Abstract
The presence of microsatellite repeat expansions within genes is associated with >30 neurological diseases. Of interest, (GGGGCC)>30-repeats within C9orf72 are associated with amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD). These expansions can be 100’s to 1000’s of units long. Thus, it is perplexing how RNA polymerase II (RNAPII) can successfully transcribe them. Recent investigations focusing on GGGGCC-transcription have identified specific, canonical complexes that may promote RNAPII-transcription at these GC-rich microsatellites: the DSIF and PAF1 complexes. These complexes may be important for resolving the unique secondary structures formed by GGGGCC-DNA during transcription. Importantly, this process can produce potentially toxic repeat-containing RNA that can encode potentially toxic peptides, impacting neuron function and health. Understanding how transcription of these repeats occurs has implications for therapeutics in multiple diseases.
Keywords: Transcription, C9orf72, ALS/FTD, Drosophila, PAF1 Complex, DSIF complex
REPEAT EXPANSION DISEASES
A growing group of diseases are associated with nucleotide repeat expansions (microsatellites) within the genome. Although many of these diseases are neurodegenerative, they also include developmental diseases. Many of these microsatellites are GC-rich, including GGGGCC-repeats, termed G4C2, found in amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD), CAG-repeats found in polyglutamine diseases, and CTG-repeats found in myotonic dystrophy 1 (DM1)[1,2]. How these microsatellites confer toxicity is a captivating field of study while the underlying mechanism(s) vary based on the location of the microsatellite within afflicted genes as they can be found in promotors, 5’UTRs, introns, exons, and 3’ UTRs. Based on their location, the repeats can confer gain-of-function toxic effects and/or disrupt the normal function of the gene and gene product.
Diseases containing microsatellites within the gene body require RNA-polymerase II (RNAPII) to transcribe through the repeat sequence. This is thought to be challenging, given that these repeats likely assume unusual secondary structures that would need to be resolved during transcription. Normal, duplexed DNA preferentially forms right-handed double helices (e.g. B-form DNA). However, GC-rich nucleic acid strands have the propensity to form non-canonical secondary structures including G-quadruplexes and R-loops[3,4]. These unique, stable structures form as a result of strong interactions between the multiple guanines (G) and/or cytosines (C) within the DNA and RNA. Recent evidence suggests that unique transcriptional machinery, specifically the DRB-Sensitivity-Inducing Factor (DSIF) and Polymerase Associated Factor 1 (PAF1) complexes, may be required to promote RNAPII activity as it moves through these microsatellites. Here we discuss the role of these factors with a focus on GGGGCC-repeats in ALS/FTD. Given this function, these complexes present as intriguing targets to limit expression of toxic expanded repeat proteins in patients.
C9ORF72-ASSOCIATED ALS/FTD
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) represent opposite ends of a spectrum for the same neurodegenerative disorder. In ALS, loss of motor neurons within the central nervous system causes motor deficits, paralysis, and death within 2–5 years[5]. ALS afflicts ~2–5/100,000 people annually, making it the most common motor neuron disease. FTD results from cortical neuron loss with death 5–10 years post-disease onset[6]. FTD occurs in ~3–15/100,000 individuals. Patients with ALS and FTD develop both motor and cognitive deficits. Pathologically, Tar DNA-binding protein 43 (TDP-43) is depleted from nuclei and can form cytoplasmic inclusions, which are hallmarks of ALS, ALS and FTD, and a subset of FTD[7]. It is unclear what causes an individual to develop ALS versus FTD, with individuals from the same pedigree, or unrelated patients harboring the same mutation, presenting with varying degrees of motor versus cognitive disabilities[8]. Recently, a number of disease modifiers have been reported that may influence presentation, including Transmembrane protein 106B (TMEM106B) and Ataxin-2[9–12]. Due to this complexity, we reference this disease as “ALS/FTD” throughout the text while we note that individual patients may fall on either end of the spectrum and present with only ALS, only FTD, or both.
Multiple mutations have been identified in patients with familial ALS and FTD, including in TARBP, ATXN2, and FUS (please see[13,14]). In particular, a hexanucleotide repeat expansion of >30 (GGGGCC)n-repeats within the first intron of C9orf72 is the most prevalent mutation known to date (Fig. 1A)[8,15]. The maximum number of repeats is estimated to range in the thousands. Intermediate (2530) repeats have been associated with neuropsychiatric symptoms and potentially other movement disorders[16,17] and may be toxic based on data in flies and mice[18,19].
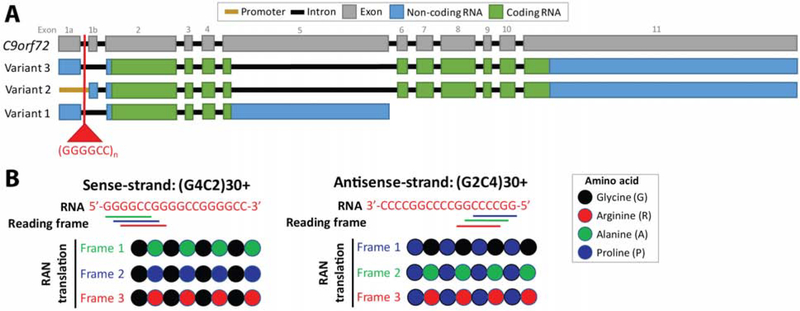
Figure 1: A G4C2 hexanucleotide repeat expansion is found within C9orf72 in ALS/FTD.
A. A hexanucleotide expansion of >30 (G4C2)n repeats is found within the C9orf72 gene in ~40% of familial ALS/FTD cases and ~7% of sporadic ALS/FTD cases. Three mRNA variants are produced from C9orf72, with the repeat expansion occurring within intron 1 of mRNA Variants 1 and 3. For mRNA Variant 2, it is in the promoter. B. Bidirectional transcription of mutant C9orf72 produces two RNA species containing the hexanucleotide repeat expansion: a sense- and an antisense-strand. These RNAs can undergo repeat-associated non-AUG translation to produce five aggregate-prone dipeptides: GA, GR, GP, PA, PR. (GP is produced by both RNA strands.) As peptides are detected in disease tissue, the RNAs for both strands containing the repeats must be made and accumulate.
How the expanded G4C2 repeats within C9orf72 contribute to disease is under intensive investigation with accumulating evidence supporting gain-of-function mechanisms as major components. Due to the G4C2-expansion, mutant C9orf72 can aberrantly produce repeat-containing RNA with bidirectional transcription producing two species, sense-G4C2 and antisense-G2C4 (termed G4C2∥G2C4) (Fig. 1B). Both species can accumulate to form foci in patient tissue[15] and may sequester RNA-binding proteins, leading to deleterious effects (see[2,20]). These RNAs can also be translated, in all three reading frames, into dipeptides which form potentially toxic aggregates[21,22]. One mechanism for translation is repeat-associated non-AUG (RAN) translation which is a field of growing interest[1,22]. Studies in model systems and with supportive data from patient-derived samples have revealed numerous pathways that may be disrupted by the expression of the G4C2-RNA and its dipeptide products, including protein quality control, translation, and nuclear-cytoplasmic transport (see[2,20]). Despite toxicity associated with specific dipeptides in model systems (Fig. 1B), particularly poly-GR (poly-glycine-arginine) and poly-PR (poly-proline-arginine), there is an ongoing debate as to their contribution to disease based on pathological patient data (see[2,21]). Cumulative protein aggregate frequency does not correlate with disease severity or tissue, although new studies strongly support a pathogenic role for individual dipeptides[23,24]. In contrast to gain-of-function mechanisms, the G4C2-mutation also disrupts expression of the C9orf72 gene product, leading to decreased protein; the contribution of gene downregulation to disease is a field of interest[2,21].
UNIQUE SECONDARY STRUCTURES OF G4C2 DNA AND RNA
Whether gain-of-function or loss-of-function mechanisms are driving toxicity associated with mutated C9orf72 in ALS/FTD, initial steps toward toxicity would require RNAPII-driven transcription of the G4C2-containing repeat. Unusual secondary structures formed by this repeat may lead to dependence on specific proteins to aid transcription.
In vitro investigations into sense G4C2-repeat DNA/RNA have shown that short (≤5) repeats fold into chair-type G-quadruplexes that exist in equilibrium with hairpin structures (Fig. 2A–B)[25,26]. G4C2-DNA seems to prefer an antiparallel topology[27–30] and G4C2-RNA a parallel topology[27,31–33]. G-quadruplexes can form within one DNA/RNA strand (intermolecular) or between multiple strands (intramolecular). Further, G4C2-DNA and nascent RNA can interact, forming hybrid secondary structures termed R-loops (Fig. 2C)[27,34,35]. These maybe stabilized by quadruplex formation on the complementary strand[36].
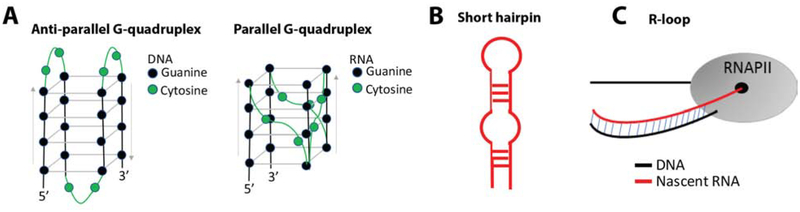
Figure 2: PAF1C impacts toxic G4C2-expression in C9orf72-associated disease.
A. (G4C2)4 DNA and RNA can form chair-type G-quadruplex secondary structures in vitro. G-quadruplexes involve the formation of G-tetrads between 4 guanines – a square planar structure held together by Hoogsteen hydrogen bonds. These G-tetrads form stacks that are stabilized by cations: particularly potassium (K+) and, to a lesser extent, sodium (Na+) then lithium (Li+). The cytosines in G4C2-structures create edgewise loops connecting the stacked G-tetrads. DNA prefers an anti-parallel conformation and RNA prefers a parallel conformation. Which structures form in vivo and with >30 G4C2 repeats still needs investigation. B. G-quadruplex structures formed by G4C2 D/RNA can coexist with another secondary structure, a short hairpin. These structures have been implicated in RAN-translation when found in repeat-containing RNA. C. During RNAPII-driven transcription of G4C2-DNA, unique DNA:RNA hybrids known as R-loops can form between the ssDNA and nascent RNA. R-loops can promote transcription while their accumulation can inhibit RNAPII, cause genomic instability, and cause DNA damage.
While less studied, the formation of quadruplexes by antisense-G2C4 DNA/RNA is debated[32,36]. G2C4-sequences could form quadruplexes, termed i-motifs, which exist in equilibrium with hairpins[37]. Antisense-G2C4 DNA may also form e-motifs, a relatively unstudied secondary structure[38,39]. In contrast, G2C4 RNA may preferentially form more canonical double helices over i-motifs[40].
To date, most structural studies have depended on in vitro data and short (≤8) repeats. Multiple factors could contribute to complexity of G4C2-structures in vivo, including the number of repeats, pH, modification of G4C2∥G2C4 strands, and interactions with endogenous proteins (e.g. RNA-binding proteins). In particular, G4C2-repeat expansions are methylated in patients[41,42] which has been shown to impact resulting structures in vitro[36].
Data supporting the formation of G4C2∥G2C4 secondary structures in cells is accumulating, but primarily correlative. R-loops were directly shown to form ex vivo at the G4C2-microsatellite in C9+-derived iPS cells[43]. Another study showed that the RNA-binding protein, Nucleolin, preferentially binds G4C2-associated G-quadruplexes in vitro and may co-localize with G4C2-RNA foci in C9+-derived tissues[27]. A compelling study identified small molecules that bind G4C2-associated G-quadruplexes in vitro and reduce RNA-foci formation, dipeptide production, and toxicity in patient-derived iPS neurons (ex vivo) and in (G4C2)36-expressing Drosophila (in vivo)[44]. A recent report defined a small molecule capable of binding the unique hairpin structure of G4C2-RNA, effectively disrupting G4C2-foci formation and RNA binding protein interactions ex vivo[26]. Overall, data are consistent with the formation of unique quadruplexes, hairpins, and R-loops in disease, with more direct ex vivo and in vivo studies required to fully define their role.
G-QUADRUPLEXES AND R-LOOPS IN GENERAL TRANSCRIPTION
RNAPII-transcription has been well-studied. RNAPII activity is mediated by a number of factors that can act at multiple stages: initiation, promoter:proximal pausing, elongation, or termination[45]. Different genes require different factors that act at one or multiple stages. Throughout the process, duplexed DNA must locally unwind to expose the individual strands to transcriptional machinery. However, this leaves the ssDNA, particularly if it is GC-rich as with the G4C2-repeat, vulnerable to forming secondary structures including quadruplexes, hairpins, or R-loops[46–48]. These structures would need to be resolved, likely by transcription activators and helicases[49–51], for RNAPII to be able to progress along the gene[52,53]. Fittingly, there are an array of accessory factors available to help elongating-RNAPII that can stimulate RNAPII interactions with DNA and recruit additional machinery to locally open the chromatin.
G-quadruplexes are found throughout the human genome and tend to be located within regulatory gene regions: promoters, 5’UTR, 3’UTR, and telomeres[54–56]. Overall, it is hypothesized that the formation of quadruplexes on template ssDNA would inhibit transcription[3,49]. Their formation on non-template ssDNA could similarly block transcription, but also could promote transcription by maintaining open chromatin. While there are increasing examples of gene regulation by G-quadruplex formation, c-MYC is particularly well studied. This proto-oncogene has a G-quadruplex within its promoter that can be stabilized with small molecules, causing reduced c-MYC expression by blocking transcription factors[57]. Multiple examples of G-quadruplex formation within introns show that they can mediate alternative splicing[58,59]. Their formation within the first intron of TOP1 has been proposed to inhibit RNAPII-transcription[60], suggesting that multiple mechanisms may be disrupted when G-quadruplexes aberrantly form within a gene-body.
In addition to G-quadruplexes, R-loops also regulate RNAPII-transcription. These DNA:RNA hybrids are proposed to facilitate transcription through multiple mechanisms, including the protection of ssDNA from methylation and recruitment of transcription factors[61,62]. Studies support that R-loop formation by GC-rich DNA/RNA occurs during transcription and R-loop accumulation can disrupt polymerase activity in vitro and in vivo. Consistent with a requirement for these structures to be cleared for successful transcription, RNA-binding and accessory proteins have been identified that resolve or prevent R-loop formation. For example, loss of THO/TREX components causes R-loops to accumulate and a reduction in RNAPII elongation rates[62–64]. The oncogenes BRCA1 and BRCA2 prevent R-loop accumulation and thus promote RNAPII-driven transcription[65,66]. Of importance, BRCA2 recruits Paf1 (discussed below) to promoter:proximal gene sites and overexpression of Paf1 can rescue deficits caused by BRCA2 loss by resolving accumulated R-loops; this suggests that BRCA1/2 is not directly acting on R-loops but rather recruits factors to resolve these hybrids.
Generally, due to the complexity of transcribing GC-rich DNA and the ability for individual sequences to form unique secondary structures, it is likely that there are a number of factors available to resolve these DNA/RNA structures within cells (e.g. senataxin, whose loss of function is associated with ALS[67]).
TRANSCRIPTION OF G4C2||G2C4 REPEAT EXPANSIONS
Regardless of whether the G4C2-repeat RNA is contributing to toxicity or if dipeptide-induced toxicity is the primary contributor, a potential therapeutic approach for gain-of-function disease mechanisms would be to inhibit the transcription of the repeat expansion.
The DSIF complex: Spt4 and Spt5
The DSIF complex was recently shown to modulate expression from expanded G4C2∥G2C4 DNA[68]. This highly conserved complex is comprised of two subunits, Spt4 and Spt5, which form a heterodimer[69].
Originally described as an RNAPII-activating complex in yeast, DSIF has since been implicated in multiple steps of RNAPII transcription, including promoter:proximal pausing, elongation, and RNA processing/termination.
The involvement of DSIF in the transcription of repeat-expansions was initially described in CAG-models for Huntington’s disease (HD)[70,71]. Notably, expanded CAG-repeats can form secondary structures similar to G4C2-repeats, including hairpins and R-loops[34,62]. Depleting Spt4 reduced RNA/protein expression and toxicity associated with (CAG)≥81 in yeast and cultured neurons[70]. Mechanistic studies in yeast revealed that Spt4 deletion caused reduced RNAPII occupancy downstream of a (CAG)99-expansion inserted into a reporter gene. This effect was not seen with an inert (CAG)29-repeat, arguing repeat-length specificity. Further investigations showed that 50% downregulation of SUPT4a (mouse Spt4) in two HD-mouse models expressing expanded (CAG)≥115 repeats reduced RNA/protein expression from the transgenes in vivo[71]. Importantly, reduced expression was associated with reduced neurodegenerative effects of the mutant-Htt-Q115+.
Kramer and colleagues took a multi-disciplinary approach to extend the discovery of DSIF as a transcriptional regulator of microsatellites to G4C2-repeats in C9orf72-disease[68]. Using gain-of-function yeast, worm, and fly models that expressed (G4C2)≥49 repeats, investigators found that depletion of Spt4 orthologues could suppress RNA expression from expanded G4C2 transgenes with concomitant reductions in toxicity. These data indicate an important, conserved role of DSIF complex in regulating G4C2-expression. Arguing dose-dependence, overexpressing SUPT4H1 (human Spt4) in C. elegans caused significant increases in G4C2-RNA levels and toxicity. In Drosophila, Spt4 was uncovered in an RNAi-based genetic screen for factors that suppress G4C2-induced degenerative effects in the fly[18,68]. Translating these findings to human C9+ disease, downregulating DSIF components, SUPT4H1 and SUPT5H, in C9+ patient-derived cells caused reduced expression of G4C2-associated transcripts, number of sense-G4C2 and antisense-G2C4 foci, and levels of one of the dipeptides produced in vivo from the repeat (Fig. 1B). Interestingly, a positive correlation between the transcript levels of SUPT4H1 or SUPT5H and levels of expression of the G4C2 repeat transcript were seen in post-mortem FTD/ALS tissue, suggesting that these factors may influence expression of the repeat in the C9+ disease[68].
There is accumulating data to suggest that this complex may affect additional repeat-associated diseases. Spt4 loss in yeast was shown to impact (CTG)105, (CAA)90 and (AAA)24 expression[70]. Further, knockdown of SUPT4a and SUPT5 (mouse Spt5) in neuroblast cells reduced expression of a GGCCTG-repeat expansion associated with spinocerebellar ataxia type 36[72]. The global impact of downregulating this complex in higher organisms requires further investigation, however. While these studies indicate selective activity at repeat-expansions, others have suggested a broader impact on transcription[18,73].
The PAF1 complex: Paf1, Leo1, CDC73, Ctr9, and Rtf1
PAF1C was first defined as an RNAPII-transcriptional regulator in S.cerevisiae[74]. The core components of PAF1C are highly conserved: Paf1, Leo1, CDC73, Ctr9 and Rtf1. Metazoan PAF1C can also include a multifunctional protein, WDR61[75]. Accumulating evidence support a conserved role of PAF1C in RNAPII-transcription at multiple stages: initiation, promoter:proximal pausing, elongation, and RNA processing/termination. In yeast, studies have shown that PAF1C recruitment is dependent on Spt5 interactions with Rtf1. In metazoans, however, recruitment of the complex may not depend on DSIF. Metazoan Rtf1 is not tightly associated with PAF1C[76–78]. Further, Leo1, CDC73, and a Paf1:Ctr9 subcomplex can all recruit other PAF1C components to RNAPII[79–82]. Following recruitment of the complex, structural data shows that PAF1C directly interacts with elongating RNAPII, stimulating its activity[83–86]. PAF1C can also recruit chromatin remodeling factors that promote transcription. Current studies show that expression from only a subset of genes is impacted by PAF1C loss, suggesting selective regulation of transcription versus a global impact. Like DSIF, RNAPII-transcription of GC-rich DNA is particularly susceptible to PAF1C loss [87,88].
Evidence supports that PAF1C may be important for promoting RNAPII-transcription of G4C2||G2C4-repeat expansions in C9+ ALS/FTD. Knockdown of PAF1C components were identified as toxicity suppressors in a large-scale RNAi-based screen in Drosophila[18]. Mechanistically, downregulation of all PAF1C components reduced RNA levels and dipeptide expression from long, toxic (G4C2)>30 transgenes in multiple fly models with concomitant reduction in (G4C2)>30-associated toxicity. Data in yeast supported these findings, showing that expression from both sense-G4C2 and antisense-G2C4 transgenes were reduced upon deletion of PAF1C components: Leo1 and CDC73.
An important finding from this study was that RNAi against PAF1C components Paf1 and Leo1 did not impact RNA levels produced from short, inert G4C2 transgenes in the fly. Rather, these RNAi only impacted expression from long, toxic repeat expansions, arguing that Paf1 and Leo1 may be selective to disease-associated repeat expansions of >30 units. Supporting this finding, only these PAF1C components were found to be upregulated in post-mortem patient brains that harbored the G4C2 microsatellite. Importantly, PAF1 and LEO1 expression, but not CDC73 expression, positively correlated with expression of C9orf72 transcripts in C9+ FTD cases. Arguing that upregulation was a direct result of the microsatellite, this was not seen in FTD patients lacking the expansion. A role of Paf1 and Leo1 in transcribing the G4C2-microsatellite was further supported by findings that Leo1 protein bound intron 1 of the C9orf72 chromatin just 3’ of the G4C2 repeat expansion in C9+-derived iPS cells. These data argue that the Paf1:Leo1 heterodimer was present during transcription through the repeat.
Overall, these data implicate PAF1C as a transcriptional regulator of expanded G4C2||G2C4-DNA in C9orf72-disease, with Paf1 and Leo1 function seeming selective for the expanded repeat. Further mechanistic analyses are needed to define how PAF1C contributes to transcription of G4C2-repeats, whether it can function at other sequences that cause microsatellites in disease, and what makes Paf1 and Leo1 so unique in the disease situation.
A model for G4C2∥G2C4-repeat transcription
Current data from multiple model systems and patient samples support that DSIF and PAF1 complexes are important for regulating RNAPII-transcription of G4C2∥G2C4-repeat DNA in C9+ situations and potentially other repeat-expansion diseases. Assuming that both complexes work cooperatively to promote RNAPII-transcription along G4C2∥G2C4-repeats, this suggests a model where the PAF1C and DSIF complexes interact with RNAPII during transcription (Fig. 3). Moreover, of the components studied, only the Paf1:Leo1 heterodimer displays specificity to disease-associated repeat expansions in metazoans. Overall, present data suggest a model wherein DSIF and PAF1C loss reduces RNAPII occupancy along the expanded G4C2∥G2C4 DNA.
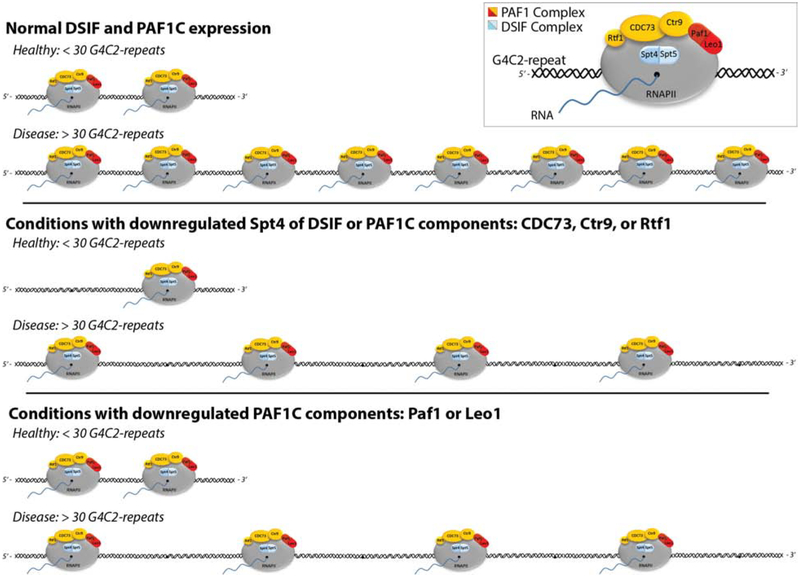
Figure 3, Key Figure: A model for G4C2||G2C4 transcription.
Top panel: Transcription elongation factors DSIF (Spt4/5) and PAF1C (Paf1/ Leo1/ CDC73/ Ctr9/ Rtf1) are recruited to, and are important for, RNAPII-transcription at G4C2-repeats – potentially acting during the elongation phase. Middle panel: in flies, depletion of Spt4, or specific Paf1C components – CDC73, Ctr9, and Rtf1 –alters expression from both inert <30 G4C2 repeats and toxic >30 G4C2 repeats. Bottom panel: by contrast, PAF1C components Paf1 and Leo1 (which form a heterodimer within PAF1C) seem most important for transcription from the toxic >30 G4C2 repeat, as their depletion only impacted G4C2-transcription in disease situations. Data indicate that PAF1C components are upregulated in response to the expression of >30 G4C2 repeats in flies, mice, and patient-derived cells[18]. Further, hSUPT4H1, hPAF1 and hLEO1, but not hCDC73, are upregulated in C9+ patient tissue, potentially the result of an increased requirement for these factors’ as they enhance transcription of the gene carrying an expanded repeat. We hypothesize that transcription of the expanded G4C2 DNA requires PAF1C and RNAPII to interact during elongation through the repeat expansion, supported by data that PAF1C is bound to G4C2-chromatin immediately 3’ of the repeat in C9+ derived patient cells[18]. DSIF may be important for recruiting PAF1C to the G4C2-repeat expansion.
Recent cryo-EM studies have investigated the structure of poised versus elongating RNAPII and its binding partners by reconstituting the human complexes in vitro[86,89]. Vos and colleagues found that DSIF was bound to RNAPII at both stages, while PAF1C was only bound to elongating-RNAPII. The specificity of PAF1C function to elongating RNAPII is consistent with ChIP studies examining binding patterns of DSIF and PAF1C components along genes[90–92]. Whether the binding pattern is the same for DSIF and PAF1C along the expanded repeat in C9+ disease needs investigation, but these findings argue that the DSIF and PAF1 complexes maybe acting at different stages of its transcription.
A surprising feature is that PAF1C and DSIF components become upregulated in response to expression of toxic (G4C2)>30 repeats[18,93]. In flies, mice, and patient-derived cells, expression of toxic ≥30 G4C2-repeats but not inert (≤30) repeats was associated with this upregulation[18]. Further, specific components of the DSIF and PAF1 complexes—SUPT4H1, PAF1, and LEO1—are upregulated in post-mortem tissue from ALS/FTD patients carrying the G4C2-expansion when compared to patients lacking this mutation[18,93]. While these findings further support that these components are of functional importance in C9+ disease, they also argue that a feed-forward loop may exist in disease that causes their upregulation. We hypothesize that G4C2-containing chromatin sequesters Spt4, Paf1, and Leo1 in C9+ expressing cells, resulting in the upregulation of these products to compensate for their depletion at other genomic loci that require their function. As these factors likely are not acting only at the G4C2-repeat, there may be additional effects caused by their upregulation. For example, repetitive elements are upregulated in C9+ post-mortem tissue while this may be impacted by DSIF and or PAF1C[93]. Overall, data suggest that misregulation of these components may have functional importance from multiple perspectives in C9+ disease, not just by impacting transcription of the G4C2-repeat.
Concluding Remarks and Future Perspectives
Extended investigations to explore if PAF1C and DSIF plays a global role in multiple repeat-associated diseases are of interest. Unique transcriptional machinery required to promote RNAPII-activity as it progresses through GC-rich repeat expansions may be common among diseases associated with microsatellites within gene bodies. The DSIF complex has been suggested to regulate RNAPII-transcription of repetitive DNA elements including (G4C2)≥8, (CAG)≥99, (CTG)105, (CAA)90, (AAA)≥24, and (GGCCTG)75 sequences[18,68,70–72]. Similar to G4C2||G2C4-repeats, DNA and RNA composed of other repeat expansions, can form quadruplexes, hairpins, and R-loops that likely need resolution during transcription[94–98]. More data to expand on described observations and provide additional mechanistic insight into how DSIF and PAF1C precisely impact RNA transcription—initiation vs elongation vs termination—are of interest.
Further, mechanistic investigations into both DSIF and PAF1C are needed to define how these complexes are functioning. Current data suggests that these complexes may help resolve the unique secondary structures formed by G4C2-repeats during transcription. Speculatively, if microsatellites can form secondary structures in vivo, as supported by accumulating data[26,27,43,44], the factors described herein may be required for resolving these aberrant structures during transcription; they may act directly or by recruiting additional factors. Of significance, DSIF may interact with GC-rich R-loops[99]. Moreover, multiple studies have implicated PAF1C components in regulating R-loops[66,99–101] and telomere repeat containing RNA (TERRA)[102–104] (a GC-rich region that is prone to R-loop and G-quadruplex formation). PAF1C is also important for c-MYC expression[105–107], a well-characterized proto-oncogene that involves a regulatory G-quadruplex within its promoter. Overall, direct investigations into DSIF and PAF1C in interactions with, and function at, R-loops and G-quadruplexes are of importance.
While we focus on the role of unique transcriptional machinery in diseases caused by gain-of-function mechanisms, this work may also impact diseases where microsatellites silence genes. For example, in Friedrich’s ataxia the presence of a GAA-repeat expansion within intron 1 of FXN silences the gene causing disease. A recent study designed synthetic transcription elongation factors that could promote expression of FXN despite the GAA-expansion[108]. These data highlight how elongation may be a critical step during RNAPII-transcription of microsatellites. In such disease situations, it would be interesting to define if increasing PAF1C and DSIF activity could promote expression of the gene in response to the presence of the microsatellite.
The concept of disrupting the expression of gain-of-function repeat expansion disease is aggressively being pursued as a therapy using reagents like small molecules and antisense oligonucleotides. While promising, there are limitations to the ASO approaches (reviewed in[109,110]). For example, as individual ASOs only target either sense- or antisense-transcripts produced from a repeat, a cocktail involving multiple ASOs would be required to effectively reduce both G4C2- and G2C4-transcripts produced by the G4C2-repeat expansion. Preventing these complementary ASOs from interacting with each other maybe difficult among other considerations. In the end, effective treatments may require combining multiple approaches, such as ASOs with antibodies targeting mutant proteins[111]. Inhibiting transcription factors that are of special importance for the expression of microsatellites could disrupt sense- and antisense-RNA production simultaneously, representing an additional/alternative approach. Moreover, data in Drosophila supports that partial inhibition of these factors maybe all that is required to significantly impact G4C2-transcripton; partial knockdown may be well tolerated (discussed in [18]). Careful considerations should be taken when targeting transcription factors, but chemotherapy drugs in cancer have shown that significant improvements for patients can be seen when targeting canonical machinery.
Highlights.
A hexanucleotide repeat expansion of >30 GGGGCC-units within C9orf72 is associated with familial ALS/FTD. Evidence supports that gain-of-function mechanisms are a primary contributor to disease.
G4C2||G2C4-DNA and RNA can form unique secondary structures, G-quadruplexes and R-loops, that likely impact transcription (and translation) of G4C2||G2C4-repeat expansions.
G-quadruplexes and R-loops formed by GC-rich DNA impact RNAPII-driven transcription under normal conditions.
Transcription of G4C2-repeat expansions in ALS/FTD requires unique machinery for RNAPII-driven expression. Breakthrough studies on the DSIF and PAF1 complexes revealed their roles in promoting expression of G4C2-repeats in disease.
Identification of unique transcriptional machinery required by GC-rich repeat expansions may highlight novel therapeutic targets universal to repeat-expansion associated diseases.
Outstanding questions.
Is G4C2 DNA/RNA able to form G-quadruplex structures in vivo? Is this tissue specific? Do these structures inhibit transcription in disease?
Can DSIF and/or PAF1 complexes mediate transcription of multiple sequences associated with repeat-expansions, implying a universal target for multiple diseases? What happens when multiple components of these complexes are simultaneously downregulated? Also, does their upregulation increase RNAPII-transcription of G4C2 repeats?
How does the PAF1 complex mediate transcription of G4C2-repeat expansions in disease? Can it help resolve G-quadruplexes and R-loops during transcription? Or is it recruiting other factors that aid in transcription of G4C2-repeat expansions?
How are the DSIF and/or PAF1 complexes recruited to G4C2-repeat expansion during RNAPII-driven transcription?
Can specific components of the PAF1 complex be targeted pharmacologically to disrupt transcription of G4C2-repeat expansions in vivo? How does this impact disease-associated phenotypes? What additional effects are seen? What is the minimal amount of inhibition required to selectively reduce transcription of repeat-expansions?
Acknowledgments
This work was funded by the Systems and Integrative Biology NIH/NIGMS training grant T32-GM07517 (to L.D.G.), NIH/NINDS R01-NS078283 (to N.M.B.), and NIH/NINDS R35-NS09727 (to N.M.B.).
Glossary
- PAF1C
polymerase associated factor 1 complex, composed of Paf1, Leo1, CDC73, Ctr9, and Rtf1
- DSIF
DRB-sensitivity-inducing factor complex, composed of Spt4 and Spt5
- G4C2
GGGGCC-repeat expansion found within the C9orf72 gene in 40% of ~familial ALS/FTD
- ALS
amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
- RAN translation
repeat associated non-AUG translation, occurs on expanded repeat sequences to produce peptide products
Footnotes
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Banez-Coronel M and Ranum LPW (2019) Repeat-associated non-AUG (RAN) translation: insights from pathology. Laboratory Investigation 99, 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook C and Petrucelli L (2019) Genetic Convergence Brings Clarity to the Enigmatic Red Line in ALS. Neuron 101, 1057–1069 [DOI] [PubMed] [Google Scholar]
- 3.Bochman ML et al. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nature Reviews Genetics 13, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushik M et al. (2016) A bouquet of DNA structures: Emerging diversity. Biochemistry and Biophysics Reports 5, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logroscino G and Piccininni M (2019) Amyotrophic Lateral Sclerosis Descriptive Epidemiology: The Origin of Geographic Difference. NED 52, 93–103 [DOI] [PubMed] [Google Scholar]
- 6.Seltman RE and Matthews BR (2012) Frontotemporal Lobar Degeneration: Epidemiology, Pathology, Diagnosis and Management. CNS Drugs 26, 841–870 [DOI] [PubMed] [Google Scholar]
- 7.Neumann M et al. (2006) Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 8.Renton AE et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagi KS et al. (2019) Meta-analysis of Genetic Modifiers Reveals Candidate Dysregulated Pathways in Amyotrophic Lateral Sclerosis. Neuroscience 396, A3–A20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pottier C et al. (2019) Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol DOI: 10.1007/s00401-019-01962-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Blitterswijk M et al. (2014) Ataxin-2 as potential disease modifier in C9ORF72 expansion carriers. Neurobiol Aging 35, 2421.e13–2421.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Blitterswijk M et al. (2014) TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol 127, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathis S et al. (2019) Genetics of amyotrophic lateral sclerosis: A review. Journal of the Neurological Sciences 399, 217–226 [DOI] [PubMed] [Google Scholar]
- 14.Pottier C et al. (2016) Genetics of FTLD: overview and what else we can expect from genetic studies. J. Neurochem 138, 32–53 [DOI] [PubMed] [Google Scholar]
- 15.DeJesus-Hernandez M et al. (2011) Expanded GGGGCC hexanucleotide repeat in non-coding region of C9ORF72 causes chromosome 9p-linked frontotemporal dementia and amyotrophic lateral sclerosis. Neuron 72, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng ASL and Tan E-K (2017) Intermediate C9orf72 alleles in neurological disorders: does size really matter? Journal of Medical Genetics 54, 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourinaris T and Houlden H (2018) C9orf72 and its Relevance in Parkinsonism and Movement Disorders: A Comprehensive Review of the Literature. Movement Disorders Clinical Practice 5, 575–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman LD et al. (2019) Expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y et al. (2016) C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron 90, 521–534 [DOI] [PubMed] [Google Scholar]
- 20.Balendra R and Isaacs AM (2018) C9orf72 -mediated ALS and FTD: multiple pathways to disease. Nature Reviews Neurology 14, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vatsavayai SC et al. (2019) C9orf72-FTD/ALS pathogenesis: evidence from human neuropathological studies. Acta Neuropathol 137, 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman LD and Bonini NM (2019) Repeat-associated non-AUG (RAN) translation mechanisms running into focus for GGGGCC-repeat associated ALS/FTD. Prog. Neurobiol DOI: 10.1016/j.pneurobio.2019.101697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saberi S et al. (2018) Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol 135, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakae N et al. (2018) Poly-GR dipeptide repeat polymers correlate with neurodegeneration and Clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathologica Communications 6, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ash PEA et al. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z-F et al. (2019) The Hairpin Form of r(G4C2)exp in c9ALS/FTD Is Repeat-Associated Non-ATG Translated and a Target for Bioactive Small Molecules. Cell Chemical Biology 26, 179–190.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haeusler AR et al. (2014) C9orf72 Nucleotide Repeat Structures Initiate Molecular Cascades of Disease. Nature 507, 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Šket P et al. (2015) Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiology of Aging 36, 1091–1096 [DOI] [PubMed] [Google Scholar]
- 29.Zhou B et al. (2015) Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Scientific Reports 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brčić J and Plavec J (2017) ALS and FTD linked GGGGCC-repeat containing DNA oligonucleotide folds into two distinct G-quadruplexes. Biochimica et Biophysica Acta (BBA) - General Subjects 1861, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 31.Fratta P et al. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy K et al. (2013) The Disease-associated r(GGGGCC)n Repeat from the C9orf72 Gene Forms Tract Length-dependent Uni- and Multimolecular RNA G-quadruplex Structures. J Biol Chem 288, 9860–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su Z et al. (2014) Discovery of a Biomarker and Lead Small Molecules to Target r(GGGGCC)-Associated Defects in c9FTD/ALS. Neuron 83, 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy K et al. (2014) Processing of double-R-loops in (CAG)·(CTG) and C9orf72 (GGGGCC)·(GGCCCC) repeats causes instability. Nucleic Acids Res 42, 10473–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diab MA et al. (2018) The G-rich Repeats in FMR1 and C9orf72 Loci Are Hotspots for Local Unpairing of DNA. Genetics 210, 1239–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamiri B et al. (2015) Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8•(GGCCCC)8 repeat: effect of CpG methylation. Nucleic Acids Res 43, 10055–10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamiri B et al. (2018) Stress-induced acidification may contribute to formation of unusual structures in C9orf72 -repeats. Biochimica et Biophysica Acta (BBA) - General Subjects 1862, 14821491. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y et al. (2017) Structure and Dynamics of DNA and RNA Double Helices Obtained from the GGGGCC and CCCCGG Hexanucleotide Repeats That Are the Hallmark of C9FTD/ALS Diseases. ACS Chem. Neurosci 8, 578–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan F et al. (2018) E-motif formed by extrahelical cytosine bases in DNA homoduplexes of trinucleotide and hexanucleotide repeats. Nucleic Acids Res 46, 942–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dodd DW et al. (2016) Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches. Biochemistry 55, 1283–1286 [DOI] [PubMed] [Google Scholar]
- 41.Bauer PO (2016) Methylation of C9orf72 expansion reduces RNA foci formation and dipeptide-repeat proteins expression in cells. Neuroscience Letters 612, 204–209 [DOI] [PubMed] [Google Scholar]
- 42.Xi Z et al. (2015) The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol. 129, 715–727 [DOI] [PubMed] [Google Scholar]
- 43.Esanov R et al. (2017) A C9ORF72 BAC mouse model recapitulates key epigenetic perturbations of ALS/FTD. Molecular Neurodegeneration 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simone R et al. (2018) G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol Med 10, 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonkers I and Lis JT (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duquette ML et al. (2004) Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18, 1618–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C et al. (2013) DNA G-quadruplex formation in response to remote downstream transcription activity: long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res 41, 7144–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng K et al. (2013) Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res 41, 5533–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armas P et al. (2016) Transcriptional control by G-quadruplexes: In vivo roles and perspectives for specific intervention. Transcription 8, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brázda V et al. (2014) DNA and RNA Quadruplex-Binding Proteins. International Journal of Molecular Sciences 15, 17493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendoza O et al. (2016) G-quadruplexes and helicases. Nucleic Acids Res 44, 1989–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jonkers I et al. (2014) Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veloso A et al. (2014) Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Research 24, 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam EYN et al. (2013) G-quadruplex structures are stable and detectable in human genomic DNA. Nature Communications 4, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chambers VS et al. (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nature Biotechnology 33, 877. [DOI] [PubMed] [Google Scholar]
- 56.Kudlicki AS (2016) G-Quadruplexes Involving Both Strands of Genomic DNA Are Highly Abundant and Colocalize with Functional Sites in the Human Genome. PLoS ONE 11, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks TA and Hurley LH (2010) Targeting MYC Expression through G-Quadruplexes. Genes Cancer 1, 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcel V et al. (2011) G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 32, 271–278 [DOI] [PubMed] [Google Scholar]
- 59.Verma SP and Das P (2018) G-quadruplex structure at intron 2 of TFE3 and its role in Xp11.2 translocation and splicing. Biochim Biophys Acta Gen Subj. 1862, 630–636 [DOI] [PubMed] [Google Scholar]
- 60.Reinhold WC et al. (2010) Exon array analyses across the NCI-60 reveals potential regulation of TOP1 by transcription pausing at guanosine quartets in the first intron. Cancer Res 70, 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crossley MP et al. (2019) R-Loops as Cellular Regulators and Genomic Threats. Molecular Cell 73, 398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim N and Jinks-Robertson S (2012) Transcription as a source of genome instability. Nat Rev Genet 13, 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huertas P and Aguilera A (2003) Cotranscriptionally Formed DNA:RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Molecular Cell 12, 711–721 [DOI] [PubMed] [Google Scholar]
- 64.Domíguez-Sánchez MS et al. (2011) Genome Instability and Transcription Elongation Impairment in Human Cells Depleted of THO/TREX. PLOS Genetics 7, e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X et al. (2017) Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nature Communications 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shivji MKK et al. (2018) BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Reports 22, 1031–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grunseich C et al. (2018) Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Molecular Cell 69, 426–437.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kramer NJ et al. (2016) Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts associated with c9FTD/ALS. Science 353, 708–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartzog GA and Fu J (2013) The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochimica et biophysica acta 1829, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu C-R et al. (2012) Spt4 Is Selectively Required for Transcription of Extended Trinucleotide Repeats. Cell 148, 690–701 [DOI] [PubMed] [Google Scholar]
- 71.Cheng H-M et al. (2015) Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown. PLoS Genet 11, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furuta N et al. (2019) Suppression of the yeast elongation factor Spt4 ortholog reduces expanded SCA36 GGCCUG repeat aggregation and cytotoxicity. Brain Research DOI: 10.1016/j.brainres.2018.12.045 [DOI] [PubMed] [Google Scholar]
- 73.Naguib A et al. (2019) SUPT4H1 Depletion Leads to a Global Reduction in RNA. Cell Reports 26, 45–53.e4 [DOI] [PubMed] [Google Scholar]
- 74.Van Oss SB et al. (2017) Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation. Trends in Biochemical Sciences 42, 788–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu B et al. (2005) The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 19, 1668–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adelman K et al. (2006) Drosophila Paf1 Modulates Chromatin Structure at Actively Transcribed Genes. Mol Cell Biol 26, 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mbogning J et al. (2013) The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast. PLoS Genet 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao Q-F et al. (2015) Characterization of the Human Transcription Elongation Factor Rtf1: Evidence for Nonoverlapping Functions of Rtf1 and the Paf1 Complex. Mol Cell Biol 35, 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dermody JL and Buratowski S (2010) Leo1 Subunit of the Yeast Paf1 Complex Binds RNA and Contributes to Complex Recruitment. J. Biol. Chem 285, 33671–33679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amrich CG et al. (2012) Cdc73 Subunit of Paf1 Complex Contains C-terminal Ras-like Domain That Promotes Association of Paf1 Complex with Chromatin. J Biol Chem 287, 10863–10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu H et al. (2012) Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J 31, 3494–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Y et al. (2018) Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation. Nature Communications 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim TS et al. (2010) RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biology 11, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chu X et al. (2013) Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Res 41, 10619–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y et al. (2017) Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nature Communications 8, 15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vos SM et al. (2018) Structure of paused transcription complex Pol II–DSIF–NELF. Nature 560, 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rondón AG et al. (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. The EMBO Journal 22, 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rondón AG et al. (2004) Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep 5, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vos SM et al. (2018) Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature 560, 607–612 [DOI] [PubMed] [Google Scholar]
- 90.Chen Y et al. (2009) DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 23, 2765–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mayer A et al. (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nature Structural & Molecular Biology 17, 1272. [DOI] [PubMed] [Google Scholar]
- 92.Van Oss SB et al. (2016) The Histone Modification Domain of Paf1 Complex Subunit Rtf1 Directly Stimulates H2B Ubiquitylation Through an Interaction with Rad6. Mol Cell 64, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prudencio M et al. (2017) Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum Mol Genet 26, 3421–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Childs-Disney JL et al. (2014) Structure of the Myotonic Dystrophy Type 2 RNA and Designed Small Molecules That Reduce Toxicity. ACS chemical biology 9, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Cruchten RTP et al. (2019) Expanded CUG repeats in DMPK transcripts adopt diverse hairpin conformations without influencing the structure of the flanking sequences. RNA 25, 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krzyzosiak WJ et al. (2012) Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res 40, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kumari D et al. (2011) Repeat Expansion Affects Both Transcription Initiation and Elongation in Friedreich Ataxia Cells. J Biol Chem 286, 4209–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin Y and Wilson JH (2011) Transcription-induced DNA toxicity at trinucleotide repeats. Cell Cycle 10, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang IX et al. (2018) Human proteins that interact with RNA/DNA hybrids. Genome Res. 28, 1405–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wahba L et al. (2011) RNase H and Multiple RNA Biogenesis Factors Cooperate to Prevent RNA:DNA Hybrids from Generating Genome Instability. Molecular Cell 44, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Landsverk HB et al. (2019) Regulation of ATR activity via the RNA polymerase II associated factors CDC73 and PNUTS-PP1. Nucleic Acids Res 47, 1797–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neidle S (2010) Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. The FEBS Journal 277, 1118–1125 [DOI] [PubMed] [Google Scholar]
- 103.Rodrigues J and Lydall D (2018) Paf1 and Ctr9, core components of the PAF1 complex, maintain low levels of telomeric repeat containing RNA. Nucleic Acids Res 46, 621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toubiana S and Selig S (2018) DNA:RNA hybrids at telomeres – when it is better to be out of the (R) loop. The FEBS Journal 285, 2552–2566 [DOI] [PubMed] [Google Scholar]
- 105.Zhi X et al. (2015) Human RNA polymerase II associated factor 1 complex promotes tumorigenesis by activating c-MYC transcription in non-small cell lung cancer. Biochemical and Biophysical Research Communications 465, 685–690 [DOI] [PubMed] [Google Scholar]
- 106.Gerlach JM et al. (2017) PAF1 complex component Leo1 helps recruit Drosophila Myc to promoters. PNAS 114, E9224–E9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sauer M and Paeschke K (2017) G-quadruplex unwinding helicases and their function in vivo. Biochemical Society Transactions 45, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 108.Erwin GS et al. (2017) Synthetic transcription elongation factors license transcription across repressive chromatin. Science 358, 1617–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.DeVos SL and Miller TM (2013) Antisense Oligonucleotides: Treating Neurodegeneration at the Level of RNA. Neurotherapeutics 10, 486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schoch KM and Miller TM (2017) Antisense oligonucleotides: Translation from mouse models to human neurodegenerative diseases. Neuron 94, 1056–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zoghbi HY (2019) Strategy to selectively remove mutant proteins could combat neurodegeneration. Nature 575, 57–58 [DOI] [PubMed] [Google Scholar]