Abstract
Tensin is a focal adhesion molecule that is known to regulate cell adhesion, migration, and proliferation. Although there are four tensin homologs (TNS1, TNS2, TNS3, and CTEN/TNS4) in mammals, only one tensin gene is found in Caenorhabditis elegans. Sequence analysis suggests that Caenorhabditis elegans tensin is slightly closer aligned with human TNS1 than with other human tensins. To establish the role of TNS1 in Caenorhabditis elegans, we have generated TNS1 knockout (KO) worms by CRISPR-Cas9 and homologous recombination directed repair approaches. Lack of TNS1 does not appear to affect the development or gross morphology of the worms. Nonetheless, defecation cycles are significantly longer in TNS1 KO worms. In addition, their pharyngeal pumping rate is markedly faster, which is likely due to a shorter pump duration in the KO worms. These findings indicate that TNS1 is not required for the development and survival of Caenorhabditis elegans but point to a critical role in modulating defecation and pharyngeal pumping rates.
Keywords: tensin, focal adhesion, defecation, pharyngeal pumping, Caenorhabditis elegans
1. Introduction
Tensin is a focal adhesion molecule that interacts with actin filaments and β integrins through its actin-binding domains and PTB (phosphotyrosine binding) domain, respectively [1] [2]. These binding activities allow tensin to anchor the actin cytoskeleton to focal adhesion sites [3]. Besides its structural role, tensin also actively engages in a variety of signaling cascades, such as those mediated through protein tyrosine kinases. With its SH2 (Src Homology 2) domain, tensin is able to recruit tyrosine-phosphorylated proteins forming a signaling complex [4]. In addition, tensin regulates small GTPase signaling pathways by interacting with DLC1 (a GTPase-activating protein) or Dock5 (a guanine nucleotide exchange factor) [5]. These interactions contribute to a group of cellular events including cell adhesion, migration, proliferation, apoptosis and differentiation [6].
The in vivo functions of tensin have been evaluated using animal models. There are four tensin homologs in mammals: TNS1, TNS2, TNS3, and Cten/TNS4 [6]. Mice lacking TNS1 develop cystic kidney diseases [7] and TNS2 null mice suffer from nephrotic syndromes in a strain-dependent manner [8]. Both mutant mice eventually die from renal failure. Loss of TNS3 in mice causes significantly stunted growth as well as developmental defects in the gut and bones [9]. Tensin genes are also found in invertebrates. Drosophila only contains one tensin gene (blistery). Tensin mutant flies lay fewer eggs which are significantly rounded [10] and the adult flies develop a wing blister phenotype [11, 12].
The nematode Caenorhabditis elegans is a bacteria-eating roundworm and contains a single tensin gene (M01E11.7). C elegans is a powerful model organism for investigating genetics, developmental biology, and animal behavior due to its short life spin, easy growth conditions, transparent body, and predetermined cell fates. In this report, we have generated TNS1 knockout worms to understand the in vivo role of TNS1 in C. elegans.
2. Materials and Methods
2.1. Sequence Analysis
Sequences were retrieved from NCBI for analysis. The sequences for CeTNS1 (NP_001305199.1), HsTNS1 (NP_072174.3), HsTNS2 (NP_736610.2), HsTNS3 (XP_011513781.1), and HsTNS4 (NP_116254.4) were then aligned using the Multiple Sequence Comparison by Log Expectation tool provided by the European Bioinformatics Institute (EBI, https://www.ebi.ac.uk/Tools/msa/muscle/). Percent identities were determined using the percent identity matrix tool included in this software. The 4 major domains of tensin (phosphatase, C2, SH2, and phosphotyrosine binding domain) were identified in each protein using the InterProScan tool, also from EBI (https://www.ebi.ac.uk/interpro/beta/). These individual domains were aligned and analyzed as described for the full-length proteins. [13]
2.2. Generation of TNS1 Knockout Worms
CRISPR-Cas9 and homologous recombination directed repair methods were employed. sgRNAs were designed to target the TNS1 gene. An oligonucleotide fragment was used to guide homologous recombination, deleting the region of TNS1 and replacing it with a 5’-TAAATAAATAAACTCGAG-3’ 3-frame stop sequence. The deletion was confirmed by PCR analysis: Primers 5’-GTTTGCACGATCGTTTAGTCCG-3’ and 5’-TCCCTTCAGTTTGACACCCTTTG-3’ were expected to amplify the modified sequence yielding a 755 bp product while giving a >15kb product from the WT genomic DNA. A second set of primers 5’-GTGTGTTGGTGTGCAGAATGAAG-3’ and 5’-TCCCTTCAGTTTGACACCCTTTG-3’ were used as a reciprocal set, giving an expected product of 587 bp from the WT sequence and no expected product from the knockout. The knockout was subsequently confirmed by sequencing. The generation and characterization of TNS1 knockout worms in N2 background were assisted by NemaMetrix (Eugene, OR).
2.3. Defecation assays
Defecation cycle activity was assayed by observing worm’s defecation behavior, which consists of a sequence of anterior and posterior contractions followed by an expulsion event. Data is displayed as average seconds per cycle. First-day adult worms, synchronized by bleaching and subsequent L1 arrest, were isolated on an agar plate containing OP50 E. coli and recorded for 10 minutes. Videos were subsequently analyzed by a blinded scorer to determine the timing of each defecation event.
2.4. Electropharyngeogram
Electrophysiological data for pharyngeal pump frequency, pump duration and pump interval were obtained over two experimental days using first day adults in 10mM 5-hydroxytryptamine (5HT) (20 – 22°C). Worms were synchronized via bleaching and subsequent L1 arrest, then plated on NMG plates seeded with OP50 E. coli until worms reached adulthood. Worms were washed from plates and incubated in 10mM 5HT prior to beginning the assay. All worms were assayed at the first day of adulthood using the NemaMetrix ScreenChip system for electrophysiological quantification, using detailed protocols posted at Nemametrix.com. Pump interval was calculated by the time from E-spike to E-spike, while pump duration was defined as the time from E-spike to R-spike on the electropharyngeogram.
3. Results
3.1. C. elegans tensin protein sequence is more similar with that of human TNS1
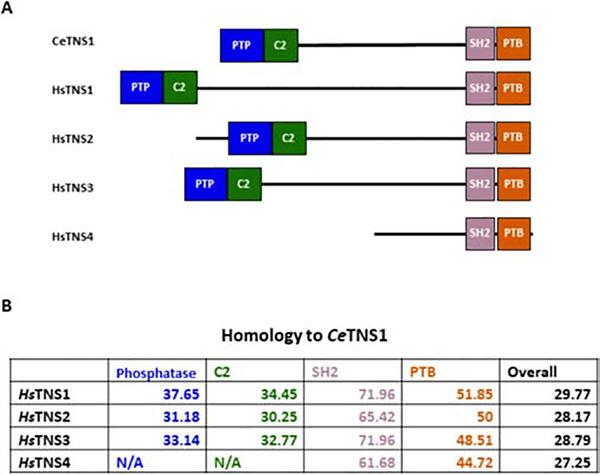
There are four tensin genes in mammals, but only one in C. elegans. To better understand the similarities between the worm and mammalian proteins, we compared the sequence of C. elegans tensin with each of the human tensin protein. All tensins contain the phosphatase domain, C2 region, SH2 domain and PTB domain except cten/TNS4, which lacks the phosphatase domain and C2 region (Figure 1A). We found that the SH2 domain is the most conserved region followed by the PTB, phosphatase domain, and then C2 regions (Figure 1B). The analysis also indicated that C. elegans tensin most closely resembles human TNS1 both overall and in its individual domains.
Figure 1. C. elegans TNS1 shares domain structure and sequence homology with Homo sapiens tensins.
(A) Domain structure of C. elegans TNS1 (CeTNS1) alongside the four human tensin proteins. The conserved domains are represented as colored boxes, while the poorly conserved central region of each protein is represented as a black line. (B) Homology of each Homo sapiens tensin (HsTNS) protein when compared with CeTNS1. HsTNS1 shows the most homology with CeTNS1, both overall and in each individual domain.
3.2. Generation of TNS1 knockout worms
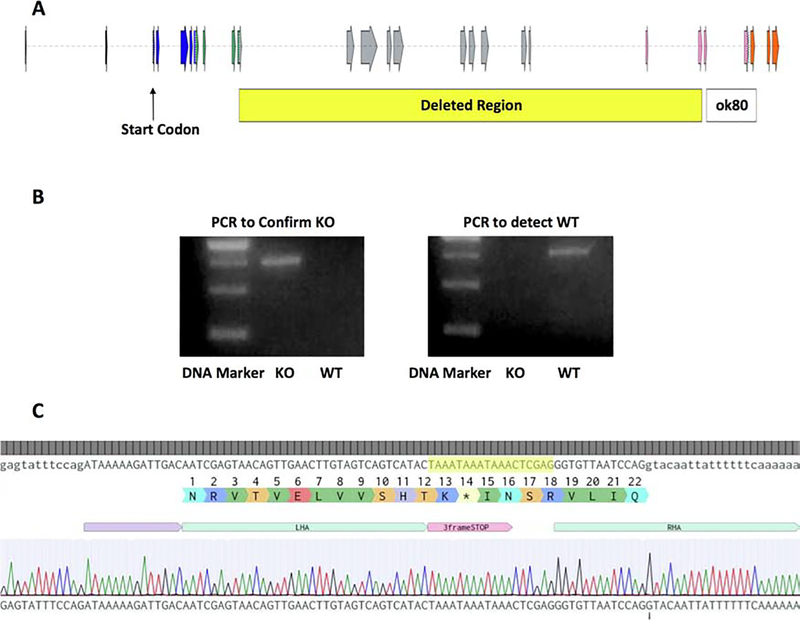
When searching available TNS1 mutant strains through the C. elegans knockout consortium data base (http://www.celeganskoconsortium.omrf.org), we found that the strain VC2621 tag-163(ok80) might be a potential knockout strain. This strain contains a 1514 bp deletion, which includes exons coding for the SH2 and PTB domains of the TNS1 gene. However, upon further analysis, TNS1 mRNA was still detected by RT-PCR assay in ok80 worms (not shown), suggesting that ok80 likely expresses a truncated form of TNS1. In order to study a complete TNS1 knockout, we have generated a 14619 bp deletion in the TNS1 gene, removing exons 11 to 21 as well as a portion of exon 10 and exon 22 by using CRISPR-Cas9 and homologous recombination directed repair technologies (Figure 2A). In place of this region we also inserted a 3 frame stop codon to ensure that no readthrough could occur between the few remaining exons. The deletion was confirmed via PCR and sequencing assays (Figure 2B & 2C). To our surprise TNS1 knockout worms are apparently normal and do not show obvious morphological or developmental defects (not shown).
Figure 2. Generation of TNS1 knockout in C. elegans.
(A) A diagram of the C. elegans TNS1 gene on chromosome 1 showing exons (arrows) in the largest splice variant. Exons coding for the phosphatase domain are shown in blue, the C2 domain in green, the SH2 domain in pink and the PTB domain in orange. A 14,619 bp region (yellow box) in the TNS1 gene is deleted and replaced with a 3-frame stop sequence in the knockout. Exons 10–22 within the deletion region were completely or partially deleted. The deletion region for the ok80 allele is shown in the white box for comparison. (B) TNS1 knockout was confirmed by PCR analysis. Genomic DNAs from the KO or WT strains were used for PCR templates. The 755-bp band in the PCR to confirm the KO indicates that the deletion has occurred, and the WT serves as a negative control (left panel). In contrast, using the primer set for the WT TNS1 gene, the 587-bp band is only detected in the WT but not the KO samples (right panel). (C) Sequencing the target region confirmed that the 3-frame stop fragment has successfully replaced the deleted region of TNS1 gene. The inserted sequence including the 3-frame stop is highlighted in yellow.
3.3. TNS1 knockout worms show slowed defecation
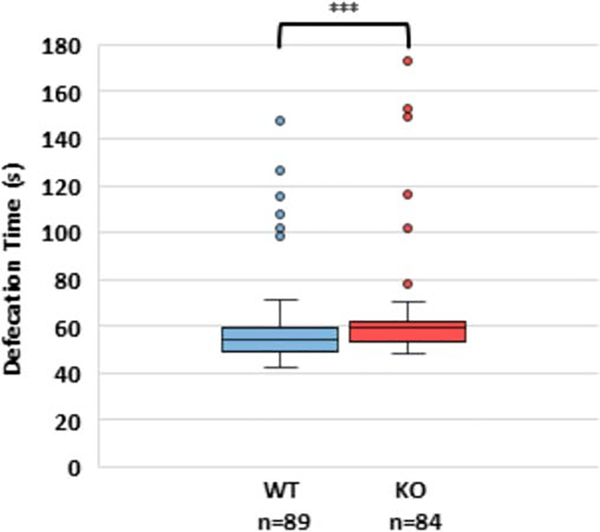
TNS1 is involved in cell adhesion processes and has been shown to localize to dense bodies, M-lines, and cell attachment sites between muscles [14], as well as to the ventral and dorsal nerve cords in C. elegans [15]. One observable process that requires coordinated muscle contractions is defecation. Therefore, we hypothesized that TNS1 knockout worms may show an altered defecation rate. We found that wild type (WT) worms’ defecation cycle lasted for 58.5 ± 1.9 seconds, which aligns closely with previously reported values of approximately 56 seconds [16]. Interestingly, the defecation cycle of TNS1 knockouts was 63.4 ± 2.3 seconds (Figure 3), which is significantly slower than that of the wild type (p < 0.001).
Figure 3: Defecation rate is slower in TNS1 knockout than wild type worms.
More than 5 defecation events from 10 different worms of each genotype were observed and the total time of the defecation cycle was calculated. Data was significant at the p<0.001 level by 2-tailed Mann-Whitney U test.
3.4. Pharyngeal pumping is faster in TNS1 knockout worms
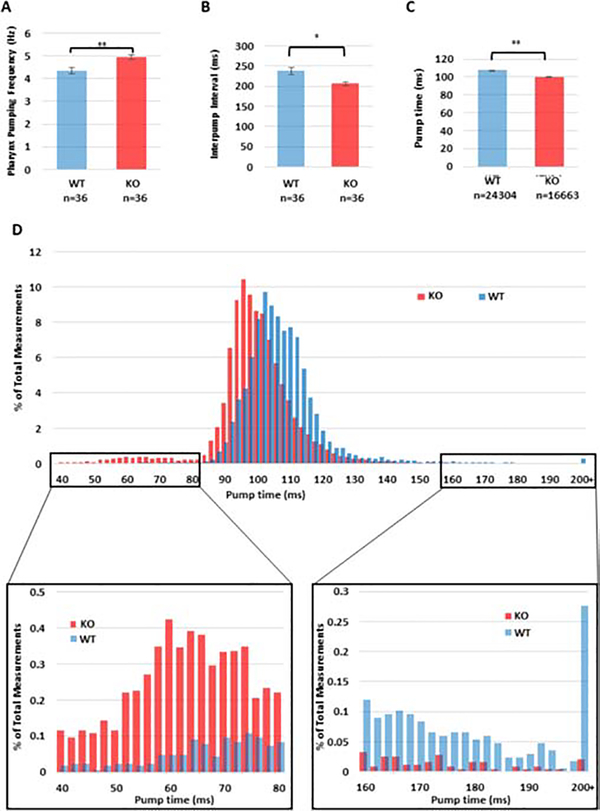
The slower defecation rate led us to test whether TNS1 KO worms were also feeding more slowly. Therefore, we investigated the pharyngeal pumping rate. The C. elegans pharynx is used to take in and process its bacterial food. This is done in a characteristic pumping mechanism, which is physiologically analogous to the mammalian heartbeat [17]. In our hands, WT worms showed a pumping frequency of 4.34 ± 0.13 Hz, which is within the previously reported rate of 200–300 pumps/minute (3.33–5 Hz) when food is present [18]. In contrast, the TNS1 KO worms had a significantly faster pumping frequency of 4.93 ± 0.10 Hz (Figure 4A, p = 0.004). The interpump interval for each pumping event was also measured. WT worms had an interpump interval of 237.4 ± 9.2 ms, while TNS1 KO was markedly reduced to 206.1 ± 5.2 (Figure 4B, p = 0.012).
Figure 4: Pharyngeal pumping is faster in TNS1 knockout worms.
Thirty-six worms of each genotype were monitored by electropharyngeogram. (A) The pumping frequency for each worm was calculated by the number of pumps over the total observation time. This showed a significant increase in pumping frequency in the TNS1 KO worms by a 2-tailed student’s t-test (p = 0.004). (B) The time interval between pump initiations (the interpump interval) was directly measured for each pump. This showed a significant reduction in interpump interval for TNS1 KO worms by a 2-tailed student’s t-test (p=0.012). (C) The amount of time spent pumping each cycle was determined to be significantly shorter for KO worms than their WT counterparts as determined by a 2-sample student’s t-test, p < 0.0001. (D) A histogram of pump times shows that the most common pumping times for KO are shorter than that observed in WT, and that a small peak of very short (<70 ms) pumps appears only in the KO mutant. The difference in very short pumps is readily apparent upon zooming in on the range from 40–80 ms. KO worms also show notable depletion of longer pumps relative to WT.
To further explore the increase in pharyngeal pumping in TNS1 mutant worms, we examined whether the phenotype was due to a shorter pump duration. We found that mean pump duration was significantly reduced from 109.11 ± 4.37 ms in WT to 99.83 ± 4.05 ms in TNS1 KO worms (Figure 4C & 4D, p < 0.0001). Furthermore, we also observed a notable increase in shorter (<75 ms) pumps (Figure 4D) and a concurrent reduction in longer (>160 ms) pumps (Figure 4D) in KO worms.
4. Discussion
In this report, we have demonstrated that TNS1 deletion in C. elegans results in a longer defecation cycle (Figure 3) and in more rapid pharyngeal pumping (Figure 4A & 4B), showing a shorter pump duration (Figure 4C & 4D). These findings demonstrate the critical role of TNS1 in C. elegans digestion.
The defecation process in C. elegans occurs in a well-defined three step process consisting of a posterior contraction, an anterior contraction, and an expulsion event [19]. Defecation rate is affected by a number of pathways, including the inositol(1,4,5)-phosphate pathway, lipid metabolism pathways, and energy metabolism pathways. Defecation rate is also highly dependent on temperature under some conditions [20] but not all [19]. Why loss of tensin causes slower defecation cycles is not immediately clear and required further investigation. It could be potentially due to changes in metabolic signaling, muscle adhesion, or even simply a consequence of lower digestive flux due to less efficient feeding.
C. elegans feeding occurs through the pharynx. This organ consists of three primary sections: the corpus, the isthmus, and the bulb [18, 21]. Muscles in the pharynx are arranged radially so that muscle contraction opens the pharynx to pull bacterial food in, and muscle relaxation closes it to expel excess water and trap the bacteria. The trapped bacteria are then moved back through the isthmus and into the bulb, where they are processed by a chitinous grinder [22] to allow the bacterial contents to be absorbed by the gut. This all happens by a characteristic neuromuscular pumping mechanism that phenotypically resembles the mammalian heartbeat [17]. The rate of pharyngeal pumping is cooperatively controlled by pharyngeal muscles and associated neurons, which are largely isolated from the somatic nervous system [23]. Defects in the system often result in abnormal feeding phenotypes [18]. All of these nerve cells can be ablated and pharyngeal pumping will still continue. However, when all neural activity in the worm is inhibited, pharyngeal pumping is also halted, indicating that some nerve signals are needed for pumping. This stands in contrast to the mammalian heart, whose cells will continue to beat independently of any neural input [24]. Interestingly, in the TNS1 knockouts we observed a marked increase in shorter than average pumps that were almost entirely absent in the WT worms (Figure 4D). It is possible that these represent abortive or poorly coordinated pumps. If this is the case, these movements could certainly impair feeding.
It is curious that C. elegans lacking TNS1 show such a mild phenotype when mice lacking even a single tensin show deleterious effects [7–9, 25], and fruit flies lacking tensin also suffer from significant defects. It is possible that TNS1 is more important for enduring stress events than for growth under optimal conditions. Kidneys, wherein TNS1 and TNS2 knockout mice show defects [7–9], are constantly bombarded with chemical stressors, so it is conceivable that the value tensin is most clearly seen under chemically stressful conditions. Similarly, drosophila tensin is required to prevent blisters during the physically stressful process of wing maturation. Therefore, it is possible that invertebrate tensin is most important in physically damaging environments. Neither of these states would be easily observed under the conditions normally used to grow C. elegans in the laboratory, which are optimized for reproducibility and rapid growth. It will be interesting to examine TNS1 KO worms under various stress conditions in the future. Interestingly, C. elegans tensin has recently been shown to be important for the regeneration of nerve axons [15]. Worms homozygous for the ok80 allele, which may still express a truncated TNS1 protein lacking the C-terminus, displayed no physiological defects under normal conditions but showed significantly less axon regrowth after injury [15]. This is very similar to our previous finding in TNS1 KO mice, where the regeneration process of skeletal muscle induced by cardiotoxin lasted significantly longer than WT mice [26]. These findings indicate that tensin plays a conserved role in the wound healing and regeneration processes from worms to mice.
Highlights.
Although four tensin members are found in mammals, C elegans only has one tensin (TNS1) gene.
TNS1 is not required for the development and survival of C elegans
Lack of TNS1 significantly extend defecation cycles in worms
The pharyngeal pumping rates in TNS1 knockout worm are markedly faster
TNS1 regulates pharyngeal pumping rates in C elegans
Acknowledgements
We thank Dr Kathryn McCormick for her helps and advice on this study. Su Hao Lo is supported in part by a NIH grant HL139473.
Abbreviations
- KO
knockout
- WT
wild type
- PTB
phosphotyrosine binding
- SH2
Src Homology 2
Footnotes
The authors disclose no potential conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chuang JZ, Lin DC, and Lin S, Molecular cloning, expression, and mapping of the high affinity actin-capping domain of chicken cardiac tensin. J Cell Biol, 1995. 128(6): 1095–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderwood DA, et al. , Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci U S A, 2003. 100(5): 2272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo SH, Tensin. Int J Biochem Cell Biol, 2004. 36(1): 31–4. [DOI] [PubMed] [Google Scholar]
- 4.Lo SH, et al. , Molecular cloning of chick cardiac muscle tensin. Full-length cDNA sequence, expression, and characterization. J Biol Chem, 1994. 269(35): 22310–9. [PubMed] [Google Scholar]
- 5.Blangy A, Tensins are versatile regulators of Rho GTPase signalling and cell adhesion. Biol Cell, 2017. 109(3): 115–126. [DOI] [PubMed] [Google Scholar]
- 6.Lo SH, Tensins. Curr Biol, 2017. 27(9): R331–r332. [DOI] [PubMed] [Google Scholar]
- 7.Lo SH, et al. , Progressive kidney degeneration in mice lacking tensin. J Cell Biol, 1997. 136(6): 1349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchio-Yamada K, et al. , Tensin2-deficient mice on FVB/N background develop severe glomerular disease. J Vet Med Sci, 2016. 78(5): 811–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang MK, et al. , Inactivation of tensin3 in mice results in growth retardation and postnatal lethality. Dev Biol, 2005. 279(2): 368–77. [DOI] [PubMed] [Google Scholar]
- 10.Cha IJ, et al. , Drosophila tensin plays an essential role in cell migration and planar polarity formation during oogenesis by mediating integrin-dependent extracellular signals to actin organization. Biochem Biophys Res Commun, 2017. 484(3): 702–709. [DOI] [PubMed] [Google Scholar]
- 11.Lee SB, et al. , blistery encodes Drosophila tensin protein and interacts with integrin and the JNK signaling pathway during wing development. Development, 2003. 130(17): 4001–10. [DOI] [PubMed] [Google Scholar]
- 12.Torgler CN, et al. , Tensin stabilizes integrin adhesive contacts in Drosophila. Dev Cell, 2004. 6(3): 357–69. [DOI] [PubMed] [Google Scholar]
- 13.Madeira F, et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res, 2019. 47(W1): W636–w641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meissner B, et al. , Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS One, 2011. 6(5): e19937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hisamoto N, et al. , C. elegans Tensin promotes axon regeneration by linking the Met-like SVH-2 and integrin signaling pathways. J Neurosci, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branicky R, et al. , Phenotypic and suppressor analysis of defecation in clk-1 mutants reveals that reaction to changes in temperature is an active process in Caenorhabditis elegans. Genetics, 2001. 159(3): 997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuler C, et al. , Arrhythmogenic effects of mutated L-type Ca 2+-channels on an optogenetically paced muscular pump in Caenorhabditis elegans. Sci Rep, 2015. 5: 14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avery L and You YJ, C. elegans feeding. WormBook, 2012: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu DW and Thomas JH, Regulation of a periodic motor program in C. elegans. J Neurosci, 1994. 14(4): 1953–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branicky R and Hekimi S, What keeps C. elegans regular: the genetics of defecation. Trends Genet, 2006. 22(10): 571–9. [DOI] [PubMed] [Google Scholar]
- 21.Albertson DG and Thomson JN, The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci, 1976. 275(938): 299–325. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. , The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Dev Biol, 2005. 285(2): 330–9. [DOI] [PubMed] [Google Scholar]
- 23.Ishita Y, Chihara T, and Okumura M, Serotonergic modulation of feeding behavior in Caenorhabditis elegans and other related nematodes. Neurosci Res, 2019. [DOI] [PubMed] [Google Scholar]
- 24.Trojanowski NF, Raizen DM, and Fang-Yen C, Pharyngeal pumping in Caenorhabditis elegans depends on tonic and phasic signaling from the nervous system. Sci Rep, 2016. 6: 22940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y, et al. , Genetic loci for resistance to podocyte injury caused by the tensin2 gene deficiency in mice. BMC Genet, 2018. 19(1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii A, and Lo SH, A role of tensin in skeletal muscle regeneration. Biochem J. 2001. 356:737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]