Abstract
As medical imaging enters its information era and presents rapidly increasing needs for big data analytics, robust pooling and harmonization of imaging data across diverse cohorts with varying acquisition protocols have become critical. We describe a comprehensive effort that merges and harmonizes a large-scale dataset of 10,477 structural brain MRI scans from participants without a known neurological or psychiatric disorder from 18 different studies that represent geographic diversity. We use this dataset and multi-atlas-based image processing methods to obtain a hierarchical partition of the brain from larger anatomical regions to individual cortical and deep structures and derive age trends of brain structure through the lifespan (3 to 96 years old). Critically, we present and validate a methodology for harmonizing this pooled dataset in the presence of nonlinear age trends. We provide a web-based visualization interface to generate and present the resulting age trends, enabling future studies of brain structure to compare their data with this reference of brain development and aging, and to examine deviations from ranges, potentially related to disease.
Keywords: MRI, Segmentation, FreeSurfer, MUSE, Brain, ROI
1. Introduction
Structural brain changes have been studied at various stages of the lifespan in relation to age and neurodegenerative diseases (Fjell and Walhovd, 2010; Habes et al., 2016), as well as to brain development (Courchesne et al., 2000; Sowell et al., 2001; Toga et al., 2006). A large number of imaging studies reported findings on age-related changes in brain structure during adolescence, early adulthood, and late adulthood (Giedd et al., 1999; Driscoll et al., 2009; Mills et al., 2016; Pfefferbaum et al., 1994; Tamnes et al., 2010; Terribilli et al., 2011). Traditionally, most neuroimaging studies have been limited to analyses on single-center datasets to minimize instrument-related variability in the data. However, in recent years there is an increasing trend towards data sharing in neuroimaging research communities, with multiple collaborative efforts for pooling existing data resources to form large, diverse samples covering a wide age range (Alfaro-Almagro et al., 2019; Thompson et al., 2014). Such collective efforts are critical for enabling development of diagnostic and prognostic biomarkers that apply across different imaging equipment as well as across the broad spectrum of demographics, which is essential for translation of neuroimaging research into clinical settings.
A number of studies have shown the importance of mega-analyses combining data from multiple cohorts. For example, data from the multi-site ENIGMA Consortium have been found to link volumetric abnormalities with post-traumatic stress disorder (Logue et al., 2018), schizophrenia (Van Erp et al., 2016), and major depressive disorder (Schmaal et al., 2016). However, there are important challenges in combining imaging data from multiple studies and sites. A major challenge is the lack of standardization in image acquisition protocols, scanner hardware, and software. Inter-scanner variability has been demonstrated to affect measurements obtained for downstream analysis such as voxel-based morphometry (Takao et al., 2011), lesion volumes (Shinohara et al., 2017), and DTI measurements (Zhu et al., 2011). Differences in sample demographics are also an important concern that should be handled carefully when combining multi-site data (LeWinn et al., 2017). For example, MR contrast may be confounded by differences in brain water content, which varies across age and diagnostic groups (Bansal et al., 2013). Additionally, the reliability of imaging-based biomarkers may be impaired by the inclusion of low-quality datasets. Although it is critical to understand and identify all sources of variability in imaging-derived measurements, assessment and optimization of reliability is typically under-appreciated in neuroscience research (Zuo et al., 2019). Finally, large-scale studies ultimately require robust and fully automated pipelines without the need to manually inspect and correct large sets of data, which is both time-consuming, subjective, and less likely to be adopted clinically.
In this paper we present a major effort designed to create the cross-sectional LIFESPAN dataset for quantitative characterization of structural age-related differences in brain anatomy through the human lifespan from age 3 to 96. For this purpose, structural brain MRI scans from 18 studies were pooled together, creating a large, and most importantly, diverse sample (N=10,477). Although our focus is on structural MRI, our methodologies are applicable to any kind of imaging data. We test the robustness of a fully automated and standardized multi-atlas labeling pipeline, namely MUSE: Multi-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters and locally optimal atlas selection (Doshi et al., 2016), which segments the brain into a set of hierarchically predefined regions of interest (ROIs) and measures the volume of each of these regions. A notable advantage of the multi-atlas segmentation methodology is that it computes the consensus labeling of a large ensemble of reference atlases, and hence simultaneously provides mechanisms for selecting atlases based on their local similarity to the target scan during the label fusion. The reference atlases represent anatomical variability across participants that span a wide age range, thus enabling a more robust segmentation across highly heterogeneous datasets.
We present a harmonization approach in this paper to address the unique challenge of combining 18 studies from diverse age ranges in the presence of nonlinear age-related differences in brain volumes. We define harmonization as the explicit removal of site-related effects in multi-site data. Through the lifespan, the brain structure changes as a result of a complex interplay between multiple maturational and neurodegenerative processes. The effect of such processes could yield large spatial and temporal variations on the brain (Toga et al., 2006). A parsimonious model of age, such as a linear or quadratic model, is unlikely to sufficiently capture the relationship between age and volume throughout the lifespan (Fjell et al., 2010; Ziegler et al., 2012). Additionally, studies in our dataset did not overlap entirely on age, making techniques based on sample matching infeasible (Karayumak et al., 2019).
In order to capture non-linearities in age-related volume differences in brain anatomy through the lifespan, we propose to fit a generalized additive model (GAM) with a penalized nonlinear term to describe age effects (Hastie and Tibshirani, 1986; Wood, 2017). Within a single model, we estimated the location (mean) and scale (variance) differences in imaging measurements across sites. In the absence of ground truth, we performed simulation experiments to evaluate the harmonization performance across various conditions of sample composition. The simulation experiments leverage a large single-scanner study covering the entire adult lifespan to serve as an estimate of ground truth. Sampling this study and using simulations, we evaluate the effects of sample demographics and relative sample sizes on the harmonization accuracy.
Other communities that handle high dimensional data-integration across multiple sites have faced the necessity of harmonization. Among the available methods, ComBat, which was originally proposed to remove batch effects in genomics data (Johnson et al., 2007), has been recently adapted to diffusion tensor imaging data (Fortin et al., 2017), cortical thickness measurements (Fortin et al., 2018), and functional connectivity matrices (Yu et al., 2018). The method was shown to remove unwanted sources of variability, specifically site differences, while preserving variations due to other biologically-relevant covariates in the data. We adopt and test ComBat in our harmonization pipeline of the LIFESPAN dataset in conjunction with GAMs, which we refer to as ComBat-GAM. We compared ComBat-GAM to no harmonization and to ComBat with a linear model, based on their performances on a multi-variate brain age prediction task.
Successful harmonization of imaging measurements enabled us to estimate age-related volume differences for each anatomical region of the LIFESPAN dataset, which we refer to as age trends. The resulting age trends are supported by the large sample size of the dataset and may serve as a reference for the neuroimaging community. We provide an interactive online tool that will allow researchers to visualize the age trends of different anatomical regions, as well as to calibrate their own data with the LIFESPAN dataset, and position user-specific data among the reference trends.
2. Material and methods
2.1. MRI datasets
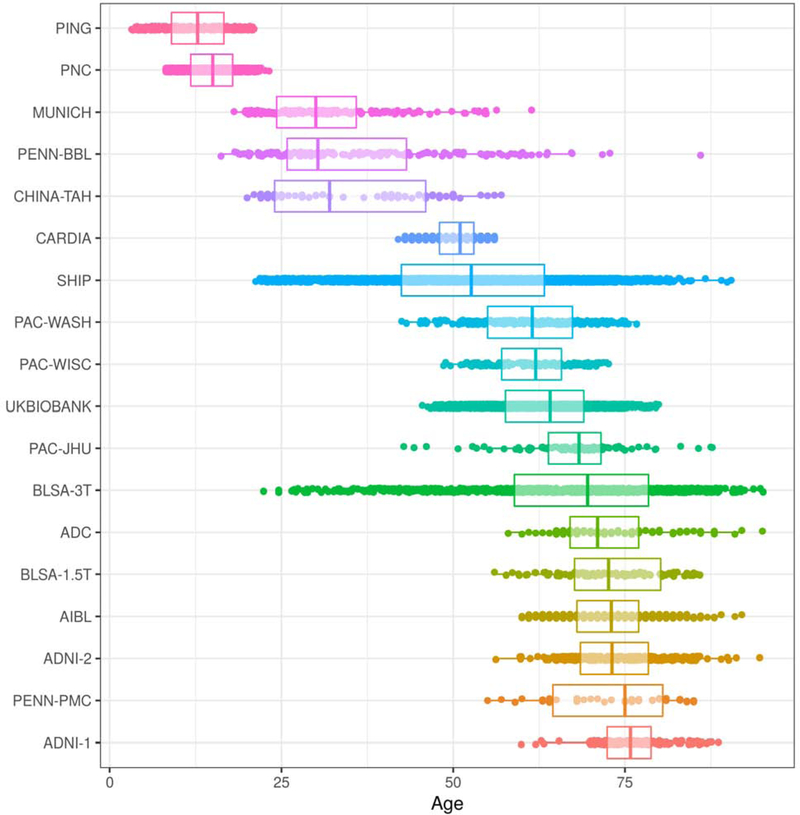
We collected structural MRI (T1) data from 18 studies. The pooled dataset included baseline scans of typically-developing and typically-aging participants from each study with available age and sex information. We defined typical development and typical aging as the absence of a known diagnosis of a neurological or psychiatric disorder. We considered multi-center imaging studies that undertook efforts to unify protocols as single studies; this includes the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Jack Jr. et al., 2008), the Baltimore Longitudinal Study of Aging (BLSA) (Armstrong et al., 2019; Resnick et al., 2003), the Coronary Artery Risk Development in Young Adults study (CARDIA) (Friedman et al., 1988), the Pediatric Imaging, Neurocognition, and Genetics study (PING) (Jernigan et al., 2016), the Philadelphia Neurodevelopmental Cohort (PNC) (Satterthwaite et al., 2016), and the UK Biobank (Alfaro-Almagro et al., 2019). Phases of ADNI (ADNI-1, ADNI-2) and BLSA (1.5T SPGR, 3T MPRAGE) were considered separate studies due to major scanner updates. A single scan was included in the LIFESPAN dataset for each ADNI and BLSA subject. Although we internally processed 21,315 scans from the UK Biobank in 10 randomized batches, we decided to include only one batch from the dataset to avoid estimating age trends that would be heavily influenced by the the UK Biobank. Table 1 shows general characteristics of the study datasets. We note the inherent demographic diversity across datasets; for example, while overall the dataset was 54% female, individual studies ranged from 38% to 69% female. The studies also cover different age ranges, though this is intended to produce a pooled dataset covering most of the human lifespan. In Supplementary Table 1 we present additional demographic diversity due to race and ethnicity for the study participants where data was available. Overall the majority of participants were white with a substantial minority of black participants; there were a small number of Chinese Asian and Hispanic participants. Figure 1 presents age distributions for each study in the LIFESPAN dataset, sorted by median age. Scanner models and acquisition protocol parameters in each dataset are given in Supplementary Table 2. Informed consent was obtained from all participants by the leading institutions of each individual study in the LIFESPAN dataset. The Ethics Committee of the leading institution of each cohort approved its study.
Table 1.
Summary characteristics of the datasets included in the LIFESPAN dataset, sorted by median age.
| Dataset Name | Country of Origin | No. Participants | No. Females (%) | Age Range (Median) |
|---|---|---|---|---|
| PING | USA | 306 | 154 (50.3) | [3, 21](12.8) |
| PNC | USA | 1,444 | 755 (52.3) | [8, 24](15) |
| MUNICH | Germany | 173 | 54 (31.2) | [18, 62](30) |
| PENN-BBL | USA | 170 | 100 (58.8) | [16, 86](30.3) |
| CHINA-TAH | China | 102 | 60 (58.8) | [20, 57](32) |
| CARDIA | USA | 719 | 377 (52.4) | [42, 56](51) |
| SHIP | Germany | 2,738 | 1,491 (54.5) | [21, 91](52.6) |
| PAC-WASH | USA | 247 | 152 (61.5) | [42, 77](61.5) |
| PAC-WISC | USA | 127 | 88 (69.3) | [48, 73](62) |
| UKBIOBANK | United Kingdom | 2,201 | 1,189 (54) | [45, 80](64.1) |
| PAC-JHU | USA | 92 | 56 (60.9) | [42, 88](68.3) |
| BLSA-3T | USA | 964 | 521 (54) | [22, 96](69.5) |
| ADC | USA | 104 | 66 (63.5) | [58, 95](71) |
| BLSA-1.5T | USA | 92 | 35 (38) | [56, 86](72.6) |
| AIBL | Australia | 446 | 249 (55.8) | [60, 92](73) |
| ADNI-2 | USA | 324 | 179 (55.2) | [56, 95](73.1) |
| PENN-PMC | USA | 39 | 21 (53.8) | [55, 85](75) |
| ADNI-1 | USA | 189 | 89 (47.1) | [59, 89](75.8) |
| LIFESPAN (total) | 10,477 | 5636 (53.8) | [3, 96](56.1) |
Figure 1:
Age distributions of studies that are part of the LIFESPAN dataset, sorted by median age. The study with youngest median age, PING, contains participants from age 3 to 21. The study with oldest median age, ADNI-1, contains participants from age 59 to 89.
2.2. MRI image processing
A fully automated processing pipeline was applied to each participant’s T1-weighted scan. Preprocessing involved correction of magnetic field intensity inhomogeneity (Tustison et al., 2010) and skull-stripping, i.e. extraction of brain tissues, using a multi-atlas method (Doshi et al., 2013). For segmenting each T1 scan into a set of pre-defined anatomical regions of interest (ROIs) we used a multi-atlas, multi-warp label-fusion method, MUSE (Doshi et al., 2016), which has obtained top accuracy in comparison to multiple benchmark methods in independent evaluations (Asman et al., 2013). In this framework, multiple atlases with semi-automatically extracted ground-truth ROI labels are first warped individually to the target image using two different non-linear registration methods. A spatially adaptive weighted voting strategy is then applied to fuse the ensemble into a final segmentation. This procedure was used to segment each image into 145 ROIs spanning the entire brain. We calculated the volumes of these 145 ROIs, as well as the volumes of 113 composite ROIs that were obtained by combining individual ROIs into larger anatomical regions following a predefined ROI hierarchy. A list of the ROIs used in the LIFESPAN dataset is given in Supplementary Table 3.
2.3. Quality control of extracted variables
A systematic quality control (QC) procedure guided by final outcome variables was conducted to identify and exclude cases of low quality. This procedure was applied on a set of 69 representative ROIs, including deep brain structures and sub-lobe level cortical parcellations, as well as the intracranial volume (ICV) (a full list of ROIs used in the QC procedure is given in Supplementary Table 4). Volumes of selected ROIs were corrected for ICV and z-score transformed independently for each dataset in order to identify data outliers. We defined outliers as volumes that were greater than three standard deviations (SD) away from the within-study sample mean of the specific ROI. All scans that included at least one outlier ROI were flagged for manual inspection.
2.4. Harmonization of imaging variables
We harmonized individual ROI volumes using a model that builds upon the statistical harmonization technique proposed in Johnson et al. (2007) for location and scale (L/S) adjustments to the data. This method estimates within a single model the location (mean) and scale (variance) differences in ROI volumes across sites, as well as variations due to other biologically-relevant covariates in the data that are intended to be preserved. Once estimated, the standardized ROI volumes can be achieved by removing location and scale effects due to site differences.
For site i, subject j, region k, a general framework for an LS-adjustment of an ROI volume, Yijk, is:
| (1) |
where fk (Xij) denotes the variation of Y captured by the biologically-relevant covariates X, gik is the estimated location effect for site site i and region k, and dik is the estimated scale effect for site site i and region k. In the linear case, fk (Xij) = ak + Xij * bk and the corresponding adjustment is:
| (2) |
In our case we substitute for fk (Xij) a Generalized Additive Model (GAM) which is a function of the covariates age, sex, and ICV, represented by xij, zij, and wij, respectively, to allow for nonlinear age trends in ROI volumes informed by the data. GAMs allow for flexible nonlinearity in xij represented using a basis expansion. Additionally, penalization in the objective function of the model fitting ensures the smoothness of fk (Xij) and avoids over-fitting to the observed data (Hastie and Tibshirani, 1986). In our design, we included a smoothed nonlinear term for age using thin plate regression splines for basis expansion as described in Wood (2003), as well as parametric terms for sex and ICV. The model was estimated based on penalized regression splines and the degree of smoothness was internally selected using the restricted maximum likelihood (REML) criterion. Accordingly, our GAM-based covariates model was estimated as:
| (3) |
We integrated the non-linear GAM model with the previously-proposed framework of ComBat (Johnson et al., 2007) for the multivariate harmonization of multiple ROIs. The main premise of ComBat is that location and scale effects for multivariate outcomes, e.g. volumes across ROIs, are drawn from a common parametric prior distribution. We assume a normal distribution as the prior for g*ik and an inverse-gamma distribution as the prior for d*ik. ComBat estimates hyperparameters of the prior distributions from the data using empirical Bayes framework. Once estimated, the hyperparameters are used to compute conditional posterior estimates of all location and scale effects, formulas for which are given in Johnson et al. (2007). ComBat adjusts an ROI volume, Y*ijk, using the conditional posterior estimates. Together with our non-linear GAM model, we have the ComBat-GAM adjustment:
| (4) |
where g*ik is the posterior estimate of the location effect for site i and region k, and d*ik is the conditional posterior estimate of the scale effect for site i and region k. We provide details of the ComBat-GAM algorithm in the supplementary materials.
2.5. Evaluation of goodness of fit with GAM versus linear and quadratic models on single-site data
We first performed a comparative evaluation of the proposed GAM structure against both linear and quadratic models on single-site data. For the comparisons we selected three large studies with different age ranges. The Philadelphia Neurodevelopmental Cohort (PNC) included 1,444 participants from ages 8 to 24 (Satterthwaite et al., 2016). The Study of Health in Pomerania (SHIP) included 2,738 participants from ages 21 to 91 (Völzke et al., 2010). The 3-Tesla cohort of the Baltimore Longitudinal Study of Aging (BLSA-3T) included 964 participants from ages 22 to 96 (Armstrong et al., 2019). For each ROI, a linear model, a quadratic model, and a GAM with a smoothed nonlinear age term were fit to predict volumes from age. In all models, sex and ICV were included as additional covariates. The regression models were applied separately on each of the three study datasets to avoid confounding with site effects. We quantified the goodness of fit by calculating the adjusted R-squared for each model. Additionally, we performed a split-sample experiment with 50% of each dataset to assess the out-of-sample fit for each model using Root Mean Square Error (RMSE). We also performed the Chisquare test to assess the hypothesis that residual sum of squares (RSS) were significantly lower using GAMs than other models.
2.6. Simulation experiments
The proposed harmonization model estimates a non-linear relationship between ROI volumes and age. The accuracy of the estimated age trend from multi-site data is a critical metric for harmonization performance. However, due to lack of ground-truth data, evaluations using real data were not possible. Therefore, we performed simulation experiments for assessing the effect of harmonization in the presence of known site effects for two different conditions. Toward this goal, we leveraged the large single-site SHIP study dataset (N=2,738) spanning ages 21 through 91.
In all experiments, we simulated volumes of the hippocampus for three hypothetical sites (named Site-A, Site-B and Site-C), using actual hippocampus volumes from SHIP. A ground truth age trend was first estimated on the entire SHIP data using a GAM model with a nonlinear term for age (sex and ICV effects were included as covariates). For each of the 3 sites, we randomly sampled data following the sample size and age range constraints imposed by each experiment. Site-specific location and scale effects were then introduced on actual hippocampus volumes to generate the simulated datasets independently for each of the two experiments.
We performed harmonization using the LS adjustment with GAM method. The error of the estimated age trend after harmonization was quantified as the mean absolute error (MAE) over 100 equallyspaced age grid-points along the estimated trend versus the ground truth trend, standardized by the mean ROI volume, to produce relative Mean Absolute Error (rMAE).
i. Effect of degree of overlap in the age ranges of data sites and sample size
Simulation Experiment I aimed to study the sensitivity of the proposed method to the amount of overlap in age ranges between harmonized datasets and the sample size of each site. For this purpose, we fixed the age ranges of Site-A and Site-C (30 to 50 and 60 to 80 years, respectively), while allowing a 30-year sliding age range for Site-B that varies from younger (30 to 60 years) to older (50 to 80 years). We also allowed the samples sizes of all sites to vary from 50 to 500. We performed a grid search over the two free parameters to identify minimum requirements for obtaining accurate age trends after harmonization.
ii. Effect of balancing sample sizes
Simulation Experiment II aimed to investigate harmonization of sites with unbalanced sample sizes. We assessed the effects of sub-sampling from a relatively larger site to create a balanced sample composition. Our main hypothesis was that leaving some data out of the harmonization in order to generate datasets balanced sample sizes might lead to more accurate alignment across studies. For this purpose, we fixed the sample size of Site-A and Site-C to 100, and varied the size of Site-B by randomly sub-sampling from 400 participants. We compared harmonization results using the complete Site-B sample (n=400) versus harmonization after sub-sampling Site-B at proportions of 25% (n=100), 50% (n=200), and 75% (n=300).
2.7. Harmonization of volumetric measurements from the LIFESPAN dataset
We applied ComBat-GAM on each of the 145 anatomical ROIs using the complete LIFESPAN sample to remove location and scale effects for each ROI.
Similar to Fortin et al. (2018), we evaluated the harmonization by assessing the accuracy on crossvalidated brain age prediction task using pre- and post-harmonized ROI volumes as features. The brain age model was constructed using a fully-connected neural network with one hidden layer. ROI volumes for the complete LIFESPAN sample were used as input features to the model, in addition to sex and ICV. Due to the redundancy between single ROIs and composite ROIs, we used only single ROIs for the feature set in the age prediction model. We performed 10-fold cross-validation, as well as leave-site-out cross validation to assess the effect of harmonization for brain age prediction on unseen sites. The network was trained with the Adam optimizer using mean squared error as the cost function with a constant learning rate of 1×10−3. The fully-connected layer consisted of 100 nodes with RELU activation functions for each node. The output layer consisted of a single node with a linear activation function. We trained separate models with 10-fold cross validation on the complete LIFESPAN dataset using unharmonized ROIs, ROIs harmonized using ComBat with a linear model and ROIs harmonized using ComBat-GAM. The predictive accuracy of each model was evaluated using mean absolute error (MAE), i.e. mean absolute difference between predicted and actual ages. We also performed leave-site-out validations, using the PNC, SHIP, and BLSA-3T studies as independent test datasets in each experiment, in order to assess the effect of harmonization in predicting the brain age for data previously unseen by the training model.
2.8. LIFESPAN age trends of ROI volumes
After harmonization, we computed lifespan volumetric trends for each anatomical ROI, using GAM to model smoothed, nonlinear age trends. Since we were primarily interested in the relationship between age and ROI volumes, we regressed-out sex and ICV. The resulting age trends are free of sex and ICV effects, and enable a comprehensive analysis of brain volumes throughout the human lifespan.
Considering the large number of ROIs, we developed an interactive application that provides the end users a practical tool for selective visualization of the computed age trends for different brain regions. The visualization application, which allows users both to display LIFESPAN age trends and to position their own data after calibration with LIFESPAN data, was created with the Shiny package (Chang et al., 2019) in the R programming language, and is hosted at the following URL: https://rpomponio.shinyapps.io/neuro_lifespan/.
3. Results
3.1. Quality control of extracted variables
We manually-inspected 1,786 images, comprising roughly 17% of the original sample. Images were assessed for overall quality and sufficient resolution. As a result, we excluded 9 cases on the basis of low overall quality. Details of the cases excluded during the QC procedure are given in Supplementary Table 5.
3.2. Evaluation of goodness of fit with GAM versus linear and quadratic models on single-site data
Compared to linear models, GAMs achieved superior goodness-of-fit based on adjusted R-square for 124/145 ROIs in PNC, 123/145 ROIs in SHIP, and 126/145 ROIs in BLSA-3T. Compared to quadratic models, GAMs achieved superior goodness-of-fit based on adjusted R-square for 101/145 ROIs in the PNC, 116/145 ROIs in SHIP, and 128/145 ROIs in BLSA-3T. A summary of the comparative evaluation is presented in Table 2, which show the clear superiority of GAMs over linear models in out-of-sample fits, and the marginal superiority of GAMs over quadratic models. Results of the Chisquare test are given in Supplementary Table 6.
Table 2.
Results of evaluation of goodness of fit with GAM versus linear and quadratic models on single-site data.
| Number (%) of ROIs in which GAM achieved superior goodness of fit based on adjusted R-Square | Number (%) of ROIs in which GAM achieved superior goodness of fit based on out-of-sample RMSE* in split-sample validation | |||
|---|---|---|---|---|
| Dataset | GAM versus Linear | GAM versus Quadratic | GAM versus Linear | GAM versus Quadratic |
| PNC (n=1,444) | 124 (85.5%) | 101 (69.7) | 105 (72.4%) | 72 (49.7%) |
| SHIP (n=2,738) | 123 (84.8%) | 116 (80%) | 103 (71.0%) | 76 (52.4%) |
| BLSA-3T (n=964) | 126 (86.9%) | 128 (88.3%) | 109 (75.2%) | 74 (51.0%) |
RMSE: Root Mean Square Error
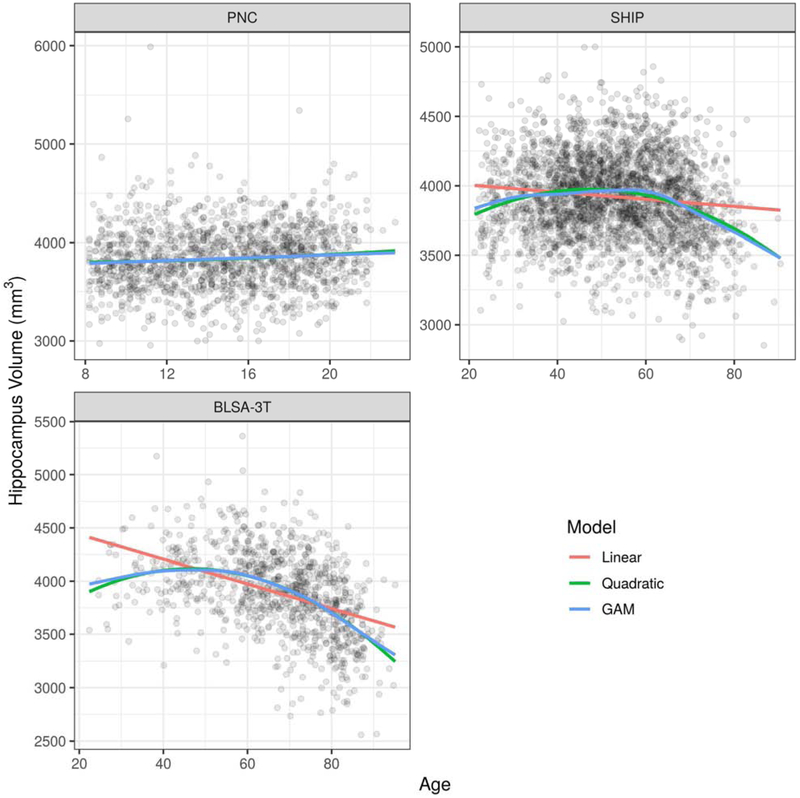
Figure 2 presents hippocampus volumes in the three selected studies with separate fits using linear models, quadratic models, and GAMs.
Figure 2:
Comparison of age trend estimates for the hippocampus volume from three studies (PNC, SHIP, and BLSA-3T) using linear models, quadratic models, and GAMs. The age trends plotted are for females and assume an average intra-cranial volume (ICV). In the top-left panel, the difference between fits is not distinguishable. In the top-right panel and the bottom-left panel, both the quadratic fit and the GAM fit exhibit clear improvement over the linear fit.
3.3. Simulation experiments
i. Effect of degree of overlap in the age ranges of data sites and sample size
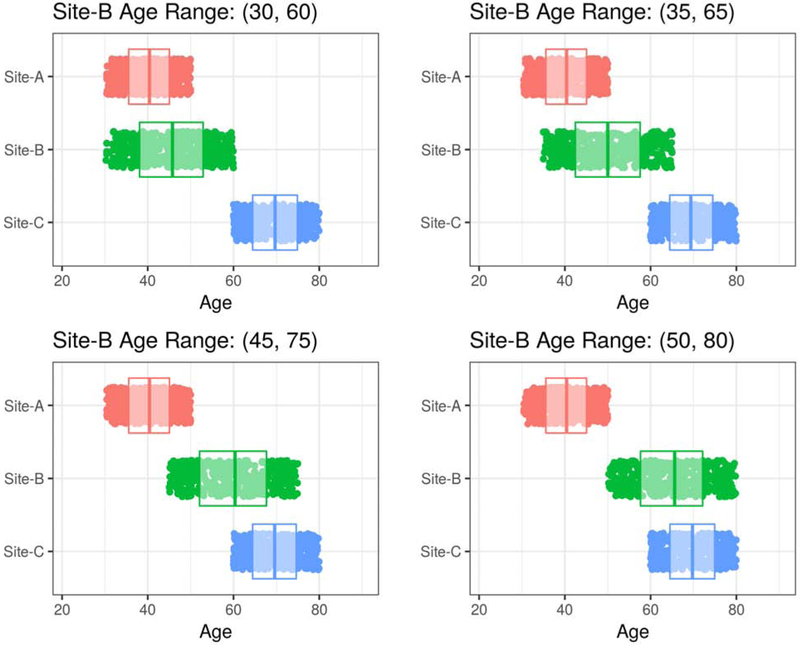
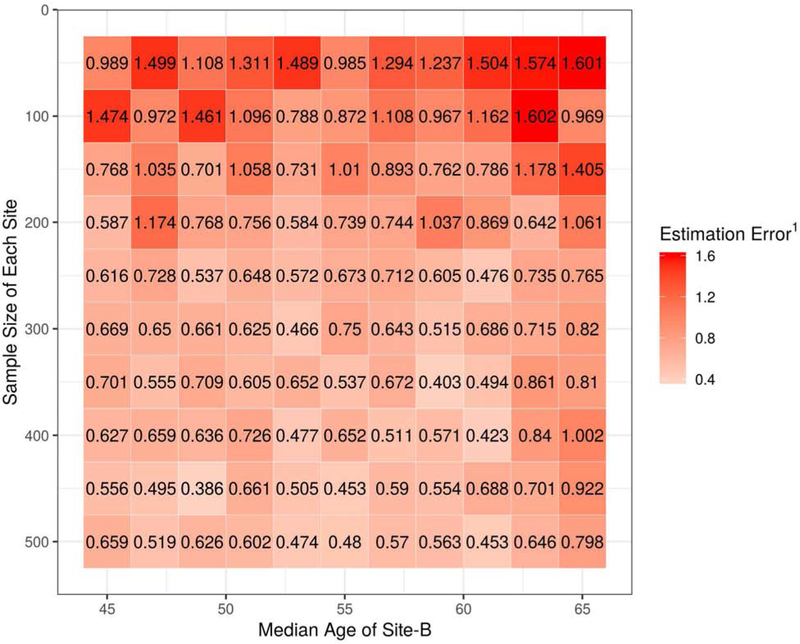
In Figure 3, we present four of the possible scenarios under the constraints of Simulation Experiment I. The age range of Site-B had a fixed width but was free to vary from younger to older ages. The sample sizes of each site were also free to vary. In Figure 4, we present the results of the grid search over the two free parameters in the simulation. Estimation error is expressed as the median relative Mean Absolute Error (rMAE) of age trend estimation across 10 randomized repetitions for each cell in the grid.
Figure 3:
Four possible scenarios under the constraints of Simulation Experiment I, which assessed the effect of different degrees of age overlap and sample size on harmonization performance. The age range of Site-B was free to vary from younger to older ages. In the upper-left panel, Site-B is overlapping only Site-A and not Site-C. In the lower-right panel, Site-B is overlapping only Site-C and not Site-A. In the upper-right panel and lower-left panel, Site-B is partially-overlapping both Site-A and Site-C.
Figure 4:
The relationship between the age trend estimation error and the two free parameters of Simulation Experiment I: the age range of Site-B and the sample size of each site. Note: 1 Estimation Error is expressed as the relative Mean Absolute Error (rMAE) of age trend estimation across 10 randomized repetitions for each cell in the grid.
Results generally improved when sample sizes were above 200 and the age overlap among sites was relatively balanced (Median age of Site-B between 50 and 60). The scope of the simulation was limited to three sites and ground-truth site effects were introduced artificially. However, we infer from the results that age range overlap is necessary for successful multi-site harmonization. In our LIFESPAN dataset, the only age ranges where a single site is present are beyond the age boundaries of 8 and 95. We caution that our age trend estimates may be less reliable at these extreme edges. In addition, we emphasize that the focus of our later analysis is not to yield strong conclusions about the age trends in developmental ranges less than 8 years old, but rather to demonstrate how to obtain age trends supported by large multi-site datasets.
ii. Effect of balancing sample sizes
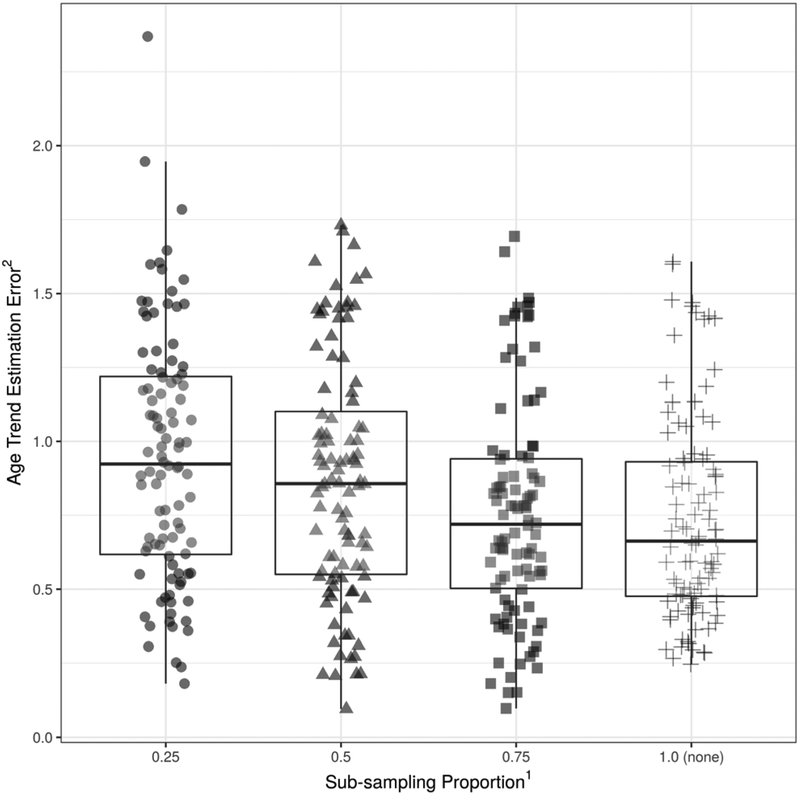
Age trend estimation errors for varying amounts of sub-sampling from the relatively large site are shown in Figure 5. The optimal performance was achieved when all data points were used, even though the relative ratio of sample sizes between sites was heavily unbalanced (n=400 vs n=100). These results suggest that the negative impact of reduced sample sizes is greater than that of unbalanced sample compositions in age trend estimation after harmonization, which is in contrast to our original hypothesis that balanced datasets would lead to better harmonization.
Figure 5:
The relationship between the age trend estimation error and the proportion of sub-sampling from Site-B in Simulation Experiment II. The original sample size of Site-B was four times larger than that of Site-A and Site-C. At 0.25, the size of Site-B after sub-sampling was equal to the size of Site-A and Site-C. At 0.5, the size of Site-B after sub-sampling was equal to the twice the size of Site-A and Site-C. Results were optimal when all data points were used. Note: 1Sub-sampling proportion was defined as the size of the sub-sampled size versus the original sample size of Site-B. 2Age Trend Estimation Error is expressed as the relative Mean Absolute Error (rMAE) of age trend estimation.
3.4. Harmonization of volumetric measurements from the LIFESPAN dataset
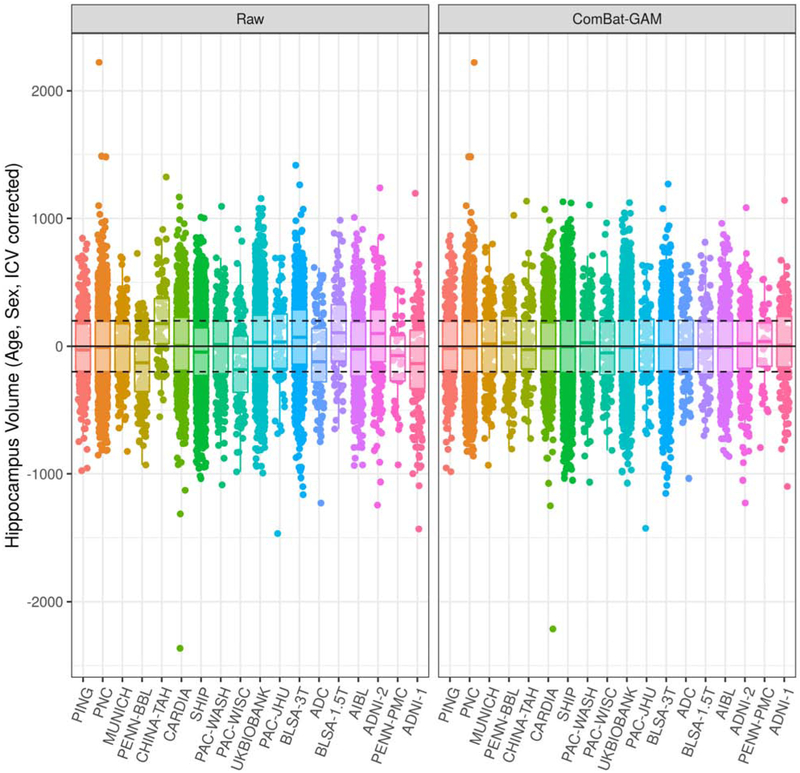
Our proposed harmonization method removed location and scale effects associated with site, after controlling for age, sex, and ICV with GAMs. Figure 6 shows the adjustments made to hippocampus volumes after harmonization. Adjustments for other important structures, as well as for total gray matter and total white matter, are shown in Supplementary Figure 1. After harmonization, the residual volumes by site are centered at zero as expected, indicating the removal of location effects. Scale effects were not as strong for the hippocampus, with the residual volumes by site showing similar variances before harmonization.
Figure 6:
Comparison of hippocampus volumes before and after harmonization, correcting for age, sex, and ICV using a GAM. Studies are ordered from youngest to oldest based on median age. In the left panel, volumes were not adjusted for site. In the right panel, volumes were adjusted with ComBat-GAM, which removes location (mean) and scale (variance) differences across sites after controlling for biological covariates. Horizontal lines are plotted at constants at 0, −200, and 200 for visual aid. Comparisons for additional ROI volumes are shown in Supplementary Figure 1.
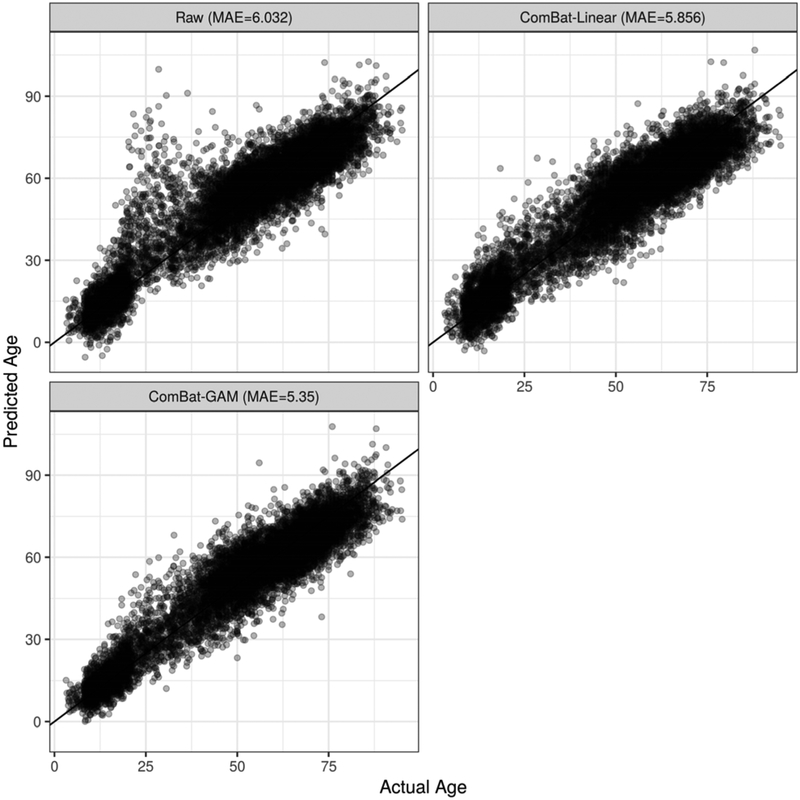
Age predictions obtained from the model trained using ROI volumes of participants with 10-fold cross validation were more accurate when the data were harmonized. Figure 7 shows predicted and actual ages for models trained on non-harmonized data, data harmonized with ComBat using a linear age model, and data harmonized using ComBat-GAM. While the application of ComBat with a linear model helped age prediction accuracy compared to no harmonization, the additional use of GAM yielded the best results of the three methods, achieving mean absolute error (MAE) of 5.35.
Figure 7:
Comparison of age prediction results using three harmonization methods and 10-fold cross validation with a fully-connected neural network using ROI Volumes as input features. MAE is the mean absolute error (i.e. actual age minus predicted age). In the top-left panel, data were unadjusted for site. In the top-right panel, data were harmonized with ComBat using a linear model. In the bottom-left panel, data were harmonized using ComBat-GAM.
In the leave-site-out validation experiments using the PNC, SHIP, and BLSA-3T as test datasets, harmonization with ComBat-GAM consistently led to improved prediction accuracy for each dataset, compared to using non-harmonized data or using data harmonized with ComBat using a linear age model (Table 3).
Table 3.
Results of leave-site-out age prediction for each harmonization method.
| MAE* obtained for each Harmonization Method | |||
|---|---|---|---|
| Dataset | Raw Data | ComBat-Linear | ComBat-GAM |
| PNC (n=1,444) | 7.418 | 7.27 | 5.412 |
| SHIP (n=2,738) | 6.737 | 6.502 | 6.151 |
| BLSA-3T (n=964) | 6.228 | 6.455 | 5.956 |
MAE: Mean absolute error, i.e. mean absolute difference between predicted and actual ages.
3.5. LIFESPAN age trends of ROI volumes
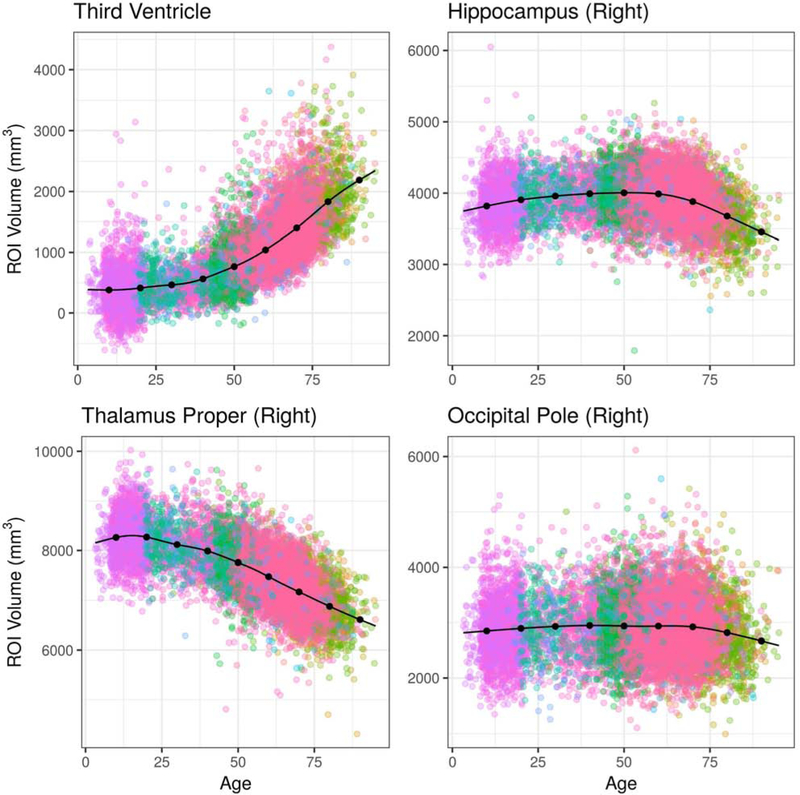
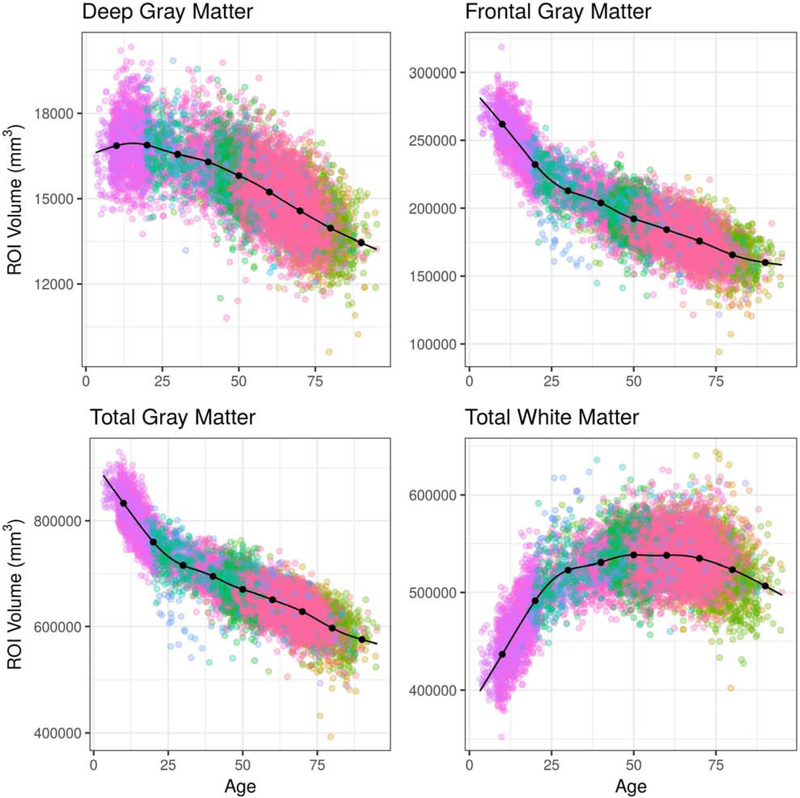
LIFESPAN age trends of the third ventricle, hippocampus, thalamus, and occipital pole are presented in Figure 8 and the age trends of 4 larger anatomical regions, total gray matter, frontal gray matter, total white matter and deep gray matter, are presented in Figure 9.
Figure 8:
Age trends for selected ROI volumes using the combined LIFESPAN dataset with 18 studies spanning the age range 3 – 96. Data were harmonized using ComBat-GAM. The age trends plotted are for females and assume an average intra-cranial volume (ICV).
Figure 9:
Age trends for selected composite ROI volumes using the combined LIFESPAN dataset with 18 studies spanning the age range 3 – 96. Composite ROI volumes were obtained by combining single ROIs into larger anatomical regions following a predefined ROI hierarchy. Data were harmonized using ComBat-GAM. The age trends plotted are for females and assume an average intra-cranial volume (ICV).
The age trends derived from the LIFESPAN data demonstrated variability at both the scales of single ROIs and composite ROIs. At the single ROI level, the hippocampus demonstrated accelerated atrophy late in the lifespan. From age 50 to 60, for example, the percentage difference in hippocampal volume declined by 0.344% over 10 years, according to the age trend. In contrast, hippocampal volume declined by 5.132% between age 70 and 80, and by 5.944% from age 80 to 90. Occipital pole volumes were relatively stable throughout the lifespan. Total gray matter volume demonstrated a period of rapid decline during adolescence, followed by more-gradual decline after age 25. Total white matter volume demonstrated growth during adolescence, stability between ages 30 and 70, and gradual decline after age 75.
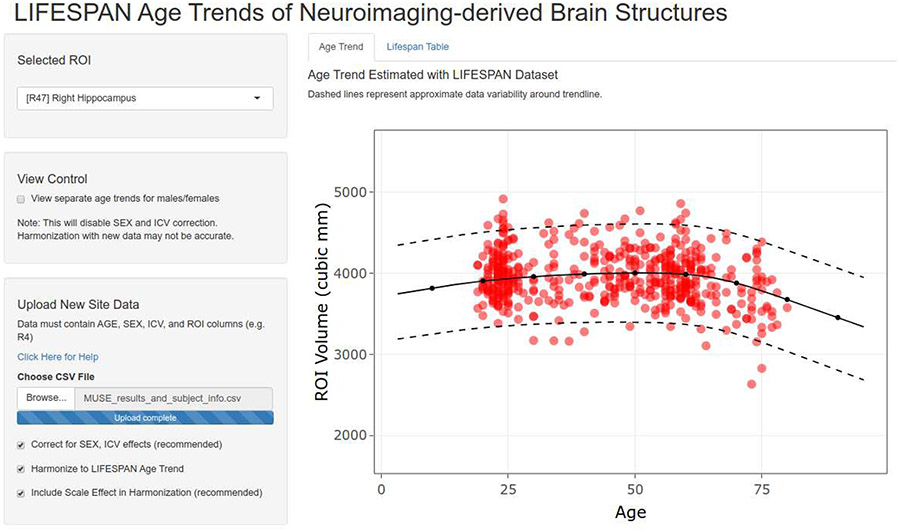
Age trends for each ROI from the harmonized dataset are made available via a web-based application hosted at the following URL: https://rpomponio.shinyapps.io/neuro_lifespan/. The application allows users to view the age trend of any ROI selected from the set of 145 ROIs harmonized in this study, as well as the 113 composite ROIs. The users may upload ROI volumes from a new study to visualize them and compare them with the presented age trends. The application also allows users to align their data to pre-calculated trends, by removing the location (mean) and scale (variance) differences between new ROI volumes and the reference dataset after controlling for age, sex, and ICV. Figure 10 shows a screenshot of the application being used to visualize the hippocampus volume for an independent dataset together with the LIFESPAN age trend for this ROI.
Figure 10:
Screenshot of the web-based application that allows visualization of the age trend for each anatomical ROI in our dataset. In red, an independent dataset has been uploaded after MUSE segmentation. New values are aligned to the LIFESPAN age trend by removing the location (mean) and scale (variance) differences between new ROI volumes and the reference dataset after controlling for age, sex, and ICV. The application is hosted at the following URL: https://rpomponio.shinyapps.io/neuro_lifespan/.
4. Discussion
We described and validated a methodology for harmonization and pooling of neuroimaging data across multiple scanners and cohorts. Using this methodology, as well as regional volumetric measures from 18 neuroimaging studies, we created a large-scale dataset of structural MRI scans covering nearly the entire human lifespan. We applied a fully-automated image processing pipeline to extract regional volumes, followed by an automated quality control procedure to ensure data integrity, and a systematic harmonization method to eliminate site effects while controlling for nonlinear age effects, with the final goal of deriving age trends of 258 brain regions at multiple resolution levels. In order to facilitate use of our methodology and data, we developed an interactive visualization and harmonization tool for displaying age trends of individual anatomical regions. This tool provides a reference frame for comparing the values of a new cohort against age trends estimated from 10,477 participants.
We proposed the use of generalized additive models (GAMs) to capture non-linearities in age-related differences in brain structure without over-fitting. Each ROI is modeled by a GAM that includes age as a nonlinear predictor and is optimized via restricted maximum likelihood with regularization to estimate a smooth function. GAMs were previously applied to capture nonlinear trends in a study of brain development in adolescents (Satterthwaite et al., 2014). In our experimental validations using three independent datasets with large sample sizes and spanning different age ranges, we demonstrated that a nonlinear model better-captured age-related differences in ROI volumes in different periods of the lifespan compared to linear and quadratic models. The superior performance of GAMs over linear models is consistent with evidence of non-linearity in various anatomical structures, such as gray matter lobes (Giedd et al., 1999), basal ganglia (Ziegler et. al., 2012), and the hippocampus in late-life participants (Allen et al., 2005; Janowitz et al., 2014).
In order to better-understand the behavior of our harmonization procedure relative to the age range covered by each study, we performed simulation experiments leveraging a single-site study in which we introduced artificial site effects. The first conclusion from these simulation experiments was that partially-overlapping age ranges were preferable to disjoint age ranges. This result was expected, as age-disjoint studies should be difficult to harmonize in the presence of nonlinear age effects. The second result suggested that using all available data was preferable to the benefit of balancing across multi-site samples.
Studies have used regional parcellation into anatomical ROIs to understand the brain morphologic changes during the lifespan as well as the effect of disease on the brain (Giedd et al., 1999; Ziegler et al., 2012). Often age has been associated with brain atrophy in various regions (Coffey et al., 1998; Habes et al., 2016), that could be linked to age-related pathologies such as neurodegenerative disorders (Dickerson et al., 2009; Whitwell et al., 2007), but also to the the normal process of aging, which was suggested to be accompanied by demyelination in the white matter and axonal loss (Hinman and Abraham, 2007). The individual’s genetic profile, lifestyle, environment, and disease-related risk factors interact together and contribute to the brain regional vulnerability to age-related changes (Janowitz et al., 2014; Rodrigue et al., 2013). Our harmonized data suggest that there is remarkable variability in the shape and nonlinearity of age trends of various ROIs, consistent with previous reports (Courchesne et al., 2000; Walhovd et al. 2011). For example, total gray matter (GM) volume decreases rapidly during late childhood and adolescence, and it continues to decrease, albeit at a much slower rate, in the adult life. We found that total brain white matter (WM) volume follows an inverted-U trend, with rapid increases throughout childhood and adolescence then assuming a downward trend around age 60, similar to the trend of Cerebral WM volume in Walhovd et al. (2005). Deep GM structures seem to be stable until early adult life, at which point volume declines.
When ROIs are used as building blocks in subsequent analyses, it is important to know the effect of harmonization on subsequently calculated biomarker indices. Toward this goal, we used predicted brain age from a model that summarizes volumetric measures across multiple ROIs as an index that captures the process of typical brain aging, and which has received increasing attention in the literature (Cole and Franke, 2017; Habes et al., 2016; Dosenbach et al., 2010; Erus et al., 2015; Franke et al., 2010). Our results indicated that harmonization has beneficial effects on the calculation of brain age by reducing the prediction error relative to unharmonized data by 11.3% based on the percentage difference in Mean Absolute Error (MAE) presented in Figure 7. This is a substantial improvement, especially since it is likely to influence the value of the residuals (brain age – age) that are typically used to flag advanced or resilient brain agers (Eavani et al., 2018).
One limitation of the current study is the lack of geographic and racial diversity of the cohorts. In part this is due to geographic biases in neuroimaging studies, which are concentrated in the USA and Europe. Asia was underrepresented, but several public imaging datasets are currently available that could augment the sample. The Consortium for Reliability and Reproducability (CoRR) is one example of a large repository of MRIs with several cohorts of Chinese participants that could be included in future analyses (Zuo et al., 2014). In addition, the Southwest University Adult Lifespan Dataset (SALD) provides images for a cross-sectional sample of healthy participants from China, covering the ages 19 to 80 (Wei et al., 2018). Beyond race, there are other sources of diversity that may affect neuroimaging, including genetic and environmental factors and subclinical pathology, which are less commonly assessed in research studies; potential neuroimaging correlates of these factors may be affected by the harmonization method. We encourage others who may have access to healthy-control datasets to use the publicly-available visualization tool we provide as a product of the LIFESPAN dataset (https://rpomponio.shinyapps.io/neuro_lifespan/). Finally, we have developed a package that enables users to apply ComBat-GAM on their own datasets (https://github.com/rpomponio/neuroHarmonize).
Our analyses have focused primarily on typically-developing and typically-aging participants, establishing age trends of brain regions for healthy controls. We included participants without neurological or psychiatric disorders; however, to harmonize studies which have a specific neurological or psychiatric disease as a focus, data from an appropriate control population is required. Patient data should then follow the same harmonization transformations, but patients should not be used in the calculation of the harmonization model. This is because the underlying assumption behind our approach is that each cohort’s measurements were drawn from the same distribution of values, albeit differing by age, sex, and intra-cranial volume (ICV). Patients with structural brain alterations could violate this assumption and, further, including them in the harmonization would attenuate disease-related effects. Hence, the age trend that we provided through the web-interface can serve as a reference based on large control population over a wide age range, and assuming a sufficient control sample is available, could assist with the harmonization task of relatively small pathologic studies, which is otherwise unfeasible.
The current study demonstrates the practical capability of pooling heterogeneous imaging datasets for downstream analysis, particularly at a large scale and in the presence of nonlinear age effects. Future efforts should focus on the application of this framework to other variables of interest and datasets, on the inclusion of patient volunteers to derive disease-specific trends, and on the extension of the current harmonization procedure to longitudinal studies.
Supplementary Material
Acknowledgements
This work was supported by the National Institute on Aging (grant number 1RF1AG054409), the National Institute of Mental Health (grant numbers 5R01MH112070; R01MH120482; R01MH112847), and the National Institutes of Health (grant number 75N95019C00022). MH was supported in part by The Allen H. and Selma W. Berkman Charitable Trust (Accelerating Research on Vascular Dementia) and the National Institutes of Health (grant number R01HL127659-04S1). TDS was supported in part by the National Institute of Mental Health (grant numbers R01MH120482, R01MH112847). DHW was supported in part by the National Institute of Mental Health (grant number R01MH113565). DAW was supported in part by the National institute on Aging (grant numbers AG010124; R01AG055005). RTS was supported in part by the National Multiple Sclerosis Society (grant number RG170728586) and National Institute of Neurological Disorders and Stroke (grant number R01NS060910). The Coronary Artery Risk Development in Young Adults Study (CARDIA) is supported by contracts HHSN268201800003I, HHSN268201800004I, HHSN268201800005I, HHSN268201800006I, and HHSN268201800007I from the National Heart, Lung, and Blood Institute (NHLBI). CARDIA was also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between NIA and NHLBI (AG0005). The Baltimore Longitudinal Study of Aging (BLSA) is supported by the Intramural Research Program, National Institute on Aging, NIH. This research has been conducted using the UK Biobank Resource under Application Number 35148. The Australian Imaging Biomakers and Lifestyle (AIBL) study was supported by funding from the Science and Industry Endowment Fund, the Dementia Collaborative Research Centres, the McCusker Alzheimer’s Research Foundation, the National Health and Medical Research Council (AUS), and the Yulgilbar Foundation, plus numerous commercial interactions supporting data collection. Details of the AIBL consortium can be found at www.AIBL.csiro.au and a list of the researchers of AIBL is provided at http://aibl.csiro.au/.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare that they have no competing interests.
References
- Fjell AM and Walhovd KB, 2010. Structural Brain Changes in Aging: Courses, Causes and Cognitive Consequences. Reviews in the Neurosciences 21 (3), 187–221. 10.1515/REVNEURO.2010.21.3.187. [DOI] [PubMed] [Google Scholar]
- Habes M, Janowitz D, Erus G, Toledo JB, Resnick SM, Doshi J, Van der Auwera S, Wittfeld K, Hegenscheid K, Hosten N, Biffar R, Homuth G, Völzke H, Grabe HJ, Hoffman W, Davatzikos C, 2016. Advanced brain aging: relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Translational Psychiatry 6, 775 10.1038/tp.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Chisum HJ, Townsend J, Cowles A, Covington J, Egaas B, Harwood M, Hinds S, Press GA, 2000. Normal Brain Development and Aging: Quantitative Analysis at in Vivo MR Imaging in Healthy Volunteers. Radiology 213 (3). 10.1148/radiology.216.3.r00au37672. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Tessner KD, Toga AW, 2001. Mapping continued brain growth and gray matter density reduction in dorsal frontal cortex: Inverse relationships during postadolescent brain maturation. Journal of Neuroscience 21 (22), 8819–8829. 10.1523/JNEUROSCI.21-22-08819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Sowell ER, 2006. Mapping brain maturation. Trends in Neurosciences 29 (3), 148–159. 10.1016/j.tins.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd J, Blumenthal J, Jeffries N, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans A, Rapoport J, 1999. Brain development during childhood and adolescence: a longitudinal MRI study. Nature Neuroscience 2, 861–863. 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Driscoll I, Davatzikos C, An Y, Wu X, Shen D, Kraut M, Resnick SM, 2009. Longitudional pattern of regional brain volume change differentiates normal aging from MCI. Neurology 72 (22), 1906–1913. 10.1212/WNL.0b013e3181a82634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KL, Goddings A, Herting MM, Meuwese R, Blakemore S, Crone EA, Dahl RE, Güroğlu B, Raznahan A, Sowell ER, Tamnes CK, 2016. Structural brain development between childhood and adulthood: Convergence across four longitudinal samples. NeuroImage 141, 273–281. 10.1016/j.neuroimage.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Mathalon DH, Sullivan EV, Rawles JM, Zipursky RB, Lim KO, 1994. A Quantitative Magnetic Resonance Imaging Study of Changes in Brain Morphology From Infancy to Late Adulthood. Archives of Neurology 51 (9), 874–887. 10.1001/archneur.1994.00540210046012. [DOI] [PubMed] [Google Scholar]
- Tamnes C, Østby Y, Fjell A, Westlye L, Tønnessen P, Walhovd K, 2010. Brain Maturation in Adolescence and Young Adulthood: Regional Age-Related Changes in Cortical Thickness and White Matter Volume and Microstructure. Cerebral Cortex 20 (3), 534–548. 10.1093/cercor/bhp118. [DOI] [PubMed] [Google Scholar]
- Terribilli D, Schaufelberger M, Duran F, Zanetti M, Curiati P, Menezes P, Scazufca M, Amaro E Jr., Leite C, Busatto G, 2011. Age-related gray matter volume changes in the brain during non-elderly adulthood. Neurobiology of Aging 32 (2), 354–368. 10.1016/j.neurobiolaging.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Vallee E, Vidaurre D, Webster M, McCarthy P, Rorden C, Daducci A, Alexander DC, Zhang H, Dragonu I, Matthews PM, Miller KL, Smith SM., 2019. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166, 400–424. 10.1016/j.neuroimage.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, et al. , 2014. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior 8 (2), 153–182. 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Roij SJH, Dennis EL, et al. , 2018. Smaller Hippocampal Volume in Posttraumatic Stress Disorder: A Multisite ENIGMA-PGC Study: Subcortical Volumetry Results From Posttraumatic Stress Dissorder Consortia. Biological Psychiatry 83 (3), 244–253. 10.1016/j.biopsych.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp TGM, Hibar DP, Rasmussen JM, et al. , 2016. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 health controls via the ENIGMA consortium. Molecular Psychiatry 21, 547–553. 10.1038/mp.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L, Veltman DJ, van Erp TGM, et al. , 2016. Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Molecular Psychiatry 21, 806–812. 10.1038/mp.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao H, Hayashi N, Ohtomo K, 2011. Effect of scanner in longitudinal studies of brain volume changes. Journal of Magnetic Resonance Imaging 32 (2), 438–444. 10.1002/jmri.22636. [DOI] [PubMed] [Google Scholar]
- Shinohara RT, Oh J, Nair G, Calabresi PA, Davatzikos C, Doshi J, Henry RG, Kim G, Linn KA, Papinutto N, Pelletier D, Pham DL, Reich DS, Rooney W, Roy S, Stern W, Tummala S, Yousuf F, Zhu A, Sicorette NL, Bakshi R, the NAIMS Cooperative, 2017. Volumetric Analysis from a Harmonized Multisite Brain MRI Study of a Single Subject with Multiple Sclerosis. American Journal of Neuroradiology 38 (8), 1501–1509. 10.3174/ajnr.A5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Hu R, Taylor M, Tso Y, Yiannoustos C, Navia B, Mori S, Ekholm S, Schifitto G, Zhong J, 2011. Quantification of accuracy and precision of multi-center DTI measurements: A diffusion phantom and human brain study. Neuroimage 56 1398–1411. 10.1016/j.neuroimage.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWinn K, Sheridan M, Keyes K, Hamilton A, McLaughlin K, 2017. Sample composition alters associations between age and brain structure. Nature Communications 8 874 10.1038/s41467-017-00908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R, Hao X, Liu F, Xu D, Liu J, Peterson BS, 2013. The effects of changing water content, relaxation times, and tissue contrast on tissue segmentation and measures of cortical anatomy in MR images. Magnetic Resonance Imaging 31 (10), 1709–1730. 10.1016/j.mri.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Xu T, Milham MP, 2019. Harnessing reliability for neuroscience research. Nature Human Behavior 3, 768–771. 10.1038/s41562-019-0655-x. [DOI] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Resnick S, Gur RC, Gur RE, Satterhwaite T, Furth S, Davatzikos C, 2016. MUSE: Multi-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally-optimal atlas selection. Neuroimage 127, 186–195. 10.1016/j.neuroimage.2015.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell A, Walhovd KB, Westlye LT, Østby Y, Tamnes CK, Jernigan TL, Ganmst A, Dale AM, 2010. When does brain aging accelerate? Dangers of quadratic fits in cross-sectional studies. Neuroimage 50 (4), 1376–1383. 10.1016/j.neuroimage.2010.01.061. [DOI] [PubMed] [Google Scholar]
- Ziegler G, Dahnke R, Jäncke L, Yotter RA, May A, Gaser C, 2012. Brain structural trajectories over the adult lifespan. Human Brain Mapping 33 (10), 2377–2389. 10.1002/hbm.21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayumak SC, Bouix S, Ning L, James A, Crow T, Shenton M, Kubicki M, Rathi Y, 2019. Retrospective harmonization of multi-site diffusion MRI data acquired with different acquisition parameters. NeuroImage 184, 180–200. 10.1016/j.neuroimage.2018.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie TJ, Tibshirani RJ, 1986. Generalized Additive Models. Statistical Science 1 (3), 297–310. 10.1214/ss/1177013604. [DOI] [Google Scholar]
- Wood S, 2017. Generalized Additive Models: An Introduction with R. Chapman & Hall/CRC; (2nd Edition). [Google Scholar]
- Johnson WE, Li C, Rabinovic A, 2007. Adjusting batch effects in mircoarray expression data using empirical Bayes methods. Biostatistics 8, 118–127. 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Fortin JP, Cullen N, Sheline Y, Taylor W, Aselcioglu I, Cook P, Adams P, Cooper C, Fava M, McGrath P, McInnis M, Phillips M, Trivedi M, Weissman M, Shinohara R., 2018. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167, 104–120. 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Parker D, Tunc B, Watanbe T, Elliott M, Ruparel K, Roalf D, Satterwaite T, Gur RC, Gur RE, Schultz R, Verma R, Shinohara R, 2017. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 161, 149–170. 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Linn K, Cook P, Phillips M, McInnis M, Fava M, Trivedi M, Weissman M, Shinohara R, Sheline Y, 2018. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Human Brain Mapping 39, 4213–4227. 10.1002/hbm.24241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr., Bernstein MA, Fox NC, et al. , 2008. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging 27 (4), 685–691. 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong NM, An Y, Beason-Held L, Doshi J, Erus G, Ferrucci L, Davatzikos C, Resnick SM, 2019. Predictors of neurodegeneration differ between cognitively normal and subsequently impaired older adults. Neurobiology of Aging 75, 178–186. 10.1016/j.neurobiolaging.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C, 2003. Longitudinal Magnetic Resonance Imaging Studies of Older Adults: A Shrinking Brain. Journal of Neuroscience 23 (8), 3295–3301. 10.1523/JNEUROSCI.23-08-03295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr, Liu K, Savage PJ, 1988. CARDIA: study design, recruitment, and some characteristics of the examined subjects. Journal of Clinical Epidemiology 41 (11), 1105–16. 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- Jernigan TL, Brown TT, Hagler DJ Jr, et al. , 2016. The Pediatric Imaging, Neruocognition, and Genetics (PING) Data Repository. Neuroimage 124, 1149–1154. https://dx.doi.org/10.1016%2Fj.neuroimage.2015.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Connolly JJ, Ruparel K, Calkins ME, Jackson C, Elliott MA, Roalf DR, Hopson R, Prabhakaran K, Behr M, Qiu H, Mentch DF, Chiavacci R, Slieman PMA, Gur RC, Hakonarson H, Gur RE, 2016. The Philadelphia Neurodevelopmental Cohort: A publicly available resource for the study of normal and abnormal brain development in youth. NeuroImahe 124 (Part B), 116–1119. 10.1016/j.neuroimage.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avantis BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: Improved N3 Bias Correction. IEEE Transactions on Medical Imaging 29 (6), 1310–1320. 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C, 2013. Multi-Atlas Skull-Stripping. Academic Radiology 20 (12), 1566–1576. 10.1016/j.acra.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asman A, Akhondi-Asl A, Wang H, Tustison N, Avants B, Warfield SK, Landman B, 2013. Miccai 2013 segmentation algorithms, theory and applications (SATA) challenge results summary. MICCAI Challenge Workshop on Segmentation: Algorithms, Theory and Applications (SATA). https://scholar.harvard.edu/akhondi-asl/publications/miccai-2013-segmentation-algorithms-theory-and-applications-sata-challenge.
- Wood S, 2003. Thin plate regression splines. Journal of the Royal Statistical Society Series B Statistical Methodology 65 (1), 95–114). 10.1111/1467-9868.00374. [DOI] [Google Scholar]
- Völzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, Aumann N, Lau K, Piontek M, Born G, et al. , 2010. Cohort Profile: The Study of Health in Pomerania. International Journal of Epidemiology 40 (2), 294–307. 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire JJ, Xie Y, McPherson J, 2019. shiny: Web Application Framework for R. R package version 1.3.2. https://cran.r-project.org/web/packages/shiny/index.html.
- Satterthwaite TD, Shinohara RT, Wolf DH, Hopson RD, Elliott MA, Vandekar SN, Ruparel K, Calkins ME, Roalf DR, Gennatas ED, Jackson C, Erus G, Prabhakaran K, Davatzikos C, Detre JA, Hakonarson H, Gur RC, Gur RE, 2014. Impact of puberty on the evolution of cerebral perfusion during adolescence. Proceedings of the National Academy of Sciences of the United States of America 111 (23), 8643–8648. 10.1073/pnas.1400178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J, Bruss J, Brown CK, Damasio H, 2005. Normal neuroanatomical variation due to age: The major lobes and a parcellation of the temporal region. Neurobiology of Aging 26 (9), 1245–1260. 10.1016/j.neurobiolaging.2005.05.023 [DOI] [PubMed] [Google Scholar]
- Janowitz D, Schwahn C, Borchardt U, Wittfeld K, Schulz A, Barnow S, Biffar R, Hoffman W, Habes M, Homuth G, Nauck M, Hegenscheid K, Lotze M, Völzke H, Freyberger HJ, Debette S, Grabe HJ, 2014. Genetic, psychosocial and clinical factors associated with hippocampal volume in the general population. Translational Psychiatry 4 10.1038/tp.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey CE, Lucke JF, Saxton JA, Ratcliff G, Jo Unitas L, Billig B, Bryan RN, 1998. Sex Differences in Brain Aging: a quantitative magnetic resonance imaging study. Archives of Neurology 55 (2). 169–179. 10.1001/archneur.55.2.169. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, et al. , 2009. The cortical Signature of Alzheimer’s Disease: Regionally Specific Cortical Thinning Relates to Symptom Severity in Very Mild to Mild AD Dementia and is Detectable in Asymptomatic Amyloid-Positive Individuals. Cerebral Cortex 19 (3), 497–510. 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve DF, Petersen RC, Jack CR Jr, 2007. 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain 130 (7), 1777–1786. 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman JD, Abraham CR, 2007. What’s Behind the Decline? The Role of White Matter in Brain Aging. Neurochemical Research 32 (12), 2023–2031. 10.1007/s11064-007-9341-x. [DOI] [PubMed] [Google Scholar]
- Rodrigue KM, Rieck JR, Kennedy KM, Devours MD, Diaz-Arrastia R, Park DC, 2013. Risk Factors for β-Amyloid Deposition in Healthy Aging: Vascular and Genetic Effects. JAMA Neurology 70 (5), 600–606. 10.1001/jamaneurol.2013.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd K, Westlye LT, Amlien I, Espeseth T, Reinvang I, Raz N, Agartz I, Salat DH, Greve DN, Fischl B, Dale AM, Fjell AM, 2011. Consistent neuroanatomical age-related volume differences across multiple scanners. Neurobiology of Aging 32 (5), 916–932. 10.1016/j.neurobiolaging.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Reinvang I, Lundervold A, Dale AM, Eilertsen DE, Quinn BT, Salat D, Makris N, Fischl B, 2005. Effects of age on volumes of cortex, white matter and subcortical structures. Neurobiology of Aging 26 (9), 1261–1270. 10.1016/j.neurobiolaging.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Cole JH, Franke K, 2017. Predicting Age Using Neuroimaging: Innovative Brain Ageing Biomarkers. Trends in Neuroscience 40 (12), 681–690. 10.1016/j.tins.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Dosenbach NUF, Nardos B, Cohen AL. Fair DA, Power JD, et al. , 2010. Prediction of Individual Brain Maturity using fMRI. Science 329 (5997), 1358–1361. 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erus G, Battapady H, Satterthwaite TD, Hakonarson H, Gur RE, Davatzikos C, Gur RC, 2015. Imaging patterns of Brain Development and their Relationship to Cognition. Cerebral Cortex 25 (6), 1676–1684. 10.1093/cercor/bht425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Ziegler G, Klöppel S, Gaser C, the Alzheimer’s Disease Neuroimaging Initiative, 2010. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. Neuroimage 50 (3), 883–892. 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Eavani H, Habes M, Satterthwaite TD, An Y, Hsieh M, Honnorat N, Erus G, Doshi J, Ferrucci L, Beason-Held LL, Resnick SM, Davatzikos C, 2018. Heterogeneity of structural and functional imaging patterns of advanced brain aging revealed via machine learning methods. Neurobiology of Aging 71, 41–50. 10.1016/j.neurobiolaging.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Anderson JS, Bellec P, et al. , 2014. An open science resource for establishing reliability and reproducability in functional connectomics. Scientific Data 1 10.1038/sdata.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Zhuang K, Ai L, Chen Q, Yang W, Liu W, Wang K, Sun J, Qiu J, 2018. Structural and functional brain scans from the cross-sectional Southwest University adult lifespan dataset. Scientific Data 5 10.1038/sdata.2018.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.