Abstract
Determining the level of consciousness in patients with disorders of consciousness (DOC) remains challenging. To address this challenge, resting-state fMRI (rs-fMRI) has been widely used for detecting the local, regional, and network activity differences between DOC patients and healthy controls. Although substantial progress has been made towards this endeavor, the identification of robust rs-fMRI-based biomarkers for level of consciousness is still lacking. Recent developments in machine learning show promise as a tool to augment the discrimination between different states of consciousness in clinical practice. Here, we investigated whether machine learning models trained to make a binary distinction between conscious wakefulness and anesthetic-induced unconsciousness would then be capable of reliably identifying pathologically induced unconsciousness. We did so by extracting rs-fMRI-based features associated with local activity, regional homogeneity, and interregional functional activity in 44 subjects during wakefulness, light sedation, and unresponsiveness (deep sedation and general anesthesia), and subsequently using those features to train three distinct candidate machine learning classifiers: support vector machine, Extra Trees, artificial neural network. First, we show that all three classifiers achieve reliable performance within-dataset (via nested cross-validation), with a mean area under the receiver operating characteristic curve (AUC) of 0.95, 0.92, and 0.94, respectively. Additionally, we observed comparable cross-dataset performance (making predictions on the DOC data) as the anesthesia-trained classifiers demonstrated a consistent ability to discriminate between unresponsive wakefulness syndrome (UWS/VS) patients and healthy controls with mean AUC’s of 0.99, 0.94, 0.98, respectively. Lastly, we explored the potential of applying the aforementioned classifiers towards discriminating intermediate states of consciousness, specifically, subjects under light anesthetic sedation and patients diagnosed as having a minimally conscious state (MCS). Our findings demonstrate that machine learning classifiers trained on rs-fMRI features derived from participants under anesthesia have potential to aid the discrimination between degrees of pathological unconsciousness in clinical patients.
Keywords: fMRI, resting-state, disorders of consciousness, anesthesia, functional connectivity, machine learning, deep learning, consciousness
Introduction
Determining the level of consciousness in patients with disorders of consciousness (DOC) remains a challenging clinical problem. The primary diagnostic tool, a behavioral assessment, is prone to erroneous conclusions (over 40% misdiagnosis rate) when relying solely on the clinician’s judgment without standardized assessment (Schnakers et al., 2009b). Though standardized behavioral exams, like the Coma Recovery Scale-Revised (CRS-R) (Giacino et al., 2004), are now widely used, misdiagnoses may also occur if patients are not assessed repeatedly within a short time window (Wannez et al., 2017). In some cases, covert consciousness (i.e., awareness without overt responsiveness) can occur due to central nervous system lesions that prevent motor activity (Fernández-Espejo et al., 2015; Monti et al., 2010; Owen et al., 2006). An analogous phenomenon, intraoperative awareness during general anesthesia, has been reported with explicit recall in 0.15% of all surgical cases (Mashour et al., 2013, 2012), and without explicit recall in 5% of all cases (Sanders et al., 2017). Further, covert consciousness was recently demonstrated by a healthy participant during propofol anesthesia using an active fMRI-based paradigm. (Huang et al., 2018b). Thus, the identification of preserved consciousness is of substantial importance in the clinical setting as the reliable detection of preserved consciousness in DOC patients can lead to an increased focus on rehabilitative efforts that may foster recovery (Fins et al., 2007; Giacino et al., 2014). Therefore, the need to establish reproducible brain markers linked to different levels of consciousness independent of behavior is paramount.
Within the last decade, there has been a surge of interest in identifying more objective techniques for measuring levels of consciousness. A wealth of previous research has explored possible neural correlates of consciousness derived from neuroimaging techniques, like functional magnetic resonance imaging (Bekinschtein et al., 2005, 2004; Chen et al., 2018; Coleman et al., 2009; Mäki-Marttunen et al., 2013) and positron emission tomography (Boly et al., 2008, 2004; Silva et al., 2010) as well as measures of neurophysiological responses to stimuli captured by electroencephalography (Bekinschtein et al., 2009; Schnakers et al., 2009a, 2008). For a review see (Laureys and Schiff, 2012; Mashour and Hudetz, 2018; Owen, 2013). Each methodological approach has unique advantages and disadvantages depending on the specific goals and application (Boly et al., 2012).
Of these techniques, resting-state fMRI (rs-fMRI)-based measurements appear especially fruitful as they are capable of providing key components in understanding the dynamic functional organization of brain activity across multiple scales (i.e., local, regional, network) that appears necessary for consciousness (Huang et al., 2018a). Accordingly, particular features of intrinsic brain activity have been associated with physiologic, pharmacologic, and pathologic states of unconsciousness (Boveroux et al., 2010; Demertzi et al., 2011; Di Perri et al., 2016; Heine et al., 2012; Roquet et al., 2016; Soddu et al., 2009). Although substantial progress has been made towards this endeavor, a robust rs-fMRI-based classification for states of consciousness is still lacking. However, recent developments in machine learning show promise as a tool to augment the discrimination between different states of consciousness in clinical practice. In the last decade, researchers have successfully built models capable of distinguishing between different degrees of awareness—locked-in syndrome, minimally conscious state (MCS), and unresponsive wakefulness syndrome/vegetative state (UWS/VS)—based on each patient’s neuroimaging data (Demertzi et al., 2019, 2015; Engemann et al., 2018; Phillips et al., 2011; Sitt et al., 2014).
Despite this progress, one persistent challenge to the study of DOC patients is the etiological heterogeneity—DOC may be induced through focal injury to neural tissues (e.g., traumatic brain injury, stroke) or more diffuse damage (e.g., Alzheimer’s disease)—each of which affects the structural integrity and functional dynamics of the brain in distinct ways (Amemiya et al., 2013; Sours et al., 2015). Taken together, the differences between DOC patients, the high misdiagnosis rate associated with behavioral assessment, and the lack of ground-truth data, pose a critical problem in establishing a robust and reproducible machine learning model. In contrast, a proposed surrogate model of study, namely anesthetic-induced unconsciousness in healthy volunteers, offers the possibility of a within-subjects design, and consequently, rigorously controlled experimental settings (Alkire et al., 2008; Mashour and Avidan, 2013). Using this paradigm, the consciousness-altering effects of a range of anesthetics have been evaluated in humans, including ketamine (Bonhomme et al., 2016), sevoflurane (Palanca et al., 2015), and propofol (Schroter et al., 2012).
The present study sought to further improve the understanding and diagnosis of DOC by systematically comparing popular machine learning approaches to classification, and by evaluating a novel source of model training data, namely the use of participants during anesthetic-induced unconsciousness. To this end, our aims were to (1) build, optimize and evaluate three distinct classes of machine learning models (i.e. support vector machine, Extra Trees, and artificial neural network) for use in distinguishing conscious wakefulness from anesthetic-induced unresponsiveness using rs-fMRI based measures, including local activity (amplitude of low-frequency fluctuations, ALFF), regional homogeneity (ReHo), and interregional functional activity. (2) Evaluate whether machine learning models trained on data collected during anesthesia make reliable generalizations to UWS/VS patients, and (3) explore the feasibility of using the above machine learning models to distinguish intermediate states of consciousness—subjects under light sedation and patients within a minimally conscious state (MCS)—from fully conscious or unconscious subjects.
Methods
Participants and fMRI Data Acquisition
The fMRI data were collected from a cohort of 83 subjects scanned at two independent research sites (Shanghai and Wisconsin). Dataset 1 involving propofol and sevoflurane anesthesia was collected in Shanghai and is hereafter referred to as Anesthesia-SHH. Dataset 2 involving propofol anesthesia was collected in Wisconsin, hereafter referred to as Anesthesia-WI. Dataset 3, hereafter referred to as DOC, had no anesthetic component, and instead included patients with disorders of consciousness, in addition to healthy controls, and was collected in Shanghai.
Dataset 1: Anesthesia-SHH
The dataset has been previously published using analyses different from those applied here (Huang et al., 2018c, 2018a, 2014). The study was approved by the Institutional Review Board (IRB) of Huashan Hospital, Fudan University. Informed consent was obtained by all the subjects to participate in the study. Thirty-two right-handed subjects were recruited (male/female: 15/17; age: 26–64 years), who were undergoing an elective trans-sphenoidal approach for resection of a pituitary microadenoma. The pituitary microadenomas were diagnosed by their size (<10 mm in diameter without growing out of the sella) based on radiological examinations and plasma endocrinal parameters. These subjects were ASA (American Society of Anesthesiologists) physical status I or II grade, with no history of craniotomy, cerebral neuropathy, vital organ dysfunction or administration of neuropsychiatric drugs. The subjects had no contraindication for an MRI examination, such as vascular clips or metallic implants. Among them, three subjects had to be excluded from the study and further data analysis because of excessive movements, resulting in 29 subjects for the following analysis.
Twenty-three subjects received propofol anesthetics with light sedation (17 out of 23) and general anesthesia (n=23), during which intravenous anesthetic propofol was infused through an intravenous catheter placed into a vein of the right hand or forearm. Propofol was administered using a target-controlled infusion (TCI) pump to obtain constant effect-site concentration, as estimated by the pharmacokinetic model (Marsh et al., 1991). Remifentanil (1.0 μg/kg) and succinylcholine (1.5 mg/kg) were administered to facilitate endotracheal intubation at general anesthesia. TCI concentrations were increased in 0.1 μg/ml steps beginning at 1.0 μg/ml until reaching the appropriate effect-site concentration. A 5-min equilibration period was allowed to ensure equilibration of propofol repartition between compartments. The TCI propofol was maintained at a stable effect-site concentration of 1.3 μg/ml for light sedation, and 4.0 μg/ml for general anesthesia of which the dose reliably induces an unconscious state (Xu et al., 2009). In addition, six subjects received sevoflurane general anesthesia. Induction was completed with 8% sevoflurane in 100% oxygen, adjusting fresh gas flow to 6 L/min, combined with remifentanil 1.0 μg/kg, succinylcholine 1.0 mg/kg and maintained with 2.6% (1.3 MAC) ETsevo in 100% oxygen, fresh gas flow at 2.0 L/min.
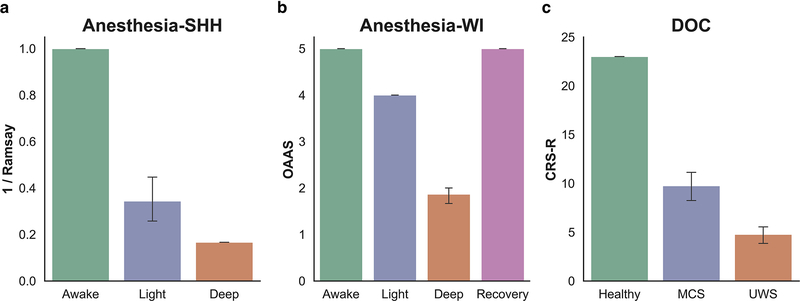
Behavioral responsiveness was assessed by the Ramsay scale (Ramsay et al., 1974) (Fig. 1a). The subjects were asked to strongly squeeze the hand of the investigator. The subject is considered fully awake if the response to verbal command (“strongly squeeze my hand!”) is clear and strong (Ramsay=1–2), in mild sedation if the response to verbal command is clear but slow (Ramsay=3–4), and in deep sedation or general anesthesia if there is no response to verbal command (Ramsay=5–6).
Fig 1.
Summary of the different behavioral responsiveness assessments used across the three included datasets. (a) The Ramsay scale (here shown as 1/Ramsay score to facilitate comparison) was applied in the Anesthesia-SHH dataset, (b) the Observer’s Assessment of Alertness/Sedation (OAAS) scale was applied in the Anesthesia-WI dataset, and the Coma Recover Scale-Revised (CRS-R) was applied in the DOC dataset.
The subjects continued to breathe spontaneously during wakefulness and light sedation. During general anesthesia, the subjects were ventilated with intermittent positive pressure ventilation, setting tidal volume at 8–10 ml/kg, respiratory rate 10–12 beats per minute, and maintaining PetCO2 (partial pressure of end-tidal CO2) at 35–45 mmHg. Two certified anesthesiologists were present throughout the study, and complete resuscitation equipment was always available. Subjects wore earplugs and headphones during the fMRI scanning.
Rs-fMRI data acquisition consisted of three 8-min scans in wakefulness baseline (n=29), light sedation (n=17) and general anesthesia (n=29), respectively. The subject’s head was fixed in the scan frame and padded with spongy cushions to minimize head movement. The subjects were asked to relax and assume a comfortable supine position with their eyes closed during scanning (an eye patch was applied). The subjects were instructed not to concentrate on anything in particular during the resting-state scan. A Siemens 3T scanner (Siemens MAGNETOM, Germany) with a standard 8-channel head coil was used to acquire gradient-echo EPI images of the whole brain (33 slices, repetition time/echo time [TR/TE]=2000/30ms, slice thickness=5mm, field of view=210mm, flip angle=90°, image matrix=64 × 64). High-resolution anatomical images were also acquired for rs-fMRI coregistration.
Dataset 2: Anesthesia-WI
The dataset has been previously published using analyses different from those applied here (Huang et al., 2018a; Liu et al., 2017a, 2017b). The Institutional Review Board of Medical College of Wisconsin (MCW) approved the experimental protocol. Fifteen healthy volunteers (male/female 9/6; 19–35 years) received propofol sedation. Four conditions of behavioral responsiveness were determined by OAAS (Observer’s Assessment of Alertness/Sedation) score (Chernik et al., 1990), namely wakefulness baseline (OAAS=5±0), propofol light sedation (OAAS=4±0), propofol deep sedation (OAAS=1.9±0.4), and recovery (OAAS=5±0). During light sedation, volunteers showed lethargic response to verbal commands, and during deep sedation volunteers showed no response to verbal commands (Fig. 1b). The corresponding target plasma concentrations vary across subjects (light sedation: 0.98 ± 0.18 μg/ml; deep sedation: 1.88 ± 0.24 μg/ml) because of the variability in individual sensitivity to anesthetics. At each level of sedation, the plasma concentration of propofol was maintained at equilibrium by continuously adjusting the infusion rate to maintain the balance between accumulation and elimination of the drug. The infusion rate was manually controlled and guided by the output of a computer simulation developed for target-controlled drug infusion (Shafer, 1996) based on the pharmacokinetic model of propofol (Marsh et al., 1991). Standard American Society of Anesthesiologists (ASA) monitoring was conducted during the experiment, including electrocardiogram, noninvasive blood pressure cuff, pulse oximetry, and end tidal carbon dioxide gas monitoring. Supplemental oxygen was administered prophylactically via nasal cannula.
Rs-fMRI data acquisition consisted of four 15-min scans in wakefulness baseline, light and deep sedation, and recovery, respectively. A 3T Signa GE 750 scanner (GE Healthcare, Waukesha, Wisconsin, USA) with a standard 32-channel transmit/receive head coil was used to acquire gradient-echo EPI images of the whole brain (41 slices, TR/TE=2000/25ms, slice thickness=3.5mm, field of view=224mm, flip angle=77°, image matrix: 64×64). High-resolution anatomical images were also acquired for rs-fMRI coregistration.
Dataset 3: DOC
The dataset has been previously published using analyses different from those applied here (Huang et al., 2018a, 2016, 2014). The study was approved by the Institutional Review Board (IRB) of Huashan Hospital, Fudan University. Informed consent was obtained from the patients’ legal representatives, and from the healthy participants. The dataset included 21 patients (male/female: 18/3) with disorders of consciousness, and 28 healthy control (HC) subjects (male/female: 14/14). The patients were assessed using a standardized behavioral exam—the Coma Recovery Scale-Revised (CRS-R) (Giacino et al., 2004)—on the day of fMRI scanning, both before and after scanning (Fig. 1c). Of those assessed, 13 patients were diagnosed as UWS/VS, and 8 were diagnosed as MCS (Table 1).
Table 1.
Clinical information for DOC
| Patient number | Gender/Age | Cause | Time of fMRI | CRS-R | Diagnosis |
|---|---|---|---|---|---|
| (days after insult) | |||||
| 1 | M/37 | TBI | 301 | 6 | UWS |
| 2 | M/78 | TBI | 211 | 7 | MCS |
| 3 | M/51 | TBI | 100 | 4 | UWS |
| 4 | M/23 | HIE | 244 | 4 | UWS |
| 5 | M/47 | SIH | 79 | 9 | MCS |
| 6 | M/48 | SIH | 78 | 6 | UWS |
| 7 | M/58 | TBI | 83 | 7 | UWS |
| 8 | M/66 | HIH | 280 | 10 | MCS |
| 9 | M/30 | TBI | 26 | 12 | MCS |
| 10 | M/8 | P-CPR | 65 | 7 | UWS |
| 11 | M/18 | TBI | 30 | 6 | MCS |
| 12 | F/32 | TBI | 73 | 12 | MCS |
| 13 | M/55 | TBI | 106 | 10 | MCS |
| 14 | M/16 | TBI | 803 | 12 | MCS |
| 15 | F/35 | TBI | 21 | 5 | UWS |
| 16 | M/46 | SIH | 18 | 2 | UWS |
| 17 | M/60 | SIH | 109 | 6 | UWS |
| 18 | M/46 | TBI | 25 | 2 | UWS |
| 19 | M/59 | SIH | 44 | 4 | UWS |
| 20 | F/52 | SIH | 51 | 4 | UWS |
| 21 | M/46 | TBI | 162 | 5 | UWS |
UWS: unresponsive wakefulness syndrome; MCS: minimally conscious state; CRS-R: Coma Recovery Scale-Revised; TBI: traumatic brain injury; SIH: spontaneous intracerebral hemorrhage; HIH: hypertensive intracerebral hemorrhage; HIE, hypoxic ischaemic encephalopathy; P-CPR: post cardiopulmonary resuscitation.
None of the healthy controls had a history of neurological or psychiatric disorders, nor were they taking any kind of medication. Of note, the labels used for classification were the patient diagnoses assigned according to their respective CRS-R scores. As mentioned earlier, diagnoses based on behavioral markers may be inaccurate, especially between MCS and USW/VS. Further, since our goal was to differentiate UWS/VS patients from healthy controls, rather than separate UWS/VS patients from MCS patients, we deemed that the CRS-R was the appropriate tool to coarsely define the groups for our classification task.
Rs-fMRI data were acquired on a Siemens 3T scanner (Siemens MAGNETOM, Germany). A standard 8-channel head coil was used to acquire gradient-echo EPI images of the whole brain (33 slices, TR/TE=2000/35ms, slice thickness=4mm, field of view=256 mm, flip angle=90°, image matrix=64 × 64). Two hundred EPI volumes (6 minutes and 40 seconds), as well as high-resolution anatomical images, were acquired.
fMRI Data Preprocessing and Feature Extraction
The following preprocessing steps were implemented in AFNI (http://afni.nimh.nih.gov/): (1) The first two frames of each fMRI run were discarded; 2) Slice timing correction; 3) Rigid head motion correction/realignment within and across runs; frame-wise displacement (FD) of head motion was calculated using frame-wise Euclidean Norm (square root of the sum squares) of the six-dimensional motion derivatives. Each frame, and the frame prior, were tagged as zeros (ones, otherwise) if the given frame’s derivative value has a Euclidean Norm above FD=0.5mm (Huang et al., 2018c) 4) Coregistration with high-resolution anatomical images; 5) Spatial normalization into Talaraich stereotactic space; 6) Using AFNI’s function 3dTproject, the time-censored data were band-pass filtered to 0.01–0.1Hz. At the same time, various undesired components (e.g., physiological estimates, motion parameters) were removed via linear regression. The undesired components included linear and nonlinear drift, time series of head motion and its temporal derivative, binarized FD time series, and mean time series from the white matter and cerebrospinal fluid; 7) Spatial smoothing with 6mm full-width at half-maximum isotropic Gaussian kernel; 8) The time-course per voxel of each run was normalized to zero mean and unit variance, accounting for differences in variance of non-neural origin (e.g., distance from head coil). Lastly, global signal regression (GSR) was not included in the following analysis as it may introduce artificial anti-correlations between regions, and therefore bias the results or interpretations (Anderson et al., 2011; Fox et al., 2009; Murphy et al., 2009, 2016; Saad et al., 2012).
Definition of Functional Networks
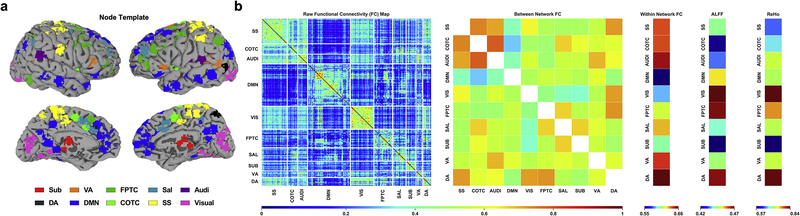
We adopted a well-established node template (Power et al., 2011) that had been slightly modified for a previous study (Huang et al., 2018a) containing 226 nodes (10mm diameter spheres, 32 voxels per sphere) within 10 functional networks: subcortical (Sub), dorsal attention (DA), ventral attention (VA), default mode (DMN), frontoparietal task control (FPTC), cinguloopercular task control (COTC), salience (Sal), sensory/somatomotor (SS), auditory (Audi), and visual networks (Visual) (Fig. 2a).
Fig 2.
Extraction of model features using fMRI-based measures of resting state activity. (a) Node template representing anatomical location of 226 seed regions of interest (ROIs) consolidated into 10 networks (Power et al., 2011): subcortical (Sub), ventral attention (VA), frontoparietal task control (FPTC), salience (Sal), auditory (Audi), dorsal attention (DA), default mode (DMN), cinguloopercular task control (COTC), sensory/somatomotor (SS), visual (Visual). (b) Raw functional connectivity map (left) generated from seed-based pairwise Pearson correlations between 226 ROIs. Activity was averaged according to network template yielding measures of between network (off-diagonal) and within network (on-diagonal) functional connectivity (middle). Two additional measures of functional segregation, the amplitude of low-frequency fluctuations (ALFF) and regional homogeneity (ReHo), were calculated independently using the network templates.
ALFF Calculation
ALFF was calculated at the voxel level by the AFNI program 3dRSFC for each subject. ALFF quantifies local resting-state signal fluctuations by measuring the integral of the signal amplitude in the frequency domain (over a low-frequency range of 0.01–0.1Hz) (Zang et al., 2007). The original approach to quantifying the ALFF was improved by calculating the ratio of the power of the low-frequency range to that of the entire frequency range resulting fractional ALFF (fALFF) (Zou et al., 2008), which was adopted in our analysis. The averaged fALFF values for each of the pre-defined 10 networks were extracted at the subject-level and separately for each condition.
ReHo Calculation
Regional homogeneity (ReHo) was calculated at the voxel level using Kendall’s coefficient of concordance (KCC) between the BOLD time series for the specified voxel and those of its 26 nearest neighbors (~2 mm radius sphere) (Zang et al., 2004). ReHo quantifies the intra-regional signal correlation. ReHo analysis was performed by AFNI program 3dReHo. As spatial smoothing could artificially enhance ReHo and reduce its reliability (Zuo et al., 2013), we calculated ReHo from non-smoothed BOLD time series. Spatial smoothing was subsequently applied, with a 6mm fullwidth at half-maximum (FWHM) Gaussian kernel, to the ReHo maps (Fisher’s Z transformed). The averaged ReHo values for each of the pre-defined 10 networks were extracted at the subject-level and separately for each condition.
FC Calculation
Inter-regional functional connectivity (FC) was calculated based on the aforementioned node template, wherein the minimal Euclidian distance between two centers of any pair of nodes is 2cm. This is notably distinct from ReHo, which reflects connectivity within an ~2mm radius sphere. We computed the Pearson correlation coefficient of the time courses between each pair of nodes, yielding a pairwise 226×226 correlation matrix (Fisher’s Z transformed). Based on this correlation matrix, the within and between network connectivity values were calculated by averaging the node-level FC values within the on-diagonal and off-diagonal components of the correlation matrix, respectively.
Model Training, Validation, & Testing
Following the above procedure, 75 features were extracted from the rs-fMRI activity: ALFF (10), ReHo (10), within network FC (10), between network FC (55) (Fig. 2b). All machine learning models were trained on the composite anesthesia dataset (n=44; n=29 from Anesthesia-SHH, n=15 from Anesthesia-WI), and subsequently evaluated for within-dataset prediction stability (i.e., reliability on the Anesthesia dataset) as well as the capacity to generalize classifications cross-dataset to pathologically unconscious patients with a DOC.
For the former, we employed a nested cross-validation strategy. First, 100 sub-samples (outer-fold) of the anesthesia dataset were generated through random sampling with replacement. Next, each outer-fold was separated into two independent datasets, an optimization dataset (80% of outer-fold) and validation dataset (20% of outer-fold). The optimization dataset was then further split using k-fold cross-validation, yielding five sub-samples (inner-folds). Each inner-fold consisted of a training dataset (80% of inner-fold) and a testing dataset (20% of inner-fold). The inner-folds were used to evaluate and optimize model hyperparameters, whereas the outer-folds were used to estimate model performance on a novel dataset. When hyperparameter optimization is used in the absence of nested cross-validation, models are more likely to overfit to the training data and overestimate performance on unseen data (Cawley and Talbot, 2010).
To quantify the external validity of the models, we used a Bootstrap sampling procedure (Efron and Tibshirani, 2007) to estimate the cross-dataset (Anesthesia to DOC) model performance; 100 sub-samples of the DOC data were generated by randomly sampling from the original data with replacement.
Across both methods, the class distributions were fixed such that there were equal numbers of both classes in the sub-samples used in model validation and testing. To provide an accurate estimate of reliability and generalizability, model performance was calculated as the mean across the 100 sub-samples. All model training and hyperparameter tuning was performed without exposing the models to the DOC data to ensure that the we did not inadvertently introduce information that would subsequently influence our analyses of generalization performance.
Model Selection
Three distinct candidate model types were evaluated within the study: support vector machine (SVM), decision tree, and artificial neural network (ANN). For a review of these commonly used supervised machine learning methods, and others, see (Caruana and Niculescu-Mizil, 2006). Both the SVM and decision tree-based models were constructed using scikit-learn (Pedregosa et al., 2011), a Python-based machine learning library popular within the neuroimaging community (Abraham et al., 2014). The ANN was built using the open source deep learning library Keras (https://keras.io) running on top of the TensorFlow platform (Dignam et al., 2016).
Support Vector Machine
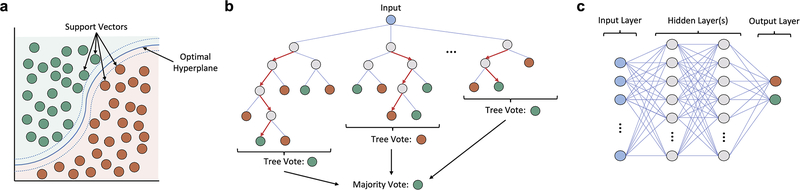
The Support vector machine (SVM) is a type of discriminative model which generates a hyperplane (i.e., decision boundary) to maximize the physical separation between two classes in N-dimensional space, where N represents the number of features (Fig. 3a). The hyperplane is defined by support vectors, the samples which lay at the boundary between classes. This technique has been widely implemented in previous neuroimaging analyses (Chennu et al., 2017; Sitt et al., 2014).
Fig 3.
Schematic representation of the three types of supervised machine learning models used in the study. (a) The Support vector machine (SVM) is a discriminative model that generates a hyperplane (i.e., decision boundary) which maximizes the separation between two classes in N-dimensional space (N = number of features). The hyperplane is defined by support vectors, the samples which lay at the boundary between classes. (b) Decision tree-based models apply a flowchart-like approach to classification wherein the input data is repeatedly split into smaller sub-groups according to some decision process until a terminal node (i.e., label) is reached. Shown is a subtype of the decision-trees class, the Random Forest, which generates many different trees from a random sample of the data, and uses bootstrap aggregation (i.e., bagging) to average the predictions across all trees. (c) Artificial Neural Networks (ANNs) represent a broad category of machine learning models which loosely imitate the physical structure of the brain. The networks are composed of individual nodes (neurons), arranged in a hierarchical structure; shown is one possible network structure, with a single input layer, two densely-connected hidden layers, an output layer with one node for each class, and only feed-forward connections throughout.
Decision Trees
Decision trees constitute a broad class of non-parametric models that visually resembles a nested tree structure. The splits (branches) of a decision tree represent points where simple decision rules are applied to parse the data until a classification is made. Decision trees seek to make high quality splits by applying metrics like Gini impurity or entropy to maximize information gain.
One particular subtype of the decision trees class, the Random Forest (Fig. 3b), is especially popular and has shown notable success in multivariate neuroimaging applications (Sarica et al., 2017). The Random Forest differs from a regular decision tree in that a multitude of trees are constructed from randomly drawn bootstrap samples of the original data. Aggregating predictions across the ensemble of structurally heterogeneous trees (i.e., bagging) helps to minimize model variance and mitigate risks of overfitting—a problem of external validity encountered often in machine learning, wherein a model is fit too tightly to the training data, and consequently, generalizes poorly when exposed to new, unseen data. The current study applied the Extra Trees (ET) variant of the Random Forest (Engemann et al., 2018; Geurts et al., 2006) which introduces additional randomness into the method for deciding split-points.
Artificial Neural Network
Artificial Neural Networks (ANN) are a class of algorithms which loosely model the neuronal structure of the brain (Fig. 3c). They are composed of an interconnected network of individual nodes (neurons) capable of adjusting the strength of their connections via a set of tunable weights and biases. The output of the neurons is defined by the application of an activation function (e.g., step function, sigmoid function). ANN’s are capable of “learning” by a process of repetition, wherein a backpropagation algorithm is repeatedly applied to automatically adjust the connection weights relative to the difference between the current prediction and expected output (Hecht-Nielsen, 1989).
We opted to construct a simple ANN with a densely-connected feedforward network structure (a.k.a multilayer perceptron), composed of: an input layer, two hidden layers, and single node (sigmoid) output layer. To address the risk of overfitting, we applied dropout to both hidden layers (20% and 50%, respectively) during training. To speed up the training process, we used the widely-popular rectified linear units (ReLU) activation function for nodes within the hidden layers (Lecun et al., 2015). Adaptive moment estimation (Adam) was chosen as the model optimizer (Kingma, Diederik and Ba, 2015) with binary cross-entropy serving as the loss metric.
Hyperparameter Optimization
Prior to training a machine learning model, a set of “hyperparameters” must be chosen. These hyperparameters represent settings that constrain the model’s behavior during training (e.g., the number of decision trees in a Random Forest model). The combination of hyperparameters chosen can cause wide variations in model performance and must be tailored to the task demands as there are no universally optimal set of hyperparameters across all applications (Thornton et al., 2012).
In practice, appropriate model hyperparameters are most often chosen by either the grid search method (systematically evaluating a range of possible combinations) or the random search method (repeatedly evaluating random combinations). The computational demands of performing a grid search rise exponentially as the number of model hyperparameters increases, therefore, the random search method has been preferred for most applications (Bergstra and Bengio, 2012).
However, given the methodology underlying grid search and random search, neither approach guarantees that the optimal combination of hyperparameters will be identified. Consequently, there has been increased interest in the development of automated hyperparameter optimization algorithms to aid in the tuning process; see (Luo, 2016) for a review.
We chose to use the Python library Hyperopt-Sklearn (Bergstra et al., 2015; Komer et al., 2018) for automated hyperparameter optimization given its ease of integration with the scikit-learn library. The Hyperopt-Sklearn library applies an optimization algorithm (i.e., Tree-Structured Parzen Estimator) to navigate a pre-defined space of hyperparameters by iteratively evaluating different combinations and subsequently modeling the likelihood probability of achieving high performance with other combinations. To improve the computational efficiency, we defined a constrained search space composed of the following tunable hyperparameters: SVM (gamma, C), ET (max tree depth, max number of features considered at each split, number of trees, decision criterion). Given the large number of tunable hyperparameters for the ANN, and high computational demands of repeated training, hyperparameter optimization was not performed on the ANN.
The default hyperparameters for the scikit-learn SVM and ET were used to compare model performance before and after hyperparameter optimization. As there is no default network structure for the Keras ANN, we chose an appropriate number of nodes for each layer through the application of the algorithmic approach recommended for two-hidden-layer feedforward networks defined in (Huang, 2003). Accordingly, the default ANN was constructed with 25 neurons in layer one, and 5 neurons in layer two.
Feature Pruning
Using the pipeline described above, we extracted 75 rs-fMRI-based features. Though we expect some of these features will be far more informative than others, much remains to be discovered about the specific biomarkers of consciousness. For this reason, we evaluated models trained on both the full set of 75 features, and models trained on a smaller subset of features isolated through feature pruning. To test the latter, we included only the features with significant differences between the awake and unresponsive states (deep sedation and general anesthesia) in the Anesthesia-SHH and Anesthesia-WI dataset. This method yielded a smaller subset of 32 features: ALFF (3), within network FC (8), between network FC (21).
Model Stress Tests
To further distinguish the models used in our analysis, we performed additional computational stress tests to evaluate whether the model classifications were robust to perturbation. To this end, we applied (1) a random drop-out of increasing fractions of the model features, and (2) a gradually reduced the signal to noise (SNR) ratio by adding increasing amounts of noise to the features. Both stress tests were conducted by making modifications solely to the DOC dataset used for testing.
To investigate how the models responded to a diffuse, nonspecific reduction in test dataset information, we randomly dropped increasing fractions of model features from the test dataset (from 0% to 100%). Features were “dropped” from the DOC dataset by setting the value for that feature, across all subjects, to zero; zeroing was necessary, rather than pure removal, to ensure that the number of features in the training dataset and testing dataset were equivalent, as required by the models.
To decrease the signal to noise ratio (SNR), we systematically introduced noise into the test dataset. For each feature, a Gaussian distribution of values was generated according to the calculated mean and variance across all subjects. The noise was added at the subject-level by randomly sampling a value on a Gaussian distribution around each feature, multiplying that sampled value by some scaling factor (ranging from 1x-100x), and finally adding the noise back to the original subject-level feature. The noised feature was then rescaled to match the original pre-noised mean and variance of the feature.
To provide a stable estimate of the effects, we employed the same, previously described bootstrap sampling procedure (B=100) in evaluating the model performance before and after each stress test.
Intermediate States
To evaluate the feasibility of discriminating intermediate states of consciousness, we applied the same preprocessing and feature-extraction procedure on data collected from three novel groups not included in the primary analyses: subjects during light propofol sedation (Light, n=15), subjects during recovery from propofol sedation (Rec, n=15), and clinical patients in a minimally conscious state (MCS, n=8).
For subjects in each of the groups not included in model training, a predicted class probability was generated, serving as a measure of the model’s confidence in the classification relative to a binary decision threshold, set at 0.5. A predicted class probability at either extremum represents a strong resemblance to one of the two groups within the anesthesia dataset used for training; predicted class probabilities greater than 0.5 (more likely awake than unresponsive) were classified as awake, whereas values less than 0.5 (more likely unresponsive than awake) were classified as unresponsive.
Statistical Analyses
A two-sample t-test was applied to analyze differences between the distribution of values across each feature for subjects during wakefulness and unresponsiveness, whereas paired t-tests were used to analyze differences in model performance before and after hyperparameter optimization as well as model performance before and after perturbation. Our analysis of each model’s predicted classification probabilities was conducted first via a one sample t-test comparing the group distributions to the binary decision threshold, set at 0.5, followed by a two sample t-test comparing the intermediate states to the two states used in training (i.e., awake, unresponsive).
Before performing the multivariate analysis, we sought to determine whether reliable classifications could be made at the single-feature level within-dataset (Anesthesia cross-validation) and cross-dataset (Anesthesia to DOC). This univariate analysis was conducted in order to explore whether using a more complex multivariate model-based approach was necessary and to further our knowledge of particular biomarkers highly related to the level of consciousness.
To quantify classification performance, receiver operating characteristic (ROC) curves were generated by first analyzing the accuracy of the predictions obtained from the different classifiers, and subsequently plotting their associated true positive rate against the false positive rate. Using the ROC curves, the area under the curve (AUC) was calculated, which served as the metric used throughout in measuring classification performance (AUC scores range from 0–1, where 0 is totally inaccurate, 1 is fully accurate, and 0.5 represented chance-level performance).
For the analyses of univariate performance and pre-post hyperparameter optimization, a Bonferroni-correction at alpha <0.05 was applied to control for the increased risk of false positives when making multiple statistical comparisons. Given that our analysis of intermediate states had a much smaller sample size, no correction was applied.
Data and code availability statement
The resting-state fMRI feature data and code for the above machine-learning pipeline are accessible at https://github.com/Justin-Campbell/ML-Anes-DOC.
Results
Univariate Performance
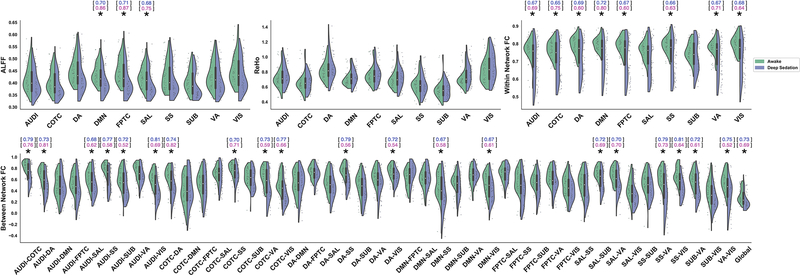
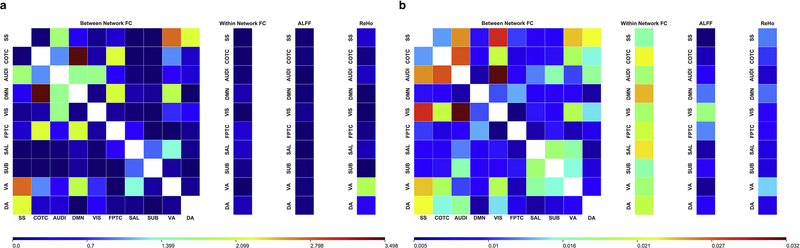
As expected, we observed several features with significant differences between the awake (n=44) and unresponsive (deep sedation/anesthesia; n=44) groups (paired sample t-test, Bonferroni-corrected p<0.05) (Fig. 4).
Fig 4.
Single feature comparisons between awake and deep sedation groups across anesthesia datasets. (a) Distribution of values for ALFF (upper-left), ReHo (upper-middle), within network FC (upper-right), and between network FC (bottom). * indicates Bonferroni-corrected p < 0.05. The ability of single features to discriminate between the two groups was evaluated using a univariate model-free analysis. The within-dataset (Anesthesia → Anesthesia; blue) and cross-dataset (Anesthesia → DOC; pink) AUC is listed above the features with significant group differences.
A subsequent analysis of the area under the ROC curves (AUC) generated from the features with group differences revealed a wide-range of univariate model-free classification performances within-dataset (AUC: 0.65–0.81) and cross-dataset (AUC: 0.52–0.87). In rare cases where a feature had an AUC of <0.50, indicating an anti-correlation with state of consciousness, the associated AUC was rectified (|AUC−0.50|+0.50) using a previously described procedure to improve interpretability (Engemann et al., 2018).
Although the above chance-level performance of univariate classifiers indicates that some features may be strongly related to different states of consciousness, the performance was not always consistent within- and cross-dataset (e.g., the dorsal attention and somatosensory networks’, DA-SS, connectivity feature had an AUC of 0.79 and 0.56, respectively). This suggests that inconsistent features may instead be closely associated with some unique aspect of anesthetic-induced unconsciousness, but does not necessarily entail information generalizable between the two.
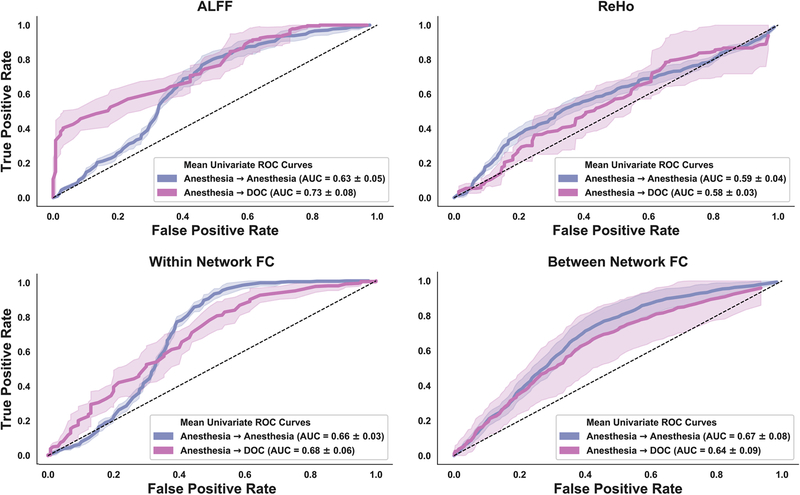
To approximate the overall performance within the four types of features (i.e., ALFF, ReHo, within network FC, between network FC), we quantified a representative ROC curve within each feature type as the mean across all its associated univariate ROC curves (Fig. 6). An analysis of the AUC from the representative ROC curves within-dataset revealed that the strongest overall performance came from between network FC features (M=0.67, SD=0.08), followed by within network FC (M=0.66, SD 0.03), ALFF (M=0.63, SD=0.05), and ReHo (M= 0.59, SD=0.04). In contrast, ALFF-based features showed the strongest overall performance cross-dataset (M=0.73, SD=0.08), followed by within network FC (M=0.68, SD=0.06), between network FC (M=0.64, SD=0.09), and ReHo (M=0.58, SD=0.03). Across both datasets, the ReHo-derived features performed the weakest, suggesting an overlap between groups as can be seen by examination of the ReHo value distributions (Fig. 4a middle).
Fig 6.
A receiver operating characteristic (ROC) curve, which plots a classifier’s true positive rate against the false positive rate, was calculated for each feature independently, both for within dataset classification (Anesthesia →Anesthesia; blue) and cross-dataset classification (Anesthesia → DOC; pink). The univariate ROC curves were subsequently averaged to yield a representative univariate ROC curve within each of the four analyses of functional connectivity. The representative ROC curve was used to determine the area under the curve (AUC), which served as the quantitative measure of univariate classifier performance. The dashed line represents chance-level performance. Shaded areas represent ± 1SD.
To ensure that the observed performance was not being driven by non-neural activity that may confound the BOLD signal we performed an analogous model-free univariate analysis using 13 features derived from head motion (standard deviation of head motion in 12 directions, Euclidean norm of all head motion parameters). Although the motion-based features performed slightly above chance-level within-dataset (M=0.66, SD=0.15), they had notably low performance cross-dataset (M=0.20, SD=0.08).
Model Performance
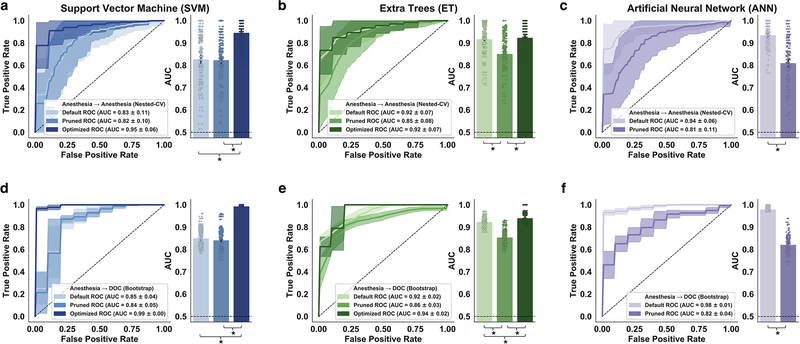
The three models all showed strong classification performance prior to feature pruning and hyperparameter optimization (Default) (within-dataset; cross-dataset): SVM (M=0.83, SD=0.11; M=0.85, SD=0.04; Fig. 7a,d), ET (M=0.92, SD=0.07; M=0.92, SD=0.02; Fig. 7b,e), ANN (M=0.94, SD=0.06; M=0.98, SD=0.01; Fig. 7c,f).
Fig 7.
Support vector machine (SVM), Extra Trees (ET), and artificial neural network (ANN) performance without hyperparameter optimization or feature selection (Default), with feature pruning only (Pruned), and with hyperparameter optimization only (Optimized). (a,b,c) Within-dataset reliability (Anesthesia → Anesthesia) for each model was evaluated using 100×5 nested cross-validation. (d,e,f) Cross-dataset generalizability (Anesthesia → DOC) was evaluated by testing the fully-trained models on 100 bootstrap samples of the DOC data. The solid lines represent the mean ROC’s across 100 evaluations. Shaded areas represent ± 1SD. The dashed line represents chance-level performance (AUC = 0.50). * indicates Bonferroni-corrected p < 0.05.
Two of the models showed significantly reduced classification performance within- and cross-dataset following feature pruning (Pruned) (within-dataset; cross-dataset): ET (t(99)=5.83, p<0.001; t(99)=16.55, p<0.001), ANN (t(99)=10.01, p<0.001; t(99)=38.10, p<0.001). In contrast, feature pruning did not appear to meaningfully affect the SVM model.
The two models which participated in hyperparameter optimization achieved a statistically significant increase in cross-dataset classification performance (Optimized): SVM (t(99)=3 3.51, p<0.001), ET (t(99)=5.48, p<0.001). Whereas hyperparameter optimization also improved within-dataset performance for the support vector machine (t(99)=8.55, p<0.001), no significant difference was observed with the ET model.
Overall, the SVM was most affected by the hyperparameter optimization, showing a marked increase in Post-Optimization vs Pre-Optimization performance (M:+0.12 within-dataset; M:+0.14 cross-dataset) and reduction in performance variability (SD:−0.05 within-dataset; SD:−0.04 cross-dataset).
Taken together, our results suggest that careful hyperparameter optimization is an essential step in constructing a robust machine learning classifier, particularly when using the SVM, and that automated methods for choosing appropriate hyperparameters (e.g., Hyperopt-Sklearn) may offer an effective, less-biased approach altogether more preferable than other manual tuning methods. Moreover, our results also suggest that pruning features based on observed group differences in the training dataset may worsen, rather than improve, classification performance within- and cross-dataset for some models.
Stress Tests
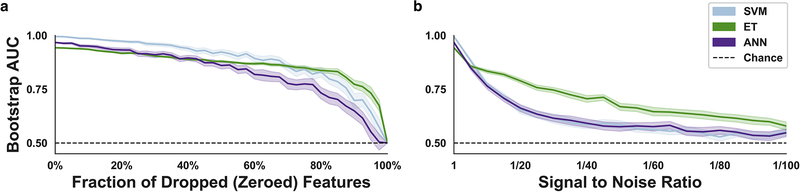
All models achieved near-optimal performance (AUC>0.95) both within- and cross-dataset following hyperparameter optimization. For this reason, we applied computational stress tests to explore which of the models continued to perform well when presented with sub-optimal data. As expected, classification performance (AUC) steadily declined as increasing numbers of features were randomly dropped from the test dataset (DOC). All three models preserved a relatively strong mean AUC (>0.80) until the number of features dropped (zeroed) exceeded 60–80% (Fig. 8a).
Fig 8.
Computational stress tests and analysis of feature importance. (a) Variable fractions of the functional connectivity features (0%−100%) were randomly dropped (zeroed) in the test dataset. The effect of random dropping was quantified using a mean area under the curve (AUC) analysis across 100 bootstrap samples of the DOC data before and after removal. (b) Performance across variable signal-to-noise ratios (1/1–1/100) was quantified using the previously described DOC sampling and testing procedure. Dotted line represents chance-level performance (AUC=0.50). Shaded areas represent ± 1 SD.
The second computational stress test, namely a systematic reduction of the signal to noise ratio (SNR), allowed us to simulate how each model responded to poor data quality (high levels of noise) (Fig. 8b). This analysis showed that the ET model retained the highest mean AUC across decreasing SNR’s, whereas the SVM and ANN models declined more rapidly to around chance-level performance: 1/25 (ET:~0.76; ANN:~0.63; SVM:~0.63), 1/50 (ET:~0.67; ANN:~0.58; SVM:~0.59), 1/100 (ET:~0.58; ANN:~0.55; SVM:~0.55).
The results of the computational stress tests suggest that the ET model is somewhat better-equipped to manage sub-optimal data; perhaps as a consequence of the model’s unique method of constructing numerous heterogeneous trees which introduce randomness into the model and subsequent averaging of predictions through the use of bootstrap aggregation.
Feature Importance
In order to better understand the particular features driving model performance, we performed an exploratory analysis of feature importance on both the SVM and ET models. Given that the optimized SVM was linear, we were able to quantify relative importance by examining the coefficients of the linear hyperplane (Fig. 9a); in line with previous recommendations, the coefficients of the hyperplane were squared (Guyon et al., 2002). Feature importance within the ET model is a readily accessible attribute of the model <sklearn.ensemble.ExtraTreesClassifier.feature_importances_> that represents how much a single feature contributes to decreasing the Gini impurity at each split (Fig. 9b).
Fig 9.
Exploratory post-hoc analysis of feature importance for the optimized support vector machine (SVM) and Extra Trees (ET) models. (a) Since the optimized SVM was linear, feature importance was quantified by squaring the weights of the coefficients used by the model. (b) Within the ET model, feature importance corresponded to how much each feature decreased the Gini impurity. Across both models, larger values (red) are associated with higher feature importance relative to features with lower values (blue).
This analysis indicated that the network-level analyses of functional connectivity, namely between network FC and within network FC, were the most informative features for the classification task across both models. Moreover, the ET model appeared to have used a wider set of features compared to the SVM.
Intermediate States
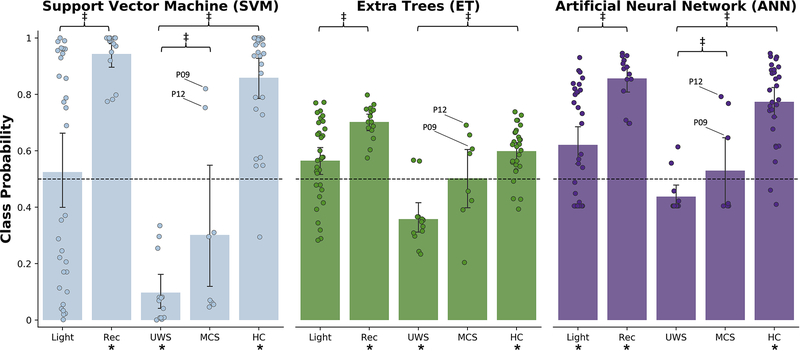
Across all three models, a similar pattern emerged with respect to the non-intermediate states (Fig. 10). Namely, the anesthesia recovery group (Rec) and healthy controls in the DOC dataset (HC) were reliably classified as awake; Rec (SVM: t(14)=20.18, p<0.001; ET: t(14)=12.86, p<0.001; ANN: t(14)=15.62, p<0.001), HC (SVM: t(27)=9.97, p<0.001; ET: t(27)=5.77, p<0.001; ANN: t(27)=9.99, p<0.001). In addition, the UWS/VS were generally classified as unresponsive; UWS/VS (SVM: t(12)=12.29, p<0.001; ET: t(12)=5.06, p<0.01; ANN: not significant), whereas the MCS classifications were indeterminate (SVM: not significant; ET: not significant; ANN: not significant). See Table 2 for a confusion matrix.
Fig 10.
Class assignment probability across models for subjects not included in the training data, from left to right: light anesthetic sedation (Light), recovery from anesthetic sedation (Rec), UWS/VS, MCS, healthy controls (HC) from the DOC dataset. Models were trained on the anesthesia datasets, such that 0 mapped to an unresponsive state and 1 mapped to an awake state. The predicted classification probabilities for each group were compared to binary decision threshold set at 0.5 to identify groups reliably classified as awake or unresponsive. A secondary analysis was performed to identify differences between the MCS and UWS/VS groups, the MCS and Wake groups, and the Light and Rec groups. * indicates uncorrected p < 0.05 for one sample t-test vs binary decision threshold. ‡ indicates uncorrected p < 0.05 for two sample t-test.
Table 2.
Confusion matrix for machine learning models
| Model Predictions | Actual Behavioral States | ||||
|---|---|---|---|---|---|
| SVM | Light | Rec | UWS | MCS | HC |
| Awake | 16 | 15 | 0 | 2 | 27 |
| Unresponsive | 16 | 0 | 13 | 6 | 1 |
| ET | Light | Rec | UWS | MCS | HC |
| Awake | 22 | 15 | 2 | 4 | 24 |
| Unresponsive | 10 | 0 | 11 | 4 | 4 |
| ANN | Light | Rec | UWS | MCS | HC |
| Awake | 19 | 15 | 2 | 3 | 26 |
| Unresponsive | 13 | 0 | 11 | 5 | 2 |
The value in each cell represents the number of subjects classified as Awake or Unresponsive across the groups not included in model training.
Secondary analyses revealed significant differences between the MCS group and the healthy controls in the DOC dataset across all three models (SVM: t(34)=5.76, p<0.001; ET: t(34)=5.62, p<0.001; ANN: t(34)=2.76, p<0.05), though significant differences between the MCS group and the UWS/VS group were only identified by the ANN model (t(19)=2.54, p<0.05). Further, we identified significant group differences between the subjects present in both the light anesthetic sedation and recovery from sedation groups for the ET and ANN models only (ET: t(14)=2.70, p<0.05; ANN: t(14)=2.98, p<0.05). Given that our analysis of intermediate states had a much smaller sample size, the p values reported in this section were uncorrected.
Upon examination of the predicted class probabilities, we identified two MCS subjects classified as awake by all three models (patient 9 and patient 12). After reviewing the Coma Recovery Scale-Revised (CRS-R) scores for each subject we discovered that these two subjects were among the highest scoring (CRS-R = 12); high scores are associated with a greater level of consciousness. Notably, patient 9 reportedly recovered (CRS-R = 23) two months after the scanning session.
Discussion
We demonstrated that the pipeline we developed—rs-fMRI feature extraction, model selection, hyperparameter optimization, and cross-validation in a pharmacologic state of unconsciousness—is sufficient for constructing a robust classifier that can be applied to pathologic states of unconsciousness. Further, our finding that MCS patients were classified as significantly different from the healthy controls suggests that there exist detectable differences in rs-fMRI activity during this intermediate state, and that, in principle, future models can be trained on these same rs-fMRI features to make graded distinctions between different levels of consciousness.
The examination of group differences at the level of single features derived from rs-fMRI activity revealed three primary conclusions. First, in line with previous studies showing the functional importance of various networks — including default-mode (Amico et al., 2014; Boly et al., 2009; Boveroux et al., 2010; Demertzi et al., 2014; Fernández-Espejo et al., 2012; Greicius et al., 2008; Huang et al., 2014; Kasahara et al., 2010; Monti et al., 2010; Norton et al., 2012; Roquet et al., 2016), frontoparietal (Boveroux et al., 2010), and salience (Guldenmund et al., 2013; Qin et al., 2015) — on the level of consciousness, we observed significantly reduced amplitude of low frequency fluctuations in those networks for both anesthesia and DOC data. We also identified consciousness-dependent breakdown of functional connectivity involving various cross-network functional connectivities. This is consistent with a role for cross-modal connectivity in consciousness via multisensory integration and top–down processes (Demertzi et al., 2015). Second, several features performed well as a model-free univariate classifier, discriminating between awake and unresponsive groups most of the time with a high degree of accuracy (e.g., connectivity between the cinguloopercular task control network and dorsal attention network, COTC-DA; within-dataset AUC: 0.74, cross-dataset AUC: 0.82), whereas others performed near chance-level (i.e., most ReHo-based features). Third, we discovered many features that performed inconsistently between the anesthesia and DOC datasets. This observed pattern of inconsistency suggests that some features may not be generalizable across datasets or linked to unconsciousness, per se, but rather might be an indicator of some other detectable change in neural activity during anesthetic-induced unconsciousness (or pathologically-induced unconsciousness).
In the past few years, an increasing cohort of studies has applied machine learning methods to examine the diagnostic value of imaging data in patients suffering from disorders of consciousness. A range of neuroimaging techniques have been utilized in this research area, including fMRI (Demertzi et al., 2015), fluorodeoxyglucose positron emission tomography (FDG-PET) (Phillips et al., 2011), and EEG (Chennu et al., 2017; Engemann et al., 2018; Sitt et al., 2014; van den Brink et al., 2018). It is noteworthy that, among the studies mentioned, resting state network-based fMRI could achieve a high discriminative accuracy (>80%) when distinguishing MCS from UWS patients (Demertzi et al., 2015). In our study, instead of training the classifier to make distinctions between MCS and UWS patients, we tested whether pharmacologic states of unconsciousness could have predictive value that generalized to pathologic states of unconsciousness (i.e., UWS patients). Accordingly, our classifiers were successful in separating conscious from unconscious subjects (>90%), a level of performance analogous to a prior FDG-PET study that reported a 100% classification accuracy when distinguishing locked-in patients from UWS patients (Phillips et al., 2011). Taken together, it seems feasible that machine learning approaches can be harnessed as tools to distinguish conscious from unconscious states. However, the classification of intermediate states (e.g., light sedation, MCS) remains challenging for several reasons. First, intermediate states of consciousness are ill defined if one conceives of consciousness as an all-or-none phenomenon. Second, the considerable inter-subject variability observed during sedation and MCS may necessitate larger sample sizes used for training machine learning models. Here, we applied a different strategy to test intermediate state classification, namely training models to distinguish consciousness from unconsciousness, and making predictions on unseen intermediate state data. Although our results of intermediate states classification were exploratory, they suggest that this pipeline could have clinical relevance if developed further.
The three candidate machine learning models we evaluated in the study—support vector machine (SVM), Extra Trees (ET), and an artificial neural network (ANN)—were chosen because of their growing popularity within the neuroimaging community and markedly distinct approach to classification. After training, each of the models tested achieved a notably high level of performance (AUC>0.95, both within- and cross-dataset). Thus, we find it reasonable to conclude that any of these models would likely be a suitable classifier for similar tasks.
Of interest, we observed near-identical performance on the validation dataset (DOC) compared to the training dataset (Anesthesia). The high performance observed across both datasets suggests that distinguishing conscious from unconscious states (using rs-fMRI features) was a relatively simple, straightforward classification. Our analysis of feature-level differences between these two states shows an often clear separation between these two groups (Fig. 4), an observation further supported by the high performance of the univariate classifiers—achieving an AUC as high as 0.87 on the DOC data (i.e., FPTC ALFF) and 0.81 on the Anesthesia data (i.e., SS-VIS between network connectivity).
There are, however, important considerations that may influence the process of model selection. First, the deep-learning based ANN was by far the most computationally demanding when it came to model training (a consequence of the backpropagation algorithm, which involves many repeated train-test epochs; see Keras documentation for a thorough review, https://keras.io), and most likely to overfit given a limited sample size. In contrast, whereas the SVM was simplest and efficient to construct, our analysis of the models before and after hyperparameter optimization revealed that the SVM was also most sensitive to hyperparameter choice. Though SVM is often used because of its relative simplicity, this illustrates that care should be taken to observe the ways in which SVM performance may change dramatically as a result of how it is constructed prior to training.
For these reasons, we believe the ET model to be a good compromise between the two—offering a good balance of computational efficiency, ease of construction, and general reliability. Finally, there is an added advantage for decision-tree-based models in particular, namely the ability to perform a post-hoc analysis of feature importance, which may help to inform feature selection in future studies. Our recommendation of this particular model is in line with other related research evaluating the ET’s ability to classify DOC patients by analyzing a wide range of distinct EEG-derived features (Engemann et al., 2018).
As part of our machine learning pipeline, we explored the relatively novel approach to hyperparameter tuning, namely automated optimization via Hyperopt-sklearn (Bergstra et al., 2015; Komer et al., 2018). Given that such methods are designed to reduce user bias in hyperparameter selection, avoid the time-intensive nature of manual hyperparameter search methods, and also provide strong gains in performance relative to default hyperparameter settings, we believe it to be a very valuable tool that will appreciate a growing application as others adopt these emerging techniques.
Contrary to our expectation, the feature pruning on the basis of observed group differences in the anesthesia dataset generally lowered performance. Here, we suspect that many of the features excluded from the pruned sub-set of features contained meaningful information used by the models. Given that careful feature selection remains a key step in constructing a machine learning model, our results suggest that elimination of redundant or non-informative features is better-achieved through methods like recursive feature elimination, in which model performance is iteratively tested with and without specific features.
Taken together, our analysis of single features, and the post-hoc exploration of SVM and ET feature importance, provides converging evidence that network-level measures of rs-fMRI activity (i.e., within network FC, between network FC) are especially relevant biomarkers for studying unconsciousness; the network-level measures tended to have high univariate model-free classification performance within- and cross-dataset, and were also identified as among the most highly important features within the ET model. Much of the recent research on neural correlates of consciousness similarly emphasizes the importance of long-range connectivity (Mashour and Hudetz, 2018) and network-level features (Amico et al., 2017; Crone et al., 2014; Fernández-Espejo et al., 2012; Fischer et al., 2016; Kotchoubey et al., 2013; Qin et al., 2015; Rosazza et al., 2016). Interestingly, though the network-level measures were generally what most separated conscious and unconscious states, we did not identify any particular networks that were universally different between the two. We propose two possible explanations for this observation: 1) the network features were derived from pre-defined network template (226 nodes), reduced the original spatial resolution from tens of thousands of voxels to hundreds. This relatively coarse estimation of brain activity may inevitably introduce inaccurate network assignment for different individuals due to inter-subject variability. 2) unconsciousness (whether induced pathologically or by anesthetics) may entail spatially diffuse, rather than focal, changes to network activity. Both explanations highlight the importance of using multivariate analyses; multivariate approaches help to address inter-subject variability (potentially, heterogeneity in DOC population) while also capturing the information from large-scale brain activity.
There are a few methodological limitations worth noting. First, during our analyses of within-and cross-dataset performance, we observed near-optimal performance both within- and cross-dataset across all three models. Though this was a positive result, it made subsequent comparisons between the three models difficult, as we did not observe a clear winner, or loser, among the three. Given that single features were, in some cases, also high performing univariate classifiers, we suspect that the discrimination task performed by the models was relatively straightforward—that is, there was usually clear separation between the two groups. This may also explain why we observed near-identical cross-dataset performance; in most machine learning applications, model performance is generally expected to decline when generalizing to novel data (relative to performance during training).
It is possible that, due to the simplicity of the classification, we achieved a sort of ceiling effect that obscured meaningful differences in how the three models would have performed on a more challenging task. Though we attempted to further delineate the models by application of computational stress tests, it is important to note the difficulty of assessing whether the differences observed are due to actual variation in model robustness, or rather, a consequence of how the different models make classifications.
Additionally, although our exploration of intermediate states indicated that the models treated the MCS group differently than the UWS/VS group and the healthy controls, only limited conclusions can be drawn. For one, since the models were not trained to execute a true multi-label classification, we cannot say that such a model would incontrovertibly achieve a similarly high level of performance when discriminating between MCS and UWS/VS or between MCS and healthy controls. Our analysis of the data collected during light anesthetic-sedation offered a preliminary indication that this group may serve as a future analog to MCS, however, as multi-label classification was not the primary goal of the study, that hypothesis was not explicitly tested and needs to be explored further.
In sum, our study both validates the use of anesthetic-induced unconsciousness as a surrogate model of study for pathologically induced unresponsiveness and establishes a pipeline for the use of rs-fMRI-based multivariate machine learning approaches to classification. In doing so, we hope to help pave the way towards a large sample verification study and the routine application of machine-learning in the clinical context.
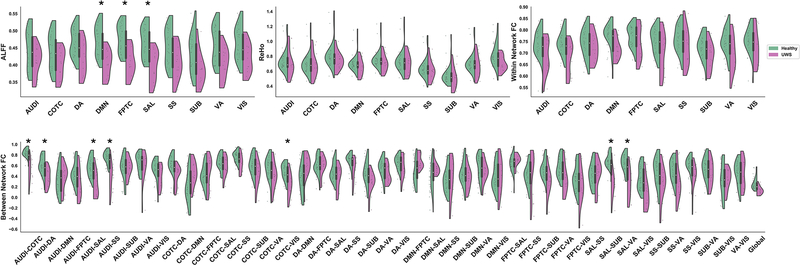
Fig 5.
Single feature comparisons between healthy controls and UWS/VS groups within DOC dataset. (a) Distribution of values for ALFF (upper-left), ReHo (upper-middle), within network FC (upper-right), and between network FC (bottom). * indicates Bonferroni-corrected p < 0.05.
Acknowledgements
This study was supported by a grant of the National Institute of General Medical Sciences of the National Institutes of Health under Award R01-GM103894, and by the Center for Consciousness Science, University of Michigan Medical School. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
We declare no conflict of interest for all authors.
References
- Abraham A, Pedregosa F, Eickenberg M, Gervais P, Mueller A, Kossaifi J, Gramfort A, Thirion B, Varoquaux G, 2014. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform 8, 14. doi: 10.3389/fninf.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkire MT, Hudetz AG, Tononi G, 2008. Consciousness and anesthesia. Science 322, 87680. doi : 10.1126/science.1149213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya S, Kunimatsu A, Saito N, Ohtomo K, 2013. Cerebral Hemodynamic Impairment: Assessment with Resting-State Functional MR Imaging. Radiology 270, 548–555. doi: 10.1148/radiol.13130982 [DOI] [PubMed] [Google Scholar]
- Amico E, Gomez F, Di Perri C, Vanhaudenhuyse A, Lesenfants D, Boveroux P, Bonhomme V, Brichant JF, Marinazzo D, Laureys S, 2014. Posterior cingulate cortex-related co-activation patterns: A resting state fMRI study in propofol-induced loss of consciousness. PLoS One 9, e100012. doi: 10.1371/journal.pone.0100012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amico E, Marinazzo D, Di Perri C, Heine L, Annen J, Martial C, Dzemidzic M, Kirsch M, Bonhomme V, Laureys S, Goñi J, 2017. Mapping the functional connectome traits of levels of consciousness. Neuroimage 148, 201–211. doi : 10.1016/j.neuroimage.2017.01.020 [DOI] [PubMed] [Google Scholar]
- Anderson JS, Druzgal TJ, Lopez-Larson M, Jeong EK, Desai K, Yurgelun-Todd D, 2011. Network anticorrelations, global regression, and phase-shifted soft tissue correction. Hum. Brain Mapp. 32, 919–934. doi: 10.1002/hbm.21079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein T, Niklison J, Sigman L, Manes F, Leiguarda R, Armony J, Owen A, Carpintiero S, Olmos L, 2004. Emotion processing in the minimally conscious state. J. Neurol. Neurosurg. Psychiatry 75, 788–793. doi: 10.1136/jnnp.2003.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein T, Tiberti C, Niklison J, Tamashiro M, Ron M, Carpintiero S, Villarreal M, Forcato C, Leiguarda R, Manes F, 2005. Assessing level of consciousness and cognitive changes from vegetative state to full recovery. Neuropsychol. Rehabil 15, 307–322. doi: 10.1080/09602010443000443 [DOI] [PubMed] [Google Scholar]
- Bekinschtein TA, Dehaene S, Rohaut B, Tadel F, Cohen L, Naccache L, 2009. Neural signature of the conscious processing of auditory regularities. Proc. Natl. Acad. Sci. U. S. A 106, 1672–1677. doi: 10.1073/pnas.0809667106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstra J, Bengio Y, 2012. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res 13, 281–305. doi: 10.1162/153244303322533223 [DOI] [Google Scholar]
- Bergstra J, Komer B, Yamins D, Eliasmith C, Cox DD, 2015. Hyperopt: A Python library for model selection and hyperparameter optimization. Comput. Sci. Discov 8. doi: 10.1088/1749-4699/8/1/014008 [DOI] [Google Scholar]
- Boly M, Faymonville M-E, Peigneux P, Lambermont B, Damas P, Del Fiore G, Degueldre C, Franck G, Luxen A, Lamy M, Moonen G, Maquet P, Laureys S, 2004. Auditory Processing in Severely Brain Injured Patients: Differences Between the Minimally Conscious State and the Persistent Vegitative State. Arch. Neurol 61, 233–238. doi: 10.1001/archneur.61.2.233 [DOI] [PubMed] [Google Scholar]
- Boly M, Faymonville M-E, Schnakers C, Peigneux P, Lambermont B, Phillips C, Lancellotti P, Luxen A, Lamy M, Moonen G, Maquet P, Laureys S, 2008. Perception of pain in the minimally conscious state with PET activation: an observational study. Lancet Neurol. 7, 1013–1020. doi: 10.1016/S1474-4422(08)70219-9 [DOI] [PubMed] [Google Scholar]
- Boly M, Massimini M, Garrido MI, Gosseries O, Noirhomme Q, Laureys S, Soddu A, 2012. Brain Connectivity in Disorders of Consciousness. Brain Connect. 2, 1–10. doi: 10.1089/brain.2011.0049 [DOI] [PubMed] [Google Scholar]
- Boly M, Massimini M, Tononi G, 2009. Theoretical approaches to the diagnosis of altered states of consciousness. Prog. Brain Res. doi: 10.1016/S0079-6123(09)17727-0 [DOI] [PubMed] [Google Scholar]
- Bonhomme V, Vanhaudenhuyse A, Demertzi A, Bruno MA, Jaquet O, Bahri MA, Plenevaux A, Boly M, Boveroux P, Soddu A, Brichant JF, Maquet P, Laureys S, 2016. Resting-state Network-specific Breakdown of Functional Connectivity during Ketamine Alteration of Consciousness in Volunteers. Anesthesiology 125, 873–888. doi: 10.1097/ALN.0000000000001275 [DOI] [PubMed] [Google Scholar]
- Boveroux P, Vanhaudenhuyse A, Bruno M-A, Noirhomme Q, Lauwick S, Luxen A, Degueldre C, Plenevaux A, Schnakers C, Philips C, Brichant JF, Bonhomme V, Maquet P, Greicius MD, Laureys S, Boly M, 2010. Breakdown of within- and between-network Resting State Functional Magnetic Resonance Imaging Connectivity during Propofol-induced Loss of Consciousness. Anesthesiology 113, 1038–1053. [DOI] [PubMed] [Google Scholar]
- Caruana R, Niculescu-Mizil A, 2006. An Empirical Comparison of Supervised Learning Algorithms. Proc. 23rd Int. Conf. Mach. Learn. - ICML ‘06. doi: 10.1145/1143844.1143865 [DOI] [Google Scholar]
- Cawley GC, Talbot NLC, 2010. On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. J. Mach. Learn. Res 11, 2079–2107. [Google Scholar]
- Chen P, Xie Q, Wu X, Huang H, Lv W, Chen L, Guo Y, Zhang S, Hu H, Wang Y, Nie Y, Yu R, Huang R, 2018. Abnormal Effective Connectivity of the Anterior Forebrain Regions in Disorders of Consciousness. Neurosci. Bull 647–658. doi: 10.1007/s12264-018-0250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennu S, Annen J, Wannez S, Thibaut A, Chatelle C, Cassol H, Martens G, Schnakers C, Gosseries O, Menon D, Laureys S, 2017. Brain networks predict metabolism, diagnosis and prognosis at the bedside in disorders of consciousness. Brain 140, 21202132. doi: 10.1093/brain/awx163 [DOI] [PubMed] [Google Scholar]
- Chernik DA, Gillings D, Laine H, Hendler J, Silver JM, Davidson AB, Schwam EM, Siegel JL, 1990. Validity and reliability of the Observer’s Assessment of Alertness/Sedation Scale: Study with Intravenous Midazolam. J Clin Psychopharmacol. [PubMed] [Google Scholar]
- Coleman MR, Davis MH, Rodd JM, Robson T, Ali A, Owen AM, Pickard JD, 2009. Towards the routine use of brain imaging to aid the clinical diagnosis of disorders of consciousness. Brain 132, 2541–2552. doi: 10.1093/brain/awp183 [DOI] [PubMed] [Google Scholar]
- Crone JS, Soddu A, Höller Y, Vanhaudenhuyse A, Schurz M, Bergmann J, Schmid E, Trinka E, Laureys S, Kronbichler M, 2014. Altered network properties of the frontoparietal network and the thalamus in impaired consciousness. NeuroImage Clin. 4, 240–248. doi: 10.1016/j.nicl.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demertzi A, Antonopoulos G, Heine L, Voss HU, Crone JS, De Los Angeles C, Bahri MA, Di Perri C, Vanhaudenhuyse A, Charland-Verville V, Kronbichler M, Trinka E, Phillips C, Gomez F, Tshibanda L, Soddu A, Schiff ND, Whitfield-Gabrieli S, Laureys S, 2015. Intrinsic functional connectivity differentiates minimally conscious from unresponsive patients. Brain 138, 2619–2631. doi: 10.1093/brain/awv169 [DOI] [PubMed] [Google Scholar]
- Demertzi A, Gómez F, Crone JS, Vanhaudenhuyse A, Tshibanda L, Noirhomme Q, Thonnard M, Charland-Verville V, Kirsch M, Laureys S, Soddu A, 2014. Multiple fMRI system-level baseline connectivity is disrupted in patients with consciousness alterations. Cortex 52, 35–46. doi: 10.1016/j.cortex.2013.11.005 [DOI] [PubMed] [Google Scholar]
- Demertzi A, Soddu A, Faymonville M-E, Bahri MA, Gosseries O, Vanhaudenhuyse A, Phillips C, Maquet P, Noirhomme Q, Luxen A, Laureys S, 2011. Hypnotic modulation of resting state fMRI default mode and extrinsic network connectivity. Prog. Brain Res. 193, 309–322. doi: 10.1016/B978-0-444-53839-0.00020-X [DOI] [PubMed] [Google Scholar]
- Demertzi A, Tagliazucchi E, Dehaene S, Deco G, Barttfeld P, Raimondo F, Martial C, Fernández-Espejo D, Rohaut B, Voss HU, Schiff ND, Owen AM, Laureys S, Naccache L, Sitt JD, 2019. Human consciousness is supported by dynamic complex patterns of brain signal coordination. Sci. Adv 5, eaat7603. doi: 10.1126/sciadv.aat7603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Perri C, Bahri MA, Amico E, Thibaut A, Heine L, Antonopoulos G, Charland-Verville V, Wannez S, Gomez F, Hustinx R, Tshibanda L, Demertzi A, Soddu A, Laureys S, 2016. Neural correlates of consciousness in patients who have emerged from a minimally conscious state: A cross-sectional multimodal imaging study. Lancet Neurol. 15, 830–842. doi: 10.1016/S1474-4422(16)00111-3 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG, 2016. TensorFlow: A System for Large-Scale Machine Learning, in: Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ‘16) pp. 265–283. doi: 10.1016/0076-6879(83)01039-3 [DOI] [Google Scholar]
- Efron B, Tibshirani R, 2007. Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Stat. Sci 1, 54–75. doi: 10.1214/ss/1177013817 [DOI] [Google Scholar]
- Engemann DA, Raimondo F, King J-R, Rohaut B, Louppe G, Faugeras F, Annen J, Cassol H, Gosseries O, Fernandez-Slezak D, Laureys S, Naccache L, Dehaene S, Sitt JD, 2018. Robust EEG-based cross-site and cross-protocol classification of states of consciousness. Brain 141, 3179–3192. doi: 10.1093/brain/awy251 [DOI] [PubMed] [Google Scholar]
- Fernández-Espejo D, Rossit S, Owen AM, 2015. A Thalamocortical Mechanism for the Absence of Overt Motor Behavior in Covertly Aware Patients. JAMA Neurol. 72, 1442–50. doi: 10.1001/jamaneurol.2015.2614 [DOI] [PubMed] [Google Scholar]
- Fernández-Espejo D, Soddu A, Cruse D, Palacios EM, Junque C, Vanhaudenhuyse A, Rivas E, Newcombe V, Menon DK, Pickard JD, Laureys S, Owen AM, 2012. A role for the default mode network in the bases of disorders of consciousness. Ann. Neurol 72, 335–343. doi: 10.1002/ana.23635 [DOI] [PubMed] [Google Scholar]
- Fins JJ, Schiff ND, Foley KM, 2007. Late recovery from the minimally conscious state: Ethical and policy implications. Neurology 68, 304–307. doi: 10.1212/01.wnl.0000252376.43779.96 [DOI] [PubMed] [Google Scholar]
- Fischer DB, Boes AD, Demertzi A, Evrard HC, Laureys S, Edlow BL, Liu H, Saper CB, Pascual-Leone A, Fox MD, Geerling JC, 2016. A human brain network derived from coma-causing brainstem lesions. Neurology 87, 1–8. doi: 10.1212/WNL.0000000000003404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME, 2009. The Global Signal and Observed Anticorrelated Resting State Brain Networks Michael. J. Neurophysiol 101, 3270–3283. doi: 10.1152/jn.90777.2008.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts P, Ernst D, Wehenkel L, 2006. Extremely randomized trees. Mach. Learn 63, 3–42. doi: 10.1007/s10994-006-6226-1 [DOI] [Google Scholar]
- Giacino JT, Fins JJ, Laureys S, Schiff ND, 2014. Disorders of consciousness after acquired brain injury: the state of the science. Nat. Rev. Neurol 10, 99–114. doi: 10.1038/nrneurol.2013.279 [DOI] [PubMed] [Google Scholar]
- Giacino JT, Kalmar K, Whyte J, 2004. The JFK Coma Recovery Scale-Revised: Measurement characteristics and diagnostic utility. Arch. Phys. Med. Rehabil 85, 2020–2029. doi: 10.1016/j.apmr.2004.02.033 [DOI] [PubMed] [Google Scholar]
- Greicius MD, Kiviniemi V, Tervonen O, Vainionpää V, Alahuhta S, Reiss AL, Menon V, 2008. Persistent default-mode network connectivity during light sedation. Hum. Brain Mapp. 29, 839–47. doi: 10.1002/hbm.20537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldenmund P, Demertzi A, Boveroux P, Boly M, Vanhaudenhuyse A, Bruno M-A, Gosseries O, Noirhomme Q, Brichant J-F, Bonhomme V, Laureys S, Soddu A, 2013. Thalamus, brainstem and salience network connectivity changes during propofol-induced sedation and unconsciousness. Brain Connect. 3, 273–85. doi: 10.1089/brain.2012.0117 [DOI] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V, 2002. Gene Selection for Cancer Classification using Support Vector Machines. Mach. Learn 46, 389–422. doi: 10.1002/bip.360320308 [DOI] [Google Scholar]
- Hecht-Nielsen R, 1989. Theory of the Backpropagation Neural Network, in: International 1989 Joint Conference on Neural Networks pp. 593–605. doi: 10.1016/0893-6080(88)90469-8 [DOI] [Google Scholar]
- Heine L, Soddu A, Gómez F, Vanhaudenhuyse A, Tshibanda L, Thonnard M, Charland-Verville V, Kirsch M, Laureys S, Demertzi A, 2012. Resting state networks and consciousness Alterations of multiple resting state network connectivity in physiological, pharmacological, and pathological consciousness states. Front. Psychol 3. doi: 10.3389/fpsyg.2012.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. Bin, 2003. Learning Capability and Storage Capacity of Two-Hidden-Layer Feedforward Networks, in: IEEE Transactions on Neural Networks. pp. 274–281. doi: 10.1109/TNN.2003.809401 [DOI] [PubMed] [Google Scholar]
- Huang Z, Liu X, Mashour GA, Hudetz AG, 2018a. Timescales of intrinsic BOLD signal dynamics and functional connectivity in pharmacologic and neuropathologic states of unconsciousness. J. Neurosci 38, 2545–17. doi: 10.1523/JNEUROSCI.2545-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Vlisides PE, Tarnal VC, Janke EL, Keefe KM, Collins MM, McKinney AM, Picton P, Harris RE, Mashour GA, Hudetz AG, 2018b. Brain imaging reveals covert consciousness during behavioral unresponsiveness induced by propofol. Sci. Rep 8, 13195. doi: 10.1038/s41598-018-31436-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Wang Z, Zhang JJ, Dai R, Wu J, Li Y, Liang W, Mao Y, Yang Z, Holland G, Zhang JJ, Northoff G, 2014. Altered temporal variance and neural synchronization of spontaneous brain activity in anesthesia. Hum. Brain Mapp. 35, 5368–5378. doi: 10.1002/hbm.22556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhang JJ, Wu J, Qin P, Wu X, Wang Z, Dai R, Li Y, Liang W, Mao Y, Yang Z, Zhang JJ, Wolff A, Northoff G, 2016. Decoupled temporal variability and signal synchronization of spontaneous brain activity in loss of consciousness: An fMRI study in anesthesia. Neuroimage 124, 693–703. doi: 10.1016/j.neuroimage.2015.08.062 [DOI] [PubMed] [Google Scholar]
- Huang Z, Zhang Jun, Wu J, Liu X, Xu J, Zhang Jianfeng, Qin P, Dai R, Yang Z, Mao Y, Hudetz AG, Northoff G, 2018c. Disrupted neural variability during propofol-induced sedation and unconsciousness. Hum. Brain Mapp. doi: 10.1002/hbm.24304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Menon DK, Salmond CH, Outtrim JG, Taylor Tavares JV, Carpenter TA, Pickard JD, Sahakian BJ, Stamatakis EA, 2010. Altered functional connectivity in the motor network after traumatic brain injury Background: a large proportion of survivors of traumatic brain injury (TBI) have persistent cognitive. Neurology 75, 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma Diederik, P., Ba JL, 2015. Adam: A Method for Stochastic Optimization, in: ICLR 2015. pp. 1–15. doi: 10.1063/1.4902458 [DOI] [Google Scholar]
- Komer B, Bergstra J, Eliasmith C, 2018. Hyperopt-Sklearn: Automatic Hyperparameter Configuration for Scikit-Learn, in: Proceedings of the 13th Python in Science Conference. SciPy, pp. 32–37. doi: 10.25080/majora-14bd3278-006 [DOI] [Google Scholar]
- Kotchoubey B, Merz S, Lang S, Markl A, Müller F, Yu T, Schwarzbauer C, 2013. Global functional connectivity reveals highly significant differences between the vegetative and the minimally conscious state. J. Neurol 260, 975–983. doi: 10.1007/s00415-012-6734-9 [DOI] [PubMed] [Google Scholar]
- Laureys S, Schiff ND, 2012. Coma and consciousness: Paradigms (re)framed by neuroimaging. Neuroimage 61, 478–491. doi: 10.1016/j.neuroimage.2011.12.041 [DOI] [PubMed] [Google Scholar]
- Lecun Y, Bengio Y, Hinton G, 2015. Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- Liu X, Lauer KK, Douglas Ward B, Roberts C, Liu S, Gollapudy S, Rohloff R, Gross W, Chen G, Xu Z, Binder JR, Li SJ, Hudetz AG, 2017a. Propofol attenuates low-frequency fluctuations of resting-state fMRI BOLD signal in the anterior frontal cortex upon loss of consciousness. Neuroimage 147, 295–301. doi: 10.1016/j.neuroimage.2016.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lauer KK, Ward BD, Roberts CJ, Liu S, Gollapudy S, Rohloff R, Gross W, Xu Z, Chen G, Binder JR, Li S-J, Hudetz AG, 2017b. Fine-grained parcellation of brain connectivity improves differentiation of states of consciousness during graded propofol sedation. Brain Connect. 7, 373–381. doi: 10.1089/brain.2016.0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, 2016. A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw. Model. Anal. Heal. Informatics Bioinforma. 5. doi: 10.1007/s13721-016-0125-6 [DOI] [Google Scholar]
- Mäki-Marttunen V, Cortes JM, Villarreal MF, Chialvo DR, 2013. Disruption of transfer entropy and inter-hemispheric brain functional connectivity in patients with disorder of consciousness. Front. Neuroinform 7, 1–11. doi: 10.1186/1471-2202-14-s1-p83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh B, Morton N, Kenny GNC, 1991. Pharmacokinetic model driven infusion of propofol in children. Br. J. Anaesth 67, 41–48. doi: 10.1093/Bja/67.1.41 [DOI] [PubMed] [Google Scholar]
- Mashour GA, Avidan MS, 2013. Capturing covert consciousness. Lancet (London, England) 381, 271–2. doi: 10.1016/S0140-6736(13)60094-X [DOI] [PubMed] [Google Scholar]
- Mashour GA, Hudetz AG, 2018. Neural Correlates of Unconsciousness in Large-Scale Brain Networks. Trends Neurosci. 41, 150–160. doi: 10.1016/j.tins.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashour GA, Kent C, Picton P, Ramachandran SK, Tremper KK, Turner CR, Shanks A, Avidan MS, 2013. Assessment of Intraoperative Awareness with Explicit Recall. Anesth. Analg 116, 889–891. doi: 10.1213/ane.0b013e318281e9ad [DOI] [PubMed] [Google Scholar]
- Mashour GA, Shanks A, Tremper KK, Kheterpal S, Turner CR, Ramachandran SK, Picton P, Schueller C, Morris M, Vandervest JC, Lin N, Avidan MS, 2012. Prevention of intraoperative awareness with explicit recall in an unselected surgical population. Anesthesiology 117, 717–725. doi: 10.1097/ALN.0b013e31826904a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti MM, Vanhaudenhuyse A, Coleman MR, Boly M, Pickard JD, Tshibanda L, Owen AM, Laureys S, 2010. Willful modulation of brain activity in disorders of consciousness. N. Engl. J. Med 362, 579–589. doi: 10.1056/NEJMoa0905370 [DOI] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA, 2009. The impact of global signal regression on resting state correlations: Are anti-correlated networks introduced? Neuroimage 44, 893–905. doi: 10.1016/j.neuroimage.2008.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy PR, Boonstra E, Nieuwenhuis S, 2016. Global gain modulation generates time-dependent urgency during perceptual choice in humans. Nat. Commun 7, 1–14. doi: 10.1038/ncomms13526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton L, Hutchison RM, Young GB, Lee DH, Sharpe MD, Mirsattari SM, 2012. Disruptions of functional connectivity in the default mode network of comatose patients. Neurology 78, 175–181. doi: 10.1212/WNL.0b013e31823fcd61 [DOI] [PubMed] [Google Scholar]
- Owen AM, 2013. Detecting consciousness: a unique role for neuroimaging. Annu. Rev. Psychol 64, 109–33. doi: 10.1146/annurev-psych-113011-143729 [DOI] [PubMed] [Google Scholar]
- Owen AM, Coleman MR, Boly M, Davis MH, Laureys S, Pickard JD, 2006. Detecting awareness in the vegetative state. Science (80-. ). 313, 1402. doi: 10.1126/science.1130197 [DOI] [PubMed] [Google Scholar]
- Palanca BJA, Mitra A, Larson-Prior L, Snyder AZ, Avidan MS, Raichle ME, 2015. Resting-state functional magnetic resonance imaging correlates of sevoflurane-induced unconsciousness. Anesthesiology 123, 346–356. doi: 10.1097/ALN.0000000000000731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E, 2011. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res 12, 2825–2830. doi: 10.1007/s13398-014-0173-7.2 [DOI] [Google Scholar]
- Phillips CL, Bruno MA, Maquet P, Boly M, Noirhomme Q, Schnakers C, Vanhaudenhuyse A, Bonjean M, Hustinx R, Moonen G, Luxen A, Laureys S, 2011. “Relevance vector machine” consciousness classifier applied to cerebral metabolism of vegetative and locked-in patients. Neuroimage 56, 797–808. doi: 10.1016/j.neuroimage.2010.05.083 [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE, 2011. Functional network organization of the human brain. Neuron 72, 665–678. doi: 10.1016/j.neuron.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Wu Xuehai, Huang Z, Duncan NW, Tang W, Wolff A, Hu J, Gao L, Jin Y, Wu Xing, Zhang Jianfeng, Lu L, Wu C, Qu X, Mao Y, Weng X, Zhang Jun, Northoff G, 2015. How are different neural networks related to consciousness? Ann. Neurol 78, 594–605. doi: 10.1002/ana.24479 [DOI] [PubMed] [Google Scholar]
- Ramsay MA, Savege TM, Simpson BR, Goodwin R, 1974. Controlled sedation with alphaxalone-alphadolone. Br. Med. J 2, 656–9. doi: 10.1136/bmj.2.5920.656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquet D, Foucher JR, Froehlig P, Renard F, Pottecher J, Besancenot H, Schneider F, Schenck M, Kremer S, 2016. Resting-state networks distinguish locked-in from vegetative state patients. NeuroImage Clin. 12, 16–22. doi: 10.1016/j.nicl.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosazza C, Andronache A, Sattin D, Bruzzone MG, Marotta G, Nigri A, Ferraro S, Sebastiano DR, Porcu L, Bersano A, Benti R, Leonardi M, D’Incerti L, Minati L, Coma Research Centre (CRC) - Besta Institute, 2016. Multimodal study of default-mode network integrity in disorders of consciousness. Ann. Neurol 79, 841–853. doi: 10.1002/ana.24634 [DOI] [PubMed] [Google Scholar]
- Saad ZS, Gotts SJ, Murphy K, Chen G, Jo HJ, Martin A, Cox RW, 2012. Trouble at Rest: How Correlation Patterns and Group Differences Become Distorted After Global Signal Regression. Brain Connect. 2, 25–32. doi: 10.1089/brain.2012.0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RD, Gaskell A, Raz A, Winders J, Stevanovic A, Rossaint R, Boncyk C, Defresne A, Tran G, Tasbihgou S, Meier S, Vlisides PE, Fardous H, Hess A, Bauer RM, Absalom A, Mashour GA, Bonhomme V, Coburn M, Sleigh J, 2017. Incidence of Connected Consciousness after Tracheal Intubation: A Prospective, International, Multicenter Cohort Study of the Isolated Forearm Technique. Anesthesiology 126, 214–222. doi: 10.1097/aln.0000000000001479 [DOI] [PubMed] [Google Scholar]
- Sarica A, Cerasa A, Quattrone A, 2017. Random Forest Algorithm for the Classification of Neuroimaging Data in Alzheimer’s Disease: A Systematic Review. Front. Aging Neurosci. 9, 329. doi: 10.3389/fnagi.2017.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnakers C, Ledoux D, Majerus S, Damas P, Damas F, Lambermont B, Lamy M, Boly M, Vanhaudenhuyse A, Moonen G, Laureys S, 2008. Diagnostic and prognostic use of bispectral index in coma, vegetative state and related disorders. Brain Inj. 22, 926–931. doi: 10.1080/02699050802530565 [DOI] [PubMed] [Google Scholar]
- Schnakers C, Perrin F, Schabus M, Hustinx R, Majerus S, Moonen G, Boly M, Vanhaudenhuyse A, Bruno MA, Laureys S, 2009a. Detecting consciousness in a total locked-in syndrome: An active event-related paradigm. Neurocase 15, 271–277. doi: 10.1080/13554790902724904 [DOI] [PubMed] [Google Scholar]
- Schnakers C, Vanhaudenhuyse A, Giacino J, Ventura M, Boly M, Majerus S, Moonen G, Laureys S, 2009b. Diagnostic accuracy of the vegetative and minimally conscious state: Clinical consensus versus standardized neurobehavioral assessment. BMC Neurol. 9, 35. doi: 10.1186/1471-2377-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroter MS, Spoormaker VI, Schorer A, Wohlschlager A, Czisch M, Kochs EF, Zimmer C, Hemmer B, Schneider G, Jordan D, Ilg R, 2012. Spatiotemporal Reconfiguration of Large-Scale Brain Functional Networks during Propofol-Induced Loss of Consciousness. J. Neurosci 32, 12832–12840. doi: 10.1523/jneurosci.6046-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer S, 1996. STANPUMP User’s Manual. Stanford Univ; Stanford, CA. [Google Scholar]
- Silva S, Alacoque X, Fourcade O, Samii K, Marque P, Woods R, Mazziotta J, Chollet F, Loubinoux I, 2010. Wakefulness and loss of awareness: Brain and brainstem interaction in the vegetative state. Neurology 74, 313–320. doi: 10.1212/WNL.0b013e3181cbcd96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitt JD, King JR, El Karoui I, Rohaut B, Faugeras F, Gramfort A, Cohen L, Sigman M, Dehaene S, Naccache L, 2014. Large scale screening of neural signatures of consciousness in patients in a vegetative or minimally conscious state. Brain 137, 2258–2270. doi: 10.1093/brain/awu141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soddu A, Boly M, Nir Y, Noirhomme Q, Vanhaudenhuyse A, Demertzi A, Arzi A, Ovadia S, Stanziano M, Papa M, Laureys S, Malach R, 2009. Reaching across the abyss: recent advances in functional magnetic resonance imaging and their potential relevance to disorders of consciousness. Prog. Brain Res. doi: 10.1016/S0079-6123(09)17718-X [DOI] [PubMed] [Google Scholar]
- Sours C, Zhuo J, Roys S, Shanmuganathan K, Gullapalli RP, 2015. Disruptions in resting state functional connectivity and cerebral blood flow in mild traumatic brain injury patients. PLoS One 10, 1–20. doi: 10.1371/journal.pone.0134019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C, Hutter F, Hoos HH, Leyton-Brown K, 2012. Auto-WEKA: Combined Selection and Hyperparameter Optimization of Classification Algorithms.
- van den Brink RL, Nieuwenhuis S, van Boxtel GJM, van Luijtelaar G, Eilander HJ, Wijnen VJM, 2018. Task-free spectral EEG dynamics track and predict patient recovery from severe acquired brain injury. NeuroImage Clin. 17, 43–52. doi: 10.1016/j.nicl.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannez S, Heine L, Thonnard M, Gosseries O, Laureys S, 2017. The Repetition of Behavioral Assessments in Diagnosis of Disorders of Consciousness. Ann. Neurol 81, 883–889. doi: 10.1002/ana.24962 [DOI] [PubMed] [Google Scholar]
- Zang Y, Jiang T, Lu Y, He Y, Tian L, 2004. Regional homogeneity approach to fMRI data analysis. Neuroimage 22, 394–400. doi: 10.1016/j.neuroimage.2003.12.030 [DOI] [PubMed] [Google Scholar]
- Zang YF, Yong H, Chao-Zhe Z, Qing-Jiu C, Man-Qiu S, Meng L, Li-Xia T, Tian-Zi J, Yu-Feng W, 2007. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 29, 83–91. doi: 10.1016/j.braindev.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Zou Q-H, Zhu C-Z, Yang Y, Zuo X-N, Long X-Y, Cao Q-J, Wang Y-F, Zang Y-F, 2008. An improved approach to detectipon of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J. Neurosci. Methods 172, 1–11. doi: 10.3174/ajnr.A1256.Functional [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X-N, Jiang L, Yang Z, Cao X-Y, He Y, Zang Y-F, Castellanos FX, Milham MP, 2013. Toward reliable characterization of functional homogeneity in the human brain: Preprocessing, scan duration, imaging resolution and computational space Xi-Nian. Neuroimage 65, 374–386. doi: 10.1016/j.pestbp.2011.02.012.Investigations [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The resting-state fMRI feature data and code for the above machine-learning pipeline are accessible at https://github.com/Justin-Campbell/ML-Anes-DOC.