Abstract
Electron cryo-microscopy (cryo-EM) is increasingly being used to determine 3D structures of a broad spectrum of biological specimens from molecules to cells. Anticipating this progress in the early 2000s, an international collaboration of scientists with expertise in both cryo-EM and structure data archiving was established (EMDataResource, previously known as EMDataBank). The major goals of the collaboration have been twofold: to develop the necessary infrastructure for archiving cryo-EM-derived density maps and models, and to promote development of cryo-EM structure validation standards. We describe how cryo-EM data archiving and validation have been developed and jointly coordinated for the Electron Microscopy Data Bank and Protein Data Bank archives over the past two decades, as well as the impact of evolving technology on data standards. Just as for X-ray crystallography and nuclear magnetic resonance, engaging the scientific community via workshops and challenging activities has played a central role in developing recommendations and requirements for the cryo-EM structure data archives.
INTRODUCTION
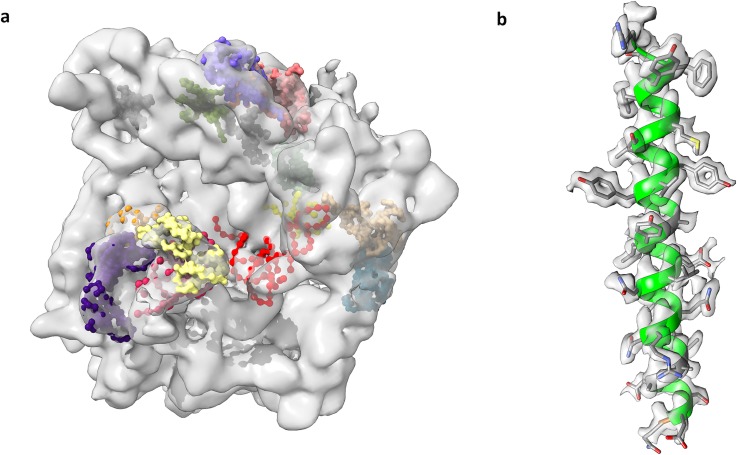
Electron cryo-microscopy (cryo-EM) has very recently become a mainstream area of structural biology and medicine, enabling 3D visualization of a wide variety of biologically important complexes that were previously inaccessible to science. Early cryo-EM 3D density maps typically lacked atomic detail, yielding only the overall molecular shape, but could still sometimes be interpreted at a “pseudo-atomic” level via fitting of previously known coordinates or homology models [Fig. 1(a)].1 Recent major technological advances now make it increasingly possible to directly visualize atomic details [Fig. 1(b)].2,3 These achievements were recognized by the award of the 2017 Chemistry Nobel Prize to cryo-EM pioneers Dubochet, Frank, and Henderson.4
FIG. 1.
Cryo-EM: contrasting early (2000) vs recent (2019). (a) Cryo-EM structure of the E. coli 70S ribosome determined by the Frank group at 11.5 Å, one of the first maps deposited in the EMDB archive (EMD-1003). It is shown here superimposed with manually fitted components deposited to PDB (1eg0).37 (b) Helical segment of the 1.8 Å apoferritin map used as a target in the 2019 model challenge,36 with a fitted model.
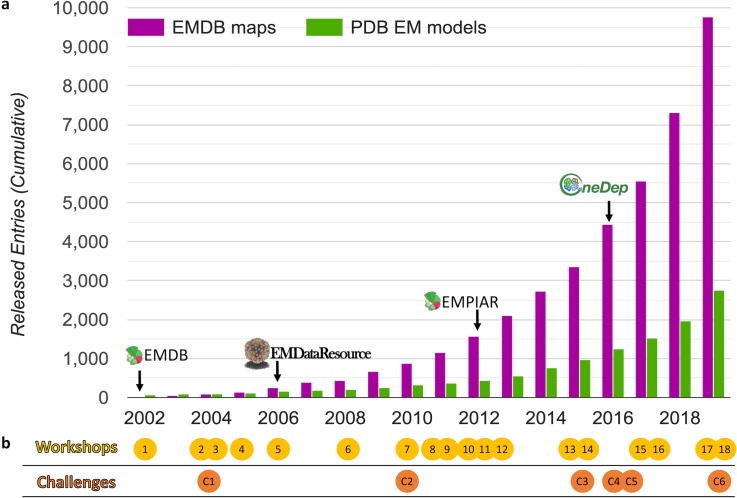
The development of cryo-EM is directly reflected by the growth of cryo-EM structure depositions contributed worldwide to public data archives [Fig. 2(a)]. The archiving systems and underlying data standards supporting deposition, annotation, release, and validation of cryo-EM structures and the associated metadata describing cryo-EM experiments have been developed over time to support this growth.5 We outline here the history of these systems and describe the process by which data standards have been developed, highlighting the role of engaging the scientific community to develop recommendations and requirements. The archiving systems and standards continue to evolve as technology drives the need for new descriptors and validation metrics.
FIG. 2.
Growth of data archives and community activity timeline. (a) Released map entries in EMDB and released EM model coordinate entries in PDB are shown, cumulative by year. Milestones (indicated with arrows) are described in the main text. Plot source: emdataresource.org. (b) Workshops (yellow circles) and challenges (orange circles) related to data standards and validation development are plotted according to the year they were held. Numbers within the circles correspond to Tables II and III rows.
CRYO-EM STRUCTURE DATA ARCHIVING
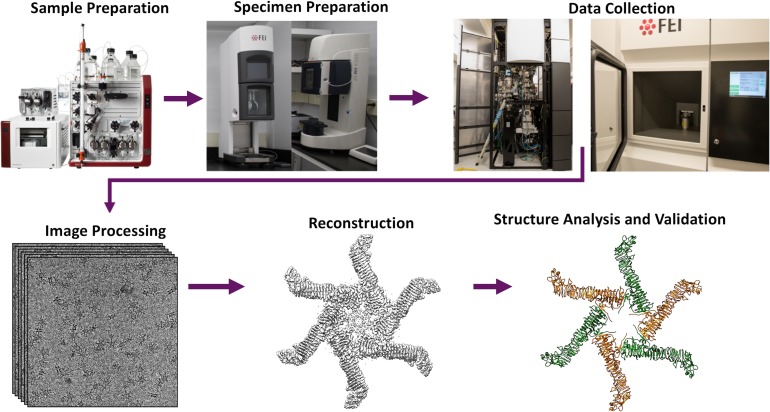
The Protein Data Bank (PDB), established in 1971 as a public archive for atomic coordinates of biological structures derived from X-ray crystallography,6 began accepting models derived from nuclear magnetic resonance spectroscopy (NMR) in 1988,7 and from electron microscopy (EM) and electron crystallography (EC) in 1990.8 In recognition of the fact that publicly available 3D density maps could accelerate discovery in structural biology and medicine, the Electron Microscopy Data Bank (EMDB) at the European Bioinformatics Institute (EBI) was launched in 2002 with support from the European Union.9 EMDB's launch was quickly followed by a pair of editorials in Structure and Nature Structural Biology encouraging electron microscopists to deposit their density maps.10,11 Similar to BioMagResBank (BMRB), which archives experimental data from NMR,12 the EMDB accepts maps determined using any cryo-EM method, including single particle reconstruction with any symmetry, helical filament reconstruction, subtomogram averaging, tomography, and electron crystallography, along with metadata describing the full experimental workflow (Fig. 3).
FIG. 3.
Cryo-EM experimental workflow. The major steps for determining a structure using cryo-EM single particle reconstruction are shown. The specimen shown is Helicobacter pylori vacuolating cytotoxin A oligomer.38
In 2006, scientists in the UK (EMDB) and USA [Research Collaboratory for Structural Bioinformatics (RCSB) and the National Center for Macromolecular Imaging (NCMI)] initiated a collaboration funded by National Institutes of Health (NIH) aimed to ensure that data archiving and validation standards for cryo-EM maps and models would be coordinated internationally.13,14 The project, formerly known as EMDataBank, was recently rebranded as EMDataResource (EMDR; emdataresource.org). The EMDR project website serves as a global resource for cryo-EM structure data archiving and retrieval, news, events, software tools, data standards, validation methods, and community challenges.
EMDep, designed and implemented at EBI, was the first system designed to collect and annotate maps and associated metadata for EMDB.9 In 2008, the EMDR team created a joint map+model deposition system for cryo-EM structures by connecting EMDep with AutoDep and ADIT (AutoDep Input Tool), the PDB data collection systems at the EBI and RCSB sites.13 The system that was implemented enabled a one stop shop for cryo-EM model and map depositions. Joint curation ensured that maps and models were deposited at the same physical scale and in the same coordinate frame. Journals that publish cryo-EM structures began to require authors to deposit maps to EMDB and models to PDB. This system supported the processing and release of nearly 4000 maps and 1000 models over a nine-year period (2008–2015).14
In 2012, the Electron Microscopy Public Image Archive (EMPIAR) was established at EBI.15 Supported by the UK Medical Research Council and UK Biotechnology & Biological Sciences Research Council, EMPIAR enables cryo-EM scientists to archive and share raw images and intermediate data files associated with their maps deposited to EMDB. Making recently collected image data broadly available has multiple benefits, including accelerating development of reconstruction software, and enriching resources for cryo-EM scientists in training. EMPIAR has its own deposition and curation system, but accesses metadata from the related EMDB entry. Individual entry storage sizes can be up to 15 TB. Approximately 4% of EMDB entries deposited since 2012 have associated EMPIAR entries.
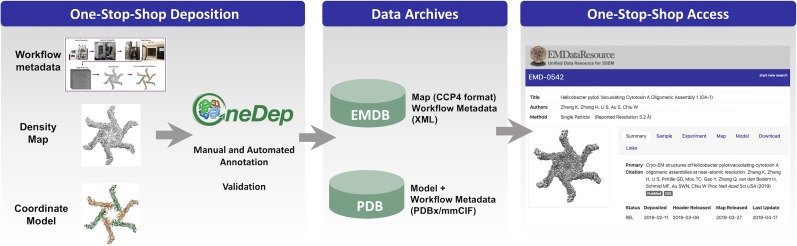
The Worldwide PDB (wwPDB) is the global organization that manages the PDB archive.16 In 2016, deposition, annotation, and release of cryo-EM structure maps and models were migrated to the wwPDB OneDep system (Fig. 4), using requirements that were initiated and developed by EMDR.17 At that time, it became mandatory to deposit maps to EMDB for all cryo-EM models deposited to PDB. In addition, structure validation reports, which can be provided by depositors in an official PDF format to journal editors and reviewers as part of manuscript review, began to be produced for all cryo-EM structures.18
FIG. 4.
Current systems for deposition, archiving, and accessing cryo-EM structures. Worldwide, every cryo-EM structure (map, experimental metadata, and optionally coordinate model) is deposited and processed through the wwPDB OneDep system (deposit.wwpdb.org), following the same annotation and validation workflow also used for X-ray crystallography and NMR structures.17,18 Map-only depositions yield an EMDB entry, while joint map+model depositions yield both EMDB and PDB entries. Workflow metadata collected in OneDep are passed to both EMDB and PDB. EMDB holds all workflow metadata while PDB holds a subset of the metadata; see Table I. The PDB and EMDB archives are accessible by FTP and rsync at wwPDB mirror sites in the US, UK, and Japan. Released cryo-EM structure data from both archives can be accessed via EMDataResource, EMDB, and wwPDB partner websites.
CREATING A DATA DICTIONARY
The foundation of any data repository is its data representation scheme. Based in part on the International Union of Crystallography dictionary for small molecule crystallography Crystallographic Information File (CIF),19 the Macromolecular Crystallographic Information File (mmCIF) was developed in the 1990s to support rich data content for the macromolecular crystallographic experiment and its results, with precise data type definitions, logical groupings for related data items, explicit parent-child relationships, enumerations for controlled vocabulary, extensibility, and many other features embedded in a computer-readable format.20 This dictionary is now the Master Format for the PDB. Particularly relevant for cryo-EM, very large complexes are readily represented, since mmCIF has no limits on the number of atoms or polymer chains.
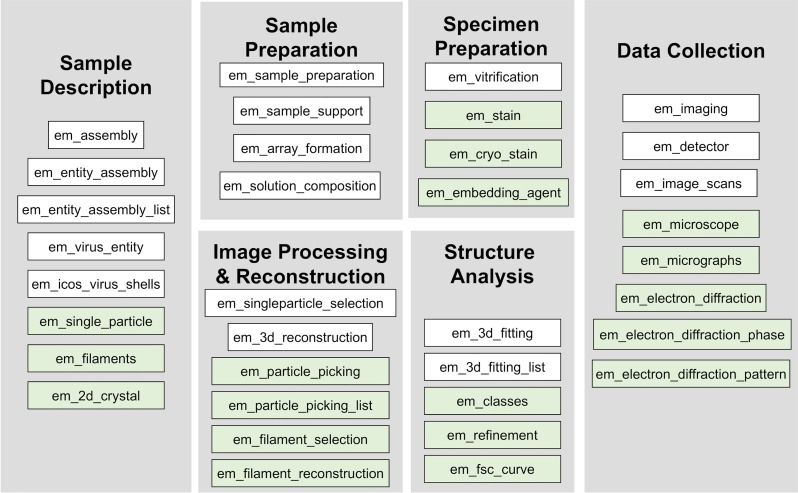
Following the lead of the crystallographic community, an mmCIF extension dictionary containing data terms for cryo-EM experiments was drafted jointly in the early 2000s based on requirements provided by the cryo-EM community. The dictionary was vetted and expanded by the scientific community via multiple workshops, and subsequently integrated by EMDR into the PDBx/mmCIF dictionary for use in the hybrid joint map+model deposition system.13 In 2015, based on feedback from additional workshops, the EMDR team further modified and expanded the dictionary in several ways. Hierarchical descriptions of complex specimens were enabled, and experimental descriptions for each of the cryo-EM methods were extended.5 The >500 term EM dictionary (Table I) is now the basis for cryo-EM depositions to both EMDB and PDB in the wwPDB OneDep system. The dictionary continues to be updated regularly to support the evolving needs of the scientific community.
TABLE I.
PDBx/mmCIF EM Dictionary used by wwPDB OneDep. As described in Fig. 4, all workflow metadata categories are collected by the OneDep system. Most categories are archived in both PDB and EMDB; asterisked categories are archived only in EMDB.
| Top level | Sample/specimen preparation | Image processing & reconstruction | Experimental data |
| em_experiment | em_buffer | em_3d_reconstruction | |
| em_software | em_buffer_component | em_image_processing | em_map* |
| em_crystal_formation | em_particle_selection | em_structure_factors* | |
| Sample description | em_embedding | em_volume_selection | em_layer_lines* |
| em_entity_assembly | em_sample_support | em_ctf_correction | |
| em_entity_assembly_molwt | em_specimen | ||
| em_entity_assembly_naturalsource | em_staining | em_2d_crystal_entity | |
| em_entity_assembly_recombinant | em_vitrification | em_3d_crystal_entity | |
| em_virus_entity | em_helical_entity | ||
| em_virus_natural_host | em_single_particle_entity | ||
| em_virus_shell | em_fiducial_markers* | ||
| em_focused_ion_beam* | em_euler_angle_assignment* | ||
| Data collection | em_grid_pretreatment* | em_final_classification* | |
| em_diffraction | em_high_pressure_freezing* | em_start_model* | |
| em_diffraction_shell | em_shadowing* | ||
| em_diffraction_stats | em_support_film* | Structure analysis | |
| em_image_recording | em_tomography* | em_3d_fitting | |
| em_image_scans | em_tomography_specimen* | em_3d_fitting_list | |
| em_imaging | em_ultramicrotomy* | em_fsc_curve* | |
| em_imaging_optics |
GATHERING COMMUNITY REQUIREMENTS
Developing a trusted scientific data repository requires careful attention to the interplay among science, technology, and community interest.21 Workshops and Challenges are two types of community outreach activities that are effective in bringing these three elements together; both have been employed multiple times to move EM data and validation standard development forward [Fig. 2(b)]. Workshops (typically 2–3 days) enable groups of experts to review current practices and develop recommendations, while Challenges (taking place over several months to a year) provide forums for experts to exercise and demonstrate current workflows and test novel procedures. Challenges can incorporate one or more workshops for planning or results review. Tables II and III list and summarize goals and outcomes of 18 international workshops and six challenges held over the past two decades. Below we provide additional descriptions of selected activities, as well as a montage of workshop photos (Fig. 5).
TABLE II.
Cryo-EM community data archiving and validation workshops.
| # | Year | Title/location | Organizers | Description | Key outcomes |
|---|---|---|---|---|---|
| 1 | 2002 | IIMS Workshop, UK | Kim Henrick, José-María Carazo, Stephen Fuller | Promote software development in the field of 3DEM molecular structure determination | Guidelines and release policies for the new EMDB archive.10,11,39 Deposition system to collect EMDB data and maps9 |
| 2 | 2004 | 3DEM Developers workshop, UK | Kim Henrick | Developer review of tools and software practices used in the field of cryoEM | Priorities developed for EMDB including electron tomography, PDB-EMDB cross-referencing, lossless map conversion, review of community map standards and conventions |
| 3 | 2004 | Cryo-EM Structure Deposition Workshop, NJ, USA | Helen Berman, Wah Chiu, Michael Rossmann | Develop community consensus on data items needed for deposition of maps and atomic models derived from cryoEM | Need for deposition one-stop-shop articulated. Recommendations incorporated in extended EM data dictionary13 |
| 4 | 2005 | 3DEM Developers Workshop, UK | Kim Henrick | Introduced EM data dictionary to software developers to facilitate its integration into 3DEM packages and electronic notebooks | Agreement to adopt a common set of conventions for maps22 |
| 5 | 2006 | CryoEM Standards Task Force, TX, USA | Wah Chiu, David Belnap, José-María Carazo | Gather cryoEM map conventions and formats with associated metadata used by different developers | Key conventions (e.g., Euler angles) were evaluated for interoperability and conversion tools were created40,41 |
| 6 | 2008 | Electron Crystallography Data Model Workshop, CA, USA | Wah Chiu, Cathy Lawson | Gather expert advice on metadata items in the EM dictionary pertaining to electron crystallography | Recommendations incorporated into EM data dictionary13 |
| 7 | 2010 | EM Validation Task Force, NJ, USA | Helen Berman, Wah Chiu, Gerard Kleywegt, Cathy Lawson | Expert review of potential validation standards for maps and models produced by 3DEM reconstruction | Recommendations summarized in white paper,25 and implemented in OneDep validation reports14,18 |
| 8 | 2011 | Model Challenge Workshop, HI, USA | Steve Ludtke, Cathy Lawson, Gerard Kleywegt, Helen Berman, Wah Chiu | Computational groups described and compared tools they used to model a selected set of published cryoEM structures | Results published in Biopolymers journal special issue42 |
| 9 | 2011 | Data Management Challenges in 3DEM, UK | Ardan Patwardhan, Gerard Kleywegt, Jason Swedlow | Gather expert advice on key topics in data archiving and validation for 3DEM data, including data model, validation, raw-data archiving | Recommendations summarized in white paper.23 Web-based visualization tools developed.28 EMPIAR raw data archive created.15 Extended data model implemented17 |
| 10 | 2012 | 3DEM Modeling Workshop, TX, USA | Wah Chiu | Current challenges in creating and validating coordinate models built into cryo-EM maps | Recommendations gathered for modeling and validation standards and future model challenges |
| 11 | 2012 | 3D Cellular Context for the Macromolecular World, UK | Ardan Patwardhan, Gerard Kleywegt, Jason Swedlow | Discussions on data archiving and validation for emerging 3D cellular imaging techniques | Recommendations summarized in white paper24 |
| 12 | 2012 | Instruct Image Processing Center Developer Workshop, Spain | José-María Carazo | Effort to standardize information exchange in single particle reconstruction and to establish algorithm benchmarking | CTF benchmarking challenge43 and EMX convention for CTF and single-particle parameters developed44 |
| 13 | 2015 | CryoEM Model Validation Workshop, MA, USA | Wah Chiu, Cathy Lawson, Paul Adams | Modeling experts met to present and discuss challenges in modeling into cryoEM maps | Gathered recommendations5 directly used in development of 2016 Model Challenge |
| 14 | 2015 | Building Bridges between Cellular and Molecular Structural Biology, UK | Ardan Patwardhan, Gerard Kleywegt | Expert discussions on how to integrate structural data from a diverse range of public archives covering cellular and molecular structural biology | Recommendations to develop tools/file formats for map segmentation, and tools to support biological structure annotation described in white paper45 |
| 15 | 2017 | Model Challenge Assessment, LA, USA | Wah Chiu, Cathy Lawson, Paul Adams | First pass analyses of models and data submitted to the 2016 Model Challenge | Recommended metrics implemented on model challenge comparison website46 |
| 16 | 2017 | CryoEM Structure Joint Challenges Workshop, CA, USA | Cathy Lawson, Wah Chiu | Joint review of the 2016 Map and Model Challenge activities | Results described in Journal of Structural Biology special issue;30 also featured in Nature Methods editorial47 |
| 17 | 2019 | Frontiers in cryo-EM Validation, UK | Gerard Kleywegt, Garib Murshudov, Elena Orlova, Ardan Patwhardhan, Alan Roseman, Peter Rosenthal, Maya Topf, Martyn Winn | Discuss current and future community needs/challenges for validation tools to support maps and models from single-particle analysis | Meeting featured in 2018 Nature editorial.48 Recommendations white paper is in preparation |
| 18 | 2019 | Model Metrics Workshop, CA, USA | Cathy Lawson, Wah Chiu | Review modeling processes and assessment results of the 2019 Model Metrics Challenge and plan for future events | Recommendations editorial and full event manuscript are in preparation |
TABLE III.
Cryo-EM community challenge activities.
| # | Year | Event | Organizing group(s) | Description | Key outcomes |
|---|---|---|---|---|---|
| C1 | 2004 | Particle Picking Challenge | National Resource for Automated Molecular Microscopy (La Jolla) | Compare diverse particle selection algorithms | Algorithms from 12 developer groups were compared and contrasted49 |
| C2 | 2010 | Model Challenge | EMDataResource, NCMI | Computational groups were asked to apply their tools to a selected set of published cryoEM structures. | Results published in Biopolymers journal special issue42 |
| C3 | 2015 | CTF Challenge | Instruct Image Processing Center (Madrid), NCMI | Evaluate community/algorithm abilities to estimate key parameters of EM Contrast Transfer Function for a wide set of experimental conditions | CTF benchmarking challenge summary and results published43 |
| C4 | 2016 | Map Challenge | EMDataResource | Establish benchmark datasets, develop best practices, evolve criteria for validation of 3DEM reconstructions | Results described in Journal of Structural Biology special issue;30 also featured in Nature Methods Editorial47 |
| C5 | 2016 | Model Challenge | EMDataResource | Establish benchmark datasets, develop best practices, evolve criteria for validation of 3DEM map-derived models | Results described in Journal of Structural Biology special issue;30 also featured in Nature Methods Editorial47 |
| C6 | 2019 | Model Metrics Challenge | EMDataResource | Identify metrics most suitable for evaluating/comparing fit of atomic coordinate models into cryo-EM maps in 1.8–3.0 Å resolution range | Recommendations editorial and full event manuscript are in preparation |
FIG. 5.
Workshop Participant Photos. (a) 2004 Cryo-EM structure deposition workshop; (b) 2010 EM Validation Task Force (EM VTF) Workshop; (c) 2011 model challenge workshop; (d) 2017 joint challenges workshop. Image in (d) Reprinted with permission from C. L. Lawson and W. Chiu, J. Struct. Biol. 204(3), 523–526 (2018). Copyright 2018 Elsevier.
EM extension dictionary development
The main goal of the 2004 Cryo-EM Structure Deposition Workshop [Fig. 5(a)], attended by ∼30 scientists including cryo-EM, image processing, crystallography, database, funding agency, and journal representatives, was to develop a global community consensus on data items for deposition of density maps and atomic models derived from cryo-EM studies. Terms were reviewed category-by-category in two focus groups, and recommendations for revisions and extensions were obtained (Fig. 6). Furthermore, participants unanimously requested a “one-stop shop” for deposition and retrieval of the cryo-EM map and model data. Following the workshop, the dictionary was further revised with follow-up input from attendees. The resulting dictionary was presented at the 2005 3DEM Gordon Research Conference, and EMDR's project website became the requested one-stop-shop portal.
FIG. 6.
Overall structure of the EM extension data dictionary circa 2005. New categories of data items recommended by participants of the 2004 Cryo-EM Structure Deposition Workshop are shown in green.
The EM extension dictionary was next reviewed by software developers at the 2005 3DEM Developers Workshop to facilitate its integration with major 3DEM packages and electronic notebook systems. There were two important outcomes: (a) the draft dictionary was unanimously accepted by the participants and (b) a set of proposed conventions for describing EM micrographs and density maps was developed.22 The conventions enable a standardized approach to image interpretation and presentation, with recommended units for common parameters, rotation and symmetry notations, and common sense principles such as “objects should have overall positive density” (early image correction procedures sometimes generated objects darker than their background depending on image processing and display software). The conventions were subsequently incorporated into the EM extension dictionary to facilitate representation of map-related data items in PDB and EMDB.
Data standards for cryo-EM structures were further addressed at the 2011 Data Management Challenges in 3D Electron Microscopy Workshop23 and the 2015 Building Bridges between Cellular and Molecular Structural Biology Workshop.24 Needs for hierarchical sample description as well as extensions to cryo-EM experimental sub-method descriptions were recognized. A future archival segmentation file format, for which requirements were gathered at the 2015 meeting, will make use of the hierarchy, enabling map regions to be connected with biological annotations.5,24
Developing validation standards
At the 2010 EM Validation Task Force (EM VTF) Workshop [Fig. 5(b)], an international group of experts explored how to assess cryo-EM maps, models, and other data deposited into EMDB and PDB. For maps, participants recognized a critical need to develop standards for assessing map resolution and accuracy. They recommended establishing two fully independent image datasets at the outset for evaluating resolution by Fourier Shell Correlation (FSC); at the time, this was not typically done, but it is now the standard procedure. However, they also advised that maps still be carefully inspected to ensure that the resolution estimate by FSC is in accordance with the map's visible features.
The EM VTF's 2012 white paper notably called for the scientific community to develop new criteria for the evaluation of maps and for the evaluation of fit of the model to the experimental map density.25 In contrast, in 2011 the VTF for X-ray crystallography published a comprehensive and detailed set of recommendations to validate structures and experimental data determined using X-ray crystallography.26 The difference reflects the fact that cryo-EM is still a rapidly evolving field.
Validation standards and raw image data archiving were additional topics of discussion at the 2011 Data Management Challenges in 3D Electron Microscopy Workshop.23 Several services were developed and implemented at EBI in response to workshop recommendations. The EMPIAR raw data archive was created,15 and stand-alone FSC and tilt-pair servers were developed for depositors to validate their cryo-EM maps.5,27 In addition, Visual Analysis web pages were designed to display an informative series of images and plots for every EMDB entry, and to help users assess data quality of released cryo-EM maps and models.28,29
Two EMDR-sponsored challenges subsequently aimed to address the 2010 EM VTF's call for improved metrics to evaluate both maps and fit of models to experimental data (2016 Map and Model Challenges). Following the 2017 Joint Challenges Workshop at Stanford, which had over 90 participants [Fig. 5(d)]; key results and recommendations were collated into a virtual special issue of the Journal of Structural Biology published in December 2018.30
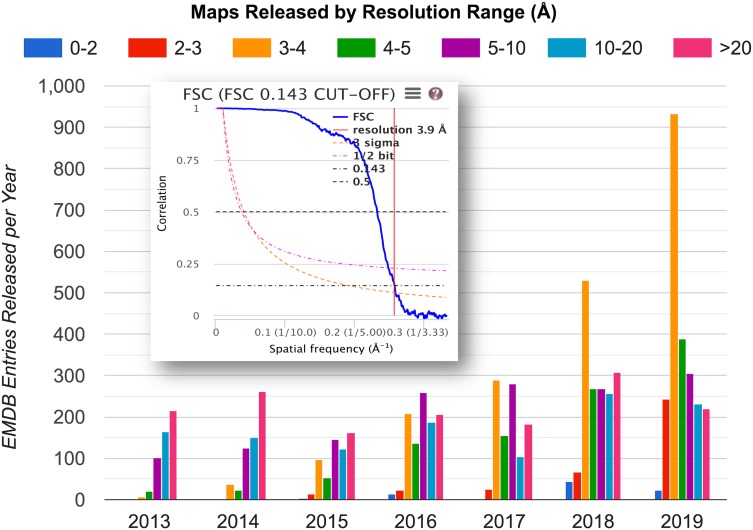
The Map Challenge provided a unique forum for critically evaluating the standard method for estimating map resolution by FSC (Fig. 7, inset). A key observation was that as currently practiced, the procedure is not sufficiently standardized: a number of different variables (e.g., map box size, voxel size, filtering and masking practice, and threshold value for interpretation) can substantially impact the outcome.31 As a result, different expert practitioners can arrive at different resolution estimates for the same level of map details. For example, two of the apoferritin maps submitted to the challenge had practitioner-estimated resolutions of 3.1 Å and 3.5 Å, respectively, though they were indistinguishable by eye. A direct conclusion is that any “reported-resolution”-based search or ranking for maps or associated models will have limited reliability. In follow-up discussions at the 2019 Frontiers in Cryo-EM Validation Workshop, one suggestion made was to have the archives independently estimate resolution by FSC from deposited unmasked, minimally filtered half-maps. This procedure would likely make comparisons between maps less susceptible (though not completely impervious) to variations in practitioner practice.
FIG. 7.
Changing cryo-EM resolution landscape. Annual distribution of depositor-reported resolution for map entries released into EMDB. The sharp increase at 2–4 Å resolution is a direct consequence of the recent advances in image detection and processing.3 Inset: example Fourier Shell Correlation (FSC) plot, which is the current standard for estimating map resolution.25 The correlation between two independent half-map reconstructions (blue curve) falls with decreasing spatial frequency; the resolution estimate (in this case 3.9 Å) is read at FSC = 0.143 (dash-dotted horizontal line). Plot source: emdataresource.org. Inset FSC plot source: EMDB visual analysis.28
The 2017 Joint Challenges Workshop also sparked lively discussions about the potential for model-based metrics to estimate not only model quality, but also to provide one or more independent measures of map resolvability. Several procedures of this type have been proposed and tested. EMRinger evaluates whether density peaks in the map fall within the possible rotameric configuration for the carbon-β atom in a side chain.32 Other procedures have been developed to measure map quality. For example, Z-scores capture how much larger the cross-correlations score (CCS) is for atoms in such features at their placed location compared to the CCS at displaced positions.33–35 Another recently devised experimental metric, Q-score, measures resolvability of the individual atom(s) in reference to the model.36
Changing validation goals
Looking at the distribution of reported resolution of maps released into EMDB annually over the past few years (Fig. 7), one can readily see a striking sharp recent increase in maps in the 2–4 Å range. This development is a direct result of recent technological improvements, and it changes the “goal-posts” for developing validation methods, adding urgency to the need for metrics to validate structures at near-atomic to atomic resolution.
The 2019 Model Metrics Challenge and associated 2019 Model Metrics Workshop were designed with the goal of evaluating metrics for map-model fit of moderately high-resolution maps (3.1–1.8 Å). A full write-up will be published elsewhere, and we note two findings here. First, the new metrics that by some means combine both model and map quality (e.g., EMRinger and Q-score) appear to be quite useful for ranking sets of structures. Second, refined Atomic Displacement Parameters (ADPs), which were included in about half of the models submitted by challenge participants, could modestly improve fit of the model to the map, particularly for the highest resolution (1.8 Å) target map. The meaning of refined ADPs/B-factors in the context of a cryo-EM density map is less clear. Best practices (e.g., to avoid overfitting) will need to be investigated.
WHERE WE ARE, WHAT'S NEXT
The initial EM validation report format released in 2016 focused on assessment of model geometry for PDB entries.18 As will be reported in more detail in a future publication, additional sections covering map analysis and visualization and map-model fit analysis and visualization will become available to EMDB and PDB depositors by early 2020. The Visual Analysis web pages hosted at EBI since 201228 have served as a test-bed for the development of the new features, which will include (a) several types of orthogonal images of the deposited map and map superimposed with model; (b) FSC curves to support depositor-reported map resolution; and (c) map-model fit statistics via “atom inclusion,” the percentage of modeled atoms falling inside a map at its recommended contour level. The new features will enable scientists (depositors, annotators, journal editors, and manuscript reviewers) to make initial assessments of map features, map quality, and map-model fit, bypassing the need to first download/view files in a graphics program.
A planned meeting in January 2020 at EBI organized by wwPDB will bring together cryo-EM and data archiving experts to discuss the current state of data archiving for cryo-EM structures derived from the single-particle reconstruction method, and to solicit recommendations on what data should be included and/or made mandatory in depositions and associated validation reports. The following points might be considered as part of the deliberations:
-
•
Can estimation of map resolution be better standardized across the community? This would enable fairer comparisons among maps determined in different laboratories and using different software packages.
-
•
Additional metrics (beyond atom inclusion) are available that describe map-model fit, including several novel procedures that effectively yield a joint assessment of map and model quality in a broad resolution range. How should map-model fit be reported as part of a structure determination and in a joint map+model deposition?
-
•
What best practice recommendations can be made for refinement of ADPs in cryo-EM models at different resolutions?
-
•
How should we evaluate multiple structures determined from a single specimen that may have variable quality and resolution?
ACKNOWLEDGMENTS
EMDataResource is funded by the U.S. National Institutes of Health/National Institute of General Medical Science, No. R01GM079429-12. We thank current and past EMDR colleagues for their contributions to data standards development, with special recognition to former and current directors of the EMDB archive including Kim Henrick, Gerard Kleywegt, and Ardan Patwardhan. We are also tremendously grateful to the cryo-EM community for their enthusiastic participation and support for data standards development.
Note: This article is part of the Special Issue: Transactions from the 69th Annual Meeting of the American Crystallographic Association: Data Best Practices: Current State and Future Needs.
References
- 1. Chiu W., Baker M. L., Jiang W., Dougherty M., and Schmid M. F., “ Electron cryomicroscopy of biological machines at subnanometer resolution,” Structure 13(3), 363–372 (2005). 10.1016/j.str.2004.12.016 [DOI] [PubMed] [Google Scholar]
- 2. Vinothkumar K. R. and Henderson R., “ Single particle electron cryomicroscopy: Trends, issues and future perspective,” Q. Rev. Biophys. 49, e13 (2016). 10.1017/S0033583516000068 [DOI] [PubMed] [Google Scholar]
- 3. Kuhlbrandt W., “ Biochemistry. The resolution revolution,” Science 343(6178), 1443–1444 (2014). 10.1126/science.1251652 [DOI] [PubMed] [Google Scholar]
- 4. Cheng Y., Glaeser R. M., and Nogales E., “ How cryo-EM became so hot,” Cell 171(6), 1229–1231 (2017). 10.1016/j.cell.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patwardhan A. and Lawson C. L., “ Databases and archiving for CryoEM,” Methods Enzymol. 579, 393–412 (2016). 10.1016/bs.mie.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.“Crystallography: Protein data bank,” Nat. New Biol. 233(42), 223–223 (1971).
- 7. Driscoll P. C., Gronenborn A. M., Beress L., and Clore G. M., “ Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing,” Biochemistry 28(5), 2188–2198 (1989). 10.1021/bi00431a033 [DOI] [PubMed] [Google Scholar]
- 8. Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., and Downing K. H., “ Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy,” J. Mol. Biol. 213(4), 899–929 (1990). 10.1016/S0022-2836(05)80271-2 [DOI] [PubMed] [Google Scholar]
- 9. Henrick K., Newman R., Tagari M., and Chagoyen M., “ EMDep: A web-based system for the deposition and validation of high-resolution electron microscopy macromolecular structural information,” J. Struct. Biol. 144(1–2), 228–237 (2003). 10.1016/j.jsb.2003.09.009 [DOI] [PubMed] [Google Scholar]
- 10. Fuller S. D., “ Depositing electron microscopy maps,” Structure 11(1), 11–12 (2003). 10.1016/S0969-2126(02)00942-5 [DOI] [PubMed] [Google Scholar]
- 11.“Editorial, A database for ‘em’,” Nat. Struct. Biol. 10(5), 313 (2003). [DOI] [PubMed]
- 12. Ulrich E. L., Akutsu H., Doreleijers J. F., Harano Y., Ioannidis Y. E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C. F., Tolmie D. E., Wenger R. K., Yao H., and Markley J. L., “ BioMagResBank,” Nucl. Acids Res. 36, D402–408 (2007). 10.1093/nar/gkm957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawson C. L., Baker M. L., Best C., Bi C., Dougherty M., Feng P., van Ginkel G., Devkota B., Lagerstedt I., Ludtke S. J., Newman R. H., Oldfield T. J., Rees I., Sahni G., Sala R., Velankar S., Warren J., Westbrook J. D., Henrick K., Kleywegt G. J., Berman H. M., and Chiu W., “ EMDataBank.org: Unified data resource for CryoEM,” Nucl. Acids Res. 39, D456–464 (2011). 10.1093/nar/gkq880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lawson C. L., Patwardhan A., Baker M. L., Hryc C., Garcia E. S., Hudson B. P., Lagerstedt I., Ludtke S. J., Pintilie G., Sala R., Westbrook J. D., Berman H. M., Kleywegt G. J., and Chiu W., “ EMDataBank unified data resource for 3DEM,” Nucl. Acids Res. 44(D1), D396–403 (2016). 10.1093/nar/gkv1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iudin A., Korir P. K., Salavert-Torres J., Kleywegt G. J., and Patwardhan A., “ EMPIAR: A public archive for raw electron microscopy image data,” Nat. Methods 13(5), 387–388 (2016). 10.1038/nmeth.3806 [DOI] [PubMed] [Google Scholar]
- 16.wwPDB Consortium, “ Protein Data Bank: The single global archive for 3D macromolecular structure data,” Nucl. Acids Res. 47(D1), D520–D528 (2019). 10.1093/nar/gky949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Young J. Y., Westbrook J. D., Feng Z., Sala R., Peisach E., Oldfield T. J., Sen S., Gutmanas A., Armstrong D. R., Berrisford J. M., Chen L., Chen M., Costanzo L. Di, Dimitropoulos D., Gao G., Ghosh S., Gore S., Guranovic V., Hendrickx P. M. S., Hudson B. P., Igarashi R., Ikegawa Y., Kobayashi N., Lawson C. L., Liang Y., Mading S., Mak L., Mir M. S., Mukhopadhyay A., Patwardhan A., Persikova I., Rinaldi L., Sanz-Garcia E., Sekharan M. R., Shao C., Swaminathan G. J., Tan L., Ulrich E. L., van Ginkel G., Yamashita R., Yang H., Zhuravleva M. A., Quesada M., Kleywegt G. J., Berman H. M., Markley J. L., Nakamura H., Velankar S., and Burley S. K., “ OneDep: Unified wwPDB system for deposition, biocuration, and validation of macromolecular structures in the PDB archive,” Structure 25(3), 536–545 (2017). 10.1016/j.str.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gore S., Sanz Garcia E., Hendrickx P. M. S., Gutmanas A., Westbrook J. D., Yang H., Feng Z., Baskaran K., Berrisford J. M., Hudson B. P., Ikegawa Y., Kobayashi N., Lawson C. L., Mading S., Mak L., Mukhopadhyay A., Oldfield T. J., Patwardhan A., Peisach E., Sahni G., Sekharan M. R., Sen S., Shao C., Smart O. S., Ulrich E. L., Yamashita R., Quesada M., Young J. Y., Nakamura H., Markley J. L., Berman H. M., Burley S. K., Velankar S., and Kleywegt G. J., “ Validation of structures in the Protein Data Bank,” Structure 25(12), 1916–1927 (2017). 10.1016/j.str.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall S. R., Allen F. H., and Brown I. D., “ The crystallographic information file (Cif)—A new standard archive file for crystallography,” Acta Crystallogr., Sect. A 47, 655–685 (1991). 10.1107/S010876739101067X [DOI] [Google Scholar]
- 20. Fitzgerald P. M. D., Westbrook J. D., Bourne P. E., McMahon B., Watenpaugh K. D., and Berman H. M., in International Tables for Crystallography G. Definition and Exchange of Crystallographic Data, edited by Hall S. R. and McMahon B. ( Springer, Dordrecht, The Netherlands, 2005), pp. 295–443. [Google Scholar]
- 21. Berman H. M., Lawson C. L., Vallat B., and Gabanyi M. J., “ Anticipating innovations in structural biology,” Q. Rev. Biophys. 51, e8 (2018). 10.1017/S0033583518000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heymann J. B., Chagoyen M., and Belnap D. M., “ Common conventions for interchange and archiving of three-dimensional electron microscopy information in structural biology,” J. Struct. Biol. 151(2), 196–207 (2005). 10.1016/j.jsb.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 23. Patwardhan A., Carazo J. M., Carragher B., Henderson R., Heymann J. B., Hill E., Jensen G. J., Lagerstedt I., Lawson C. L., Ludtke S. J., Mastronarde D., Moore W. J., Roseman A., Rosenthal P., Sorzano C. O., Sanz-Garcia E., Scheres S. H., Subramaniam S., Westbrook J., Winn M., Swedlow J. R., and Kleywegt G. J., “ Data management challenges in three-dimensional EM,” Nat. Struct. Mol. Biol. 19(12), 1203–1207 (2012). 10.1038/nsmb.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patwardhan A., Ashton A., Brandt R., Butcher S., Carzaniga R., Chiu W., Collinson L., Doux P., Duke E., Ellisman M. H., Franken E., Grunewald K., Heriche J. K., Koster A., Kuhlbrandt W., Lagerstedt I., Larabell C., Lawson C. L., Saibil H. R., Sanz-Garcia E., Subramaniam S., Verkade P., Swedlow J. R., and Kleywegt G. J., “ A 3D cellular context for the macromolecular world,” Nat. Struct. Mol. Biol. 21(10), 841–845 (2014). 10.1038/nsmb.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henderson R., Sali A., Baker M. L., Carragher B., Devkota B., Downing K. H., Egelman E. H., Feng Z., Frank J., Grigorieff N., Jiang W., Ludtke S. J., Medalia O., Penczek P. A., Rosenthal P. B., Rossmann M. G., Schmid M. F., Schroder G. F., Steven A. C., Stokes D. L., Westbrook J. D., Wriggers W., Yang H., Young J., Berman H. M., Chiu W., Kleywegt G. J., and Lawson C. L., “ Outcome of the first electron microscopy validation task force meeting,” Structure 20(2), 205–214 (2012). 10.1016/j.str.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Read R. J., Adams P. D., Arendall W. B., Brunger A. T., Emsley P., Joosten R. P., Kleywegt G. J., Krissinel E. B., Lutteke T., Otwinowski Z., Perrakis A., Richardson J. S., Sheffler W. H., Smith J. L., Tickle I. J., Vriend G., and Zwart P. H., “ A new generation of crystallographic validation tools for the Protein Data Bank,” Structure 19(10), 1395–1412 (2011). 10.1016/j.str.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wasilewski S. and Rosenthal P. B., “ Web server for tilt-pair validation of single particle maps from electron cryomicroscopy,” J. Struct. Biol. 186(1), 122–131 (2014). 10.1016/j.jsb.2014.02.012 [DOI] [PubMed] [Google Scholar]
- 28. Lagerstedt I., Moore W. J., Patwardhan A., Sanz-Garcia E., Best C., Swedlow J. R., and Kleywegt G. J., “ Web-based visualisation and analysis of 3D electron-microscopy data from EMDB and PDB,” J. Struct. Biol. 184(2), 173–181 (2013). 10.1016/j.jsb.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abbott S., Iudin A., Korir P. K., Somasundharam S., and Patwardhan A., “ EMDB web resources,” Curr. Protoc. Bioinf. 61(1), 5.10.11–15.10.12 (2018). 10.1002/cpbi.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lawson C. L. and Chiu W., “ Comparing cryo-EM structures,” J. Struct. Biol. 204(3), 523–526 (2018). 10.1016/j.jsb.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heymann J. B., Marabini R., Kazemi M., Sorzano C. O. S., Holmdahl M., Mendez J. H., Stagg S. M., Jonic S., Palovcak E., Armache J. P., Zhao J., Cheng Y., Pintilie G., Chiu W., Patwardhan A., and Carazo J. M., “ The first single particle analysis map challenge: A summary of the assessments,” J. Struct. Biol. 204(2), 291–300 (2018). 10.1016/j.jsb.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barad B. A., Echols N., Wang R. Y., Cheng Y., DiMaio F., Adams P. D., and Fraser J. S., “ EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy,” Nat. Methods 12(10), 943–946 (2015). 10.1038/nmeth.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pintilie G. and Chiu W., “ Assessment of structural features in Cryo-EM density maps using SSE and side chain Z-scores,” J. Struct. Biol. 204(3), 564–571 (2018). 10.1016/j.jsb.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mendez J. H. and Stagg S. M., “ Assessing the quality of single particle reconstructions by atomic model building,” J. Struct. Biol. 204(2), 276–282 (2018). 10.1016/j.jsb.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. M. A. Herzik, Jr. , Fraser J. S., and Lander G. C., “ A multi-model approach to assessing local and global cryo-EM map quality,” Structure 27(2), 344–358.E3 (2019). 10.1016/j.str.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pintilie G., Zhang K., Su Z., Li S., Schmid M. F., and Chiu W., “ Measurement of atom resolvability in cryo-EM maps with Q-scores,” Nat. Methods (in press) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gabashvili I. S., Agrawal R. K., Spahn C. M., Grassucci R. A., Svergun D. I., Frank J., and Penczek P., “ Solution structure of the E. coli 70S ribosome at 11.5 A resolution,” Cell 100(5), 537–549 (2000). 10.1016/S0092-8674(00)80690-X [DOI] [PubMed] [Google Scholar]
- 38. Zhang K., Zhang H., Li S., Pintilie G. D., Mou T. C., Gao Y., Zhang Q., van den Bedem H., Schmid M. F., Au S. W. N., and Chiu W., “ Cryo-EM structures of Helicobacter pylori vacuolating cytotoxin A oligomeric assemblies at near-atomic resolution,” Proc. Natl. Acad. Sci. U. S. A. 116(14), 6800–6805 (2019). 10.1073/pnas.1821959116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tagari M., Newman R., Chagoyen M., Carazo J. M., and Henrick K., “ New electron microscopy database and deposition system,” Trends Biochem. Sci. 27(11), 589 (2002). 10.1016/S0968-0004(02)02176-X [DOI] [PubMed] [Google Scholar]
- 40. Heymann J. B. and Belnap D. M., “ Bsoft: Image processing and molecular modeling for electron microscopy,” J. Struct. Biol. 157(1), 3–18 (2007). 10.1016/j.jsb.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 41. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., and Ludtke S. J., “ EMAN2: An extensible image processing suite for electron microscopy,” J. Struct. Biol. 157(1), 38–46 (2007). 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 42. Ludtke S. J., Lawson C. L., Kleywegt G. J., Berman H., and Chiu W., “ The 2010 cryo-EM modeling challenge,” Biopolymers 97(9), 651–654 (2012). 10.1002/bip.22081 [DOI] [PubMed] [Google Scholar]
- 43. Marabini R., Carragher B., Chen S., Chen J., Cheng A., Downing K. H., Frank J., Grassucci R. A., Bernard Heymann J., Jiang W., Jonic S., Liao H. Y., Ludtke S. J., Patwari S., Piotrowski A. L., Quintana A., Sorzano C. O., Stahlberg H., Vargas J., Voss N. R., Chiu W., and Carazo J. M., “ CTF challenge: Result summary,” J. Struct. Biol. 190(3), 348–359 (2015). 10.1016/j.jsb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marabini R., Ludtke S. J., Murray S. C., Chiu W., de la Rosa-Trevin J. M., Patwardhan A., Heymann J. B., and Carazo J. M., “ The electron microscopy exchange (EMX) initiative,” J. Struct. Biol. 194(2), 156–163 (2016). 10.1016/j.jsb.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patwardhan A., Brandt R., Butcher S. J., Collinson L., Gault D., Grunewald K., Hecksel C., Huiskonen J. T., Iudin A., Jones M. L., Korir P. K., Koster A. J., Lagerstedt I., Lawson C. L., Mastronarde D., McCormick M., Parkinson H., Rosenthal P. B., Saalfeld S., Saibil H. R., Sarntivijai S., Solanes Valero I., Subramaniam S., Swedlow J. R., Tudose I., Winn M., and Kleywegt G. J., “ Building bridges between cellular and molecular structural biology,” Elife 6, e25835 (2017). 10.7554/eLife.25835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kryshtafovych A., Adams P. D., Lawson C. L., and Chiu W., “ Evaluation system and web infrastructure for the second cryo-EM model challenge,” J. Struct. Biol. 204(1), 96–108 (2018). 10.1016/j.jsb.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.“Editorial, Challenges for cryo-EM,” Nat. Methods 15, 985 (2018). [DOI] [PubMed]
- 48. Baker M., “ Cryo-electron microscopy shapes up,” Nature 561, 565–567 (2018). 10.1038/d41586-018-06791-6 [DOI] [PubMed] [Google Scholar]
- 49. Zhu Y., Carragher B., Glaeser R. M., Fellmann D., Bajaj C., Bern M., Mouche F., de Haas F., Hall R. J., Kriegman D. J., Ludtke S. J., Mallick S. P., Penczek P. A., Roseman A. M., Sigworth F. J., Volkmann N., and Potter C. S., “ Automatic particle selection: Results of a comparative study,” J. Struct. Biol. 145(1–2), 3–14 (2004). 10.1016/j.jsb.2003.09.033 [DOI] [PubMed] [Google Scholar]