Natural history collections provide resources to investigate specimens from diverse and distant places, shedding light on trait variation and distributions of species at scales not possible for individual researchers in the field. Additionally, insights can be gained on the biotic interactions of wild species, among which infectious diseases are known to have a major influence on the ecology and evolution of natural populations (Dobson and Crawley, 1994; Burdon and Thrall, 2008; Ghelardini et al., 2016). However, our knowledge of disease distributions, in terms of spatial, temporal, and host‐specific patterns, remains significantly lacking outside of humans and agriculturally important organisms. Wild plant populations often occur across a broader range of densities, environmental gradients, and ecological communities than do crop plants. Understanding disease distributions in these populations can yield critically important information on pathogen transmission modes (Alexander et al., 1996, 2007; Bruns et al., 2018) and factors leading certain host species to be more or less heavily impacted (Burdon and Chilvers, 1982; Eastburn et al., 2011).
Multiple studies have utilized museum collections to investigate wild plant–pathogen systems (Ristaino et al., 2001; Li et al., 2006; Alexander et al., 2007; Saville et al., 2016; Andrew et al., 2018), among other plant ecological interactions (e.g., plant–animal interaction, Beauvais et al., 2017, or phytosociology, Hanan‐A et al., 2016). There are two types of natural history collections relevant to such disease‐related studies: pathogen‐centered collections that directly target and preserve the disease‐causing organism and broader plant‐centered collections that nonetheless may retain evidence of the disease occurrence and cause. While diverse phytopathogenic fungi are documented in fungaria (i.e., mycological herbaria) (Brock et al., 2009; Heberling and Burke, 2019), surveys of general plant collections for symptoms and signs of pathogens (Alexander et al., 2007; Yoshida et al., 2014) can additionally reveal the patterns of infection incidence based on the proportions of plants affected (Hood et al., 2010). Such ecological insights beyond the immediate intention of collectors (exaptations, sensu Heberling et al., 2017) have been highlighted as an important yet underutilized benefit of natural history collections (Heberling et al., 2017; Meineke et al., 2018a, b).
Despite the tremendous value of herbarium specimens, depositions of new specimens have substantially declined over the last several decades (Prather et al., 2004; Lavoie and Lachance, 2006; Renner and Rockinger, 2016; Beauvais et al., 2017; Romberg and Rivera, 2017), limiting our ability to assess whether historic distributions are impacted by ongoing environmental changes or to find and study extant populations (Applequist et al., 2007; Meineke et al., 2018a; Lang et al., 2019). The rise in citizen science platforms, such as iNaturalist (http://www.inaturalist.org), eBird (http://www.ebird.org), or Zooniverse (http://www.zooniverse.org), holds new potential for assessing the ecology of wildlife species. iNaturalist alone currently has over 10 million plant observations, primarily from within the past decade; other online platforms (e.g., calflora.org) add to the available data. Several recent studies have emphasized the value of such platforms as complementary to museum collections for documenting species distributions (e.g., Heberling and Isaac, 2018). Some citizen science projects have targeted diseases (e.g., Meentemeyer et al., 2015; Brown et al., 2017), which are particularly informative in the case of emerging diseases that threaten domestic or wildlife populations. Such data are similar to the pathogen‐centered natural history collections mentioned above in reporting the occurrence of disease. Diseases are less often a desired subject of citizen scientists documenting general biodiversity inventories, but a similar strategy of quantitatively surveying online plant observations for telltale signs of pathogens may be feasible and could contribute valuable and current information on how often plants are affected by of these important ecological interactions. Such insights are especially important in this era of strong environmental change when access to information at broad geographic scales is urgently needed.
Here, we aimed to assess the utility of recent citizen‐driven natural history observations for gaining insights into the ecological interactions of wild plant populations. We compared different sources of information on disease occurrence, illustrated by using and advancing knowledge on anther‐smut disease affecting a diverse range of plant taxa. We compared data on the distribution of anther smut from fungaria, surveys of general collections in plant herbaria, and surveys of plant observations deposited to the iNaturalist online platform. We show the effectiveness of the new online resources for quantifying variation in disease incidence across members of the Caryophyllaceae host family and demonstrating species‐specific results for disease occurrence across temporal (i.e., phenological) and spatial scales (i.e., within native and introduced ranges) that complement prior studies. While pointing to other diseases where similar approaches are reasonable, we show that the tremendous efforts in citizen‐driven observations can be extended beyond their original aims regarding plant distributions, enriching our understanding of important ecological interactions and integrating with approaches based on herbarium collections.
ANTHER SMUT AS A MODEL FOR BROADLY DISTRIBUTED DISEASES
Anther smut has been a particularly informative model for pathogen surveys in general herbarium collections. This disease is caused by fungi that replace the host's pollen with dark‐colored spores (Schäfer et al., 2010). Although anther smut can be clearly ascertained upon examination of the anthers in comparison to healthy flowers (Fig. 1), multiple studies have noted the general lack of recognition of the disease by plant collectors (Antonovics et al., 2003; Hood and Antonovics, 2003; Rabeler and Hartman, 2005; Bueker et al., 2016) or the assumption that appearance is part of the plant species natural phenotypic variation (Hood and Antonovics, 2003; Antonovics and Hood, 2018). This lack of recognition by collectors has allowed for comparison of disease rates based upon the proportions of specimens with anther smut for a species (Hood et al., 2010) or comparisons of disease incidence across hosts’ geographic distributions or time periods of collections (Antonovics et al., 2003; Bueker et al., 2016).
Figure 1.

Silene virginica observations showing signs of anther‐smut disease as dark, powdery, spore‐filled anthers. (A) Herbarium sample of S. virginica (Collector: M. Nee, 1983; image courtesy of the C. V. Starr Virtual Herbarium, http://sweetgum.nybg.org/science/vh/; barcode NY 331579). (B) iNaturalist observation of diseased S. virginica (observer: M. Rung, ‘wildflowerenthusiast5’, 2014; http://www.inaturalist.org/observations/1084300). (C). iNaturalist observation of healthy S. virginica (observer: J. Gallagher, ‘judygva’, 2016; https://www.inaturalist.org/observations/4283886).
Herbarium surveys for this disease have included broad groups of host taxa (Hood et al., 2010) and targeted host species (Antonovics et al., 2003; Rabeler and Hartman, 2005; Bueker et al., 2016). A nearly global disease distribution has been revealed, which often extends to the limits of individual host species ranges and is associated with particular host traits (e.g., perennial life history; Thrall et al., 1993; Hood et al., 2010). Herbarium surveys for anther smut are facilitated by the main symptom being in the flowers, which are often the target of preservation due to their importance in taxonomic determination. Similarly, photographs of flowers are the focus of natural history observations of plants recently made available through online platforms.
The best studied of these anther‐smut pathogens are members of the basidiomycete genus Microbotryum (subphylum Pucciniomycotina) occurring on plants in the carnation family, the Caryophyllaceae. The fitness consequences of infection are large because the hosts are sterilized and the infection is systemic and persistent (Alexander and Maltby, 1990). Sporulation in the anthers can facilitate pathogen spread via pollinators, and anther smut has been studied as a model of frequency‐dependent and vector‐borne disease transmission (Alexander and Antonovics, 1988; Bucheli and Shykoff, 1999). The most affected members of the Caryophyllaceae include the tribes Sileneae and Caryophylleae (Thrall et al., 1993), but with wide variation in occurrence among the species even within plant genera (Hood et al., 2010).
Microbotryum species can also occur on several other plant families (Piątek et al., 2005; Kemler et al., 2009) and, in some cases, cause other forms of smut diseases (e.g., ovary smut, whole‐flower smut or leaf smut) (Kashefi and Vánky, 2004; Lui et al., 2009). Additionally, there are a few instances of convergence for the anther‐smut disease syndrome, caused by distantly related fungi in the subphylum Ustilagomycotina (Bauer et al., 2008; Roets et al., 2008).
SURVEYS OF NATURAL HISTORY OBSERVATIONS OLD AND NEW
The availability of rapidly accumulating online natural history observations presents the opportunity to compare them with independent data sets based upon specimen collections. These include specimens of Microbotryum (formerly Ustilago violacea) in fungarium collections, prior published herbarium surveys of general plant collection, and a survey of plant observations deposited in the iNaturalist website.
Fungarium data were retrieved from the Mycology Collections data Portal (MyCoPortal; http://mycoportal.org; accessed 18 June 2019), integrating records from ca. 120 institutions, and from examination of collections at the Royal Botanic Gardens, Kew (K). Specimen details (i.e., date, locality, host species) were used to remove duplicate depositions; a few recognized instances of anther smut in agricultural greenhouses (Conners, 1933; Spencer and White, 1951) and experimental inoculation studies (Kniep, 1919) were also removed. A total of 1394 specimens remained, of which 846 contained decipherable and informative location data.
Data from prior herbarium surveys of general plant collections included studies that focused on the tribe Sileneae, representing ca. 39,000 specimens from a dozen major herbaria (Hood et al., 2010) and targeted surveys of the North American native host species Silene virginica and Silene caroliniana from regional collections (Antonovics et al., 2003).
iNaturalist observations within the Caryophyllaceae (n = 79,801) were examined (by M. E. Hood) for anther smut as the presence of dark‐colored fungal spores in place of pollen (Fig 1) (http://inaturalist.org; accessed 2 July 2017 through 24 June 2019). Plant identifications are often evaluated by the online community, and those ascertained at least to the level of tribe were used for quantifying disease incidence (Table 1). Summary statistics on observation numbers per species, geolocation data, and phenology were collected from iNaturalist main webpages for each species. An iNaturalist project, called Wildflower Health Watch, was created for adding observations that included anther smut; a small number of observations did not permit assembly into projects and were not included in further analyses. iNaturalist entries explicitly for Microbotryum species were few and were not included in this study. Plant species in other families known to harbor anther smut were also examined to assess detectability of the disease among iNaturalist observations.
Table 1.
Observations of anther‐smut disease in the Caryophyllaceae among iNaturalist data.
| Family | Subfamily | Tribe | Total count | Diseased count | Proportion diseased |
|---|---|---|---|---|---|
| Caryophyllaceae | 79,801 | 380 | 0.00476 | ||
| Caryophylloideae | 38,011 | 357 | 0.00939 | ||
| Sileneae | 26,946 | 257 | 0.00954 | ||
| Caryophylleae | 11,065 | 100 | 0.00904 | ||
| Alsinoideae | 35,205 | 23 | 0.00065 | ||
| Alsineae | 27,102 | 11 | 0.00041 | ||
| Arenarieae | 2,472 | 12 | 0.00485 | ||
| Sagineae | 2,884 | 0 | 0.00000 | ||
| Sclerantheae | 2,117 | 0 | 0.00000 | ||
| Eremogoneae | 630 | 0 | 0.00000 | ||
| Paronychioideae | 6,585 | 0 | 0.00000 | ||
| Sperguleae | 2,986 | 0 | 0.00000 | ||
| Polycarpeae | 2,366 | 0 | 0.00000 | ||
| Paronychieae | 1,233 | 0 | 0.00000 | ||
| Corrigioleae | 0 | 0 | 0.00000 |
Tribe‐level data are summed to produce subfamily‐ and family‐level statistics.
This iNaturalist survey produced a quantity of anther smut observations comparable to those from the fungaria or the prior survey of herbarium general plant collections. However, this quantity of observations has been achieved in iNaturalist over the much narrower observation timespan of a decade, compared to two centuries for the specimen collections, and in a period when the number of observations in the other databases has markedly declined (Fig. 2). The number of diseased species in the Caryophyllaceae found in the iNaturalist observations (n = 43) is smaller than in the other databases (fungaria, N = 97; herbarium general plant collection, N = 111) (Appendix S1). Still, 13 host–pathogen combinations in the iNaturalist survey had not been recorded in either the fungarium data or the general plant herbarium collection survey (Table 2). Overall, the incidence of disease in iNaturalist is very similar to herbarium survey of the Sileneae specimens (iNaturalist disease incidence = 0.95%; herbarium survey = 0.89%; χ 2 = 0.689, df = 1, P = 0.407).
Figure 2.
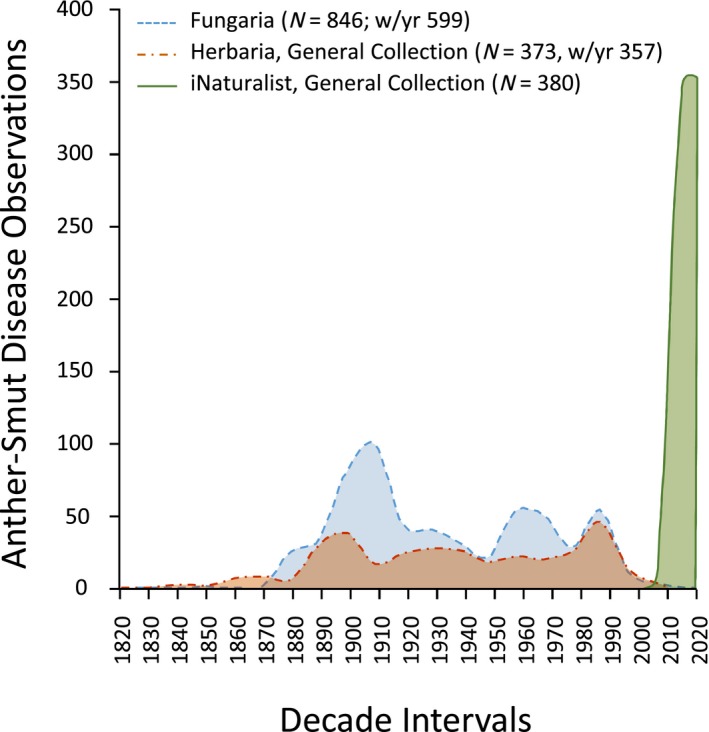
Anther‐smut disease observations over time and by data collection type. The blue dashed line represents fungaria (i.e., mycological herbaria) for hosts in the Caryophyllaceae (N = 846, number of observations with year data [w/yr] = 599), red dash‐dotted line represents the survey of herbarium general collections of hosts primarily in the Sileneae from Hood et al. (2010) (N = 373, w/yr = 357), and green solid line represents the survey of iNaturalist observations of hosts in the Caryophyllaceae (N = 361, w/yr = 361). Herbarium general collection survey includes data obtained before the last interval from 2010 to 2020.
Table 2.
Caryophyllaceae species with anther‐smut disease among iNaturalist observations.
| Host species | Tribe | Total count | Diseased count | Proportion diseased | Deviation | Binomial distribution probability |
|---|---|---|---|---|---|---|
| Moehringia lateriflora | Ar | 463 | 12 | 0.02592 | high | 0.0000 |
| Myosoton aquaticum | Al | 452 | 7 | 0.01549 | high | 0.0000 |
| Stellaria borealis | Al | 34 | 1 | 0.02941 | high | 0.0219 |
| Mononeuria groenlandica | Al | 195 | 1 | 0.00513 | low | 0.9926 |
| Dianthus carthusianorum | C | 485 | 24 | 0.04948 | high | 0.0000 |
| Saponaria pumila | C | 10 | 3 | 0.30000 | high | 0.0001 |
| Dianthus arenarius † | C | 39 | 4 | 0.10256 | high | 0.0005 |
| Dianthus superbus | C | 175 | 7 | 0.04000 | high | 0.0014 |
| Dianthus pavonius | C | 29 | 3 | 0.10345 | high | 0.0025 |
| Dianthus pygmaeus † | C | 31 | 3 | 0.09677 | high | 0.0031 |
| Dianthus graniticus † | C | 1 | 1 | 1.00000 | high | 0.0094 |
| Dianthus saxicola † | C | 3 | 1 | 0.33333 | high | 0.0279 |
| Dianthus hyssopifolius † | C | 28 | 2 | 0.07143 | high | 0.0284 |
| Saponaria ocymoides | C | 253 | 6 | 0.02372 | high | 0.0335 |
| Dianthus acicularis † | C | 4 | 1 | 0.25000 | high | 0.0370 |
| Dianthus capitatus † | C | 9 | 1 | 0.11111 | high | 0.0814 |
| Dianthus sylvestris | C | 113 | 3 | 0.02655 | high | 0.0910 |
| Dianthus repens † | C | 11 | 1 | 0.09091 | high | 0.0986 |
| Saponaria officinalis | C | 2492 | 17 | 0.00682 | low | 0.1062 |
| Dianthus gallicus † | C | 13 | 1 | 0.07692 | high | 0.1154 |
| Dianthus monspessulanus | C | 63 | 2 | 0.03175 | high | 0.1185 |
| Dianthus seguieri † | C | 16 | 1 | 0.06250 | high | 0.1401 |
| Dianthus deltoids | C | 632 | 7 | 0.01108 | high | 0.3831 |
| Gypsophila repens | C | 74 | 1 | 0.01351 | high | 0.5025 |
| Dianthus chinensis † | C | 201 | 2 | 0.00995 | low | 0.7073 |
| Silene virginica | S | 1723 | 119 | 0.06907 | high | 0.0000 |
| Viscaria vulgaris | S | 345 | 14 | 0.04058 | high | 0.0000 |
| Silene flos‐cuculi | S | 1577 | 2 | 0.00127 | low | 0.0000 |
| Silene laciniata | S | 1916 | 4 | 0.00209 | low | 0.0001 |
| Silene uniflora | S | 462 | 13 | 0.02814 | high | 0.0005 |
| Silene dioica | S | 3240 | 47 | 0.01451 | high | 0.0030 |
| Silene latifolia | S | 4741 | 28 | 0.00591 | low | 0.0053 |
| Silene acaulis | S | 927 | 17 | 0.01834 | high | 0.0079 |
| Silene atropurpurea † | S | 3 | 1 | 0.33333 | high | 0.0279 |
| Silene italica | S | 32 | 2 | 0.06250 | high | 0.0363 |
| Silene vallesia | S | 4 | 1 | 0.25000 | high | 0.0370 |
| Silene grayi | S | 5 | 1 | 0.20000 | high | 0.0461 |
| Silene saxifraga | S | 12 | 1 | 0.08333 | high | 0.1070 |
| Silene parryi | S | 41 | 1 | 0.02439 | high | 0.3208 |
| Silene fulgens † | S | 50 | 1 | 0.02000 | high | 0.3761 |
| Silene caroliniana | S | 195 | 1 | 0.00513 | low | 0.4525 |
| Silene baccifera | S | 74 | 1 | 0.01351 | high | 0.5025 |
| Silene lemmonii | S | 106 | 1 | 0.00943 | low | 0.7375 |
| Silene vulgaris * | S | 2727 | 0 | 0.00000 | low | 0.0000 |
Species are ranked within tribes according to the significance of the deviation of observed diseased observations from expectations based upon subfamily diseased proportions (Table 1). Deviation indicates high or low rates of disease relative to tribe‐level proportions. *Silene vulgaris is also included as an example of statistically assessing disease absence. †Indicates host–pathogen combinations not found in prior surveys of herbarium general collections or fungaria data. Tribes are abbreviated as Ar = Arenarieae, Al = Alsinoideae, C = Caryophylloideae, S = Sileneae. Bold binomial distribution probabilities indicate statistical significance at the level of alpha = 0.05 following the Bonferroni corrected threshold of 0.0014.
Geographic distributions of anther‐smut observations differed somewhat among three sources of data (Fig. 3). The fungarium collections were focused mostly in western Europe, with the largest numbers from Germany, Sweden, and Austria (Fig. 3A), likely reflecting the intense activity of fungal biologists in Germany, especially coinciding with the peak in collections during the pre‐war period of the 20th century (Ainsworth, 1976). The survey of general plant collections displayed the broadest distribution among continents, and indeed, the underlying aim of that study was to cover herbaria from distant regions (Hood et al., 2010) (Fig. 3B). The iNaturalist distribution was more restricted in geographic coverage, with fewer records from southern or extreme far northern latitudes (Fig. 3C).
Figure 3.
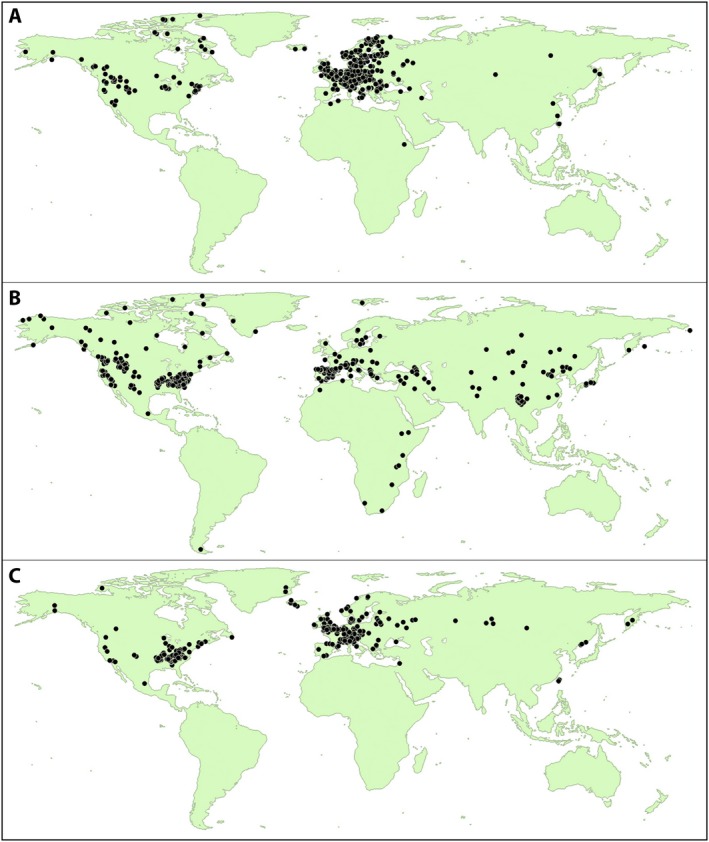
Geographic distribution of anther‐smut disease observations. (A) Fungaria (i.e., mycological herbaria), (B) survey of herbarium general collections, and (C) survey of iNaturalist observations.
The differences seen between the iNaturalist and historic collections may be due to differing goals of the observers. In general, herbaria contain specimens primarily collected to be representative of diversity from a comprehensive range of geographic locations. On the other hand, iNaturalist data are being generated by citizen scientists, probably more locally and without particular research aims in mind. As a result, observed distributions from any particular source of data represent where the observers tend to go, and they do not necessarily reflect unbiased ranges of the organisms at broad geographic scales.
RESOLVING AMONG‐SPECIES VARIATION IN DISEASE INCIDENCE
Pathogens are expected to cause disease on a fraction of their host species, and thus surveys of disease distributions are facilitated by large numbers of host observations. Indeed, on the iNaturalist platform, anther smut was infrequent overall, occurring in 0.48% of 79,801 Caryophyllaceae observations (Table 1). However, the disease incidence varied greatly among taxonomic divisions within this plant family. Among subfamilies, incidence of disease ranged from 0.00% to 0.95%. Disease incidence was highest in the Caryophylloideae, low in the Alsinoideae, and no disease was recorded in the Paronchioideae, consistent with a prior review of the literature (Thrall et al., 1993). These differences among subfamilies were highly significant (χ 2 = 329, df = 2, P < 0.001). Within the Caryophylloideae subfamily, the Sileneae and Caryophylleae tribes were nearly equal in disease incidence at 0.95% and 0.90% (χ 2 = 0.211, df = 1, P = 0.646), including in these tribes the genera Silene and Dianthus, respectively, that are known to be commonly diseased (Thrall et al., 1993). Results so similar to prior herbarium surveys lend confidence in the ability of citizen observations to be informative of real differences among host groups.
Among the 43 plant species in the Caryophyllaceae where anther smut was found in the iNaturalist survey (Table 2), all have perennial life histories, consistent with conclusions of prior studies (Thrall et al., 1993; Hood et al., 2010). There was large variation among plant species in the proportions of observations with anther smut, and disease‐rich species were not restricted to one section of the family (Table 2). Several species were found to have significantly higher disease incidence than the average for their subfamily, most notably including (in descending order of the significance of the binomial probabilities, analyzed like in the herbarium survey of Hood et al., 2010): Silene virginica (Sileneae), Moehringia lateriflora (Arenarieae), and Dianthus carthusianorum (Caryophylleae). Of particular note was S. virginica, with 6.91% of 1723 plants observed being diseased, and these constituted nearly one third of all disease observations within the family. This level of disease was not statistically different from the 8.32% among 1022 herbarium specimens surveyed in a prior study (Antonovics et al., 2003) (χ 2 = 1.855, df = 1, P < 0.173). The statistical significance of the absence of disease was not assessed for all perennial species (see Hood et al., 2010), although some species known to be diseased in nature were notable for the significantly low rate of disease among iNaturalist observations, including Silene flos‐cuculi, Silene laciniata, and Silene vulgaris as examples (Table 2).
The disease incidence among herbarium general collections and among iNaturalist data was remarkably similar despite being recorded by different sets of observers and at different times. This similarity in disease incidence is consistent with prior indications that anther smut generally goes unrecognized by a botanist when making field collections for herbarium use (Antonovics et al., 2003), and thus disease proportion among total observations can be a relatively objective measure for comparisons of disease incidence among host taxa. Flower size and color have been evaluated for potentially influencing disease recognition by collectors, but no significant relationship was found between these two traits and disease incidence among host species (Hood et al., 2010). Differing incidence of disease between very similar‐appearing species support the neutrality of general collections with regard to anther smut. A striking example is Silene uniflora and S. vulgaris that are so similar as to previously be considered conspecific forms (Marsden‐Jones and Turrill, 1957), but the iNaturalist survey showed statistically significant higher and lower disease incidence, respectively, for these two hosts compared to the other Sileneae taxa. Finally, multiple aspects of anther smut were confirmed by the iNaturalist data, supporting this application to understanding the ecology of disease interactions. For instance, the subfamily differences in incidence were consistent with disease incidence based on a review of previous publications (Thrall et al., 1993). There was also a consistent association of the disease with perennial host life histories across data sets as previously reported (Hood et al., 2010).
WITHIN‐SPECIES VARIATION IN DISEASE INCIDENCE
Spatial and temporal patterns provide further information about the dynamics of disease as threats to natural populations and could be detected within some species having a larger number of iNaturalist observations. Within‐species patterns included assessment of disease incidence caused by Microbotryum lychnidis‐dioicae in the native (Europe) and introduced (North America) ranges of Silene latifolia. In North America, only one specimen was diseased among 2727 observations, significantly fewer than the 27 of 1923 observations in the native range (χ 2 = 35.6, df = 1, P < 0.001). This difference is consistent with the escape from enemies hypothesis, which says there should be lower disease incidence in the introduced range (Wolfe, 2002).
Two comparisons were performed for the heavily diseased species, S. virginica, to assess distributions obtained from the iNaturalist survey relative to distributions reported in prior studies. The first comparison included geographic distributions of diseased hosts among all host observations of this species (Fig. 4). The current (iNaturalist) distribution is remarkably similar to that obtained from historic collections (Antonovics et al., 2003) (the overall disease incidence between the two data sets was also similar as noted above). The common occurrence of anther smut near the margins of the host distribution was also consistent with the observations of Antonovics et al. (2003). Using the same spatial statistical approach as in that prior study, the iNaturalist data similarly showed a lack of a statistically significant difference in local host densities between observations of diseased hosts versus healthy hosts (mean total observation numbers in 50‐km radius from diseased hosts = 40.6 and healthy hosts = 40.2, t‐test F 234 = 1.284, P = 0.258; mean total observation numbers in 100 km radius from diseased hosts = 94.6 and healthy hosts = 89.4; t‐test F 234 = 0.114, P = 0.736). Because anther smut has a pollinator‐mediated (i.e., frequency‐dependent) component to disease transmission, it has been suggested that the disease may persist even at the margins of host ranges where density can be too low to maintain purely density‐dependent transmission (Bruns et al., 2018). The lack of positive association of anther smut in S. virginica with regional measures of host density is consistent with findings of prior studies on this and other species (Antonovics et al., 2003; Bueker et al., 2016).
Figure 4.
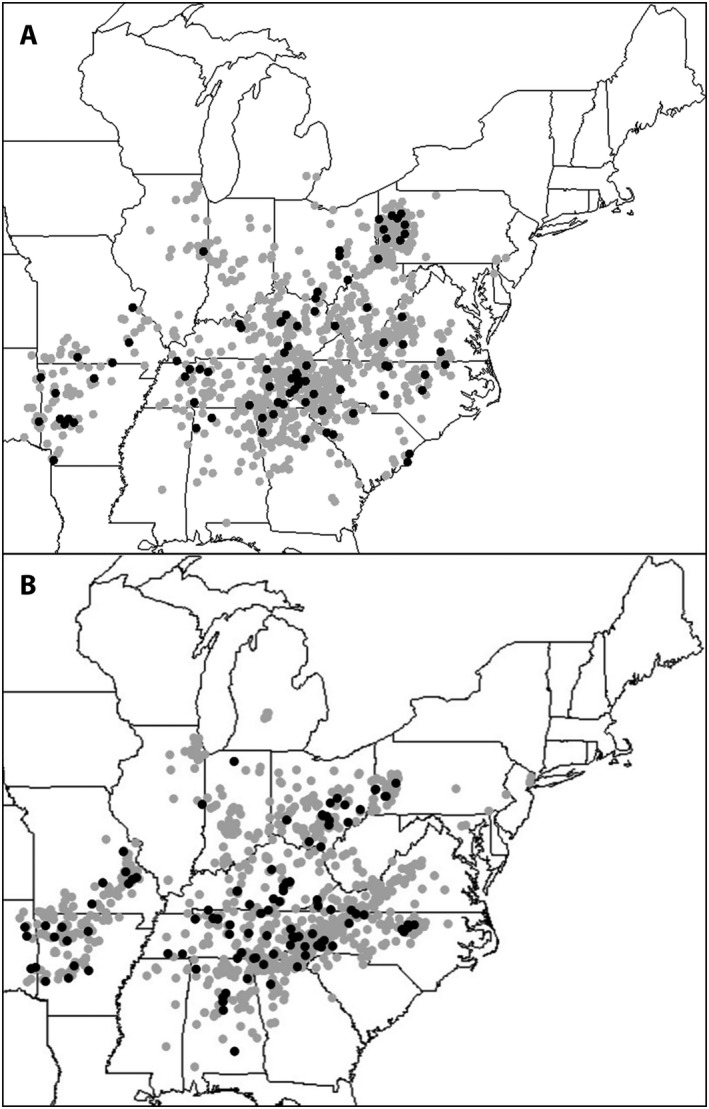
Distribution of anther‐smut disease within the geographic range of Silene virginica. (A) Survey of general plant collections; image reproduced from Antonovics et al. (2003) (total herbarium sheets = 1022, diseased = 85). Mapped positions for the general plant collection survey are randomly generated locations within the county of the collection's origin. (B) Survey of iNaturalist observations (total observations = 1723, diseased = 119). Black dots indicate observations of anther‐smut disease, and gray dots indicate observations of healthy hosts.
The second comparison was of phenological patterns of diseased versus healthy hosts of S. virginica. The temporal distribution of flowering observations of diseased S. virginica significantly shifted toward the earlier period of the season relative to the distribution of healthy hosts (Fig. 5) (two‐sample K‐S test, D max = 0.328, P < 0.001). This result is consistent with the earlier flowering of diseased plants documented in field‐based studies of other species (Alexander and Antonovics, 1988; Shykoff and Kaltz, 1998; Carlsson and Elmqvist, 1992; Tang et al., 2019) and likely indicates an important role of the flowering timing for pathogen dispersal and epidemiology.
Figure 5.
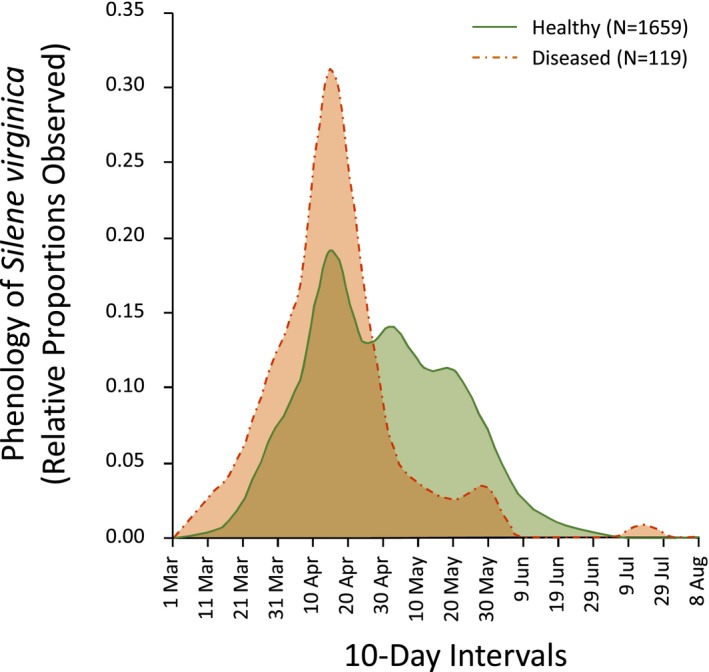
Phenology of Silene virginica from iNaturalist data showing observations with anther‐smut disease and healthy observations. The red dash‐dotted line represents diseased observations (N = 119), and green solid line represents healthy observations (N = 1659).
FURTHER UTILITY OF ONLINE NATURAL HISTORY PLATFORMS
Investigation with the anther‐smut disease has demonstrated informative and novel ways that online platforms for citizen observations like iNaturalist can add to the resources available in natural history collections, including insights on disease distributions and details of the host–pathogen interactions. Noting that herbaria have faced recent declines in submissions (Prather et al., 2004; Lavoie and Lachance, 2006; Renner and Rockinger, 2016), here we can see this impact as the general absence of anther‐smut observations in the last few decades in data we summarize from both mycological collections and surveys of general plant collections. We show that citizen observations can be valuable for filling a widening gap in our knowledge of recent species distributions and that such online platforms present opportunities for deeper research that quantifies contemporary disease incidence over spatial, temporal, and host‐specificity scales.
While the occurrence of anther smut on the Caryophyllaceae provided quantitative insights to several aspects of disease ecology, the approach is not limited to this host–pathogen system (Appendix S2). Anther smut was also detected in the iNaturalist platform for targeted searches of species in other plant families previously known to harbor the fungus. Thus, Microbotryum was found infecting Pinguicula alpina (Lentibulariaceae), Calandrinia acaulis, and Calandrinia affinis (Montiaceae) (Kemler et al., 2009; Elvebakk et al., 2015; Ziegler et al., 2018). Based upon similar floral morphology symptoms, anther smut caused by Ustilagomycotina fungi was also detected in several Scilla species (fungus Antherospora) (Bauer et al., 2008) and Oxalis species (fungus Thecaphora) (Roets et al., 2008).
Extending to other types of plant disease, phyllody symptoms in goldenrod (Solidago canadensis) characteristic of phytoplasma infection, fungal anthracnose symptoms on the petals of flowering dogwood (Cornus florida), and ergot sclerotia (Claviceps fungi) on inflorescences of wild quack grass (Elymus repens) can each be found readily in quick surveys of iNaturalist observations of the host plants (Appendix S2). The rates and facility of finding diseased observations will depend upon the particulars of the host–pathogen system, including how conspicuous and unambiguous the symptoms might be. Still, there is strong potential for identifying individual occurrences for a broad range of interactions, especially those that produce observable phenotypes in floral structures. Diseases of leaves or particularly fruits also present likely possibilities, such as Gymnosporangium rust disease of hawthorns (Crataegus sp.) that is easily found among the online postings. In a broader context, observation may be available in sufficient numbers to provide meaningful insights for rare variants in other, non‐disease traits (e.g., polymorphism of floral color, or sex ratios in gynodioecious species).
An important benefit of the rise in citizen‐driven observations is information on disease in extant populations, whether to be used for comparison with historic data or for directing future field research. While herbarium records have indicated changes in plant distributions within the two centuries of accumulated observations (Antonovics et al., 2003; Meineke et al., 2018a), such cumulative distributions, even partitioned over short time intervals, can sometime not accurately reflect field observations of extant populations (Antonovics et al., 2001; Applequist et al., 2007). Moreover, a rising threat from disease emergence has resulted from increasing rates of climate change and the redistribution of species, by natural and anthropogenic means (Smith et al., 2014; see also Lafferty and Mordecai, 2016). Monitoring the tempo and spatial patterns of disease spread during this period of changes in host geographic ranges would provide critical information to inform conservation and intervention strategies.
We have also found that the ability to quickly contact an observer online is especially helpful in obtaining additional details about disease occurrence or even biological specimens. Particularly for rare or obscure organisms, guidance to extant populations can fundamentally alter the prospect for field studies. Equally as important, this information can be used to target the acquisition of new material for preservation in herbarium collections. The usefulness of herbarium specimens from many years ago for DNA isolation and analysis (e.g., Malmstrom et al., 2007; Andrew et al., 2018; Heberling and Burke, 2019) means that locating extant populations also makes it possible to compare past and current genetic structure. Heberling and Isaac (2018) suggested using iNaturalist as a direct supplement to new herbarium specimens by linking the online observations to the herbarium sample. The reverse also holds potential, connecting new observations to bioinformatics resources on preserved specimens. Readily accessible means for annotating specimens and accessing an assembly of observations as online resources for particular research projects hold great potential to advance studies in natural plant populations.
Finally, the disease surveys from general collections can identify host species deserving of further targeted study, even where rates of infection may not have risen to statistical significance here due to small sample sizes. For example, our field research in the maritime Alps (e.g., Bruns et al., 2017; Petit et al., 2017) was prompted by the observation of several diseased Silene campanula specimens among relatively few herbarium sheets at the Muséum National d'Histoire Naturelle (P). Similarly, Dianthus pavonius had 3 among 29 total iNaturalist observations showing anther smut symptoms, which is suggestive of a heavy disease impact. Confirmation of this suggestion was provided by a recent study by Bruns et al. (2018), which included numerous transects in natural populations of D. pavonius where the product of disease incidence among populations (0.46) and average infection prevalence within diseased populations (0.18) are entirely consistent with the high disease rate among iNaturalist observations.
This study shows that natural history observations currently being generated in high volumes for online platforms can yield valuable insights into the ecological interactions of wild plant species. Where observers are unaware that symptoms are also being recorded, quantitative assessments of disease incidence can also be achieved. This approach is similar to surveys of museum collections. Yet, while herbaria provide essential information on long‐term distributions and direct access to specimens for trait measurements, the iNaturalist observations help fill a widening gap in very recent distributions at a time when deposition of new herbarium materials has declined. As citizen‐driven plant observations accumulate, there is great potential to discover new and diverse host–pathogen associations and to direct field studies to extant populations. Therefore, the impact of online observations intended to record plant occurrence can be enhanced by mining for disease interactions that are important drivers of plant ecology and evolution.
AUTHOR CONTRIBUTIONS
M.H. originated and designed the project. A.K. and M.H. contributed to data collections and analysis, and both contributed to drafting and revising the manuscript.
Supporting information
APPENDIX S1. Data on hosts observed with anther smut (Microbotryum fungi) on plants in the Caryophyllaceae.
APPENDIX S2. Example iNaturalist observations showing symptoms for a range of disease and host types.
ACKNOWLEDGMENTS
We thank the U.S. National Institutes of Health for support (award NIGMS‐R01GM122061). Thanks is also given to Dr. Michael Nee, Matt Rung, and Judy Gallagher for the use of herbarium and diseased and healthy iNaturalist images, respectively, and to Dr. Janis Antonovics for helpful comments on an earlier version of the manuscript. We also thank the editors and anonymous reviewers, whose comments greatly helped to improve this work.
Kido, A. and Hood M. E.. 2020. Mining new sources of natural history observations for disease interactions. American Journal of Botany 107(1): 3–11.
LITERATURE CITED
- Ainsworth, G. C. 1976. Introduction to the history of mycology. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Alexander, H. M. , and Antonovics J.. 1988. Disease spread and population dynamics of anther‐smut infection of Silene alba caused by the fungus Ustilago violacea . Journal of Ecology 76: 91–104. [Google Scholar]
- Alexander, H. M. , and Maltby A.. 1990. Anther‐smut infection of Silene alba caused by Ustilago violacea: factors determining fungal reproduction. Oecologia 84: 249–253. [DOI] [PubMed] [Google Scholar]
- Alexander, H. M. , Thrall P. H., Antonovics J., Jarosz A. M., and Oudemans P. V.. 1996. Population dynamics and genetics of plant disease: a case study of anther‐smut disease. Ecology 77: 990–996. [Google Scholar]
- Alexander, H. M. , Price S., Houser R., Finch D., and Tourtellot M.. 2007. Is there reduction in disease and pre‐dispersal seed predation at the border of a host plant's range? Field and herbarium studies of Carex blanda . Journal of Ecology 95: 446–457. [Google Scholar]
- Andrew, C. , Diez J., James T. Y., and Kauserud H.. 2018. Fungarium specimens: a largely untapped source in global change biology and beyond. Philosophical Transactions of the Royal Society, B, Biological Sciences 374: 20170392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics, J. , and Hood M. E.. 2018. Linnaeus, smut disease, and living contagion. Archives of Natural History 45: 213–232. [Google Scholar]
- Antonovics, J. , Newman T. J., and Best B. J.. 2001. Spatially explicit studies on the ecology and genetics of population margins In Silvertown J. and Antonovics J. [eds.], Integrating ecology and evolution in a spatial context, 97–116. Blackwell Science, Oxford, UK. [Google Scholar]
- Antonovics, J. , Hood M. E., Thrall P. H., Abrams J. Y., and Duthie G. M.. 2003. Herbarium studies on the distribution of anther‐smut fungus (Microbotryum violaceum) and Silene species (Caryophyllaceae) in the eastern United States. American Journal of Botany 90: 1522–1531. [DOI] [PubMed] [Google Scholar]
- Applequist, W. L. , McGlinn D. J., Miller M., Long Q. G., and Miller J. S.. 2007. How well do herbarium data predict the location of present populations? A test using Echinacea species in Missouri. Biodiversity and Conservation 16: 1397–1407. [Google Scholar]
- Bauer, R. , Lutz M., Begerow D., Piatek M., Vánky K., Bacigálová K., and Oberwinkler F.. 2008. Anther smut fungi on monocots. Micological Research 112: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Beauvais, M. P. , Pellerin S., Dubé J., and Lavoie C.. 2017. Herbarium specimens as tools to assess the impact of large herbivores on plant species. Botany 95: 153–162. [Google Scholar]
- Brock, P. M. , Döring H., and Bidartondo M. I.. 2009. How to know unknown fungi: the role of a herbarium. New Phytologist 181: 719–724. [DOI] [PubMed] [Google Scholar]
- Brown, N. , van den Bosch F., Parnell S., and Denman S.. 2017. Integrating regulatory surveys and citizen science to map outbreaks of forest diseases: acute oak decline in England and Wales. Proceedings of the Royal Society, B, Biological Sciences 284: 20170547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns, E. , Antonovics J., Carasso V., and Hood M. E.. 2017. Transmission and temporal dynamics of anther‐smut disease (Microbotryum) on alpine carnation (Dianthus pavonius). Journal of Ecology 105: 1413–1424. [Google Scholar]
- Bruns, E. , Antonovics J., and Hood M. E.. 2018. Is there a disease‐free halo at species range limits? The co‐distribution of anther‐smut disease and its host species. Journal of Ecology 107: 1–11. [Google Scholar]
- Bucheli, E. , and Shykoff J. A.. 1999. The influence of plant spacing on density‐dependent versus frequency‐dependent spore transmission of the anther smut Microbotryum violaceum . Oecologia 119: 55–62. [DOI] [PubMed] [Google Scholar]
- Bueker, B. , Eberlein C., Gladieuz P., Schaefer A., Snirc A., Bennett D. J., Begerow D., et al. 2016. Distribution and population structure of the anther smut Microbotryum silenes‐acaulis parasitizing an arctic‐alpine plant. Molecular Ecology 25: 811–824. [DOI] [PubMed] [Google Scholar]
- Burdon, J. J. , and Chilvers G. A.. 1982. Host density as a factor in plant disease ecology. Annual Review of Phytopathology 10: 143–166. [Google Scholar]
- Burdon, J. J. , and Thrall P. H.. 2008. Pathogen evolution across agro‐ecological interface: implications for disease management. Evolutionary Applications 1: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, U. , and Elmqvist T..1992. Epidemiology of anther-smut disease (microbotryum violaceum) and numeric regulation of populations of silene dioica . Oecologia 90: 509–517. [DOI] [PubMed] [Google Scholar]
- Conners, I. L. 1933. Carnation (Dianthus). Canadian Plant Disease Survey 13: 65. [Google Scholar]
- Dobson, A. , and Crawley M.. 1994. Pathogens and the structure of plant communities. Trends in Ecology & Evolution 9: 393–398. [DOI] [PubMed] [Google Scholar]
- Eastburn, D. M. , McElrone A. J., and Bilgin D. D.. 2011. Influence of atmospheric and climatic change on plant–pathogen interactions. Plant Pathology 60: 54–69. [Google Scholar]
- Elvebakk, A. , Flores A. R., and Watson J. M.. 2015. Revisions in the South American Calandrinia caespitosa complex (Montiaceae). Phytotaxa 203: 1–23. [Google Scholar]
- Ghelardini, L. , Pepori A. L., Luchi N., Capretti P., and Santini A.. 2016. Drivers of emerging fungal disease of forest trees. Forest Ecology and Management 281: 235–246. [Google Scholar]
- Hanan‐A, A. M. , Vibrans H., Cacho N. I., Villaseñor J. L., Ortiz E., and Gómez‐G V. A.. 2016. Use of herbarium data to evaluate weediness in five congeners. AOB Plants 8: plv144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberling, J. M. , and Burke D. J.. 2019. Utilizing herbarium specimens to quantify historical mycorrhizal communities. Plant Journal 7: e01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberling, J. M. , and Isaac B. L.. 2017. Herbarium specimens as exaptations: new uses for old collections. American Journal of Botany 104: 963–965. [DOI] [PubMed] [Google Scholar]
- Heberling, J. M. , and Isaac B. L.. 2018. iNaturalist as a tool to expand research value of museum specimens. Applications in Plant Sciences 6: e1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E. , and Antonovics J.. 2003. Plant species descriptions show signs of disease. Proceedings of the Royal Society, B, Biological Sciences 270: S156–S158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E. , Mena‐Alí J. I., Gibson A. K., Oixelman B., Giraud T., Yockteng R., Arroyo M. T. K., et al. 2010. Distribution of the anther‐smut pathogen Microbotryum on species of the Caryophyllaceae. New Phytologist 187: 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashefi, J. , and Vánky K.. 2004. Microbotryum scolymi, a rare smut fungus new for Greece. Journal of Plant Pathology 86: 157–159. [Google Scholar]
- Kemler, M. , Lutz M., Göker M., Oberwinkler F., and Begerow D.. 2009. Hidden diversity in the non‐caryophyllaceous plant‐parasite members of Microbotryum (Picciniomycotina: Microbotryales). Systematics and Biodiversity 7: 297–306. [Google Scholar]
- Kniep, H. 1919. Untersuchungen uber den Antherenbrand (Ustilago violacea Pers.), Ein Beitrag zum Sexualitatsproblem. Zeitschrift für Botanik 11: 257–284. [Google Scholar]
- Lafferty, K. D. , and Mordecai E. A.. 2016. The rise and fall of infectious disease in a warmer world. F1000Research 5: 2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, P. L. , Willems F. M., Scheepens J. F., Burbano H. A., and Bossdorf O.. 2019. Using herbaria to study global environmental change. New Phytologist 221: 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, C. , and Lachance D.. 2006. A new herbarium‐based method for reconstructing the phenology of plant species across large areas. American Journal of Botany 93: 512–516. [DOI] [PubMed] [Google Scholar]
- Li, W. , Brlansky R. H., and Hartung J. S.. 2006. Amplification of DNA of Xanthomonas axonopodis pv. citri from historic citrus canker herbarium specimens. Journal of Microbiological Methods 65: 237–246. [DOI] [PubMed] [Google Scholar]
- Lui, T. , Tian H., He S., and Guo L.. 2009. Microbotryum scorzonerae (Microbotryaceae), new to China, on a new host plant. Mycotaxon 108: 245–247. [Google Scholar]
- Malmstrom, C. M. , Shu R., Linton E. W., Newton L. A., and Cook M. A.. 2007. Barley yellow dwarf viruses (BYDVs) preserved in herbarium specimens illuminate historical disease ecology of invasive and native grasses. Journal of Ecology 95: 1153–1166. [Google Scholar]
- Marsden‐Jones, E. M. , and Turrill W. B.. 1957. The bladder campions (Silene maritima and S. vulgaris). Ray Society, London, UK. [Google Scholar]
- Meentemeyer, R. K. , Dorning M. A., Vogler J. B., Schmidt D., and Garbelotto M.. 2015. Citizen science helps predict risk of emerging infectious disease. Frontiers in Ecology and the Environment 13: 189–194. [Google Scholar]
- Meineke, E. K. , Davis C. C., and Davies T. J.. 2018a. The unrealized potential of herbaria for global change biology. Ecological Monographs 88: 505–525. [Google Scholar]
- Meineke, E. K. , Davies T. J., Daru B. H., and Davis C.C.. 2018b. Biological collections for understanding biodiversity in the Anthropocene. Philosophical Transactions of the Royal Society, B, Biological Sciences 374: 20170386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, E. , Silver C., Cornille A., Gladieux P., Rosenthal L., Bruns E., Yee S., et al. 2017. Co‐occurrence and hybridization of anther‐smut pathogens specialized on Dianthus hosts. Molecular Ecology 26: 1877–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piątek, M. , Mułenko W., Piątek J., and Bacigálová K.. 2005. Taxonomy and distribution of Microbotryum pinguiculae, a species of smut fungi new for the Carpathians. Polish Botanical Journal 50: 153–158. [Google Scholar]
- Prather, L. A. , Alvarez‐Fuentes O., Mayfield M. H., and Ferguson C. J.. 2004. Implications of the decline in plant collecting for systematic and floristic research. Systematic Botany 29: 216–220. [Google Scholar]
- Rabeler, R. K. , and Hartman R. L.. 2005. Caryophyllaceae In Flora of North America Editorial Committee [ed.],Flora of North America north of Mexico, 3–215. Oxford University Press, NY, NY, USA. [Google Scholar]
- Renner, S. S. , and Rockinger A.. 2016. Is plant collecting in Germany coming to an end? Willdenowia 46: 93–97. [Google Scholar]
- Ristaino, J. B. , Groves C. T., and Parra G. R.. 2001. PCR amplification of the Irish potato famine pathogen from historic specimens. Nature 411: 695–697. [DOI] [PubMed] [Google Scholar]
- Roets, F. , Dreyer L. L., Wingfield M. J., and Begerow D.. 2008. Thecaphora capensis sp. nov., an unusual new anther smut on Oxalis in South Africa. Persoonia 21: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg, M. , and Rivera Y.. 2017. Voucher specimens? Yes please! NPDN News USDA APHIS 12: 1–2. [Google Scholar]
- Saville, A. C. , Martin M. D., and Ristaino J. B.. 2016. Historic late blight outbreaks caused by a widespread dominant lineage of Phytophthora infestans (Mont.) de Bary. PLoS ONE 11: e0168381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, A. M. , Kemler M., Bauer R., and Begerow D.. 2010. The illustrated life cycle of Microbotryum on the host plant Silene latifolia . Botany 88: 875–885. [Google Scholar]
- Shykoff, J. A. , and Kaltz O.. 1998. Phenotypic changes in host plants diseased by Microbotryum violaceum: parasite manipulation, side effects, and trade‐offs. International Journal of Plant Sciences 159: 236–243. [Google Scholar]
- Smith, K. F. , Goldberg M., Rosenthal S., Carlson L., Chen J., Chen C., and Ramachandran S.. 2014. Global rise in human infectious disease outbreaks. Journal of The Royal Society Interface 11: 20140950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, J. L. , and White H. E.. 1951. Anther smut of carnation. Phytopathology 41: 291–299. [Google Scholar]
- Tang, H. , Hood M. E., Ren Z.‐X., Li D.‐Z., Wolfe L. M., and Wang H.. 2019. Specificity and seasonal prevalence of anther‐smut disease Microbotryum on sympatric Himalayan Silene species. Journal of Evolutionary Biology 32: 451–462. [DOI] [PubMed] [Google Scholar]
- Thrall, P. H. , Biere A., and Antonovics J.. 1993. Plant life‐history and disease susceptibility—the occurrence of Ustilago violacea on different species within the Caryophyllaceae. Journal of Ecology 81: 489–498. [Google Scholar]
- Wolfe, L. M. 2002. Why alien invaders succeed: support for the escape‐from‐enemy hypothesis. American Naturalist 160: 705–711. [DOI] [PubMed] [Google Scholar]
- Yoshida, K. , Burbano H. A., Krause J., Thines M., Weigel D., and Kamoun S.. 2014. Mining herbaria for plant pathogen genomes: back to the future. PLOS Pathogens 10: e1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, R. , Lutz M., Piątek J., and Piątek M.. 2018. Dismantling a complex of anther smuts (Microbotryum) on carniverous plants in the genus Pinguicula . Mycologia 110: 361–374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Data on hosts observed with anther smut (Microbotryum fungi) on plants in the Caryophyllaceae.
APPENDIX S2. Example iNaturalist observations showing symptoms for a range of disease and host types.


