Abstract
Purpose
This study aimed to investigate the regulatory effects and mechanisms of long non-coding RNA (LncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) on gastric cancer (GC) cells.
Methods
The expression of MALAT1 was detected in GC tissues and two GC cell lines (SGC-7901 and BGC-823). MALAT1 was overexpressed and silenced in GC cells by the transfection of pcDNA-MALAT1 and siRNA-MALAT1, respectively. The proliferation and apoptosis of transfected cells, as well as the tumor volume and weight in mice injected with transfected cells were determined. After identifying the interaction between microRNA-22-3p (miR-22-3p) and MALAT1/epidermal growth factor receptor 3 (ErbB3), the effects of miR-22-3p/ErbB3 silencing on the proliferation and apoptosis of GC cells were evaluated.
Results
MALAT1 was significantly upregulated in GC tissues and cells and negatively associated with the survival of GC patients. Overexpression of MALAT1 significantly promoted the proliferation and inhibited the apoptosis of SGC-7901 cells, while silencing of MALAT1 exerts contrary effects on BGC-823 cells. Silencing of MALAT1 also significantly inhibited the tumor growth in mice. In addition, MALAT1 negatively regulated its target miR-22-3p. Silencing of miR-22-3p reversed the anti-tumor effects of MALAT1 silencing on GC cells. MiR-22-3p negatively regulated its target ErbB3. Silencing of ErbB3 reversed the tumor-promoting effects of miR-22-3p silencing on GC cells.
Conclusion
Silencing of MALAT1 inhibited the proliferation and promoted the apoptosis of GC cells through upregulating miR-22-3p and downregulating ErbB3.
Keywords: long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1, gastric cancer, MicroRNA22-3p, epidermal growth factor receptor 3
Introduction
Gastric cancer (GC) is a gastrointestinal cancer that occurs in the inside lining of the stomach.1 Surgical resection, as well as chemotherapy, and radiotherapy provide great advantages in the treatment of GC in clinical practice. However, the recurrence, metastasis, and resistance greatly limit the therapeutic outcomes.2 The 5-year overall survival rate is <30% for GC and <10% for metastatic GC worldwide.3 Since molecular-based targeting therapy has become a promising therapeutic strategy for GC,4 novel therapeutic targets against GC are urgently needed to be discovered.
Long non-coding RNAs (LncRNAs) play key regulatory roles in diverse cellular processes, such as proliferation, apoptosis, differentiation, invasion, and migration.5 Increasing pieces of evidence have proved that lncRNAs are associated with the occurrence and development of GC, such as H19,6 HULC,7 LINC00152,8 HOTAIR,9 CCAT2,10 and MEG3.11 LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is an oncogene that associated the tumorigenesis and metastasis of GC.12 High plasma MALAT1 is independently related with the poor prognosis of GC patients and is considered as a metastasis biomarker.13 MALAT1 knockdown inhibits the proliferation of SGC-7901 cells through arresting cells in G0/G1 phase.14 Knockdown of MALAT1 markedly reduces the migration, invasion, tumorigenicity, metastasis, and vasculogenic mimicry of BGC823 and SGC7901 cells.15
LncRNAs can modulate the translation and degradation of mRNAs through interacting with microRNAs (miRs).16 Previous studies have proved that the regulatory role of MALAT1 on GC is realized by targeting specific miRs, such as miR-122,13 −1297,17 −124,18 and −23b-3p.19 MiR-22 is a tumor suppressor in GC that positively associated with the outcomes of GC patients.20 Overexpression of miR-22 markedly suppresses the proliferation, invasion, and migration of SGC-7901 cells.21 It is noteworthy that miR-22-3p is the target of MALAT1. Tang et al showed that MALAT1 protects the endothelium against ox-LDL-induced dysfunction via up-regulating miR-22-3p.22 Li et al proved that MALAT1 influences the proliferation and migration of renal cell carcinoma cells by targeting miR-22-3p.23 However, whether the regulatory role of MALAT1 on GC is associated with miR-22 remains unclear.
Receptor tyrosine-protein kinase 3 (ErbB3), also known as HER3 is an oncogene that positively associated with the poor survival of GC patients.24 Knockdown of ErbB3 markedly inhibits the proliferation of GC cells and the growth of tumor tissues in mice.25 Previous studies have proved that ErbB3 is a target of miR-22. Ling et al showed that miR-22 inhibits the proliferation and invasion of lung cancer cells through post-transcriptional regulation of ErbB3.26 Up-regulation of miR-22 promotes the apoptosis of pancreatic acinar cells by repressing RrbB3 in acute edematous pancreatitis.27 However, research on the regulatory role of miR-22/ErbB3 interaction in GC is limited.
Here, the regulatory effects of MALAT1 on the proliferation and apoptosis of GC cells, as well as tumor growth in mice were evaluated. In addition, the potential regulatory mechanisms of MALAT1 relating with miR-22-3p/ErbB3 were determined. Our findings may reveal a novel therapeutic target against GC and open up new insights into the underlying mechanisms.
Materials and Methods
Tissue Samples
GC patients (N = 37) were collected in our hospital from April 2012 to February 2013. GC was staged in accordance with the UICC/AJCC staging system in 2010. Patients who had undergone preoperative radiotherapy, chemotherapy, and/or immunotherapy were excluded from this study. The clinical parameters of GC patients are shown in Table 1. Histopathologically confirmed GC tissues and adjacent normal tissues were collected through surgical resection, and immediately frozen in liquid nitrogen. This study was approved by the local Institutional Review Board, and informed consents were obtained.
Table 1.
The Clinical Parameters of Patients with Gastric Cancer (GC)
| Clinical Parameters | ||
|---|---|---|
| Age | Mean ± SD | 59 ± 10 |
| < 60 (N) | 14 | |
| ≥ 60 (N) | 23 | |
| Gender | Male (N) | 24 |
| Female (N) | 13 | |
| Lymph node metastasis | N0 (N) | 9 |
| N1 (N) | 8 | |
| N2 (N) | 11 | |
| N3 (N) | 9 | |
| Distant metastasis | M0 (N) | 33 |
| M1(N) | 4 | |
| Invasion depth | T1 (N) | 11 |
| T2(N) | 9 | |
| T3(N) | 11 | |
| T4 (N) | 6 | |
| TNM stage | I (N) | 12 |
| II (N) | 4 | |
| III (N) | 14 | |
| IV (N) | 7 | |
| Histological type | Poorly differentiated, undifferentiated adenocarcinoma | 27 |
| Tubular adenocarcinoma | 1 | |
| Mucinous adenocarcinoma | 4 | |
| Signet ring cell carcinoma | 5 |
Cell Culture
Human GC cell lines, SGC-7901 (moderately differentiation) and BGC-823 (poorly differentiation), as well as a normal human gastric mucosal epithelial cell line, GES-1 were purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). Cells were cultured in DMEM containing 10% FBS at 37°C with 5% CO2.
Cell Transfection
PcDNA-MALAT1, pcDNA negative control (pcDNA-NC), siRNA-MALAT1 (si-MALAT1), siRNA-MALAT1 NC (si-MALAT1 NC), siRNA-ErbB3 (si-ErbB3), and siRNA-ErbB3 NC (si-ErbB3 NC) were purchased from Jima (Shanghai, China). MiR-22-3p mimics, miR-22-3p mimics NC (mimics NC), miR-22-3p inhibitors, and miR-22-3p inhibitors NC (inhibitors NC) were purchased from Ribobio (Guangzhou, China). When reaching 80% confluence, cells were transfected with the above agents for 48 h by using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from frozen tissues and cells using TRIzol agent, and reverse-transcribed into cDNA using a cDNA Reverse Transcription Kit (Invitrogen). qRT-PCR was carried out on ABI PRISM 7300 (ABI, Foster City, CA, USA) with specific primers (Table 2). GAPDH and U6 were used as internal controls. The PCR program included 95°C for 10 s, 40 cycles at 95°C for 5 s, 60°C for 15 s, and 72°C for 31 s. Relative mRNA expression level was calculated according to the 2−ΔΔCt method.
Table 2.
The Primers Sequences Used in Quantitative Real-Time PCR
| Primers | Sequences |
|---|---|
| miR-22-3p F | 5ʹ-AAGCTGCCAGTTGAAGAACTGTA-3’ |
| miR-22-3p R | 5ʹ-TTACCTAGCGTATCGTTGAC-3’ |
| MALAT1 F | 5ʹ-AAAGCAAGGTCTCCCCACAAG-3’ |
| MALAT1 R | 5ʹ-GGTCTGTGCTAGATCAAAAGGCA-3’ |
| ErbB3 F | 5ʹ-CAAGATTCCAGTCTGCATTAAAGTC-3’ |
| ErbB3 R | 5ʹ-CAGCATATGATCTGTCACAGCTTG-3’ |
| GAPDH F | 5ʹ-AGAAGGCTGG GGCTCATTTG-3’ |
| GAPDH R | 5ʹ-AGGGGCCATC CACAGTCT TC-3’ |
| U6 F | 5ʹ-CTCGCTTCGGCAGCACA-3’ |
| U6 R | 5ʹ-AACGCTTCACGAATTTGCGT-3’ |
Dual Luciferase Reporter Gene (DLR) Assay
For detecting the interaction between miR-22-3p and MALAT1, cells were co-transfected with luciferase plasmid pGL3 (Promega, Madison, WI, USA) carrying MALAT1-wildtype (MALAT1-WT)/MALAT1-mutant (MALAT1-MUT) (Ribobio) and miR-22-3p mimics/mimics NC by using lipofectamine 3000 (Invitrogen). For detecting the interaction between miR-22-3p and ErbB3, cells were co-transfected with pGL3 carrying ErbB3-wildtype (ErbB3-WT)/ErbB3-mutant (ErbB3-MUT) (Ribobio) and miR-22-3p mimics/mimics NC. After 48 h of incubation, the fluorescence was visualized by using Dual Luciferase Reporter Assay Kit (Promega) on a Microplate Reader (Invitrogen).
CCK-8 and Cloning Assay
The proliferation of transfected GC cells was detected by CCK-8 and cloning assay. Simply, at 0, 24, 48 and 72 h post-culturing, cells were incubated with CCK-8 solution (Sigma) for 4 h. After 10 min of incubation with DMSO, the OD 450nm was detected by a Microplate Reader (Invitrogen). In addition, cells were cultured for 14 days in 6-well plates. The formed clones were stained with 0.1% crystal violet for 30 min. Positively stained clones (>50 cells) were counted under a microscope (Olympus, Japan).
Cell Apoptosis Assay
The cell apoptosis was detected by using Annexin V-propidium iodide (PI) kit (keygen, Jiangsu, China). Simply, cells were stained with Annexin V-EGFP and PI for 10 min under darkness. The apoptosis rate was analyzed on a Flow Cytometry.
Western Blot
The proteins were isolated from cells/tissues by RIPA Lysis buffer, separated by 10% SDS-PAGE, and transferred to polyvinylidene fluoride membrane. The membrane was blocked with 5% skim milk for 2 h, and incubated with primary antibodies, including anti-ErbR3 (#12708, 1:1000, Cell Signaling, Boston, MA, USA), -Ki67 (ab16667, 1:1000, Abcam, Cambridge, England), -PCNA (#13110, 1:1000, Cell Signaling), -Bax (#5023, 1:1000, Cell Signaling), -Bcl-2 (#4223, 1:1000, Cell Signaling), and -Cleaved Caspase-3 (#9661, 1:1000, Cell Signaling) overnight at 4°C. β-actin was used as an internal control (ab8227, 1:2000, Abcam). Followed by 1 h of incubation with HRP-conjugated secondary antibody (SN134, 1:5000, SunShine, Nanjing, China) at 25°C, the protein bands were visualized using an HRP development kit (Invitrogen).
Establishment of Tumor Model in Mice
A total of 24 mice were purchased from Hfkbio (Beijing, China), and fed at 22°C with free access to water and food. Mice were randomly divided into two groups, including si-MALAT1 and si-MALAT1 NC group (N = 12, each group). SGC-7901 cells transfected with si-MALAT1 or si-MALAT1 NC (100 mL, 1×106 cells/mL) were injected into the right axilla of mice. After injection, the tumor volume was measured every 1 week by vernier caliper. After the injection for 5 weeks, mice were killed by cervical dislocation. The tumor tissues were separated from mice and then weighted by using an analytical balance. All animal assays were performed in accordance with the guidelines of the Public Health Service Policy on Care and Use of Laboratory Animals.
Statistical Analyses
Each assay was performed for at least 3 times. Data were expressed as mean ± standard deviation (SD). Statistical analysis was performed by SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). Comparison was determined by Student’s t-test (two groups) or one-way ANOVA (> two groups), followed by Tukey’s multiple comparisons test (two groups). The correlation between miR-22-3p and MALAT1/ErbB3 was determined by Pearson correlation assay. A P-value < 0.05 represented significantly different.
Results
MALAT1 Was Upregulated in GC Tissues and Cells
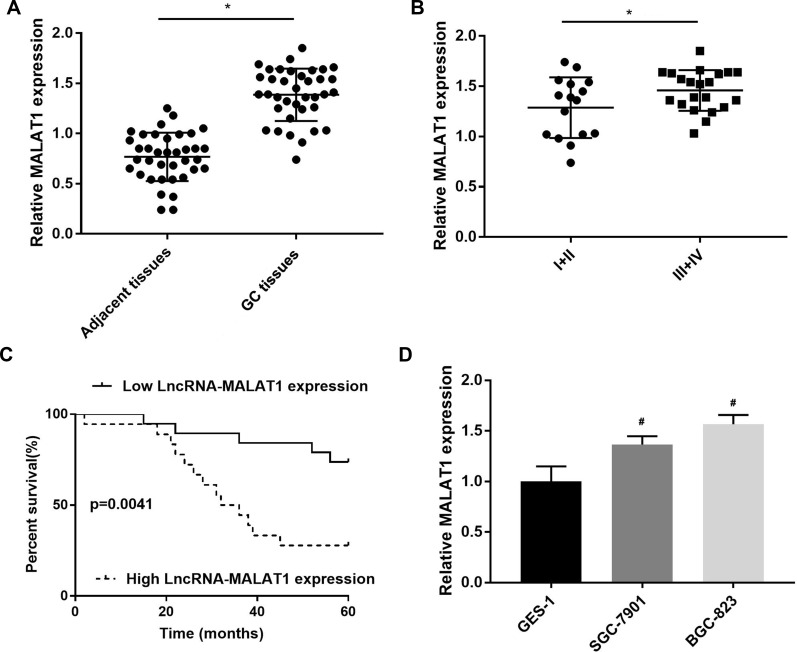
MALAT1 expression was first detected in GC tissues. MALAT1 expression was significantly upregulated in GC tissues at the mRNA level when compared with adjacent normal tissues (P < 0.05) (Figure 1A). MALAT1 expression was significantly higher in GC tissues at Ⅲ+Ⅳ stage than in GC tissues at Ⅰ+Ⅱ stage at the mRNA level (P < 0.05) (Figure 1B). The upregulation of MALAT1 in GC tissues was negatively associated with the survival of GC patients (P = 0.0041) (Figure 1C). In addition, MALAT1 expression was significantly upregulated in SGC-7901 and BGC-823 cells at the mRNA level, when compared with GES-1 cells (P < 0.05) (Figure 1D).
Figure 1.
The expression of LncRNA MALAT1 in gastric cancer (GC) tissues and cells. (A) Relative MALAT1 expression in GC tissues and adjacent normal tissues at the mRNA level (N = 37); (B) Relative MALAT1 expression in GC tissues at Ⅲ+Ⅳ and Ⅰ+Ⅱ stage at the mRNA level (N = 37); (C) The relationship between MALAT1 expression and the percent survival of GC patients (N = 37); (D) Relative MALAT1 expression in GC cell lines, SGC-7901 and BGC-823 and a normal human gastric mucosal epithelial cell line, GES-1 at the mRNA level. Each experiment (qRT-PCR) was performed in five replicates. *P < 0.05 vs adjacent tissues; #P < 0.05 vs GES-1.
MALAT1 Promoted the Proliferation and Inhibited the Apoptosis of GC Cells
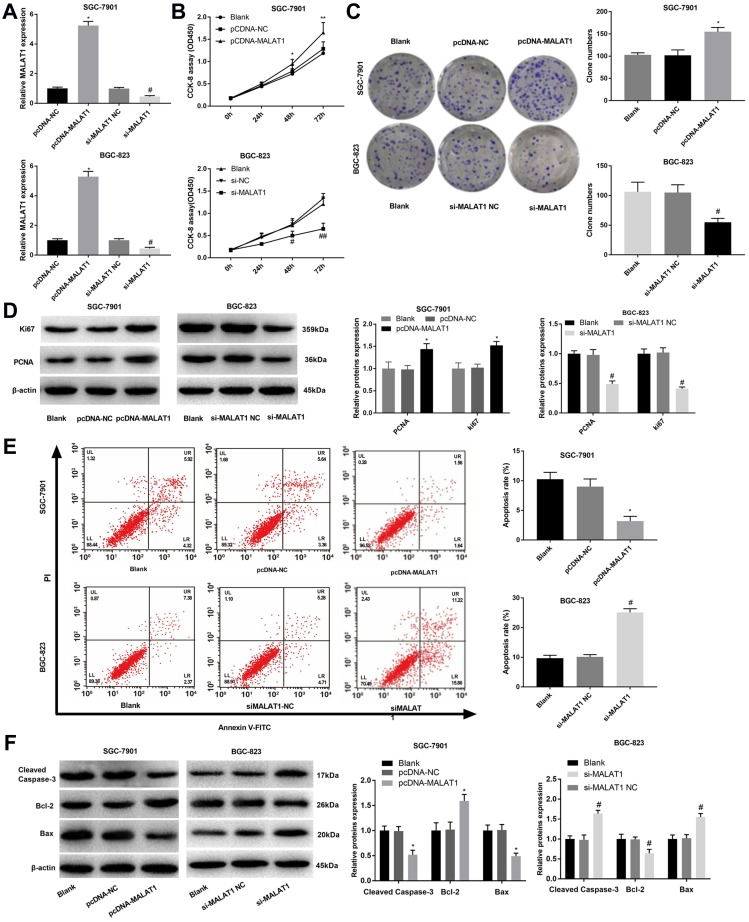
In order to reveal the regulatory roles of MALAT1 on GC cells, MALAT1 was overexpressed and silenced in SGC-7901 and BGC-823 cells by the transfection of pcDNA-MALAT1 and si-MALAT1, respectively (P < 0.05) (Figure 2A). MALAT1 overexpression significantly increased the OD450 value of SGC-7901 cells at 48 and 72 h post-culturing (P < 0.05) (Figure 2B), as well as the clone numbers of SGC-7901 cells (P < 0.05) (Figure 2C). The overexpression of MALAT1 also significantly upregulated two proliferation markers, Ki67 and PCNA in SGC-7901 cells at the protein level (P < 0.05) (Figure 2D). On the contrary, the silencing of MALAT1 significantly decreased the OD450 value, the clone number, as well as the expression Ki67 and PCNA in BGC-823 cells (P < 0.05) (Figure 2A–D). Overexpression of MALAT1 also significantly reduced the apoptosis rate of SGC-7901 cells (P < 0.05) (Figure 2E), decreased the expression of Cleaved Caspase-3 and Bax, and increased the expression of Bcl-2 in SGC-7901 cells at the protein level (P < 0.05) (Figure 2F). The silencing of MALAT1 exhibited contrary results on BGC-823 cells with the overexpression of MALAT1 (P < 0.05) (Figure 2E and F).
Figure 2.
The regulatory effects of LncRNA MALAT1 on the proliferation and apoptosis of SGC-7901 and BGC-823 cells. (A) Relative MALAT1 expression at the mRNA level; (B) OD450 values; (C) Clone numbers; (D) Relative protein expression of Ki67 and PCNA; (E) Apoptosis rate; (F) Relative protein expression of Cleaved Caspase-3, Bax, and Bcl-2. Black, normal cells without transfection; pcDNA-NC, pcDNA-MALAT1 negative control; si-MALAT1, siRNA-MALAT1; si-MALAT1 NC, siRNA-MALAT1 negative control. Each experiment was performed in three replicates. *P < 0.05 vs Black and pcDNA-NC; #P < 0.05 vs Black and si-MALAT1 NC.
miR-22-3p Was a Target of MALAT1
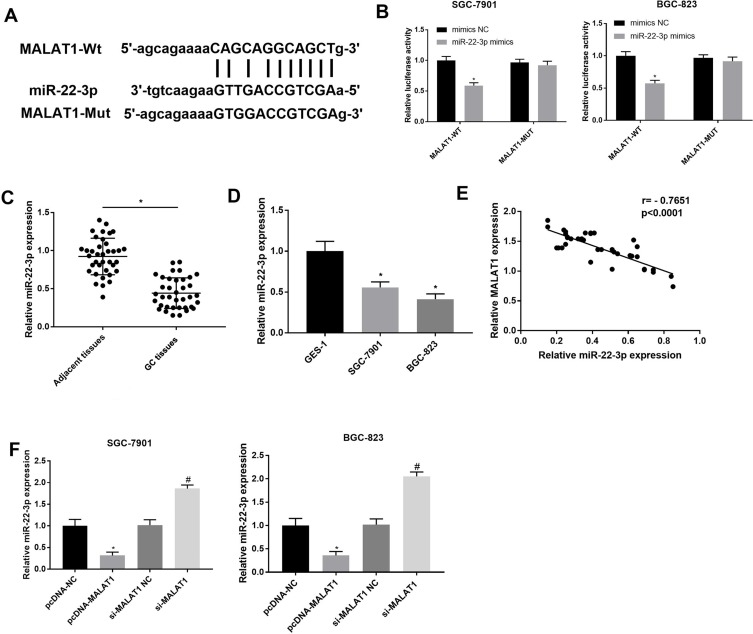
A binding site at miR-22-3p was predicated on MALAT1 by StarBase (http://starbase.sysu.edu.cn/) (Figure 3A). Because miR-22-3p is a tumor suppressor in GC, the regulatory relationship between MALAT1 and miR-22-3p was further analyzed. DLR assay identified that miR-22-3p was a target of MALAT1 (P < 0.05) (Figure 3B). MiR-22-3p expression was significantly downregulated in GC tissues at the mRNA level when compared with adjacent normal tissues (P < 0.05) (Figure 3C), and also significantly downregulated in SGC-7901 and BGC-823 cells at the mRNA level when compared with GES-1 cells (P < 0.05) (Figure 3D). In GC tissues, MALAT1 expression was negatively associated with miR-22-3p expression (P < 0.0001) (Figure 3E). In addition, the overexpression and silencing of MALAT1 significantly downregulated and upregulated miR-22-3p in SGC-7901 and BGC-823 cells, respectively (P < 0.05) (Figure 3F).
Figure 3.
Interaction between LncRNA MALAT1 and miR-22-3p. (A) A binding site predicated by StarBase (http://starbase.sysu.edu.cn/); (B) A target interaction identified by DLR assay; (C) Relative miR-22-3p expression in gastric cancer (GC) tissues and adjacent normal tissues at the mRNA level (N = 37); (D) Relative MALAT1 expression in GC cell lines, SGC-7901 and BGC-823 and a normal human gastric mucosal epithelial cell line, GES-1 at the mRNA level; (E) Correlation analysis of the relationship between MALAT1 and miR-22-3p in GC tissues (N = 37); (F) Relative miR-22-3p expression in transfected SGC-7901 and BGC-823 cells. pcDNA-NC, pcDNA-MALAT1 negative control; si-MALAT1, siRNA-MALAT1; si-MALAT1 NC, siRNA-MALAT1 negative control. Each experiment was performed in three replicates. *P < 0.05 vs mimics NC (B), adjacent tissues (C), GES-1 (D), and pcDNA-NC (F); #P < 0.05 vs si-MALAT1 NC.
Silencing of miR-22-3p Reversed the Anti-Tumor Effects of si-MALAT1 on GC Cells
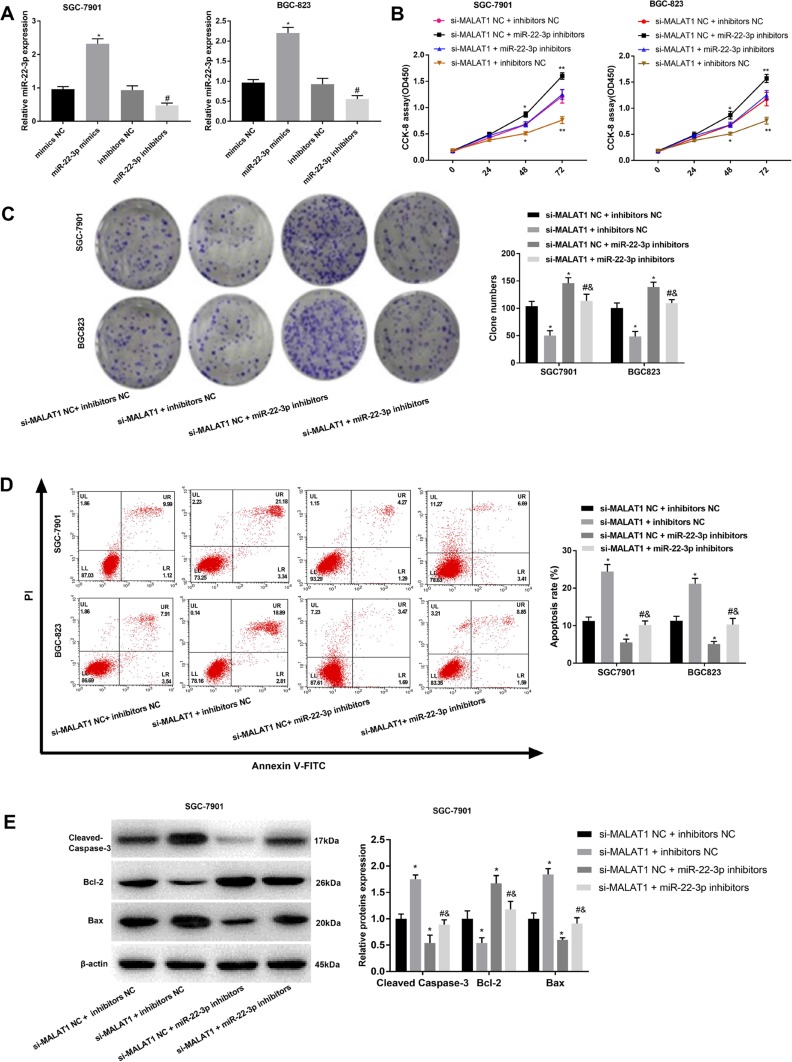
MiR-22-3p was silenced in SGC-7901 and BGC-823 cells by the transfection of miR-22-3p inhibitors (P < 0.05) (Figure 4A). Silencing of miR-22-3p significantly increased the OD450 value and clone number of SGC-7901 and BGC-823 cells (P < 0.05) (Figure 4B and C). Silencing of miR-22-3p also significantly decreased the apoptosis rate, decreased the expression of Cleaved Caspase-3 and Bax, and increased the expression of Bcl-2 in SGC-7901 and BGC-823 cells (P < 0.05) (Figure 4D and E). Noteworthily, the silencing of miR-22-3p significantly reversed the anti-proliferation and pro-apoptosis effects of si-MALAT1 on GC cells (P < 0.05) (Figure 4B–E).
Figure 4.
The regulatory effects of miR-22-3p on the proliferation and apoptosis of SGC-7901 and BGC-823 cells. (A) Relative MALAT1 expression at the mRNA level; (B) OD450 values; (C) Clone numbers; (D) Apoptosis rate; (E) Relative protein expression of Cleaved Caspase-3, Bax, and Bcl-2. Mimics NC, miR-22-3p mimics negative control; inhibitors NC, miR-22-3p inhibitors negative control; si-MALAT1, siRNA-MALAT1; si-MALAT1 NC, siRNA-MALAT1 negative control. Each experiment was performed in three replicates. *P < 0.05 vs mimics NC (A), and si-MALAT1 NC + inhibitors NC (B–E); #P < 0.05 vs si-MALAT1 + inhibitors NC; &P < 0.05 vs si-MALAT1 NC + miR-22-3p inhibitors.
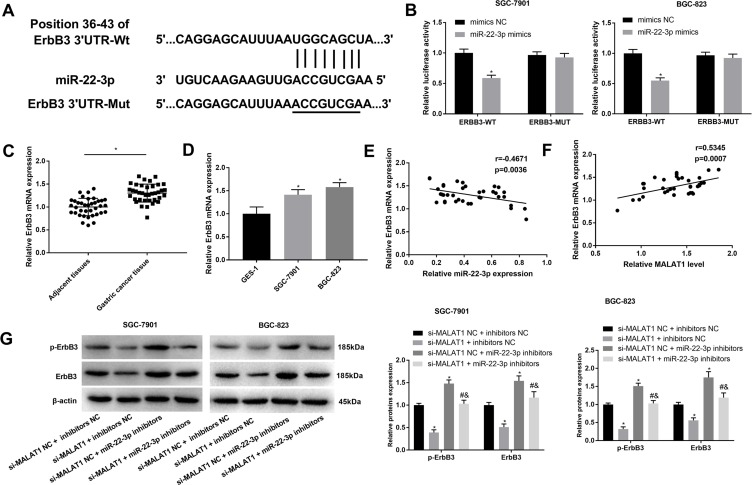
ErbB3 Was a Target of miR-22-3p
A binding site at 3ʹURT of ErbB3 was predicated on miR-22-3p by Target Scan (http://www.targetscan.org/) (Figure 5A). Because ErbB3 is a tumor promoter in GC, the regulatory relationship between miR-22-3p and ErbB3 was further analyzed. DLR assay identified that ErbB3 was a target of miR-22-3p (P < 0.05) (Figure 5B). ErbB3 expression was significantly upregulated in GC tissues when compared with adjacent normal tissues and significantly upregulated in SGC-7901 and BGC-823 cells when compared with GES-1 cells at the mRNA level (P < 0.05) (Figure 5C and D). In GC tissues, ErbB3 expression was negatively associated with miR-22-3p expression (P = 0.0036), and positively associated with MALAT1 expression (P = 0.0007) (Figure 5E and F). In addition, the silencing of MALAT1 significantly downregulated ErbB3 and p-ErbB3 in SGC-7901 and BGC-823 cells (P < 0.05). MiR-22-3p overexpression significantly reversed the inhibitory role of si-MALAT1 on the expression of ErbB3 and p-ErbB3 (P < 0.05) (Figure 5G).
Figure 5.
Interaction between ErbB3 and miR-22-3p. (A) A binding site predicated by Target Scan (http://www.targetscan.org/); (B) A target interaction identified by DLR assay; (C) Relative ErbB3 expression in gastric cancer (GC) tissues and adjacent normal tissues at the mRNA level (N = 37); (D) Relative ErbB3 expression in GC cell lines, SGC-7901 and BGC-823 and a normal human gastric mucosal epithelial cell line, GES-1 at the mRNA level; (E) Correlation analysis of the relationship between ErbB3 and miR-22-3p in GC tissues (N = 37); (F) Correlation analysis of the relationship between ErbB3 and MALAT1 in GC tissues (N = 37); (G) Relative ErbB3 and p-ErbB3 expression in transfected SGC-7901 and BGC-823 cells. si-MALAT1, siRNA-MALAT1; si-MALAT1 NC, siRNA-MALAT1 negative control; inhibitors NC, miR-22-3p inhibitors negative control. Each experiment was performed in three replicates. *P < 0.05 vs mimics NC (B), adjacent tissues (C), GES-1 (D), and si-MALAT1 NC + inhibitors NC (G); #P < 0.05 vs si-MALAT1 + inhibitors NC; &P < 0.05 vs si-MALAT1 NC + miR-22-3p inhibitors.
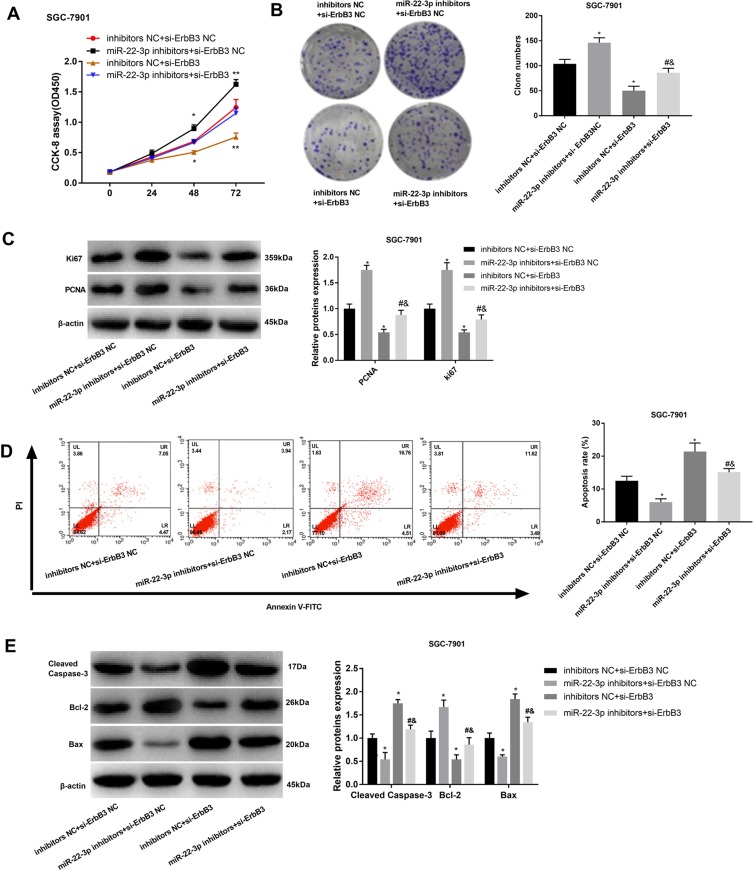
Silencing of ErbB3 Reversed the Tumor-Promoting Effects of miR-22-3p Inhibitors on GC Cells
In order to further identify the regulatory roles of ErbB3 on GC cells, ErbB3 was silenced in SGC-7901 cells by the transfection of si-ErbB3. CCK-8 and cloning assay showed that the silencing of ErbB3 significantly decreased the OD450 value and clone number of SGC-7901 cells (P < 0.05) (Figure 6A and B). The silencing of ErbB3 also significantly downregulated PCNA and Ki67 in SGC-7901 cells (P < 0.05) (Figure 6C). Silencing of miR-22-3p significantly increased the apoptosis rate, upregulated Cleaved Caspase-3 and Bax, and downregulated Bcl-2 in SGC-7901 cells (P < 0.05) (Figure 6D and E). Noteworthily, the silencing of miR-22-3p significantly reversed the anti-apoptosis and pro-proliferation effects of miR-22-3p inhibitors on SGC-7901 cells (P < 0.05) (Figure 6A–E).
Figure 6.
The regulatory effects of ErbB3 on the proliferation and apoptosis of SGC-7901 cells. (A) OD450 values; (B) Clone numbers; (C) Relative protein expression of Ki67 and PCNA; (D) Apoptosis rate; (E) Relative protein expression of Cleaved Caspase-3, Bax, and Bcl-2. si-ErbB3, siRNA-ErbB3; si-ErbB3 NC, siRNA-ErbB3 negative control; inhibitors NC, miR-22-3p inhibitors negative control. Each experiment was performed in three replicates. *P < 0.05 vs inhibitors NC + si-ErbB3 NC; #P < 0.05 vs miR-22-3p inhibitors + si-ErbB3 NC; &P < 0.05 vs inhibitors NC + si-ErbB3.
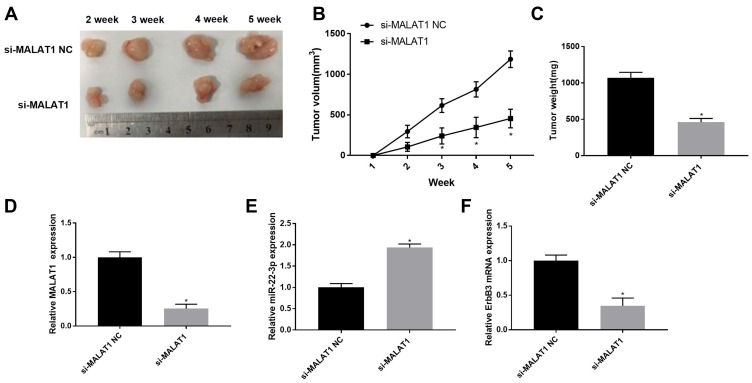
Silencing of MALAT1 Inhibited the Tumor Growth in Mice
The effects of MALAT1 silencing on the tumor growth in vivo were further analyzed. The tumor volume was significantly increased in mice injected with SGC-7901 cells in a time-dependent manner. The tumor volume was significantly lower in mice injected with si-MALAT1-transfected SGC-7901 cells than those injected with si-MALAT1 NC-transfected cells beginning from the second week post-injection (P < 0.05) (Figure 7A and B). Five weeks later, the tumor weight was significantly lower in mice injected with si-MALAT1-transfected SGC-7901 cells than those injected with si-MALAT1 NC-transfected cells (P < 0.05) (Figure 7C). In addition, MALAT1 silencing significantly upregulated miR-22-3p and downregulated MALAT1 and ErbB3 in tumor tissues (P < 0.05) (Figure 7D–F).
Figure 7.
The tumor growth in mice injected with SGC-7901 cells. (A) Separated tumor tissue; (B) Tumor volume (N = 10); (C) Tumor weight (N = 10); (D) Relative MALAT1 expression in tumor tissues at the mRNA level (N = 5); (E) Relative miR-22-3p expression in tumor tissues at the mRNA level (N = 5); (F) Relative ErbB3 expression in tumor tissues at the mRNA level (N = 5). Each experiment was performed in three replicates. si-MALAT1, siRNA-MALAT1; si-MALAT1 NC, siRNA-MALAT1 negative control. *P < 0.05 vs si-MALAT1 NC.
Discussion
GC is a severe gastrointestinal malignancy that accompanied with high mortality.3 In recent years, molecular-based targeting therapy has become a promising therapeutic strategy for GC.28 MALAT1 is an important lncRNA that acts as an oncogene, as well as a therapeutic target in cancer. MALAT1 is upregulated in diverse types of cancers, such as ovarian cancer,29 prostate cancer,30 lung cancer,12 colorectal cancer,31 and GC.13 In consistent with previous studies, MALAT1 expression was significantly upregulated in GC tissues when compared with adjacent normal tissues, and significantly upregulated in SGC-7901 and BGC-823 when compared with GES-1 cells. Previous studies have proved that high plasma MALAT1 is positively associated with poor prognosis, and high MALAT1 expression is positively associated with metastasis, and TNM stage.13,17 Here, MALAT1 expression was significantly upregulated in GC tissues at Ⅲ+Ⅳ stage when compared withⅠ+Ⅱ stage. MALAT1 overexpression was negatively associated with the survival of GC patients. Our findings are just consistent with previous studies, and further indicate that MALAT1 is a potential diagnostic and prognostic factor for GC.
A large number of researches have proved that MALAT1 is important in the proliferation and apoptosis of GC cells. For example, MALAT1 knockdown inhibits the proliferation of SGC-7901 cells via blocking cells in G0/G1 phase.14 MALAT1 knockdown inhibits the proliferation ability of MKN45 and MKN28 cells.17 MALAT1 knockdown promotes G0/G1 phase arrest, as well as the apoptosis of MKN45 and CTC141 cells.13 In this study, we found that the silencing of MALAT1 significantly increased the OD450 value, the clone number, Ki67, PCNA and Bcl-2 expression, and decreased the apoptosis rate, and Cleaved Caspase-3 and Bax expression in SGC-7901 cells. Noteworthily, silencing of MALAT1 exhibited contrary results on BGC-823 cells with the overexpression of MALAT1. In consistent with previous studies, these findings indicate silencing of MALAT1 can inhibit the proliferation and promote the apoptosis of GC cells in vitro. A previous study has proved that siRNA-mediated suppression of MALAT1 suppresses the tumor formation and growth in mice.32 Here, MALAT1 silencing significantly reduced the tumor volume and weight in mice, which further illustrate that the MALAT1 silencing exerts great anti-tumor potential on GC.
The regulatory effects of MALAT1 on GC are closely associated with the interaction with diverse miRs, such as miR-122-IGF-1R,13 miR-1297,17 and miR-23b-3p.19 In this study, miR-22-3p was identified as a target of MALAT1. MiR-22 is a tumor suppressor that downregulated in diverse cancers, including breast cancer,33 colon cancer,34 liver cancer,35 and GC.21 In consistent with previous studies, miR-22-3p expression was significantly downregulated in GC tissues and cells. A previous study has proved that miR-22 overexpression decreased the proliferation ability of SGC-7901 cells, and the tumor growth in mice.21 Similarly, we found that the silencing of miR-22-3p significantly promotes the proliferation, and inhibits the apoptosis of GC cells. Since MALAT1 expression was negatively associated with miR-22-3p expression, miR-22-3p upregulation may contribute to the anti-tumor effects of MALAT1 silencing on GC cells. This hypothesis is further illustrated by that miR-22-3p silencing reversed the anti-proliferation and pro-apoptosis effects of si-MALAT1.
ErbB3, a member of the epidermal growth factor receptor family plays an important regulatory role in the growth, chemotherapeutic resistance, and metastasis of cancer.36 In this study, ErbB3 was identified as a target of miR-22-3p, which is just consistent with a previous research that miR-22 downregulated ErbB3 through binding to ErbB3.21 In addition, a previous study has proved that ErbB3 knockdown inhibits the proliferation and promotes the apoptosis of GC cells, and promotes the tumor growth in vivo.25 In consistent with this study, we found that the silencing of ErbB3 significantly inhibits the proliferation, and promotes the apoptosis of GC cells. Since ErbB3 was negatively associated with miR-22 in GC, we suspect that ErbB3 downregulation may contribute to the tumor-promoting effects of miR-22-3p inhibitors on SGC-7901 cells. This hypothesis is further illustrated by that the silencing of ErbB3 significantly reversed the tumor-promoting effects of miR-22-3p inhibitors on SGC-7901 cells. The anti-tumor effects of MALAT1 silencing on GC cells may attribute to miR-22-3p-induced downregulation of ErbB3.
Conclusions
In conclusion, lncRNA MALAT1 expression was upregulated in GC tissues and cells and could be used as a potential diagnostic and prognostic factor for GC. Silencing of MALAT1 inhibited the proliferation and promoted the apoptosis of GC cells through upregulating miR-22-3p and downregulating ErbB3. Since silencing of MALAT1 also inhibited the tumor growth in mice, it may be a promising therapeutic target for GC. However, the efficacy and safety of MALAT1 silencing in the treatment of GC are still unclear. Further researches in these fields are still needed.
Ethics Approval and Consent to Participate
This study was conducted after obtaining local ethical committee approval of Hebei Medical University and Hebei General Hospital. All patients signed informed consent, and this was conducted in accordance with the Declaration of Helsinki. All animal experiments were conducted after obtaining the approval of Hebei Medical University and Hebei General Hospital’s Animal Ethics Committee. All animal assays were performed in accordance with the guidelines of the Public Health Service Policy on Care and Use of Laboratory Animals.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Carcas LP. Gastric cancer review. J Carcinog. 2014;13(1):14. doi: 10.4103/1477-3163.146506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski W. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong HS, Smithers BM. Epidemiology of gastric cancer. Cancer Rev. 2012;02(1):1–7. [Google Scholar]
- 4.Fontana E, Smyth EC. Novel targets in the treatment of advanced gastric cancer: a perspective review. Ther Adv Med Oncol. 2016;8(2):113–125. doi: 10.1177/1758834015616935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Sun J, Wang J, et al. Long noncoding RNAs in gastric cancer: functions and clinical applications. Onco Targets Ther. 2016;9:681–697. doi: 10.2147/OTT.S95412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang F, Bi J, Xue X, et al. Up‐regulated long non‐coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279(17):3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31(1):358–364. doi: 10.3892/or.2013.2850 [DOI] [PubMed] [Google Scholar]
- 8.Pang Q, Ge J, Shao Y, et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumor Biol. 2014;35(6):5441–5447. doi: 10.1007/s13277-014-1709-3 [DOI] [PubMed] [Google Scholar]
- 9.Endo H, Shiroki T, Nakagawa T, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8(10):e77070. doi: 10.1371/journal.pone.0077070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C, Hua L, Yao K, Chen J, Zhang J, Hu J. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8(1):779–785. [PMC free article] [PubMed] [Google Scholar]
- 11.Peng W, Si S, Zhang Q, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34(1):79. doi: 10.1186/s13046-015-0197-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol. 2015;8(12):15903–15910. [PMC free article] [PubMed] [Google Scholar]
- 13.Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7(35):56209–56218. doi: 10.18632/oncotarget.v7i35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Su L, Chen X, et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68(5):557–564. doi: 10.1016/j.biopha.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wu Z, Yuan J, et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035 [DOI] [PubMed] [Google Scholar]
- 16.Wu T, Qu L, He G, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7(10):11533–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 2017;17(1):44. doi: 10.1186/s12935-017-0408-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang TH, Liang LZ, Liu XL, et al. Long non-coding RNA MALAT1 interacts with miR-124 and modulates tongue cancer growth by targeting JAG1. Oncol Rep. 2017;37(4):2087–2094. doi: 10.3892/or.2017.5445 [DOI] [PubMed] [Google Scholar]
- 19.Yiren H, Y Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. doi: 10.1186/s12943-017-0743-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Li F, Zhang Y, Tu Y, Gao X. Reduced expression of miR-22 in gastric cancer is related to clinicopathologic characteristics or patient prognosis. Diagn Pathol. 2013;8(1):102. doi: 10.1186/1746-1596-8-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo Q, Cao LY, Yu T, et al. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6(11):e2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y, Jin X, Xiang Y, et al. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. 2015;589(20 Pt B):3189–3196. doi: 10.1016/j.febslet.2015.08.046 [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Ma Z, Xu X. Long noncoding RNA MALAT1 correlates with cell viability and mobility by targeting miR223p in renal cell carcinoma via the PI3K/Akt pathway. Oncol Rep. 2019;41(2):1113–1121. doi: 10.3892/or.2018.6853 [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Yuan H, Li Y, Han Y. The role of HER3 in gastric cancer. Biomed Pharmacother. 2014;68(6):809–812. doi: 10.1016/j.biopha.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Chen Y, Li G, et al. Her3 is associated with poor survival of gastric adenocarcinoma: her3 promotes proliferation, survival and migration of human gastric cancer mediated by PI3K/AKT signaling pathway. Med Oncol. 2014;31(4):903. doi: 10.1007/s12032-014-0903-x [DOI] [PubMed] [Google Scholar]
- 26.Ling B, Wang GX, Long G, Qiu JH, Hu ZL. Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. J Cancer Res Clin Oncol. 2012;138(8):1355–1361. doi: 10.1007/s00432-012-1194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin T, Fu Q, Pan YF, et al. Expressions of miR-22 and miR-135a in acute pancreatitis. J Huazhong Univ Sci Technolog Med Sci. 2014;34(2):225–233. doi: 10.1007/s11596-014-1263-7 [DOI] [PubMed] [Google Scholar]
- 28.Cidon EU, Ellis SG, Inam Y, Adeleke S, Zarif S, Geldart T. Molecular targeted agents for gastric cancer: a step forward towards personalized therapy. Cancers. 2013;5(1):64–91. doi: 10.3390/cancers5010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu L, Wang X, Guo Y. Long non-coding RNA MALAT1 is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp Ther Med. 2017;13(6):3055–3060. doi: 10.3892/etm.2017.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebastian A, Hum NR, Hudson BD, Loots GG. Cancer–osteoblast interaction reduces sost expression in osteoblasts and up-regulates lncRNA MALAT1 in prostate cancer. Microarrays. 2015;4(4):503–519. doi: 10.3390/microarrays4040503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thoracic Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac [DOI] [PubMed] [Google Scholar]
- 33.Xiong J, Yu D, Wei N, et al. An estrogen receptor α suppressor, microRNA‐22, is downregulated in estrogen receptor α‐positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277(7):1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x [DOI] [PubMed] [Google Scholar]
- 34.Li B, Song Y, Liu T, et al. miRNA-22 suppresses colon cancer cell migration and invasion by inhibiting the expression of T-cell lymphoma invasion and metastasis 1 and matrix metalloproteinases 2 and 9. Oncol Rep. 2013;29(5):1932–1938. doi: 10.3892/or.2013.2300 [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Yang Y, Yang T, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103(8):1215–1220. doi: 10.1038/sj.bjc.6605895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Huang X, Lee CK, Liu B. Elevated expression of erbB3 confers paclitaxel resistance in erbB2-overexpressing breast cancer cells via upregulation of Survivin. Oncogene. 2010;29(29):4225–4236. doi: 10.1038/onc.2010.180 [DOI] [PubMed] [Google Scholar]