Abstract
Recent advances in image reconstruction techniques have enabled high resolution MRI studies of fetal brain development in human subjects. Rhesus macaques (Macaca mulatta) are valuable animal models for use in studies of fetal brain development due to the similarities between this species and humans in brain development and anatomy. There is a need to develop fetal brain templates for the rhesus macaque to facilitate the characterization of the normal brain growth trajectory and departures from this trajectory in rhesus models of neurodevelopmental disorders. Here we have developed unbiased population-based anatomical T2-weighted, fractional anisotropy (FA) and apparent diffusion coefficient (ADC) templates for fetal brain from MR images scanned at 3 time points over the second and third trimesters of the 168 day gestational term. Specifically, atlas images are constructed for brains at gestational ages of 85 days (G85, N = 18, 9 females), 110 days (G110, N = 10, 7 females) and 135 days (G135, N = 16, 7 females). We utilized this atlas to perform segmentation of fetal brain MR images and fetal brain volumetric and microstructure analysis. The T2-weighted template images facilitated characterization of the growth within six fetal brain regions. The template images of diffusion tensor indices provided information related to the maturation of white matter tracts. These growth trajectories are referenced to human studies of fetal brain development. Similarities in the temporal and regional patterns of brain growth over the corresponding periods of central nervous system development are identified between the two species. Atlas images are available online as a reference for registration, reconstruction, segmentation, and for longitudinal analysis of early fetal brain growth over this unique time window.
1. Introduction
In primates, the fetal brain undergoes dramatic changes in size, shape, and magnetic resonance image (MRI) contrast during the second half of gestation. As a consequence, MRI can be used to characterize the normal trajectory of development in vivo, and provide a sensitive method for detecting anatomical abnormalities associated with neurodevelopmental disorders, over this time range. Historically, fetal motion has precluded the acquisition of high-resolution 3D brain images for quantitative analyses of development. However, advances in retrospective motion correction methods have addressed this problem. It is now possible to reconstruct high-resolution 3D images of the moving fetal brain from multiple 2D image series (Rousseau et al., 2006; Jiang et al., 2007; Gholipour et al., 2010; Kim et al., 2010a, 2010b, 2011; Kuklisova--Murgasova et al., 2012; Kainz et al., 2015; Tourbier et al., 2015), and it is also possible to assess anisotropy in water diffusion in utero using these methods (Huang et al., 2009; Jiang et al., 2009; Fogtmann et al., 2014). Quantitative characterization of macroscopic changes in the fetal brain, as well as the analysis of water diffusion anisotropy, can therefore be used to monitor the cellular-level development of white matter (Scola et al., 2016) and cortical gray matter (Wang et al., 2017) tissue in the developing brain.
Rhesus macaques are important animal models for study of fetal brain development in the contexts of neurodevelopmental disorders. In contrast to rodent species, which are born at early stages of central nervous system development, nonhuman primates are born at a similar stage of brain maturation as humans (Workman et al., 2013). In addition, rhesus macaque and human brains share anatomical similarities of potential relevance to functional and behavioral deficits observed in individuals affected by neurodevelopmental disorders, such as the large intracranial volume fraction occupied by white matter (WM), and the gyral/sulcal folding patterns of the cerebral cortex. It is easier to control experimental variables such as MRI scan parameters and environmental factors, such as gestational age at the time of the MRI procedure, in studies of animals compared to human subjects. Normative template images are important tools in the analysis of brain volume and microstructural properties as assessed through measurements of water diffusion. Multiple templates of human fetal development have been reported (Habas et al., 2010; Kuklisova-Murgasova et al., 2011; Serag et al., 2012; Zhan et al., 2013; Dittrich et al., 2014; Gholipour et al., 2017; Khan et al., 2019), but to date, template images are not available for rhesus macaque fetal brains. The purpose of this work is therefore to provide normative templates of the fetal rhesus macaque brain over the second half of the 168-day gestational term.
High resolution, 3D, isotropic T2-weighted, apparent diffusion coefficient (ADC), and fractional anisotropy (FA) images were reconstructed from 2D slice series’s at 0.5 × 0.5 × 0.5 mm3 sized voxels, or 0.75 × 0.75 × 0.75 mm3 sized voxels for the T2-weighted images, and diffusion parameter maps, respectively. Template images are generated for fetal rhesus macaque brains at gestational ages (G)85, G110, and G135. At the earliest of these time points, approximately halfway through the gestational term, cerebral cortical folds have not yet formed, and laminar pattern is the dominant organizing feature of the developmentally transient germinal matrix zones, the intermediate zone, the subplate, and the cortical plate. At subsequent time points, maturation of cerebral WM gives rise to defined tracts with high FA. In order to quantify volumes of the developing structures, age specific label maps were constructed with reference to the T2-weighted images. Similarly, WM regions of interest (ROIs), based on a subset of regions delineated in the UW-DTIRMAC271 atlas (Zakszewski et al., 2014) for adult rhesus macaques, are delineated on FA parameter maps. These data are made available to the research community (www.nitrc.org). It is anticipated that this normative dataset for fetal brain development in the rhesus macaque will be of value as a reference dataset for characterizing brain development in this species, and for comparisons to findings in fetal brains in animal model studies of neurodevelopmental disorders.
2. Materials and methods
2.1. Subjects
All animal procedures were approved by the Oregon National Primate Research Center (ONPRC) and were conducted in full accordance with Public Health Service Policy on Human Care and Use of Laboratory Animals. A total of 24 dams produced 28 fetuses that contributed to the template images in this study (four dams generated two fetuses each, as separate pregnancies). These were derived from control animals in three separate studies that adopted different experimental designs. Dams 1–5 (Table 1) were part of a longitudinal study of normative development of the fetal cerebral cortex (Wang et al., 2017) and were singly-housed throughout pregnancy. Dams 6–16 consist of the control animals from a cross-sectional study of the effect of fetal alcohol exposure on brain development, and detailed rearing procedures are described in (Jimenez et al., 2019). Dams 17–24 were the control animals in a longitudinally-designed study of the effect of maternal protein restriction on fetal growth and development. These animals were group-housed and experimental procedures are detailed in (Roberts et al., 2018). The gestational ages in which fetal MRI data were acquired for each fetus are listed in Table 1. All MRI procedures were performed within two days of three gestational ages of the rhesus macaque 168 day gestational term: gestational day (G)85, G110, and G135. Several of the fetuses studied were scanned at more than one time point. Thus, brains of 17 fetuses (8 females) were scanned at a gestational age of 85 ± 2 days, brains of 10 fetuses (7 females) were scanned at a gestational age of 110 ± 1 days, and brains of 16 fetuses (7 females) were scanned at a gestational age of 135 ± 2 days.
Table 1.
Fetal gestaion age (days) at MRI scan
| Fetal # | Sex | GA at scan (days) | GA at scan (days) | GA at scan (days) |
|---|---|---|---|---|
| 1 | M | 83 | 111 | 133 |
| 2 | M | 85 | 111 | 133 |
| 3 | F | 85 | 111 | 135 |
| 4 | F | 87 | 110 | |
| 5 | F | 87 | 111 | |
| 6 | M | 84a | ||
| 7 | M | 87a | ||
| 8 | F | 83a | ||
| 9 | F | 110a | ||
| 10 | F | 110a | ||
| 11 | F | 110a | ||
| 12 | F | 110a | ||
| 13 | M | 110a | ||
| 14 | F | 135a | ||
| 15 | F | 134a | ||
| 16 | M | 135a | ||
| 17 | F | 137a | ||
| 18 | M | 133a | ||
| 19 | M | 84a | 134a | |
| 20 | F | 86a | 136a | |
| 21a | M | 83a | 137a | |
| 21b | M | 83a | 136a | |
| 22a | M | 133a | ||
| 22b | F | 85a | 134a | |
| 23a | F | 85a | ||
| 23b | F | 86a | 135 | |
| 24a | M | 85a | 135a | |
| 24b | M | 84a |
Fetuses in which DTI data were acquired.
2.2. Fetal MRI acquisition and brain image reconstruction
Previously described data acquisition and image reconstruction procedures were followed (Fogtmann et al., 2014; Wang et al., 2015, 2017). For all MRI procedures, sedation was induced with 10 mg/kg of ketamine and subjects were intubated for maintenance of anesthesia with 1% isoflurane. Images were acquired using a Siemens 3T Tim Trio equipped with a quadrature transmit, 15-channel receive human “extremity” radiofrequency coil (QED, Cleveland, OH). The fetal head position was determined using a series of half-Fourier acquisition single-shot turbo spin-echo (HASTE) images. Subsequently, T2-weighted images were acquired using HASTE with the settings TR = 1200 ms, TE = 102 ms, a generalized autocalibrating partially parallel acquisition (GRAPPA) factor = 2, an echo train length of 220, and effective echo spacing of 3.26 ms. Nine contiguous 2D image stacks with in-plane resolution of 0.5 × 0.5 mm2, and 1 mm slice thicknesses were acquired along the maternal axial, sagittal, and coronal axes in an interleaved manner. Each contiguous stack was acquired in 66 s, and so the total acquisition time for the T2-weighted images was 9 min and 54 s. This facilitates reconstruction of a 3D volume with isotropic resolution (Fogtmann et al., 2014). Subsequent to acquiring the T2-weighted images, diffusion-weighted data were acquired using a diffusion-weighted, 2D spin-echo-based echo planar imaging (EPI) sequence (b = 0, and 20 diffusion-weighted volumes with b = 500 s/mm2, TR = 5000 ms; TE = 93 ms; GRAPPA factor = 2; EPI factor = 78; echo spacing = 1.09 ms, in-plane resolution = 1.13 × 1.13 mm2; slice thickness = 3 mm along the maternal axial, sagittal, and coronal axes, separately). A total of 27 images images were acquired, separated into groups of 9 images oriented along three orthogonal planes, as with the T2-weighted images, to facilitate the reconstruction of 3D images with isotropic resolution. Each image required 2 min and 2 s to acquire, resulting in a total diffusion MRI acquisition time of 54 min and 54 s.
For the T2-weighted 2D image stacks, motion estimation and reconstruction (SLIMMER) procedures (Kim et al., 2011) were used for slice position and orientation registration. The 3D volume was reconstructed using an iterative deconvolution of the slice profiles in the orthogonal slice planes with 0.5 mm isotropic resolution (Fogtmann et al., 2014).
For fetal DTI calculations, the iterative reconstruction method used for the T2-weighted images was followed to generate a 3D “b0” image with isotropic resolution consisting of (0.75mm)3 voxels. Inter-slice motion estimation of all slices in the diffusion-weighted images was performed using an optimization model-based framework based on refinement of a rigid-body registration. Finally, an imaging model of the slice profile and in-plane point spread function in fixed anatomical coordinates was used to create 3D DTI diffusion tensor maps with a spatial resolution sequentially refined to (0.75 mm)3 (Fogtmann et al., 2014). Following high-resolution motion estimation and diffusion tensor estimation at the highest resolution, parameter maps, including fractional anisotropy (FA) and the apparent diffusion coefficient (ADC), were calculated.
2.3. Preprocessing of reconstructed images
Reconstructed 3D images of fetal heads generated following the above procedures underwent a series of subsequent image preprocessing operations to generate brain images to serve as input for the template construction procedure. Preprocessing includes (a) skull-stripping, (b) bias field correction and intensity normalization, and (c) co-registration to a common template coordinate space. These steps are illustrated in Fig. S1.
a). Skull-stripping
For each of the three gestational ages, the T2-weighted image of a randomly selected subject served as a reference, and manual brain segmentation was performed using ITK-SNAP software (Yushkevich et al., 2006). Next, all other T2-weighted images at the same gestational age were nonlinearly registered to the selected reference fetal image with symmetric diffeomorphic deformable transformation method (SyN (Klein et al., 2009),), as implemented in the “antsRegistrationSyN.sh” function of the Advanced Normalization Tools package (ANTS) (Avants et al., 2008). The SyN registration procedure is invertible, and therefore the reference brain mask can be mapped to each individual subject using the transformation parameters obtained from the registration, with nearest neighbor interpolation. The transformed masks were used to skull strip the T2-weighed images. In order to perform skull-stripping on the DTI images, the b0 image was nonlinearly registered to the T2-weighted image acquired from the same individual, and the mask of the T2-weighted image was mapped to the DTI reference frame. All brain masks were verified visually and modified manually if necessary.
b). Bias field correction and intensity normalization
Bias intensity correction was performed on skull-stripped brains using the “N4Bias-FieldCorrection” function in ANTS (Tustison et al., 2010). The intensity of each corrected brain image was then divided by the mean intensity of non-zero voxels in the brain-masked image to achieve the intensity normalization. This preprocessing step was only implemented for the T2-weighted images.
c). Registration to a common space
All normalized brain images were registered to a standardized AC-PC orientation through a rigid body registration. To do this, the images from an arbitrarily chosen fetus that was scanned at all thee time points (Fetus #1 in Table 1) were manually AC-PC aligned using standard functions in 3D Slicer (http://www.slicer.org ) (Fedorov et al., 2012). Subsequently, all other brain images from the same gestational age were rigid-body-transformed to the AC-PC aligned reference image.
2.4. T2-weighted image template construction
Template images were constructed following previously described procedures (Guimond et al., 2000) with minor modifications. Specifically, as detailed below, all subjects were transformed to a single subject’s reference frame, and the linear components of these transformations were averaged to generate template images that are average in size for each gestational age group. The specific steps of the procedure are outlined in Table 2. For a given gestational age, the group of N preprocessed images is denoted I0, I1 …, IN−1. One of these was randomly selected to serve as an initial reference, IR, and the following iterative procedure was performed.
Table 2.
Algorithm to generate template T2-weighted images.
| 1. For each Ii, solve Eq. (1) to calculate , Ai and Øi |
| 2. Calculate the average of all by ; |
| 3. Calculate the average of all by , and then ; |
| 4. Calculate the average of all by , and then ; |
| 5. Repeat 1 to 4 with updated reference by to achieve convergence. |
- Step 1. Determine the linear (affine) transform Ai and nonlinear warp field, Øi that registers preprocessed image Ii to IR by solving
(1)
This step was implemented by SyN registration in ANTS (Avants et al., 2014). Due to the invertible property of SyN registration, it is possible to obtain the inverse nonlinear warp field and affine matrix , which transforms the reference IR to image Ii.
Step 2. Average all transformed images to produce the mean intensity image . This image represents the average intensity from all images at each voxel location, but the brain has shape and size properties that are biased by the selection of IR.
Step 3. The mean inverse deformation field, , contains information related to the average shape of the brains in the preprocessed images Ii. The deformed mean image, , which is the result of applying the mean inverse deformation field to , is unbiased with regard to brain shape (Guimond et al., 2000).
Step 4. The mean inverse affine transformation, , contains linear information such as brain size within the preprocessed images. The transformed image , which is the result of applying the mean inverse affine transformation to , is unbiased with regard to brain size.
- Step 5. Replace IR in Step 1 with , obtained in Step 4, and repeat all Steps until converges to a stable result, at which point is deemed the template image. Convergence was assessed by evaluating the average displacement distance (ADD) of the mean deformation field
in which xj is the set of coordinates that overlap the brain mask, NV is the number of such coordinates, and is a 3D vector expressing the average displacement for voxel xj. We found that the difference of ADD from iteration 1 to iteration 2, for all three templates, was at least 10-fold larger than the difference in ADD from iteration 2 to iteration 3. The differences in ADD from iterations 2 and 3 was always less than 0.01 voxel (or 0.005 mm). Subsequent iterations did not result in further reduction in ADD. A total of 5 iterations was performed for the G85 template, and 3 iterations were performed for the G110 and G135 templates. Plots of ADD as a function of iteration number are shown in Fig. S2.(2)
2.5. DTI template construction
DTI data is available for a subset of the scans included in T2-weighted template (Table 1superscript a). In order to provide a representation of the FA and ADC images in the T2-weighted template image space, the skull-stripped “b0” images were nonlinearly registered to the T2-weighted template image of corresponding gestational age. The resulting registration parameters were then applied to transform the FA image to the T2-weighted template space. The voxel-based intensity average of all transformed FA images was used as the initial reference for FA template construction, which was refined through the iterative Steps 1–4, above. The number of iterations was chosen by visually inspecting the contrast between brain region boundaries in the FA map, yielding 5 iterations for the G85 images, and 3 iterations for G110 and G135 (plots of ADD vs. iteration number plots are shown in Fig. S2). The registration parameters in the final iteration for each gestational age were applied to transform the ADC images to the FA template space and then the voxel-based intensity average of all transformed ADC images was considered as the ADC template.
2.6. Data analysis and statistics
All statistical analyses were performed using the R software platform (http://www.R-project.org).
2.6.1. Analysis of rhesus brain regional volume changes with gestational age
Volumes within the cortical plate, the developing white matter, the striatum, the thalamus, the cerebellum, and the brainstem were analyzed as a function of gestational age. The measured sets of volumes were found to not be normally distributed, nor homoscedastic across gestational ages (Shapiro-Wilk test, p < 0.05 and Fligner-Killen test, p < 0.05, respectively). Therefore, the logarithm of brain region volumes were analyzed, as this transformation resulted in the data being normally distributed for all structures except the ventricles at G110 (p = 0.01) and homoscedastic across gestational age except for the CSF (p = 0.025). The log-transformed data were then analyzed in two stages by linear mixed effect modeling using the R function lmer in the R package “lmerTest” (Kuznetsova et al., 2017). The reported p-values were computed via Satterthwaite’s degrees of freedom method. The first stage considered 1 random effect (Fetus) and 4 fixed effects: the gestational age (Age), the brain structure (Structure), the experimental cohort in which the animal belongs (Cohort), and the fetal sex (Sex) with the formula
| (3) |
2.6.2. Analysis of rhesus macaque fetal brain growth
In addition to the above analysis of brain volume change, regional growth rates were explicitly analyzed. As elaborated in the Results and Discussion, this analysis was peformed to remove the effect of size in comparisons between brain regions: larger brain regions usually undergo larger changes in volume per unit time whereas the growth rate is normalized to the brain region size. The growth rate over a given interval is the rate of change (volume per day) divided by the average volume over the interval. Three fetuses (fetuses 1–3, Table 1) were scanned at all three gestational ages, which enables assessments of growth over the two intervals G85 to G110, and G110 to G135 for these animals. These data were analyzed with the formula
| (4) |
2.6.3. Analysis of human fetal brain growth
Brain growth was computed for the human fetal brain, for the same set of structures analyzed in the rhesus macaque, using data obtained from (Gholipour et al., 2017), in which segmented images are available for each week of gestation over an age range that spans 21–37 weeks. The human equivalent gestational ages for rhesus macaque G85, G110, and G135 are 21 weeks, 31 weeks, and 42 weeks (or typically, the second week of postnatal life), respectively (Workman et al., 2013). Growth rates for each structure in the human brain, over the period corresponding to the rhesus brain from G85 to G110, were obtained by fitting the volume of each structure to a linear function over the 21 weeks to 31 weeks age range, and dividing the slope (volume per unit time) by the estimated volume at 26 weeks of age (the average volume). It was not possible to estimate the growth rate for the period exactly corresponding to G110 to G135, because data were not available in (Gholipour et al., 2017) for ages older than 37 weeks, and the G135 human equivalent is 42 weeks. Therefore, brain region volumes from 31 weeks to 37 weeks were used to approximate growth over the 31–42 week age range by dividing the slope of a linear fit over the 31–37 weeks age range by the estimated volume at 36.5 weeks.
2.6.4. Analysis of diffusion tensor imaging parameters
As with the brain region volumes, the parameters FA and ADC are not normally distributed (Shapiro-Wilk test, p < 0.05). However, in contrast to the brain volume data, no DTI parameter was analyzed for a given fetus at more than 1 gestational age. Therefore, Wilcoxon rank sum tests were used to compare white matter FA and ADC values between gestational ages G110 and G135. To adjust for multiple comparisons among the 10 WM fiber tracts, a Bonferroni corrected p-value of α/10 = 0.005 was used as the threshold for statistical significance.
3. Results
Age-specific label maps were generated by manual segmentation of the T2-weighted template images, consulting histology-based adult rhesus macaque atlases (Saleem and Logothetis, 2012), fetal human brain atlases (Bayer and Altman, 2005) (Bayer and Altman, 2002), and MRI studies of human and non-human primate fetal brains (Kinoshita et al., 2001; Huang et al., 2006; Scott et al., 2011; Rohlfing et al., 2012; Calabrese et al., 2015; Wang et al., 2015; Andescavage et al., 2017; Gholipour et al., 2017). Manual delineation of anatomical structures was carried out by one author (X.W.), primarily in the coronal plane and modified on the axial and sagittal planes iteratively. The set of tissue categories delineated on the T2-weighted templates are summarized in Table 3 and Figs. 1–3. During the second half of gestation, the fetal brain undergoes rapid maturation as transient fetal zones disappear, new structures emerge, and many structures differentiate into resolvable sub-structures. As a result, the set of tissue categories differs for the three gestational ages (see Table 3).
Table 3.
Tissue categories of T2W templates.
| Tissue categories | G85 | G110 | G135 | ||
|---|---|---|---|---|---|
| Cerebrospinal fluid (CSF) | ✓ | ✓ | ✓ | ||
| Cortical plate (CP) | ✓ | ✓ | ✓ | ||
| Subplate (SP) | WM | ✓ | ✓ | ✓ | |
| Germinal and fibrous layers (GFL) | ✓ | ||||
| Ventricles (VENT) | ✓ | ✓ | ✓ | ||
| Striatum | Caudate (Ca) | ✓ | ✓ | ✓ | |
| Lentiform nucleus | Putamen (Pu) | ✓ | ✓ | ||
| Globus pallidus (GP) | ✓ | ||||
| Thalamus | ✓ | ✓ | ✓ | ||
| Hypothalamus | ✓ | ✓ | |||
| Brainstem | ✓ | ✓ | ✓ | ||
| Cerebellum | ✓ | ✓ | ✓ | ||
| Corpus Callosum (CC) | ✓ | ✓ | ✓ | ||
| Hippocampus | ✓ | ✓ | |||
| Amygdala | ✓ | ✓ | |||
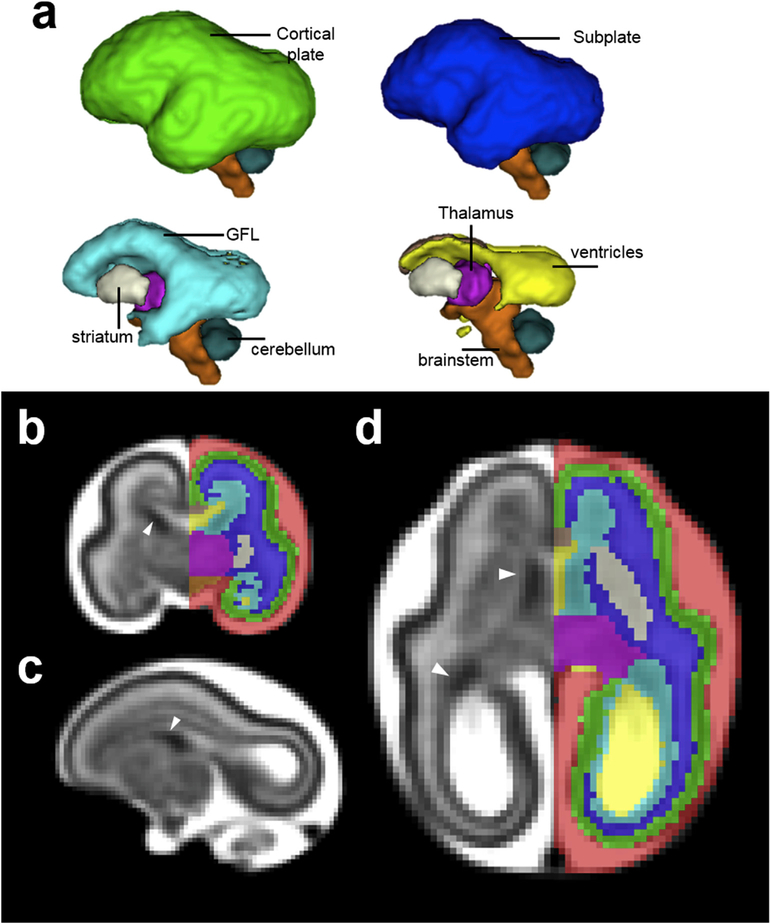
Fig. 1.
(a) 3D, (b) coronal, (c) sagittal, and (d) axial views of the G85 template and tissue segmentation. The T2-weighted template has a stratified pattern (c-d), cortical plate (CP) is the cell-dense outermost layer with the lowest T2-weighted intensity (green) and the subplate (SP) is the intermediate layer with relatively high T2-weighted intensity (blue). The germinal and fibrous layers (GFL), the hypointense layer (cyan) lining the ventricles, contains extends into ganglionic eminence rostrally (white arrow heads), and includes the ventricular zone, inner subventricular zone, inner fiber layer, outer subventricular zone and outer fiber layer (b-d). Adequate contrast also exists among deep gray nuclei, allowing us to further differentiate striatum (cream) from thalamus (magenta) (b and d). The brainstem (orange), which has slightly lower T2-weighted intensity than the adjacent thalamus, can also be delineated (b).
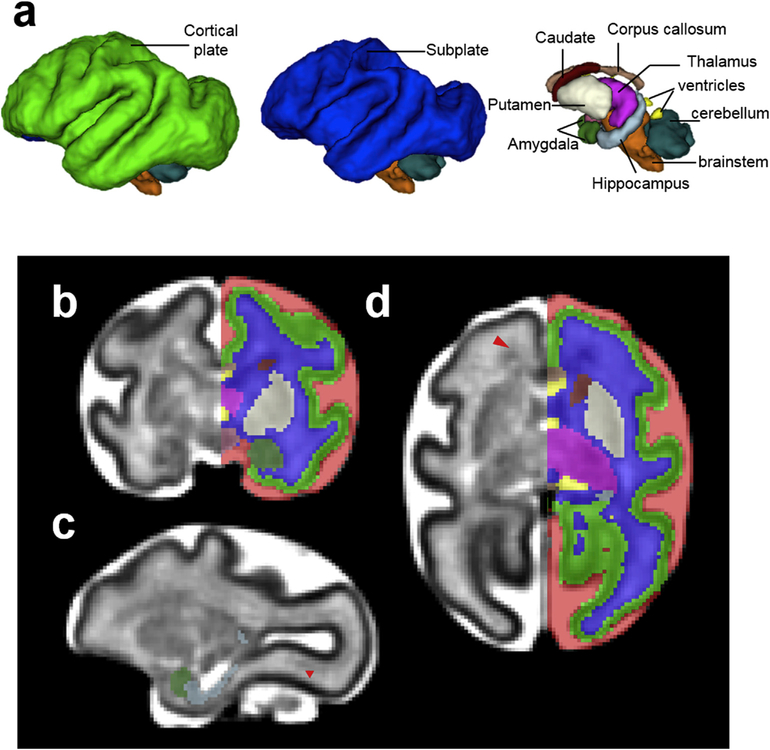
Fig. 3.
(a) 3D, (b) coronal, (c) sagittal, and (d) axial views of the G135 template and tissue segmentation. At G135, the lentiform nucleus can be further segmented into the putamen (cream) and the Globus pallidus (yellow) because of their increased contrast on T2-weighted images (b and d). At this developmental stage, the caudate nucleus appears hypointense compared to the putamen on the T2-weighted template (b and d). Although heterogeneity within thalamus can be observed (b-d), it is not further segmented into different nuclei. Likewise, brainstem is not further segmented into midbrain, pons, or medulla despite T2-weighted intensity differences (c).
3.1. Age-specific label maps at G85, G110, and G135
At G85, the T2-weighted template has a typical stratified pattern as seen in human (Scott et al., 2011) and non-human primates (Wang et al., 2015) of comparable gestational age. As shown in Fig. 1b, the cell-dense cortical plate (CP) is the outermost layer with the lowest T2-weighted intensity (green). The G85 cortical plate is lissencephalic, and gyri and sulci emerge over the G110 and G135 time points. A detailed description of cortical folding in the rhesus macaque over this age range is provided in (Wang et al., 2017). The subplate (SP) is subjacent to the CP, and has relatively high image intensity (blue). The ventricular zone, inner and outer subventricular zones, and inner and outer fibrous layers cannot be distinguished with in vivo MRI resolution (Smart et al., 2002), and all contribute to a hypointense germinal and fibrous layer (Wang et al., 2015) (GFL, cyan). In addition, the GFL contains the ganglionic eminences (white arrow heads, Fig. 1b–d). Note that tissue classification used here is slightly different from that adopted in a study of human fetal development of comparable age duration (Scott et al., 2011) where those authors combined subplate and IZ, reserving GMAT as a label only for the neural stem cell rich zone adjacent to lateral ventricles. Adequate contrast also exists among deep gray nuclei, allowing us to further segment them into striatum (cream) and thalamus (magenta) (Fig. 1b,c, and d). The rostral border of the brainstem (orange) was distinguished from the neighboring thalamus and WM (as well as hypothalamus at G110 and G135) by a subtle but distinct difference in T2-weighted intensity in which the brainstem was consistently hypointense (Fig. 1b).
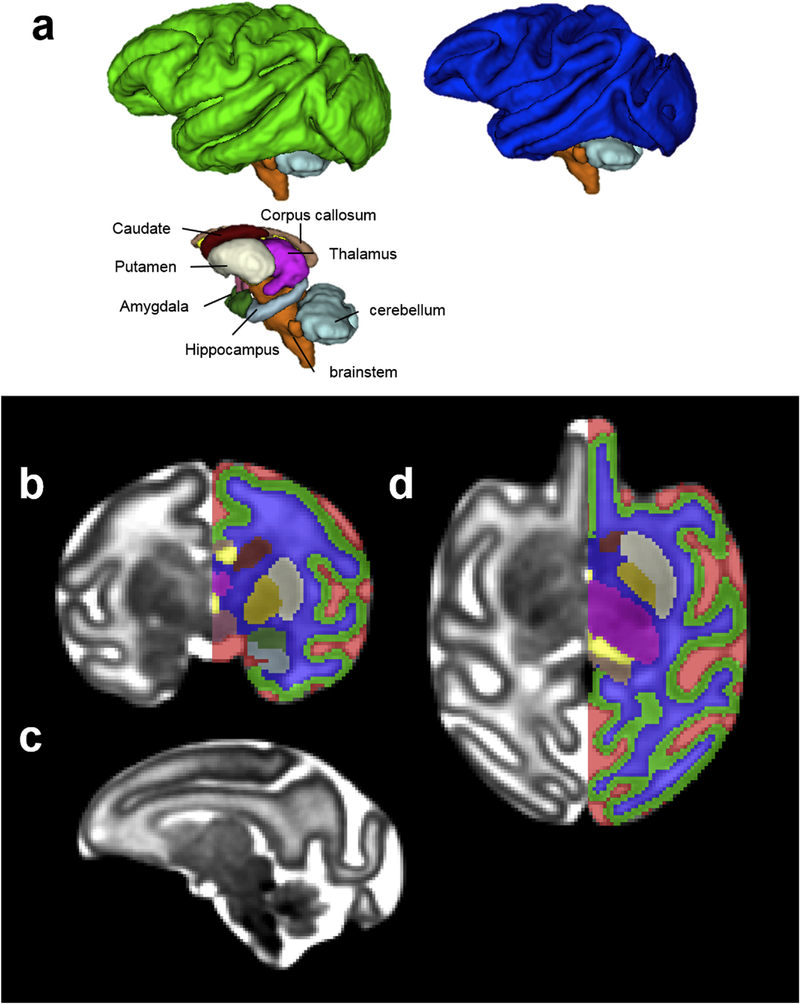
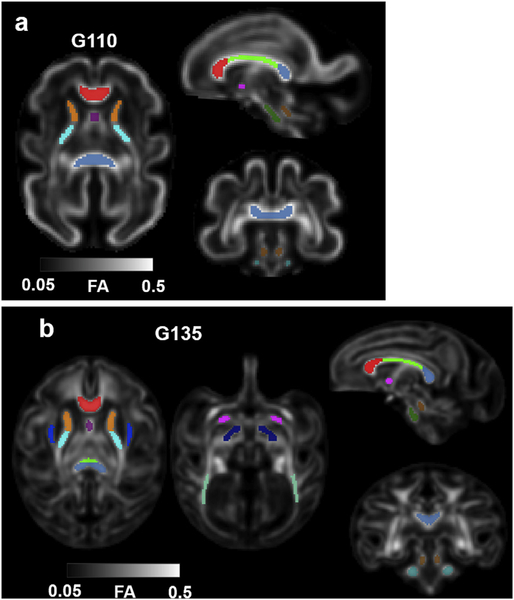
By G110, the GFL has decreased in its relative size compared to that at G85 and can only be identified at frontal and occipital levels (Fig. 2d, red arrowheads) on the T2-weighted template. Due to its similar image intensity to adjacent tissue zones, and diffuse boundary, GFL is not segmented at this gestation age (Fig. 2b and d). At G110, the striatum can be further segmented into caudate (maroon) and lentiform nucleus (cream) (Fig. 2b and d). Due to their similar T2-weighted image intensity at this developmental stage, the anterior limb of the internal capsule (ALIC), which is readily identifiable on the FA map, was used as the boundary between the two nuclei (Fig. 2b and d). In addition, hippocampus (light blue) and amygdala (dark green) can also be identified and delineated at this gestational age on the T2-weighted template (Fig. 2c). Hypothalamus (rose), located ventral to the thalamus and rostral to the brain stem (Fig. 2b and d), can also be segmented. By G135, the GFL has decreased dramatically in size and cannot be identified in our images. The lentiform nucleus, on the other hand, can be further segmented into the putamen (cream) and the Globus pallidus (yellow) because of the increased contrast on T2-weighted images at this gestational age (Fig. 3a and d). At this developmental stage, the caudate nucleus appears hypointense compared to the putamen on the T2-weighted template. Although heterogeneity within the thalamus can be observed (Fig. 3d), it is not further segmented into different nuclei. Likewise, brainstem is not further segmented into midbrain, pons, or medulla despite some T2-weighted intensity differences between these structures (Fig. 3c).
Fig. 2.
(a) 3D, (b) coronal, (c) sagittal, and (d) axial views of the G110 template and tissue segmentation. The GFL can only be identified at the frontal and occipital levels (d, red arrowhead). The striatum can be further segmented into caudate (maroon) and lentiform nucleus (cream) (b and d) at this age. Hippocampus (light blue) and amygdala (dark green) can also be identified and delineated at this gestational age on the T2-weighted template (c). The hypothalamus (rose), located ventral to the thalamus and rostral to the brain stem (b), can also be segmented.
3.2. Analysis of brain structure volume changes with gestation
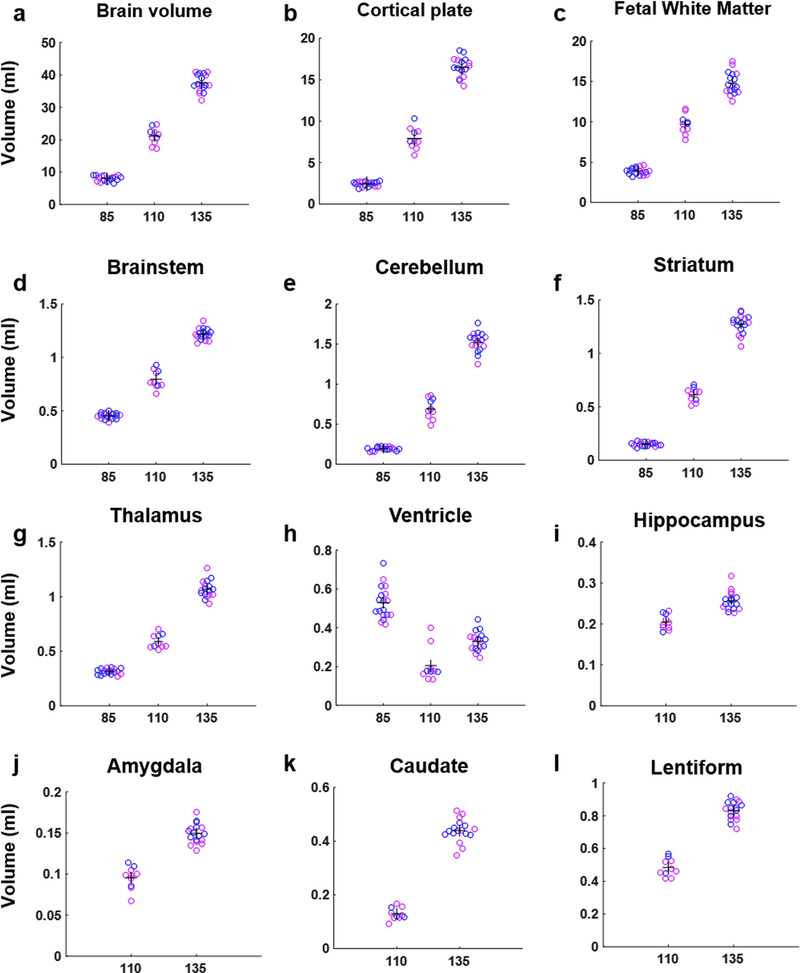
In order to quantify the sizes of each brain structure from G85 to G135, the tissue labels of the templates at each gestational age were transformed to each individual brain’s space using the inverse of warp field and affine tranform generated during registering the individual to the template. Visual inspection was carried out to evaluate the robustness of non-linear transformation processes. Manual adjustments were performed on only 2 of the 43 brains, both of which were at the G110 time point in which the automatic registration procedures resulted in the lateral ventricles being labeled as subplate/fetal white matter. Volumes of each anatomical structure were then extracted for each fetal brain, and are plotted in Fig. 4 (pink data points correspond to females and blue data points correspond to males). A statistical analysis was performed on the volumes of six brain structures that were measured at all three gestational ages. These were the CP, WM (the sum of SP and GFL for G85), brainstem, cerebellum, striatum, and thalamus. Log-transformed brain region volumes were analyzed using a linear mixed effect model, and this revealed significant effects of age, brain region, and an age-by-region interaction (for all three effects, p < 10−6). No statistically significant effects of sex (p = 0.27) or cohort (p = 0.93) were observed. In addition, exploratory analyses including interaction terms between sex and age did not reveal significant effects (data not shown). Expansion was also observed in the structures that could only be delineated at G110 and G135, which includes the hippocampus, amygdala, caudate and lentiform nuclei (Fig. 4i–l). The volumes (in mL) of each brain structure from each scan are listed in Table S1.
Fig. 4.
From G85 to G135, the volume of the whole brain, CP, WM (sum of SP and GFL for G85), brainstem, cerebellum, striatum, and thalamus all increase (a-g). Ventricular volume first decreases from G85 to G110 and then increases from G110 to G135 (h). Among the structures that can only be delineated at G110 and G135, expansion was observed in the hippocampus, amygdala, caudate and lentiform nuclei (i-l). Blue dots represent males and pink dots represent females.
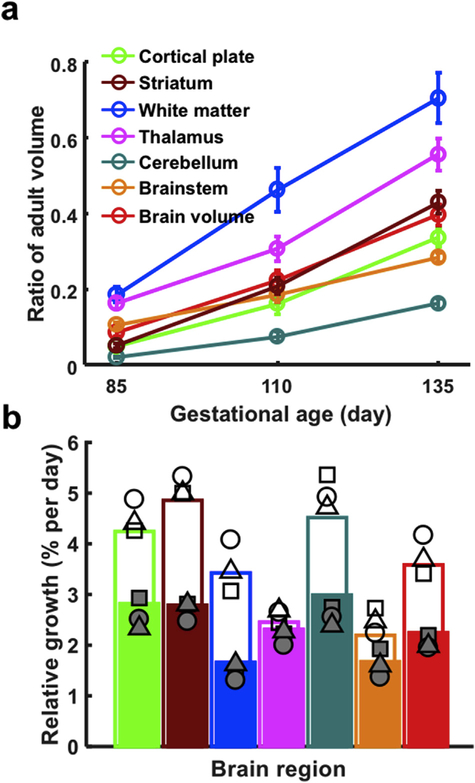
Fig. 5 compares growth of different brain structures over the two intervals examined (G85 to G110, and G110 to 135). In Fig. 5a, the brain region volumes are illustrated relative to their volumes at maturity. The INIA19 brain atlas (Rohlfing et al., 2012) provides a template image of the mature rhesus macaque brain. In a recent study that characterized brain size changes from late adolescence through middle-aged adulthood in 71 (13 female) rhesus macaques (Shnitko et al., 2019), the set of the 6 brain regions analyzed above were delineated on the INIA19 atlas. The volumes of these brain structures derived from the INIA19 template were found to be representative of the volume for each structure in the mature rhesus macaque. Ratios of volume at maturity, are plotted as a function of gestational age for each structure in Fig. 5a. Error bars represent the standard deviation in brain region volume at each age. At G85, all structures are less than 20% of their volumes at maturity, with the white matter being the highest percentage (18.6% of its adult volume) and cerebellum being the lowest (2.0% of its adult volume). The fetal white matter remained the highest ratio of its adult volume thoughout the study’s gestational age range (46.3% at G110 and 70.5% at G135), followed by thalamus (16.3 at G85, 30.7% at G110, and 55.6% at G135). The cerebellum remains the structure exhibiting the smallest ratio of its adult volume from G85 to G135 (7.4% at G110 and 16.2% at G135).
Fig. 5.
(a) Growth trajectories of different structures, and (b) relative growth rates over the G85-G110 (open bars) and G110-G135 intervals (closed bars). In (a), at G85, all structures are less than 20% of their volumes at maturity. By G135, a wide range of volumes are observed, the fetal white matter possessing highest percentage of its adult volume (70.5%) while the cerebellum exhibits the smallest percentage of its adult volume (16.2%). Error bars are standard deviations for each brain region at each age. In (b), relative brain growth (%/day) is universally faster between G85 and G110 than between G110 and G135. Growth is faster in telencephalic structures (cortical plate, WM, and the striatum between G85 and G110) than in other structures (thalamus, brainstem), with the notable exception of the cerebellum. Growth rates from individual fetuses 1–3 in Table 1 (fetus 1 is represented as a circle, fetus 2 is represented as a square, and fetus 3 is represented as a triangle) are overlaid on growth rates calculated from average brain volumes (colored bars) to illustrate the scale of variability in the growth measures.
Growth rates calculated from the average volumes measured for the six brain regions are shown in Fig. 5b for the G85 to G110 interval (open bars) and the G110 to G135 interval (closed bars). In contrast to the absolute change in volume with time (e.g. Figs. 4 and 5a), which in most cases is larger for larger structures, the brain growth rate reflects processes such as cell division, and increase in cell volume, independent of the size of the brain structure being analyzed. As shown in Fig. 5b, growth appears faster between G85 and G110 than between G110 and G135. Brain regional volumes were measured at all three gestational ages for three fetuses (fetuses 1–3, Table 1), and these are plotted as open symbols for the G85 to G110 interval, and shaded symbols for the G110 to G135 interval (fetus 1 is represented with circles, fetus 2 is represented with squares, and fetus 3 is represented with triangles). Note that the three fetuses with longitudinal measurements are only a subset of the animals used to compute average growth rates, and therefore the average of the three data points does not necessarily coincide exactly with the overall mean, represented by bars in Fig. 5b. Linear mixed effects modeling was performed on this subset of brains, and this analysis reveals main effects of interval, brain region, as well as in interval-by-brain region interaction (for all three effects, p < 10−6). As shown in Fig. 5b, growth appears to be more rapid in telencephalic structures (cortical plate and WM, and the striatum between G85 and G110) than in other structures (thalamus, brainstem), with the notable exception of the cerebellum, which continues rapid growth over a protracted period, extending into postnatal life (Tiemeier et al., 2010).
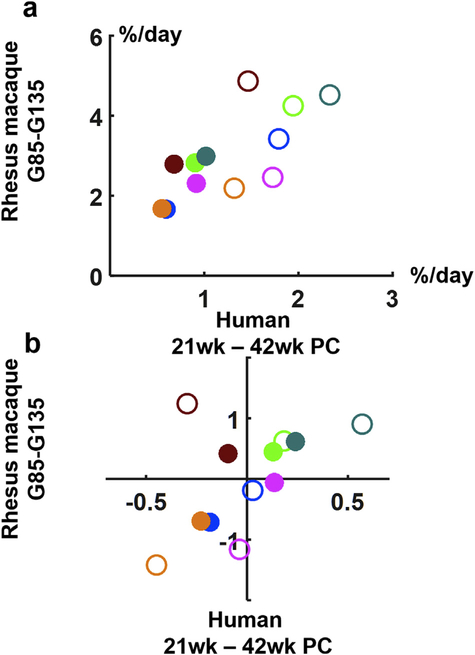
Given the observed dependence of growth rate on gestational age and brain region, it was of interest to determine whether human brain development exhibits similar dependencies. Growth rates for rhesus macaque (bars, Fig. 5b) are plotted as a function of growth rates for the same structures measured in humans (Gholipour et al., 2017), in Fig. 6a. This comparision yields three noteworthy findings. First, overall brain growth is approximately 2.4 times faster in rhesus monkeys than in humans over the two intervals evaluated (it is 2.1 faster over the interval from G85 to G110, and 2.6 faster from G110 to G135). Second, the growth rate is approximately 1.7 times faster (1.5 times faster in monkeys and 1.9 times faster in humans) in the first interval compared to the second growth interval. Last, the dependence of growth rate on brain region in humans is similar, but not perfectly coincident with the pattern observed in rhesus monkeys. In order to isolate the effects of brain region from the effects of developmental stage, Fig. 6b shows the residual difference in growth rates from the average brain growth rate for each of the two intervals. The correlation falls short of statistically significant (r = 0.54, p = 0.07), largely due to the striatum undergoing more rapid growth relative to average brain growth in monkeys, compared to humans, at both developmental intervals. For example, the residual growth rates for all structures except for the striatum are highly correlated (r = 0.9, p = 0.0005).
Fig. 6.
Relative growth rates for rhesus macaque fetal brain structures for G85 to G110 and human 21–31 weeks (open symbols) and rhesus G110 to G135 and human 31–37 weeks (closed symbols). In (a) rhesus brain region growth is plotted as a function of relative growth rates for corresponding human fetal brain structures obtained from (Gholipour et al., 2017). Each brain structure is color-coded as in Fig. 5. In (b), the residuals of brain growth rates are plotted following subtraction of the average brain growth rate for rhesus and human over the first interval (open symbols) and subtraction of the average brain growth rate over the second interval (closed symbols).
3.3. White matter tracts at G110 and G135
To study the regional trends in white matter maturation from G110 to G135, white matter tracts were identified on the G110 and G135 FA templates and the ROIs listed in Table 4 were manually defined within those tracts (Fig. 7). The specific set of tracts was selected based on two criterea: 1) the tract possesses sufficiently high FA (>0.2) in the macaque fetal template, and 2) the tract was included among those characterized in a human fetal FA atlas that spans a comparable developmental period (21–39 weeks (Khan et al., 2019),). In addition, the anterior commissure (AC) and the superior cerebellar peduncle (SCP), which were not included in the above-mentioned human fetal template, were also included in our atlas. The former is a interhemispheric fiber bundle, and the latter is a pair of white matter structure that connects the cerebellum to the pons. Both of these tracts can be identified as early as G110, with FA > 0.2. To guide the delineation of these ROIs, we used the adult rhesus macaque white matter atlas UW_DTIRMAC271 (Zakszewski et al., 2014) as a reference, in which white mater labels are overlaid on a FA template.
Table 4.
White Matter ROIs on DTI templates.
| White Matter categories | G110 | G135 |
|---|---|---|
| Genu of the Corpus Callosum (GCC) | ✓ | ✓ |
| Body of the Corpus Callosum (BCC) | ✓ | ✓ |
| Splenium of the Corpus Callosum (SCC) | ✓ | ✓ |
| Anterior commissure (AC) | ✓ | ✓ |
| Anterior limb of the internal capsule (ALIC) | ✓ | ✓ |
| Posterior limb of the internal capsule (PLIC) | ✓ | ✓ |
| External capsule (EC) | ✓ | |
| Fornix | ✓ | ✓ |
| Cerebrospinal tract (CST) | ✓ | ✓ |
| Post thalamic radiation (PTR) | ✓ | |
| Cerebral peduncle (CP) | ✓ | |
| Superior cerebellar peduncle (SCP) | ✓ | ✓ |
| Middle cerebellar peduncle (MCP) | ✓ | ✓ |
Fig. 7.
White matter ROIs on (a) G110 and (b) G135 FA templates. As early as G110, major white matter tracts such as corpus callosum (CC) (further divided into genu (GCC), body (BCC), and splenium (SCC)) (red, green, and blue, respectively), fornix (purple), anterior (orange) and posterior (cyan) limbs of the internal capsule (ALIC and PLIC, respectively), anterior commissure (AC) (magenta), cerebral spinal tract at the midbrain (CST) (olive green), superior cerebral peduncle (SCP), and medial cerebral peduncle (MCP) can be identified and segmented on the FA template (a). At G135, the external capsule (EC), post thalamic radiation (PTR), and cerebral peduncle (CP), can be identified and delineated on the FA template (b).
3.4. White matter maturation
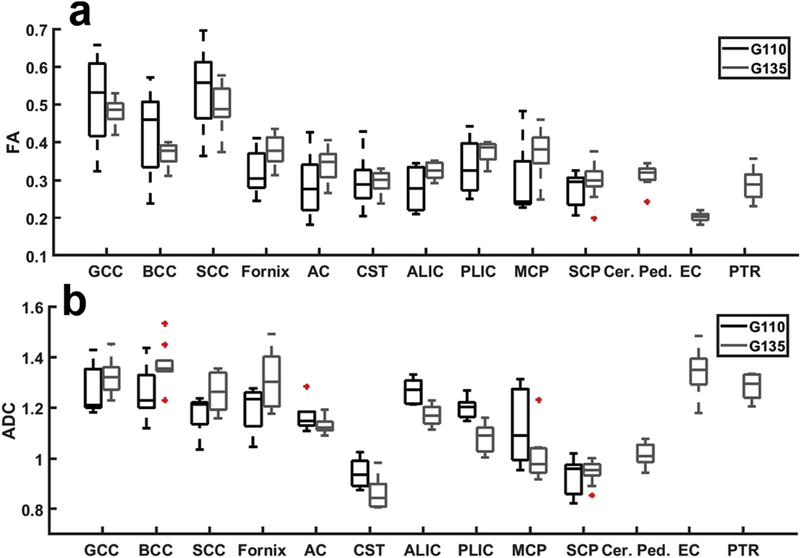
To assess white matter maturation, white matter ROIs identified on the G110 and G135 FA templates were projected to individual FA spaces. Careful visual inspection was carried out and no manual editing was needed. ROI-averaged FA values were extracted from each subject (Fig. 8). A Wilcoxon rank sum test was used to compare white matter FA between G110 and G135 and no significant differences were detected in any of the featured tracts (Fig. 8a). The same test was also performed to compare the water ADC within the same set of white matter ROIs. Significant reductions in ADC were detected in the ALIC (p = 0.00023) and PLIC (p = 0.00023). The FA and ADC values of each white matter ROI from each scan listed in Tables S2 and S3, respectively.
Fig. 8.
Boxplots show the average (a) FA and (b) ADC values for each white matter ROI across subjects. For each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. Outlier points (red + symbols) are defined as those differing by more than three standard deviations from the mean. Whiskers reflect the range of all non-outlier data values.
4. Discussion
4.1. Rhesus macaque fetal brain development
The fetal brain templates and label maps described here provide normative data for rhesus brain development at gestational ages G85, G110, and G135. Over this developmental period, the fetal brain is a highly dynamic structure. The volumes and shapes of each brain structure change substantially, and the sets of resolvable brain regions differ between the three time points. At G85, the dominant structural feature is the stratified cerebrum. At 0.5 mm3 isotropic resolution, three resloved lamina, herein termed the GFL, SP, and CP, span the ventricular zone to the pial surface. By G110, the appearance of gyral and sulcal folds is accompanied by the transition of the GFL to differentiating tracts. In addition, the hippocampus and amygdala can be delineated, and the lentiform nucleus and caudate can be discerned within the striatum. The mature folding pattern of the cerebral cortex is apparent in the G135 brain (Wang et al., 2017), and at this gestational age, additional WM fiber systems are apparent, and the lentiform nucleus can be further subdivided into the globus pallidus and putamen. As a consequence of the dramatic anatomical changes that occur over this interval, studies using rhesus macaques as animal models of neurodevelopmental disorders must control for gestational age, to distinguish treatment effects from age effects on brain structure.
A recent advance in the construction of human fetal brain templates and labelmaps is the development of methods to create continuous 4D images that represent the brain over a range of gestational ages (Gholipour et al., 2017). This is accomplished by generating a general procedure for constructing a template at any given gestational age using a combination of images of similar stages of maturation. In order to construct such a template, it is necessary to have a relatively uniform sampling of fetal brains across the gestational age range of interest. Due to this requirement, as a limitation of this study, it was not feasible to construct a 4D template because the set of available images were all obtained from fetuses very close to specified ages (within two gestational days). However, one strength of this study is the accuracy with which gestational age is known. As detailed previously (Jimenez et al., 2019), daily determinations of plasma estradiol concentration enabled the day of conception to be estimated with precision of one day. This is considerably more precise than estimating post-conceptional age based on the day of last menses, as is common in human studies. Therefore, the variance observed in the measures quantified herein reflect variance in brain anatomy, rather than uncertainty in gestatinal age.
A recent longitudinal neuroimaging study of rhesus macaque brain development from 1 week to 5 years postnatal age demonstrated that the female rhesus brain is smaller than males (Scott et al., 2011). For example, the one-week-old female brain is 59.9 mL compared to 62.4 mL for males. In spite of this, statistically significant sex differences in brain size or rate of maturation were not observed in the current study of fetal brain development. At G135, the average female and male brain sizes were 37.2 ± 3.3 mL and 38.0 ± 2.2 mL, respectively. Although it is possible that studies with more animals would statistically establish sex differences in fetal brain volumes, MRI-based brain biometry studies of large samples sizes in humans (N = 128) report only limited sex differences, such as in ventricular volume (Kyriakopoulou et al., 2017). An additional study involving human 166 fetuses did not find differences in brain volume between male and females (Andescavage et al., 2017). Therefore it is likely that sex differences in brain size are more subtle during the phase of fetal development characterized here than they are postnatally.
The animals studied here consist of the control groups from three separate experiments. As a result, some cohort differences exist between animals in environmental factors, such as housing conditions, and differences in experimental design, which influenced the number of scans performed per animal. Fetuses 1–18 (Table 1) were derived from singly-housed dams, whereas the dams of the other fetuses were group-housed. In addition, some systematic differences between animal care staff, and conventions adopted for the control diets exist. Another potential confound was the need to sedate pregnant dams during MRI procedures. Fetal exposure to anesthesia is associated with increased programmed cell death in neurons and glial cells (Brambrink et al., 2012; Creeley et al., 2014). Therefore, animals that underwent multiple imaging procedures experienced higher levels of exposure to isoflurane. In order to assess whether such differences could have influenced the fetal brain growth parameters reported here, analyses were performed to determine whether the different cohorts exhibited differences in fetal brain structure. No such differences were observed. The lack of statistically significant effects of cohort indicates that differences in maternal rearing did not influence fetal brain development in a manner that was detectable in this study.
4.2. Relationship to human development
Interconversion between rhesus and human gestational ages requires consideration of the differences in gestational term length (168 days for rhesus compared to 270 days for human) and the fact that rhesus macaques are born at a later stage of central nervous system development than humans. Systematic comparative analysis of CNS development across many mammals have lead to the development of a model (Workman et al., 2013) that associates G85, G110, and G135 in rhesus macaques with human ages of 21, 31, and 42 weeks post conception, respectively. The work of Gholipour et al. (2017) provides volume measurements of the same six structures quantified in rhesus macaques herein, over the gestational age range from 21 to 37 weeks, with a temporal resolution of 1 measurement per week. This data set therefore spans the interval that corresponds to G85 to G110 in rhesus macaques. The G110 to G135 interval is only partially sampled with the 31 through 37 week time points in humans. It would be difficult to span the entire 31–42 week period because human term is 40 weeks, and to our knowledge there are no studies that implement consistent data acquisition and analysis procedures for quantifying bain development during the relevant fetal and postnatal periods in normal term pregnancies. Therefore development from G110 to G135 is compared to the 31–37 week period in humans, with the acknowledged caveats that, in addition to the relatively few numbers of fetal resus brains in which data are available, neurodevelopmental events occuring between 37 and 42 weeks do not contribute to the analysis of human data peformed here.
Rhesus brain growth, quantified in percent change in volume per day, is approximately 2.4 times faster in rhesus macaques than it is in humans. This difference relates to the underlying cellular biological processes that give rise to macroscopic growth, which are not well-characterized. For example, comparisons between rhesus macaque and human neural progenitor cell cycle kinetics have not revealed large differences between these two species (Breunig et al., 2011). Further study of growth rates, incorporating information from additional species, could be of value for linking developmental processes on the cellular scale to brain growth. In turn, such knowledge would serve to facilitate mechanistic interpretations of abnormal growth trajectories observed in individuals affected by neurodevelopmental disorders in the future.
In spite of the difference in brain growth rates, monkeys and humans exhibit similarities in changes in growth rate with development and regional patterns of growth. In both species growth decreases by a factor of 1.7 between the first and second interval studied here. As shown in Fig. 5b, larger relative growth rates were observed in the telencephalic gray matter and cerebellum, with lower rates observed in WM, thalamus and brainstem, for both species. One possible exception to the overall similar regional pattern of growth is the striatum, which deviates from the monkey to human growth rate difference of 2.4, particularly over the G85 to G110 interval (Fig. 6b). Further study will be necessary to establish whether the striatal growth rate difference observed here results from species differences in development, or from inconsistencies in delineating striatal area boundaries between time points and/or between species.
Several similarities also exist between the patterns of WM maturation observed in the rhesus macaque and those reported in human studies. In humans, WM volume expansion with age is unique compared to other structures quantified, in that its rate of volume expansion decreases over the second half of gestation (Andescavage et al., 2017). This is similar in rhesus macaques (Fig. 5a), where the volume change in WM is slightly lower between G110 and G135 than it was between G85 and G110. Regional differences in FA maturation are also similar with humans. Changes in FA are minor over the second half of gestation in both the human (Khan et al., 2019) and macaque (Fig. 8a) corpus callosum. In contrast, although differences in FA are not statistically significant, the fornix, internal capsules, and MCP show the largest differences in the mean values (Fig. 8), and these tracts undergo the greatest developmental changes in humans (Khan et al., 2019). We note that larger sample sizes (N ≈ 15 per gestational age) would be necessary to be powered at a level of 0.8 to detect differences in FA on the order observed in these structures (about 0.05), assuming a sample standard deviation of 0.05.
5. Conclusion
Over the period from G85 to G135, the rhesus macaque fetal brain transforms from a lissencephalic, radially-organized structure to a folded shape resembling the mature brain, with several WM structures and deep gray matter nuclei apparent in magnetic resonance images with (0.5 mm)3 isotropic resolution. Gestation-age-specific labelmaps have been delineated on template T2-weighted images and DTI parameter maps. Volume and diffusion parameter measurements reflecting normal maturation over this developmental period are presented. The template images, labelmaps, and data reflecting each of the identified structures have been made publicly available to the research community (www.nitrc.org). Due to the cost of generating rhesus macaques at specific stages of gestation, it is hoped that the data described here will be a valuable resource to supplement the power of rhesus fetal brain measurements conducted by others.
Supplementary Material
Acknowledgements
This work was supported by NIH grants R01AA021981 (C.D.K.), OD011092 (C.D.K. and K.A.G) R01EB017133 (C.S.), R01NS055064 (C.S.), and Bill and Melinda Gates Foundation grant OPP1110865 (C.D.K.).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuroimage.2019.116310.
References
- Andescavage NN, du Plessis A, McCarter R, Serag A, Evangelou I, Vezina G, Robertson R, Limperopoulos C, 2017. Complex trajectories of brain development in the healthy human fetus. Cerebr. Cortex 27, 5274–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC, 2014. The Insight ToolKit image registration framework. Front. Neuroinf 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, Altman J, 2002. The Human Brain during the Third Trimester. CRC Press, Boca Raton. [Google Scholar]
- Bayer SA, Altman J, 2005. The Human Brain during the Second Trimester. CRC Press, Boca Raton. [Google Scholar]
- Brambrink AM, Evers AS, Avidan MS, Farber NB, Smith DJ, Martin LD, Dissen GA, Creeley CE, Olney JW, 2012. Ketamine-induced neuroapoptosis in the fetal and neonatal rhesus macaque brain. Anesthesiology 116, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, Haydar TF, Rakic P, 2011. Neural stem cells: historical perspective and future prospects. Neuron 70, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Shi Y, Styner MA, Johnson GA, 2015. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage 117, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creeley CE, Dikranian KT, Dissen GA, Back SA, Olney JW, Brambrink AM, 2014. Isoflurane-induced apoptosis of neurons and oligodendrocytes in the fetal rhesus macaque brain. Anesthesiology 120, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich E, Riklin Raviv T, Kasprian G, Donner R, Brugger PC, Prayer D, Langs G, 2014. A spatio-temporal latent atlas for semi-supervised learning of fetal brain segmentations and morphological age estimation. Med. Image Anal 18, 9–21. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R, 2012. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogtmann M, Seshamani S, Kroenke C, Xi C, Chapman T, Wilm J, Rousseau F, Studholme C, 2014. A unified approach to diffusion direction sensitive slice registration and 3-D DTI reconstruction from moving fetal brain anatomy. IEEE Trans. Med. Imaging 33, 272–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Estroff JA, Warfield SK, 2010. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. IEEE Trans. Med. Imaging 29, 1739–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholipour A, Rollins CK, Velasco-Annis C, Ouaalam A, Akhondi-Asl A, Afacan O, Ortinau CM, Clancy S, Limperopoulos C, Yang E, Estroff JA, Warfield SK, 2017. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Sci. Rep 7, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond A, Meunier J, Thirion J-P, 2000. Average brain models: a convergence study. Comp. Vis. Image Underst. 77, 192–210. [Google Scholar]
- Habas PA, Kim K, Corbett-Detig JM, Rousseau F, Glenn OA, Barkovich AJ, Studholme C, 2010. A spatiotemporal atlas of MR intensity, tissue probability and shape of the fetal brain with application to segmentation. Neuroimage 53, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Xue R, Zhang J, Ren T, Richards LJ, Yarowsky P, Miller MI, Mori S, 2009. Anatomical characterization of human fetal brain development with diffusion tensor magnetic resonance imaging. J. Neurosci 29, 4263–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Wakana S, Zhang W, Ren T, Richards LJ, Yarowsky P, Donohue P, Graham E, van Zijl PC, Mori S, 2006. White and gray matter development in human fetal, newborn and pediatric brains. Neuroimage 33, 27–38. [DOI] [PubMed] [Google Scholar]
- Jiang S, Xue H, Counsell S, Anjari M, Allsop J, Rutherford M, Rueckert D, Hajnal JV, 2009. Diffusion tensor imaging (DTI) of the brain in moving subjects: application to in-utero fetal and ex-utero studies. Magn. Reson. Med 62, 645–655. [DOI] [PubMed] [Google Scholar]
- Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV, 2007. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Trans. Med. Imaging 26, 967–980. [DOI] [PubMed] [Google Scholar]
- Jimenez VA, Wang X, Newman N, Walter NAR, Gonzales S, Lo JO, Ford MM, Cuzon Carlson VC, Grant KA, Kroenke CD, 2019. Detecting neurodevelopmental effects of early-gestation ethanol exposure: a nonhuman primate model of ethanol drinking during pregnancy. Alcohol Clin. Exp. Res 43, 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainz B, Steinberger M, Wein W, Kuklisova-Murgasova M, Malamateniou C, Keraudren K, Torsney-Weir T, Rutherford M, Aljabar P, Hajnal JV, Rueckert D, 2015. Fast volume reconstruction from motion corrupted stacks of 2D slices. IEEE Trans. Med. Imaging 34, 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Habas P, Rajagopalan V, Scott J, Corbett-Detig J, Rousseau F, Glenn O, Barkovich J, Studholme C, 2010a. Non-iterative relative bias correction for 3D reconstruction of in utero fetal brain MR imaging. In: Conf Proc IEEE Eng Med Biol Soc 2010, pp. 879–882. [DOI] [PubMed] [Google Scholar]
- Khan S, Vasung L, Marami B, Rollins CK, Afacan O, Ortinau CM, Yang E, Warfield SK, Gholipour A, 2019. Fetal brain growth portrayed by a spatiotemporal diffusion tensor MRI atlas computed from in utero images. Neuroimage 185, 593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Habas PA, Rajagopalan V, Scott JA, Corbett-Detig JM, Rousseau F, Barkovich AJ, Glenn OA, Studholme C, 2011. Bias field inconsistency correction of motion-scattered multislice MRI for improved 3D image reconstruction. IEEE Trans. Med. Imaging 30, 1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C, 2010b. Intersection based motion correction of multislice MRI for 3-D in utero fetal brain image formation. IEEE Trans. Med. Imaging 29, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita Y, Okudera T, Tsuru E, Yokota A, 2001. Volumetric analysis of the germinal matrix and lateral ventricles performed using MR images of postmortem fetuses. AJNR (Am. J. Neuroradiol.) 22, 382–388. [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV, 2009. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46, 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklisova-Murgasova M, Aljabar P, Srinivasan L, Counsell SJ, Doria V, Serag A, Gousias IS, Boardman JP, Rutherford MA, Edwards AD, Hajnal JV, Rueckert D, 2011. A dynamic 4D probabilistic atlas of the developing brain. Neuroimage 54, 2750–2763. [DOI] [PubMed] [Google Scholar]
- Kuklisova-Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA, 2012. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Med. Image Anal 16, 1550–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB, 2017. lmerTest package: tests in linear mixed effects models. J. Stat. Softw 82, 1–26. [Google Scholar]
- Kyriakopoulou V, Vatansever D, Davidson A, Patkee P, Elkommos S, Chew A, Martinez-Biarge M, Hagberg B, Damodaram M, Allsop J, Fox M, Hajnal JV, Rutherford MA, 2017. Normative biometry of the fetal brain using magnetic resonance imaging. Brain Struct. Funct 222, 2295–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts VHJ, Lo JO, Lewandowski KS, Blundell P, Grove KL, Kroenke CD, Sullivan EL, Roberts CT Jr., Frias AE, 2018. Adverse placental perfusion and pregnancy outcomes in a new nonhuman primate model of gestational protein restriction. Reprod. Sci 25, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing T, Kroenke CD, Sullivan EV, Dubach MF, Bowden DM, Grant KA, Pfefferbaum A, 2012. The INIA19 template and NeuroMaps atlas for primate brain image parcellation and spatial normalization. Front. Neuroinf 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich JA, Studholme C, 2006. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Acad. Radiol 13, 1072–1081. [DOI] [PubMed] [Google Scholar]
- Saleem KS, Logothetis NK, 2012. A Combined MRI and Histology Atlas of the Rhesus Monkey Brain in Stereotaxic Coordinates. Academic Press, London. [Google Scholar]
- Scola E, Sirgiovanni I, Avignone S, Cinnante CM, Biffi R, Fumagalli M, Triulzi F, 2016. Fetal development of the corpus callosum: insights from a 3T DTI and tractography study in a patient with segmental callosal agenesis. Neuroradiol. J 29, 323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Habas PA, Kim K, Rajagopalan V, Hamzelou KS, Corbett-Detig JM, Barkovich AJ, Glenn OA, Studholme C, 2011. Growth trajectories of the human fetal brain tissues estimated from 3D reconstructed in utero MRI. Int. J. Dev. Neurosci 29, 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serag A, Aljabar P, Ball G, Counsell SJ, Boardman JP, Rutherford MA, Edwards AD, Hajnal JV, Rueckert D, 2012. Construction of a consistent highdefinition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage 59, 2255–2265. [DOI] [PubMed] [Google Scholar]
- Shnitko TA, Liu Z, Wang X, Grant KA, Kroenke CD, 2019. Chronic alcohol drinking slows brain development in adolescent and young adult nonhuman primates. eNeuro 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H, 2002. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebr. Cortex 12, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN, 2010. Cerebellum development during childhood and adolescence: a longitudinal morphometric MRI study. Neuroimage 49, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourbier S, Bresson X, Hagmann P, Thiran JP, Meuli R, Cuadra MB, 2015. An efficient total variation algorithm for super-resolution in fetal brain MRI with adaptive regularization. Neuroimage 118, 584–597. [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pettersson DR, Studholme C, Kroenke CD, 2015. Characterization of laminar zones in the mid-gestation primate brain with magnetic resonance imaging and histological methods. Front. Neuroanat 9, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Studholme C, Grigsby PL, Frias AE, Cuzon Carlson VC, Kroenke CD, 2017. Folding, but not surface area expansion, is associated with cellular morphological maturation in the fetal cerebral cortex. J. Neurosci 37, 1971–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL, 2013. Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci 33, 7368–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G, 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zakszewski E, Adluru N, Tromp do PM, Kalin N, Alexander AL, 2014. A diffusiontensor-based white matter atlas for rhesus macaques. PLoS One 9, e107398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Dinov ID, Li J, Zhang Z, Hobel S, Shi Y, Lin X, Zamanyan A, Feng L, Teng G, Fang F, Tang Y, Zang F, Toga AW, Liu S, 2013. Spatial-temporal atlas of human fetal brain development during the early second trimester. Neuroimage 82, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.