Abstract
Li-Fraumeni syndrome (LFS) is a rare autosomal dominant disorder associated with TP53 germline mutations and an increased lifetime risk of multiple primary cancers (MPC). Penetrance estimation of time to first and second primary cancer within LFS remains challenging due to limited data and the difficulty of characterizing the effects of a primary cancer on the penetrance of a second primary cancer. Using a recurrent events survival modeling approach that incorporates a family-wise likelihood to efficiently integrate the pedigree structure, we estimated the penetrance for both first and second primary cancer (PC) diagnosis from a pediatric sarcoma cohort at MD Anderson Cancer Center (MDACC, number of families=189; single primary cancer [SPC]=771; MPC=87). Validation of the risk prediction performance was performed using an independent MDACC clinical cohort of TP53 tested individuals (SPC=102; MPC=58). Findings showed that an individual diagnosed at a later age was more likely to be diagnosed with a second PC. Additionally, TP53 mutation carriers had a hazard ratio of 1.65 (95%CI: 1.1, 2.5) for developing a second PC versus SPC. The area under the ROC curve (AUC) for predicting individual outcomes of MPC versus SPC was 0.77. In summary, we provide the first set of penetrance estimates for SPC and MPC for TP53 germline mutation carriers, and demonstrate its accuracy for cancer risk assessment.
Keywords: LFS, multiple primary cancers, pedigree based survival analysis, mathematical model, risk prediction
Introduction
Li-Fraumeni syndrome (LFS) is a familial cancer syndrome associated with germline TP53 mutations and predisposing to a high lifetime probability of developing a wide spectrum of cancers(1). Clinical management of families affected with TP53 mutations remains a significant challenge. With the increasing success of treating cancer patients, many TP53 mutation carriers survive their first primary cancers only to develop additional primary cancers throughout their lives. Multiple primary cancers (MPC) are typically defined as cancers of different histologic type than another cancer in the same individual. It has been estimated that the risk for a second primary diagnosis can be as high as 50% for germline TP53 mutation carriers(2) and multiple malignancies have been previously observed in 43% of TP53 carriers(3). Many of these patients are aware that they are at increased risk of a second primary diagnosis, however the age of onset for a second primary is yet un-characterized in a statistically rigorous manner. Use of age-specific penetrance estimates of TP53 mutation carriers may have implications for surveillance and clinical management. A rigorous cancer surveillance program for LFS at the University of Texas MD Anderson Cancer Center (MDACC) currently follows advanced screening protocols(4). Counseling patients and family members could be further enhanced with a comprehensive cancer risk prediction tool that would enable the patients to understand or quantify their risk, lower anxiety for the unknown and improve adherence to screening.
Penetrance refers to the proportion of individuals carrying a deleterious variant of a disease predisposing gene (genotype) that also express clinical symptoms (phenotype) by a certain age. Penetrance can be incomplete and age-related, and precisely modeling it is of great importance to personalized risk assessment in medical genetics. Penetrance is further influenced by characteristics of the genotype, such as the specific impact that a variant has on a gene function and modifier factors(5–8). Additionally, age-of-onset penetrance estimation is also important for characterizing genetic effects of a disease. The goal of this paper is to present a penetrance, estimated with information from all individuals from genetic pedigrees, and to independently validate risk for both first and second primary cancer diagnosis.
Methods
Model development
We defined the penetrance of primary cancer as the cumulative probability of developing the next primary cancer by a certain age given the mutation status of disease susceptibility variant and prior cancer history (e.g., previous primary cancer occurrence and diagnosis age). In this study, we estimated the penetrance specific to the first and the second primary cancer. The model for penetrance estimation is built based on survival modeling of recurrent events and on Mendelian inheritance property of genotypes, which allows us to model data from pedigree studies. The underlying theory for the statistical model has been previously developed(9). In brief, we considered the multiple primary cancer occurrence in a randomly selected individual as a non-homogenous Poisson process (NHPP) and built the model with the following two major components: 1. Recurrent events modeling, which was devised to estimate the time varying hazard that fully characterizes the primary cancer occurrence process. We used a proportional hazard function where the baseline is a function of current age and the exponential component can incorporate covariates of interest. The model can thus consider effects from current age, cancer history or genetic factors when estimating the risk for next primary cancer development.
For the LFS data, we incorporate a covariate X(t) = {G,S,G × S,D(t),G × D(t)}T into the NHPP model, where D(t) is a time-dependent, but periodically fixed MPC variable that is coded as t > T1 and 0 otherwise. We propose the following multiplicative model for the conditional intensity function given X (t) as
| (1) |
where β denotes the coefficient parameter that controls effects of covariate X(t) on the intensity and λ0(t) is a baseline intensity function. And 2. Unknown genotypes among family members were imputed via the Elston-Stewart algorithm(10), which significantly increases the statistical power for parameter estimation using all available cancer outcomes in these families. This approach improves computational efficiency by exploiting the Mendelian inheritance property when inferring missing genotypes in pedigrees. We also corrected for ascertainment bias due to selection of families though cancer-affected individuals (Supplementary Figure 1) and finally made inference on model parameters via a Markov chain Monte Carlo method. All 95% confidence intervals are 95% confidence bands derived from posterior distributions.
We specified the model with three main effects (TP53 genotype (0 for non-carrier and 1 for carrier), sex (0 for female and 1 for male) and current cancer status (0 for no cancer diagnosis and 1 for one primary cancer diagnosis) and their interactions. We then computed the deviance information criterion (DIC) to identify the best set of covariates. We compare five different combinations of G, S and D(t). We observe that the simplest model with {G,S,D(t)} achieves the minimum DIC value. However, we decided to select the second best model in terms of the DIC, with {G,S,G × S,D(t),G × D(t)} as our final model since it has been reported that cancer status has different effects on cancer risk for mutation carriers and non-carriers(2,11). Our model assumes similar penetrance of all cancer types due to limited number of patients for estimating penetrance for each cancer type separately. We considered death from any other cause or last follow-up as censoring events when estimating penetrance from cancer-free survival.
Model training population and validation study population
We used a MDACC pediatric sarcoma cohort data to train the mode(12,13). The data were collected based on probands with sarcoma diagnosed before age 16 and with at least 3 years after-diagnosis survival. The data collection was extended to the probands’ blood relatives, which includes the probands’ grandparents, parents, parental siblings, siblings and offspring and pedigrees could be further extended through affected relatives using a sequential sampling scheme(14). For each individual, the sex and the diagnoses of any malignant cancer except the non-melanoma skin cancer were recorded from the date of birth until the date of death or last contact date. Death certificates and medical records were requested for all reported deaths and cancer(12–14). Only confirmed invasive cancers, confirmed by either records or validated through multiple family members, were included in the MDACC pediatric sarcoma cohort(12–14). A summary of the MDACC pediatric sarcoma cohort is presented in Table 1. The primary cancer diagnoses were determined based on the histology and site information recorded for each cancer event (Supplementary Figures 2, 3). Bilateral breast cancers were found in this population, however, they were considered to be a single primary cancer diagnosis. Benign tumor information was collected but was not used in model training. Mutation carrier status in this study was defined by PCR screening of exons 2–11 of the TP53 gene from peripheral-blood cell samples. More information about mutation testing can be found elsewhere(15). For the MDA pediatric sarcoma cohort, which began collecting data in the 1980’s, blood samples tested for a germline TP53 mutation were compared to the available literature for a suggestion of pathogenicity. If the mutation had not been reported then available family data was used to observe if direct transmission of the same TP53 mutation was present and if cancers segregated as established LFS phenotypes. This method was used by the study investigator to annotate the mutations due to lack of functional studies at the time. After the IARC database was established, the study investigator used the database as a resource to review prior pathogenicity determinations and update the patients if necessary. Supplementary Table 1 lists all deleterious TP53 mutations that were observed in the MDACC cohorts. The final data was comprised 189 families and a total of 3,706 individuals, with a total of 964 (26.0%) individuals genotyped for TP53 mutations status (Table 1). Among 570 (15.4%) primary cancer patients identified, 52 (1.4%) patients developed multiple primary cancer during follow-up. In this data set we have approximately equal number of cancer patients or healthy individuals for each sex (Table 1).
Table 1:
Number of primary cancer patients by the TP53 mutation status and sex in the training dataset: the MDACC pediatric sarcoma cohort data. Abbreviations: SPC, single primary cancer patients; MPC, multiple primary cancer patients.
| Wildtype | Mutation | Unknown | ||
|---|---|---|---|---|
| Female | 344 | 8 | 1214 | |
| Female | 118 | 21 | 96 | |
| Female | 3 | 19 | 9 | |
| Total number of individuals | 868 | 96 | 2742 |
For model prediction performance validation, we used an independent MDACC data set of prospectively followed families that were selected based off of clinical LFS criteria(3,16,17) that includes both TP53 germline mutation carrier families and wild-type families. Specifically, appropriately trained personnel in the Department of Genetics identify potentially eligible subjects primarily using 1) surgery schedules, 2) ClinicStation, 3) patient census, 4) patient clinics, 5) patient-study areas and referrals from inside and outside the institution, and 6) self-referrals. The list of potentially eligible subjects is then sent to the protocol’s principal investigator for approval. Patients were then contacted, with approval of treating physician, to determine interest in the study. Data collection was performed similarly to the pediatric sarcoma study. The number of primary cancers in this data is summarized in Table 2. We only used the individuals with available genotype information for validation (Table 2).
Table 2:
Number of primary cancer patients by the TP53 mutation status and Sex in the validation dataset with probands removed to address ascertainment bias: the MDACC prospective clinical cohort. Abbreviations: SPC, single primary cancer patients; MPC, multiple primary cancer patients.
| Wildtype | Mutation | ||
|---|---|---|---|
| Female | 136 | 30 | |
| Female | 12 | 43 | |
| Female | 2 | 34 | |
| Total number of individuals | 281 | 199 |
Prevalence and de novo mutation rate
We assumed the TP53 mutation follows Hardy-Weinberg equilibrium but this could be changed when homozygous genotype information is published. The mutation prevalence for pathogenic TP53 mutations was specified as 0.0006 for LFSPRO, which was derived in our previous study(18). The assumed frequencies for the three genotypes (homozygous reference, heterozygous and homozygous variant) were 0.9988, 0.0005996 and 3.6e-07, respectively. We used 0.00012 as a default value of de novo mutation rate when evaluating the familywise likelihood(19).
Validation study design
We evaluated the model prediction performance on primary cancer risk using the average annual risk computed with our TP53 penetrance estimates. The risk was calculated as the cumulative probability of developing the next primary cancer divided by the follow-up time. The receiver operating characteristic (ROC) curve was used to evaluate the sensitivity and specificity of predicting a primary cancer incidence using the estimated risk probability at various cutoffs. For Kaplan-Meier (KM) method-based risk prediction, we obtained KM survival functions for the time from date of birth to first primary cancer. These survival probabilities were then converted to penetrance estimate to compute the average annual risk. We used Jackknife to compute the standard errors of prediction performances(20,21). In brief, each subsample was generated by omitting the ith family and the average under the curve (AUC) was calculated for this subsample as previously described. The standard error (se) was calculated using the Jackknife technique,
where n is the number of families, and is the mean estimate of AUC values among all Jackknife subsamples.
Results
Age-of-onset penetrance curves for single- and multiple-primary cancers
Table 1 provides a summary of the pediatric sarcoma cohort used to train our model. There are a total of 96 known TP53 mutation carriers, among which 48 were diagnosed with one primary cancer and 31 were diagnosed with more than one primary cancer. There are 2,742 individuals who were not tested for TP53 mutations, among which 244 were diagnosed with one primary cancer and 15 were diagnosed with more than one primary cancer. Table 3 provides the coefficient estimates for all covariates including sex, genotype, cancer diagnosis and the interaction terms. As expected, the risk of the first and second primary cancers are strongly associated the TP53 mutation status with a hazard ratio (HR) of eβ =27 (95% CI: 18, 40). Importantly, as illustrated in Figure 1, the TP53 mutation carriers with a primary cancer diagnosis present a sharper increase in risk of having another primary cancer diagnosis over age than carriers who are still disease-free (HR=1.65, 95%CI: 1.10, 2.48). Such effect was not observed in the non-carriers (HR=0.82, 95%CI: 0.40, 1.48).
Table 3:
Summary of covariate coefficients and their 95% confidence intervals estimated by our model.
| Covariate | Coefficient Estimate | 95% CI | Hazard Ratios |
|---|---|---|---|
| Genotype | 3.288 | (2.871, 3.687) | 26.782 |
| Sex | 0.027 | (−0.187,0.241) | 1.027 |
| Genotype × Sex | −0.354 | (−0.817,0.106) | 0.702 |
| Cancer status | −0.197 | (−0.929,0.389) | 0.821 |
| Genotype × Cancer status | 0.700 | (−0.033,1.548) | 2.014 |
| Cancer status + Genotype ×Cancer status | 0.502 | (0.091, 0.908) | 1.652 |
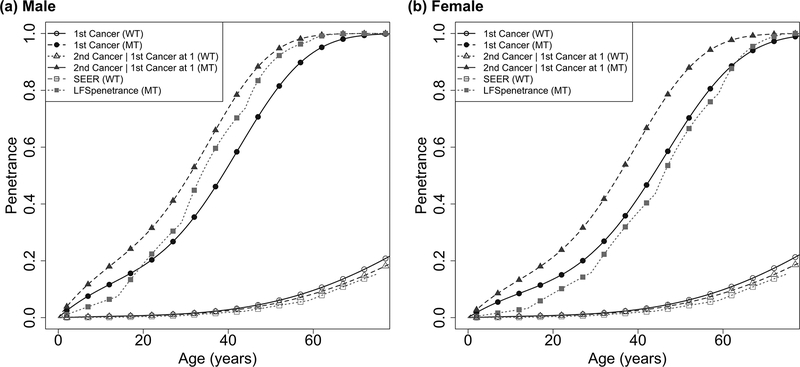
Figure 1:
Penetrance estimates over time to the first cancer and time to the second cancer given the first cancer diagnosed at age 1, for A) male and B) female. Previously reported penetrance estimates for time to the first cancer are also shown for comparison: the SEER estimate for non-carriers, and the LFS penetrance for the first primary cancer for carriers by Wu et al., 201028.
Using our model, we have obtained an accurate estimate of the onset of the first primary cancer by including cancer cases without genotype information from the family data. Among females, the HR for mutation carriers as compared to non-carriers is 26.8 (95%CI: 17.62, 39.88), while the HR among males for carriers versus non-carriers is 19.26 (95%CI: 13.14, 27.95). Consistent with previous results(15), the number of mutation carriers are similar in males and females in this study (Table 1). However, the estimated cancer risks are different in early ages between males and females (cumulative risk by age 30 at 0.239 for male, and 0.317 for female), possibly due to the early onset of breast cancer in females (Figure 1A, 1B).
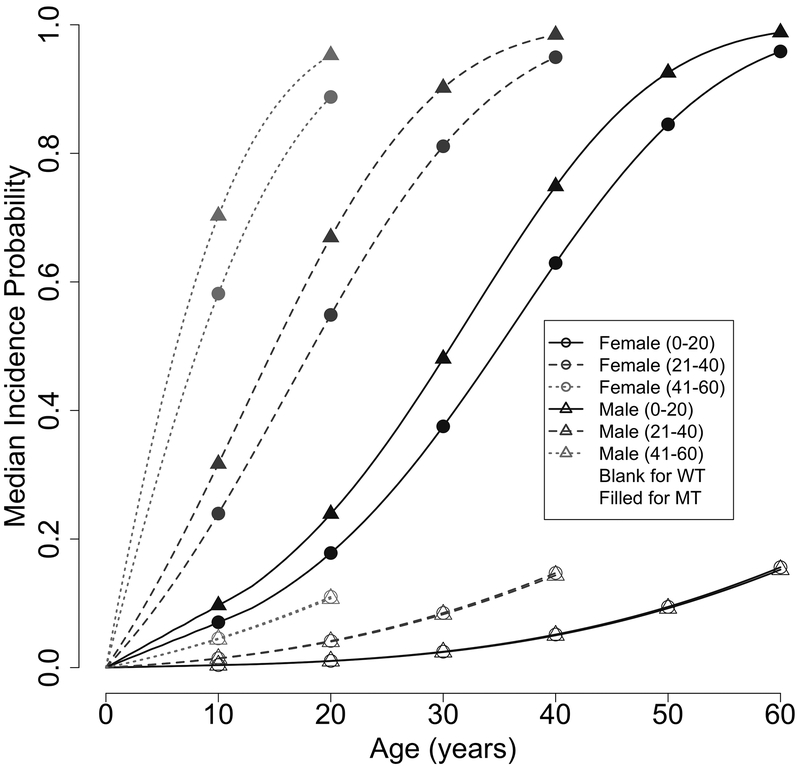
We have further obtained a novel set of penetrance estimates for the age-of-onset for second primary cancers among individuals who have had been diagnosed with a primary cancer. As presented in our hazard model (Table 3), this set of penetrance is dependent on the age of diagnosis (AoD) of the first primary cancer (PC). To illustrate the dynamics with AoD, Figure 2 shows the median age-of-onset risk for MPC within an interval of 20 for the age of diagnosis of the prior SPC: 0–20, 21–40 and 41–60. Here we observe a sharper increase in risk of developing a second primary cancer over age for individuals who have a later age-of-onset in their first primary cancer. Correspondingly, Table 4 shows our estimated median (at 50% probability) time-to-a second cancer diagnosis for the 20-year age intervals, for both females and males, e.g., for a female carrier, the median times are 29 years for the early age group (0–20), 14 years for the middle age group (21–40) and 6 years for the late age group (41–60). Interestingly, similar observations can be made with non-carriers (Figure 2). Therefore our novel SPC/MPC penetrance estimates allows us to observe an age-dependent effect in the diagnosis of MPC in our cohort.
Figure 2:
Illustration of the effect of second primary cancer on age-dependent penetrance estimates using median incidence probabilities in time windows of 20 years: 0–20, 21–40, 41–60. The x-axis denotes gap time, which is the number of years from the onset of the first primary cancer.
Table 4:
Median second primary cancer-free times (in years) since the first primary cancer diagnosis age and their 95% confidence intervals (in parenthesis) estimated for TP53 mutation carriers, stratified by sex and age of diagnosis for the first primary cancer.
| Median time to Second Cancer | ||
|---|---|---|
| Age of diagnosis of the first primary cancer | Female | Male |
| 0 – 20 | 32 (27–37) | 36 (31–41) |
| 21 – 40 | 16 (13–20) | 19 (16–24) |
| 41 – 60 | 7 (6–10) | 9 (7–12) |
Validation of risk prediction for first and second primary cancers
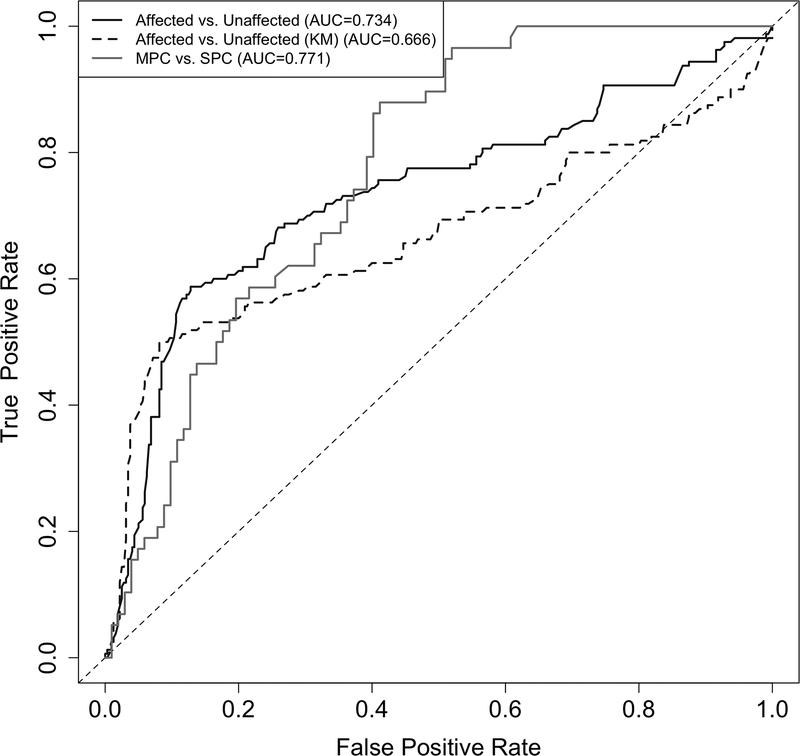
Table 2 shows an overview of the validation population used for assessing the cancer risk prediction performance of the estimated penetrance. This dataset is not used for model development or model parameter estimation and hence serves as an independent test for risk prediction performance. We used individuals with known mutation status and cancer outcomes: 74 SPC and 55 MPC among the carriers, and 28 SPC and 3 MPC among the non-carriers. We evaluated the performance of our penetrance estimates by two types of risk assessment: 1). We evaluated the risk probability estimated for first primary cancer diagnosis; and 2) we evaluated the risk of second primary cancer diagnosis among individuals who have already had a first primary cancer diagnosis. As shown in Figure 3, our penetrance estimates achieved AUCs of 0.73 (standard error: 0.031) and 0.77 (standard error: 0.040) when predicting the first or the second primary cancer diagnosis, respectively. Our prediction for the first primary cancer diagnosis outperformed the commonly used KM method with a corresponding AUC of 0.67 (standard error: 0.036).
Figure 3:
Comparison of validation performance between our multiple primary cancer-specific penetrance and those estimated from Kaplan-Meier (KM) method in predicting the first or the second primary cancer occurrence using the MDACC prospective data. Sample size: n(Affected)=160, n(Unaffected)=320, n(MPC)=58, n(SPC)=102.
Discussion
This study presents a new set of penetrance for the first or second primary cancer diagnosis in patients with LFS and validated its risk prediction performance through an independent LFS dataset. In contrast with previous studies(2,15), our NHPP model allowed us to utilize information from all family members, like sex, genotype if available, and age of diagnosis of the first primary cancer, by properly exploiting the family structure, using patient data with or without mutation test results. Including all individuals, regardless of testing, increased our training data sample size from 311 tested cancer patients to 570 total cancer patients, which substantially increased the statistical power for more accurate parameter estimation. Our final penetrance estimates for second diagnosis are age-of-diagnosis-dependent and varied with TP53 genotype and sex. Based on the new penetrance estimates, we observed the risk of second cancer diagnosis increased with older age of first cancer diagnosis. This observation appears similar to a generic age effect wherein older people get more cancers and more often however, our observations remain significant because we have been able to quantify the risk of TP53 mutation carriers which is higher for getting a second cancer than the first cancer (HR=1.65, 95%CI: 1.1, 2.5), after adjusted for a baseline age effect. Our penetrance estimates are able to characterize this observation as a continuum rather that a “early life” and “late life” comparison of diagnosed primaries and we were able to perform external validation for our curves. The penetrance of our model demonstrated a better risk prediction performance as compared to classical nonparametric methods, such as KM, for survival outcomes, as shown in the validation results. We have integrated the new penetrance estimates in our risk prediction software LFSPRO as LFSPR_2.0.0 to provide risk estimates, which is freely available at https://bioinformatics.mdanderson.org/public-software/lfspro/.
Stringent surveillance recommendations for TP53 mutation carriers have been established that includes annual whole body MRI (WB-MRI) and brain MRI, among other screening exams, for early detection of tumors(4). Studies have shown that this intensive cancer surveillance protocol has led to the early detection of primary cancers usually only requiring resection instead of chemotherapy and/or radiation, both of which have potential for contributing to treatment related late effects(2). Once identified early, treatment for carriers with a new primary can be quickly assessed increasing the likelihood of a positive outcome after early diagnosis for participants, which has been stated as a key benefit for continued screening(22,23). However, clinical studies of this rigorous screening protocol have reported to have psychosocial drawbacks(22,23). Nevertheless, early detection and peace of mind after results disclosure are noted as benefits gained through the screening process that outweigh the drawbacks(23). Psychosocial studies assessed after long term participation in surveillance programs are not yet available to determine if burnout is an issue. Implementation of age-specific penetrance estimation in LFS screening programs could give genetic counselors and clinicians an opportunity to provide a more complete picture of predicted risk to time of first or secondary primary to their patients. Since secondary primaries are estimated to occur in 50% of carriers(2), patients are encouraged to maintain the rigorous LFS screening protocol. The open-source R package, LFSPRO, which estimates the probability of an individual being a TP53 germline mutation carrier, has been expanded to also estimate risk to either first or second primary cancer (lfspro.mode function with mode= “mpc”) within 5, 10, 15 and 20 years. We are currently acquiring feedback on LFSPRO’s utility within the MDACC Li-Fraumeni Education and Early Detection (LEAD) program(23,24) which consists of a multidisciplinary team that works together to perform LFS screening protocols, analyze screening results and discuss future recommendations with the patients. Our goal is for LFSPRO to be used by genetic counselors and LFS clinicians as a tool to tailor their discussion of early cancer risks with their patients. However, at this time, our R package does not provide prediction beyond the second primary cancer or the recurrence of a primary cancer.
Our MPC penetrance estimation for the MDACC LFS cohorts is the first step in forming penetrance estimation within the established LFS community. Before becoming a clinical tool there are many factors that still need to be considered. First, we collapsed all cancer types into a single one and did not investigate the cause-specific penetrance for second primary cancer. Previous studies have shown the risk estimates varied among different cancer types, with breast cancer risk being dominant among female carriers(2,25). Second, we did not take into account the effects of allelic heterogeneity. The different point mutations in TP53 are hypothesized to contribute to different effects of the syndrome(26). However, due to rarity of the syndrome, limited sample size of individual mutations within the initial training data set (Supplementary Table 1) and lack of a standard on which to group the different mutations, it was not possible to include allelic specific information into our multiple primary modeling at this time. Third, we did not incorporate the effect of treatment in our modeling because of limited treatment information in the pediatric sarcoma cohort data, which is a prevalent issue in most datasets collected to date. Previous studies(27,28) have shown that cancer treatment may be a risk modifier for time to next cancer. Radiation and/or chemotherapy were used for the majority of cancer cases in this study. The focus on 2nd primary cancers in the LFS populations is limited due to the lack of treatment data regarding radiation use/dose, generally contraindicated in this population due to its association with treatment-induced second neoplasms. Also, treatment of patients differs over the span of the cohort study due to technological innovation making it more difficult to capture in one study. A complete collection of treatment information continues to be a challenging task for family studies, but remains essential for future studies on multiple primary cancers in LFS. When such information is available, it can be incorporated into a penetrance model such as proposed here. Though treatment effects are currently not estimated, they are implicitly accounted for in terms of risk prediction, with the other parameter estimates absorbing the effect of treatments. This explains the good predictive performance of our current model as the independent validation set was also collected at MDACC. One potential drawback is a direct application of our penetrance to other study populations may not fit.
In summary, our study provides age-specific penetrance estimates for first or second primary cancer in individuals with LFS and has successfully validated its discrimination power between primary cancer patients and cancer-free individuals through another LFS data set. These estimates have the potential to provide a more accurate primary cancer risk assessment for patients with LFS, especially for cancer survivors who desire a better risk management of any future cancer development.
Supplementary Material
Significance:
Findings present an open-source R package LFSPRO that could be used for genetic counseling and health management of individuals with Li-Fraumeni syndrome as it estimates the risk of both first and second primary cancer diagnosis.
Acknowledgments:
S.J. Shin, E.B. Dodd and W. Wang were supported in part by the Cancer Prevention Research Institute of Texas through grant number RP130090. S.J. Shin was supported in part by National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) through grant number 2019R1A4A1028134. E.B. Dodd and W. Wang were supported in part by the U.S. NCI through grant numbers 1R01 CA183793, 1R01 CA174206, 2R01 CA158113. W. Wang is supported in part by U.S. NCI P30CA016672. F. Gao and J. Chen are supported by U.S. NCI 1R01 CA183793. J. Bojadzeiva, X. Kong and L.C. Strong were supported in part by the U.S. NIH through grant P01CA34936. J. Ning was funded in part by the National Cancer Institute R01 (R01CA193878) and the Andrew Sabin Family Fellowship. C.I. Amos was supported by the U.S. NCI 1U01CA196386, CA196386S1, and 1R01CA186566. C.I. Amos would also like to thank the CPRIT RR170048 grant and the U.S. NCI U19CA203654 and U19CA203654S1 grants for their additional funding. We thank Dr. Jialu Li for his contribution to this project and we thank Dr. Banu Arun and Jessica Ross for their helpful comments.
Footnotes
Disclaimer: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Malkin D Li-Fraumeni Syndrome. Vol. 2, Genes & Cancer; 2011. p. 475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer [Internet]. 2016. December 1;122(23):3673–81. Available from: http://doi.wiley.com/10.1002/cncr.30248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bougeard G, Renaux-Petel M, Flaman J-M, Charbonnier C, Fermey P, Belotti M, et al. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J Clin Oncol [Internet]. 2015. July 20;33(21):2345–52. Available from: http://ascopubs.org/doi/10.1200/JCO.2014.59.5728 [DOI] [PubMed] [Google Scholar]
- 4.Villani A, Shore A, Wasserman JD, Stephens D, Kim RH, Druker H, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016;17(9):1295–305. [DOI] [PubMed] [Google Scholar]
- 5.Beutler E, Felitti VJ. The clinical penetrance of hereditary hemochromatosis [1]. Hepatology. 2003;37(3):711. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Wang W, Broman KW, Katki HA, Parmigiani G. Bayesmendel: an R environment for Mendelian risk prediction. Stat Appl Genet Mol Biol. 2004;3(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marroni F, Aretini P, D’Andrea E, Caligo MA, Cortesi L, Viel A, et al. Penetrances of breast and ovarian cancer in a large series of families tested for BRCA1/2 mutations. Eur J Hum Genet [Internet]. 2004. September 1;12:899 Available from: 10.1038/sj.ejhg.5201256 [DOI] [PubMed] [Google Scholar]
- 8.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The Risk of Cancer Associated with Specific Mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med [Internet]. 1997;336(20):1401–8. Available from: 10.1056/NEJM199705153362001 [DOI] [PubMed] [Google Scholar]
- 9.Shin SJ, Li J, Ning J, Bojadzieva J, Strong LC, Wang W. Bayesian estimation of a semiparametric recurrent event model with applications to the penetrance estimation of multiple primary cancers in Li-Fraumeni syndrome. Biostatistics [Internet]. 2018;00:1–16. Available from: https://academic.oup.com/biostatistics/advance-article-abstract/doi/10.1093/biostatistics/kxy066/5183544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elston RC, Stewart J. A General Model for the Genetic Analysis of Pedigree Data. Hum Hered. 1971;21(6):523–42. [DOI] [PubMed] [Google Scholar]
- 11.van Eggermond AM, Schaapveld M, Lugtenburg PJ, Krol ADG, de Boer JP, Zijlstra JM, et al. Risk of multiple primary malignancies following treatment of Hodgkin lymphoma. Blood [Internet]. 2014. July 17;124(3):319 LP–327. Available from: http://www.bloodjournal.org/content/124/3/319.abstract [DOI] [PubMed] [Google Scholar]
- 12.Strong LC, Stine M, Norsted TL. Cancer in survivors of childhood soft tissue sarcoma and their relatives. J Natl Cancer Inst. 1987;79(6):1213–20. [PubMed] [Google Scholar]
- 13.Lustbader ED, Williams WR, Bondy ML, Strom S, Strong LC. Segregation analysis of cancer in families of childhood soft-tissue-sarcoma patients. Am J Hum Genet. 1992/08/01. 1992;51(2):344–56. [PMC free article] [PubMed] [Google Scholar]
- 14.Bondy ML, Lustbader ED, Strom SS, Strong LC. Segregation analysis of 159 soft tissue sarcoma kindreds: comparison of fixed and sequential sampling schemes. Genet Epidemiol. 1992/01/01. 1992;9(5):291–304. [DOI] [PubMed] [Google Scholar]
- 15.Hwang S-J, Lozano G, Amos CI, Strong LC. Germline p53 Mutations in a Cohort with Childhood Sarcoma: Sex Differences in Cancer Risk. Am J Hum Genet [Internet]. 2003. April;72(4):975–83. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0002929707606181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li FP, Fraumeni JFJ, Mulvihill JJ, Blattner WA, Dreyfus MG, Tucker MA, et al. A cancer family syndrome in 24 kindreds. Cancer Res [Internet]. 1988;48:5358–62. Available from: http://cancerres.aacrjournals.org/content/canres/48/18/5358.full.pdf [PubMed] [Google Scholar]
- 17.Chompret A, Abel A, Stoppa-Lyonnet D, Brugieres L, Pages S, Feunteuns J, et al. Sensitivity and predictive value of criteria for p53 germline mutation screening. J Med Genet. 2001;38:43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng G, Bojadzieva J, Ballinger ML, Li J, Blackford AL, Mai PL, et al. Estimating TP53 Mutation Carrier Probability in Families with Li–Fraumeni Syndrome Using LFSPRO. Cancer Epidemiol Biomarkers Prev [Internet]. 2017. June;26(6):837–44. Available from: http://cebp.aacrjournals.org/lookup/doi/10.1158/1055-9965.EPI-16-0695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez KD, Buzin CH, Noltner KA, Gu D, Li W, Malkin D, et al. High frequency of de novo mutations in Li-Fraumeni syndrome. J Med Genet [Internet]. 2009. October 1;46(10):689–93. Available from: http://jmg.bmj.com/cgi/doi/10.1136/jmg.2008.058958 [DOI] [PubMed] [Google Scholar]
- 20.Efron B, Stein C. The Jackknife Estimate of Variance. Ann Stat. 1981;9(3):586–96. [Google Scholar]
- 21.Efron B The Jackknife, the Bootstrap, and Other Resampling Plans. Philadelphia, Pa.: Society for Industrial and Applied Mathematics, 1982.; 1982. [Google Scholar]
- 22.McBride KA, Ballinger ML, Schlub TE, Young M-A, Tattersall MHN, Kirk J, et al. Psychosocial morbidity in TP53 mutation carriers: is whole-body cancer screening beneficial? Fam Cancer [Internet]. 2017. July 25;16(3):423–32. Available from: http://link.springer.com/10.1007/s10689-016-9964-7 [DOI] [PubMed] [Google Scholar]
- 23.Ross J, Bojadzieva J, Peterson S, Noblin SJ, Yzquierdo R, Askins M, et al. The psychosocial effects of the Li-Fraumeni Education and Early Detection (LEAD) program on individuals with Li-Fraumeni syndrome. Genet Med [Internet]. 2017. September 16;19(9):1064–70. Available from: http://www.nature.com/doifinder/10.1038/gim.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bojadzieva J, Amini B, Day SF, Jackson TL, Thomas PS, Willis BJ, et al. Whole body magnetic resonance imaging (WB-MRI) and brain MRI baseline surveillance in TP53 germline mutation carriers: experience from the Li-Fraumeni Syndrome Education and Early Detection (LEAD) clinic. Fam Cancer [Internet]. 2018;17(2):287–94. Available from: 10.1007/s10689-017-0034-6 [DOI] [PubMed] [Google Scholar]
- 25.Shin SJ, Yuan Y, Strong LC, Bojadzieva J, Wang W. Bayesian Semiparametric Estimation of Cancer-Specific Age-at-Onset Penetrance With Application to Li-Fraumeni Syndrome. J Am Stat Assoc [Internet]. 2018;0(0):1–12. Available from: 10.1080/01621459.2018.1482749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heymann S, Delaloge S, Rahal A, Caron O, Frebourg T, Barreau L, et al. Radio-induced malignancies after breast cancer postoperative radiotherapy in patients with Li-Fraumeni syndrome. Radiat Oncol [Internet]. 2010. November;5(1):104 Available from: 10.1186/1748-717X-5-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inskip PD, Curtis RE. New malignancies following childhood cancer in the United States, 1973–2002. Int J Cancer [Internet]. 2007;121(10):2233–40. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/ijc.22827 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.