Abstract
Pluripotent stem cells recapitulate many aspects of embryogenesis in vitro. Here, we established a novel culture system to differentiate human embryonic stem cell aggregates (HESCA), and evaluated its utility for teratogenicity assessment. Culture of HESCA with modulators of developmental signals induced morphogenetic and molecular changes associated with differentiation of the paraxial mesoderm and neuroectoderm. To examine impact of teratogenic exposures on HESCA differentiation, 18 compounds were tested, for which adequate information on in vivo plasma concentrations is available. HESCA treated with each compound were examined for gross morphology and transcript levels of 15 embryogenesis regulator genes. Significant alterations in the transcript levels were observed for 94% (15/16) of the teratogenic exposures within 5-fold margin, whereas no alteration was observed for 92% (11/12) of the non-teratogenic exposures. Our study demonstrates that transcriptional changes in HESCA serve as predictive indicator of teratogenicity in a manner comparable to in vivo exposure levels.
Keywords: Human embryonic stem cell, 3-dimensional aggregate, in vitro assay, teratogenic exposure, Daston list, paraxial mesoderm, neuroectoderm, embryogenesis regulators
1. Introduction
Gestational exposure to certain chemicals can adversely affect embryogenesis and cause birth defects. To identify such teratogenic chemical exposures, several investigative approaches have been explored, including human epidemiologic studies, experimentations with pregnant animals, and in vitro assays using embryonic stem (ES) cells. ES cells are pluripotent, and can be induced to differentiate into a variety of cell types to model embryogenesis in vitro. In ES cell-based tests, teratogenic potential is estimated according to inhibitory effects of chemical exposures on cell proliferation and differentiation [1,2]. To date, various formats of ES cell-based assays have been reported, differing in types of differentiating cells (e.g., cardiomyocytes, neurons and osteoblasts) and in endpoints of analyses (e.g., morphological assessment and gene expression profiling) [3–6]. In vitro assays, such as ES cell-based tests, offer numerous benefits to teratology research, because they are generally fast, economical, expandable for high-throughput screening, amenable for mechanistic interrogations, and devoid of ethical issues associated with animal experimentations. Nonetheless, ES cell-based tests have not yet become a standard practice for the purpose of regulatory assessment of teratogenic chemical exposures.
To employ ES cell-based tests effectively for teratogenicity assessment, it is critical to identify strengths and weaknesses of each assay format. In each format, ES cells are manipulated to recapitulate only limited aspects of embryogenesis with respect to developmental stages and structures. It is highly unlikely that a single assay format can detect all kinds of teratogenic exposures in a sensitive and specific manner. Therefore, multiple formats of assays would be required to compensate for deficiencies in each other. Various test formats have been validated for their capability and limitation using known teratogenic and non-teratogenic chemical exposures as references. In many of those studies, reference chemicals are classified into either teratogens (developmental toxicants) or non-teratogens (non-toxicants). However, such dichotomous classification of chemicals is not realistic, because their teratogenicity depends on dose and timing of exposure [7–9]. For example, a given chemical can be teratogenic at one exposure level, but non-teratogenic at another lower exposure level. In this regard, Daston and colleagues set forth the concept of exposure-based assessment, and compiled a list of exposures, i.e., types of chemicals with specific concentrations, that exhibit teratogenic effects, or lack thereof, based on in vivo studies in rats [10]. This list of chemical exposures (coined as the Daston list) may be used to validate effectiveness of in vitro teratogenicity screening assays, including ES cell-based tests, in a manner relevant for in vivo exposure levels.
Additionally, some of pharmaceutical compounds used for therapeutic purposes in human may also serve as suitable references to validate in vitro assays. For many pharmaceutical compounds, pharmacokinetics data have been collected through preclinical and clinical studies, which provide crucial information on exposures, such as therapeutic ranges of compound concentrations in blood plasma [11]. Furthermore, epidemiologic studies have made statistically sound linkages between increased incidence of birth defects and consumption of certain medications during pregnancy, including thalidomide [12] and valproic acid [13]. Such investigations, along with data from model animal studies, have helped to classify human medications into those that are contraindicated for use during pregnancy due to increased risks of malformations or into those that pose no risk, even though pregnancy risks of many medications are still unclear due to lack of sufficient human data [14]. These contraindicated and no risk medications may be used for validation of in vitro screening assays in reference to their therapeutic plasma concentrations, which may further reveal strengths and weaknesses of each assay.
The goal of the present study is to develop a new simple culture protocol to differentiate human ES cells into embryonic tissues that are relevant for birth defects research, and also to evaluate the effectiveness of the protocol as an assay platform for assessment of teratogenic chemical exposures. We first explored three different culture conditions for ES cell differentiation by using small molecule modulators of the WNT, NODAL and retinoic acid signaling pathways, all of which are the key regulators of body patterning during early embryogenesis. Based on morphological and molecular impact of these conditions, we formulated a specific culture protocol to induce 3-dimensional cell aggregates that exhibit characteristics of the axial skeleton and central nervous system precursors. Then, we used these human ES cell aggregates (HESCA) to examine effects of 18 chemical compounds, including those from the Daston list and human medications with known pregnancy risks, by quantitatively measuring expression levels of embryogenesis regulator genes.
2. Materials and methods
2.1. Maintenance and aggregation culture of human embryonic stem cells
H9 line (WA09, National Institutes of Health registration number 0062) of human ES cells was obtained commercially (WiCell Research Institute, Madison, WI). Cells were maintained in the feeder-free maintenance culture medium mTeSR1 (Stemcell Technologies, Vancouver, BC) with 40 units/mL penicillin and 40 μg/mL streptomycin, and passaged every other day. Cells were detached and dissociated in TrypLE Express (Thermo Fisher Scientific, Waltham, MA), washed in Basal Medium (Component #85851 of mTeSR1), and transferred to a flask that had been pre-coated with iMatrix-511 (Takara Bio, Mountain View, CA) and filled with mTeSR1 (Figure 1A). iMatrix-511 is a recombinant laminin-511 E8 fragment, which supports adhesion, survival and expansion of dissociated human ES cells [15]. mTeSR1 was refreshed one day after passaging. For differentiation, dissociated cells were suspended at a density of 20 cells/μL in the Aggregation Culture Medium (ACM; 80% Minimum Essential Medium Alpha with nucleosides and GlutaMAX [Thermo Fisher Scientific] and 20% 5X Supplement [Component #85852 of mTeSR1]). ACM was supplemented with 10 μM CHIR99021 (Calbiochem, La Jolla, CA), 2 μM SB431542 (Stemcell Technologies), 2 nM retinoic acid (Sigma-Aldrich, St. Louis, MO) and test compounds (Table 1) at varying concentrations, depending on the experiments. 50 μL of cell suspension was placed in each well of 96-well plates (ultra-low attachment, round bottom; Corning, Corning, NY). Plates were centrifuged at 1,000 rpm (180 × g) for 5 minutes at ambient temperature to facilitate cell aggregation (Figure 1A), and were incubated at 37°C in humidified air with 4.5% CO2 for up to 5 days. More details of the maintenance and aggregation culture protocols are provided in Supplementary Data. Note that antibiotics (penicillin and streptomycin) were included in the maintenance culture medium to reduce risks of bacterial contamination during routine passaging, which is in line with common practice for general cell culture. However, antibiotics were not included in ACM during differentiation culture to avoid potential influences on the effects of test compounds. Such precaution may be of significance in light of a recent study showing that penicillin and streptomycin alter expression levels of various genes in a human liver cell line [16]. Nonetheless, in our assay using human embryonic stem cell aggregates (HESCA; Supplementary Figure), antibiotics at the same concentrations as used in the maintenance medium (i.e., 40 units/mL penicillin and 40 μg/mL streptomycin) did not significantly alter the transcript levels of the embryogenesis regulator genes (Table 2).
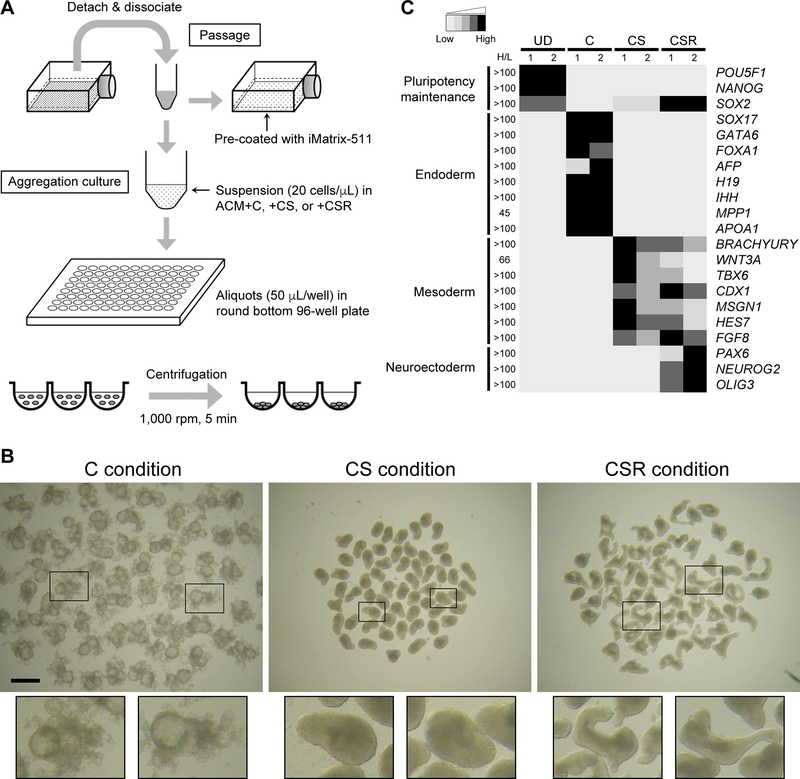
Figure 1.
Differentiation of human embryonic stem cell aggregates (HESCA). A, Diagram depicting the procedures for maintenance and aggregation culture. B, Images of HESCA that have been cultured for 5 days under three different conditions: C (10 μM CHIR99021 [WNT signaling activator]), CS (10 μM CHIR99021 and 2 μM SB431542 [NODAL signaling inhibitor]) and CSR (10 μM CHIR99021, 2 μM SB431542 and 2 nM retinoic acid). Scale bar = 1 mm. Enlarged images of representative aggregates are shown in framed boxes for each condition at the bottom. C, Grayscale representation of the transcript levels of genes associated with pluripotency maintenance and differentiation of endoderm, mesoderm and neuroectoderm. A total of 8 samples, i.e., two independent sets (1 and 2) of undifferentiated cells (UD) and Day 5 HESCA cultured under C, CS and CSR conditions, were subjected to RNA-seq analyses. Normalized transcript levels for each gene were compared among the 8 samples. Relative transcript amounts of individual samples were assigned with 5 levels of grayscales from the lowest to the highest, and the ratio of the highest to the lowest levels is indicated as H/L for each gene. Detailed transcript amounts are shown in Supplementary Data.
Table 1.
Compounds tested in the present study
| Compound name | CAS number | Vendor* (catalog number) | Stock concentration (solvent) | Comments** |
|---|---|---|---|---|
| Abacavir | 188062–50–2 | Sigma-Aldrich (SML0089) | 50 mM (water) | Daston (P: 80 μM, N: 18 μM) |
| All-trans retinoic acid | 302–79–4 | Sigma-Aldrich (R2625) | 100 μM (DMSO) | Daston (P: 200 nM, N: 1.7 nM) |
| Caffeine | 58–08–2 | Sigma-Aldrich (C0750) | 100 mM (water) | Daston (P: 325 μM, N: 7.7 μM) |
| Captopril | 62571–86–2 | Santa Cruz (sc-200566) | 50 mM (DMSO) | Contraindicated (0.23–2.3 μM) |
| Dabigatran | 211915–06–9 | Santa Cruz (sc-351724) | 20 mM (DMSO) | Daston (P: 7 μM, N: 1 μM) |
| Doxylamine | 562–10–7 | Sigma-Aldrich (D3775) | 50 mM (DMSO) | Safe (0.18–0.74 μM) |
| Ethylene glycol | 107–21–1 | Sigma-Aldrich (324558) | 18 M (100%) | Daston (P: 57 mM, N: 1.4 mM) |
| Fluorouracil | 51–21–8 | Sigma-Aldrich (F6627) | 5 mM (DMSO) | Contraindicated (0.38–2.3 μM) |
| Glycolic acid | 79–14–1 | Sigma-Aldrich (G8284) | 1 M (water) | Daston (P: 5 mM, N: 275 μM) |
| Hydroxyurea | 127–07–1 | Sigma-Aldrich (H8627) | 10 mM (water) | Control for proliferation assay |
| Isotretinoin | 4759–48–2 | Cayman (21648) | 40 μM (DMSO) | Contraindicated (3.3–6.7 nM) |
| MEHP | 4376–20–9 | Sigma-Aldrich (796832) | 500 mM (DMSO) | Daston (P: 146 μM, N: 1 μM) |
| Methanol | 67–56–1 | Fisher Scientific (A412P-4) | 24.7 M (100%) | Daston (P: 270 mM, N: 22 μM) |
| Methylmercury | 22967–92–6 | Sigma-Aldrich (442534) | 10 mM (DMSO) | Control for proliferation assay |
| Metoclopramide | 7232–21–5 | Sigma-Aldrich (M0763) | 50 mM (DMSO) | Safe (0.17–0.5 μM) |
| Nilotinib | 641571–10–0 | Cayman (10010422) | 20 mM (DMSO) | Daston (P: 28 μM, N: 2 μM) |
| Phenytoin | 57–41–0 | Cayman (24037) | 50 mM (DMSO) | Contraindicated (19.8–59.7 μM) |
| Ramelteon | 196597–26–9 | Cayman (20389) | 200 mM (DMSO) | Daston (P: 81 μM, N: 19 nM) |
| Thalidomide | 50–35–1 | Santa Cruz (sc-201445) | 10 mM (DMSO) | Contraindicated (1.9–5.8 μM) |
| Valproic acid | 1069–66–5 | Sigma-Aldrich (P4543) | 100 mM (water) | Contraindicated (277–693 μM) |
CAS: Chemical Abstracts Service, DMSO: dimethyl sulfoxide, MEHP: mono-2-ethylhexyl phthalate
Sigma-Aldrich (St. Louis, MO), Santa Cruz Biotechnology (Dallas, TX), Cayman Chemical (Ann Arbor, MI), Fisher Scientific (Waltham, MA)
Daston: compounds in the Daston list with P (positive) and N (negative) plasma concentrations (Daston et al., 2014). Contraindicated: human medications contraindicated for use during pregnancy. Safe: human medications safe for use during pregnancy. Therapeutic plasma concentrations of these human medications are indicated in parentheses (Schulz et al., 2012).
Table 2.
Genes examined in the present study
| Gene symbol | Descriptions* | Primer sequences (5’ ➔ 3’) |
|---|---|---|
| ACTB | Cytoplasmic actin; House keeping; Expressed broadly and constitutively (used as a loading control for normalization) | F: TACAGGAAGTCCCTTGCCATCCTA R: ACATCTCAAGTTGGGGGACAAAAA |
| ALDH1A2 | Retinal dehydrogenase; Involved in synthesis of retinoic acid; Expressed in somites | F: GTCTGTCCCTCTCTGCTTTCTCT R: TCCTCCTCCCTTTATCCCACTTTC |
| APOA1 | Apolipoprotein A; Involved in lipid metabolism; Expressed in endodermal tissues, such as liver, intestine and yolk sac | F: TGTGTACGTGGATGTGCTCAAAGA R: CTCCTGCCACTTCTTCTGGAAGTC |
| BRACHYURY | T-box transcription factor; Associated with initiation of gastrulation; Expressed in early primitive streak | F: ATCCTCATCCTCAGTTTGGAGGTG R: ACACAGGTGTCCATGAGGCTATGA |
| CDX1 | Homeodomain transcription factor; Regulates anterior-posterior axial patterning; Expressed in primitive streak | F: TCAGAGCTGGCTGCCAATC R: TGGAACCAGATCTTCACCTGC |
| EYA1 | EYA (eyes absent) transcriptional coactivator; Involved in mesoderm patterning; Expressed in mesoderm | F: CAGTCTTGACCTCTGCCTTTGTG R: AGGTTTGCTGTATTGGAGAAGCTG |
| FGF8 | Ligand for FGF signaling; Regulates mesoderm patterning and behavior; Expressed in primitive streak | F: AGCAGAGTTCGAGTCCGAGGAG R: CAGCGCTGTGTAGTTGTTCTCCA |
| FOXA1 | Forkhead transcription factor; Regulates endoderm differentiation; Expressed in endoderm lineages | F: CAAACCAAACCGTCAACAGCATAA R: GGGCAAGGAAGGAGGAGAATTT |
| FOXC2 | Forkhead transcription factor; Involved in mesoderm patterning; Expressed in paraxial mesoderm | F: GTCTGTGAAGAGCGCAGGTAACTT R: GGGCTCGCTATGGGATTTGGTC |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase; House keeping; Expressed broadly and constitutively | F: TAGAAAAACCTGCCAAATATGATG R: GTCTCTCTCTTCCTCTTGTGCTCT |
| GATA6 | GATA transcription factor; Involved in endoderm specification; Expressed in endoderm lineages | F: AACTACCACCTTATGGCGCAGAAA R: CCCATCTTGACCCGAATACTTGAG |
| HES7 | HLH transcription factor; Involved in somite segmentation; Expressed in presomitic mesoderm | F: ACCCGGGATCGAGCTGAGAATAG R: ACTCGCGGAAACCGGACAAGTAG |
| HOXB1 | HOX transcription factor; Involved in anterior-posterior axial patterning; Expressed in a gradient along body axis | F: CGGGCCTTCTCAGTACTACCCTCT R: TCTGGAACCAAATCTTGACCTGTG |
| HOXB3 | HOX transcription factor; Involved in anterior-posterior axial patterning; Expressed in a gradient along body axis | F: GGGATGGTGAGAGATCCTGAAAGA R: CTCGTGTCACCAACTGCTTTCTGT |
| HOXB5 | HOX transcription factor; Involved in anterior-posterior axial patterning; Expressed in a gradient along body axis | F: GCCAATTTCACCGAAATAGACGAG R: CCAAACGAAATATCGTAACACAAGG |
| HOXB7 | HOX transcription factor; Involved in anterior-posterior axial patterning; Expressed in a gradient along body axis | F: GGAGGGAGAAAGGAGAAATTTTGG R: GGTTAGTCCAGACCCACAGTCCAC |
| HOXB9 | HOX transcription factor; Involved in anterior-posterior axial patterning; Expressed in a gradient along body axis | F: GAGCCTTGACAATGTTGTCCTCCT R: ACGGAAGGTGGAGGGATTTTTCTA |
| IHH | Ligand for Hedgehog signaling; Involved in body patterning; Expressed in the primitive endoderm | F: CCCGTCGTGGTGTAGTCATAGAG R: CCCAGGGAATTTAGCAGCATCAA |
| MEOX1 | Homeodomain transcription factor; Regulates somite segmentation; Expressed in somites | F: GGGCTGAACAACTAGGAGCTGAAA R: GAGGGATTTTCTTGTCCCCTTTTG |
| MESP2 | HLH transcription factor; Involved in somite segmentation; Expressed in unsegmented paraxial mesoderm | F: GTAGCTTGGGTGCCTCCTTATTTG R: CCCTTCTTAGAGCAGCTCAGACT |
| MPP1 | Palmitoylated membrane protein; Expressed in endoderm tissues, such as liver and intestine | F: GAATTCCACCCGCCTTGCTTTA R: TTGCTGTGCATCACCCTTGATTAG |
| MSGN1 | HLH transcription factor; Regulates somite patterning; Expressed in unsegmented paraxial mesoderm | F: GCCTGGTAGAGGTGGACTACAATA R: TGAGTGTCTGGATCTTGGTGAGAG |
| NANOG | Homeodomain transcription factor; Regulates pluripotency maintenance; Expressed in epiblast | F: TGAAGCATCCGACTGTAAAGAATC R: CATTGCTATTCTTCGGCCAGTTGT |
| NEUROG2 | HLH transcription factor; Regulates neural differentiation; Expressed in neural tube | F: TCCTCGGTTGTTTCTTGCATTTCT R: AGATCACAGGAACCAGTTGCATTC |
| NODAL | Ligand for Nodal signaling; Inducer of endoderm specification; Expressed in primitive endoderm and primitive streak | F: GACATCATCCGCAGCCTACAGG R: GACCTGGGACAAAGTGACAGTG |
| OLIG3 | HLH transcription factor; Involved in neural patterning; Expressed in neural tube | F: AGAGATCTCAGACTCCACTTGACC R: CACAGAGTCAGAGAAAGCCAACTC |
| PAX3 | Paired-box transcription factor; Involved in somite and neural patterning; Expressed in somites and neural tube | F: GTACACCAAAGCACGATTCCTT R: GCTATAGGTGGGTGGACAGTAG |
| PAX6 | Paired-box transcription factor; Involved in neural specification and patterning; Expressed in neuroectoderm | F: TGTCTTCCCTAGAAATCCTCAGAATGA R: ACAAAATGATTGGACCGTGAACAG |
| POU5F1 | POU-domain transcription factor; Regulates pluripotency maintenance; Expressed in epiblast | F: TGGTGCCGTGAAGCTGGAGAAGGA R: CTGCTTGATCGCTTGCCCTTCTGG |
| SOX17 | SRY-box transcription factor; Involved in endoderm specification; Expressed in endoderm lineages | F: CAAAGAAATGTTGTCCTGGGTGTG R: GTTCACCCTTTTCGAGGATGAGAA |
| SOX2 | SRY-box transcription factor; Regulates pluripotency maintenance; Expressed in epiblast and neural tube | F: CCCCCGGCGGCAATAGCA R: TCGGCGCCGGGGAGATACAT |
| TBX6 | T-box transcription factor; Involved in paraxial mesoderm specification; Expressed in caudal end | F: ATCTCCGTGACAGCCTACCAGAAC R: AGTACATGGGTTTGGAGCCCACAT |
| TCF15 | HLH transcription factor; Regulates patterning of paraxial mesoderm; Expressed in somites | F: CCATCTGCACCTTCTGCCTCAG R: CCCGGTCCCTACACAAAGAAGG |
| UNCX | Homeodomain transcription factor; Involved in segmentation of paraxial mesoderm; Expressed in somites | F: CTCAGGCTCCGACTCACGCAAC R: CTCTCTTGGGAGGAGGAGGGTCTC |
| WNT3A | Ligand for Wnt signaling; Regulates paraxial mesoderm specification; Expressed in late primitive streak | F: AGTGACACGCTCATGTGCAGAA R: GAAGGAGCCCGTCTCAGGGTTG |
| WNT5A | Ligand for Wnt signaling; Regulates axial elongation; Expressed at caudal end | F: CTACCGCTTTGCCAAGGAGTTC R: CGTACTTCTCCTTCAGGGCATCA |
Molecular features, functional roles and expression patterns during early embryogenesis are largely based on studies in the mouse(Mouse Genome Informatics: www.informatics.jax.org).
2.2. RNA-seq
RNA extraction, library preparation and sequencing were performed with the assistance of the Genomics and Bioinformatics Shared Resource at the University of Hawaii Cancer Center. A total of 8 samples, i.e., 2 independent sets of 4 samples each (undifferentiated cells and Day 5 aggregates cultured under the C [ACM supplemented with CHIR99021], CS [ACM supplemented with CHIR99021 and SB431542] and CSR [ACM supplemented with CHIR99021, SB431542 and retinoic acid] conditions; Figure 1), were subjected to RNA-seq analyses. Total RNA was extracted from each sample using the Direct-zol RNA Microprep Kit (Zymo Research, Irvine, CA), according to the manufacturer’s instructions, including the optional DNase I treatment. 400 ng of RNA per sample was processed for library preparation using the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, Ipswich, MA). Libraries were sequenced using the NextSeq 500 System with a High Output Flow Cell (400 M reads, 150 cycles) (Illumina, San Diego, CA). Raw sequencing reads were first trimmed for adapters, low quality bases on both the 5’ and 3’ ends (Phred quality score Q < 20), and short reads filtered (< 20 bases) using Cutadapt [17]. Trimmed reads were aligned to human genome reference build hg38 using STAR [18]. Gene expression was quantified using Partek Flow software (Partek, St. Louis, MO), and normalized to account for differences in sequencing depth among samples using DESeq2 [19]. An Excel spreadsheet file containing the normalized gene expression data for all 8 samples is provided in Supplementary Data.
2.3. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from HESCA using the Direct-zol RNA Microprep Kit, and processed for cDNA synthesis using oligo-dT (18) primer and M-MLV Reverse Transcriptase (Promega, Madison, WI). PCR was performed using the CFX96 Real-Time PCR Detection System and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) with the condition described previously [20]. Primer sequences and brief descriptions of the genes examined are summarized in Table 2. Data files were opened in CFX Manager software (Bio-Rad) and Ct values were transferred to the Excel program for analyses. ACTB, which encodes β-actin, was used as a housekeeping gene to normalize the expression levels of other genes. For each experiment, three independent sets of samples using different collections of cell suspensions were examined as biological replicates. For the time-course study (data presented in Figure 2), each set consisted of 10 samples: Day 0 (before aggregation), Days 3, 4 and 5 of the three different conditions (C, CS and CSR), all of which were originated from the same cell suspension. For the chemical exposure study (data presented in Figure 3), each set consisted of 6 samples: control (vehicle only) and 5 different concentrations of a given compound, all of which were originated from the same cell suspension and cultured in the same 96-well plate. Note that control and all compound-treated groups in each set contained the same final concentration of dimethyl sulfoxide (DMSO), which was used as solvent to prepare stocks of certain compounds (Table 1). Relative transcript levels were calculated for each set of experiment, as previously described [20], and the means and standard deviations of the three replicates were presented. Fifteen specific genes were selected to evaluate impact of compound exposures, based on the following two criteria: (1) transcript levels were up-regulated progressively towards Day 5 in control aggregates cultured under the CSR condition (data presented in Figure 2), and (2) null mutations of the homologous genes cause distinct malformations in the mouse embryo (Table 3), indicating their critical roles as embryogenesis regulators.
Figure 2.
Time course of transcript levels in HESCA that were cultured under the C (blue), CS (red) and CSR (green) conditions, and examined by quantitative RT-PCR. Vertical axes of graphs are relative transcript levels in arbitrary unit, and mean ± standard deviation (n=3) are shown with the line graphs.
Figure 3.
Transcript levels of the 15 embryogenesis regulator genes in Day 5 HESCA-CSR that have been treated with test compounds, and examined by quantitative RT-PCR. Vertical axes of graphs are relative transcript levels in arbitrary unit, and mean ± standard deviation (n=3) are shown with the bar graphs. Asterisks denote transcript levels that were more than 2-fold different from the corresponding no compound control with statistical significance (P < 0.05). ATRA: all-trans retinoic acid, MEHP: mono-ethylhexyl phthalate. Note that the concentrations of ATRA indicated here are in addition to 2 nM retinoic acid that was already included in the CSR culture medium.
Table 3.
Functional significance of the 15 embryogenesis regulator genes that were examined in the present study to assess molecular impact of teratogenic chemical exposures
| Gene symbol | Phenotype of homozygous null mutant mouse* |
|---|---|
| Aldh1a2 | Failure to turn and elongate along the anterior-posterior body axis; lack of limb buds; heart abnormalities |
| Eya1 | Abnormal kidneys; agenesis of thymus and parathyroid; thyroid hypoplasia |
| Fgf8 | Cardiovascular defects; abnormal left-right axis determination; impaired limb and craniofacial development |
| Foxc2 | Cardiac abnormalities; skeletal defects in the neurocranium and spine |
| Hoxb7 | First and second rib defects |
| Hoxb9 | Rib fusion; abnormal rib attachment to the sternum |
| Meox1 | Hemi-vertebrae; rib and vertebral fusions |
| Mesp2 | Absence of segmented somites; fused vertebral column and dorsal root ganglia; impaired sclerotomal polarity |
| Neurog2 | Neuronal differentiation defects; abnormal retinal and spinal cord interneuron development |
| Olig3 | Impaired development of class A neurons in the dorsal spinal cord |
| Pax3 | Malformations of neural tube, spinal ganglia, heart, vertebral column, hindbrain and limb musculature |
| Pax6 | Small eyes; cataract; craniofacial and forebrain defects |
| Tcf15 | Disrupted somites; patterning defects of the axial skeleton, peripheral nerves, and skeletal muscles |
| Uncx | Severe skeletal defects, including absence of pedicles, transverse processes and proximal ribs |
| Wnt5a | Caudal truncation with shortened anterior-posterior axis; truncation of the snout, tongue and mandible; short fore- and hindlimbs; absent genital tubercle and lung abnormalities |
Information is based on the Mouse Genome Informatics database (www.informatics.jax.org) and references therein.
2.4. Test compounds
Descriptions of test compounds, i.e., name, Chemical Abstract Service (CAS) number, vendor, stock concentration and solvent, are shown in Table 1. The Daston list contains 28 compounds, among which 11 are designated with both positive and negative concentrations, corresponding to maternal plasma Cmax at a dose level that produces significant teratogenicity or embryo lethality and to Cmax that does not cause developmental toxicity, respectively [10]. By contrast, the remaining 17 compounds were designated with only one concentration (either positive or negative). In the present study, the Daston compounds with both positive and negative concentrations were specifically tested, as they are most suited to evaluate sensitivity of assays. Note that SB-209770, one of the 11 compounds with both positive and negative concentrations was not commercially available, and therefore not tested. For 8 of the human medications with known teratogenicity (or lack thereof), a range of the therapeutic plasma concentrations [11] is indicated. Transcriptional effects of each compound were tested at 5 different concentrations of 2- to 4-fold dilution series, as described in Supplementary Data. Appropriate dilution series were selected through pilot experiments to include both negative (absence of impact on gene transcription) and positive (presence of impact) concentrations, except for captopril, metoclopramide and nilotinib. The highest exposures tested for captopril and metoclopramide were 100 μM to keep the amount of the solvent DMSO in the culture medium no more than 0.26%, although they were negative concentrations. The intention of keeping a low DMSO concentration was to minimize unexpected effects of excess solvent on gene expression profiles. The lowest exposure tested for nilotinib was 0.25 μM, which is already 8 times lower than the in vivo non-teratogenic level [10] but was a positive concentration in the present study. Pilot experiments also implicated that high concentrations of certain compounds were evidently detrimental for growth or survival of cell aggregates, which were not used for further analyses. Namely, aggregates did not survive or grow robustly in 160 μM abacavir, 14.6 μM glycolic acid and 8 μM nilotinib. Methanol was not tested at concentrations above 1,024 mM, which was equivalent to 4% volume in the culture medium. This was to minimize excessive dilution of the culture medium by methanol, which may significantly change the osmotic pressure and cause unexpected effects on gene expression profiles.
2.5. ALERT score for assessment of gene expression changes by compound exposures
To assess overall impact of compound exposure, ALERT (Altered Level of Embryogenesis Regulator Transcript) score was formulated as a total number of genes whose transcript levels were significantly altered. An alteration in the transcript level was deemed significant when difference in the mean values between control and exposed groups was more than 2-fold (either increase or decrease) with statistical significance (P < 0.05). The reason for selecting more than 2-fold differences was because malformations are often evident only in homozygous, but not in heterozygous, mutant mice of many embryogenesis regulator genes, implicating that a reduction in expression by less than 2-fold is insufficient to cause birth defects. Because a total of 15 embryogenesis regulators were examined, the highest possible ALERT score is 15 (i.e., all genes are significantly altered) whereas the lowest is 0 (i.e., none is altered). In the present study, all positive ALERT scores (1–15) were considered as adverse impact because any one of the 15 embryogenesis regulators is indispensable for normal embryogenesis. However, higher ALERT scores, i.e., alterations in more genes, implicate potentially severer impact than lower ALERT scores.
2.6. Cell aggregate morphology
HESCA were grouped together and photographed using DSC-70 digital camera (Sony Electronics, San Diego, CA) connected to Stemi 305 dissection microscope (Carl Zeiss, Thornwood, NY). Image files were opened in ImageJ (http://rsb.infonihgov/ij), and morphological parameters (area and circularity) of individual HESCA were measured by tracing their circumference manually using the polygon selection tool. Measurements were exported to Microsoft Excel, where Elongation Distortion Index (= 1/circularity - 1) was calculated. As described previously, area was used as proxy for the size of cell aggregates, whereas EDI was used to gauge the extent of elongation or distortion of aggregates [21]. Area and EDI of individual cell aggregates were normalized against the average of control cell aggregates, and defined as relative area and relative EDI, respectively, which are expressed in percentages (i.e., averages of relative area and relative EDI of control cell aggregates are 100%). Data from three replicates were compiled, and their averages are shown with standard deviation. Thus, a total of 29–30 cell aggregates were scored for each exposure (1 aggregate was occasionally lost or damaged during photography).
2.7. Cell proliferation assay
The impact of test compounds on cell proliferation and viability was evaluated using the CyQUANT Cell Proliferation Assay Kit, which measures the amount of DNA using a specific fluorescence probe as a quantitative proxy for the number of cells (ThermoFisher Scientific). H9 cells were seeded in 96-well plates (pre-coated with iMatrix-511) at a density of 5 × 104 cells/well, and were cultured in mTeSR (100 μL/well) supplemented with each of the test compounds at 5 different concentrations of 2- to 4-fold dilution series or vehicle only as a control. After 2 days of culture, cells were processed for DNA quantitation using Quantus Fluorometer (Promega) according to the manufacturer’s instructions. Cell seeding density was optimized through a series of pilot experiments to confirm that cell numbers at the end of 2 days of culture were proportionate to intensities of fluorescence. Relative DNA amount was calculated based on ratio of the fluorescence intensity in compound-treated cells to that in the control (set as 100%), and the means and standard deviations of 3 sets of biological replicates were presented. In addition to the teratogenic and non-teratogenic compounds listed above (2.4.), methylmercury and hydroxyurea were also tested as positive controls for the assay to detect cytotoxic agents.
2.8. Statistical analyses
The means of relative transcript levels, morphological parameters and relative DNA amount were first examined by the one-way analysis of variance (ANOVA) to determine whether there were any significant differences among their means, which was then followed by post-hoc two-sample t-test to compare between control and compound-exposed groups. For transcript level comparisons, P-values of less than 0.05 were considered statistically significant. For comparisons of morphological parameters, P-values of less than 0.01 were considered statistically significant. The morphological criteria that were previously established for the morphology-based assessment of developmental toxicity using the mouse P19C5 stem cell aggregates [22] were also applied in the present study. Specifically, a compound exposure was considered as having a morphological impact when it caused a reduction in the mean relative area by more than 20% compared to control aggregates and/or a decrease or increase in the mean relative EDI by more than 40% compared to control aggregates. For the cell proliferation assay, a compound exposure was considered as having an adverse impact on cell proliferation or viability when it caused a reduction in the mean relative DNA amount by more than 20% compared to control with P-values of less than 0.05.
3. Results
3.1. Differentiation of human embryonic stem cell aggregates
Human embryonic stem cell aggregates (HESCA) were cultured for 5 days in three different conditions that we formulated (Figure 1A), and the states of differentiation were assessed by gene expression profiling. In the first condition, HESCA were cultured in the Aggregation Culture Medium (ACM) supplemented with 10 μM CHIR99021, a pharmacological agent that activates canonical WNT signaling (designated as C condition hereafter), which is essential for gastrulation in the mouse embryo [23]. By Day 5 of culture, HESCA under C condition (HESCA-C) exhibited distinct morphology with numerous cystic structures (Figure 1B). RNA-seq analysis showed that transcripts associated with the endoderm lineages, namely SOX17, GATA6, FOXA1, AFP, H19, IHH, MPP1 and APOA1, were markedly elevated in HESCA-C compared to undifferentiated cells (Figure 1C). Time course of gene expression changes were further examined in HESCA-C by qRT-PCR. Transcripts encoding the pluripotency maintenance factors (POU5F1, NANOG and SOX2) were down-regulated by Day 3 (Figure 2). Transcript levels of the endoderm lineage-associated genes (SOX17, GATA6, FOXA1, IHH, MPP1 and APOA1) were all up-regulated by Day 3, and further increased or maintained until Day 5 (Figure 2). By contrast, transcripts associated with the mesoderm lineage (BRACHYURY, WNT3A, TBX6, CDX1, MSGN1, HES7 and FGF8) were elevated only transiently at Day 3, but were down-regulated by Day 5 (Figure 2). This suggests that differentiation of endoderm, but not mesoderm, is promoted in HESCA-C. Note that the NODAL transcript was strongly up-regulated under C condition (Figure 2), raising the possibility that NODAL/ACTIVIN signaling, which is responsible for endoderm induction [24], is enhanced as a consequence of WNT signaling activation.
For the second culture condition, the excessive activation of NODAL/ACTIVIN signaling was counteracted by supplementing the C medium with 2 μM SB431542, an antagonist of the NODAL receptors (designated as CS condition). The morphology of HESCA-CS was distinct from HESCA-C, such that the surface was smooth with no cystic structure (Figure 1B). Transcripts associated with the mesoderm lineage (BRACHYURY, WNT3A, TBX6, CDX1, MSGN1, HES7 and FGF8) were all up-regulated in HESCA-CS, much more so than in HESCA-C, whereas up-regulation of the endoderm-associated genes, including NODAL, was suppressed (Figure 2). Furthermore, expressions of genes involved in caudal elongation (WNT5A and ALDH1A2), somite development (PAX3, TCF15, FOXC2, EYA1, MESP2, MEOX1 and UNCX) and axial patterning (HOXB1, HOXB3, HOXB5, HOXB7 and HOXB9) (Table 2) were also up-regulated in HESCA-CS. This suggests that CS condition allows differentiation of mesoderm, particularly the paraxial mesoderm. However, transcripts associated with neural development, such as SOX2, PAX6, NEUROG2 and OLIG3 (Table 2), were not up-regulated in HESCA-CS (Figure 2).
Retinoic acid (RA) has been employed experimentally to induce neural differentiation in both mouse and human pluripotent stem cells [25–27]. Hence, for the third condition, a low concentration of RA (2 nM) was added to the CS medium (designated as CSR condition). Morphologically, HESCA-CSR was more distorted and elongated compared to HESCA-CS, implicating occurrence of morphogenetic process, although the overall shape was variable among aggregates (Figure 1B). Transcripts encoding SOX2, PAX6, NEUROG2 and OLIG3 were all up-regulated in HESCA-CSR, suggesting that neuroectoderm development is promoted under CSR condition (Figure 1C). Note that all of the mesoderm-associated transcripts described above were also robustly up-regulated in HESCA-CSR, even though the levels were slightly lower than in HESCA-CS for several genes (e.g., TBX6, MSGN1, HES7, FOXC2, MEOX1 and UNCX) (Figure 2). This suggests that CSR condition induces differentiation of the neuroectoderm and paraxial mesoderm.
3.2. Impact of teratogenic exposures on gene expression profiles
Many human birth defects involve malformations in the axial skeleton and central nervous system, which are embryologically derived from the paraxial mesoderm and neuroectoderm, respectively [28,29]. HESCA-CSR expressed molecular characteristics of these embryonic tissues, and therefore was explored as a platform to assess teratogenicity of chemical exposures. A total of 18 chemicals (Table 1) were chosen for assessment, for which there are adequate in vivo data on plasma concentrations and teratogenicity. Ten of the chemicals (abacavir, all-trans retinoic acid [ATRA], caffeine, dabigatran, ethylene glycol, glycolic acid, methanol, mono-ethylhexyl phthalate [MEHP], nilotinib and ramelteon) are from the Daston list [10], for which both positive (teratogenic) and negative (non-teratogenic) plasma concentrations have been determined in rats. Eight of the chemicals (captopril, doxylamine, fluorouracil, isotretinoin, metoclopramide, phenytoin, thalidomide and valproic acid [VPA]) are human medications, for which therapeutic plasma concentrations and epidemiologic evidence for teratogenicity or lack thereof have been established in human (Table 1).
Cell aggregates were cultured in the CSR medium containing test chemicals, and examined at Day 5 for the transcript levels of 15 embryogenesis regulator genes (Table 3). Transcriptional changes by chemical exposures were unidirectional for ALDH1A2 (decrease), EYA1 (decrease), FOXC2 (decrease), HOXB7 (increase), HOXB9 (increase), PAX3 (decrease) and UNCX (decrease) (Figure 3). By contrast, responses of other genes, namely FGF8, MEOX1, MESP2, NEUROG2, OLIG3, PAX6, TCF15 and WNT5A, were decrease or increase depending on exposures. For example, OLIG3 was decreased by fluorouracil at 2–4 μM, but increased by thalidomide at 1–16 μM (Figure 3). Thus, gene expression profiles in HESCA-CSR were differentially and uniquely affected by individual compounds in a concentration-dependent manner.
ALERT scores (see Materials and methods) were determined for each exposure to assess overall impact on gene expressions. In the case of the Daston list compounds, ALERT scores were largely consistent with teratogenicity of in vivo plasma concentrations. For 6 compounds (abacavir, ATRA, caffeine, dabigatran, MEHP and ramelteon), the lowest concentrations to yield positive ALERT scores (≥1) fell right between the teratogenic and non-teratogenic levels (Figure 4A), indicating that HESCA-CSR responded to these compounds in a dose-dependent manner comparable to in vivo. For 3 compounds (ethylene glycol, glycolic acid and methanol), 1.5 to 4.5 times higher concentrations than in vivo teratogenic levels were required to produce positive ALERT scores, suggesting that molecular responses of HESCA-CSR to these compounds may be less sensitive than in vivo. By contrast, nilotinib yielded a positive ALERT score even at non-teratogenic level (2 μM), suggesting heightened sensitivity of HESCA-CSR to this compound (Figure 4A).
Figure 4.
Molecular impact of chemical exposures on HESCA-CSR, represented by the ALERT (Altered Level of Embryogenesis Regulator Transcript) score. A, Impact of 10 compounds from the Daston list. Numbers next to open and solid arrows at the bottom of each graph correspond to the non-teratogenic and teratogenic plasma concentrations, respectively, according to Daston et al. (2014). B, Impact of 8 human medications with known pregnancy risks. Numbers with gray bars at the bottom of each graph indicate the therapeutic plasma concentration ranges of the medications in human, according to Schulz et al. (2012).
Out of the 8 human medications tested, 6 have been linked to causing birth defects, and therefore are contraindicated for use during pregnancy (Table 1). Five of these medications (fluorouracil, isotretinoin, phenytoin, thalidomide and VPA) yielded positive ALERT scores at concentrations at or close to (within 2-fold range) the therapeutic plasma levels (Figure 4B). Captopril, one of the contraindicated medications, however, did not produce positive ALERT scores even at 100 μM, which is more than 40 times higher than therapeutic plasma levels. Two human medications that are considered safe during pregnancy yielded no ALERT score even at 34 times (for doxylamine) or 200 times (for metoclopramide) higher concentrations than therapeutic levels (Figure 4B). Thus, HESCA-CSR responded to 7 out of 8 medications in a manner reflecting in vivo teratogenicity or lack thereof in human.
3.3. Impact of teratogenic exposures on aggregate morphology
During 5 days of culture, HESCA-CSR progressively grew in size (Figure 5A). HESCA-CSR were initially round until Day 2 of culture, but gradually transformed into more elongated and distorted shape towards Day 5 (Figure 5A). These morphological changes were quantifiable using morphological parameters, specifically area and Elongation Distortion Index (EDI) (Figure 5B). Many embryogenesis regulators involved in axial patterning and morphogenesis were progressively up-regulated after Day 3, such as ALDH1A2, FGF8, HOXB9, MEOX1 and WNT5A (Figure 2), which coincided with the timing of shape change. Thus, to examine whether alterations of their expression levels by teratogenic exposures are accompanied by changes in morphogenesis, morphological parameters were compared between compound-exposed and control HESCA-CSR groups.
Figure 5.
Morphogenesis of HESCA-CSR. A, Images of HESCA-CSR at different day points of culture that were originated from the same cell suspension. Enlarged images of representative aggregates are shown in framed boxes to highlight the round (Days 1 and 2) and slightly distorted shapes (Day 3). Scale bar = 500 μm. B, Quantitation of morphological parameters. Means ± standard deviations (n=20) are shown for relative area and Elongation Distortion Index (EDI) of HESCA-CSR that are shown in A. Asterisks denote statistical significance (P < 0.01).
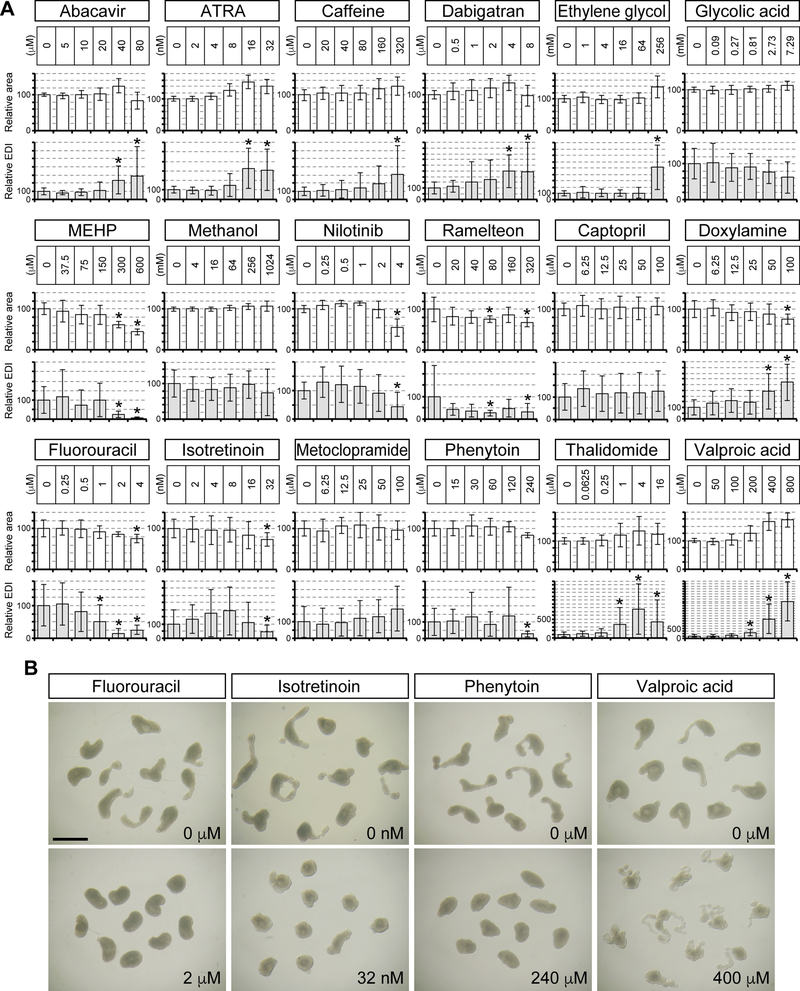
Some of the exposures that yielded positive ALERT scores significantly affected HESCA-CSR morphology, whereas others did not (Figure 6A, Table 4). Note that relative area was not significantly reduced by many of the exposures with positive ALERT scores, such as abacavir at 40 μM, ATRA at 32 nM, caffeine at 320 μM, glycolic acid at 7.29 mM, methanol at 1,024 mM, fluorouracil at 2 μM, isotretinoin at 8 nM, phenytoin at 60 μM, thalidomide at 1 μM and valproic acid at 200 μM. These exposures also appeared to lack in general cytotoxicity, as they did not significantly reduce cell proliferation or viability in monolayer culture (Figure 7). None of the non-teratogenic exposures significantly affected HESCA-CSR morphology (Table 5). Some of the teratogenic exposures caused either a decrease or increase in relative EDI, depending on exposures. Shown in Figure 6B are examples of Day 5 aggregates that were exposed to the teratogenic human medications. Elongation and distortion of HESCA-CSR were significantly less when exposed to fluorouracil at 2 μM, isotretinoin at 32 nM, and phenytoin at 240 μM, compared to the corresponding controls. By contrast, aggregates exposed to VPA at 400 μM exhibited more skinny and convoluted appearance, contributing to increased relative EDI (Figure 6A, B). Nonetheless, several of the teratogenic exposures that yielded positive ALERT scores did not significantly change relative area or relative EDI, namely glycolic acid at 7.29 mM, MEHP at 75 μM, methanol at 1,024 mM, isotretinoin at 8 nM and phenytoin at 60 μM. This implicates that molecular assays based on the transcript levels may be more sensitive to detect teratogenic chemical exposures than measurement of the morphometric parameters.
Figure 6.
Impact of teratogenic exposures on HESCA-CSR morphogenesis. A, Morphological parameters of Day 5 HESCA-CSR that were treated with test compounds. Means ± standard deviations are shown for relative area and relative EDI. Asterisks denote morphological changes, which are defined as reduction in mean relative area by >20% or deviation in mean relative EDI by >40% compared to controls with statistical significance (P < 0.01). B, Images of representative Day 5 HESCA-CSR that were exposed to the human medications that are teratogenic. Scale bar = 1 mm.
Table 4.
Effects of teratogenic exposures on morphology and gene expression in HESCA-CSR
| Teratogenic exposure level *1 | Test concentration *2 | Morphological effects | ALERT score | Concordance (conc. margin) *3 |
|---|---|---|---|---|
| Abacavir (80 μM) | 80 μM | EDI ↑ | 14 | Yes |
| All-trans retinoic acid (200 nM) | 32 nM | EDI ↑ | 11 | Yes |
| Caffeine (325 μM) | 320 μM | EDI ↑ | 7 | Yes |
| Dabigatran (7 μM) | 4 μM | EDI ↑ | 8 | Yes |
| Ethylene glycol (57 mM) | 256 mM | EDI ↑ | 10 | Yes (< 5-fold) |
| Glycolic acid (5 mM) | 7.29 mM | Not significant | 3 | Yes (< 2-fold) |
| MEHP (146 μM) | 75 μM | Not significant | 3 | Yes |
| Methanol (270 mM) | 1,024 mM | Not significant | 3 | Yes (< 4-fold) |
| Nilotinib (28 μM) | 4 μM | Area ↓, EDI ↓ | 13 | Yes |
| Ramelteon (81 μM) | 80 μM | Area ↓, EDI ↓ | 2 | Yes |
| Captopril (0.23–2.3 μM) | 100 μM | Not significant | 0 | No |
| Fluorouracil (0.38–2.3 μM) | 2 μM | EDI ↓ | 5 | Yes |
| Isotretinoin (3.3–6.7 nM) | 8 nM | Not significant | 5 | Yes (< 2-fold) |
| Phenytoin (19.8–59.7 μM) | 60 μM | Not significant | 5 | Yes (< 2-fold) |
| Thalidomide (1.9–5.8 μM) | 1 μM | EDI ↑ | 12 | Yes |
| Valproic acid (277–693 μM) | 200 μM | EDI ↑ | 11 | Yes |
HESCA-CSR: Human embryonic stem cell aggregate cultured under the CSR condition, MEHP: mono-2-ethylhexyl phthalate, EDI: Elongation Distortion Index, ALERT: Altered Level of Embryogenesis Regulator Transcript
Positive effect (teratogenic) msternal plasma concentration in the Daston list (Daston et al., 2014) and therapeutic plasma concentrations of human medications with teratogenic potential (Schulz et al., 2012).
Concentrations exposed to HESCA-CSR.
Yes indicates that positive ALERT score was associated with the teratogenic exposure level (within a concentration margin indicated in parenthesis for several compounds).
Figure 7.
Impact of compound exposures on human ES cell proliferation. Relative DNA amount, as proxy for relative cell number, was measured after 2 days of monolayer culture in the maintenance medium supplemented with test compounds. Relative values are calculated as a percentage of the corresponding control value (set as 100), and means ± standard deviations (n=3) are shown. Asterisks denote reductions in the relative DNA amount by >20% compared to controls with statistical significance (P < 0.05).
Table 5.
Effects of non-teratogenic exposures on morphology and gene expression in HESCA-CSR
| Non-teratogenic exposure level *1 | Test concentration *2 | Morphological effects | ALERT score | Concordance *3 |
|---|---|---|---|---|
| Abacavir (18 μM) | 20 μM | Not significant | 0 | Yes |
| All-trans retinoic acid (1.7 nM) | 2 nM | Not significant | 0 | Yes |
| Caffeine (7.7 μM) | 20 μM | Not significant | 0 | Yes |
| Dabigatran (1 μM) | 1 μM | Not significant | 0 | Yes |
| Ethylene glycol (1.4 mM) | 4 mM | Not significant | 0 | Yes |
| Glycolic acid (0.275 mM) | 0.81 mM | Not significant | 0 | Yes |
| MEHP (1 μM) | 37.5 μM | Not significant | 0 | Yes |
| Methanol (22 μM) | 4 mM | Not significant | 0 | Yes |
| Nilotinib (2 μM) | 2 μM | Not significant | 4 | No |
| Ramelteon (19 nM) | 20 μM | Not significant | 0 | Yes |
| Doxylamine (0.18–0.74 μM) | 6.25 μM | Not significant | 0 | Yes |
| Metoclopramide (0.17–0.5 μM) | 6.25 μM | Not significant | 0 | Yes |
HESCA-CSR: Human embryonic stem cell aggregate cultured under the CSR condition, MEHP: mono-2-ethylhexyl phthalate, EDI: Elongation Distortion Index, ALERT: Altered Level of Embryogenesis Regulator Transcript
Negative effect maternal plasma concentrations in the Daston list (Daston et al., 2014) and therapeutic plasma concentrations of human medications with no teratogenic risk (Schulz et al., 2012)
Concentrations exposed to HESCA-CSR
Yes when ALERT score was zero at or higher than the non-teratogenic exposure levels
4. Discussion
Our long-term goal is to develop an effective in vitro platform for teratogenicity screening by establishing a novel culture protocol for the differentiation of human ES cell aggregates. To date, various in vitro assays have been devised in efforts to detect teratogenic exposures, many of which utilize mouse or human ES cells [4,30–33]. Inclusion of human ES cells is particularly critical in light of certain chemicals, such as thalidomide, impacting human but not mouse embryogenesis [12]. Accordingly, various assay formats using human ES cells have been reported, which differ in types of cell differentiation and endpoint analyses [34–40]. In the present study, we developed a new protocol employing a highly simple culture method (Figure 1A), which efficiently and consistently generated 3-dimensional cell aggregates (coined as HESCA-CSR). HESCA-CSR exhibited morphogenetic and molecular changes associated with differentiation of the paraxial mesoderm and neuroectoderm within 5 days, which are the precursors of the axial skeleton and central nervous system, respectively. This underscores that our differentiation protocol is teratologically relevance, because malformations of the axial skeleton and nervous system are found in various human birth defects, such as hemivertebra, rib fusion, spina bifida and rachischisis [28,29]. Another crucial aspect of the present study is that effectiveness of the assay was evaluated based on exposure levels of chemicals, the validation strategy that corresponds to the real world of teratogenicity testing [7,10]. Many of the chemicals tested in the present study quantifiably altered transcript levels of embryogenesis regulators in a manner consistent with in vivo teratogenic concentrations. Specifically, significant alterations in the transcript levels were observed for 94% (15/16) of the teratogenic exposures within 5-fold margin (Table 4), whereas no alteration was observed for 92% (11/12) of the non-teratogenic exposures (Table 5). These results suggest that HESCA-CSR can serve as an effective assay platform for teratogenicity screening, although it also revealed a few seemingly nonconcordant cases, which may help future efforts to improve human ES cell-based assays to detect a wide range of teratogenic chemicals in a specific and sensitive fashion.
Responses of HESCA-CSR to several chemical exposures were seemingly nonconcordant with their in vivo teratogenicity. The most conspicuous case was captopril, a drug contraindicated during pregnancy, which did not affect the transcript levels at all concentrations tested, even at 40 times higher than the therapeutic levels (Figure 4). This implicates that the HESCA-CSR platform may not be suitable for detecting teratogenicity of this chemical. Captopril is an antihypertensive drug of the angiotensin-converting enzyme (ACE) inhibitor class, whose intake during pregnancy is linked to an increased incidence of certain birth defects, such as renal tubular dysgenesis and hypocalvaria [41]. Notably, the adverse effects of ACE inhibitors are mostly caused by exposure during the fetal period (> eighth week of development) rather than the embryonic period (≤ eighth week) [42]. HESCA-CSR appeared to recapitulate only up to the fourth week of development based on the gene expression profile. This suggests a limitation of the assay in detecting teratogenic insults that occur at later stages, specifically in mid-to-late gestation. Additionally, our assay showed that three chemicals (ethylene glycol, glycolic acid and methanol) needed higher concentrations than the in vivo levels to affect HESCA-CSR. Although only 1.5 times higher was sufficient for glycolic acid to yield a positive ALERT score, 4 to 5 times more were required for the other two, indicating that the assay is less sensitive in detecting teratogenicity of these chemicals. In in vivo situation, ethylene glycol and methanol are oxidized mainly by the liver into glycolic acid and formic acid, respectively, both of which are more potent in teratogenicity than the precursors [43,44]. Lack of sufficient metabolic activities may contribute to the reduced sensitivity of many in vitro assay platforms, including HESCA-CSR, against those chemicals that become teratogenic only after metabolic conversions. However, absence of metabolic activities can provide valuable investigative opportunities to interrogate which chemical structures and metabolic intermediates are responsible for teratogenic effects. Lastly, our HESCA-CSR assay appeared to be hypersensitive to nilotinib, which yielded positive ALERT scores even at the non-teratogenic concentrations. Nilotinib is a tyrosine kinase inhibitor used for the treatment of leukemias. It is currently unknown whether our assay platform is hypersensitive to other types of tyrosine kinase inhibitors. However, nilotinib also affects the morphology-based assay with mouse P19C5 stem cell aggregates at non-teratogenic concentrations [22], raising the possibility that hypersensitivity against this compound is a general feature of stem cell-based in vitro tests. Further studies are necessary to examine whether the state of nilotinib is considerably different in the blood plasma versus in the culture medium (e.g., due to metabolic modifications or binding to serum proteins), which may diminish teratogenic potential of the drug in vivo compared to in vitro.
To assess adverse effects of chemical exposures, 15 genes were specifically chosen for expression profiling, all of which are likely to encode key regulators for human embryogenesis. In principle, profiling of more genes by RNA-seq, as explored in previous studies [45,46], should provide expansive data to thoroughly evaluate molecular impact of chemical exposures, although the cost would be prohibitively high to analyze multiple compounds at multiple concentrations in multiple biological replicates. Nonetheless, such whole transcriptome analysis may reveal significant effects of teratogenic exposures that could have been missed by profiling of the 15 genes. It should be noted, however, that not all genes are equally important for embryogenesis, because losses of some genes cause severe birth defects, whereas others result in no overt phenotype [47,48]. For this reason, transcriptomic profiles need to be carefully evaluated in accordance with functional significance of individual genes that are relevant for the etiology of birth defects. Furthermore, the nature and extent of alterations in transcript levels may matter in interpreting the impact of chemical exposures. In the present assay, we scored either decrease or increase by >2-fold as a significant alteration. However, functional consequences of alterations are likely to be different between decrease and increase in transcript levels. Many mouse studies have examined effects of loss-of-function mutations, largely based on gene knockout experiments, which provide valuable insights into how a reduction in the transcript level affects embryogenesis. By contrast, much fewer studies have addressed how overexpression or gain-of-function mutation of specific genes would affect embryo development. As a result, current knowledge on possible impact of increased gene expressions is scarce. Nonetheless, there are some examples that underscore the significance of excessive gene activity in birth defects, such as constitutive activation of FGF signaling that causes various congenital skeletal malformations in human [49]. Therefore, criteria to select genes for profiling and also to define embryologically relevant adverse effects need to be prudently appraised in order to enhance in vitro assessment of teratogenic exposures.
In the present study, any positive ALERT score, whether 1 or 15, was considered as an adverse impact on the embryogenesis regulator expression. For many compounds (abacavir, ATRA, caffeine, dabigatran, nilotinib, ramelteon, isotretinoin, thalidomide and valproic acid), the level of ALERT scores tended to be higher for HESCA-CSR that were exposed to higher concentrations (Figure 4). This implicates that the level of ALERT score correlates with teratogenic intensity of exposures. However, it is unclear whether a compound that yields a higher ALERT score is more of a concern than that which gives a lower score (e.g., valproic acid and fluorouracil yielded 13 and 5, respectively, at the therapeutic concentrations; Figure 4). Intuitively, compound exposures that alter expressions of greater numbers of embryogenesis regulators (i.e., higher ALERT score) may be expected to affect more embryonic structures (i.e., more severe teratogenicity). ALERT scores are based on specific changes (>2-fold decrease or increase) in the transcript levels of the selected 15 genes, which may be too restricted to be reflective of a wide spectrum of birth defects in human. The practical utility of the ALERT score needs to be more vigorously evaluated in future studies with additional information on in vivo concentrations and teratogenic severity (including types of embryonic structures affected) for a broad range of compounds.
Many common birth defects are not only due to impaired cell proliferation or differentiation, but are also caused by misregulation of morphogenesis, i.e., shape changes of embryonic tissues. Examples of such birth defects are neural tube defects, heart septal defects, gastroschisis, and cleft lip and palate [28,29]. Hence, inclusion of morphogenesis aspects should reinforce ES cell-based tests to detect a wide spectrum of teratogenic exposures [40,50]. Morphogenesis of embryonic body axis elongation, comparable to gastrulation, can be recapitulated in 3-dimensional aggregates of mouse P19/P19C5 embryonal carcinoma stem cells [51,52] and mouse ES cells [53,54]. In particular, in vitro axial morphogenesis of P19C5 cell aggregates has been evaluated as a platform to assess teratogenic exposures using the Daston list [22] and human medications with known pregnancy risks [21], in which morphometric measurement of aggregates was effectively applied for teratogenicity assessment. In the present study, HESCA exhibited morphological transformation from spherical to highly distorted shapes under the CSR condition (Figure 5A). Whether the morphological transformation of HESCA-CSR is comparable to the gastrulation-like morphogenesis of mouse P19C5 cell aggregates is yet to be investigated. However, only 10 out of the 16 teratogenic exposures significantly altered morphogenesis of HESCA-CSR (Table 4), suggesting that morphological assessment is not as sensitive as the ALERT score. Nonetheless, visual assessment of aggregate morphology may still be useful as a preliminary test to gauge potential impact of chemical exposures. Morphological assessment can provide approximate estimation of adverse or toxic concentration ranges (e.g., relative area can be used to assess general cytotoxicity), and may be practical and economical before investing on more laborious and costly gene expression profiling. In addition, certain teratogenic chemical exposures may primarily affect morphogenesis without significantly altering transcript levels, such as through direct impact on cytoskeletal or cell polarity regulator proteins. Therefore, while the present study highlighted the efficacy of transcript profiling, morphogenetic aspects of the assay may still be valuable and should not be dismissed in future screening endeavors.
The CSR culture condition was specifically explored in the present study, although other conditions may also be useful for assessment of teratogenic exposures. Transcripts associated with the endoderm lineages, such as SOX17, FOXA1 and GATA6, were enriched in HESCA-C, but were underrepresented in HESCA-CS and HESCA-CSR (Figure 1C). If there are teratogenic exposures that specifically impair development of endoderm but not of mesoderm or neuroectoderm, those exposures may not be detected using HESCA-CSR. Existence of such endoderm-specific teratogenic exposures is currently unknown. However, there are human birth defects, in which endoderm-derived tissues are affected, such as pyloric stenosis and annular pancreas [28,29]. Assays using HESCA-C, which exhibited unique morphology and gene expression profile, may yield valuable insights into effects of chemical exposures on endoderm development, and compensate for potential deficiencies of HESCA-CSR. It would be important, however, to further characterize what types of endoderm are developing in HESCA-C. There are two distinct lineages of endoderm that emerge during embryogenesis: the primitive and definitive endoderm lineages. The primitive endoderm gives rise to extraembryonic tissues that provide nutrient support and inductive signals for embryo patterning, whereas the definitive endoderm contributes to the inner lining of the respiratory and digestive tracts. Many genes, including SOX17, FOXA1 and GATA6, are expressed in both the primitive and definitive endoderm lineages at early stages [55]. A recent RNA-seq analysis of the first-trimester human yolk sac, a derivative of the primitive endoderm, has revealed that the tissue is highly enriched with transcripts encoding AFP, APOA1, APOA2, COL1A3, FGB, H19 and TTR [56]. All of these genes were strongly up-regulated in HESCA-C (Supplementary Data), suggesting that the C culture condition promotes differentiation of the primitive endoderm lineage. Thus, HESCA-C can be further explored as an in vitro model to study development of human primitive endoderm, which may be of particular interest in light of the studies that implicate the yolk sac as the primary target of hyperglycemia in the pathogenesis of diabetic embryopathy [57,58].
Exposure-based assessment using the Daston list is a valuable strategy to validate individual in vitro assays for teratogenicity screening [10], and was employed to evaluate the HESCA-CSR platform in the present study. The Daston list was also applied to evaluate the morphology-based assay using the mouse P19C5 gastrulation model [22], the zebrafish development assay [59], and also another assay platform of human ES cells, which measures changes in specific metabolite levels as an indication of teratogenicity [38,60]. Exposure-based assessment of more assay platforms should delineate their applicability and limitation, and help assemble the most effective combination of assays to detect a wide range of teratogenic exposures. Note, however, that the Daston list was derived from studies using animals, specifically rats [10]. Because the HESCA-CSR platform is based on human cells, it is yet to be determined how effectively it can respond to teratogenic and non-teratogenic exposures that are elucidated using animal models. Given that the ultimate goal is to develop screening assays to detect teratogenic exposures for human, it may be crucial to build a new list, which is comparable to the Daston list in concept but is based on pharmacokinetics and teratogenicity data in human.
In theory, a single platform would be sufficient for teratogenicity screening, if it were to recapitulate the whole aspects of embryogenesis faithfully. Several recent studies report that various morphological and molecular events of early embryogenesis can be recreated in vitro in 3-dimensional aggregates of mouse and human stem cells [52–54,61–64]. This implicates a future possibility of stem cells being used to create embryo-like entities, and ethical issues of such synthetic embryos have been debated [65–68]. From a practical point of view, however, teratogenicity research would benefit more from the use of multiple platforms, each of which represents only limited aspects of embryogenesis. This is because recapitulation of specific aspects is more likely to be achieved easily, consistently and economically, which are pivotal for conducting reliable screening of many compounds at various concentrations. Also, without the ethical concern of creating embryo-like entities, limited but complementary platforms of multiple ES cell-based assays are more likely to move forward without obstruction, while circumventing another ethical issue, i.e., animal experimentations.
Supplementary Material
Highlights.
Human ES cell aggregates exhibited morphogenesis under a new differentiation protocol
Transcripts associated with paraxial mesoderm and neuroectoderm were up-regulated
Transcript levels in aggregates were significantly altered by most teratogenic exposures
Size and shape of aggregates were quantifiably altered by many teratogenic exposures
ACKNOWLEDGEMENTS
We are grateful to Dr. Karolina Peplowska at the Cancer Center Genomics Shared Resource and Dr. Vedbar Khadka at the JABSOM Bioinformatics Core for their technical assistance with RNA-seq analysis.
FUNDING
This work was supported by grants from the Johns Hopkins Center for Alternatives to Animal Testing (CAAT), the Alternatives Research & Development Foundation (ARDF), the Harold K.L. Castle Foundation and the National Institutes of Health (R03 HD088970) to Y.M. RNA-seq analysis was performed at the University of Hawaii John A. Burns School of Medicine Bioinformatics Core, which was supported by grants from the Hawaii Community Foundation, Bears Care Foundation and the National Institutes of Health (R01 CA223490, R21 CA164764) to Y.D., and (P20 GM103466, P30 GM114737, U54 MD007601) to M.M.
Footnotes
COMPETING INTERESTS
The authors have no competing interest to report.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Riebeling C, Hayess K, Peters AK, Steemans M, Spielmann H, Luch A, Seiler AE, Assaying embryotoxicity in the test tube: current limitations of the embryonic stem cell test (EST) challenging its applicability domain. Crit. Rev. Toxicol 42 (2012) 443–464. [DOI] [PubMed] [Google Scholar]
- [2].Theunissen PT, Piersma AH, Innovative approaches in the embryonic stem cell test (EST). Front. Biosci 17 (2012) 1965–1975. [DOI] [PubMed] [Google Scholar]
- [3].Kuske B, Pulyanina PY, zur Nieden NI, Embryonic stem cell test: stem cell use in predicting developmental cardiotoxicity and osteotoxicity. Methods Mol. Biol 889 (2012) 147–179. [DOI] [PubMed] [Google Scholar]
- [4].Luz AL, Tokar EJ, Pluripotent stem cells in developmental toxicity testing: a review of methodological advances. Toxicol. Sci 165 (2018) 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Seiler AE, Buesen R, Visan A, Spielmann H, Use of murine embryonic stem cells in embryotoxicity assays: the embryonic stem cell test. Methods Mol. Biol 329 (2006) 371–395. [DOI] [PubMed] [Google Scholar]
- [6].Theunissen PT, Pennings JL, van Dartel DA, Robinson JF, Kleinjans JC, Piersma AH, Complementary detection of embryotoxic properties of substances in the neural and cardiac embryonic stem cell tests. Toxicol. Sci 132 (2013) 118–130. [DOI] [PubMed] [Google Scholar]
- [7].Daston GP, Chapin RE, Scialli AR, Piersma AH, Carney EW, Rogers JM, Friedman JM, A different approach to validating screening assays for developmental toxicity. Birth Def. Res. B Dev. Reprod. Toxicol 89 (2010) 526–530. [DOI] [PubMed] [Google Scholar]
- [8].Friedman JM, The principles of teratology: are they still true? Birth Def. Res. A Clin. Mol. Teratol 88 (2010) 766–768. [DOI] [PubMed] [Google Scholar]
- [9].Jelínek R, The contribution of new findings and ideas to the old principles of teratology. Reprod. Toxicol 20 (2005) 295–300. [DOI] [PubMed] [Google Scholar]
- [10].Daston GP, Beyer BK, Carney EW, Chapin RE, Friedman JM, Piersma AH, Rogers JM, Scialli AR, Exposure-based validation list for developmental toxicity screening assays. Birth Def. Res. B Dev. Reprod. Toxicol 101 (2014) 423–428. [DOI] [PubMed] [Google Scholar]
- [11].Schulz M, Iwersen-Bergmann S, Andresen H, Schmoldt A, Therapeutic and toxic blood concentrations of nearly 1,000 drugs and other xenobiotics. Crit. Care 16 (2012) R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vargesson N, Thalidomide-induced teratogenesis: history and mechanisms. Birth Def. Res. C Embryo Today 105 (2015) 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jentink J, Loane MA, Dolk H, Barisic I, Garne E, Morris JK, de Jong-van den Berg LT, Valproic acid monotherapy in pregnancy and major congenital malformations. N. Engl. J. Med 362 (2010) 2185–2193. [DOI] [PubMed] [Google Scholar]
- [14].Sahin L, Nallani SC, Tassinari MS, Medication use in pregnancy and the pregnancy and lactation labeling rule. Clin. Pharmacol. Ther 100 (2016) 23–25. [DOI] [PubMed] [Google Scholar]
- [15].Miyazaki T, Futaki S, Suemori H, Taniguchi Y, Yamada M, Kawasaki M, Hayashi M, Kumagai H, Nakatsuji N, Sekiguchi K, Kawase E, Laminin E8 fragments support efficient adhesion and expansion of dissociated human pluripotent stem cells. Nat. Commun 3 (2012) 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ryu AH, Eckalbar WL, Kreimer A, Yosef N, Ahituv N, Use antibiotics in cell culture with caution: genome-wide identification of antibiotic-induced changes in gene expression and regulation. Sci Rep. 7 (2017) 7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17 (2011) 10–12. [Google Scholar]
- [18].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (2013) 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Warkus ELL, Marikawa Y, Fluoxetine inhibits canonical Wnt signaling to impair embryoid body morphogenesis: potential teratogenic mechanisms of a commonly used antidepressant. Toxicol. Sci 165 (2018) 372–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Warkus EL, Yuen AA, Lau CG, Marikawa Y, Use of in vitro morphogenesis of mouse embryoid bodies to assess developmental toxicity of therapeutic drugs contraindicated in pregnancy. Toxicol. Sci 149 (2016) 15–30. [DOI] [PubMed] [Google Scholar]
- [22].Warkus ELL, Marikawa Y, Exposure-based validation of an in vitro gastrulation model for developmental toxicity assays. Toxicol. Sci 157 (2017) 235–245. [DOI] [PubMed] [Google Scholar]
- [23].Marikawa Y, Wnt/beta-catenin signaling and body plan formation in mouse embryos. Semin. Cell Dev. Biol 17 (2006) 175–184. [DOI] [PubMed] [Google Scholar]
- [24].Jaremko KL, Marikawa Y, Regulation of developmental competence and commitment towards the definitive endoderm lineage in human embryonic stem cells. Stem Cell Res. 10 (2013) 489–502. [DOI] [PubMed] [Google Scholar]
- [25].Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI, Embryonic stem cells express neuronal properties in vitro. Dev. Biol 168 (1995) 342–357. [DOI] [PubMed] [Google Scholar]
- [26].Guan K, Chang H, Rolletschek A, Wobus AM, Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell Tissue Res. 305 (2001) 171–176. [DOI] [PubMed] [Google Scholar]
- [27].Verrier L, Davidson L, Gierliński M, Dady A, Storey KG, Neural differentiation, selection and transcriptomic profiling of human neuromesodermal progenitor-like cells in vitro. Development 145 (2018) dev166215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Moore K, Persaud TVN, Torchia M, The Developing Human 10th Edition: Clinically Oriented Embryology. Saunders Co.; Philadelphia, (2015). [Google Scholar]
- [29].Sadler TW, Langman’s Medical Embryology 13th edition. Lippincott Williams & Wilkins, Philadelphia: (2014). [Google Scholar]
- [30].Augustine-Rauch K, Zhang CX, Panzica-Kelly JM, A developmental toxicology assay platform for screening teratogenic liability of pharmaceutical compounds. Birth Def. Res. B Dev. Reprod. Toxicol 107 (2016) 4–20. [DOI] [PubMed] [Google Scholar]
- [31].Buck K, zur Nieden NI, Risk assessment using human pluripotent stem cells: recent advances in developmental toxicity screens In: Rasmussen TP (Ed.), Stem Cells in Birth Defects Research and Developmental Toxicology. Wiley, New York, NY, (2018) 91–118. [Google Scholar]
- [32].Kugler J, Huhse B, Tralau T, Luch A, Embryonic stem cells and the next generation of developmental toxicity testing. Expert Opin. Drug Metab. Toxicol 13 (2017) 833–841. [DOI] [PubMed] [Google Scholar]
- [33].Rezvanfar MA, Hodjat M, Abdollahi M, Growing knowledge of using embryonic stem cells as a novel tool in developmental risk assessment of environmental toxicants. Life Sci. 158 (2016) 137–160. [DOI] [PubMed] [Google Scholar]
- [34].Flamier A, Singh S, Rasmussen TP, A standardized human embryoid body platform for the detection and analysis of teratogens. PLoS One 12 (2017) e0171101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kameoka S, Babiarz J, Kolaja K, Chiao E, A high-throughput screen for teratogens using human pluripotent stem cells. Toxicol. Sci 137 (2014) 76–90. [DOI] [PubMed] [Google Scholar]
- [36].Mayshar Y, Yanuka O, Benvenisty N, Teratogen screening using transcriptome profiling of differentiating human embryonic stem cells. J. Cell Mol. Med 15 (2011) 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mehta A, Konala VB, Khanna A, Majumdar AS, Assessment of drug induced developmental toxicity using human embryonic stem cells. Cell Biol. Int 32 (2008) 1412–1424. [DOI] [PubMed] [Google Scholar]
- [38].Palmer JA, Smith AM, Egnash LA, Conard KR, West PR, Burrier RE, Donley EL, Kirchner FR, Establishment and assessment of a new human embryonic stem cell-based biomarker assay for developmental toxicity screening. Birth Def. Res. B Dev. Reprod. Toxicol 98 (2013) 343–363. [DOI] [PubMed] [Google Scholar]
- [39].West PR, Weir AM, Smith AM, Donley EL, Cezar GG, Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol 247 (2010) 18–27. [DOI] [PubMed] [Google Scholar]
- [40].Xing J, Cao Y, Yu Y, Li H, Song Z, Yu H, In vitro micropatterned human pluripotent stem cell test (μP-hPST) for morphometric-based teratogen screening. Sci. Rep 7 (2017) 8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pryde PG, Sedman AB, Nugent CE, Jr M Barr, Angiotensin-converting enzyme inhibitor fetopathy. J. Am. Soc. Nephrol 3 (1993) 1575–1582. [DOI] [PubMed] [Google Scholar]
- [42].Barr M, Angiotensin-converting enzyme inhibitor fetopathy In: Kavlock RJ, and Daston GP (Ed.), Drug Toxicity in Embryonic Development II Advances in Understanding Mechanisms of Birth Defects: Mechanistic Understanding of Human Developmental Toxicants. Springer, New York, NY, (1997) 265–294. [Google Scholar]
- [43].Andrews JE, Ebron-McCoy M, Kavlock RJ, Rogers JM, Developmental toxicity of formate and formic acid in whole embryo culture: a comparative study with mouse and rat embryos. Teratology 51 (1995) 243–251. [DOI] [PubMed] [Google Scholar]
- [44].Neeper-Bradley TL, Tyl RW, Fisher LC, Kubena MF, Vrbanic MA, Losco PE, Determination of a no-observed-effect level for developmental toxicity of ethylene glycol administered by gavage to CD rats and CD-1 mice. Fundam. Appl. Toxicol 27 (1995) 121–130. [DOI] [PubMed] [Google Scholar]
- [45].Chen M, Zhang M, Borlak J, Tong W, A decade of toxicogenomic research and its contribution to toxicological science. Toxicol. Sci 130 (2012) 217–228. [DOI] [PubMed] [Google Scholar]
- [46].McHale CM, Zhang L, Thomas R, Smith MT, Analysis of the transcriptome in molecular epidemiology studies. Environ. Mol. Mutagen 54 (2013) 500–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Barbaric I, Miller G, Dear TN, Appearances can be deceiving: phenotypes of knockout mice. Brief. Funct. Genomic. Proteomic 6 (2007) 91–103. [DOI] [PubMed] [Google Scholar]
- [48].Nowak MA, Boerlijst MC, Cooke J, Smith JM, Evolution of genetic redundancy. Nature 388 (1997) 167–171. [DOI] [PubMed] [Google Scholar]
- [49].Burke D, Wilkes D, Blundell TL, Malcolm S, Fibroblast growth factor receptors: lessons from the genes. Trends Biochem. Sci 23 (1998) 59–62. [DOI] [PubMed] [Google Scholar]
- [50].Belair DG, Wolf CJ, Moorefield SD, Wood C, Becker C, Abbott BD, A three-dimensional organoid culture model to assess the influence of chemicals on morphogenetic fusion. Toxicol. Sci 166 (2018) 394–408. [DOI] [PubMed] [Google Scholar]
- [51].Lau CG, Marikawa Y, Morphology-based mammalian stem cell tests reveal potential developmental toxicity of donepezil. Mol. Reprod. Dev 81 (2014) 994–1008. [DOI] [PubMed] [Google Scholar]
- [52].Marikawa Y, Tamashiro DA, Fujita TC, Alarcón VB, Aggregated P19 mouse embryonal carcinoma cells as a simple in vitro model to study the molecular regulations of mesoderm formation and axial elongation morphogenesis. Genesis 47 (2009) 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Beccari L, Moris N, Girgin M, Turner DA, Baillie-Johnson P, Cossy AC, Lutolf MP, Duboule D, Arias AM, Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562 (2018) 272–276. [DOI] [PubMed] [Google Scholar]
- [54].van den Brink SC, Baillie-Johnson P, Balayo T, Hadjantonakis AK, Nowotschin S, Turner DA, Martinez Arias A, Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141 (2014) 4231–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zorn AM, Wells JM, Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol 25 (2009) 221–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cindrova-Davies T, Jauniaux E, Elliot MG, Gong S, Burton GJ, Charnock-Jones DS, RNA-seq reveals conservation of function among the yolk sacs of human, mouse, and chicken. Proc. Natl. Acad. Sci. USA 114 (2017) E4753–E4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dong D, Reece EA, Lin X, Wu Y, AriasVillela N, Yang P, New development of the yolk sac theory in diabetic embryopathy: molecular mechanism and link to structural birth defects. Am. J. Obstet. Gynecol 214 (2016) 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Reece EA, Pinter E, Homko C, Wu YK, Naftolin F, The yolk sac theory: closing the circle on why diabetes-associated malformations occur. J. Soc. Gynecol. Investig 1 (1994) 3–13. [PubMed] [Google Scholar]
- [59].Cassar S, Beekhuijzen M, Beyer B, Chapin R, Dorau M, Hoberman A, Krupp E, LeConte I, Stedman D, Stethem C, van den Oetelaar D, Tornesi B, A multi-institutional study benchmarking the zebrafish developmental assay for prediction of embryotoxic plasma concentrations from rat embryo-fetal development studies. Reprod. Toxicol 86 (2019) 33–44. [DOI] [PubMed] [Google Scholar]
- [60].Palmer JA, Smith AM, Colwell MR, Smart BJ, Ludwig MA, Burrier RE, Donley ELR, Kirchner FR, Prediction of developmental toxicity potential dictated by in vivo exposures with a biomarker-based human pluripotent stem cell assay. Birth Def. Res 109 (2017) 658. [Google Scholar]
- [61].Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka-Goetz M, Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356 (2017) 6334. [DOI] [PubMed] [Google Scholar]
- [62].Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset JC, Korving J, Vivié J, Truckenmüller RK, van Oudenaarden A, van Blitterswijk CA, Geijsen N, Blastocyst-like structures generated solely from stem cells. Nature 557 (2018) 106–111. [DOI] [PubMed] [Google Scholar]
- [63].Shao Y, Taniguchi K, Townshend RF, Miki T, Gumucio DL, Fu J, A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun 8 (2017) 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sozen B, Amadei G, Cox A, Wang R, Na E, Czukiewska S, Chappell L, Voet T, Michel G, Jing N, Glover DM, Zernicka-Goetz M, Self-assembly of embryonic and two extraembryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol 20 (2018) 979–989. [DOI] [PubMed] [Google Scholar]
- [65].Aach J, Lunshof J, Iyer E, Church GM, Addressing the ethical issues raised by synthetic human entities with embryo-like features. Elife 6 (2017) e20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hyun I, Engineering ethics and self-organizing models of human development: opportunities and challenges. Cell Stem Cell 21 (2017) 718–720. [DOI] [PubMed] [Google Scholar]
- [67].Munsie M, Hyun I, Sugarman J, Ethical issues in human organoid and gastruloid research. Development 144 (2017) 942–945. [DOI] [PubMed] [Google Scholar]
- [68].Rivron N, Pera M, Rossant J, Martinez Arias A, Zernicka-Goetz M, Fu J, van den Brink S, Bredenoord A, Dondorp W, de Wert G, Hyun I, Munsie M, Isasi R, Debate ethics of embryo models from stem cells. Nature 564 (2018) 183–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.