Abstract
Mutations in mitochondrial DNA (mtDNA) have been linked to risk, progression, and treatment response of head and neck squamous cell carcinoma (HNSCC). Due to their clonal nature and high copy number, mitochondrial mutations could serve as powerful molecular markers for detection of cancer cells in bodily fluids, surgical margins, biopsies and lymph node (LN) metastasis, especially at sites where tumor involvement is not histologically apparent. Despite a pressing need for high-throughput, cost-effective mtDNA mutation profiling system, current methods for library preparation are still imperfect for detection of low prevalence heteroplasmic mutations. To this end, we have designed an ultra-deep amplicon-based sequencing library preparation approach that covers the entire mitochondrial genome. We sequenced mtDNA in 28 HNSCCs, matched LNs, surgical margins and bodily fluids, and applied multiregional sequencing approach on 14 primary tumors. Our results demonstrate that this quick, sensitive and cost-efficient method allows obtaining a snapshot on the mitochondrial heterogeneity, and can be used for detection of low frequency tumor-associated mtDNA mutations in LNs, sputum and serum specimens. These findings provide the foundation for using mitochondrial sequencing for risk assessment, early detection, and tumor surveillance.
Introduction
Head and neck squamous cell carcinoma (HNSCC) accounts for 650,000 new cases worldwide [1–5] and over 50,000 cases in the US alone [6]. HNSCC prognosis is poor, with a 5-year overall survival of ~50% [7–9]. The prognosis and treatment regiments vary dramatically depending on the presence or absence of lymph node (LN) metastasis. Detection of regional LN metastasis is usually based on the clinical examination and imaging, followed by post-operational histological analysis. Unfortunately, current tools for detection of tumor cells in surgically resected LN may, in some cases, miss the presence of micrometastasis. Novel strategies for detection of micrometastasis based on the analysis of tumor-derived genetic aberrations by next generation sequencing (NGS) offer new hope for improved risk assessment and better selection of the treatment regimen [10, 11]. However, detection of mutations in minuscule metastatic lesions comprised of only few neoplastic cells may still fall below the detection threshold of these highly sensitive techniques. Further, early disease and micrometastatic lesions have lower levels of neoplastic cells and therefore more likely to yield false negative results.
Current theories of tumorigenesis hold that HNSCCs are driven by the acquisition of genetic alterations, including changes in mitochondrial DNA (mtDNA) [12–16]. Due to the lack of protective histones and limited DNA repair mechanisms, mtDNA is susceptible to damage by environmental carcinogens as well as endogenous reactive oxygen species, a byproduct of the oxidative phosphorylation system [17]. As a result, the mutation rate in mtDNA is approximately 10 times higher than in nuclear genomic DNA [13, 17]. Numerous somatic mutations in both the coding and control regions of mtDNA have been extensively examined in a broad range of primary human cancers by us [13, 17–21] and others [22, 23]. These data suggest that some mtDNA alterations may be directly involved in tumorigenesis and not merely epiphenomena [24, 25]. By virtue of their clonal nature, higher mutation rate and copy number, assessing tumor-specific mtDNA mutations in histologically clean LN and surgical margins may provide a more sensitive diagnostic tool and eventually reduce the false negative rate in patients. Additionally, due to its circular configuration, mtDNA thought to be more stable than genomic DNA, and may therefore be suitable for sequencing when isolated from low quality formalin-fixed paraffin-embedded (FFPE) or body fluid-derived (e.g. plasma, serum) samples.
Using an array-based sequencing platform (MitoChip) for high-throughput mtDNA sequencing [26], our group has showed that mtDNA alterations are valuable markers of head and neck tumorigenesis, and that mtDNA mutations and copy number increase with histopathologic grade in premalignant and malignant HNSCC lesions [27, 28]. While mitochondrial mutations could be extracted from whole-genome nuclear DNA sequencing data [29, 30], most current methods for mtDNA library preparation require pre-amplification and are imperfect for detection of low prevalence and heteroplasmic variants, especially in very small and highly degraded samples. Although recently reported PCR-based library construction approaches have increased the capacity to assess mutation across mitochondrial genome and demonstrate a substantial improvement in sequencing of the fresh frozen and FFPE specimens [31, 32], lack of multiplexing and workflow complexities increase their overall turnaround times and costs, posing a practical barrier to clinical translation. While several studies used early generation methods such as Sanger sequencing and Affymetrix MitoChip assays to map mtDNA mutations in head and neck neoplasms over the last decade, to the best of our knowledge, only two recent works have leveraged NGS sequencing to achieve higher sequencing resolution. One study extracted the mtDNA sequences from the off-target mitochondrial reads derived from the whole-exome sequencing (WES) of HNSCC samples and matched lymphocytes obtained from TCGA database [30]. However, as mentioned earlier, the lack of adequate mtDNA coverage depth in WES data limits detection of minor alleles of lower frequency. A second study used long-range PCR enrichment and Illumina HiSeq 2500 sequencing on 28 tumors, but without matched normal controls. Lack of matched controls confounds somatic mutation variant calling since mutations identified as putatively somatic may represent low-level germline heteroplasmies [33].
To address these limitations, we have developed a novel amplicon-based approach for ultra-deep sequencing of the entire mitochondrial genome specifically suited for micro-sized low quality specimens. This multiplexing amplicon-based library preparation approach allows running up to 48 samples (for both sequencing and copy number analysis) in a single amplification reaction, allowing for efficient and accurate library preparation. Using this approach, we performed whole mtDNA sequencing of 28 HNSCC tumors and multiple matched metastatic or histologically clean lymph nodes (LN). For a subset of patients, we also sequenced matched sputum, serum and histological clean surgical margins. Furthermore, to assess intra-tumor mitochondrial heterogeneity, we have applied a multiregional sequencing approach on 14 primary tumor specimens. This method allowed us to obtain a snapshot on the extent of mitochondrial heterogeneity of these lesions, including detection of low frequency tumor-associated mtDNA mutations in LN, surgical margins and bodily fluids.
Materials and Methods
Schematic of workflow for sequencing and analysis of somatic mitochondrial mutations in HNSCC samples
Samples and DNA extraction
Samples were collected from the Johns Hopkins Head & Neck Database (HAND) following Institutional Review Boards (IRB)-approved protocols. Informed written consent was obtained from all patients before sampling. All samples were reviewed by a senior pathologist to reconfirm the diagnosis and were processed for analysis by the Johns Hopkins Pathology Core. Lesions with a low neoplastic cellularity (<70%) were additionally macrodissected to remove contaminating normal cells before DNA extraction. Neoplastic cellularity was estimated from the sequential slides, which highly reflect cellularity of the section used for DNA sequencing. Genomic DNA was isolated from fresh frozen samples (tumors, margins and lymph nodes) by the QIAamp DNA Kit (Qiagen) and quantified with the Nanodrop system (Thermo Scientific). As a control, matched lymphocyte DNA were used in each case. Plasma and sputum DNA was extracted by digestion with 50 μg ml−1 proteinase K (Boehringer Mannheim) in the presence of 1% SDS at 48 °C overnight followed by phenol/chloroform extraction and ethanol precipitation.
Assay design and mitochondrial genome amplification
Amplicon-based library preparation approach, consisting of 148 primers pairs that cover the entire mitochondrial genome (with 86.2% of the mt-genome regions are covered by two different amplicon sets) was designed to be used on Fluidigm Access Array platform (Supplementary Table 1). To enhance the coverage of degraded material the amplicon size was limited to ~ 170 base pairs. Access Array System creates amplicon libraries using a unique tagging protocol, in which primers attach sample-specific barcodes and sequencer-specific tags to each PCR product, allowing to run 48 samples in one multiplex sequencing reaction. In brief, 10ng of genomic DNA was used for mtDNA amplification using a microfluidic chip according to the manufacturer’s instructions. Each sample was combined with primer pairs in a microfluidic chip and thermal cycling on a Fluidigm FC1 Cycler was performed. PCR products were then collected using the IFC controller and transferred to a 96-well plate. In a separate PCR, Illumina sequence-specific adaptors and barcodes were attached. Raw NGS data has been uploaded to the NCBI Sequence Read Archive repository. Accession code for deposited data is PRJNA592200. Any interested reader is also welcome to contact us for receiving the original raw data as FASTQ-files.
Sequencing and variant calling
Pooled and indexed PCR products were sequenced on the Illumina MiSeq instrument following standard protocols with the following modifications: Illumina-specific sequencing primers were substituted with a mixture of two Fluidigm-specific primers pairs (FL1 and FL2). The sequences were aligned to the revised Cambridge Reference Sequence (rCRS) using Bowtie2. Assessment of quality control (using Phred scores) and detection of somatic mutations was performed using MitoSeek [34]. Tumor samples, margins, lymph nodes, sputum and serum were compared pairwise with lymphocyte controls. Preparation of pairwise sample input and batch processing of output data was performed using R. Prior to statistical analyses, the data cleaning was performed by: i) checking for missing values, ii) calculation and verification of minor allele frequencies and iii) removing mutations called using insufficient (<100) reads. The following conditions were applied for each position: an event was identified as a somatic mutation only when (i) the position is covered with at least 100 reads in both the tumor and normal; (ii) distinct paired reads contain the mutation in the tumor; (iii) the number of distinct paired reads containing a particular mutation in the tumor is at least 3% of the total distinct read pairs and (iv) the mismatched base is not present in >0.5% of the reads in the matched normal sample. For TCGA-HNSC WES data the following variant calling criteria were used: an event was identified as a somatic mutation when (i) the position is covered with at least 10 reads in both the tumor and normal; (ii) at least 3 unique reads supporting variants in tumor sample with at least 3% variant allele fraction; (iii) the mismatched base is not present in the matched normal sample.
Copy number estimation
For copy number estimation, primer pairs targeting three nuclear genes (RPP30, RPPH1 and TERT) were incorporated into the Access Array mtDNA primers set, and relative rate of amplification of mtDNA encoded TRNL1 sequence was compared to a reference sequence of each one of these three nuclear genes. For each sample, the consensus sequences from each of the paired end reads were obtained using flash. The consensus sequences was then aligned to the genome consisting only of the three genes (RPP30, RPPH1, TERT) using bowtie2. Finally, the total number of reads aligning to either of the three genes was calculated using bedtools. Relative abundance of mitochondrial DNA was measured as a ratio of mtDNA/nDNA (calculated by dividing the mtDNA signal for TRNL1 gene by each one of the corresponding nuclear reference genes).
ddPCR for mutation validation:
Four selected somatic variants identified by NGS were technically validated by ddPCR using BioRad QX100 platform. All ddPCR assays used in this study were designed and optimized to work in the ddPCR system by Bio-Rad. Cross-reactivity with nuclear DNA was considered upon assays design. The Bio-Rad assays IDs are summarized in Supplementary Table 13. Briefly, the ddPCR mixture contains 8 μl of 2 X ddPCR Supermix (Bio-Rad), 400nM of forward and reverse primers, 125nM mutant and wild-tube probe and 2 μl template DNA (10 to 20ng) in each 20 μl reaction. The reactions were loaded into a droplet cartridge and placed in the droplet generator, where vacuum is applied to the cartridge. This draws both the PCR reagents and oil through a flow-focusing nozzle where around 20,000 individual droplets ≈ 1 nl in size are formed, suspended in an emulsion. The emulsion was transferred into a 96 well plate, sealed using a thermal plate sealer and cycled using a C-1000 thermal cycler (Bio-Rad) under the manufacturer’s recommended conditions. Annealing temperature was optimized for each assay. After amplification, the plate was transferred to a droplet reader from which raw fluorescence amplitude data is extracted to the Quantasoft software for downstream analysis.
Results
mtDNA sequencing assay design
For tracking mitochondrial mutagenesis across body sites and within tumor samples, an ultra-deep PCR-based next-generation sequencing (NGS) of the entire mitochondrial genome was established (see Methods section). A schematic of workflow for sequencing and data analysis is shown in Supplementary Figure 1. Primers pairs (149) were designed to cover the entire mitochondrial genome with a dual coverage of 86.2% (Figure 1A, Supplementary Table 1). We used this method to sequence mtDNA from 28 primary HNSCC samples (Table 1). These samples were obtained from patients who did not receive chemotherapy or radiation prior to tumor biopsy. For each patient, matched lymphocyte DNA was used as a control. To assess intra-tumor heterogeneity and spatial distribution of mtDNA mutations in HNSCCs, a multiregional sequencing approach was applied to different zones of histologic progression (superficial zone; mid-zone; deep invasive tumor front) in 14 of the 28 tumor specimens where such specimens were available. (Supplementary Table 2). Overall, we obtained over 97% coverage with a median average depth of 7903 × and 5133 × for HNSCCs or lymphocyte DNA respectively (Supplementary Table 3). A custom bioinformatics workflow (based on MitoSeek [34]) was designed with the following variant calling criteria: an event was identified as a somatic mutation only when (i) the position is covered with at least 100 reads in both the tumor and normal; (ii) the number of reads containing a particular mutation in the tumor is at least 3% of the total distinct read pairs and (iii) the mismatched base is not present in >0.5% of the reads in the matched normal sample. Mutations arising from misplaced genome alignments, including paralogous sequences, were identified and excluded by searching the reference genome. Indels were removed from the analysis, as detecting heteroplasmic indels is unreliable using short-read sequencing [35].
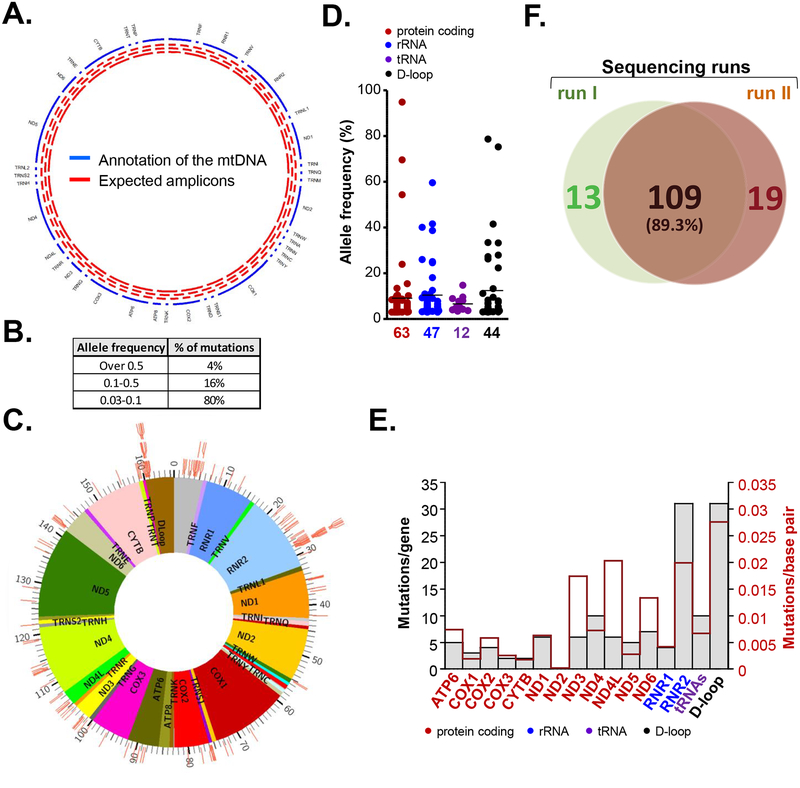
Figure 1. Somatic mutations of the mitochondrial genome in primary HNSCC tumors.
A. Schematic presentation of the assay design. Tracks on the circos plot: blue – annotation of the mtDNA genes, red -expected amplicons. B. Table depicting percentage of detected somatic mtDNA mutations with indicated allelic frequency. C. Circos plot showing the landscape of detected somatic mtDNA mutations across the mitochondrial genome. Each tick represents a mutation at a specific genome location. D. Dot plot depicting minor allele frequency across the functional elements of mt genome. E. Number of somatic mutations in functional regions of the mitochondrial genome (grey bars - absolute numbers; red line - mutations per base pair). F. Venn diagram summarizing mtDNA mutations concurrently detected by two independent sequencing runs.
Table 1.
Socio-demographic data of patients used in this study. Patient’s age, gender, tumor site, TNM classification and HPV status.
| Sample ID | Age at Diagnosis | Gender | Site | Race | Subsite | Stage | T classification | N classification | Tumor HPV (by PCR) | Smoking status (Current, Former or Never) | Pack years | Drinking Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 34 | Male | OC | W | Lateral tongue | III | T2 | N1 | Neg | Never | NA | Nondrinker |
| 2 | 58 | Male | OP | W | Base of tongue | IV | T2 | N2 | Pos | Former | 40-pack-years | Nondrinker |
| 3 | 54 | Male | OC | W | Buccal mucosa | IV | T2 | N2 | Neg | Occasional smoker (since teenage years) | Unknown | Unknown |
| 4 | 67 | Male | L | W | Glottic | III | T3 | N0 | Neg | Former | 10-pack-years | Former infrequent drinker |
| 5 | 57 | Male | OP | W | Soft palate | II | T2 | N0 | Pos | Former | 5 pack-years | Former infrequent drinker |
| 6 | 70 | Male | OC | W | Mandibular alveolus | IV | T4a | N0 | Neg | Current | 60 pack-years | Current heavier drinker |
| 7 | 50 | Male | OC | W | Lateral tongue | I | T1 | N0 | Neg | Former | >15 pack-years | Nondrinker |
| 8 | 86 | Male | OC | Tongue | IV | T1 | N3 | Neg | Former | NA | NA | |
| 9 | 59 | Male | OP | W | Base of tongue | III | T2 | N1 | Pos | Current | 6 pack-years | Nondrinker |
| 10 | 82 | Male | OC | W | Tongue | IV | T2 | N2 | Neg | Former | 75 pack-years | Former regular drinker |
| 11 | 61 | Male | OC | W | Tongue | IV | T4 | N2 | Neg | Former | 20 pack-years | Former regular drinker |
| 12 | 60 | Male | L | W | Supraglottis | IV | T4a | N2 | Neg | Former | 40 pack-years | Current infrequent drinker |
| 13 | 69 | Male | OC | W | Retromolar trigone | IV | T4a | N0 | Neg | Current | 50 pack-years | Current heavier drinker |
| 14 | 68 | Male | L | W | Supraglottis | IV | T3 | N2 | Neg | Former | 90 pack-years | Former regular drinker |
| 15 | 43 | Female | OC | O | Tongue | III | T3 | N0 | Neg | Never | n/a | Nondrinker |
| 16 | 58 | Male | L | W | Supraglottis | IV | T3 | N2 | Neg | Current | 30 pack-years | Current heavier drinker |
| 17 | 47 | Male | OP | W | Tonsil | III | T3 | N2 | Pos | Current | Unknown | Former regular drinker |
| 18 | 84 | Male | OC | B | Floor of mouth | III | T3 | N1 | Neg | Former | >50 pack-years | Nondrinker |
| 19 | 51 | Female | OC | W | Floor of mouth alveolar ridge | II | T2 | N0 | Neg | Current | 16 pack-years | Current heavier drinker |
| 20 | 51 | Male | OC | W | Lateral tongue | I | T1 | N0 | Neg | Never | NA | Current infrequent drinker |
| 21 | 64 | Female | OP | W | Base of tongue with involvement of larynx | IV | T4a | N2 | Neg | Current | >50 pack-years | Current light drinker |
| 22 | 59 | Female | OC | W | Lateral tongue | III | T2 | N1 | Neg | Current | >50 pack-years | Current heavier drinker |
| 23 | 56 | Male | OC | B | Mandibular alveolus | IV | T4a | N2 | Neg | Current | 10 pack-years | Current moderate drinker |
| 24 | 63 | Male | OC | A | Retromolar trigone | I | T1 | N0 | Neg | Former cigarette smoker/chewed betel for 40 years | Unknown | Former regular drinker |
| 25 | 54 | Female | OC | W | Buccal mucosa/retromolar trigone | IV | T2 | N2 | Neg | Former | Unknown | Current light drinker |
| 26 | 76 | Male | OC | B | Floor of mouth | III | T2 | N1 | Neg | Current | 20 pack-years | Current moderate drinker |
| 27 | 48 | Male | OC | W | Oral cavity with invasion to mandible | IV | T4 | N2 | Neg | Never | NA | Nondrinker |
| 28 | 77 | Female | OC | W | Lateral tongue | I | T1 | N0 | Neg | Current | Unk nown | Unknown |
OC = oral cavity, OP = oropharynx, H = hypopharynx, L = larynx, and NA = not available
A = Asian/Pacific Islander, B = Black/African-American, W = White, O = Other
Occasional smoker: An adult who has smoked at least 100 cigarettes in his or her lifetime, who smokes now, but does not smoke every day. CDC/National Center for Health Statistics https://www.cdc.gov/nchs/nhis/tobacco/tobacco_glossary.htm
Lifetime abstainer – Fewer than 12 drinks in lifetime.
Former infrequent drinker – Fewer than 12 drinks in any one year and no drinks in past year.
Former regular drinker – At least 12 drinks in any one year in lifetime but no drinks in past year.
Current infrequent drinker – 1–11 drinks in past year.
Current light drinker – At least 12 drinks in the past year but 3 drinks or fewer per week, on average over the past year.
Current moderate drinker – More than 3 drinks but no more than 7 drinks per week for women and more than 3 drinks but no more than 14 drinks per week for men, on average over the past year.
Current heavier drinker – More than 7 drinks per week for women; more than 14 drinks per week for men, on average over the past year.
CDC/National Center for Health Statistics https://www.cdc.gov/nchs/nhis/alcohol/alcohol_glossary.htm
Nondrinker: An adult who does not currently drink or have a known history of drinking.
Somatic mutations of the mitochondrial genome in primary HNSCC tumors
166 somatic point mutation (132 unique) were detected in 14 (50%) of 28 tumors (Supplementary Table 4). Sixty one of the codon specific mutations detected in this study were reported in various hematopoietic and solid malignancies including HNSCC by several large pan-cancer NGS analyses (Supplementary Table 5), supporting their status as legitimate somatic mutations with potential functional implications [30, 36, 37]. In agreement with the previous report, most of the mutations were G > A (47%) and C > T (37%) base transitions [33]; these mtDNA substitutions are known to be common in solid malignancies and are associated with DNA damage due to increased mitochondrial reactive oxygen species (mtROS) (Supplementary Table 6) [29, 38]. The mutations were largely heteroplasmic in nature; allelic frequency greater than 50% was only observed in 4% of mutations (Figure 1B). Although the distribution of the mutations spanned the entire mitochondrial genome and no obvious hot spots were detected (Figure 1C), we identified 10 specific positions (5 in RNR2 and 5 in the D-Loop) mutated in two (7%) patients each (Supplemental Table 7). Eight of these mutations were not reported as known polymorphisms in mtDB and mitomap databases and 4 were previously identified in multiple solid neoplasms (Supplemental Table 7). While further validation in larger HNSCC cohorts is required, this observation may suggest that at least some of these revolving clonal events are not random, but influenced by cancer specific mutational process. Within functional regions, 63, 47, 12 and 44 mutations were located in protein coding (68% non-synonymous mutations including 2 stop codons), rRNA coding, tRNA coding and regulatory regions (D-loop) respectively (Figure 1D, Supplementary Figure 2). Within the coding area, higher mutation accumulation was found in RNR2, followed by ND4, ND6, ND1, ND3, ND4L and ATP6. Taking the different region lengths into account, mutation to base pair ratio was highest in the D-Loop, RNR2 and ND4L, followed by ND3 (Figure 1E).
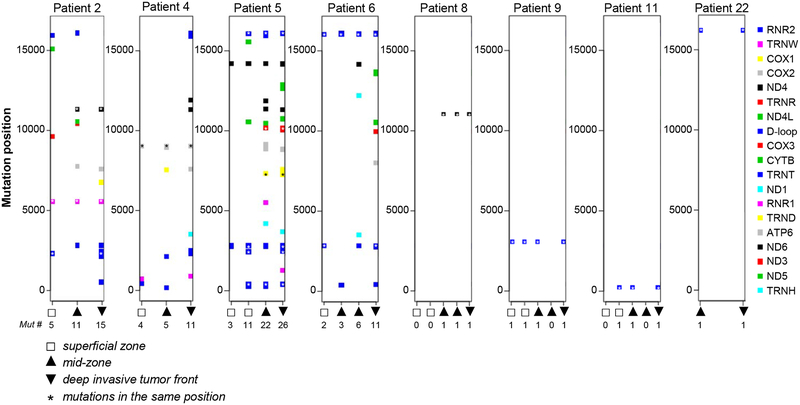
To validate the reproducibility of variant calling, new libraries were prepared from DNA collected from all zones of the 4 tumors with high (patients 2, 4, 5 and 6) and 1 tumor with low (patient 3) number of mutations. These libraries were sequenced and mtDNA variants were assessed independently. The analysis has confirmed 89.3 % of the unique variants, and the prevalence of the mutant reads were very consistent between the two independent sequencing runs, supporting the reproducibility of this high-throughput library preparation approach (Figure 1F, Supplementary Table 8). Out of 14 tumors with multiregional sequencing, mtDNA aberrations were detected in 10 neoplasms. Interestingly, while sequencing of samples collected from different zones within the same HNSCC tumor revealed a large degree of heterogeneity among mutated mitochondrial foci, clonal events (mutations present in more than one zone) were detected in 8 patients with multi-regional collections (Figure 2, Supplementary Table 4), suggesting that they may occur early in tumor evolution, likely even before tumorigenesis. Interestingly, tumors with highest mutations number (such as patient 2, 4, 5 and 6) showed a progressive increase in mutation numbers from superficial to deep invasive tumor zone, with one zone demonstrably sub-clonal to the other (Figure 2).
Figure 2. Intratumor heterogeneity of mtDNA mutations in primary HNSCC lesions.
Eight tumors with multi-regional sequencing (3 or 5 zones per tumor) in which clonal events (mutations present in more than one zone) were detected are shown. Each color quadrant represents somatic mutation detected in a particular gene (see color code on the right side of the panel). Genomic position of each mutation in indicated on axis Y. Axis X - white quadrants indicate tissues collected form superficial zone, up-pointed triangles indicate tissues collected from the tumor mid zone, down-pointed triangles indicate specimens collected from the deep invasive front. Number of mutations detected in each specimen are indicated beneath each zone. Asterisks indicate mutations detected in the same genomic position.
Comparing the landscape of heteroplasmic mtDNA mutations with TCGA-HNSC dataset
Although most of the currently available whole-exome capture kits do not target mtDNA genes, it was previously reported that due to the abundance of mtDNA in human cells, off-target reads derived from the mitochondrial genome could be used to identify mtDNA mutations in WES data obtained from TCGA [30]. To validate the distribution of mutations across genetic regions observed in our study, we analyzed off-target mtDNA sequences extracted from the WES data of tumors and matched lymphocytes obtained from TCGAHNSC database. Confirming previous report [30], the off-target coverage was not uniform across the mitochondrial genome in exome studies (mean coverage - 69%) and significantly lower than coverage from our in-house platform (>95%) (Supplementary Figure 3A). Expectedly, the average read depth was quite low (67X) compared to our targeted sequencing approach. Thus, the variant calling criteria was modified for WES analysis. An event was identified as a somatic mutation when (i) the position is covered with at least 10 reads in both the tumor and normal; (ii) at least 3 unique reads supporting variants in tumor sample with at least 3% variant allele fraction; (iii) the mismatched base is not present in the matched normal sample (see Methods for more details). Using these criteria, we have detected 376 somatic mutations in 60 of 521 TCGA-HNSC cases. Although the direct comparison of the WES and targeted sequencing data should be approached with caution, the dispersion of somatic mutations across the mitochondrial functional regions and base transitions in our samples was similar to that seen in TCGA tumors by our (Supplementary Figure 3B and C) and other’s [33, 37] analysis, providing validation of our local cohort results. That said, it is important to note that decreased depth in the WES data may also lead to some degree of MAF overestimation; lower number of total reads would cause large changes in MAF even with small increases in mutant allele read number. We believe that this further emphasizes the need for the new high-depth mtDNA sequencing technologies that may accurately detect low-level heteroplasmies.
Spatial distribution of mtDNA abundance in HNSCC
Although changes in mtDNA content is a useful clinical biomarker for various types of cancers including HNSCC [27, 39, 40], results are controversial as several analytical factors can affect accurate copy number (CN) measurement. mtDNA is commonly quantified by calculating a ratio between a target mitochondrial gene and a reference nuclear gene using quantitative real time PCR. Often, these measurements are made in separate experiments. To overcome these technical limitations, we validated and optimized primer pairs for three nuclear housekeeping genes located at a highly conserved region of the genome (RPP30, RPPH1 and TERT) and incorporated these primers to the mtDNA primers set, allowing for quantitative copy number analysis. To test the reproducibility of the three targets, relative abundance of mtDNA was calculated in 28 HNSCC cases by dividing the signal for mtDNA encoded TRNL1 (MT-TL1) gene by each one of the corresponding nuclear reference genes (mtDNA/nDNA ratio). The TRNL1 was chosen because of the rarity of deletions in this region and absence of nuclear pseudogenes [41–44]. Since all samples have been processed in identical manner, the sample with a higher mtDNA/nDNA value contains more copies of mtDNA. This approach eliminates the variability that results from assaying nuclear and mitochondrial targets separately. In our HNSCC dataset, all 3 ratios produced consistent values in each individual sample and comparable distribution of relative mtDNA abundance (Figure 3A), suggesting that these assays are equally suitable for assessing mtDNA copy number changes. Based on these results, the average of 3 mtDNA/nDNA ratios was used for further analysis.
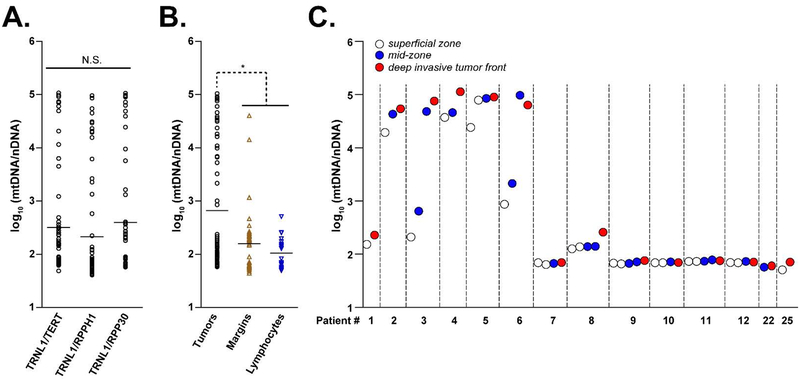
Figure 3. Spatial distribution of mtDNA abundance in HNSCC.
A. Comparison of mtDNA copy number index calculated using three nuclear-encoded housekeeping genes. All 3 ratios produced comparable (non-significant) distribution of relative mtDNA abundance. B. Relative mtDNA abundance (calculated as average of 3 mtDNA/nDNA ratios) in tumor samples (black circles), matched histologically negative surgical margin specimens (brown up-pointed triangles) and lymphocytes (blue down-pointed triangles). C. mtDNA copy number index in 14 HNSCC tumors with multiregional sequencing. White circles indicate tissues collected form superficial zone, blue circles indicate tissues collected from the tumor mid zone, red circles indicate specimens collected from the deep invasive front.
We next applied this method to estimate the relative mtDNA abundance in 42 matched histologically negative surgical margin specimens available for 18 of the 28 tumors analyzed. We observed a significant increase in mtDNA/nDNA ratios in invasive tumors compared to margins/lymphocytes (Figure 3B), supporting previous reports that increase in mitochondrial content is associated with HNSCC progression [28]. Although analysis of spatial heterogeneity in 14 tumors with multiregional sequencing revealed that mtDNA abundance did not differ substantially between the different zones in most of the patients (besides patient 3 and 6), a trend towards mtDNA content increase was seen in deep invasive tumor front section in 8 (# 1, 2, 3, 4, 5, 6, 8, and 25) of 14 patients (Figure 3C).
Detection of tumor-specific mtDNA alterations in matched LNs, margins and bodily fluids
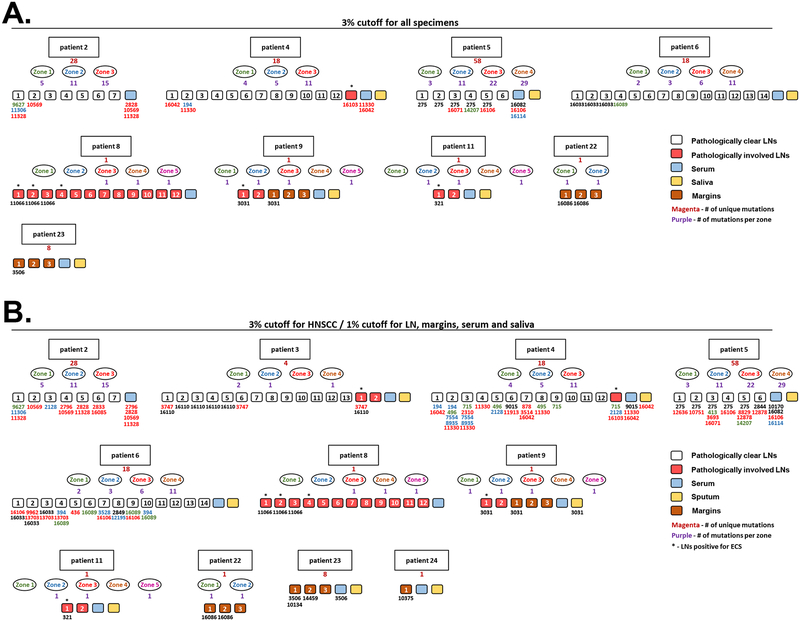
High copy number per cell and low degradation level make mtDNA mutations a promising tool for assessing the presence of tumor-specific genetic aberrations in non-cancerous tissues such as surgical margins, LNs and bodily fluids. As most of the tumor-specific mtDNA mutations are heteroplasmic in nature, new high-throughput approaches that may accurately detect low-level heteroplasmic variants are eminently needed. To deploy our mtDNA sequencing workflow to non-cancerous specimens, we carefully reviewed the surgical pathology files at Johns Hopkins Hospital and identified matching LNs, surgical margins and bodily fluids collected at the time of surgery from 14 patients with at least one mtDNA mutation in their primary tumor. We used the same library preparation platform and mutation calling criteria to sequence mtDNA from 73 LNs (21 histologically metastatic and 52 clear nodes) isolated from 10 patients, 15 histologically uninvolved margins from 7 patients, as well as 10 sputum and 12 serum specimens from 10 and 12 patients respectively (Supplementary Table 9). Similarly to the primary tumor samples, we have focused on variants called with an allele fraction of ≥3%, to reduce potential false-positive calls of mtDNA somatic mutations. Overall, 98% of reads mapped to the target mtDNA genome and average depth was 3396X across all samples tested, ranging from 4146X for LNs and 1556 for sputum specimens (Supplementary Table 3). The somatic mutations detected in these samples are summarized in Supplementary Table 10. Tumor-specific mtDNA mutations were detected in 6 (29%) histologically metastatic LNs resected from 4 patients (Figure 4A,Supplementary Table 11). Notably, of these nodes, 5 (83%) were positive for extracapsular spread (ECS), whereas only 3 of 15 (20%) postoperative metastatic LN specimens in which tumor-specific mtDNA was not detected were positive for ECS, suggesting that increased spreading of the tumor cells may contribute to higher detection rate of the tumor associated mutations. As routine analyses for metastases within LNs from neck dissection specimens is typically limited to histologic examination of one or only a few sections of each node, the failure to detect presence of tumor-associated mutations in all involved lymph nodes may be due to irregular dissemination of tumor cells in the specimens available for the analysis. Four patients carried tumor-specific mutations in 19 (36%) LNs (often multiple) that were histologically free of atypical cells (Figure 4A, Supplementary Table 11), suggesting that involvement of these specimens could have been undetected by routine diagnostic methods. Furthermore, tumor-derived mutations were detected in 4 (27%) histologically uninvolved margins from 3 patients (even though margins specimens were only available for patients with low number of mtDNA mutations in their primary malignancy), as well as in 3 (25%) serum specimens. No tumor-associated mutations were detected in sputum specimens, which may be attributed to the presence of high-DNase activity and subsequent DNA degradation in the sputum [45]. Of note, somatic mutations that were not seen in the primary tumors were detected in a subset of the sequenced LNs, serum and sputum specimens Supplementary Table 10. While the nature of these mutations remains to be investigated, these variants could have been missed due to the undersampling of the heterogeneous primary tumor, or may result from the clonal evolution within the secondary site. Notably, we have detected a substantially higher number of tumor-specific mutations when detection threshold for somatic variants in LNs, margins and serum specimens was lowered to 1% (Figure 4B, Supplementary Table 12). Additionally, tumor specific mutations were detected in 2 sputum samples. While it is possible that these mutations represent alignment artifacts and workflow errors, given the high average sequencing depth of mtDNA (4146X in LNs), a 1% minor allele frequency represents a mutant allele read count of at least 40. Thus, we hypothesize that these mutations are likely true and not a sequencing artifact.
Figure 4: Detection of tumor-specific mtDNA alterations in matched LNs, margins and bodily fluids.
A. Figure summarizes cancer-associated mutations among different lesions collected form the same patient (only tumors in which cancer associated mutations were found in LNs, margins or bodily fluids are shown). From top to bottom - rectangular shapes indicate patient’s number, magenta number beneath the rectangular shape indicates a number of unique somatic mutations detected in this HNSCC malignancy, ovals represent different zones collected for this patient, purple numbers beneath the ovals depict the amount of mutations detected in each distinct tumor zone. Small quadrats indicate different specimens collected from the same patients: white – pathologically clear LNs, red – pathologically involved LNs, cyan – serum, yellow – sputum, brown – surgical margins. Numbers beneath the small quadrats indicate the positions of tumor associated mutation detected in this specimens (the color code indicates the zonal origin of this mutation. Black numbers indicate clonal mutations). Asterisk indicates metastatic LNs positive for extracapsular spread. 3% detection cutoff was used for mutations calling in all samples (primary HNSCC tumors, LNs, margins, serum and sputum. B. Same as (A), however 1% cutoff was used for variants calling in LN, margins, serum and sputum.
Validation of selective mtDNA mutations with ultra-sensitive ddPCR
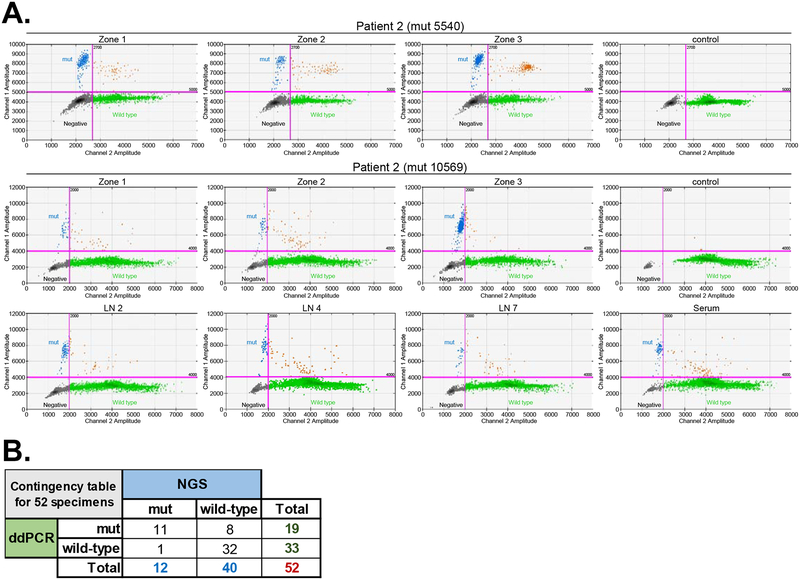
As the detection of true heteroplasmic variants requires a high degree of sensitivity, we next utilized ultra-sensitive ddPCR to further validate the accuracy of the mtDNA sequencing approach. Due to its high sensitivity, ddPCR can detect heteroplasmies with minor allele frequency (MAF) > 0.2% [46], and might be considered as a gold-standard method for detecting the tumor-associated mutations in research settings [47]. Four ddPCR assays were designed by Bio-Rad laboratories to probe cancer-associated mtDNA substitutions detected by NGS in 3 different patients (substitution at the positions 5540 and 10569 in patient 2, position 16103 in patient 4 and position 16082 in patient 5) (Supplementary Table 13). MAF of the selected mutation detected in these patients by NGS ranged from 1.3% to 94.8%. Eleven of 12 (91.6%) of the mtDNA mutations detected by NGS (including variants reported in LNs and one serum sample) were correspondingly confirmed by ddPCR (Figure 5A and 5B, Supplemental Table 14). Altogether, of the 52 samples assessed by both NGS and ddPCR, 44 (84.6%) were detected as either positive or negative by both platforms. Notably, despite the high concordance (Figure 5B), ddPCR detected low-frequency mtDNA mutations in 8 additional specimens that were considered wild-type by NGS analysis, probably due to the overall higher sensitivity of the ddPCR assay.
Figure 5. Validation of selective mtDNA mutations with ultra-sensitive ddPCR.
A. ddPCR validation of 2 mutations (at positions 2240 and 10569) detected in primary tumor of patient #2 is shown as an example. DNA samples extracted from all specimens collected form patient #2 (3 distinct primary tumor zones, 7 LNs, serum and lymphocytes) were probed with two different ddPCR assays specifically designed for detecting each of the above substitutions. Only specimens were mutation was detected are shown. Blue dot clusters indicate mutation detected by the specific assay. Black dot cluster indicates empty droplets. Green clusters indicate droplets containing wild-type alleles. Brown clusters indicate droplets containing both wild-type and mutant alleles. B. Contingency table depicts the concordance between selected mutation detection by NGS and ddPCR.
Discussion
mtDNA mutations have been increasingly reported in a wide variety of cancers, including HNSCC [13, 17–21]. While the potential role of cancer associated somatic mutations of the mitochondrial genome is poorly understood, it is becoming apparent that some genetic alterations in the mtDNA contribute to tumorigenesis. Moreover, multiple studies indicated that alterations in mtDNA copy number also correlate with increased cancer risk [27, 48, 49], contributing to a growing acknowledgment of cancer as a mitochondrial metabolic disease.
Due to their clonal nature, high copy number, and circular structure, mitochondrial mutations could provide a powerful molecular marker for detection of cancer cells in surgical margins, and other minute cellular samples such as biopsies and micrometastatic lesions. Detecting micrometastasis and margin involvement is a major challenge using traditional histopathologic examination, and inadequate detection of tumor in these samples may be contributing to increased morbidity/mortality due to inadvertent down-staging of the underlying tumor. The small volume of the biopsied tissue and limited number of histopathological samples might reduce the rate of accurate diagnosis even by highly sensitive molecular technologies. Therefore, there is a pressing need for a rapid and cost-effective mtDNA mutation profiling system in patient specimens, which would also allow tracing the changes in cell-free mitochondrial DNA (cfmtDNA) in bodily fluids during the course of the disease or cancer treatment. Although recently emerged NGS-based approaches offer promising capabilities in detecting heteroplasmic mtDNA changes, their use as a modality for routine application has recognized limitations. Enrichment of mtDNA by long-range PCR generates large amplicons and impedes the analysis of fragmented specimens such as FFPE and bodily-fluids samples [50]. Amplification of amplicon libraries with several primer pools in separate reactions increases workflow complexity and elevates the cost [31, 32]. Whereas probe capture enrichment methodology is more expensive, labor-intensive and requires longer turnaround time [51]. These challenges motivated us to develop an ultra-deep amplicon-based multiplex approach for rapid sequencing of the entire mitochondrial genome and simultaneous assessment of mtDNA copy number that overcomes these complications and enables analysis of micro-sized biopsies and highly degraded material (see Materials and Method).
To test this technology, we applied it on DNA extracted from the 165 tissue samples (including primary tumors, metastatic and histologically clear lymph nodes, resection margins, blood and sputum) collected from 28 HNSCC cancer patients, disease notorious for poor prognosis and high locoregional recurrence rate [52]. DNA extracted from matched frozen lymphocytes was used as a control. Overall, 98% of reads mapped to the mtDNA reference sequence and average depth was over 2000X across all tested specimens. Notably, sequencing of the paraffin-embedded specimens that were available for 2 HNSCC neoplasms, demonstrated only a slight elevation in mutation load compared to the DNA extracted from the fresh frozen material (not shown). Furthermore, nearly 98% of mutations were concurrently detected in two parallel sequencing runs, supporting the high reproducibility of this library preparation method.
Analysis of large datasets, such as the International Cancer Genome Consortium and TCGA, suggested that approximately 60% of all solid tumors bear at least one mtDNA mutation [30]. Consistently with these observations, we have identified mtDNA mutation in 50% of HNSCC tumors sequenced in our study. Of the 166 somatic mutations detected across patients in this study, the majority (38%) were located in protein coding area and were enriched for the non-synonymous aberrations, supporting previous observations in multiple cancer types including HNSCC [29, 33]. These pathogenic coding alterations may potentially result in respiratory chain deficiency, promoting tumorigenesis [29, 53]. Indeed, somatic mutations in multiple mitochondrial genes have been strongly linked with tumor formation [13, 15, 18, 21, 54–56]. For instance, the majority of somatic mutations in the coding regions in this work and other studies occurred in subunits of complex-I (NADH dehydrogenase; genes ND1–6 and ND4L) [57, 58]. This complex serves as an entry point for electrons donated from NADH to the electron transport chain (ETC), and is critical for proper ATP production via oxidative phosphorylation. Loss of ND function may promote mtROS production and decrease electron flux via the ETC, which has been associated with an oncogenic phenotype [57–59]. Similarly, mutations in the cytochrome oxidase and ATP synthase proteins found in our dataset could also be effecting tumorigenesis by exacerbating mitochondrial dysfunction via loss of ETC efficiency [57]. However the exact functional role these mutations play in cancerogenesis is still incomplete. This is in part due to technical limitations; the efficacy of nucleic acid delivery into the mitochondria is low [60], making manipulation of the mitochondrial genome very difficult. Several recent approaches developed to improve DNA import into mammalian mitochondria [60] or to manipulate mitochondrial genes inside the organelle (such as mitochondria-adapted CRISPR/Cas9 platform) [61] are promising but still require validation and optimization. As the current technical limitations of these methods are overcome [62], our understanding of the biological role of mtDNA mutations in tumorigenesis will grow. Somatic mutations in non-coding regulatory D-loop region (which play role in mitochondrial replication) and RNA coding genes (which play role in translation of all 13 proteins encoded by the mitochondrial genome) may also result in altered mitochondrial function and have an impact on head and neck cancer progression [21, 28, 63, 64]; however their clinicopathological significance is still controversial [12, 63, 65–68] and the exact mechanism of how alterations in these domains contribute to malignant transformation remains to be elucidated.
Heteroplasmic somatic changes in mitochondrial genome might also serve as a sensitive tool for deciphering spatial tumor evolution. Several recent studies have used multiregional sequencing approach to analyze the spatial heterogeneity of mtDNA in lung, hepatocellular and colorectal cancer [69, 70]. Similar to the observations obtained in our study, there was a substantial inter-tumor variability, with the majority of discovered somatic mutations restricted to specific areas of the heterogeneous lesions, and only a fraction of variants detected across all segments collected from the same tumor. Though limited by sample size, an association between higher mutational burden and mtDNA content in invasive tumor front sections was observed in our study. This is supportive of the branched evolutionary growth of HNSCC [71]. It is also important to note that the majority of prior studies investigating cancer associated mtDNA mutations have used data derived from the bulk tumor sequencing, which may result in potential underestimation of the true mutational spectrum in heterogeneous lesions. Larger confirmatory studies using longitudinal samples (that allow tracking a clonal population over time) and precise calculation of the percentage of abnormal cells containing each variant are warranted to validate our observations and provide further insight into mechanisms of evolutionary tumor growth.
Our group and others have reported that mtDNA-CN levels, a critical component of overall mitochondrial health, was increased with stages of HNSCC progression and negatively correlated with patient survival [27, 40]. While these observations suggest that mtDNA-CN is a useful clinical biomarker for disease risk stratification, the current methods for relative mtDNA content assessment employ calculation of a ratio between mitochondrial and nuclear encoded genes measured in two separate PCR experiments, which can affect the accurate quantification. In contrast, the method optimized in this study allows sequencing of the entire mtDNA genome as well as simultaneous quantification of target and reference genes in a single amplification reaction, thus improving correct quantification and ensuring scalability and translational ability of the assay. In agreement with previous reports [28], our method revealed higher level of relative mtDNA content in invasive HNSCC tumors compared to matched histologically negative surgical margin specimens, confirming the success of our multifunctional sequencing approach. Interestingly, in a subset of samples with multi-regional sequencing we have observed a trend towards progressive increase in relative mtDNA-CN level from superficial to deep invasive tumor zone, which coincided with a trend towards higher mutational load in these specimens. While we could not conduct statistically meaningful association analyses due to the relatively small number of samples with multiregional sequencing, it is tempting to hypothesize that defects in the intrinsic DNA repair machinery or loss of efficient clearance of disease mitochondria (i.e. mitophagy) could lead to increased copies of defective mtDNA nucleoids per cell, thus increasing copy number. Alternatively, a higher mtDNA-CN may also be a consequence of cell compensation for defective oxidative phosphorylation and lower ATP production per mitochondria. While our results may be indicative of the potential role of mtDNA content increase in the transition to invasive disease, further studies in larger patients’ cohorts are necessary to confirm the biological relevance of this observation.
In order to accurately call low-level somatic mtDNA variants, paired lymphocyte DNA has been used as a source of normal DNA control for the analysis of our “in house” and TCGA HNSC samples, since these cells are assumed to be the authentic germline mitochondrial genome free of tumor-specific genetic changes. Matched tumor adjacent tissues have been used as ‘normal DNA’ reference in several recent studies analyzing mitochondrial genome extracted from TCGA [30, 37]. However such histologically normal material may sustain somatic mtDNA changes (field cancerization), thus confounding somatic mutation calling. For instance, using lymphocyte controls, our analysis detected evidence of somatic mutations in several histologically “clean” samples. Longitudinal studies are needed to determine whether detection of such mutations in histologically normal specimens portends recurrence at nearby sites at a future timepoint. Therefore, comparison to the mtDNA profile of a distant, uninvolved source (blood lymphocytes) is necessary to accurately call somatic heteroplasmy mutations. Supporting previous reports in various solid malignancies including HNSCC [30, 33], only a fraction of the somatic variants among the tumor samples in our cohort were high-level heteroplasmies. As the current hypothesis is that the emergence of discernible effect on mitochondrial physiology requires mutant allelic frequency to exceed 50%, only a minority of somatic mtDNA variants in HNSCC are likely to be bona fide cancer drivers. Although the phenotypic consequences of low-level heteroplasmies is controversial and remains to be experimentally validated, recent evidence indicate that accumulation of low level heteroplasmic mutations may be associated with tumorigenesis [37, 72]. Furthermore, due to their high number, presence in bodily fluids and rapid development of highly sensitive sequencing techniques, these low-level tumor-specific mtDNA mutations may serve as an early indication of potential cancer development and may represent a means for tracking tumor progression and monitoring treatment response non-invasively [72].
Ultra-sensitive ddPCR method was recently used for detecting rare heteroplasmic mutations in mtDNA [46, 73]. Therefore, to further evaluate the NGS accuracy we used ddPCR assays to assess 52 specimens collected from 3 patients for the presence of 4 mtDNA mutations detected by NGS. While ddPCR confirmed presence or absence of the mutant allele with 82.7% concordance, NGS approach appears to be more prone to false negatives, as 8 specimens (15.4%) discovered as positive by ddPCR were reported as wild-type by NGS. As both assays were performed using the same DNA extraction aliquot to minimize effects of intratumoral heterogeneity, these differences may be attributed to the higher sensitivity of the ddPCR approach. However, despite its very high sensitivity, ddPCR platforms include two fluorescence filters which support only duplex reactions. While multiplexing up to 10-plex is possible with some optimization and specific reaction setups, ddPCR platform is not yet suitable for screening of a large number of mtDNA mutations due to its high cost and difficulties associated with assays design for multiplex purposes. On the other hand, the NGS method described here allows rapid sequencing of the 48 samples in one multiplex reaction at relatively low cost. Furthermore, we have recently adjusted our protocol for ultra-deep mtDNA sequencing using the Fluidigm Juno microfluidics platform, allowing parallel sequencing of up to 192 samples. Although further analyses using larger sample cohorts are warranted to better assess sensitivity, specificity and reproducibility of the assay as a function of the heteroplasmy level, these results demonstrate the potential of amplicon based NGS method to detect low-prevalence heteroplasmies in primary tumors, LNs and bodily fluids down to the 1% level.
There is evidence that lower levels of heteroplasmy of certain mtDNA mutations modulates DNA replication, glucose metabolism and lifespan in mice [74] and significantly increases the risk of age-related chronic diseases [75, 76]. However, to date, little is known about the role of mtDNA heteroplasmies in human cancer. Current research on mtDNA heteroplasmy is significantly hindered by the lack of sensitive high throughput methods capable to accurately detect low-level mtDNA variants. Therefore, development of sensitive sequencing approaches for heteroplasmy detection are necessary for studying the association between mitochondrial mutations and malignant diseases.
Taken together, our results indicate the potential of the amplicon-based NGS platform described in this study as an accurate, simple, rapid and cost-effective method suitable for high throughput mtDNA analysis in primary tumor tissues, lymph nodes as well as in highly degraded bodily fluids. These observations provide a foundation for further advancing its development for risk assessment, early cancer detection, and tumor surveillance in HNSCC and other solid malignancies.
Supplementary Material
Supplementary Table 4. Detailed list of all somatic mtDNA mutations found in primary HNSCC specimens.
Supplementary Table 13. Summary of ddPCR assays designed by Bio-Rad.
Supplementary Table 14. Summary of the ddPCR analysis results in specimens collected from patients 2, 4 and 5.
Supplementary Table 1. List of 148 primer pairs designed and used in this study for mtDNA amplification. Table summarizes the GC-content, length distribution and location of the Access Array amplicons.
Supplementary Table 2. Table summarizes the number of zones collected and sequenced for each HNSCC lesion.
Supplementary Table 3. Table lists average coverage and sequencing depth for indicated specimen types.
Supplementary Table 5. List of somatic mtDNA mutations found in HNSCC tumors that have been previously observed in solid tumors (including HNSCC).
Supplementary Table 6. Spectrum of base-pair substitutions in HNSCC tumors.
Supplementary Table 7. Table summarizes mutations detected in more than one HNSCC patient.
Supplementary Table 8. List of somatic mtDNA mutations detected by two independent sequencing runs in patients 2, 3, 4, 5 and 6. Red – mutations detected in both runs, black – mutations detected by only one sequencing run, blue – mutations that were detected by the first run but were found in additional tumor zones upon resequencing.
Supplementary Table 9. Table summarizes all match specimens collected from the same patient.
Supplementary Table 10. List of all somatic mtDNA mutations detected in LNs, margins, serum and sputum specimens. Tumor-associated mutations (variants that were also detected in the match primary tumor specimens) are highlighted in bold.
Supplementary Table 11. Detailed list of tumor-associated mtDNA mutations detected in LNs, margins and serum specimens at 3% variant calling threshold.
Supplementary Table 12. Detailed list of tumor-associated mtDNA mutations detected in LNs, margins and serum specimens at 1% variant calling threshold.
Supplementary Figure 1. Schematic of workflow for sequencing and analysis of somatic mitochondrial mutations in HNSCC samples. Briefly, gDNA from samples and matched control specimens are extracted and amplified as described in Methods. Following library preparation and next-generation sequencing, the reads are aligned. The mitochondrial (MT)-targeted reads are extracted for further analysis. CN calculations are performed using the nuclear reads as described in Methods. Additional quality control steps are taken prior to somatic mutation discovery using MitoSeek. Data cleaning including: calculation of MAF using mutant allele read counts, assessment of adequate depth, annotation of D-Loop and non-coding region reads based on base position, and removal of duplicate entries was performed in R. Subsequent to this, somatic mutations from each sample (and matched control) was associated with the proper clinical sample to obtain group level data and perform statistics.
Supplementary Figure 2. Box plot represents distributions of mutant allele frequencies (MAFs) of somatic mutations detected in primary HNSCC tumors across the functional mtDNA regions.
Supplementary Figure 3. Analysis of WES data derived from TCGA-HNSC dataset. A. Average coverage. B. Mutational distribution across the mitochondrial genome. C. Frequency of base pair transitions.
Highlights:
We develop an ultra-deep approach for sequencing of the mitochondrial genome.
We sequenced 28 HNSCC tumors and matched lymph nodes, margins, blood and sputum.
Somatic mutation were detected in 50% of the tumors.
Clonal events were detected in patients with multi-regional sequencing.
mtDNA content was increased in invasive tumor front section in a subset of patients.
Tumor-specific mutations were detected in matched LNs, margins and bodily fluids.
Selective mtDNA mutations were validated with ultra-sensitive ddPCR.
Acknowledgements
This work was supported by NIDCR/NIH grant R01DE027809 (E.I.) and Specialized Programs of Research Excellence in Human Cancers (SPORE) P50DE019032 (E.I.). A.D.S. was supported by the Swiss Cancer League grant BIL KLS-3649-02-2015.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflict of interest.
References
- [1].Kamangar F, Dores GM, Anderson WF, Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world, J Clin Oncol, 24 (2006) 2137–2150. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Siegel R, Xu J, Ward E, Cancer statistics, 2010, CA Cancer J Clin, 60 (2010) 277–300. [DOI] [PubMed] [Google Scholar]
- [3].Parkin DM, Pisani P, Ferlay J, Global cancer statistics, CA Cancer J Clin, 49 (1999) 33–64, 31. [DOI] [PubMed] [Google Scholar]
- [4].Argiris A, Karamouzis MV, Raben D, Ferris RL, Head and neck cancer, Lancet, 371 (2008) 1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].John Andrew Ridge RM, Miriam N Lango, Steven Feigenberg, Head and Neck Tumors, Haller Daniel G., Wagman Lawrence D., Camphausen Kevin A., Hoskins William J. (Eds) Cancer Management: A Multidisciplinary Approach., ed. 2013. (2013). [Google Scholar]
- [6].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2015, CA Cancer J Clin, 65 (2015) 5–29. [DOI] [PubMed] [Google Scholar]
- [7].Leemans CR, Braakhuis BJ, Brakenhoff RH, The molecular biology of head and neck cancer, Nature reviews. Cancer, 11 (2011) 9–22. [DOI] [PubMed] [Google Scholar]
- [8].Hunter KD, Parkinson EK, Harrison PR, Profiling early head and neck cancer, Nature reviews. Cancer, 5 (2005) 127–135. [DOI] [PubMed] [Google Scholar]
- [9].Bettendorf O, Piffko J, Bankfalvi A, Prognostic and predictive factors in oral squamous cell cancer: important tools for planning individual therapy?, Oral oncology, 40 (2004) 110–119. [DOI] [PubMed] [Google Scholar]
- [10].Prado K, Zhang KX, Pellegrini M, Chin AI, Sequencing of cancer cell subpopulations identifies micrometastases in a bladder cancer patient, Oncotarget, 8 (2017) 45619–45625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meder L, Konig K, Fassunke J, Ozretic L, Wolf J, Merkelbach-Bruse S, Heukamp LC, Buettner R, Implementing amplicon-based next generation sequencing in the diagnosis of small cell lung carcinoma metastases, Experimental and molecular pathology, 99 (2015) 682–686. [DOI] [PubMed] [Google Scholar]
- [12].Challen C, Brown H, Cai C, Betts G, Paterson I, Sloan P, West C, Birch-Machin M, Robinson M, Mitochondrial DNA mutations in head and neck cancer are infrequent and lack prognostic utility, British journal of cancer, 104 (2011) 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dasgupta S, Koch R, Westra WH, Califano JA, Ha PK, Sidransky D, Koch WM, Mitochondrial DNA mutation in normal margins and tumors of recurrent head and neck squamous cell carcinoma patients, Cancer prevention research, 3 (2010) 1205–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Koc EC, Haciosmanoglu E, Claudio PP, Wolf A, Califano L, Friscia M, Cortese A, Koc H, Impaired mitocondrial protein synthesis in head and neck squamous cell carcinoma, Mitochondrion, 24 (2015) 113–121. [DOI] [PubMed] [Google Scholar]
- [15].Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA, Mitochondrial mutations contribute to HIF1alpha accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenease kinase 2 in head and neck squamous cell carcinoma, Clinical cancer research: an official journal of the American Association for Cancer Research, 15 (2009) 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mithani SK, Taube JM, Zhou S, Smith IM, Koch WM, Westra WH, Califano JA, Mitochondrial mutations are a late event in the progression of head and neck squamous cell cancer, Clinical cancer research: an official journal of the American Association for Cancer Research, 13 (2007) 4331–4335. [DOI] [PubMed] [Google Scholar]
- [17].Chatterjee A, Dasgupta S, Sidransky D, Mitochondrial subversion in cancer, Cancer prevention research, 4 (2011) 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, Lam W, Zhang W, Gazdar AF, Fisher PB, Sidransky D, Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation, Journal of cellular physiology, 227 (2012) 2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jakupciak JP, Maragh S, Markowitz ME, Greenberg AK, Hoque MO, Maitra A, Barker PE, Wagner PD, Rom WN, Srivastava S, Sidransky D, O’Connell CD, Performance of mitochondrial DNA mutations detecting early stage cancer, BMC cancer, 8 (2008) 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dasgupta S, Yung RC, Westra WH, Rini DA, Brandes J, Sidransky D, Following mitochondrial footprints through a long mucosal path to lung cancer, PloS one, 4 (2009) e6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou S, Kachhap S, Sun W, Wu G, Chuang A, Poeta L, Grumbine L, Mithani SK, Chatterjee A, Koch W, Westra WH, Maitra A, Glazer C, Carducci M, Sidransky D, McFate T, Verma A, Califano JA, Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck, Proceedings of the National Academy of Sciences of the United States of America, 104 (2007) 7540–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tan DJ, Bai RK, Wong LJ, Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer, Cancer research, 62 (2002) 972–976. [PubMed] [Google Scholar]
- [23].Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O’Sullivan J, Salk JJ, Bielas JH, Decreased mitochondrial DNA mutagenesis in human colorectal cancer, PLoS genetics, 8 (2012) e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC, mtDNA mutations increase tumorigenicity in prostate cancer, Proceedings of the National Academy of Sciences of the United States of America, 102 (2005) 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S, Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis, Cancer research, 65 (2005) 1655–1663. [DOI] [PubMed] [Google Scholar]
- [26].Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A, The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection, Genome research, 14 (2004) 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, Califano JA, Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions, Clinical cancer research: an official journal of the American Association for Cancer Research, 10 (2004) 8512–8515. [DOI] [PubMed] [Google Scholar]
- [28].Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, Sidransky D, Califano JA, Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation, Clinical cancer research: an official journal of the American Association for Cancer Research, 8 (2002) 2260–2265. [PubMed] [Google Scholar]
- [29].Larman TC, DePalma SR, Hadjipanayis AG, Cancer N, Protopopov A, Zhang J, Gabriel SB, Chin L, Seidman CE, Kucherlapati R, Seidman JG, Spectrum of somatic mitochondrial mutations in five cancers, Proceedings of the National Academy of Sciences of the United States of America, 109 (2012) 14087–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, Davies HR, Papaemmanuil E, Gundem G, Shlien A, Bolli N, Behjati S, Tarpey PS, Nangalia J, Massie CE, Butler AP, Teague JW, Vassiliou GS, Green AR, Du MQ, Unnikrishnan A, Pimanda JE, Teh BT, Munshi N, Greaves M, Vyas P, El-Naggar AK, Santarius T, Collins VP, Grundy R, Taylor JA, Hayes DN, Malkin D, Group IBC, Group ICMD, Group IPC, Foster CS, Warren AY, Whitaker HC, Brewer D, Eeles R, Cooper C, Neal D, Visakorpi T, Isaacs WB, Bova GS, Flanagan AM, Futreal PA, Lynch AG, Chinnery PF, McDermott U, Stratton MR, Campbell PJ, Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer, eLife, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Amer W, Toth C, Vassella E, Meinrath J, Koitzsch U, Arens A, Huang J, Eischeid H, Adam A, Buettner R, Scheel A, Schaefer SC, Odenthal M, Evolution analysis of heterogeneous non-small cell lung carcinoma by ultra-deep sequencing of the mitochondrial genome, Scientific reports, 7 (2017) 11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kazdal D, Harms A, Endris V, Penzel R, Kriegsmann M, Eichhorn F, Muley T, Stenzinger A, Pfarr N, Weichert W, Warth A, Prevalence of somatic mitochondrial mutations and spatial distribution of mitochondria in non-small cell lung cancer, British journal of cancer, 117 (2017) 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kloss-Brandstatter A, Weissensteiner H, Erhart G, Schafer G, Forer L, Schonherr S, Pacher D, Seifarth C, Stockl A, Fendt L, Sottsas I, Klocker H, Huck CW, Rasse M, Kronenberg F, Kloss FR, Validation of Next-Generation Sequencing of Entire Mitochondrial Genomes and the Diversity of Mitochondrial DNA Mutations in Oral Squamous Cell Carcinoma, PloS one, 10 (2015) e0135643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guo Y, Li J, Li CI, Shyr Y, Samuels DC, MitoSeek: extracting mitochondria information and performing high throughput mitochondria sequencing analysis, Bioinformatics, 29 (2013) 1210–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Daber R, Sukhadia S, Morrissette JJ, Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets, Cancer genetics, 206 (2013) 441–448. [DOI] [PubMed] [Google Scholar]
- [36].Li D, Du X, Guo X, Zhan L, Li X, Yin C, Chen C, Li M, Li B, Yang H, Xing J, Site-specific selection reveals selective constraints and functionality of tumor somatic mtDNA mutations, Journal of experimental & clinical cancer research: CR, 36 (2017) 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Grandhi S, Bosworth C, Maddox W, Sensiba C, Akhavanfard S, Ni Y, LaFramboise T, Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection, Human molecular genetics, 26 (2017) 2912–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kauppila JH, Stewart JB, Mitochondrial DNA: Radically free of free-radical driven mutations, Biochimica et biophysica acta, 1847 (2015) 1354–1361. [DOI] [PubMed] [Google Scholar]
- [39].Hu L, Yao X, Shen Y, Altered mitochondrial DNA copy number contributes to human cancer risk: evidence from an updated meta-analysis, Scientific reports, 6 (2016) 35859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cheau-Feng Lin F, Jeng YC, Huang TY, Chi CS, Chou MC, Chin-Shaw Tsai S, Mitochondrial DNA copy number is associated with diagnosis and prognosis of head and neck cancer, Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals, 19 (2014) 269–274. [DOI] [PubMed] [Google Scholar]
- [41].Weerts MJ, Sieuwerts AM, Smid M, Look MP, Foekens JA, Sleijfer S, Martens JW, Mitochondrial DNA content in breast cancer: Impact on in vitro and in vivo phenotype and patient prognosis, Oncotarget, 7 (2016) 29166–29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bai RK, Wong LJ, Simultaneous detection and quantification of mitochondrial DNA deletion(s), depletion, and over-replication in patients with mitochondrial disease, The Journal of molecular diagnostics: JMD, 7 (2005) 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dimmock D, Tang LY, Schmitt ES, Wong LJ, Quantitative evaluation of the mitochondrial DNA depletion syndrome, Clinical chemistry, 56 (2010) 1119–1127. [DOI] [PubMed] [Google Scholar]
- [44].Weerts MJA, Hollestelle A, Sieuwerts AM, Foekens JA, Sleijfer S, Martens JWM, Low Tumor Mitochondrial DNA Content Is Associated with Better Outcome in Breast Cancer Patients Receiving Anthracycline-Based Chemotherapy, Clinical cancer research: an official journal of the American Association for Cancer Research, 23 (2017) 4735–4743. [DOI] [PubMed] [Google Scholar]
- [45].Nadano D, Yasuda T, Kishi K, Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method, Clinical chemistry, 39 (1993) 448–452. [PubMed] [Google Scholar]
- [46].Rebolledo-Jaramillo B, Su MS, Stoler N, McElhoe JA, Dickins B, Blankenberg D, Korneliussen TS, Chiaromonte F, Nielsen R, Holland MM, Paul IM, Nekrutenko A, Makova KD, Maternal age effect and severe germline bottleneck in the inheritance of human mitochondrial DNA, Proceedings of the National Academy of Sciences of the United States of America, 111 (2014) 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huggett JF, Cowen S, Foy CA, Considerations for digital PCR as an accurate molecular diagnostic tool, Clinical chemistry, 61 (2015) 79–88. [DOI] [PubMed] [Google Scholar]
- [48].Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, Sukumar S, Sidransky D, Tumor-specific changes in mtDNA content in human cancer, International journal of cancer. Journal international du cancer, 116 (2005) 920–924. [DOI] [PubMed] [Google Scholar]
- [49].Bonner MR, Shen M, Liu CS, Divita M, He X, Lan Q, Mitochondrial DNA content and lung cancer risk in Xuan Wei, China, Lung cancer, 63 (2009) 331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Viertler C, Groelz D, Gundisch S, Kashofer K, Reischauer B, Riegman PH, Winther R, Wyrich R, Becker KF, Oelmuller U, Zatloukal K, A new technology for stabilization of biomolecules in tissues for combined histological and molecular analyses, The Journal of molecular diagnostics: JMD, 14 (2012) 458–466. [DOI] [PubMed] [Google Scholar]
- [51].Shih SY, Bose N, Goncalves ABR, Erlich HA, Calloway CD, Applications of Probe Capture Enrichment Next Generation Sequencing for Whole Mitochondrial Genome and 426 Nuclear SNPs for Forensically Challenging Samples, Genes, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bourhis J, Le Maitre A, Baujat B, Audry H, Pignon JP, N.C.C.G. Meta-Analysis of Chemotherapy in Head, N.C.G. Meta-Analysis of Radiotherapy in Carcinoma of Head, G. Meta-Analysis of Chemotherapy in Nasopharynx Carcinoma Collaborative, Individual patients’ data meta-analyses in head and neck cancer, Current opinion in oncology, 19 (2007) 188–194. [DOI] [PubMed] [Google Scholar]
- [53].Lai CH, Huang SF, Liao CT, Chen IH, Wang HM, Hsieh LL, Clinical significance in oral cavity squamous cell carcinoma of pathogenic somatic mitochondrial mutations, PloS one, 8 (2013) e65578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Koshikawa N, Hayashi J, Nakagawara A, Takenaga K, Reactive oxygen species-generating mitochondrial DNA mutation up-regulates hypoxia-inducible factor-1alpha gene transcription via phosphatidylinositol 3-kinase-Akt/protein kinase C/histone deacetylase pathway, J Biol Chem, 284 (2009) 33185–33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J, ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis, Science, 320 (2008) 661–664. [DOI] [PubMed] [Google Scholar]
- [56].Yuan Y, Wang W, Li H, Yu Y, Tao J, Huang S, Zeng Z, Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma, BMC cancer, 15 (2015) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hertweck KL, Dasgupta S, The Landscape of mtDNA Modifications in Cancer: A Tale of Two Cities, Front Oncol, 7 (2017) 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kurelac I, MacKay A, Lambros MB, Di Cesare E, Cenacchi G, Ceccarelli C, Morra I, Melcarne A, Morandi L, Calabrese FM, Attimonelli M, Tallini G, Reis-Filho JS, Gasparre G, Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumorigenesis that correlate with low genomic instability in pituitary adenomas, Human molecular genetics, 22 (2013) 226–238. [DOI] [PubMed] [Google Scholar]
- [59].Park JS, Sharma LK, Li H, Xiang R, Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J, Bai Y, A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis, Human molecular genetics, 18 (2009) 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jang YH, Lim KI, Recent Advances in Mitochondria-Targeted Gene Delivery, Molecules, 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jo A, Ham S, Lee GH, Lee YI, Kim S, Lee YS, Shin JH, Lee Y, Efficient Mitochondrial Genome Editing by CRISPR/Cas9, BioMed research international, 2015 (2015) 305716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gammage PA, Moraes CT, Minczuk M, Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR Ized, Trends Genet, 34 (2018) 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lin JC, Wang CC, Jiang RS, Wang WY, Liu SA, Impact of somatic mutations in the D-loop of mitochondrial DNA on the survival of oral squamous cell carcinoma patients, PloS one, 10 (2015) e0124322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Stewart JB, Alaei-Mahabadi B, Sabarinathan R, Samuelsson T, Gorodkin J, Gustafsson CM, Larsson E, Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers, PLoS genetics, 11 (2015) e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lievre A, Blons H, Houllier AM, Laccourreye O, Brasnu D, Beaune P, Laurent-Puig P, Clinicopathological significance of mitochondrial D-Loop mutations in head and neck carcinoma, British journal of cancer, 94 (2006) 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mondal R, Ghosh SK, Talukdar FR, Laskar RS, Association of mitochondrial D-loop mutations with GSTM1 and GSTT1 polymorphisms in oral carcinoma: a case control study from northeast India, Oral oncology, 49 (2013) 345–353. [DOI] [PubMed] [Google Scholar]
- [67].Zhang R, Wang R, Zhang F, Wu C, Fan H, Li Y, Wang C, Guo Z, Single nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinoma, Journal of experimental & clinical cancer research: CR, 29 (2010) 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Smith PM, Elson JL, Greaves LC, Wortmann SB, Rodenburg RJ, Lightowlers RN, Chrzanowska Lightowlers ZM, Taylor RW, Vila-Sanjurjo A, The role of the mitochondrial ribosome in human disease: searching for mutations in 12S mitochondrial rRNA with high disruptive potential, Human molecular genetics, 23 (2014) 949–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kazdal D, Harms A, Endris V, Penzel R, Oliveira C, Kriegsmann M, Longuespee R, Winter H, Schneider MA, Muley T, Pfarr N, Weichert W, Stenzinger A, Warth A, Subclonal evolution of pulmonary adenocarcinomas delineated by spatially distributed somatic mitochondrial mutations, Lung cancer, 126 (2018) 80–88. [DOI] [PubMed] [Google Scholar]
- [70].Li X, Guo X, Li D, Du X, Yin C, Chen C, Fang W, Bian Z, Zhang J, Li B, Yang H, Xing J, Multi-regional sequencing reveals intratumor heterogeneity and positive selection of somatic mtDNA mutations in hepatocellular carcinoma and colorectal cancer, International journal of cancer, 143 (2018) 1143–1152. [DOI] [PubMed] [Google Scholar]
- [71].Burrell RA, McGranahan N, Bartek J, Swanton C, The causes and consequences of genetic heterogeneity in cancer evolution, Nature, 501 (2013) 338–345. [DOI] [PubMed] [Google Scholar]
- [72].He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA Jr., Kinzler KW, Vogelstein B, Papadopoulos N, Heteroplasmic mitochondrial DNA mutations in normal and tumour cells, Nature, 464 (2010) 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sofronova JK, Ilinsky YY, Orishchenko KE, Chupakhin EG, Lunev EA, Mazunin IO, Detection of Mutations in Mitochondrial DNA by Droplet Digital PCR, Biochemistry. Biokhimiia, 81 (2016) 1031–1037. [DOI] [PubMed] [Google Scholar]
- [74].Hirose M, Schilf P, Gupta Y, Zarse K, Kunstner A, Fahnrich A, Busch H, Yin J, Wright MN, Ziegler A, Vallier M, Belheouane M, Baines JF, Tautz D, Johann K, Oelkrug R, Mittag J, Lehnert H, Othman A, Johren O, Schwaninger M, Prehn C, Adamski J, Shima K, Rupp J, Hasler R, Fuellen G, Kohling R, Ristow M, Ibrahim SM, Low-level mitochondrial heteroplasmy modulates DNA replication, glucose metabolism and lifespan in mice, Scientific reports, 8 (2018) 5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sobenin IA, Mitrofanov KY, Zhelankin AV, Sazonova MA, Postnov AY, Revin VV, Bobryshev YV, Orekhov AN, Quantitative assessment of heteroplasmy of mitochondrial genome: perspectives in diagnostics and methodological pitfalls, BioMed research international, 2014 (2014) 292017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kang E, Wang X, Tippner-Hedges R, Ma H, Folmes CD, Gutierrez NM, Lee Y, Van Dyken C, Ahmed R, Li Y, Koski A, Hayama T, Luo S, Harding CO, Amato P, Jensen J, Battaglia D, Lee D, Wu D, Terzic A, Wolf DP, Huang T, Mitalipov S, Age-Related Accumulation of Somatic Mitochondrial DNA Mutations in Adult-Derived Human iPSCs, Cell stem cell, 18 (2016) 625–636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 4. Detailed list of all somatic mtDNA mutations found in primary HNSCC specimens.
Supplementary Table 13. Summary of ddPCR assays designed by Bio-Rad.
Supplementary Table 14. Summary of the ddPCR analysis results in specimens collected from patients 2, 4 and 5.
Supplementary Table 1. List of 148 primer pairs designed and used in this study for mtDNA amplification. Table summarizes the GC-content, length distribution and location of the Access Array amplicons.
Supplementary Table 2. Table summarizes the number of zones collected and sequenced for each HNSCC lesion.
Supplementary Table 3. Table lists average coverage and sequencing depth for indicated specimen types.
Supplementary Table 5. List of somatic mtDNA mutations found in HNSCC tumors that have been previously observed in solid tumors (including HNSCC).
Supplementary Table 6. Spectrum of base-pair substitutions in HNSCC tumors.
Supplementary Table 7. Table summarizes mutations detected in more than one HNSCC patient.
Supplementary Table 8. List of somatic mtDNA mutations detected by two independent sequencing runs in patients 2, 3, 4, 5 and 6. Red – mutations detected in both runs, black – mutations detected by only one sequencing run, blue – mutations that were detected by the first run but were found in additional tumor zones upon resequencing.
Supplementary Table 9. Table summarizes all match specimens collected from the same patient.
Supplementary Table 10. List of all somatic mtDNA mutations detected in LNs, margins, serum and sputum specimens. Tumor-associated mutations (variants that were also detected in the match primary tumor specimens) are highlighted in bold.
Supplementary Table 11. Detailed list of tumor-associated mtDNA mutations detected in LNs, margins and serum specimens at 3% variant calling threshold.
Supplementary Table 12. Detailed list of tumor-associated mtDNA mutations detected in LNs, margins and serum specimens at 1% variant calling threshold.
Supplementary Figure 1. Schematic of workflow for sequencing and analysis of somatic mitochondrial mutations in HNSCC samples. Briefly, gDNA from samples and matched control specimens are extracted and amplified as described in Methods. Following library preparation and next-generation sequencing, the reads are aligned. The mitochondrial (MT)-targeted reads are extracted for further analysis. CN calculations are performed using the nuclear reads as described in Methods. Additional quality control steps are taken prior to somatic mutation discovery using MitoSeek. Data cleaning including: calculation of MAF using mutant allele read counts, assessment of adequate depth, annotation of D-Loop and non-coding region reads based on base position, and removal of duplicate entries was performed in R. Subsequent to this, somatic mutations from each sample (and matched control) was associated with the proper clinical sample to obtain group level data and perform statistics.
Supplementary Figure 2. Box plot represents distributions of mutant allele frequencies (MAFs) of somatic mutations detected in primary HNSCC tumors across the functional mtDNA regions.
Supplementary Figure 3. Analysis of WES data derived from TCGA-HNSC dataset. A. Average coverage. B. Mutational distribution across the mitochondrial genome. C. Frequency of base pair transitions.