Abstract
Mitochondrial function in white adipose tissue (WAT) is an important yet understudied aspect in adipocyte biology. Here, we report a role for amyloid precursor protein (APP) in compromising WAT mitochondrial function through a high-fat diet (HFD)-induced, unconventional mis-localization to mitochondria that further promotes obesity. In humans and mice, obese conditions significantly induce APP production in WAT and its enrichment in mitochondria. Mechanistically, a HFD-induced dysregulation of signal recognition particle subunit 54c is responsible for the mis-targeting of APP to adipocyte mitochondria. Mis-localized APP blocks the protein import machinery, leading to mitochondrial dysfunction in WAT. Adipocyte-specific and mitochondria-targeted APP overexpressing mice display increased body mass and reduced insulin sensitivity, along with dysfunctional WAT due to a dramatic hypertrophic program in adipocytes. Elimination of adipocyte APP rescues HFD-impaired mitochondrial function with significant protection from weight gain and systemic metabolic deficiency. Our data highlights an important role of APP in modulating WAT mitochondrial function and obesity-associated metabolic dysfunction.
Introduction
A better understanding of the processes that cause adipose tissue (AT) dysfunction is essential for understanding the pathophysiology of obesity, and could identify novel targets for preventing and treating obesity-associated comorbidities1, 2. Weight gain causes an expansion of AT that involves adipocyte hypertrophy3, 4, which contributes to impaired adipocyte function and subsequent AT inflammation and fibrosis5. Large adipocytes are resistant to insulin’s action on lipolysis and glucose uptake, and show impaired secretion of adiponectin, an insulin sensitizing hormone5. Hypertrophic adipocytes are at increased risk of hypoxia, which drives a fibrotic program that limits healthy AT expansion6, 7. Clinical studies have correlated enlarged adipocytes to AT dysfunction, whole-body metabolic defects and systemic insulin resistance8. Increased subcutaneous adipocyte size is a predictor of obesity-related comorbidities, such as type 2 diabetes9. Accordingly, targeting adipocyte hypertrophy has the potential to improve metabolic function in people with obesity. However, the driving forces for adipocyte enlargement have yet to be identified.
Another characteristic of metabolically unhealthy adipocytes is mitochondrial dysfunction10, 11, which affects adipogenesis, adipokine secretion, lipogenesis, and lipolysis12, 13. Evidence from human and animal studies show that obesity is associated with AT mitochondrial dysfunction, manifested by reduced mitochondrial DNA (mtDNA) in white AT (WAT), decreased electron transport chain (ETC) gene expression, impaired mitochondrial oxidative capacity, and increased reactive oxygen species (ROS)14. By employing transgenic and tissue specific knockout models in rodents, several factors that modulate AT mitochondrial function have been identified, including uncoupling protein 1 (UCP1)15, peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α)16, and mitoNEET17.
Amyloid precursor protein (APP) is intensively investigated in the neuroscience field, due to its essential contribution to Alzheimer’s disease (AD) pathogenesis through the generation of toxic amyloid β (Aβ) aggregates, potentially causing neurodegeneration18. However, very few studies have been conducted to investigate the action of APP in peripheral organs. Notably, APP is expressed in both mouse and human WAT, where it is significantly induced by high-fat diets (HFD)19, 20. Zheng et al. noticed that systemic App-deficiency leads to 15%–20% reduction in body weight21, but the reasons for the lighter body mass remain unknown.
Here, we report an unconventional location for APP in adipocyte mitochondria under obese conditions, and an unexpected role of APP in regulating adipocyte mitochondrial function, lipolysis and hypertrophy. Following the observation that APP is up-regulated in AT and enriched in adipocyte mitochondria with obesity, we generated adipocyte-specific, doxycycline (Dox)-inducible App overexpressing and knockout mouse models. Enhanced presence of APP in mitochondria impairs adipocyte mitochondrial function, resulting in a suppressed lipolysis and rapid adipocyte hypertrophy. Adipocyte-specific elimination of App maintains mitochondrial respiration, protects mice from HFD-induced adipocyte hypertrophy, and promotes systemic metabolic health.
Results
APP is increased in WAT in obese human and mice and accumulates in adipocyte mitochondria.
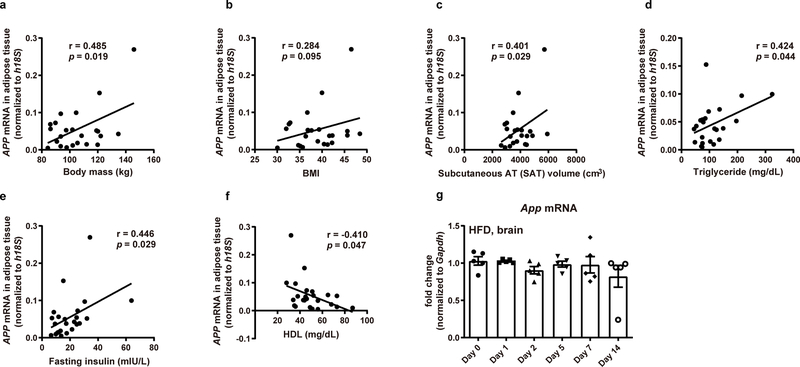
We aimed to examine the alterations in APP levels in human and mouse WAT, particularly in the context of obesity. We found an inverse relationship between sWAT APP mRNA levels and insulin sensitivity in people with obesity, as judged by the glucose infusion rate (GIR) during a hyperinsulinemic-euglycemic clamp procedure (Fig. 1a). In addition, human sWAT APP mRNA levels also show positive correlations with body mass (Extended Data Fig. 1a), body mass index (BMI) (Extended Data Fig. 1b), subcutaneous adiposity (Extended Data Fig. 1c), plasma triglycerides (Extended Data Fig. 1d), and fasting insulin levels (Extended Data Fig. 1e), but are inversely correlated with high-density lipoprotein (HDL)-cholesterol (Extended Data Fig. 1f). These findings demonstrate that increased sWAT APP expression is associated with obesity, insulin resistance, and atherogenic dyslipidemia in humans. Furthermore, we evaluated public human genomic data resources to gather more evidence demonstrating the clinical relevance of APP changes in human obesity. In a report by Pietilainen et al.22, global gene expression analysis (E-MEXP-1425) was performed utilizing sWAT samples from lean and obese monozygotic twin pairs, and APP levels are significantly increased in the obese co-twins (Fig. 1b). In another study conducted in 20 non-obese and 19 obese Pima Indians (GSE2508)23, we found the expression of APP in sWAT is higher in obese than in non-obese groups (Fig. 1c). One study (GSE27951)24 that reported adipocyte size in sWAT demonstrated that APP expression progressively increases with enhancements in mean adipocyte diameter (Fig. 1d).
Fig. 1. APP is increased in WAT in obese human and mice and accumulates in adipocyte mitochondria.
(a) Relationship between sWAT APP mRNA levels and insulin sensitivity, assessed as the glucose infusion rate divided by plasma insulin concentration (M/I) during a hyperinsulinemic-euglycemic clamp procedure in people with obesity (n=23). FFM: fat free mass. (b-d) APP expression extracted from human genomic databases: APP levels in sWAT from lean and obese co-twins (b, Accession #: E-MEXP-1425, n=11 subjects per group), from lean and obese female (F) or male (M) Pima Indians (c, Accession #: GSE2508, n=10 (lean F, obese F and lean M) or 9 (obese M) subjects per group), and from human subjects with a range of subcutaneous adipocyte sizes (d, Accession #: GSE27951, n=12 (115μm), 9 (121μm) or 11 (125μm) subjects per group. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range). (e-h) App mRNA levels in different tissues from Ob/Ob and wild-type mice (e, n=5), in 30-week HFD and chow fed mice (f, n=5), in adipocytes (n=5) and stromal vascular fractions (SVF, n=6) from sWAT (left) and eWAT (right) in 30-week HFD and chow fed mice (g) and App mRNA levels from sWAT (left) and eWAT (right) in acute HFD challenged wild-type mice for 0, 1, 2, 5, 7 and 14 days (h, n=7 mice for Day 0 group; n=5 mice for Day 1, 2, 5, 7 and 14 groups). (i) Representative Western blotting image (left panel) for APP in mitochondrial (Mito), post-mitochondria (Post-mito, including cytosol and microsomal (ER) compartments) fractions, and whole tissue lysate from sWAT in 30-week HFD and chow fed mice and its quantification (right panel). COXIV: mitochondrial marker; β-Tubulin: cytoplasmic marker; Calreticulin: ER marker. n=3 mice per group. Images are chosen from three independent experiments. Data are presented as mean ± SEM of biologically independent samples. Pearson correlation analysis for correlation coefficient (r) and two-tailed p-value (a); Two-tailed Student’s t-test (b-c, e-g, i); One-way ANOVA test followed by a Tukey post-test (d, h). See also Extended Data Fig. 1.
In agreement with above human studies, we demonstrated that App mRNA levels are significantly up-regulated in both the ob/ob mice (Fig. 1e) and HFD-induced obese mice (Fig. 1f). The increase of APP expression is much greater in WAT than in other metabolically active organs examined, such as brown AT (BAT) and liver. We further demonstrated that in both sWAT and epididymal WAT (eWAT), the transcription of App is enhanced specifically in adipocytes, but not in the stromal vascular fractions (SVF) (Fig. 1g). In addition to long-term (30 weeks) HFD insults, we also fed HFD for shorter periods, and showed that an acute HFD challenge significantly induces App levels in both sWAT and eWAT (Fig. 1h), but not in the brain (Extended Data Fig. 1g). Exposing mice to HFD for 24 hrs, we noticed a ~100-fold increase of App mRNA levels in sWAT, suggesting that App induction is among the earliest adverse effects of a HFD insult.
The mechanistic details as to how higher levels of APP cause cellular changes are not known. To facilitate defining the function of APP, we explored the intracellular localization of the upregulated APP protein in WAT. Naturally, the majority of APP is targeted to the endoplasmic reticulum (ER) and subsequently enter the secretory pathway25. Surprisingly, we observed that in addition to an increase in the cytoplasmic fractions, APP protein is also significantly enriched in adipocyte mitochondria (Fig. 1i). This unconventional localization to mitochondria prompted us to further pursue studying the impact of APP on adipose biology in relationship with adipocyte mitochondrial function.
Adipocyte-specific APP overexpressing mice are more sensitive to diet induced obesity.
To directly investigate the role of APP in white adipocytes in a tissue/cell-specific manner, we generated a mouse model carrying a tetracycline responsive element (TRE)-driven and mitochondria-targeted APP transgenic cassette (TRE-mito-APP). By crossing TRE-mito-APP mice with the adiponectin promoter-driven rtTA expressing mice (Adipo-rtTA, APP-), we achieved adipocyte-specific, Dox-inducible APP overexpression (Adipo-APP, APP+) (Fig. 2a). After HFD/Dox feeding, we assessed App mRNA levels in different fat pads and other tissues, and there is a significant induction in App mRNA in sWAT and eWAT (Fig. 2b). However, there is no increase in App expression observed in mesenteric WAT (mWAT) and perirenal WAT (pWAT), or the liver and the brain (Fig. 2b and Extended Data Fig. 2a). APP transgenic mice display significantly higher levels of APP protein in sWAT (Fig. 2c). Additionally, we also validated the substantial increase of APP expression and co-localization with mitochondrial marker TIM23 in sWAT in transgenic mice (Fig. 2d). The over-expressed APP significantly accumulates in mitochondria, as suggested by a much higher level of APP protein detected in mitochondria purified from APP transgenic mouse sWAT (Extended Data Fig. 2b), and further supported by a significant co-localization of APP and TIM23 in the TRE-mito-APP transfected HEK293T cells (Extended Data Fig. 2c).
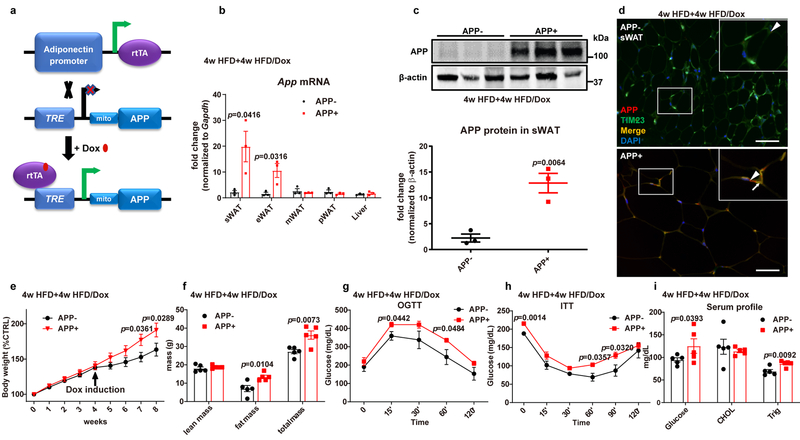
Fig. 2. Adipocyte-specific APP overexpressing mice are more sensitive to diet induced obesity.
(a) Schematic illustration of the adipocyte-specific, Dox-inducible APP transgenic mouse model. A mitochondrial (mito) pre-sequence is connected to APP695 cDNA to target overexpressed APP into adipocyte mitochondria. (b-d) Validation of APP overexpression in WAT of transgenic mice fed with 4-week HFD plus 4-week HFD/Dox 600mg/kg: (b) App mRNA levels in different tissues from control (APP-) and APP overexpressing (APP+) mice (n=4 mice per group); (c) Representative Western blotting image (upper panel) for APP protein levels in sWAT and its quantification (lower panel) (n=3 mice per group); (d) Immunofluorescence staining for APP (red) and TIM23 (green) in sWAT, DAPI (blue) for nuclear labeling, bar = 60 μm. Triangles indicate over-expressed APP signals, and arrows indicate the merge between APP and TIM23 signals. Representative images in c and d are chosen from three independent experiments. (e-i) Both control and APP transgenic mice are subject to the following metabolic analyses: (e) Relative body weight for 8 weeks; (f) Body composition data obtained via NMR system; Glucose levels at different time-points from (g) OGTT and (h) ITT experiments; (i) Fasting glucose, cholesterol and triglyceride levels in serum samples from both groups. n=8 mice (e, g, h) or n=5 mice (f, i) per group. For all the statistics: data are presented as mean ± SEM of biologically independent samples. Two-tailed Student’s t-test (b-c, f, i); Two-way ANOVA followed by a Tukey post-test (e, g-h). See also Extended Data Fig. 2.
We aimed to evaluate the metabolic consequences of mitochondrial APP in response to HFD feeding. Body weights start to diverge almost as soon as the transgene is induced by Dox, and reach a difference with statistical significance around week 7 or 8 (Fig. 2e). The body weight increase is primarily due to an increase in fat mass (Fig. 2f). Concomitant with the obese phenotype, the oral glucose tolerance is impaired in APP overexpressing mice (Fig. 2g). We also measured circulating insulin levels during the OGTT and found that both basal and glucose-stimulated insulin concentrations are markedly increased in APP overexpressing mice (Extended Data Fig. 2d). Consistent with the results from the OGTT, an insulin tolerance test (ITT) also demonstrates decreased insulin sensitivity in APP transgenic mice (Fig. 2h). Beyond the effects on carbohydrate metabolism, we also observed a difference in circulating lipid profiles. Although cholesterol levels remain unchanged, serum triglycerides are significantly elevated in APP transgenic mice (Fig. 2i). Taken together, increased APP in WAT increases HFD induced body weight gain, glucose intolerance, insulin resistance, and dyslipidemia.
APP overexpression causes dysfunctional WAT and liver steatosis.
To understand how the local APP induction results in profound metabolic changes, we examined the detailed temporal events during AT expansion. sWAT from APP transgenic mice displays a dysfunctional expansion phenomenon with significantly enlarged adipocytes and enhanced crown-like structures (Fig. 3a). Similarly, we also observed substantially larger adipocyte size and dramatically increased infiltration of inflammatory cells in eWAT (Fig. 3b). Gene expression analysis validates an enhanced inflammatory program in both sWAT and eWAT after APP overexpression (Figs. 3c and 3d). Although anti-inflammatory (M2-like) macrophage markers Il10, Mrc1 and Clec10a are unaffected, generic macrophage signature genes Adgre1 and Il6 show a significant elevation; pro-inflammatory (M1) macrophage markers such as Ifng, Tnfa and Nos2 are highly induced in sWAT upon APP overexpression. The eWAT of APP transgenic mice also demonstrates an enhanced pattern of pro-inflammatory gene expression. Obesity is frequently associated with AT inflammation that is linked to increased AT fibrosis6. Thus, we also probed for the expression of factors involved in AT fibrosis. The major driver for AT fibrosis, Hif1a, is significantly upregulated at the transcriptional level in both sWAT and eWAT of APP transgenic mice (Figs. 3c and 3d), further highlighted by significantly increased expression of Hif1a target genes, such as Col1a1, Col3a1 and Col6a1 in sWAT (Fig. 3c), as well as Lox, Col3a1 and Col6a1 in eWAT (Fig. 3d). We also tested circulating adiponectin levels and found that APP transgenic mice display significantly lower levels of adiponectin in plasma (Fig. 3e), highlighting the influence of AT dysfunction as a systemic mediator of metabolic health. Highly inflammatory and fibrotic AT is characteristic of dysfunctional fat, usually related to unfavorable changes in the liver. Adipo-APP transgenic mice rapidly develop liver steatosis, while control mice remain mostly protected from HFD-induced lipid accumulation (Fig. 3f). Lipid profiling further shows a significant elevation in liver triglycerides levels (Fig. 3g). By demonstrating significantly elevated AT inflammation/fibrosis and decreased adiponectin, we link adipocyte-specific APP overexpression to a dysfunctional AT expansion, which leads to impaired whole-body metabolism.
Fig. 3. APP overexpression causes dysfunctional WAT and liver steatosis.
(a-b) Representative H&E staining images from three independent cohorts of sWAT (a) and eWAT (b) sections from 4-week HFD plus 4-week HFD/Dox 600mg/kg fed control and APP overexpressing mice. Bar = 60 μm. (c-d) Inflammatory and fibrosis related gene mRNA levels assessed by qPCR in sWAT (c) and eWAT (d) of control and APP transgenic mice. General inflammatory (MΦ, macrophages) markers: Adgre1 (F4/80), Il6 (IL-6); anti-inflammatory macrophage (M2 MΦ, M2 macrophages) markers: Mrc1 (CD206), Il10 (IL-10), Clec10a (CD301); pro-inflammatory macrophage (M1 MΦ, M1 macrophages) markers: Ifng, Tnfa, Nos2; fibrotic markers: Hif1a, Lox, Col1a1, Col3a1, Col6a1 and Tgfb1. n=5 mice per group. (e) Circulating adiponectin immunoblot image (upper panel) and quantification (lower panel) in control and APP transgenic mice. n=3 mice per group. Image is chosen from three independent experiments. (f) Representative H&E staining images of liver tissues from both groups, chosen from three independent staining experiments. Bar = 161 μm. (g) Cholesterol and triglyceride levels in liver samples from both groups. n=5 mice per group. For all the statistics: data are presented as mean ± SEM of biologically independent samples. Two-tailed Student’s t-test (c-e, g).
APP overexpression leads to adipocyte hypertrophy by impairing stimulated lipolysis.
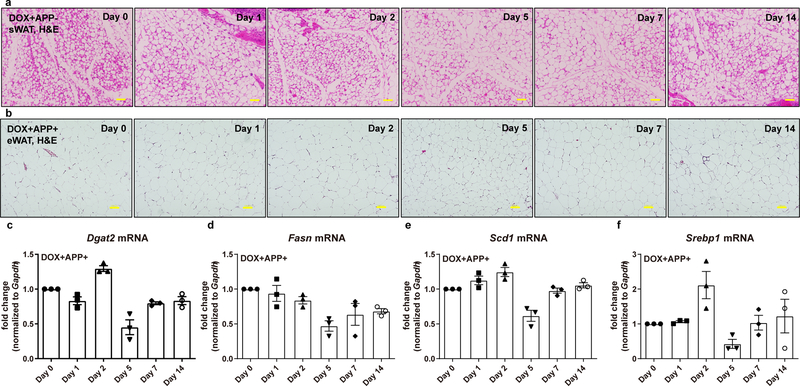
The driving force for AT dysfunction after APP induction is unclear. Next, we took advantage of the Dox-inducibility of the APP transgenic mouse model to examine the very initial changes in AT by subjecting mice to acute Dox feeding. With the acute induction, APP overexpressing sWAT displays a rapid enlargement in adipocyte size (Fig. 4a), yet without a significant difference on total body weight. Not surprisingly, the average adipocyte size in sWAT from control mice remains stable (Extended Data Fig. 3a). This demonstrates that APP overexpression per se, even in the absence of HFD, is sufficient to induce adipocyte hypotrophy. In addition, the presence of perilipin-1 is largely maintained, suggesting that adipocytes stayed viable (Fig. 4b). By performing image-based quantitative analysis to map the distribution of adipocyte cell size for each time-point, we saw a marked and progressive shift from smaller to larger adipocytes in sWAT over time (Fig. 4c). The increase in average adipocyte size starts within 24 hours of APP induction, and continues to progress from day 2 until day 14 (Fig. 4d). In contrast to sWAT, we did not observe a comparable increase in adipocyte size in eWAT (Extended Data Fig. 3b), suggesting that the sWAT is the primary tissue affected upon APP overexpression. Indeed, APP expression is significantly higher in sWAT compared to eWAT (Fig. 2b).
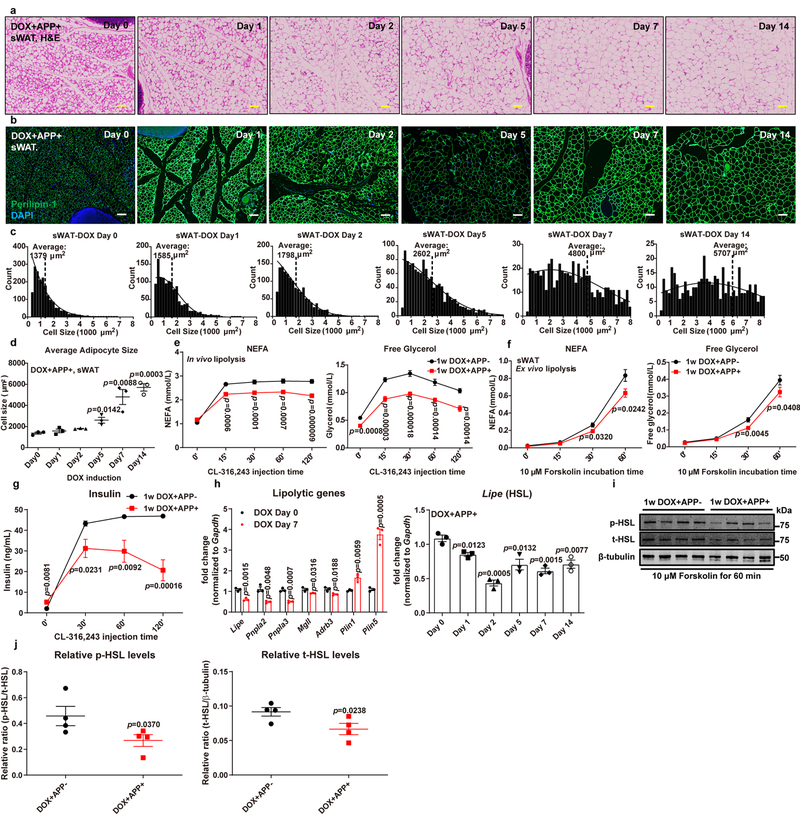
Fig. 4. APP overexpression leads to adipocyte hypotrophy by impairing stimulated lipolysis.
(a-d) Rapid enlargement of subcutaneous adipocytes in APP transgenic mice under acute Dox 600mg/kg induction for 0, 1, 2, 5, 7 and 14 days: Representative images for H&E staining (a) and perilipin-1 (Green) immunofluorescence staining (b) in sWAT sections from different time-points, chosen from two independent experiments. Bar = 161 μm; (c) Cell count distribution according to adipocyte cell size at different time-points, replicated in two independent cohorts; (d) Average adipocyte size in sWAT at different time-points. n=3 mice per time point. (e-g) Both control and APP transgenic mice induced by 1-week Dox 600mg/kg feeding are subject to in vivo and ex vivo lipolysis analysis: (e) Serum NEFA (left), free glycerol (right) and (g) insulin levels at different time-points in both groups after β3-adrenoceptor agonist CL-316,243 (1 mg/kg) injection, n=8 mice per group; (f) NEFA (left) and free glycerol (right) levels in the mediums obtained from ex vivo cultured sWAT fat pads at different time points of 10 μM forskolin incubation, n=12 tissues per group. (h) Lipolytic gene expressions in 7-day Dox 600mg/kg fed control and APP overexpressing mice (left panel, n=3 mice per group) and a time-course change for Lipe (gene name for HSL) transcriptions in sWAT of APP transgenic mice after Dox induction (right panel, n=3 mice per time point). (i-j) (i) Representative immunoblot analysis of phosphorylated HSL (p-HSL) and total HSL (t-HSL) levels in the sWAT fat pads from both groups after ex vivo lipolysis assays, and images are chosen from three independent experiments; (j) Quantification for relative p-HSL and t-HSL expression, n=4 tissues per group. For all statistics: data are shown as mean ± SEM of biologically independent samples. One-way ANOVA followed by a Tukey post-test (d, right panel in h); Two-way ANOVA followed by a Tukey post-test (e-g); Two-tailed Student’s t-test (left panel in h, j). See also Extended Data Fig. 3.
We aimed to dissect the underlying mechanism(s) as to how adipocytes can undergo such a rapid onset of hypotrophy upon APP induction. Mechanistically, adipocyte size is controlled by the balance between lipid storage and mobilization, which is mainly driven by lipogenesis and lipolysis26. We did not find any significant changes for genes that are critical for fatty acid synthesis in sWAT after acute induction of APP (Extended Data Fig. 3c–f), suggesting that de novo lipogenesis is not the primary reason for APP-induced adipocyte hypertrophy. Thus, we put our focus on investigating the impact of APP on lipolytic processes. The increase in serum non-esterified fatty acid (NEFA) and free glycerol levels in APP overexpression mice is blunted upon injection of the selective β3-adrenergic receptor agonist CL-316,243 (Fig. 4e). Consistently, we also confirmed that sWAT fat pads taken from APP transgenic mice demonstrate an impaired response to forskolin-induced NEFA and glycerol release (Fig. 4f). Furthermore, we noticed that while control mice show enhanced circulating insulin levels, APP overexpressing mice fail to trigger a marked release of insulin in response to the CL-316,243 (Fig. 4g). In addition to functional read outs, we detected changes in lipolysis-related gene transcription, and demonstrated that expression levels for critical enzymes involved in fatty acid breakdown are significantly decreased, such as Lipe (HSL), Pnpla2 (ATGL) and Pnpla3; meanwhile, Plin1 and Plin5, which negatively control adipocyte lipolysis, are markedly upregulated (Fig. 4h). Notably, as a key rate-limiting factor in lipolysis, Lipe displays a time-dependent downregulation after APP induction (Fig. 4h). At the protein level, APP overexpression not only decreases total HSL expression, but also attenuates the phosphorylation of HSL (Figs. 4i–j), in support of HSL suppression playing an important role in APP-mediated inhibition of lipolysis. We conclude that acute APP induction in adipocytes causes hypertrophy of sWAT at least in part due to impaired stimulated lipolysis, though we cannot rule out other mechanisms during regular feeding/fasting conditions in these mice.
APP impairs adipocyte mitochondrial function due to defective mitochondrial protein import.
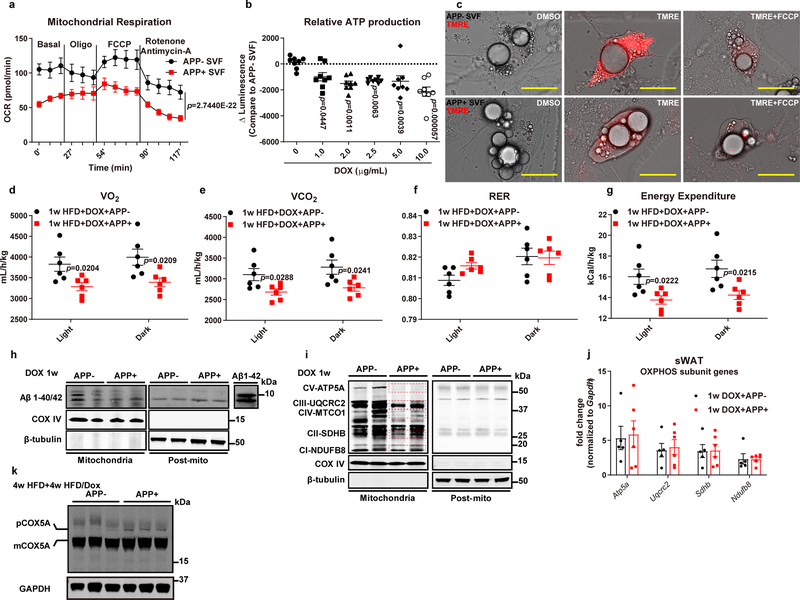
To test whether the inhibited adipocyte lipolysis by APP is associated with mitochondrial dysfunction, we performed a time-course study to investigate the impact of APP induction on mitochondrial respiration in adipocytes. APP overexpression in adipocytes dramatically impairs mitochondrial function time-dependently (Fig. 5a). Notably, the mitochondrial dysfunction in sWAT happens within one day after Dox, in line with the APP-induced rapid adipocyte enlargement. We also performed mitochondrial respiration assays on in vitro differentiated adipocytes, and consistent with the ex vivo decline in mitochondrial respiration, we observed a significant impairment in basal oxygen consumption, ATP-linked respiration, and maximal respiratory capacity in APP overexpressing adipocytes (Extended Data Fig. 4a). In addition, adipocytes derived from APP transgenic mice have a significantly diminished membrane potential, as judged by the loss in mitochondrial fluorescence in the presence of TMRE (Extended Data Fig. 4c). Further, APP overexpression results in a decline in ATP production in vitro (Extended Data Fig. 4b). To demonstrate that the mitochondrial dysfunction in adipocytes influences whole body energy expenditure, we performed metabolic cage studies. APP transgenic mice have lower levels of oxygen consumption and CO2 production during both light and dark cycles (Fig. 5b, Extended Data Fig. 4d–e). Although the respiratory exchange ratio (RER) is identical between genotypes (Extended Data Fig. 4f), energy expenditure in APP overexpressing mice is significantly suppressed (Extended Data Fig. 4g). In addition to functional changes, we also assessed the morphological alterations in adipocyte mitochondria. Mitochondria in APP overexpressing adipocytes show elongation, and typical mitochondrial cristae structures disappear (Fig. 5c), consistent with APP impairing mitochondrial respiration.
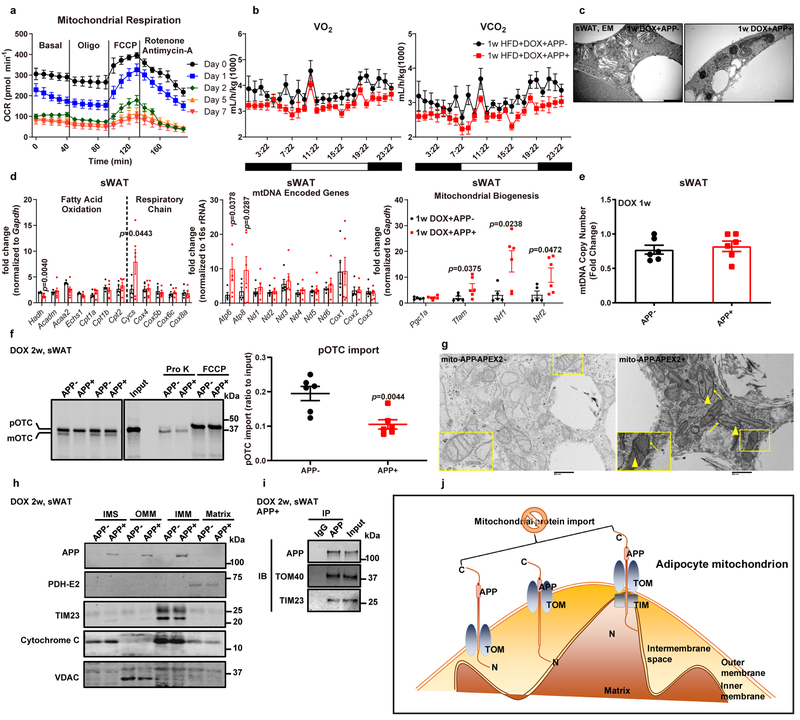
Fig. 5. APP impairs adipocyte mitochondrial function due to defective mitochondrial protein import.
(a) Ex vivo mitochondrial respiration in sWAT fat pads from APP transgenic mice fed with different periods of Dox 600 mg/kg diet. Oxygen consumption rate (OCR) at basal level and post Oligomycin (Oligo), FCCP and Rotenone/Antimycin-A injection is shown. n=5 tissues each time point. (b) Oxygen consumption (VO2, left) and carbon dioxide production (VCO2, right) measured from indirect calorimetry of control and APP overexpressing mice under 1-week HFD/Dox challenge. n=6 mice per group. Representative images for one dark-night cycle are shown. (c) EM images for sWAT sections from both groups under 1-week Dox 600mg/kg induction. Bar = 1,000 nm. Images are chosen from two independent biological samples. (d-e) Mitochondrial function related gene expressions (d) and mtDNA copy number (e) in 1-week Dox fed control and APP transgenic mice. For mtDNA copy number, fold change is normalized to the control with the highest copy number value. n=5 mice in APP- group and 6 mice in APP+ group. (f) Representative autoradiography image (left) and statistics (right) of pOTC import assessed in isolated mitochondria (incubation for 30 minutes) from sWAT of 2-week Dox fed control or APP transgenic mice. 25% of [35S]pOTC added to each reaction is loaded as input. 0.4 mg/mL protease K (Pro K) is added to digest unimported pOTC proteins. 40 μmol/L FCCP inhibits pOTC import via depolarizing the mitochondria (negative control). n=6 mice per group. Images are chosen from three independent experiments. (g) EM images for differentiated adipocytes from SVF isolated from control (left) or APP-APEX2 transgenic mice (right). Arrows: staining associated with the mitochondrial outer membrane; arrowheads: dark contrast in the cristae. Bar = 0.5 μm. Images are chosen from two independent biological samples. (h) Representative Western blotting image for APP in mitochondrial sub-localization of purified mitochondria from sWAT in control and APP transgenic mice. PDH-E2: mitochondrial matrix; TIM23: inner mitochondrial membrane (IMM); Cytochrome C: intermembrane space (IMS); VDAC: outer mitochondrial membrane (OMM). (i) Representative Western blotting image of co-immunoprecipitation (IP) analysis using either IgG or anti-APP antibody incubated with isolated mitochondria from APP transgenic mice fed with 2-week Dox diet. The input (20% of lysate) and IP samples are subject to immunoblot (IB) with APP, TOM40, and TIM23 primary antibodies. Images are chosen from three independent experiments in h and i. (j) The proposed “clogging” model. For all statistics: data are shown as mean ± SEM of biologically independent samples. Two-tailed Student’s t-test (d-f). See also Extended Data Fig. 4–5.
How does APP enhancement cause dysfunctional mitochondria? At the transcriptional level, we found that the majority of mitochondrial genes involved in fatty acid oxidation and respiratory chain function remain unchanged, along with unaltered levels of most mitochondrial DNA-encoded genes (Fig. 5d). Furthermore, several key mitochondrial components, such as Cycs, Atp6 and Atp8, and mitochondrial biogenesis regulators Tfam, Nrf1 and Nrf2 show significantly increases, perhaps as a counter-regulatory response to mitochondrial dysfunction (Fig. 5d). We additionally found that APP overexpression does not lead to changes in mtDNA copy number (Fig. 5e). We conclude that the APP-mediated mitochondrial dysfunction is not related to a transcriptional phenomenon. Previous reports suggested that the accumulation of amyloid-β 40/42 (Aβ 40/42) plays a toxic role for multiple aspects of neuronal mitochondrial function27. However, we did not observe an accumulation of Aβ levels in isolated mitochondria after APP induction (Extended Data Fig. 4h), suggesting that in our adipocyte-specific APP transgenic model, Aβ 40/42 deposition is not the major mediator of mitochondrial dysfunction.
Two additional studies have provided further insights through linking full-length APP with mitochondrial dysfunction28, 29. These groups demonstrated that in AD patients, full-length APP is observed in mitochondria, and accumulation of APP in the mitochondrial importing channels blocks protein import, since APP is likely to clog the import machinery due to one domain that is rich in acidic amino acids (residues 220–290). Although these observations are intriguing in neurons, such a mechanism has never been explored in any peripheral tissues. Does APP also induce impaired protein import into adipocyte mitochondria? Crucial components of the ETC, including ATP5A (Complex V), UQCRC2 (Complex III), SDHB (Complex II) and NDUFB8 (Complex I) are significantly suppressed by APP overexpression (Extended Data Fig. 4i). All these proteins are nuclear encoded and require transport through the mitochondrial protein importing machinery. However, we did not observe a transcriptional change in these key ETC genes (Extended Data Fig. 4j). Combined together, we showed that even though no changes in nuclear encoded mitochondrial gene transcription are observed, the levels of key components located in mitochondria are significantly decreased, supporting the notion that APP decreases mitochondrial protein import. We then moved forward to directly validate that APP mis-targeting to mitochondria inhibits adipocyte mitochondrial protein import by taking advantage a well-established in vitro protein import assay30, 31. This assay utilizes a radiolabeled ornithine transcarbamylase precursor (pOTC) protein to test the efficiency of protein import in isolated mitochondria. By incubating [35S]pOTC with mitochondria isolated from sWAT, we demonstrated that APP overexpression significantly reduces the kinetics of pOTC import, as judged by a 46% reduction in pOTC import (Fig. 5f). In vivo, APP transgenic mice show increased amounts of COX5A precursor accumulation caused by mitochondrial protein import defects (Extended Data Fig. 4k). To further establish that APP accumulation in mitochondria causes deficient protein import, we performed three independent experiments to refine the sub-mitochondrial localization of APP. The first line of evidence arose from our experiments using the ascorbate peroxidase 2 (APEX2) staining system32. Upon harvesting differentiated adipocytes from APP-APEX2 transgenic mice, we observed plenty of APEX2-positive contrast across mitochondrial outer and inner membranes, suggesting that upon targeting of APP to mitochondria, the protein will reside close to the mitochondrial protein import machinery (Fig. 5g). We also examined the in vivo mitochondrial sub-localization of APP through biochemical sub-fractionation and observed that after its mis-targeting into mitochondria, APP is distributed in the outer membrane, intermembrane space, and inner membrane fractions, with no signal in the matrix (Fig. 5h). In addition to the physical presence of APP across mitochondrial outer and inner membranes, we further performed co-immunoprecipitation assays to show that APP robustly binds to TOM40 and TIM23, the key subunits belonging to translocases of outer membrane and inner membrane, respectively (Fig. 5i). The strong interaction between APP and TOM40/TIM23 may form the molecular basis for the APP “clogging” action in the protein import pathway, thereby suppressing the pOTC and other nuclear encoded proteins translocating into adipocyte mitochondria. Collectively, upon APP induction, adipocytes develop mitochondrial dysfunction, and one of the underlying reasons is a blockade of protein import due to “clogging” of the import channels by APP (Fig. 5j).
Dysregulation of SRP54c is responsible for mistargeting of APP into adipocyte mitochondria.
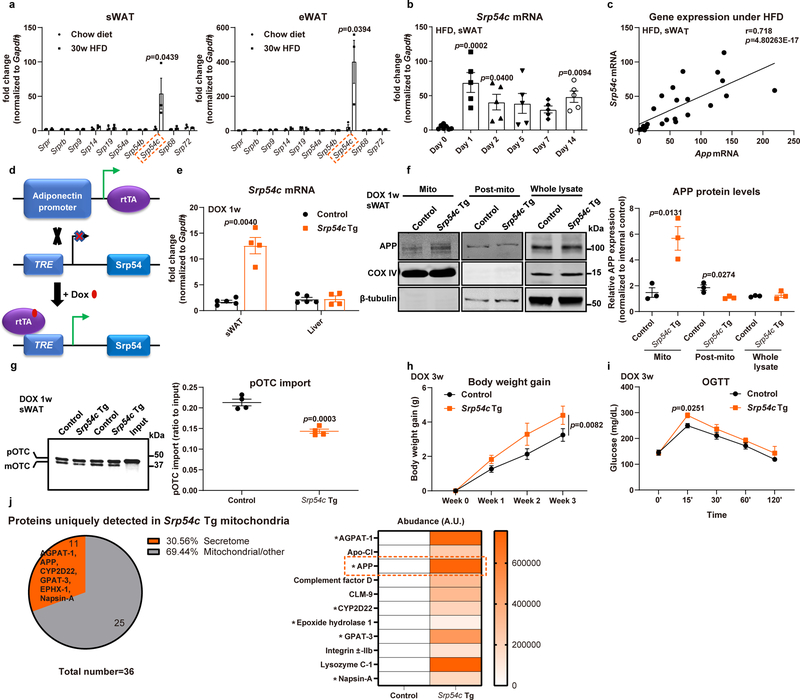
The driving force responsible for mistargeting APP to adipocyte mitochondria under obese conditions remains unclear. A recent study by Costa et al. identified an unanticipated role for the yeast signal recognition particle (SRP) in protein-targeting efficiency and specificity33, showing that auxin-induced SRP loss causes conventional ER-targeted transcripts to be mis-targeted into mitochondria, thus leading to rapid mitochondrial defects. We explored whether mis-localization of APP is mediated through SRP dysregulation. Examining expression levels of different SRP components in WAT of obese mice, we identified that one SRP subunit, SRP54c, stands out as specifically and massively upregulated in sWAT and eWAT upon prolonged HFD feeding (Extended Data Fig. 5a). Interestingly, even under an acute HFD challenge, Srp54c transcripts show a consistently increased expression pattern (Extended Data Fig. 5b) similar to App (Fig. 1h). We indeed demonstrated that sWAT Srp54c and App mRNA expressions are significantly correlated (Extended Data Fig. 5c), consistent with the idea that SRP54c is involved in the regulation of APP in adipocytes. To further dissect the in vivo function of SRP54c, we employed an adipocyte-specific, Dox-inducible overexpression model of SRP54c (Srp54c Tg) (Extended Data Fig. 5d). We confirmed the tissue-specificity and inducibility of the Srp54c transgenic mouse model, by observing a significant induction of Srp54c mRNA selectively in sWAT, but not in the liver (Extended Data Fig. 5e). We hypothesized that SRP54c overexpressing mice would effectively mimic the imbalance of SRP subunits under diet-induced obesity and possibly affect the APP localization. Indeed, Extended Data Fig. 5f demonstrates that one-week of overproduction of SRP54c in adipocytes is sufficient to induce mis-targeting of APP into mitochondria, leading to a 6-fold increase of APP accumulation in mitochondria in Srp54c transgenic mice without affecting total APP levels. By rapidly and dramatically inducing the mis-localization of APP, Srp54c transgenic mice also display a 33% impairment in pOTC import compared to control mice (Extended Data Fig. 5g), phenocopying the mitochondrial protein import inhibition effects seen in APP transgenic mice. Metabolically, we observed that SRP54c overexpression results in enhanced body weight gain under chow diet (Extended Data Fig. 5h) and accelerated onset of glucose intolerance at earlier time-points (Extended Data Fig. 5i). To further gain mechanistic insights into how overproduced SRP54c contributes to adipocyte mitochondrial dysfunction, we subjected highly purified mitochondria to unbiased proteomic analysis through mass-spectrometry. Upon retrieving the data, we particularly focused on the 36 proteins uniquely detected in mitochondria from sWAT of Srp54c Tg mice. Among these, a large portion (31%) of the polypeptides belongs to the secretome (Extended Data Fig. 5j). Notably, we observed a prominent presence of APP (Extended Data Fig. 5j), suggesting that mis-targeting of APP plays an important role mediating the mitochondrial dysfunction effects by SRP54c. Several additional proteins containing well-defined ER targeting sequence are also found in these mitochondrial fractions, such as AGPAT-1, CYP2D22, EPHX-1, GPAT-3, and Napsin-A. Collectively, our data demonstrate that HFD induces a dysregulation of SRP54c, which drives APP mis-targeting and the excessive SRP54c is sufficient to phenocopy APP transgenic mice. This is the first set of observations that recognizes SRP54c as a key mediator responsible for mis-targeting of APP into mitochondria under obese conditions, triggering the subsequent profound metabolic dysfunction.
App AKO protects mice from obesity with enhanced adipocyte mitochondrial function.
Our gain-of-function studies suggest that depleting APP in adipocytes may exert a protective role against HFD-induced obesity and insulin resistance. Thus, we first determined whether halting the overproduction of APP through removal of Dox would reverse the metabolic dysregulation in APP transgenic mice. We demonstrated that by removing Dox, mice gain weight significantly more slowly than mice continuously exposed to HFD/Dox diets (Extended Data Fig. 6a). Without the APP overexpression, adipocyte mitochondrial respiration recovers, as judged by analysis in ex vivo sWAT (Extended Data Fig. 6b) and in isolated mitochondria (Extended Data Fig. 6g). In addition, OGTTs and ITTs both show an improvement in glucose intolerance and insulin resistance without APP overexpression. In contrast, mice continuing to overexpress APP show impaired glucose metabolism and insulin resistance (Extended Data Fig. 6c–d). Importantly, these changes are not due to the removal of Dox per se, since control mice under the same conditions do not show any of these changes upon removal of Dox, including identical body weight gain, unchanged mitochondrial respiration, and comparable glucose tolerance assessed by an OGTT (Extended Data Fig. 6e–h). We observed that mice without Dox have smaller adipocytes (Extended Data Fig. 6i), suggesting that hypotrophy of sWAT adipocytes is attenuated upon cessation of APP overexpression. We also noticed that sWAT from mice withdrawn from Dox diets show fewer inflammatory cells. These improvements in AT upon withdrawing Dox trigger improvements in the liver, which displays reduced lipid accumulation (Extended Data Fig. 6j). In the same experimental settings, control mice show similar adipocyte size and inflammatory cell infiltration in sWAT as well as identical liver steatosis with or without Dox feeding (Extended Data Fig. 6i–j). Thus, upon eliminating APP overexpression in adipocytes by removing Dox, we demonstrate that mitochondrial function is recovered, functional adipocytes are restored, and a significantly improved whole-body metabolism is seen.
To directly examine the impact of endogenous APP on diet-induced obesity and insulin resistance, we eliminated App inducibly and specifically in adipocytes (App AKO) (Fig. 6a). We validated the inducibility and tissue-specificity for App elimination by demonstrating a significant decrease in App mRNA levels in both sWAT and eWAT from App AKO mice (Fig. 6b). Notably, App transcripts are also remarkably downregulated in the BAT from App AKO mice, while its expression is unaffected in the liver (Fig. 6b). We further confirmed that APP protein is significantly reduced in the App AKO mouse sWAT (Fig. 6c). Upon demonstrating a substantial reduction of App selectively in adipocytes, we moved forward to probe for the impact of adipocyte-specific App depletion on whole-body metabolism under a HFD challenge. In contrast to the obese phenotype observed for APP overexpressing mice, App AKO mice show reduced weight gain (Fig. 6d). We noticed an immediate divergence of the weight gain after adding Dox. App AKO mice are not only leaner, but also maintain better glucose tolerance and higher insulin sensitivity, as judged by both OGTT (Fig. 6e) and ITT (Fig. 6f), respectively. We further speculated that a prolonged HFD/Dox exposure allowing a significant induction of mitochondrial dysfunction in adipocytes could fully uncover the protective role of App depletion from obesity. Thus, we performed a 12-week HFD/Dox feeding experiment on control and App AKO mice. We found that App AKO mice are significantly protected from HFD-induced body weight gain (Fig. 6g). App AKO mice also display much better glucose tolerance at later time-points (Fig. 6h), and maintain drastically lower circulating insulin levels during OGTT (Extended Data Fig. 7a), suggesting an insulin sensitive phenotype, which is further validated by a significant improvement in ITT (Extended Data Fig. 7b).
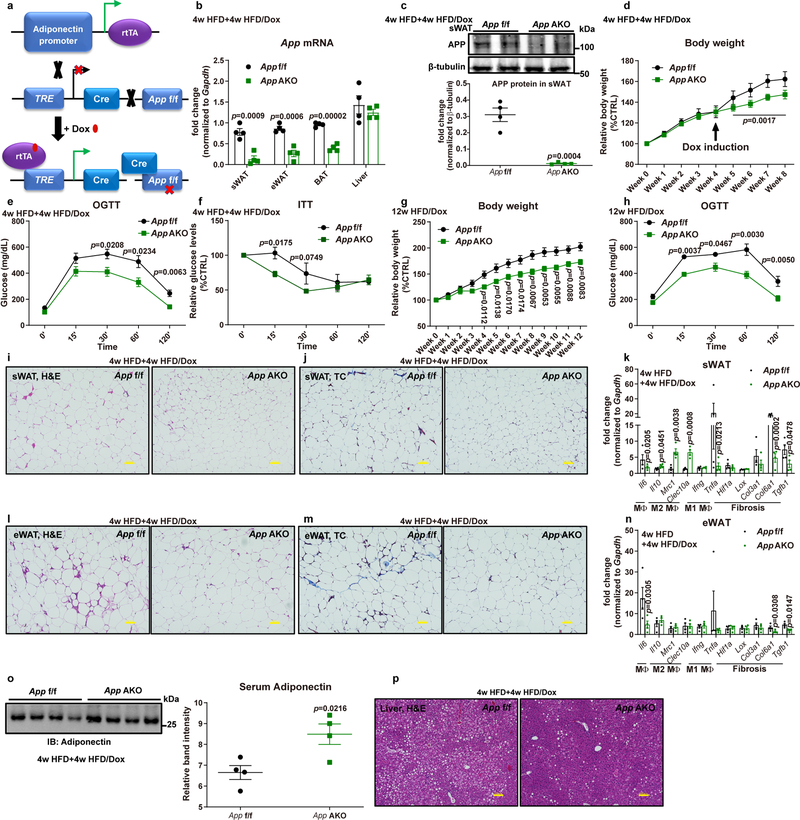
Fig. 6. App AKO protects mice from obesity.
(a) Schematic illustration of the adipocyte-specific, Dox-inducible App knockout mouse model. (b-c) Validation of App deletion in adipocyte-specific App knock-out mice (App AKO) compared to control mice (App f/f) fed with 4-week HFD plus 4-week HFD/Dox 600mg/kg: (b) App mRNA levels in different tissues from App f/f and App AKO mice (n=4 mice per group); (c) Representative western blotting image (upper panel) for APP protein levels in sWAT and its quantification (lower panel, n=4 mice per group.). Images are representative of three independent experiments. (d-h) Both App f/f and App AKO mice are subject to the following metabolic analyses: (d) Relative body weight for 8 weeks (n=8 mice per group); Glucose levels at different time-points from (e) OGTT and (f) ITT experiments (n=8 mice per group); (g) Relative body weights for 12 weeks of HFD/Dox feeding; (h) Glucose levels at different time-points of OGTT assays (n=8 mice per group in g and h). (i-n) Inflammatory and fibrotic phenotypes in sWAT and eWAT from App f/f and App AKO mice: Representative H&E staining images for (i) sWAT and (l) eWAT; Representative trichrome (TC) staining images for (j) sWAT and (m) eWAT; Inflammation and fibrosis related gene expressions in (k) sWAT and (n) eWAT of mice from both groups (images are chosen from four independent experiments). Bar = 100 μm. (o) Circulating adiponectin immunoblot (left panel) and quantification (right panel) in both groups. n=4 mice per group. (p) Representative H&E staining pictures of liver tissues from both groups (images are chosen from four independent experiments). Bar = 100 μm. For all statistics: data are shown as mean ± SEM of biologically independent samples. Two-tailed Student’s t-test (b-c, k, n-o); Two-way ANOVA followed by a Tukey post-test (d-h). See also Extended Data Fig. 6 and 7.
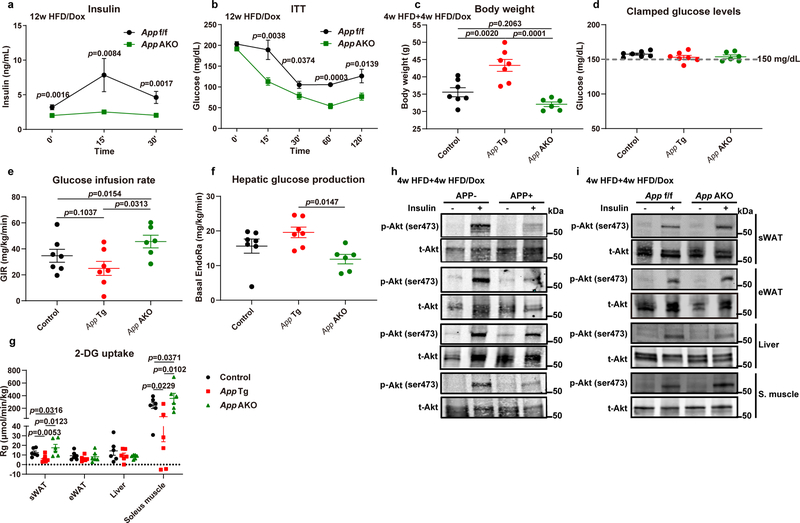
To further strengthen our conclusion and dissect which tissue(s) are responsible for changes in insulin sensitivity, we performed hyperinsulinemic-euglycemic clamp studies in both the APP transgenic (App Tg) and App AKO mouse models. Consistent with our previous results, we showed that APP overexpressing mice gain significantly more body weight compared to control and App AKO mice (Extended Data Fig. 7c). During the clamp studies, we managed to maintain clamped glucose levels at ~150 mg/dL in all three groups (Extended Data Fig. 7d). The GIR needed to maintain euglycemia is lower in APP transgenic mice and dramatically higher in App AKO mice (Extended Data Fig. 7e). Basal endogenous glucose output (EndoRa) is more efficiently suppressed in App AKO mice, but only shows a trend towards an increase in APP transgenic mice (Extended Data Fig. 7f), suggesting that the liver is not a major organ contributing to whole-body insulin sensitivity changes. We then measured the radiolabeled glucose (2-DG) uptake in different metabolic tissues and demonstrated that in both sWAT and soleus muscle, APP transgenic mice display a greatly diminished insulin-stimulated glucose uptake. In contrast, App AKO mice show significantly enhanced glucose uptake (Extended Data Fig. 7g). To complement the clamp studies, we examined the response of individual metabolic tissues to insulin, as assessed by the phosphorylation of Akt (pAkt). In APP overexpressing mice, pAkt levels are clearly lower in sWAT, eWAT and soleus muscle compared to control mice; in contrast, App AKO mice show an enhanced pAkt signal upon insulin stimulation in sWAT, eWAT and muscle (Extended Data Fig. 7h–i).
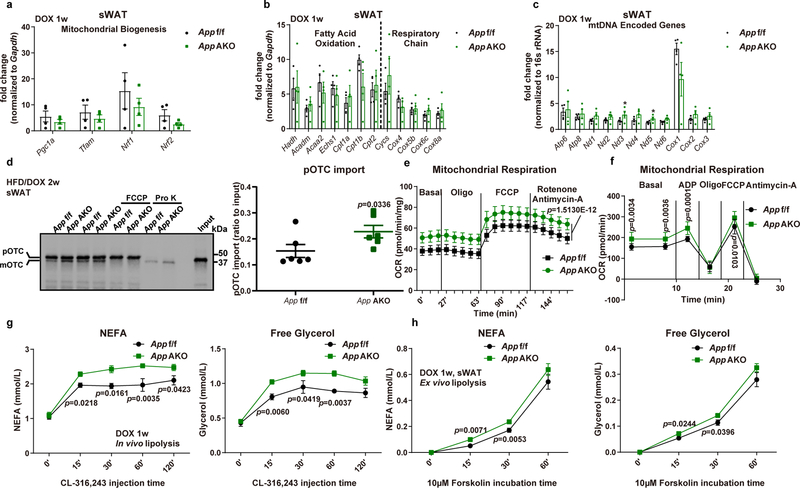
Fig. 7. App AKO enhances adipocyte mitochondrial function.
(a-c) (a) Mitochondrial biogenesis related gene expressions, (b) genes involved in fatty acid oxidation and respiratory chain component genes, and (c) mtDNA encoded gene transcription in App f/f and App AKO mice after 1-week Dox induction are shown, n=4 mice per group. (d) Representative autoradiography image (left) and statistics (right) of pOTC import assessed in isolated mitochondria (incubation for 30 minutes) from sWAT of 2-week HFD/Dox fed control or App AKO mice. 25% of [35S]pOTC added to each reaction is loaded as input. Pro K: 0.4 mg/mL; FCCP: 40 μmol/L. n=6 mice per group. Image is chosen from three independent experiments (e-f) (e) Ex vivo mitochondrial respiration in sWAT fat pads and (f) respiration measurements in isolated mitochondria from App f/f and App AKO mice fed with Dox containing diet for 1 week, n=10 per group. (g-h) Both control and App AKO mice induced by 1-week Dox 600mg/kg feeding are subject to in vivo and ex vivo lipolysis analysis: (g) Serum NEFA (left), free glycerol (right) at different time-points in both groups after β3-adrenoceptor agonist CL-316,243 (1 mg/kg) injection, n=6 mice per group; (h) NEFA (left) and free glycerol (right) levels in the mediums obtained from ex vivo cultured sWAT fat pads at different time points of 10 μM forskolin incubation, n=12 tissues per group. For all statistics: data are shown as mean ± SEM of biologically independent samples. Two-tailed Student’s t-test (a-d); Two-way ANOVA followed by a Tukey post-test (e-h).
We observed a remarkable improvement in App AKO mice in the sWAT histology, displaying significantly decreased numbers of hypertrophic adipocytes (Fig. 6i), reduced inflammatory cell infiltration and fibrotic areas (Fig. 6j). We further validated that App AKO mice are protected from sWAT inflammation and fibrosis by measuring expression levels of some critical genes involved in these processes (Fig. 6k). Similarly, we showed that in eWAT of App AKO mice, the average adipocyte size remains identical compared to control mice, but a dramatic reduction in inflammatory cell infiltration is seen (Fig. 6l). The healthier phenotype is also supported by decreased fibrosis (Fig. 6m). Transcriptionally, markers for AT inflammation/fibrosis are significantly downregulated in eWAT of App AKO mice (Fig. 6n). In agreement with the overall insulin sensitive phenotype, App AKO mice show elevated circulating adiponectin levels (Fig. 6o), reflecting better functionality of adipocytes and an enhanced whole-body metabolic homeostasis. Accompanying the morphological and functional improvements in WAT, hepatic lipid accumulation is significantly attenuated through the adipocyte-specific depletion of App (Fig. 6p), further underlining that App AKO mice maintain functional AT and consequently improve whole-body metabolism.
Next, we investigated whether the protective effects against diet-induced obesity in App AKO mice are related to improved adipocyte mitochondrial function. The transcription of genes involved in mitochondrial biogenesis and function are largely unaffected (Figs. 7a–c). In contrast to APP overexpressing mice, we observed an enhancement in pOTC import activity due to adipocyte-specific deletion of App, as suggested by a 50% increase in pOTC import (Fig. 7d). We further examined ex vivo mitochondrial respiration and demonstrated a significantly augmented mitochondrial functionality in App AKO mice by showing increased basal respiration and FCCP-induced maximal respiratory capacity in sWAT fat pads (Fig. 7e), as well as enhanced respiration in isolated mitochondria (Fig. 7f). Then we put our focus on investigating whether adipocyte App deletion can enhance lipolytic processes. Upon injection of CL-316,243, we noticed that elimination of App dramatically induces lipolysis, as indicated by a significant enhancement in serum NEFA and free glycerol levels (Fig. 7g). Consistently, we also confirmed that sWAT fat pads taken from App AKO mice demonstrate an augmented response to forskolin-induced NEFA and glycerol release (Fig. 7h). Taken together, the lack of App sustains fully functional adipocytes and preserves metabolic function system-wide, likely driven by improvements along the axis of mitochondrial function and lipolysis.
Discussion
In the present study, we demonstrate that APP, a critical modulator for AD in the brain, also contributes to adipocyte dysfunction during weight gain and obesity. The potential clinical relevance of the findings from our rodent models is supported by our findings that obesity, insulin resistance and dyslipidemia are associated with upregulation of APP gene expression in AT in patients. Consistently, Puig et al. report that App-deficient mice show improved glucose tolerance and reduced insulin resistance upon HFD feeding34. Mutant APP is shown to exacerbate insulin resistance and macrophage infiltration in HFD challenged AT35. In debating, Czeczor et al. suggest that systemic App knockout mice develop glucose intolerance under HFD, with a mechanism linking it to impaired insulin secretion from β-cells36. APP is expressed in many different cell types, such as neuronal cells, immune cells, β-cells, and adipocytes. The interpretation of the metabolic characterization of congenital and ubiquitous loss-of-function models is therefore very challenging. Our present study utilizes mouse models with adipocyte-specific manipulations, which provides direct evidence uncovering the crucial role of APP in adipocyte physiology and obesity.
Few studies have investigated the relationship between full-length APP and mitochondrial function. APP accumulates in mitochondria in neuronal cells and impairs capacity to import nuclear-encoded components due to “clogging” by APP28, and APP is highly enriched and further impairs mitochondrial function in isolated mitochondria from affected brain regions of AD patients29. A recent study confirms that APP overexpression in neurons causes its presence in mitochondria-associated membranes37. We, for the first time, demonstrate an accumulation of APP in adipocyte mitochondria during HFD feeding. We further show that this is not just an epiphenomenon, but that APP impairs nuclear-encoded protein import into mitochondria, thereby suppressing mitochondrial function; while App knockout in adipocytes prevents mitochondrial dysfunction. Thus, we provide evidence for the mis-location of APP in mitochondria of white adipocytes and its crucial role for inducing mitochondrial dysfunction and promoting obesity. A recent in vitro study shows that knocking down App in differentiated adipocytes enhances mitochondrial respiration38. Furthermore, we speculate that this process of mitochondrial mis-localization may not be limited to neuronal cells and white adipocytes, but could expand to other peripheral cell types, such as brown adipocytes, pancreatic β-cells, and others.
The induced mitochondrial dysfunction by full-length APP relies on its translocation from the conventional secretory pathway to mistargeting into mitochondria. Our data suggest a misinterpretation of the signal sequence of APP as a mitochondrial pre-sequence. This raises several unresolved questions, particularly on the mechanism(s) driving APP translocating into mitochondrial fractions. At the level of the targeting sequence, previous work suggests that three positively charged residues near the amino-terminal region are responsible for its mitochondrial targeting in neuronal cells29. We will examine whether these residues also mediate mis-targeting of APP into mitochondria in white adipocytes in our future efforts. Post-translational modifications are also potential mediators for protein mistargeting, as indicated by phosphorylation of P4502B1, an ER-targeted component, promoting its translocation into mitochondria39. Our present study further explores the contribution of the SRP for the mis-localization of APP, based on recent observations which identify an unanticipated role for yeast SRP in protein-targeting efficiency and specificity33. Our data demonstrate that HFD specifically induces a dysregulation of SRP54c, and SRP54c induction is sufficient to increase APP translocation into mitochondria and reproduce the obese and glucose intolerant phenotypes in mitochondria-targeted APP transgenic mice. Furthermore, we speculate that in response to disproportionate overexpression of SRP54c, certain proteins can undergo mis-localization from ER to mitochondria. To identify the proteins susceptible to mis-localization and to investigate their relation to obesity requires future studies.
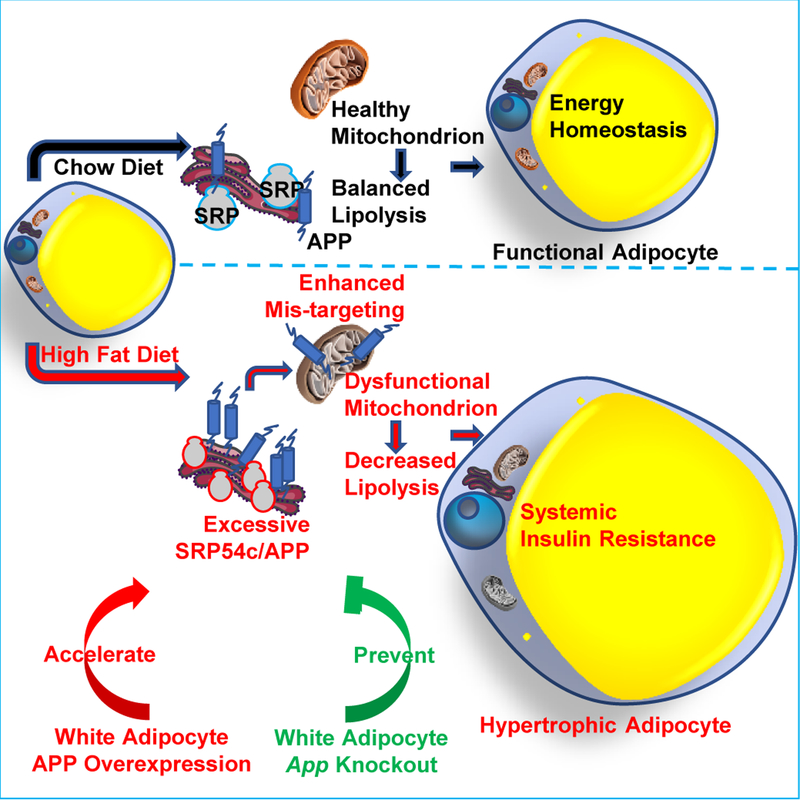
In summary, we highlight a modulating role of APP in adipocyte mitochondrial function, lipolysis and hypertrophy in the context of obesity (Fig. 8). We believe this role of APP as a mediator for adipocyte mitochondrial dysfunction could expand the role of APP as a target for neurodegenerative disease into obesity and associated comorbidities.
Fig. 8. A modulating role of APP in adipocyte mitochondrial function, lipolysis and hypertrophy in the context of obesity.
HFD challenge induces white adipocyte APP overexpression and subsequent mis-targeting into mitochondria, leading to impaired mitochondrial functions. The mitochondrial dysfunction further decreases catecholamine-induced lipolysis, resulting in rapid hypertrophy in adipocytes, followed by a significant obese phenotype and systemic insulin resistance. Adipocyte-specific overexpression of APP recapitulates and further accelerates the AT dysfunction and obese problems, while the specific elimination of App in white adipocytes prevents mice from diet induced metabolic deficiency.
Methods
Study subjects
Twenty-four men and women with obesity participated in this study (Supplementary Table 1). All subjects completed a screening history and physical examination, standard blood tests and an oral glucose tolerance test. No subject had diabetes or other serious illnesses, were taking medications that could interfere with insulin action, consumed excessive alcohol (>14 drinks/week for women and >21 drinks/week for men), or smoked tobacco products. Written informed consent was obtained from all subjects before their participation in this study, which was approved by the Institutional Review Board of Washington University School of Medicine in St. Louis, MO. The trial is registered at ClinicalTrials.gov website ().
Animal models
All performed procedures on animals have been approved by the Institutional Animal Care and Use Committee (IACUC) of UT Southwestern Medical Center at Dallas (APN# 2015–101207). All experiments were conducted using littermate-controlled male mice and were started at the age of 8–10 weeks, except that SVF were isolated from mice at the age of 4–6 weeks. All experimental animals were maintained on a C57BL/6 background and housed in a barrier specific pathogen free animal facility with 12-hour dark-light cycle with free access to water and food (Chow diet or special diet, see details as followed). Standard chow diet (#5058, LabDiet) was regularly fed to mice. For HFD only studies, regular HFD with 60% caloric from fat (D12492, Research Diets) were fed to mice. During HFD challenge experiments together with Dox induction, mice were fed with a paste diet containing 60% caloric from fat supplemented with 600mg/kg Dox (S7067, Bio-Serv). Dox containing pellet diet (600 mg/kg, S4107, Bio-Serv) was utilized for Dox induction. For the experimental cohorts performed on special diets, both control and transgenic or knock-out mice were fed with the same kind of diet to eliminate the impacts from food.
Inducible adipocyte-APP overexpression mouse model: we generated a mouse model which is Dox inducible, adipocyte-specific APP overexpression by crossing the tetracycline responsive element (TRE)-mito-APP transgenic mice with adiponectin promoter driven- reverse tetracycline-dependent transcriptional activator (rtTA) transgenic mice (Adipo-rtTA as previously reported4). For the TRE-mito-APP construct, we replaced the original ER targeting signaling sequence for mouse APP695 with the COX8 mitochondrial signaling sequence (5’ -ATGTCTGTCCTGACGCCACTGCTGCTGAGGAGCCTGACCGGCTCGGCCCGGCGGCTCATGGTGCCG CGGGCTCAGGTCCACTCG - 3’) to facilitate the APP protein targeting to mitochondria.
Inducible adipocyte-SRP54c overexpression mouse model: we generated a mouse model which is Dox inducible, adipocyte-specific SRP54c overexpression by crossing the TRE-SRP54c transgenic mice with Adipo-rtTA transgenic mice. TRE-mito-APP, TRE-SRP54c, and Adipo-rtTA mice were generated in the UT Southwestern Medical Center Transgenic Core facility.
Inducible adipocyte-App knock-out mouse model: with the courtesy of Dr. Hui Zheng from Balor College of Medicine, an adipocyte-specific App knock-out mouse model was generated by crossing Adipo-rtTA and TRE-Cre mice (B6.Cg-Tg(tetO-cre)1Jaw/J, No. 006234) together with Appflox/flox (App f/f) mice provided by Dr. Zheng, in which the depletion of App gene was controlled by Dox feeding.
C57BL/6J Wild-type and Ob/Ob Mice: male C57BL/6J mice (No. 000664) and Ob/Ob mice (B6.Cg-Lepob/J, No. 000632) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA).
HEK293T-rtTA cell line
A stable HEK293T-rtTA cell line was generated from our laboratory following previously reported protocols40 by a selection from embryonic kidney epithelial HEK293T cells (ATCC# CRL-3216) transfected with CMV enhancer/promoter-driven CMV-rtTA. HEK293T-rtTA cells were cultured in DMEM high glucose media (#11965–092, Gibco, ThermoFisher Scientific, Waltham, MA, USA) containing 10% FBS (TMS-013-B, Millipore Sigma, St. Louis, MO, USA) and penicillin-streptomycin (30–001-CI, Corning, Corning, NY, USA), which was for in vitro Dox-inducible targeted gene expressions.
Stromal vascular fraction (SVF) differentiated adipocytes
SVF were isolated from mouse sWAT and cultured in DMEM/F12 media containing glutamax (#10565–018, Gibco, ThermoFisher Scientific, Waltham, MA, USA) supplemented with 10% FBS, penicillin-streptomycin and gentamicin (#15750060, Gibco, ThermoFisher Scientific, Waltham, MA, USA) in a humid incubator with 10% CO2 at 37 °C. After SVF cells grew toward over 95% confluency, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1μM dexamethasone, 5μg/ml Insulin and 1 μM rosiglitazone were added into the culture media to initiate in vitro differentiation. After 48 hours of incubation, media containing only 5μg/ml Insulin was replaced to maintain the differentiation of SVF derived adipocytes.
Antibodies and Reagents
Anti APP antibody (#101336) for western blotting and immunofluorescence staining was purchased from GeneTex (Irvine, CA, USA). Antibodies against β-actin (A3854) and amyloid β 1–40/42 (AB5076) as well as amyloid β protein fragment 1–42 (A9810) were from Millipore Sigma (St. Louis, MO, USA). Anti COXIV antibody (ab14744), PDH-E2 (ab110332) antibody, and total OXPHOS rodent WB antibody cocktail (ab110413) were purchased from Abcam (Cambridge, MA, USA). Anti-TIM23 (sc-514463), anti-TOM40 (sc-365467), anti-COX5A (sc-376907) and anti-VDAC (sc-98708) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Perilipin-1 antibody (20R-PP004) was obtained from Fitzgerald, Inc. (Acton, MA, USA). Primary antibodies against β-tubulin (#86298S), calreticulin (#12238T), phosphorylated HSL (pHSL, Ser660, #4126S), total HSL (#4107), cytochrome C (#4272), phosphorylated Akt (pAkt, Ser473, #9271S) and total Akt (#2920S) came from Cell Signaling Technology, Inc. (Beverly, MA, USA). Unless specifically indicated, all other reagents were obtained from Millipore Sigma Corporation (St. Louis, MO, USA).
Body composition measurements
For the human study, body fat mass and fat-free mass were determined by using dual-energy x-ray absorptiometry, and intrahepatic triglyceride content was determined by using magnetic resonance imaging.
The precise measurements of mouse whole body compositions including total body fat and lean mass were performed through the Bruker Minispec mq10 system (Bruker Corporation, Billerica, MA, USA).
Co-immunoprecipitation assay
The assay was performed as previously described41. Briefly, isolated mitochondria from sWAT were lysed using Pierce IP lysis buffer (#87787, ThermoFisher Scientific) containing protease inhibitor cocktail (Calbiochem, San Diego, CA, USA). Following an overnight incubation with anti-APP antibody or IgG (negative control) at 4°C, the lysates were mixed with Protein G Sepharose 4 Fast Flow (#17–0618-01, GE Healthcare Life Sciences, Pittsburgh, PA, USA) at room temperature for 1 hour to capture immune complexes. After three sequential washes with Pierce IP lysis buffer, samples were eluted, resuspended in 2× loading buffer, and subject to immunoblotting with indicated antibodies.
Histological analysis and adipocyte size quantification
Mice adipose tissue and liver paraffin-embedded sections were cut at 4μm and stained with hematoxylin and eosin (H&E) as well as Masson’s Trichrome C staining, performed by the University of Texas Southwestern Medical Center Histology Core. Images (100× or 200× magnification) were recorded by the FSX100 Inverted Microscope (Olympus, Waltham, MA, USA), and representative histological images were shown.
For adipocyte size quantification, bright-field H&E staining images were taken through a Keyence BZ-X710 microscope (Keyence, Itasca, IL, USA). Adipocyte size analysis was conducted according to previously validated protocols42. Incorporated Keyence BZ-X Analyzer software was utilized for analyzing and calculating the area for each adipocyte. At least 600 adipocytes were quantified for each individual mouse.
Hyperinsulinemic-euglycemic clamp procedure and adipose tissue biopsies.
The hyperinsulinemic-euglycemic clamp procedure and AT biopsy were performed in people with obesity as previously described43. Subjects were admitted to the Clinical and Translational Research Unit at Washington University School of Medicine in the evening before the procedure. At 1900 h subjects were served a standard meal and then fasted until study completion the next day. At 0600 h the following morning, one catheter was inserted into a forearm vein to infuse stable isotopically labeled glucose tracer, dextrose and insulin, and a second catheter was inserted into a radial artery in the contralateral wrist to obtain blood samples. At 0700h, a primed (22.5 μmol/kg), continuous (0.25 μmol/kg/min), infusion of [6,6-2H2] glucose (Cambridge Isotope Laboratories) was started. After infusing the glucose tracer for 3.5 hours, insulin was infused at a rate of 50 mU·m−2 body-surface area·min−1 (initiated with a priming dose of 200 mU·m−2·min−1 for 5 min and then 100 mU·m−2·min−1 for 5 min) for 3.5 hours (3.5 to 7.0 h). The infusion of glucose tracer was stopped during insulin infusion to account for the expected decrease in endogenous glucose production. Euglycemia (~100 mg/dL) during insulin infusion was maintained by variable rate infusion of 20% dextrose, which was enriched to 2.5% with [6,6-2H2] glucose to help ensure a constant glucose tracer-to-tracee ratio (TTR). Blood samples were collected before beginning the tracer infusion to determine background plasma glucose TTR, and every 10 min during the final 30 min of the basal period and insulin infusion to determine glucose and insulin concentrations and glucose kinetics. Adipose tissue biopsies were obtained during the basal stage of the clamp procedure at ~0700h. After anesthetizing the skin and underlying tissues by percutaneous injection of lidocaine, a small skin incision was made, and periumbilical abdominal subcutaneous adipose tissue was aspirated through a 4-mm liposuction cannula connected to a 30-cc syringe. Tissue samples were immediately rinsed with ice-cold saline, and frozen in liquid nitrogen, before being stored at −80°C for subsequent RNA extraction.
Hyperinsulinemic-euglycemic clamp experiments and radiolabeled 2-Deoxy-D-glucose ([3H] 2-DG, NET328A001MC, PerkinElmer, Houston, TX, USA) uptake assays on conscious, unrestrained mice were performed by the Metabolic Phenotyping Core as previously described17, 44. Briefly, jugular implantation surgeries were performed on mice four days before clamps. On the clamping day, food was removed at 0800 h for 4hr fast. The primed 3-[3H]-D-glucose (Cat. NET331C001MC, PerkinElmer, Houston, TX, USA) was infused at the rate of 0.05 uCi/min for 90 minutes before clamping. During the 2-hour clamping, continuous insulin infusion (4mU/kg/min) and 3-[3H]-D-glucose infusion (0.1 uCi/min) were maintained. Variable infusion of 50% glucose were performed to maintain plasma glucose at ~150 mg/dl. For 2-[14C]-deoxyglucose (Cat. NET495001MC, PerkinElmer) uptake, a bolus (13 uCi) was injected and followed by a 50 ul saline injection. After 25 minutes, mice were dissected, and targeted tissues were collected for 2-DG uptake assay.
Immunofluorescence staining
Paraffin-embedded slides were prepared by the University of Texas Southwestern Medical Center Histology Core for immunostaining. Briefly, deparaffinized sections were stained with anti-APP (1:200), anti-TIM23 (1:500) or anti-perilipin-1 (1:500) primary antibodies and incubated overnight at 4 °C. Then the corresponding fluorescent labeled secondary antibodies (Life Technologies, Carlsbad, CA, USA) were added and finally the sections were counterstained with DAPI before adding coverslips. The fluorescence images were acquired with an Olympus FSX100 Microscope.
Indirect ATP detection assay
Cellular ATP levels were detected through the Mitochondrial ToxGlo Assay (G8000) supplied by Promega (Madison, WI, USA). Briefly, SVF derived from sWAT of different mice were seeded into 96-well plates and then differentiated into adipocytes. Then ATP detection reagents from the kit were added according to the instructions, and the plate was subject to luminescence signaling reading through a FLUOstar Omega microplate reader (BMG LABTECH Inc., Cary, NC, USA).
Isolation of stromal vascular fraction (SVF) and adipocyte differentiation
SVF isolation and in vitro differentiation of adipocytes were performed following the previously established protocols45. Briefly, subcutaneous and epididymal adipose tissues were harvested, rinsed with PBS, and then completely minced. Tissues were digested for two hours at 37 ˚C with standard digestion buffer (100 mM HEPES PH 7.4, 120 mM NaCl, 50 mM KCl, 5 mM glucose, 1 mM CaCl2, 1.5% bovine serum albumin (BSA), and 1 mg/mL collagenase D (Roche)) in a water-bath shaker. After digestion, the mixture was passed through 100 μm cell strainers and then centrifuged at 600g for 5 minutes. After collecting the floating adipocytes for future use, the pelleted stromal vascular cells were re-suspended in PBS with 2% FBS. The cells were filtered with 40 μm cell strainers and centrifuged at 600g for 5 minutes, and then the re-suspended SVF cells in growing media DMEM/F12 containing 10% FBS were seeded into cell culture dishes.
For in vitro differentiation, SVF cells isolated by collagenase digestion were plated onto collagen-coated dishes and cultured in 10% CO2 at 37 °C until confluency. Confluent cultures were stimulated with adipogenic cocktail (growth media supplemented with 5 μg/ml insulin, 1 μM dexamethasone, 0.5 mM IBMX and 1 μM rosiglitazone) for 48 h. Subsequently, cells were maintained in growth media supplemented with 5 μg/ml insulin until further experiments.
Lipolysis Assays
For in vivo lipolysis assay, mice were administrated with β3-adrenergic receptor agonist CL-316, 243 (C5976, Millipore Sigma) at dose of 1 mg/kg body weight, and then blood was collected from tail veins at each indicated time point. Blood samples were centrifuged at 6,000 rpm for 15 minutes and stored for further measurements. For ex vivo lipolysis assay, ~20 mg of sWAT fat pads was placed into 200 μl low glucose DMEM (#10–014-CV, Corning, Corning, NY, USA) containing 2% fatty acid free BSA, and then incubated with 10 μM Forskolin at 37 °C in a humid incubator containing 5% CO2 for 60 min. Medium was collected and stored for measurements.
Both NEFA and free glycerol levels in serum and culture medium were measured through commercially available colorimetric assays, respectively (NEFA: # 999–34691, FUJIFILM Wako Diagnostics U.S.A., Mountain View, CA, USA; free glycerol: F6428, Millipore Sigma, St. Louis, MO, USA).
Metabolic cage studies
The metabolic cage studies were performed by The University of Texas Southwestern Medical Center Metabolic Phenotyping Core facility. Briefly, the mice were maintained on a 12-hour dark/light cycle at room temperature. Metabolic parameters including oxygen consumption, carbon dioxide generation, food intake and water consumption were monitored and recorded continuously using the TSE calorimetric system (TSE System, Germany). During the whole process, all transgenic mice and their littermate controls were single housed in the metabolic chambers and kept on Dox 600 mg/kg diets and water ad libitum.
Mitochondrial DNA (mtDNA) copy number
mtDNA copy number was determined as previously described using quantitative real-time PCR of extracted genomic DNA from mouse sWAT46, 47. Here, we chose the mitochondrial-encoded gene cytochrome c oxidase 3 (Mt-Co3) and compared to the nucleus gene NADH dehydrogenase (ubiquinone) flavoprotein 1 (Ndufv1).
Mitochondrial isolation and Percoll gradient-based purification
Mitochondria and cytosol containing microsomes were isolated and purified from sWAT using Percoll gradient fractionation protocol as previously reported48. Briefly, tissues were harvested and lysed by Dounce homogenization in Sucrose isolation buffer (0.25 M sucrose in 10 mM HEPES, pH 7.4) adding protease inhibitor cocktails. After centrifugation at 10,300g at 4 °C for 15 minutes, the total cytoplasm fractions including microsomes (ER, Golgi apparatus and plasma membrane) and crude mitochondrial fraction were isolated. The obtained crude mitochondrial fraction was further purified through a Percoll gradient (40% Percoll and 0.25 M sucrose in 10 mM HEPES, pH 7.4) using a Sorvall Discovery M150 SE (ThermoFisher Scientific) ultracentrifuge to spin at 288,000 rpm at 4 °C for 40 minutes. The diffuse mitochondrial band was then transferred to new tubes and centrifuged at 10,000g at 4 °C for 10 minutes to pellet the purified mitochondria. After resuspending the mitochondrial pellet, protein concentrations were determined by the bicinchoninic acid (BCA) assay kit (Thermo Scientific Pierce, Rockford, IL, USA) and subject to immunoblotting or mass-spectrometry based proteomic analysis performed by UT Southwestern Proteomics Core. Secretome was analyzed and predicted through VerSeDa (Vertebrate Secretome Database, http://genomics.cicbiogune.es/VerSeDa/index.php).
Mitochondrial membrane potential staining
Mitochondrial membrane potential (MMP) was assessed by staining the cells with a commercially available MitoStatus dye tetramethyl rhodamine ethyl ester (TMRE, # 564696) from BD Biosciences (San Jose, CA, USA). Briefly, SVF differentiated adipocytes cultured in glass bottom microwell dishes were induced by Dox for 48 hours and then incubated with 200 nM TMRE for 20 minutes at 37 °C. After a gentle wash, the cells were fixed, and fluorescence images were captured using the Keyence BZ-X710 microscope.
Mitochondrial protein import assay
The mitochondrial protein import assay was performed as previously described with a few modifications30, 31. This assay utilizes a radiolabeled ornithine transcarbamylase precursor (pOTC) protein, which represents an abundant nuclear encoded mitochondrial protein requiring translocases of the outer and inner mitochondrial membrane to be imported into its final destination in mitochondria49. This allows insights into the kinetics and the efficiency of protein import activity in isolated mitochondria. Briefly, mouse pOTC cDNA was cloned into pGEM-3Zf(+) plasmid (P2271, Promega, Madison, WI, USA), and then expressed in the in vitro TNT-coupled reticulocyte lysate system (L4610, Promega) suppled with L-[35S]methionine (NEG709A500UC, PerkinElmer). Following the translation, radiolabeled pOTC protein was incubated with isolated mitochondria from mouse fat pads at 30 °C for 30 minutes. Mitochondria with imported mature OTC (mOTC) were collected by centrifugation at 9,000g for 10 minutes and then subject to subsequent electrophoresis and membrane transfer. The radioactive OTC on the membrane were exposed to a phosphor screen (FBCA 1417, ThermoFisher Scientific), visualized through the Bio-Rad Pharos FX Plus Molecular Imager, and then quantified through the Bio-Rad Quantity One 1-D analysis software (version 4.6.6). The pOTC import was calculated as the ratio of mOTC (cleaved band, represents fully imported OTC) compared to the input (25% of [35S]pOTC amount added to each reaction).
Mitochondrial respiration measurement
Mitochondrial respiration was determined using the Seahorse XF24 Extracellular Flux Analyzer (Agilent, Santa Clara, CA, USA) following the manufacturers’ instructions. In brief, mitochondrial stress test was performed according to a manufacture recommended BOFA (Basal-Oligomycin-FCCP-Antimycin A/Rotenone) protocol17. Ex vivo and in vitro mitochondrial function were measured by utilizing 5–10 mg sWAT fat pads and SVF differentiated adipocytes, respectively. For tissues, oligomycin (2 μM), FCCP (8 μM), and antimycin A (10 μM)/ rotenone (3 μM) were added; for cultured cells, oligomycin (1 μM), FCCP (4 μM), and antimycin A (10 μM)/ rotenone (1 μM) were injected. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were recorded through the Seahorse instrument.
Mitochondrial sub-fractionation
After isolation and percoll-purification of AT mitochondria, pelleted mitochondria were subject to further sub-fractionation as previously described50. In short, 60 μg/mL water‐soluble digitonin was added to resuspended mitochondria and gently stirred for 15 minutes. Following centrifugation at 10, 000g for 10 minutes, the “mitoplasts” (inner membranes surrounding the mitochondrial matrix) were pelted and the low-speed supernatants were collected. The mitoplast pellet was subject to subtle sonication through a Diagenode Bioruptor (UCD-300, Diagenode Inc., Denville, NJ, USA). Both post-sonication mitoplasts and the supernatants were further centrifuged at 144, 000g for 1 hour to generate two sets of pellets (the inner‐membrane pellet from the mitoplasts and the outer‐membrane pellet from the low-speed supernatants) and supernatants (the matrix fraction from the mitoplasts and the intermembrane space fraction from the low-speed supernatant). The four fractions were stored at −80°C until further analysis through western blotting.
RNA extraction and real-time quantitative PCR (qPCR)
Upon sacrificing mice, depots of adipose tissues including sWAT, eWAT, mesenteric WAT, perirenal WAT and BAT, as well as liver and brain tissues were harvested and quickly frozen in liquid nitrogen for future use. For the total RNA extraction, a hybrid protocol of Trizol (Invitrogen, Carlsbad, CA, USA) reagents and the RNeasy RNA extraction kit (#74106, Qiagen, Valencia, CA, USA) was performed. Briefly, after homogenizing the tissues by using a TissueLyser (Qiagen, Valencia, CA, USA), the RNAs were isolated following the protocol according to the RNeasy kit. Then the quality and concentration of the RNA were determined through the Nanodrop Spectrophotometer (N1–1000, Thermo Scientific, Wilmington, DE, USA). A total of 1 μg RNA underwent subsequently reverse transcriptional reactions with an iScript cDNA synthesis kit (#170–8891, Bio-Rad Laboratories, Inc., Hercules, CA, USA). cDNAs were diluted and stored until being utilized for further qPCR quantification of relative gene expressions. Briefly, we utilized the SYBR Green PCR Master Mix (A25742, Life Technologies, Carlsbad, CA, USA) to perform the qPCR reactions, and the experiments were conducted on an ABI Prism 7900HT Real-Time PCR System or a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). All the primer sequences in this study were validated in previous studies and are listed in Supplementary Table 2, and human RPS18 or mouse Gapdh was used as the internal control for relative quantification.
Serum and liver chemistry
Upon sacrificing the mice, whole blood samples were obtained and centrifuged at 6,000 rpm for 15 minutes to collect the serum. Samples were analyzed for glucose, cholesterol, triglycerides and free fatty acid (NEFA) measurements through a Vitros 250 Chemistry system (Ortho Clinical Diagnostics, Raritan, NJ, USA). Similarly, after homogenizing ~100 mg of frozen liver tissues, the lysates were utilized for tissue cholesterol and triglycerides measurements after extraction.
Systemic metabolic tests
Systemic metabolic tests in this study included body weight monitoring, oral glucose tolerance test (OGTT) and insulin tolerance test (ITT). For the OGTTs, 5-hour fasting was conducted prior to administration of 2.5g/kg body weight glucose to the mice through oral gavage. Blood was collected from tail veins at each time point indicated, and then the blood samples were centrifuged at 6,000 rpm for 15 minutes and stored for further measurements. Glucose concentrations were measured by using an oxidase-peroxidase based colorimetric assay (Sigma-Aldrich, St. Louis, MO, USA). Insulin levels were measured through a commercially available ELISA kit (#80-INSMSU-E01) purchased from Alpco (Salem, NH, USA). For the ITTs, mice underwent 5-hour fasting before 1u/kg body weight of insulin was administered through intraperitoneal (i.p.) injection. Blood samples were collected at the indicated time points and utilized for glucose measurement.
Transmission Electron Microscopy and visualization of APP in mitochondria
Briefly, sWAT from different mice were fixed by perfusion with a fixation buffer (0.1 mM sodium cacodylate containing 4% paraformaldehyde and 1% glutaraldehyde) and the harvested tissue were then transferred to 2.5% glutaraldehyde in 0.1 mM sodium cacodylate buffer and cut into pieces. Then the tissue pieces were sent to University of Texas Southwestern Medical Center Electron Microscopy Core for subsequent sectioning and imaging processing. EM pictures were acquired on the JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA, USA).
APEX2 staining system is a recently developed electron microscopy approach to visualize the subcellular localization of APEX2 fusion proteins32. For visualization of APP in the mitochondria by EM, we merged APEX2 carboxy-terminally with full-length APP (APP-APEX2) and generated an APP-APEX2 transgenic mouse model enabling adipocyte specific overexpression of APEX2 fused APP to perform staining in primary cultured adipocytes. APP-APEX2 overexpressed and control adipocytes were grown on gridded glass bottom microwell dishes and underwent APEX2 staining according to the published protocol30. Briefly, after fixation by 2% glutaraldehyde for 1 hour at 4 °C and wash by sodium cacodylate buffer, fresh solution containing DAB (0.5 mg/ml) and 10 mM H2O2 in cold sodium cacodylate buffer was added to cells for incubation for 15 minutes. The DAB reaction product can be visualized and monitored by light microscopy, and all the staining procedures were performed gently and kept on ice. The post staining cells were sent for standard EM sample processing performed by the UT Southwestern EM core facility.
Western blot analysis
After frozen tissue samples were homogenized, they were lysed in RIPA buffer (1% Triton-X100, 50 mmol/L Tris-HCl (pH 8.0), 0.25 mol/L NaCl, 5 mmol/L EDTA) containing PMSF (phenylmethylsulfonyl fluoride) and protease inhibitor cocktail (Calbiochem, San Diego, CA, USA). Following centrifugation at 12,000g for 15 min at 4°C, the supernatants were collected, and protein concentrations were measured by the BCA kit. For immunoblotting, protein samples (20–60 μg) or serum (0.1 µl diluted in 10 µl Tris-buffered saline Tween (TBS-T, 20mmol/L Tris, 137 mmol/L NaCl, pH 7.6)) were loaded and separated by 4% – 12% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). Upon completion of the electrophoresis, the proteins were transferred to a nitrocellulose membrane. Non-specific binding sites were blocked by pre-incubating the membrane in 5% BSA in TBS-T. The membranes were then incubated overnight at 4°C with specific primary antibodies: anti-APP (1:1000), anti-COXIV (1:1000), anti-β-tubulin (1:1000), anti-calreticulin (1:1000), anti-β-actin (1:5000), anti-adiponectin (1:2000), anti-pHSL (1:1000), anti-HSL (1:1000), anti-amyloid β 1–40/42 (1:1000), anti-OXPHOS rodent WB antibody cocktail (1:1000), anti-PDH-E2 (1:1000), anti-TIM23 (1:500), anti-cytochrome C (1:1000), anti-VDAC (1:200), anti-TOM40 (1:400), anti-COX5A (1:500), anti-pAkt (1:1000) and anti-Akt (1:1000). After washing, fluorescent-conjugated secondary antibody (IRDye, LI-COR, Lincoln, NE, USA) were added and incubated at room temperature for 1 hour. Then post three-time wash, the Western blot bands were scanned by the LI-COR Odyssey Imager (LI-COR, Lincoln, NE, USA), and the band intensity was analyzed through the LI-COR Odyssey Imager software.
Statistical analysis
All data are presented as means ± SEM of biologically independent samples, except that data in Supplementary Table 1 are presented as mean ± SD. We utilized GraphPad Prism 7.04 (GraphPad Software, Inc., La Jolla, CA, USA) to perform the statistical analyses. For computing correlation coefficients, Pearson correlation calculations were performed. For comparisons between two independent groups, a two-tailed Student’s t-test was used. For comparisons between more than two independent groups, a one-way analysis of variance (ANOVA) followed by a Tukey post-test was used. Differences between two groups over time were determined by a two-way ANOVA followed by a Tukey post-test to compare replicate means in each time point. P < 0.05 was considered statistically significant, and exact p-values were labeled in figures.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. APP is increased in WAT in obesity and adipocyte-specific APP overexpressing mice are more sensitive to diet induced obesity (related to Figs. 1a and 1h).
(a-f) The correlation between APP mRNA levels in sWAT from obese human patients with (a) body mass, (b) BMI, (c) subcutaneous AT volume, (d) triglycerides, (e) fasting insulin and (f) HDL-cholesterol levels. n=23 (a-d) or 24 (e, f) patients. (g) App transcription in the brain in acute HFD challenged wild-type mice for 0, 1, 2, 5, 7 and 14 days. n=7 for Day 0 group; n=5 for Day 1, 2, 5, 7 and 14 groups. Data are shown as mean ± SEM of biologically independent samples. Statistics: Pearson correlation analysis for correlation coefficient (r) and two-tailed p-value (a-f); One-way ANOVA followed by a Tukey post-test, and non-significance was found (g).
Extended Data Fig. 2. Adipocyte-specific APP overexpressing mice are more sensitive to diet induced obesity (related to Figs. 2c, 2d and 2g).
(a) Representative western blotting image for APP protein levels in brain samples from control and APP transgenic mice under HFD/Dox feeding. n=3 mice per group. (b) Western blotting for APP in mitochondrial and post-mitochondrial fractions from sWAT in control and APP overexpressing mice fed with Dox for 1 week. COXIV: mitochondrial marker; β-tubulin: cytoplasmic marker. n=2 mice per group. Representative image is chosen from three independent experiments in a and b. (c) Immunofluorescence staining for APP (red), TIM23 (green) and DAPI (blue) for nuclear labeling in HEK293T cells transfected with empty vector (left) or the mito-APP construct (right). Orange colors indicate the merge between APP and TIM23. Bar = 20 μm. The staining experiments have been replicated three times. (d) Insulin levels measured in serum samples obtained from OGTT experiments. n=8 mice per group. Data are shown as mean ± SEM of biologically independent samples. Two-way ANOVA followed by a Tukey post-test (d).
Extended Data Fig. 3. APP overexpression leads to adipocyte hypotrophy by impairing stimulated lipolysis (related to Figs. 4a and 4e–j).
(a) Representative images for H&E staining in sWAT sections from control mice (APP-) at different time-points of Dox 600mg/kg induction, chosen from two independent experiments, bar = 161 μm. (b) Representative H&E staining images in eWAT sections from APP transgenic mice (APP+) at different time-points of Dox 600mg/kg induction, chosen from two independent experiments, bar = 100 μm. (c-e) Lipogenesis related gene transcriptions in sWAT of APP transgenic mice at different time points of Dox 600mg/kg feeding: (c) Dgat2, (d) Fasn, (e) Scd1 and (f) Srebp1, n=3 mice per time point. Data are shown as mean ± SEM of biologically independent samples. One-way ANOVA followed by a Tukey post-test (c-f) and no statistical significance has been found.
Extended Data Fig. 4. APP impairs adipocyte mitochondrial function due to defective mitochondrial protein import (related to Figs. 5a–b, 5f).
(a) In vitro mitochondrial respiration (OCR) in sWAT SVF differentiated adipocytes from control (APP-) and APP transgenic (APP+) mice. Cells are pre-incubated for 5.0 μg/mL Dox for 48 hours to induce APP overexpression. n=5 per group. (b) Relative in vitro ATP production changes from SVF differentiated adipocytes of APP transgenic mice compared to control mice. Different dosages of Dox have been applied to cells. n=8 per dosage. (c) Combined light and fluorescence microscopy images displaying mitochondrial membrane potential (MMP) through TMRE staining (Red) from SVF differentiated adipocytes of control and APP transgenic mice. DMSO serves as the negative control, and FCCP is a positive control to collapse MMP. Images are chosen from three independent experiments and are representative of at least 12 fields for each group. Bar = 50 μm. (d-g) Quantification for indirect calorimetry measurements in control and APP transgenic mice in light and dark cycles: (d) Oxygen consumption (VO2); (e) carbon dioxide production (VCO2); (f) respiratory exchange ratio (RER) and (g) calculated energy expenditure. n=6 mice per group. (h) Western blotting for Aβ–40/42 in mitochondrial and cytoplasmic fractions from sWAT in control and APP overexpressing mice. Aβ 1–42 protein has been loaded separately as a positive control. n=2 mice per group. Representative image is chosen from three independent experiments. (i) Western blotting for mitochondrial complex components using oxidative phosphorylation antibody cocktail in mitochondrial and cytoplasmic fractions from sWAT in control and APP overexpressing mice. n=2 mice per group. Representative image is chosen from three independent experiments. (j) Gene expressions for mitochondrial complex components measured in (i). n=5 (APP-) or 6 (APP+) mice per group. (k) Western blotting for COX5A in sWAT in control and APP overexpressing mice. n=3 mice per group. Representative image is chosen from three independent experiment. For all statistics: data are shown as mean ± SEM of biologically independent samples. Two-way ANOVA (a); One-way ANOVA followed by a Tukey post-test (b); Two-tailed Student’s t-test (d-g, j).
Extended Data Fig. 5. Dysregulation of SRP54c is responsible for mistargeting of APP into adipocyte mitochondria (related to Fig. 5).
(a-b) SRP subunit gene mRNA levels in anchoring adipocytes in 30-week HFD and chow fed mice (a, left: sWAT; right: eWAT, n=3 mice per group) and Srp54c mRNA levels from sWAT in acute HFD challenged wild-type mice for 0, 1, 2, 5, 7 and 14 days (b), n=7 mice in Day 0 group and n=5 mice in Day 1, 2, 5, 7 and 14 groups. (c) The correlation between Srp54c mRNA levels in sWAT from HFD challenged wildtype mice with App mRNA expression, n=32. (d) Schematic illustration of the adipocyte-specific, Dox-inducible SRP54c transgenic mouse model. (e) Validation of SRP54c overexpression in WAT of transgenic mice fed with 1-week Dox 600mg/kg diet by detecting Srp54c mRNA levels in different tissues from control (Control) and SRP54c overexpressing (Srp54c Tg) mice (n=4 mice per group). (f) Representative Western blotting image (left panel) for APP in purified mitochondrial, post-mito, and whole tissue lysate from sWAT in 1-week Dox fed mice and its quantification (right panel). n=3 mice per group. Images are chosen from three independent experiments. (g) Representative autoradiography image (left panel) and statistics (right panel) of pOTC import assessed in isolated mitochondria (incubation for 30 minutes) from sWAT of 1-week Dox fed control or Srp54c Tg mice. 25% of [35S]pOTC added to each reaction is loaded as input. n=4 mice per group. (h-i) Upon 3-week Dox feeding, both control and Srp54c Tg mice are subject to metabolic analysis, including (h) body weight monitoring and (i) OGTT assays. n=4 mice per group. (j) Proteomics analysis performed in purified mitochondria from WAT of control and Srp54c Tg mice: left, percentage of secretome proteins among uniquely detected proteins in Srp54c Tg mitochondria; right, heat map depicting enrichment of identified proteins belonging to secretome (* indicates a polypeptide containing a well-defined ER signaling sequence). For all statistical graphs, numeric data are presented as mean ± SEM of biologically independent samples. Two-tailed Student’s t-test (a, e-g); One-way ANOVA followed by a Tukey post-test (b); Pearson correlation analysis for correlation coefficient (r) and two-tailed p-value (c); Two-way ANOVA followed by a Tukey post-test (h-i).
Extended Data Fig. 6. Halting APP overexpression reverses the obese phenotypes (related to Fig. 6).
(a-d) APP overexpressing mice (APP+) are fed with HFD/Dox diets for 8 weeks, followed by dividing into two groups, one group continuously on HFD/Dox feeding (Keep Dox) and the other group fed with HFD without Dox (Withdraw Dox). Two groups of mice are subject to the following metabolic analyses (n=6 mice per group): (a) Body weight for 12 weeks; (b) Ex vivo mitochondrial respiration (OCR) in sWAT fat pads from both groups, n=10 tissues per group; Glucose levels at different time-points from (c) OGTT and (d) ITT experiments. (e-f, h) Control mice (APP-) are fed with HFD/Dox diets for 8 weeks, followed by dividing the cohorts into two groups, one group continuously on HFD/Dox feeding (“Keep Dox”) and the other group fed with HFD without Dox (“Withdraw Dox”). Two groups of mice were subjected to the following metabolic analyses: (e, n=6 mice per group) Body weights for the 12 week exposure; (f, n=10 tissues per group) Ex vivo mitochondrial respiration (OCR) in sWAT fat pads from both groups; Glucose levels at different time-points from (h, n=6 mice per group) OGTT experiments. (g) Combined high-resolution respirometry measured in isolated mitochondria from four groups including control or APP transgenic mice kept with or without Dox feeding, n=5 mice per group. (i-j) AT inflammatory and liver steatosis phenotypes in control or APP transgenic mice kept with or without Dox feeding: (i) Representative H&E staining images for sWAT; (j) Representative H&E staining images of liver tissues from four groups. Images are chosen from three independent experiments. For all the statistics: data are presented as mean ± SEM of biologically independent samples. Two-way ANOVA followed by a Tukey post-test (a-h), and no statistical significance was found in e, f, and h.
Extended Data Fig. 7. App AKO protects mice from obesity with enhanced adipocyte mitochondrial function (related to Figs. 6e–h).
(a-b) In 12-week HFD/Dox feeding control or App AKO mice, (a) insulin levels measured in serum samples obtained from OGTT experiments and (b) glucose levels at difference time-points during ITT assays. n=8 mice per group. (c-g) For insulin sensitivity measurement, three groups of mice including control, APP adipocyte-specific transgenic (App Tg) and App adipocyte-specific knockout (App AKO) mice are subject to hyperinsulinemic-euglycemic clamp studies: (c) Body weight, (d) Clamped glucose levels, (e) Glucose infusion rate, (f) Basal hepatic glucose production, and (g) 2-DG uptake in different metabolic tissues are shown. For c-f, n=7 mice in control and App Tg groups, n=6 mice in App AKO group. For g, n=6 mice per group. (h-i) Representative immunoblot image of phosphorylated Akt (p-Akt, Ser 473) and total Akt expression in different metabolic tissues from (h) both control and APP overexpressing mice or (i) both control and App AKO mice after saline or insulin injection (i.v.) for 5 min. For the Western blot image, n=3 mice per group, and the representative images are chosen from three independent experiments. For all statistics: data are shown as mean ± SEM of biologically independent samples. Two-way ANOVA followed by a Tukey post-test (a-b); One-way ANOVA followed by a Tukey post-test (c-g).
Supplementary Material
Acknowledgements
We are grateful to Dr. Hui Zheng at Baylor College of Medicine for generously sharing Appflox/flox mice for our study. We also sincerely thank the Transgenic Core Facility at UT Southwestern for the generation of the transgenic mouse lines and the Metabolic Phenotyping Core, Proteomics Core (Lemoff A), Pathology Core, Live Cell Imaging Core Facility and Electron Microscopy Core Facility (Luby-Phelps K) at UT Southwestern for their excellent experimental assistance. This study was supported by US National Institutes of Health grants P01-DK088761, R01-DK55758, DK56341, RR024992, and R01-DK099110, a Novo Nordisk Foundation Excellence project grant (P.E.S.) and a grant from the Pershing Square Foundation (S.K.). I.W.A. was supported by the Swedish Research Council (2012–1601 to I.W.A.).
Footnotes
Competing interests
All the authors declare no competing interests.
Data Availability
All data that support the findings of this study are included in the main figures, extended data figures, and supplementary information. Proteomics raw data has been deposited to MassIVE (MSV000084498).
References
- 1.Hruby A, Hu FB. The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics 33, 673–689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusminski CM, Bickel PE, Scherer PE. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov 15, 639–660 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Choe SS, Huh JY, Hwang IJ, Kim JI, Kim JB. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front Endocrinol (Lausanne) 7, 30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 19, 1338–1344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tandon P, Wafer R, Minchin JEN. Adipose morphology and metabolic disease. J Exp Biol 221, jeb164970 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Crewe C, An YA, Scherer PE. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J Clin Invest 127, 74–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell 156, 20–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenkula KG, Erlanson-Albertsson C. Adipose cell size: importance in health and disease. Am J Physiol Regul Integr Comp Physiol 315, R284–R295 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Lonn M, Mehlig K, Bengtsson C, Lissner L. Adipocyte size predicts incidence of type 2 diabetes in women. FASEB J 24, 326–331 (2010). [DOI] [PubMed] [Google Scholar]
- 10.de Mello AH, Costa AB, Engel JDG, Rezin GT. Mitochondrial dysfunction in obesity. Life Sci 192, 26–32 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab 23, 435–443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudina S, Graham TE. Mitochondrial function/dysfunction in white adipose tissue. Exp Physiol 99, 1168–1178 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Fassina G, Dorigo P, Gaion RM. Equilibrium between metabolic pathways producing energy: a key factor in regulating lipolysis. Pharmacol Res Commun 6, 1–21 (1974). [DOI] [PubMed] [Google Scholar]
- 14.Cedikova M, et al. Mitochondria in White, Brown, and Beige Adipocytes. Stem Cells Int 2016, 6067349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poher AL, et al. Ectopic UCP1 Overexpression in White Adipose Tissue Improves Insulin Sensitivity in Lou/C Rats, a Model of Obesity Resistance. Diabetes 64, 3700–3712 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kleiner S, et al. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proc Natl Acad Sci U S A 109, 9635–9640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusminski CM, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med 18, 1539–1549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy MP, LeVine H 3rd. Alzheimer’s disease and the amyloid-beta peptide. J Alzheimers Dis 19, 311–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YH, Tharp WG, Maple RL, Nair S, Permana PA, Pratley RE. Amyloid precursor protein expression is upregulated in adipocytes in obesity. Obesity (Silver Spring) 16, 1493–1500 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Puig KL, Floden AM, Adhikari R, Golovko MY, Combs CK. Amyloid precursor protein and proinflammatory changes are regulated in brain and adipose tissue in a murine model of high fat diet-induced obesity. PLoS One 7, e30378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H, et al. beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81, 525–531 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Pietilainen KH, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med 5, e51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YH, et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia 48, 1776–1783 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller P, et al. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord 11, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhou X, Li G, Zhang Y, Wu Y, Song W. Modifications and Trafficking of APP in the Pathogenesis of Alzheimer’s Disease. Front Mol Neurosci 10, 294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haczeyni F, Bell-Anderson KS, Farrell GC. Causes and mechanisms of adipocyte enlargement and adipose expansion. Obes Rev 19, 406–420 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Mossmann D, et al. Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab 20, 662–669 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer’s amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol 161, 41–54 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devi L, Prabhu BM, Galati DF, Avadhani NG, Anandatheerthavarada HK. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer’s disease brain is associated with mitochondrial dysfunction. J Neurosci 26, 9057–9068 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning-Krieg UC, Scherer PE, Schatz G. Sequential action of mitochondrial chaperones in protein import into the matrix. EMBO J 10, 3273–3280 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano H, et al. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci 17, 822–831 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martell JD, Deerinck TJ, Lam SS, Ellisman MH, Ting AY. Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat Protoc 12, 1792–1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa EA, Subramanian K, Nunnari J, Weissman JS. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 359, 689–692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puig KL, et al. Amyloid precursor protein modulates macrophage phenotype and diet-dependent weight gain. Sci Rep 7, 43725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freeman LR, Zhang L, Dasuri K, Fernandez-Kim SO, Bruce-Keller AJ, Keller JN. Mutant amyloid precursor protein differentially alters adipose biology under obesogenic and non-obesogenic conditions. PLoS One 7, e43193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czeczor JK, et al. APP deficiency results in resistance to obesity but impairs glucose tolerance upon high fat feeding. J Endocrinol 237, 311–322 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Pera M, et al. Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in Alzheimer disease. EMBO J 36, 3356–3371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjune JI, et al. IRX5 regulates adipocyte amyloid precursor protein and mitochondrial respiration in obesity. Int J Obes (Lond), (2018). [DOI] [PMC free article] [PubMed]
- 39.Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol 151, 277–288 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das AT, Tenenbaum L, Berkhout B. Tet-On Systems For Doxycycline-inducible Gene Expression. Curr Gene Ther 16, 156–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao M, et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab 23, 1167–1184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao M, Hepler C, Vishvanath L, MacPherson KA, Busbuso NC, Gupta RK. Fetal development of subcutaneous white adipose tissue is dependent on Zfp423. Mol Metab 6, 111–124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabbrini E, et al. Metabolically normal obese people are protected from adverse effects following weight gain. J Clin Invest 125, 787–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo JB, et al. Knockdown of ANT2 reduces adipocyte hypoxia and improves insulin resistance in obesity. Nat Metab 1, 86–97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An YA, et al. Angiopoietin-2 in white adipose tissue improves metabolic homeostasis through enhanced angiogenesis. Elife 6, e24071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotzbeck P, et al. Brown adipose tissue whitening leads to brown adipocyte death and adipose tissue inflammation. J Lipid Res 59, 784–794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreux PA, et al. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat Metab 1, 595–603 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Crewe C, et al. An Endothelial-to-Adipocyte Extracellular Vesicle Axis Governed by Metabolic State. Cell 175, 695–708 e613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfanner N, Warscheid B, Wiedemann N. Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol 20, 267–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pallotti F, Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol 80, 3–44 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.