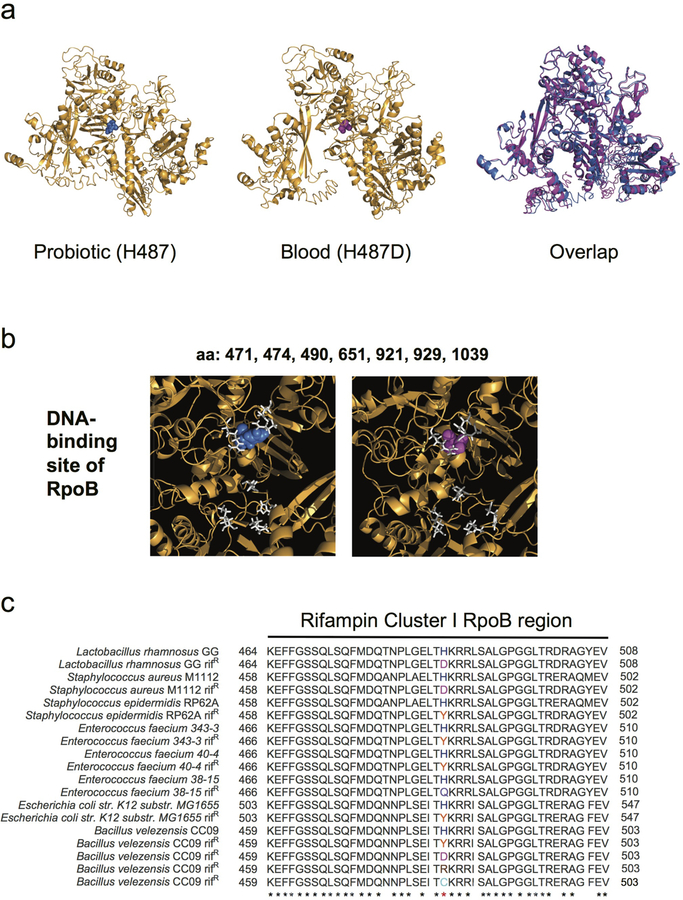
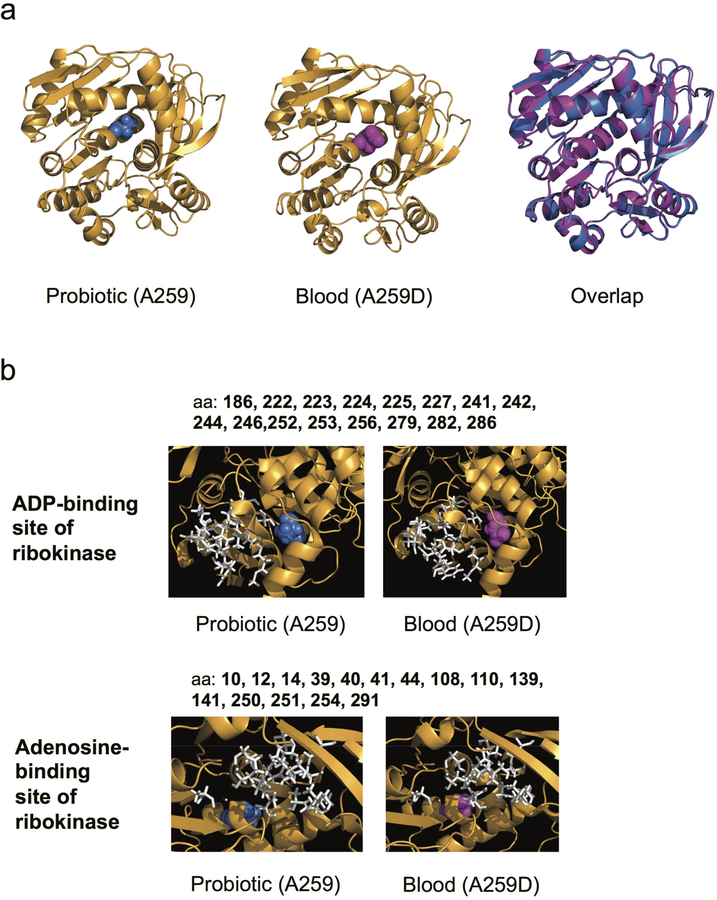
In addition to the 6 blood isolate mutations which were pre-existing in the probiotic product, we identified 5 blood isolate mutations not appearing in the isolates from the probiotic product, suggesting de novo evolution within the patient (Figure 2, Supplementary Table 5). These mutations, appearing in the blood isolates, where not found in any of the 16 genomes isolated from each of the 3 capsules. To further test for their possible existence in the product, we deep-sequenced capsules from five different batches, obtained both from the hospital and from a commercial pharmacy, and identified diverse loci (batches 2–6, Methods; Figure 2, Extended Data Figure 1, Supplementary Table 6). No pre-existing genotypic diversity was found at the loci of the 5 blood-isolate-specific mutations (Figure 2, Supplementary Table 7). One of these 5 blood-isolate-specific mutations was in an intergenic promoter mutation, 2 were nonsynonymous coding mutations (H487D in the RNA polymerase RpoB (Figure 3a, Extended Data Fig. 2) and A259D near the active site of the RbsK ribokinase (Extended Data Fig. 3)), and 2 were synonymous mutations (at G44 of the YhfS transferase and at V132 of phosphoglucomutase. These mutations, existing in the blood but not identified in the probiotic product, could represent de novo mutations selected within the patient.
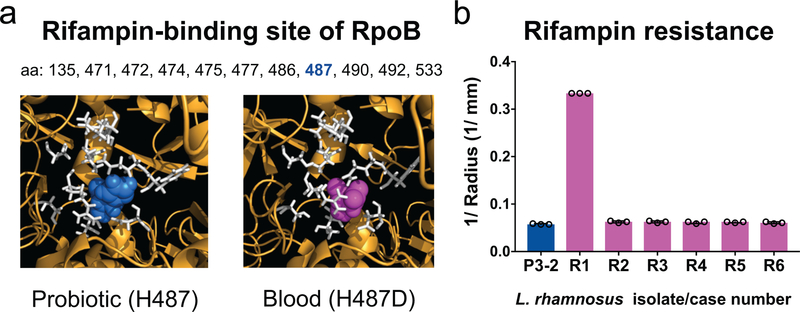
Fig 3. The Lactobacillus rhamnosus blood-isolate-specific rpoB SNP occurs at the rifampin-binding site and confers rifampin resistance.
(a) Predicted structure of L. rhamnosus GG RNA polymerase β-subunit RpoB showing the rifampin-binding site (white) with histidine 487 of the probiotic (blue, left) and aspartic acid 487 of the blood isolate from Patient R1 (magenta, right). (b) Rifampin susceptibility testing of blood isolates of each patient (R1–R6) compared to a probiotic isolate with no SNPs (P3–2). Bars depict the medians of 3 independent experiments, and error bars show the interquartile ranges. *P = 0.0021 for R1 compared to P3–2 by Kruskal- Wallis test followed by Dunn’s multiple comparisons test. The blood isolate from Patient R1 was resistant based on zone cutoffs for S. aureus (Supplementary Table 8).
The blood-isolate-specific mutation in the rpoB RNA polymerase gene (H487D) appeared in an isolate from Patient R1, who had been concurrently receiving Lactobacillus rhamnosus GG and the rifampin derivative rifaximin during the three months prior to bacteremia. This mutation, changing a specific residue in the cleft of RpoB DNA-binding site, is known to provide resistance to rifampin (Figure 3a, Extended Data Fig. 2)24–26. Antibiotic susceptibility measurements showed that this blood isolate was indeed resistant to rifampin, while all other blood isolates as well as a probiotic isolate containing no SNPs were sensitive (Figure 3b). In contrast, susceptibilities to other antibiotics were nearly identical among the blood and probiotic isolates (Supplementary Table 8), suggesting that the R1 isolate adapted specifically to resist rifampin. Interestingly, while rifampin resistance mutations at other rpoB positions typically confer decreased fitness, mutations at the H487 position can retain fitness similar to the wild-type27. Indeed, the R1 isolate carrying the rpoB mutation showed no significant fitness cost compared with the probiotic strain containing no SNPs (Supplementary Table 9). The specificity of the rpoB mutation to the patient receiving rifampin, together with its associated resistance and growth phenotypes, further suggest that the probiotic strains could acquire adaptive mutations increasing their fitness in the host environment.
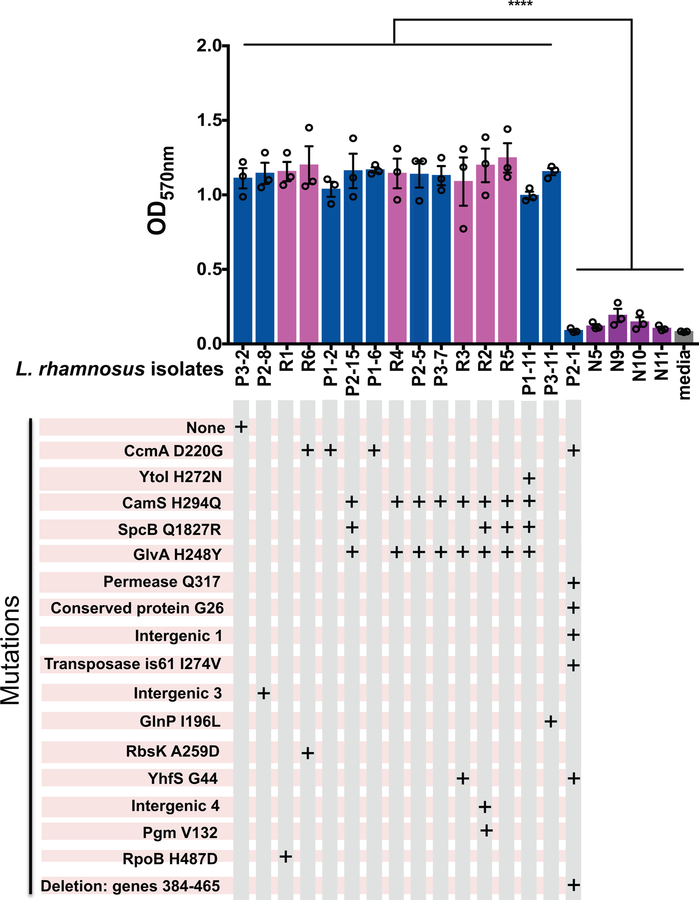
We further considered other adaptive phenotypes. While survival in serum or human whole blood was similar among the probiotic and blood isolates (Supplementary Figure 1), biofilm formation (Extended Data Figure 4), which could lead to increased adhesion to a central venous catheter and/or enhanced survival in the GI tract, was significantly higher in the L. rhamnosus LGG blood and probiotic isolates compared to the L. rhamnosus non-LGG blood isolates (from Patients N5, N9–N11) and to the probiotic isolate P2–1 containing an 82-gene deletion, which includes the spaCBA pilus genes critical for biofilm28 (Supplementary Table 10). These results suggest that biofilm is not required for bacteremia and that the LGG probiotic products can contain mutants with markedly different biofilm phenotypes.
While our patient population was critically ill, the patients developing LGG bacteremia while receiving probiotics did not have the typical risk factors for Lactobacillus bacteremia such as severe immune compromise or bowel disintegrity. Furthermore, in a case-control study comparing potential risk factors for bacteremia in these 6 cases with 16 matched control ICU patients who received probiotics but did not have bacteremia (Methods), we found no significant differences in device utilization, vasopressor support, recent surgery, diarrhea, parenteral nutrition, or antibiotic exposure (Supplementary Table 11). While the low number of patients in this case-control study may limit statistical power, our inclusion of a control group with case-control methodology represents a significant improvement over prior descriptive studies in understanding specific risk factors within the ICU. The lack of strong differences between the patients who had bacteremia and the control group that did not have bacteremia suggests that the ICU patients at risk for transmission of probiotics from product to blood may not be easily identifiable.
The exact mechanism of transmission from probiotic to blood is unclear. Nearly all of these patients had a central line, and direct contamination of the central line with a probiotic strain or with stool containing the probiotic strain could lead to the observed probiotic bacteremia. Alternatively, the probiotic bacteria could have translocated across the bowel wall. The antibiotic resistance related adaptation we observed could appear either prior to or immediately following the transmission of the bacteria to the blood. Our results suggest that these adaptive mutations are absent in the probiotic capsule and therefore evolved within the host environment, yet given possible genomic variations among batches of the probiotic product, we cannot exclude that some of these presumably blood-specific mutations were pre-existing in the specific capsules given to each patient. In any case, appearing either through rare mutations preexisting in the product or de novo during treatment, these emerging antibiotic-resistant probiotic bacteria could potentially undermine treatment efficacy. It would be interesting in future studies to explore the importance of other de novo mutations with additional in vitro phenotyping or in animal models.
In summary, our epidemiological analysis uncovered a statistically and clinically significant risk for bacteremia with probiotic Lactobacilli in the ICU, and genome-level analysis identified 6 independent cases of transmission of probiotics from capsule to blood in ICU patients treated with probiotics. Our results also provide evidence of within-host evolution of the probiotic, including acquisition of antibiotic resistance. Probiotics have shown significant benefits for acute infectious diarrhea, antibiotic-associated diarrhea, and ulcerative colitis2,10,11. Yet, our findings highlight that as ICU patients have increased risk for probiotic-associated bacteremia, these potential benefits must be weighed against this risk when considering the continued use of probiotics in the ICU.
Methods
Patient inclusion criteria and clinical data.
Eighteen cases of Lactobacillus bacteremia were identified through usual surveillance activities of the Infection Prevention and Control program at Boston Children’s Hospital from January 2009 to June 2014 (6 patients receiving probiotics; 12 patients not receiving probiotics, Supplementary Table 1). Based on pharmacy records from January 2009 through June 2014, there were 15,736 probiotic doses administered to 645 ICU patients, including 5,859 (37%) in a medical ICU; 4,080 (26%) in an intermediate care program (ICP); 3,560 (23%) in a medical-surgical ICU; 2,114 (13%) in a cardiac ICU; and 123 (0.8%) in a neonatal ICU, all at a single center (Boston Children’s Hospital). The majority (522 of 645, 81%) of the ICU patients who received probiotics, including all 6 patients who developed bacteremia, received a probiotic containing Lactobacillus rhamnosus GG (LGG). ICU patients who were prescribed probiotics received a median of 8 (IQR 3–23) doses, with a range from 1–347 doses. Twenty-four percent of doses were given by mouth, 62% by gastrostomy or jejunostomy tube, and 14% by nasogastric or nasojejunal tube. The average numbers of doses per month did not significantly change over time, with 217 doses/month in 2009, 216 doses/month in 2010, 244 doses/month in 2011, 268 doses/month in 2012, 239 doses/month in 2013, and 249 doses/month in 2014. Probiotics were administered to only 3% of ICU patients (645 ICU patients among a total of 22,174 patients admitted to these ICUs during the study period). Probiotics were most commonly prescribed because patients had been receiving them prior to ICU admission. There were no ICU-specific guidelines for probiotic administration. Nearly all patients had a central venous line (CVL) at the time of the bacteremia (only Patients R2, N1, and N5 did not), so nearly all of these bacteremias met CDC criteria for central line-associated bloodstream infection (CLABSI). We did not examine the details of probiotic doses administered to non-ICU patients. The study was approved by the Boston Children’s Hospital IRB.
Our study was not designed to assess the clinical impact of bacteremia, although we do note that these episodes of bacteremia manifested initially as clinically active infection and that nearly all the patients were treated with intravenous antibiotics directed at Lactobacillus. CVLs were removed during treatment from 2 of the 5 patients receiving probiotics who had a CVL and from 9 of the 10 patients not receiving probiotics who had a CVL. Notably, 2 of the 6 cases of LGG bacteremia (Patients R2 and R4) and 1 of the 4 cases of non-LGG L. rhamnosus bacteremia (Patient N5) were considered by their treating physicians as potential contaminants or transient bacteremias and were not specifically treated with a long course of antibiotics. However, since Patient R4 had a CVL, the bacteremia was classified as a CLABSI. None of the patients had endocarditis, and none died within 7 days of bacteremia.
Isolation of bacteria from probiotic capsules and blood.
Blood isolates were frozen at time of isolation by the clinical microbiology lab and later streaked on MRS-agar plates and the Lactobacillus species were identified by MALDI-TOF. To isolate DNA for whole-genome sequencing, single colonies were picked (16 colonies per probiotic product batch and a single colony for blood samples) to inoculate MRS broth (BD, 288130), and overnight cultures were frozen. To isolate individual bacteria from probiotic capsules, we employed two complementary techniques. For probiotic product batch 1, a capsule was streaked on a CDC Anaerobe Blood Agar plate, bacterial lawn was scraped off the plate, frozen (−80°C, glycerol) and then streaked to single colonies on MRS-agar plates (BD, 288210) incubated at 37°C. For product batches 2 and 3, capsules were re-suspended and thoroughly vortexed in phosphate-buffered saline solution, streaked on MRS-agar plates incubated at 37°C until colonies showed, and then re-streaked to purity.
Whole-genome sequencing of individual isolates and deep sequencing.
For single isolates, DNA was extracted from frozen overnight bacterial cultures derived from single colonies picked as described above (Macherey-Nagel, NucleoSpin 96 Tissue). For capsule deep sequencing (probiotic product batches 2–6, Supplementary Table 2), DNA was extracted from 100 ul (>108 cells) of resuspended capsule (same kit as above). Batch 1 was not available for deep sequencing. Nextera sequencing libraries were prepared32 and sequenced in an Illumina HiSeq 2500 machine in rapid-mode to produce 125 base paired-end reads. DNA extraction and library preparation for single-isolate deep sequencing used as control (see “Methods: Genomic Analysis of capsule deep sequencing” below) was done similarly to other single isolates, and sequencing was done at the same Illumina run as the capsule DNA deep sequencing.
Genomic data analysis of isolates.
Illumina reads were filtered to remove reads contaminated by Nextera adapter or low-quality bases (>2 bases with Phred Score<20) yielding an average 1.13M reads per sample (standard deviation=2.9·105). These reads were aligned to indicated reference genomes using Bowtie 1.2.1.1 allowing a maximum of 3 mismatches per read. The fraction of aligned reads was used to determine distance between isolates and reference genomes. Alignment to closest reference genome (Genbank FM179322) was further analyzed. Base calling was done using SAMtools and BCFtools 0.1.19. A genome position was identified as a SNP if more than a single allele was identified across isolates using a quality threshold of FQ<−80. Phylogeny was based on the identified SNPs and was determined by the PHYLIP dnapars algorithm which carries out unrooted parsimony.
Gene content analysis.
For each isolate, a “raw copy number” for each gene was calculated as the median base coverage across the gene divided by the median coverage across the genome of the isolate. To remove gene specific biases, this raw copy number was further normalized by the median raw value of the gene across all isolates – yielding the gene copy number used to identify deleted genes. For Figure 2, a similar analysis of genomic coverage was done, where for each 6Kb region the mean read coverage was divided by the median coverage across the genome and normalized by the median of this region-specific value across isolates.
Genomic analysis of capsule deep sequencing.
Reads were filtered and aligned to the reference genome as described for single isolate analysis above (GenBank: FM179322). This resulted in coverage of >97% of the reference genome. Per batch, median coverage of these positions was 372–1268X (Supplementary Table 2). As a control, a single isolate colony (batch 2, isolate 15) was also sequenced at high depth (median coverage 897X). To identify variable loci in the probiotic batches, we performed, for each probiotic batch and genomic position, a Fisher exact test comparing the number of reads calling the reference versus the alternative base in the probiotics versus the the single-isolate control. To control for multiple comparisons, a p-value of (0.05)/(genome length) = 1.66×10−8 was used for calling within batch diversity.
Data and Code Availability.
Illumina files from gene sequencing have been deposited in the public database. Accession codes will be provided prior to publication. Higher level analysis (e.g. SNP calling) is provided in supplementary materials. All other data are available from the authors upon reasonable request.
Bacterial strains and culture conditions for in vitro assays.
L. rhamnosus bacteria were grown at 37°C with 5% CO2 for 48h on Trypticase Soy Agar II with 5% sheep blood (BD Biosciences). Liquid culture was performed using MRS broth (Sigma-Aldrich) supplemented with 0.001% Tween 80 (Sigma-Aldrich, MRST) at 37°C with 5% CO2 for 24h statically. P. aeruginosa PAO1 galU mutant33 were grown at 37°C overnight on Trypticase Soy Agar II (BD Biosciences). Liquid culture was performed using LB broth Miller (Fisher BioReagents) at 37°C, at 200 rpm overnight. Modified TSB (mTSB) medium consisted of 15 g/L TSB (BD Biosciences) and 20 g/L of Bacto-proteose peptone no. 3 (BD Biosciences) was used for the biofilm assay.
Biofilm assay.
The assay for biofilm formation was based on a previous report with minor modifications34. In brief, 3×107 CFU were added in 200 μL of mTSB in three replicates in flat bottom polystyrene 96-well plates (Costar) at 37°C with 5% CO2 for 72 h. Bacteria were dumped out by inverting the plate. The plate was then washed with water, and attached bacteria were stained for 30 min with 200 μL 0.1% (wt/vol) crystal violet in an isopropanol-methanol-PBS solution (1:1:18 [vol:vol:vol]). Plates were washed with water, left to dry for 15 min, and then 150 μL of 33% glacial acetic acid was added to each well. Biofilm was measured at 570 nm (Versa max, Molecular devices).
Antibiotic resistance testing.
Disk diffusion susceptibility testing was performed by the Boston Children’s Hospital Infectious Diseases Diagnostic Laboratory using the standard methods of Staphylococcus species (specific disk diffusion methods for Lactobacillus are not available)35.
Competition assay.
The single culture and competition assays were based on a previous report, with modifications36. In brief, bacteria of the probiotic strain containing no SNPs (P3–2) and of the blood isolate R1, from overnight MRST liquid cultures were adjusted to OD600 of 0.05, either in single culture or mixed in a 1:1 ratio. During growth in MRST broth at 37°C with 5% CO2 for 24 h CFUs were determined every 2 hours by serial dilutions on MRST agar plates for the single cultures and on both MRST agar and MRST agar with 1 μg/ml rifampicin (Research Products International) for the competition cultures.
BLASTP of RpoB protein.
The protein accession numbers of the RpoB protein from the bacteria L. rhamnosus GG (CAR88393.1), S. aureus M1112 (EWR31828.1), S. epidermidis RP62A (AAW53580.1), E. faecium 343–3 (AAO00728.1), E. faecium 38–15 (AAO00731.1), E. faecium 40–4 (AAO00730.1), E. coli K-12 substr. MG1655 (NP_418414.1), B. velezensis CC09 (ANB47365.1) were used in COBALT for amino acid alignment from NCBI (https://www.stva.ncbi.nlm.nih.gov/tools/cobalt).
Whole blood killing assay.
Bacteria were grown on TSA with 5% sheep blood (BD Biosciences) and incubated for 48h at 37°C with 5% CO2. Overnight cultures in MRST medium were washed once in PBS (Boston Bio-products) and adjusted to give 106 CFU/ 50 μL. 50 μL of each strain were added to 450 μL of heparinized blood from a healthy donor. Inoculum CFUs were determined by serial dilutions on TSA with 5% sheep blood. After 1 h and 3 h of rotation at 37°C, serial dilutions were plated to determine the number of surviving CFU. In parallel, static tubes were held at 37°C as a non-phagocytosis control for all time points (0h, 1h and 3h).
Serum sensitivity assays.
Bacteria were grown on TSA with 5% sheep blood (BD Biosciences) and incubated 48 h at 37°C with 5% CO2. Static overnight cultures in MRST medium at 37°C with 5% CO2 were washed in PBS once and then diluted in PBS plus 1 mM CaCl2 and 1 mM MgCl2, and 100 μL aliquots were placed in a sterile 96-well plate to give a final inoculum of approximately 5×106 CFU per well. Pooled male, type AB human serum (Sigma-Aldrich) was diluted in PBS plus 1 mM CaCl2 and 1 mM MgCl2 to give twice the desired final concentration. Final serum concentrations used were 50% and 25%. Human serum (50%) that was heat-inactivated by incubation at 56°C for 30 min, and 0% serum served as controls. Equal volumes (100 μL) of sera and bacterial suspensions were mixed and incubated at 37°C for 1 h with gentle shaking. An aliquot from each well was serially diluted and then plated on TSA with 5% sheep blood after incubation for 48 h at 37°C with 5% CO2 for enumeration. A serum-sensitive, LPS-rough strain of Pseudomonas aeruginosa (PAO1 galU33), grown on TSA 37°C overnight and then in liquid culture in LB at 37°C overnight, was used as a positive control.
Case-control study methods.
Since all cases of Lactobacillus bacteremia in patients receiving probiotics occurred in an ICU, cases were matched with up to 3 control patients who had received probiotics in an ICU within 90 days of the case and had similar or longer length of ICU exposure prior to censoring. Controls were selected randomly using incidence density sampling. Censoring occurred at date of bacteremia or, for controls, at date of discharge, death, or transfer from the ICU. Five cases had 3 controls identified, while 1 case with a particularly long ICU stay, had only 1 possible control identified. As a result, 6 cases were compared with 16 controls in our analysis.
Patient data for the 6 cases and 16 controls were collected retrospectively by chart review onto a standardized case report form. Temperature, WBC count, C-reactive protein (CRP), and 30-day mortality were collected from the date of bacteremia or censoring. Clinical variables previously associated with either Lactobacillus bacteremia or with central-line associated bloodstream infections in general were collected for the 7 days prior to bacteremia or death/discharge37–40.
Immunodeficiency was defined as active oncologic diagnosis, solid organ or stem cell transplant, primary immunodeficiency, receipt of immunosuppressant within 6 weeks prior, or neutropenia (absolute neutrophil count (ANC) or total white blood cell (WBC) count < 500 occurring for at least two days and within three calendar days before or after the date of culture or death/discharge). Medical device data included endovascular prosthetic material, central venous catheter, tracheostomy tube, gastrostomy tube, and urinary catheter. Gastrointestinal breakdown included documentation of mucositis, diarrhea, or skin breakdown around the gastrostomy or jejunostomy insertion site. Diarrhea was identified by documentation in the physician or nursing notes or by stool output of > 20 mL/kg in a 24-hour period as per the CDC definition of mucosal barrier injury41. Antibiotic data included oral and intravenous antibiotics regardless of indication.
For the case-control study, odds ratios for continuous and categorical variables were generated by exact conditional logistic regression using SAS 9.4 (Carey, NC).
Statistics Details
Figure 3b: Kruskal-Wallis (P = 0.0297, Kruskal-Wallis statistic = 13.99) test followed by Dunn’s multiple comparisons test to P3–2 were performed. α = 0.05. Statistics Table for Figure 3b: See Supplementary Table 12a.
Extended Data Figure 4: 3 independent experiments were performed on different days. In each experiment, each bacterial isolate had 3 technical replicates. P<0.0001 by ANOVA with Tukey’s multiple comparisons test for the pairwise comparison of any of the isolates making biofilm (defined as OD570 >1) compared to either P2–1, N5, N9, N10, N11, or medium control. There were no statistically significant differences among the isolates making biofilm or among the isolates not making biofilm. F = 38.93. DF=42. Statistics Table for Extended Data Figure 4: See Supplementary Table 12b.
Supplementary Figure 1a: 2 independent experiments on different days were performed. Error bars show the interquartile range of 3 technical replicates for all apart from 50% h.i. PAO1 galU control, which had 2 technical replicates. *P = 0.0448 for PAO1 galU 50% serum versus 50% h.i. by Kruskal-Wallis test (P = 0.0297, Kruskal-Wallis statistic = 26.88) followed by Dunn’s multiple comparisons test.
Supplementary Figure 1b: 2 independent experiments on different days were performed. Error bars show SD of 3 technical replicates. The ratios t1h/t0h and t3h/t0h were used for statistical analysis. For t1h/t0h: P=0.1893 by one-way ANOVA test followed by Dunnett’s multiple comparisons test. F = 1.677. There were no statistically significant differences upon multiple pairwise comparisons against P3–2. P3–2 vs. P1–1: P = 0.7058, P3–2 vs. R1: P = 0.9998, P3–2 vs. R2: P = 0.5002, P3–2 vs. R3: P = 0.9020, P3–2 vs. R4: P = 0.9547, P3–2 vs. R5: P = 0.8192, P3–2 vs. R6: P = 0.2698. DF = 15. For t3h/t0h: P=0.1901 by one-way ANOVA test followed by Dunnett’s multiple comparisons test. F = 1.658. There were no statistically significant differences upon multiple pairwise comparisons against P3–2. P3–2 vs. P1–1: P = 0.8893, P3–2 vs. R1: P = 0.9998, P3–2 vs. R2: P = 0.8571, P3–2 vs. R3: P = 0.9998, P3–2 vs. R4: P = 0.8957, P3–2 vs. R5: P = 0.2034, P3–2 vs. R6: P = 0.9353. DF=16.
Extended Data Table 9: 3 independent experiments on different days were performed. In each experiment 3 independent bacterial cultures of each strain were used. Values shown are the median with 25% and 75% percentiles. For single culture experiment; doubling time: P > 0.9999, Mann-Whitney U = 4, no. of divisions: P > 0.9999, Mann-Whitney U = 4 by an unpaired two-tailed Mann-Whitney test. For competition culture experiment; doubling time: P = 0.1, Mann-Whitney U = 0, no. of divisions: P = 0.1, Mann-Whitney U = 0 by an unpaired two tailed Mann-Whitney test.
Please refer to the Life Sciences Reporting Summary for additional details.
Extended Data
Extended Data Figure 1.
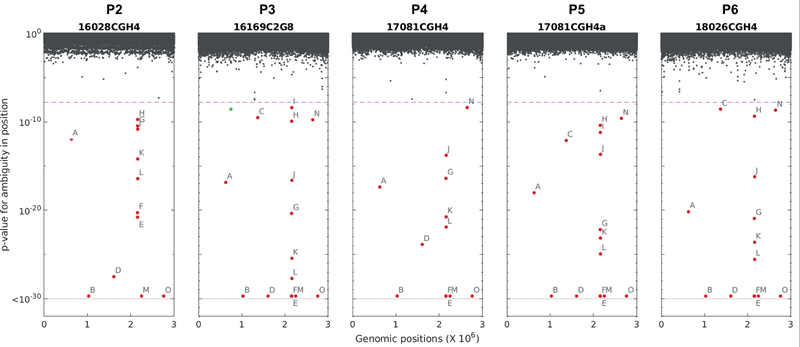
Deep sequencing identifies loci of diversity across probiotic product batches. Five probiotic batches (batches P2–P6, see Supplementary Table 2) were sequenced at high depth together with a single colony. In each batch, for each position in the reference genome, a two-sided Fisher exact test was carried out to determine differences in diversity between the batch derived sequences and the colony derived ones, and the respective p-values were plotted. Significant loci (p-value<1.66e-8) are marked with labels A-O (for details see Supplementary Table 6). A single locus of increased diversity in the colony in comparison to only one of the probiotic batches was also observed (green).
Extended Data Figure 2. The blood-isolate-specific rpoB SNP does not perturb the RpoB predicted structure but occurs near the DNA-binding site and is associated with rifampin resistance in other bacterial species.
(a) Predicted structures of L. rhamnosus GG RNA polymerase β-subunit RpoB with histidine at position 487 seen in the probiotic (blue, left), aspartic acid at position 487 seen in the blood isolate from Patient R1 (magenta, middle), and overlap (right). (b) Predicted DNA-binding site amino acids are shown in white, with the histidine (blue) of the probiotic (left) and the aspartic acid (magenta) of blood isolate from Patient R1 (right) shown compared to the DNA-binding positions. (c) Amino acid (aa) sequence alignment of the Rifampin cluster I of the RpoB protein from L. rhamnosus GG and other genera. Numbering begins and ends at the first and last aa of the cluster; asterisks depict evolutionarily conserved aa residues; red asterisk shows the conservation across species of the histidine. In magenta, aa substitution H487D of the L. rhamnosus GG rifampin-resistant isolate (Patient R1) found in this study, H481D of S. aureus M1112 rifampin-resistant isolate24, and H482D of B. velezensis rifampin-resistant isolate29; in orange, substitution H481Y of S. epidermidis RP62A rifampin-resistant isolate30, H489Y of E. faecium 343–3 rifampin-resistant isolate27, H489Y of E. faecium 40–4 rifampin-resistant isolate27, H526Y of E. coli K-12 substr. MG1655 rifampin-resistant isolate31, and H482Y of B. velezensis rifampin-resistant isolate29; in lavender, substitution H489Q of E. faecium 38–15 rifampin-resistant isolate27; in brown, substitution H482R of B. velezensis rifampin-resistant isolate29; in turquoise, substitution H482C of B. velezensis rifampin-resistant isolate29.
Extended Data Figure 3. The blood-isolate-specific ribokinase SNP does not perturb the predicted structure of ribokinase but occurs near the active site.
(a) Predicted structures of probiotic ribokinase with A259 (blue, left), blood isolate from Patient R1 with ribokinase A259D SNP (magenta, middle) and overlap (right). (b) The predicted binding site amino acids of ribokinase for adenosine are shown in white, with the alanine 259 (blue) of the probiotic (left) and the aspartic acid (magenta) of blood isolate 1 (right) shown compared to the adenosine-binding positions.
Extended Data Figure 4. Biofilm formation of probiotic and blood L. rhamnosus isolates.
Blood isolates from patients receiving (R1–R6) and those not receiving probiotics (N5, N9, N10, N11), as well as selected probiotic isolates, were tested for biofilm formation. Isolates are grouped by similar mutations, as depicted in the grid below the isolate labels. Isogenic probiotic isolates from different probiotic capsules were used as controls, if available, as were controls for mutations found in blood isolates, when available. Px-y, were x = probiotic batch number, y = probiotic isolate number. Bars depict means of three independent experiments performed on different days, with 3 technical replicates per isolate in each experiment. Error bars depict the SEM. **** P<0.0001 by ANOVA test followed by Tukey’s multiple comparisons test for the pairwise comparison of any of the isolates making biofilm (defined as OD570 >1) compared to either P2–1, N5, N9, N10, N11, or medium control. There were no statistically significant differences among the isolates making biofilm or among the isolates not making biofilm.
Supplementary Material
Supplementary Figure 1. Bacterial survival in human serum and whole blood are similar among the probiotic and blood isolates. (a) Survival in human serum of L. rhamnosus GG probiotic control strain without SNPs (P3–2) and blood L. rhamnosus isolates (R1–R6). The concentrations of serum tested were 50% vs. 50% heat-inactivated (h.i.). The bars show the medians. Error bars indicate interquartile ranges. *P = 0.0448 for 50% serum vs. 50% h.i. serum by Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Results shown are representative of 2 independent experiments. PAO1 ΔgalU is a Pseudomonas aeruginosa galU mutant, an LPS-rough, serum-sensitive strain (positive control). (b) Survival in human whole blood of probiotic strains without SNPs (P3–2) or with the CcmA SNP (P1–1), and blood isolates (R1–R6). The bars show the means of 3 technical replicates. Error bars indicate SD. P = 0.1893 (t1h/t0h) and P = 0.1901 (t3h/t0h) by ANOVA test followed by Dunnett’s multiple comparison test of survival indices at 1 and 3 hours compared to time 0, using the probiotic P3–2 as control. Results shown are representative of 2 independent experiments performed on different days using blood from two different donors. Px-y where x = probiotic batch number and y = isolate number.
Supplementary Table 1 and Supplementary Table 5.
Acknowledgements.
We thank T. Moniz who provided the probiotic administration data; J. Kinlay and P. Scanlon who noted the pattern of cases; A. Mello, who provided epidemiologic data, and R. Marshall and E. Derderian, who performed strain analysis and antibiotic susceptibility testing on the clinical isolates.
Funding. This work was funded in part by the Richard A. and Susan F. Smith President’s Innovation Award (to G.P.P.) and by funds for the Translational Research for Infection Prevention in Pediatric Anesthesia and Critical Care (TRIPPACC) Program of the Department of Anesthesiology, Critical Care and Pain Medicine at Boston Children’s Hospital (to G.P.P.), US National Institutes of Health grant R01 GM081617 (to R.K.), The Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine (to R.K.) and European Research Council FP7 ERC grant 281891 (to R.K.).
Footnotes
Competing Interests Statement
The authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the interpretation of the article.
Data Availability
Sequence data are available under BioProjectID PRJNA562050. BioSample accession numbers are: SAMN12632778-SAMN12632834. Figure 3, Extended Data Figure 4, Supplementary Table 9, and Supplementary Figure 1 have associated raw data. All other data are available from the corresponding authors upon reasonable request.
References
- 1.Sarah HY, Jernigan JA & McDonald LC Prevalence of probiotic use among inpatients: a descriptive study of 145 US hospitals. Am. J. Infect. Control 44, 548–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szajewska H What are the indications for using probiotics in children? Arch. Dis. Child 101, 398–403 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Barraud D, Bollaert P-E & Gibot S Impact of the administration of probiotics on mortality in critically ill adult patients: a meta-analysis of randomized controlled trials. Chest 143, 646–655 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Barraud D et al. Probiotics in the critically ill patient: a double blind, randomized, placebo-controlled trial. Intensive Care Med. 36, 1540–1547 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Honeycutt TCB et al. Probiotic administration and the incidence of nosocomial infection in pediatric intensive care: a randomized placebo-controlled trial. Pediatr. Crit. Care Med. 8, 452–8; quiz 464 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Suez J et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell 174, 1406–1423.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Zmora N et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 174, 1388–1405.e21 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Kunz AN, Noel JM & Fairchok MP Two cases of Lactobacillus bacteremia during probiotic treatment of short gut syndrome. J. Pediatr. Gastroenterol. Nutr 38, 457–458 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Salminen MK et al. Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. rhamnosus GG. Clin. Infect. Dis 38, 62–69 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Thomas DW & Greer FR Clinical report—probiotics and prebiotics in pediatrics. Pediatrics (2010). [DOI] [PubMed] [Google Scholar]
- 11.Ghouri YA et al. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin. Exp. Gastroenterol 7, 473–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodorakopoulou M, Perros E, Giamarellos-Bourboulis EJ & Dimopoulos G Controversies in the management of the critically ill: the role of probiotics. Int. J. Antimicrob. Agents 42 Suppl, S41–4 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Zhang G-Q, Hu H-J, Liu C-Y, Shakya S & Li Z-Y Probiotics for Preventing Late-Onset Sepsis in Preterm Neonates: A PRISMA-Compliant Systematic Review and Meta-Analysis of Randomized Controlled Trials. Medicine 95, e2581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siempos II, Ntaidou TK & Falagas ME Impact of the administration of probiotics on the incidence of ventilator-associated pneumonia: a meta-analysis of randomized controlled trials. Crit. Care Med. 38, 954–962 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Gu W-J, Wei C-Y & Yin R-X Lack of efficacy of probiotics in preventing ventilator-associated pneumonia probiotics for ventilator-associated pneumonia: a systematic review and meta-analysis of randomized controlled trials. Chest 142, 859–868 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Oudhuis GJ, Bergmans DCJJ & Verbon A Probiotics for prevention of nosocomial infections: efficacy and adverse effects. Curr. Opin. Crit. Care 17, 487–492 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Lolis N et al. Saccharomyces boulardii fungaemia in an intensive care unit patient treated with caspofungin. Crit. Care 12, 414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebeer S, Vanderleyden J & De Keersmaecker SCJ Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev 72, 728–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salminen MK et al. Lactobacillus bacteremia during a rapid increase in probiotic use of Lactobacillus rhamnosus GG in Finland. Clin. Infect. Dis 35, 1155–1160 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Chung H et al. Global and local selection acting on the pathogen Stenotrophomonas maltophilia in the human lung. Nat. Commun 8, 14078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieberman TD et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med 22, 1470–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman TD et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet 43, 1275–1280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith EE et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A 103, 8487–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y, Cui L, Katayama Y, Kozue K & Hiramatsu K Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol 49, 2680–2684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua X et al. Global Effect of rpoB Mutation on Protein Expression in Enterococcus faecium. Jundishapur Journal of Microbiology 9, (2016). [Google Scholar]
- 26.Campbell EA et al. Structural, functional, and genetic analysis of sorangicin inhibition of bacterial RNA polymerase. EMBO J. 24, 674–682 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enne VI, Delsol AA, Roe JM & Bennett PM Rifampicin resistance and its fitness cost in Enterococcus faecium. J. Antimicrob. Chemother 53, 203–207 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Lebeer S et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol 78, 185–193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai X-C et al. Rifampicin-Resistance Mutations in the rpoB Gene in Bacillus velezensis CC09 have Pleiotropic Effects. Front. Microbiol 8, 178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wi YM et al. Rifampicin resistance in Staphylococcus epidermidis: molecular characterisation and fitness cost of rpoB mutations. Int. J. Antimicrob. Agents 51, 670–677 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Xu M, Zhou YN, Goldstein BP & Jin DJ Cross-resistance of Escherichia coli RNA polymerases conferring rifampin resistance to different antibiotics. J. Bacteriol 187, 2783–2792 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baym M et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10, e0128036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priebe GP et al. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun 72, 4224–4232 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebeer S, Verhoeven TLA, Perea Vélez M, Vanderleyden J & De Keersmaecker SCJ Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol 73, 6768–6775 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical & Laboratory Standards Institute--CLSI. Performance Standards of Antimicrobial Susceptibility Testing; 27th Informational Supplement; M100–S27. (2017). [Google Scholar]
- 36.Mariam DH, Mengistu Y, Hoffner SE & Andersson DI Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 1289–1294 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wylie MC et al. Risk Factors for Central Line–Associated Bloodstream Infection in Pediatric Intensive Care Units. Infect. Control Hosp. Epidemiol 31, 1049–1056 (2010). [DOI] [PubMed] [Google Scholar]
- 38.De Groote MA, Frank DN, Dowell E, Glode MP & Pace NR Lactobacillus rhamnosus GG bacteremia associated with probiotic use in a child with short gut syndrome. Pediatr. Infect. Dis. J 24, 278–280 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Ledoux D, Labombardi VJ & Karter D Lactobacillus acidophilus bacteraemia after use of a probiotic in a patient with AIDS and Hodgkin’s disease. Int. J. STD AIDS 17, 280–282 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Gouriet F, Million M, Henri M, Fournier P-E & Raoult D Lactobacillus rhamnosus bacteremia: an emerging clinical entity. Eur. J. Clin. Microbiol. Infect. Dis 31, 2469–2480 (2012). [DOI] [PubMed] [Google Scholar]
- 41.See I et al. Mucosal barrier injury laboratory-confirmed bloodstream infection: results from a field test of a new National Healthcare Safety Network definition. Infect. Control Hosp. Epidemiol 34, 769–776 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Bacterial survival in human serum and whole blood are similar among the probiotic and blood isolates. (a) Survival in human serum of L. rhamnosus GG probiotic control strain without SNPs (P3–2) and blood L. rhamnosus isolates (R1–R6). The concentrations of serum tested were 50% vs. 50% heat-inactivated (h.i.). The bars show the medians. Error bars indicate interquartile ranges. *P = 0.0448 for 50% serum vs. 50% h.i. serum by Kruskal-Wallis test followed by Dunn’s multiple comparisons test. Results shown are representative of 2 independent experiments. PAO1 ΔgalU is a Pseudomonas aeruginosa galU mutant, an LPS-rough, serum-sensitive strain (positive control). (b) Survival in human whole blood of probiotic strains without SNPs (P3–2) or with the CcmA SNP (P1–1), and blood isolates (R1–R6). The bars show the means of 3 technical replicates. Error bars indicate SD. P = 0.1893 (t1h/t0h) and P = 0.1901 (t3h/t0h) by ANOVA test followed by Dunnett’s multiple comparison test of survival indices at 1 and 3 hours compared to time 0, using the probiotic P3–2 as control. Results shown are representative of 2 independent experiments performed on different days using blood from two different donors. Px-y where x = probiotic batch number and y = isolate number.
Supplementary Table 1 and Supplementary Table 5.
Data Availability Statement
Illumina files from gene sequencing have been deposited in the public database. Accession codes will be provided prior to publication. Higher level analysis (e.g. SNP calling) is provided in supplementary materials. All other data are available from the authors upon reasonable request.