SUMMARY
Innate behaviors involve both reflexive motor programs and enduring internal states, but how these responses are coordinated by the brain is not clear. In Drosophila, male-specific P1 interneurons promote courtship song, as well as a persistent internal state that prolongs courtship and enhances aggressiveness. However, P1 neurons themselves are not persistently active. Here we identify pCd neurons as persistently active, indirect P1 targets that are required for P1-evoked persistent courtship and aggression. Acute activation of pCd neurons alone is inefficacious, but enhances and prolongs courtship or aggression promoted by female cues. Brief female exposure induces a persistent increase in male aggressiveness, an effect abrogated by interruption of pCd activity. pCd activity is not sufficient but necessary for persistent physiological activity, implying an essential role in a persistence network. Thus, P1 neurons coordinate both command-like control of courtship song, and a persistent internal state of social arousal mediated by pCd neurons.
In Brief
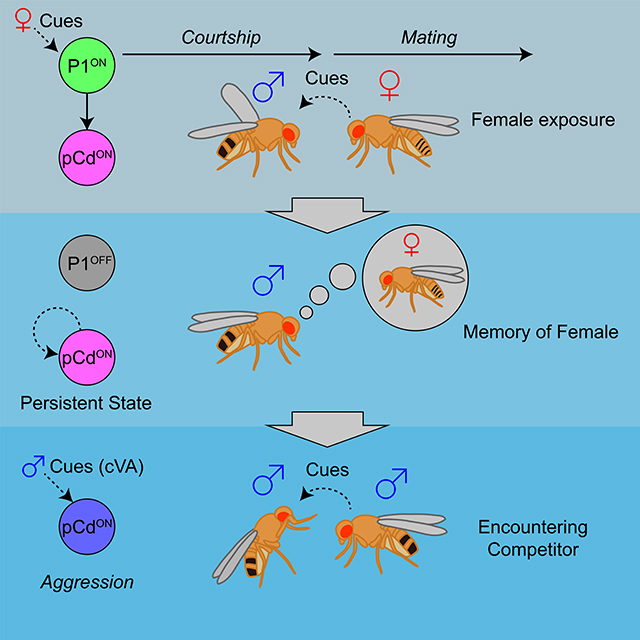
How do brains coordinate rapid reflex responses and persistent internal states underlying innate behaviors? Jung et al. describe a circuit node in male fly brains that is activated by female sensory cues, and which triggers both immediate courtship behavior and persistent arousal via parallel pathways. The persistent state induced by female exposure is maintained in part by pCd neurons, even after such exposure is terminated, and can be integrated with male-specific chemosensory cues that promote aggression.
Graphical Abstract
INTRODUCTION
Animal behaviors triggered by specific sensory cues evolve over multiple time-scales, from rapid reflex reactions to more enduring responses accompanied by changes in internal state (Tinbergen, 1951; Bargmann, 2012). The former allow quick survival reactions, while the latter afford time to integrate contextual and other influences on behavioral decisions. How these reflexive and integrative pathways are coordinated by neural circuits remains poorly understood. One useful feature of integrative responses is that they allow behaviors to persist on time-scales beyond the duration of the triggering sensory stimulus (Anderson and Adolphs, 2014). Studies in C. elegans have identified neuromodulatory circuits involving serotonin and PDF, which control persistent states of roaming vs. dwelling associated with exploration vs. exploitation of food resources (Anderson and Adolphs, 2014). In mice, transient activation of agouti-related peptide (AgRP) -expressing neurons in the arcute nucleus of the hypothalamus promotes persistent food-seeking behavior, an effect mediate by neuropeptide Y (NPY) signaling (Chen et al., 2016; Chen et al., 2019). Transient activation of steroidogenic factor 1 (SF1) neurons in the dorsomedial/central portion of the ventromedial hypothalamus (VMHdm/c) promotes persistent defensive behaviors (Kunwar et al., 2015). However the circuit-level mechanisms underlying these persistent effects are not well understood.
In Drosophila melanogaster, male-specific P1 interneurons (Yamamoto and Koganezawa, 2013) are activated by female-specific pheromones (Kohatsu et al., 2011; Clowney et al., 2015; Kallman et al., 2015), and control male courtship behaviors such as singing (Pan et al., 2011; von Philipsborn et al., 2011), as well as internal states that regulate aggression (Hoopfer et al., 2015), mating (Kohatsu and Yamamoto, 2015; Zhang et al., 2016), feeding (Zhang et al., 2018b) and sleep (Chen et al., 2017) (reviewed in Ref. (Auer and Benton, 2016)). Artificial stimulation of P1 neurons in solitary males can trigger rapid-onset courtship song (Pan et al., 2011; von Philipsborn et al., 2011; Inagaki et al., 2014). Nevertheless, singing persists for minutes after stimulation offset (Bath et al., 2014; Inagaki et al., 2014; Clowney et al., 2015). Similarly, the effect of P1 activation to promote inter-male aggressiveness endures for minutes after photostimulation offset (Hoopfer, 2016; Watanabe et al., 2017).
In contrast to these persistent behavioral effects, optogenetically evoked P1 physiological activity, measured via calcium imaging in live, head-fixed flies, returns to baseline in tens of seconds (Inagaki et al., 2014; Hoopfer et al., 2015) (although it has been reported to persist in brain explants (Zhang et al., 2018a)). These data suggest that persistent behavioral states evoked by P1 stimulation are not encoded in P1 neurons themselves, but rather in one or more of their downstream targets. We therefore sought to identify such persistently activated P1 targets, and to understand their functional role in the encoding of persistent behavioral states.
RESULTS
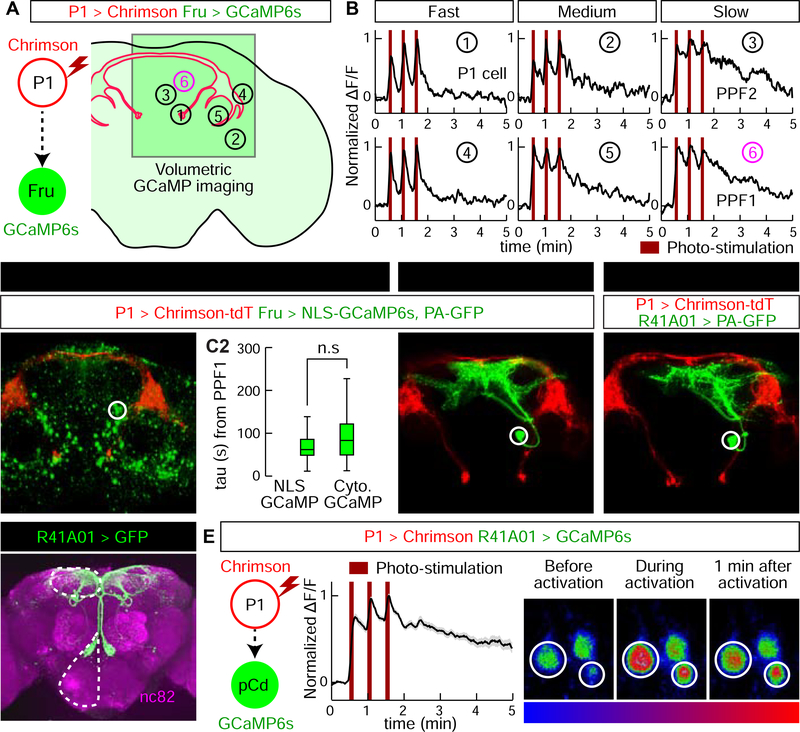
To search for P1 follower cells exhibiting persistent responses, we expressed the red-shifted opsin Chrimson (Klapoetke et al., 2014) in P1a-split GAL4 neurons (Hoopfer et al., 2015; Anderson, 2016; Hoopfer, 2016), and a calcium indicator (GCaMP6s (Chen et al., 2013)) in ~2,000 Fruitless (Fru)-LexA (Mellert et al., 2010) neurons (Figure 1A). Optogenetic stimulation was calibrated to activate P1 cells at a level comparable to that evoked in these cells by female abdomen touching in the same preparation. Fru+ cells activated by P1 stimulation were identified by volumetric imaging (30 4-μm optical sections covering a 250 μm × 250 μm × 120 μm volume; Figure S1D, E). On average, we monitored activity of 191 Fru+ cell somata and identified ~37 cells per fly that responded to P1 stimulation (≥2/3 trials evoking a peak ΔF/F response >4σ above baseline; Figure S1F), in 14 distinct brain regions. Different putative P1 follower cells showed different response durations, in a continuous distribution ranging from those similar to P1 (τ~15 s; see Methods) to those lasting much longer (Figure 1B, Figure S1G, I). We used several criteria to select cells for further study: 1) median tau value > 5-fold that of P1 (τ>~75 s); 2) persistent P1 responses detected in >75% of tested flies (n=12); 3) >2 cells/fly per hemibrain; 4) cells genetically accessible using specific GAL4 drivers.
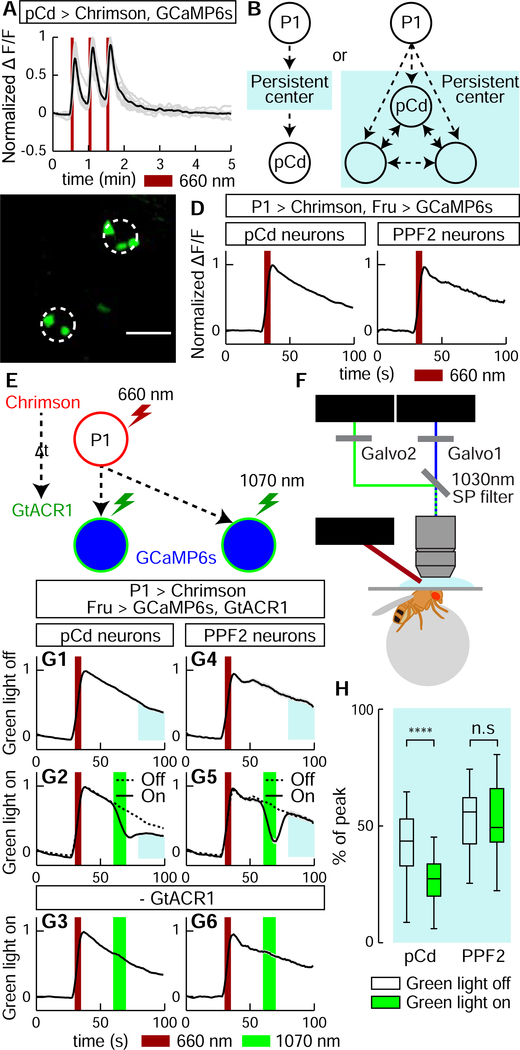
Figure 1. Identification of P1 follower cells with long-lasting responses.
(A) Experimental schematic. Green square indicates imaging field containing different putative P1 follower cells (numbered circles). (B) Representative GCaMP6s traces (normalized ΔF/F ); numbers correspond to cells in (A). PPF1 cells (➅) are pCd neurons. 655 nm light (10 Hz, 10 ms pulse-width, 25 s inter-stimulation interval) was delivered for Chrimson stimulation (dark red bars). (C1–4) Identification of GAL4 driver labeling PPF1 (pCd) neurons (See Figure S2A for details). (C1) LexAop-NLS-GCaMP expressed in Fru-LexA neurons; white circle, PPF1 somata. (C2) Comparison between NLS GCaMP6s and Cytoplasmic GCaMP6s. Decay constants (tau) were calculated by curve fitting (See Figure S1I and Methods for details). n=32 trials, 11 cells from 7 flies (NLS GCaMP), 77 cells from 12 flies (Cytoplasmic GCaMP). Statistical significance in this and in all other figures (unless otherwise indicated) was calculated using a Mann-Whitney U-test. Boxplots throughout show the median (center line), 25th and 75th percentiles (box), and 1.5 times the interquartile range (whiskers). Outliers were defined as data points falling outside 1.5x the interquartile range of the data, and were excluded from plots for clarity, but not from statistical analyses. (C3) PPF1 projections revealed by Fru-LexA>PA-GFP activation (Datta et al., 2008). (C4) PPF1 neurons labeled by R41A01-LexA>PA-GFP. Non-PPF1 PA-GFP and NLS-GCaMP basal fluorescence have been masked for clarity. All images in C1, C2, and C4 are maximum intensity z-projections of 2-pm optical sections acquired by 2-P imaging. (D) Central brain R41A01 Gal4 neurons revealed by UAS-myr::GFP reporter. Superior medial protocerebrum (SMP) and sub-esophageal zone (SEZ) are indicated by dashed outlines. (E) LexAop-GCaMP6s response of pCd neurons labeled by R41A01-LexA following P1-Gal4/UAS-Chrimson stimulation (see Supp. Table 1 for genotypes). Left, schematic; middle, normalized ΔF/F trace (n=23 trials, 15 cells from 10 flies; mean±sem); right, fluorescent images taken before, during, and 1 minute after P1 activation (averaged over 5 frames). White circles indicate two responding cells.
We identified several putative persistent P1 follower (PPF) cells, which met the first criterion. These neurons were present in ~5 distinct clusters, each containing ~1–3 PPF cells, within a relatively small brain region (see Figure 1A. Cells in one such cluster, PPF1 (Figure 1B, #6) exhibited a median τ~83 s (Figure S1G, H). Cells in three other clusters including PPF2 (Figure 1B, #3), showed a median τ>~75, but failed to meet the second and third criteria. Another cluster in addition to PPF1 met all 3 criteria, but was not genetically accessible.
To gain specific genetic access to PPF1 neurons, we first examined the anatomy of these cells by combining P1 stimulation-evoked GCaMP imaging with photo-activatable GFP (PA-GFP) labeling of responding cells (Datta et al., 2008). We generated a nuclear-localized GCaMP (NLS-GCaMP6s) to prevent cytoplasmic GCaMP signal from obscuring PA-GFP fluorescence (Figure 1C1). NLS-GCaMP6s also detected persistent responses to P1 stimulation in PPF1 cells (Figure 1C2). We then focused a 720 nm two-photon laser on the identified PPF1 cells, and revealed their projection pattern via diffusion of activated PA-GFP (Datta et al., 2008) (Figure 1C3). By comparing the morphology of PPF1 neurons with Fru-MARCM (Cachero et al., 2010; Chiang et al., 2011) and Gal4 line image databases (Jenett et al., 2012), we identified two Gal4 drivers, R41A01 and R21D06, which labeled morphologically similar neurons (Figure 1C4, D; Figure S2A–D). To verify that R41A01 and R21D06 indeed label PPF1 neurons, we performed functional imaging in R41A01>GCaMP6s or R21D06>GCaMP6s flies, and confirmed persistent responses to P1 activation in PPF1 somata (Figure 1E and Figure S2C); whether such persistent responses are present in all neurites is difficult to ascertain. Interestingly, these neurons exhibited stepwise integration of P1 input (Figure 1E); however repeated P1 stimulation trials (as done in volume imaging, 30 trials, Figure 1B) sensitized PPF1 neurons (Figure S3).
Gal4 line R41A01 labels a cell cluster called pCd, previously reported to play an important role in female sexual receptivity (Zhou et al., 2014). Analysis of marker expression indicated that pCd cells are cholinergic neurons (Diao et al., 2015) that express both Fru and Dsx (Figure S2F–I), two sex-determination factors that label neurons involved in male courtship and aggression (Manoli et al., 2005; Stockinger et al., 2005; Rideout et al., 2007; Kimura et al., 2008; Pan et al., 2011; Yamamoto and Koganezawa, 2013). pCd neurons project densely to the superior-medial protocerebrum (SMP), while extending an additional long fiber bundle ventrally to innervate the dorsal region of the subesophageal zone (SEZ; Figure 1D). Double labeling of pCd neurons with somatodendritic (Denmark-RFP (Nicolai et al., 2010)) and pre-synaptic (Syt-GFP (Zhang et al., 2002)) markers revealed that their SMP projections are mostly dendritic, while their pre-synaptic terminals are located in the SMP and the SEZ (Figure S4D). Registration of P1 pre-synaptic labeling with pCd somatodendritic labeling in a common brain template failed to reveal clear overlap (Figure S4G–I), and application of the GFP Reconstitution Across Synaptic Partner (GRASP (Feinberg et al., 2008)) technique failed to detect close proximity between P1 and pCd neurons (Figure S4J–R), suggesting that functional connectivity between these cells is unlikely to be monosynaptic.
pCd neuronal activity is required for P1-induced persistent social behaviors
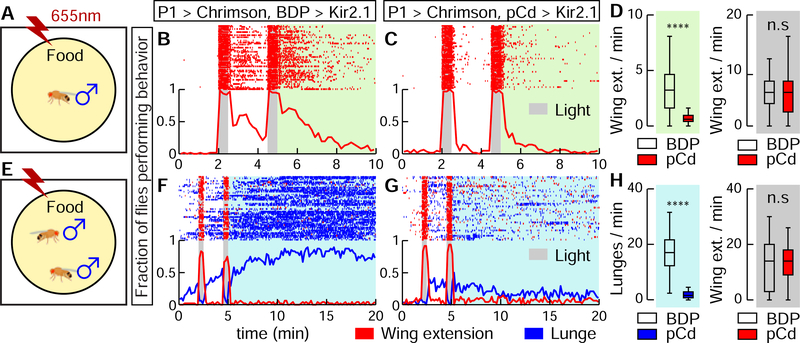
To test whether Pl-evoked persistent social behaviors require pCd activity, we silenced the latter using R41A01-LexA>LexAop-Kir2.1 while activating P1a-split GAL4 neurons using UAS-Chrimson. In solitary males (Figure 2A), silencing pCd neurons dramatically reduced persistent wing extension evoked by Chrimson activation of P1 cells (Figure 2B vs. 2C, green shading; 2D). Importantly, time-locked wing-extension during photostimulation was unaffected (Figure 2B–D, gray shading). Persistent aggression evoked by P1 activation in pairs of males (Hoopfer et al., 2015; Watanabe et al., 2017) (Figure 2E, F) was also strongly reduced by silencing pCd neurons (Figure 2G, H, blue shading), while wing-extension during photostimulation was unaffected. This result was confirmed using a more specific R41A01∩R21D06 intersectional split-GAL4 driver (Figure S2D) to silence pCd neurons, and R15A01-LexA to activate P1 cells (Figure S5). Thus pCd activity is required for enduring, but not for time-locked, behavioral responses to P1 activation.
Figure 2. Activity of pCd neurons is required for P1-induced persistent behaviors.
(A) Schematic (approximately to scale). Chrimson activation at 655 nm (Inagaki et al., 2014) was performed in solitary males on food. (B, C) Behavior of flies during (gray shading) and after (green shading) P1a (Hoopfer et al., 2015; Anderson, 2016) neuronal activation, either without (B; BDP is enhancerless LexA control driver), or with (C; pCd-LexA) Kir2.1-mediated (Baines et al., 2001) inhibition of pCd neurons. Grey bars, 30 s photostimulations (40 Hz, 10 ms pulse-width) at 2 min intervals. Upper: Wing extension raster plot (red ticks). Lower: fraction of flies performing wing extensions (red line) in 10 s time bins. n=62 (B), 63 (C). (D) Wing extension frequency per fly after (green shading) or during (grey shading) photostimulation. **** P < 0.0001. (E) As in (A), but using male pairs. (F, G) Plot properties as in (B, C). Grey bars, 30 s photostimulation periods (2 Hz, 10 ms pulse-width) at 2 min intervals. Upper: raster plot showing wing extensions (red ticks) and lunges (blue ticks). Lower: fraction of flies performing wing extensions (red line) or lunges (blue line) in 20 s time bins. n=48 for each genotypes. (H) Lunge frequency after photostimulation (light blue shading, left), and wing extension frequency during photostimulation (grey shading, right). Lunging during, and wing extension after photostimulation were ≤ 1 event/min and are omitted for clarity. Statistics as in (D).
pCd neurons amplify and prolong, but do not trigger, social behaviors
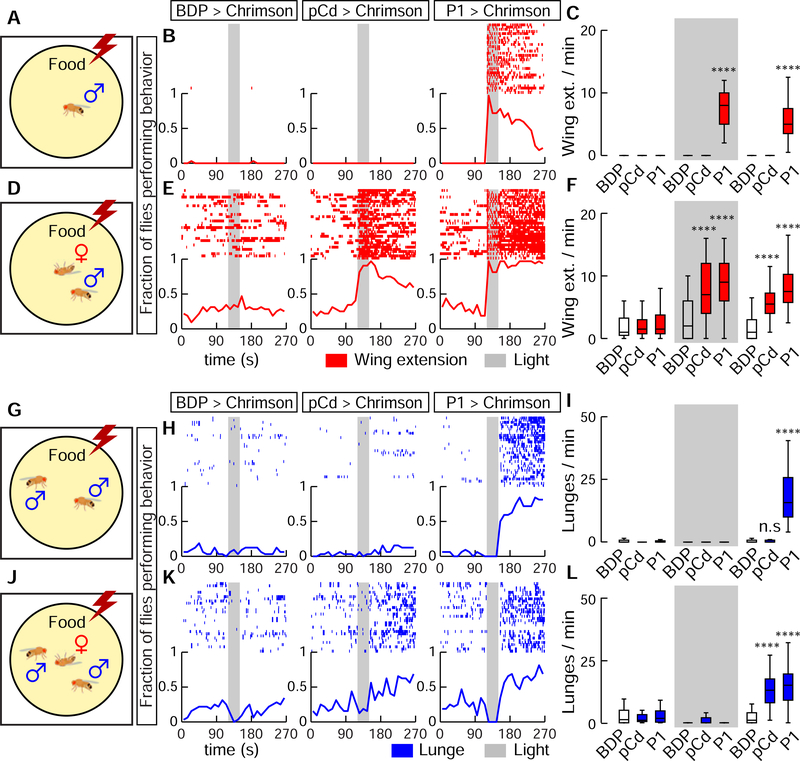
We next investigated the effect on behavior of optogenetically stimulating pCd neurons. Interestingly, optogenetic activation of pCd neurons in solitary flies had no visible effect, in contrast to optogenetic activation of P1 neurons (Inagaki et al., 2014; Clowney et al., 2015; Hoopfer et al., 2015) (Figure 3A, B). Persistent internal states can change an animal’s behavioral response to sensory cues. We reasoned that if pCd neurons promote such a persistent internal state, then their optogenetic activation, while insufficient to evoke behavior on its own, might nevertheless suffice to modify the behavioral response of the flies to an external social stimulus. To test this, we examined the effect of pCd stimulation on the behavioral response of males to female cues (Figure 3D). Activation of pCd neurons in the presence of a dead female dramatically elevated courtship behavior during photostimulation, and this effect persisted for several minutes after stimulus offset (Figure 3B vs. 3E, pCd>Chrimson; Figure 3C, F, pCd).
Figure 3. Activation of pCd neurons amplifies and extends male social behaviors induced by female cues.
(A, D, G, and H), experimental schematics illustrating optogenetic activation of pCd neurons in solitary males (A-F) or pairs of group-housed males (G-L), tested without (A-C, G-I) or with (D-F, J-L) a dead female. Raster plots and fraction of flies performing behaviors (red and blue lines, 10 s time bins) are shown in (B, E, H, and K). Plot properties same as in Figure 2. Grey bars, 30 s Chrimson activation at 655 nm (10 Hz, 10 ms pulse-width). Quantification and statistical tests shown in (C, F, I, and L). n=32 flies each. Statistical test used was a Kruskal-Wallis test. **** Dunn’s corrected P < 0.0001 for between-genotype comparisons. Courtship data are omitted in (H, K) for clarity.
Activation of pCd neurons in pairs of non-aggressive group-housed male flies did not promote aggression, unlike P1 activation (Hoopfer et al., 2015) (Figure 3G–I). But in the presence of a dead female, which produced increased baseline aggression in male flies (Lim et al., 2014), activation of pCd neurons significantly enhanced (fly aggressiveness after photostimulation, an effect not observed in photostimulated controls (Figure 3J–L). Thus, unlike P1 activation, which can substitute for the effect of dead females to trigger courtship or aggression, pCd activation alone cannot do so (Figure 3B–C, H-I). However pCd neuron activation can enhance and extend the effect of a dead female to promote these social behaviors.
pCd neurons are required for sustained courtship and aggressive drive
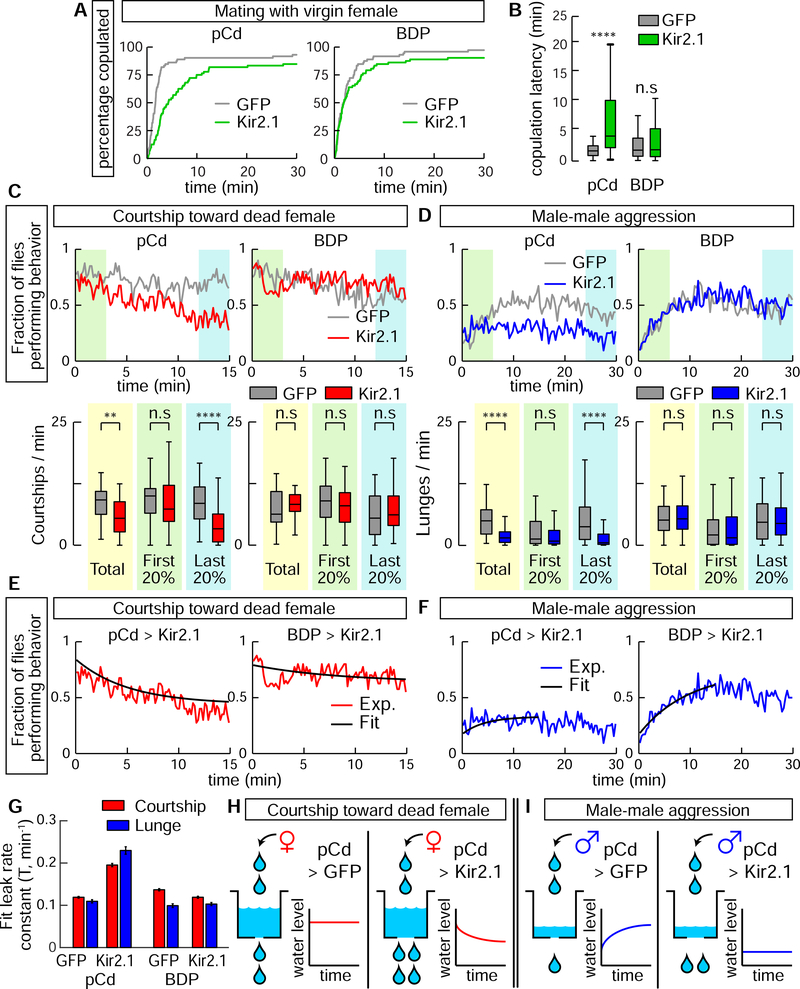
Given that pCd neuronal activity is required for optogenetic P1 activation-evoked social behavior (Figure 2), we next investigated its requirement during natural social behavior. Silencing pCd neurons significantly increased the latency to copulation (Figure 4A, B). To examine the effect of silencing on courtship per se, without rapid progression to copulation, we tested males in the presence of a freeze-killed virgin female, which induced robust unilateral wing-extensions (UWEs; courtship song (Tauber and Eberl, 2003)). In controls (BDP-GAL4> Kir2.1 or GFP), the fraction of flies exhibiting UWEs was relatively constant across the 15 min assay (Figure 4C, BDP, gray and red lines). However in pCd>Kir2.1 flies, UWEs declined significantly during that interval, in comparison to pCd>GFP controls (Figure 4C, pCd, red line, green vs blue shading).
Figure 4. Inhibition of pCd neurons increases copulation latency and reduces endurance of naturally occurring social behaviors.
(A) Individual males of the indicated genotypes were paired with a live wild-type virgin female. Cumulative percentage of flies that copulated over 30 min is shown. (B) Quantification and statistical tests for copulation latency. **** P < 0.0001 for between-genotype comparisons (Mann-Whitney U-test). (C) Solitary male flies were incubated with a dead female and courtship (unilateral wing extension bouts, UWEs) measured over 15 min. Left panels show experimental (pCd>Kir2.1, red line) and responder control (UAS-GFP, grey line) flies, right panels show enhancerless driver controls (BDP-Gal4; red and grey lines). Upper: fraction of flies performing behavior in 10 s time bins. Lower: number of UWE bouts per min per fly over entire 15 min observation (yellow shading), first (green shading) and last (blue shading) 20% (3 min) of the interval. n=40 flies per genotype. ** P < 0.01, **** P < 0.0001. (D) Pairs of single-housed males monitored over 30 min. Plot properties and statistical tests same as in (C), except blue color indicates lunging. Fraction of flies performing behavior was binned in 20 s time intervals. n=64 flies per genotypes. (E, F) Curve fitting of (E) courtship data from (C), or (F) lunging data from (D). Black lines show exponential fit curve for each experiment. Goodness of fit (MSE): courtship; 0.0042 (pCd>GFP), 0.0051 (pCd>Kir2.1), 0.0056 (BDP>GFP), 0.0058 (BDP>Kir2.1); Aggression; 0.0028 (pCd>GFP), 0.0031 (pCd>Kir2.1), 0.0045 (BDP>GFP), 0.0029 (BDP>Kir2.1). (G) Leak rate constants derived from curve fitting in (E, F); note that both courtship and lunging in pCd>Kir2.1 flies are best fit by assuming increased leak constants, relative to genetic controls. (H, I) Illustration of modeling results. Water level represents level of activity in a hypothetical leaky integrator driving behavior (Lorenz and Leyhausen, 1973). Inhibition of pCd activity with Kir2.1 increases leak rate constant of the integrator.
We next performed parallel experiments for aggression. Single-housed (SH) male flies will fight on food in the absence of females (Wang et al., 2008; Lim et al., 2014), and the intensity of fighting escalates over time (Figure 4D, BDP). However in SH pCd>Kir2.1 flies, aggression did not escalate over time, although initial levels of lunging were similar to controls (Figure 4D, pCd, blue line, green vs. blue shading). These data demonstrate a requirement for pCd neurons in escalated aggression, independent of any influence from females. Importantly, in both assays, silencing pCd neurons did not impair initiation of social behavior, consistent with the inability of pCd optogenetic stimulation to trigger these behaviors (Figure C, D); rather it influenced their amplitude and kinetics.
The effect of pCd silencing on courtship vs. aggression was subtly different: in the former case, silencing pCd neurons caused UWEs to steadily decline over time, whereas during aggression, natural escalation failed to occur (Figure 4C vs. 4D, pCd, red vs. blue lines). To investigate whether a common mechanism could explain both phenotypes, we asked whether both data could be jointly fit by a “leaky integrator” model (Chaudhuri and Fiete, 2016). Such models formalize classical “hydraulic” theories of behavioral drive (Lorenz and Leyhausen, 1973), in which the instantaneous level of activity in a neural integrator circuit determines either the rate or type of an animal’s behavior; here, we sought to fit the time-evolving rate of UWEs (Figure 4E), or of lunging (Figure 4F). Our leaky integrator model assumed that flies received sensory input from conspecifics with a rate constant R, and that the activity of the neural circuit integrating conspecific sensory cues decayed from its initial condition to steady-state with a “leak” rate constant T (min−1).
The behavioral data in each assay were well fit by models in which the only free parameter allowed to vary by genotype was T (Figure 4E–G). For UWEs, in control flies the relatively flat line reflects the fact that the initial rate of behavior is high, and already close to the steady-state where “fill” and “leak” rates are equal (Figure 4H, left). In contrast, the faster decline of UWEs in pCd>Kir2.1 flies (Figure 4E) was best fit by an increase in T (Figure 4G, red bars). During aggression, control flies exhibit escalation (Figure 4F, BDP>Kir2.1) because the initial rate of aggression is low, and the sensory input rate constant R is greater than T for this behavior (Figure 4I, left). Increasing T in pCd>Kir2.1 flies therefore converts aggression to a relatively flat line (Figure 4F; 4I, right). Thus, the superficially different courtship vs. aggression phenotypes caused by silencing pCd neurons can be explained by a common mechanism, whereby inhibition of pCd neurons increases the leak rate constant of a neural integrator, which may control a state of social arousal or drive (Anderson and Adolphs, 2014; Anderson, 2016).
pCd neurons display neural integrator properties
We next investigated whether pCd neurons display integrator properties at the level of their physiology. The observation that they exhibit stepwise summation of P1 input (Figure 1E, Figure S3A) is consistent with this idea. Surprisingly, repeated direct stimulation of PPF1 neurons did not exhibit such summation, and evoked faster-decaying responses (median τ~13.4 s) than evoked by indirect P1 activation (median τ~83 s), indicating that persistent activity cannot be triggered cell autonomously (Figure 5A). However, pCd function might be necessary, although not sufficient, for persistent activity (Figure 5B, right). If so, then persistent pCd activity should not recover from transient inhibition performed during the decay phase following P1 stimulation (Guo et al., 2017; Inagaki et al., 2019). Alternatively, if pCd cells simply “inherit” persistence passively from an upstream input (Figure 5B, left), their persistent P1 response should recover following transient inhibition. We therefore stimulated P1 neurons (5 s) while imaging from pCd cells, and after a short delay (25 s) briefly (~10 s) inhibited pCd activity using the green light-sensitive inhibitory opsin GtACR1 (Mohammad et al., 2017) and 2-photon spiral scanning (Rickgauer and Tank, 2009) at 1070 nm to restrict inhibition to pCd cells (Figure 5E, F and Methods).
Figure 5. pCd neuronal activity is required for physiological persistence.
(A) pCd response to direct optogenetic stimulation is not persistent. Gray lines depict individual pCd cell responses (n=27 from 9 flies), black line shows the mean for all cells. Dark red bars, Chrimson stimulation (655 nm light (10 Hz, 10 ms pulse-width, 25 s inter-stimulation interval). (B) Schematic illustrating alternatives tested by the experiment in (E-H). Light blue shading depicts hypothetical persistence-encoding network (“center”). If pCd neurons simply inherit persistence passively from the center (left), then persistence should rebound following transient pCd silencing. If persistence does not rebound, it implies that pCd activity is required for the center to maintain persistence (right). (C) Representative two-photon image showing cell body locations of pCd and PPF2 neurons expressing Fruitless>GCaMP6s in vivo. Dashed white circles indicate spiral scanning area for GtACR actuation in (E-H). Maximum intensity projection of 5 × 4 μm optical sections, averaged over 10 frames. (D) Normalized ΔF/F traces from pCd (left, n=36 trials from 8 flies), and PPF2 (right, n=29 trials from 5 flies) neurons upon P1 activation. Mean±sem. Dark red bar indicates P1 photostimulation (5 s, 10 Hz, 10 ms pulse-width, 660 nm LED). (E) Experimental schematic. pCd or PPF2 neuron cell bodies are locally photo-inhibited with GtACR1 (~10 s, spiral scanning, see Methods for details) after a delay (At, 25 s) following P1 activation (5 s). (F) Schematic illustrating imaging setup with 1070 nm 2-photon laser for GtACR1 photo-inhibition, and 920 nm 2-photon laser for in vivo GCaMP imaging. (G) Normalized ΔF/F from pCd neurons (G1-G3), and PPF2 neurons (G4-G6) with GtACR actuation (green bars) applied during Pl-induced persistent phase. G1 and G4: without photo-inhibition; G3 and G6, 1070 nm irradiation without GtACR1 expression. Dashed lines in G2 and G5 are mean of G1 and G4 traces, respectively. n=36 trials from 8 flies for pCd neurons, and 16 (5 flies) for PPF2 neurons. n=40 (8 pCd flies), 29 (6 PPF2 flies) for genetic controls. Mean±sem. (H) Normalized area under the curve (blue shaded regions in (G)) after photo-inhibition. **** P < 0.0001.
Actuation of GtACR1 in pCd neurons following P1 stimulation caused a rapid, ~68% decrease in ΔF/F signal, which did not recover to control levels following the offset of inhibition, but rather remained flat (Figure 5G2, blue shaded area, solid vs. dashed line and Figure 5H, pCd, green bar). This effect is not due to irreversible damage to pCd neurons by photo-inhibition, since reactivation of P1 neurons following transient pCd inhibition reliably re-evoked pCd persistent activity, and multiple cycles of P1 stimulation with or without GtACR1 actuation could be performed with consistent results (Figure S6A–B, pCd). Furthermore, 2-photon spiral scanning at 1070 nm of pCd neurons lacking GtACR1 had no effect (Figure 5G3), confirming that the decrease in GCaMP signal is due to inhibition of activity by GtACR1 and not to 2-photon irradiation. As the experiment was originally performed using Fru-LexA to label pCd cells, we confirmed the result using a pCd-specific driver (Figure S6C–E, blue shading).
As an additional control, we also performed the same manipulation on PPF2 neurons, another FruM+ population located near pCd (Figure 5C), which also showed persistent responses to P1 activation (Figure 1B, #3; Figure 5D, PPF2). In this case, following GtACR inhibition PPF2 activity quickly recovered to the level observed at the equivalent time-point in controls without 1070 nm photo-inhibition (Figure 5G5, 5H, PPF2 and Figure S6B, PPF2). Thus, PPF2 activity is not required continuously to maintain a persistent response to P1 activation. In contrast, persistence in pCd neurons requires their continuous activity. However the fact that persistent activity cannot be evoked by direct stimulation of pCd neurons alone suggests that persistence likely requires co-activation of a network comprised of multiple neurons.
pCd neurons are required for an effect of females to persistently enhance male aggressiveness
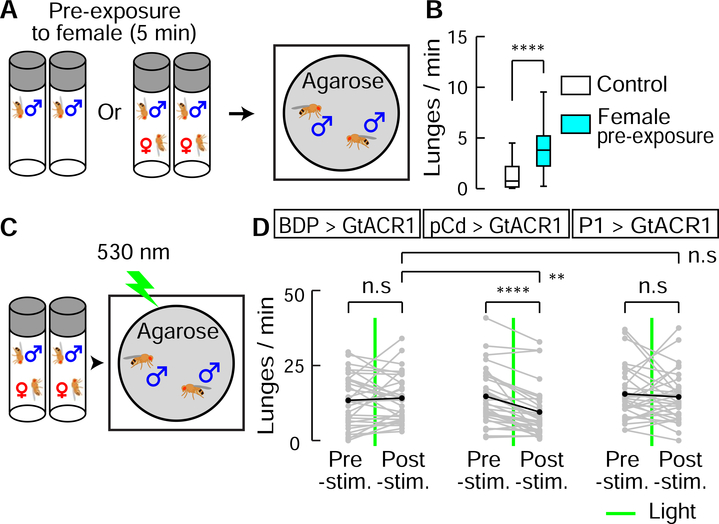
The foregoing data indicated that pCd neurons are required to maintain a P1 activation-triggered persistent internal state, which prolongs wing extension in solitary males and promotes aggression when male flies encounter another male. We next asked whether pCd neurons are similarly required for a persistent internal state triggered by naturalistic cues. Since P1 neurons are activated by female cues (reviewed in ref. (Auer and Benton, 2016)), we examined the influence of transient female exposure on male aggressive behavior. Previous studies have demonstrated that females can enhance inter-male aggression (ref. (Lim, 2014; Lim et al., 2014) and see Figure 3H vs. K), but whether this effect can persist following the removal of females was not clear. To investigate this, we pre-incubated individual male flies for 5 min with or without a live female, and then gently transferred them into an agarose-covered arena to measure their aggression (Figure 6A). Male flies pre-incubated with a female showed significantly higher levels of lunging than controls (Figure 6B), indicating a persistent influence of female exposure to enhance aggressiveness.
Figure 6. Role of pCd neurons in a female-induced enhancement of male aggressiveness.
(A) Schematic illustrating female induced inter-male aggression experiment. Single-housed male flies were pre-incubated in vial with or without (control) a virgin female for 5 min. Subsequently, pairs of pre-incubated males were placed in behavioral arenas with an agarose substrate. (B) Lunge frequency per fly after pre-incubation without (white) or with (blue) a female. n=32 flies each. Statistical test used was a Mann-Whitney U-test. **** P < 0.001. (C) Schematic of experimental design. (D) Lunge number before (pre-stim.) and after (post-stim.) GtACR1-mediated neural silencing. Green lines depict exposure to green light (530 nm, 10 Hz, 10 ms pulse-width) for 10 s. Gray points show lunge frequencies for individual flies, and black points show mean values. Statistical tests used were Wilcoxon signed test (within fly comparison) and Kruskal-Wallis test (between genotype comparison). ** Dunn’s corrected P < 0.01, **** P < 0.0001
We next asked whether this persistent influence requires continuous pCd activity. To do this, male flies expressing GtACR1 in pCd neurons were pre-incubated with females, and briefly photostimulated with green light during the aggression test (Figure 6C). Transient inhibition of pCd neurons abrogated the effect of female pre-exposure to enhance aggression (Figure 6D), mirroring the effect of such transient inhibition to disrupt persistent physiological activity in these cells (Figure 5G2). Thus, continuous pCd neuron activity is required to maintain a persistent behavioral state change induced by female presentation. Importantly, this effect was not observed when P1 neurons were transiently silenced using GtACR, although such silencing of P1 cells did transiently disrupt male courtship towards females (Figure S7), as previously reported (Zhang et al., 2018a).
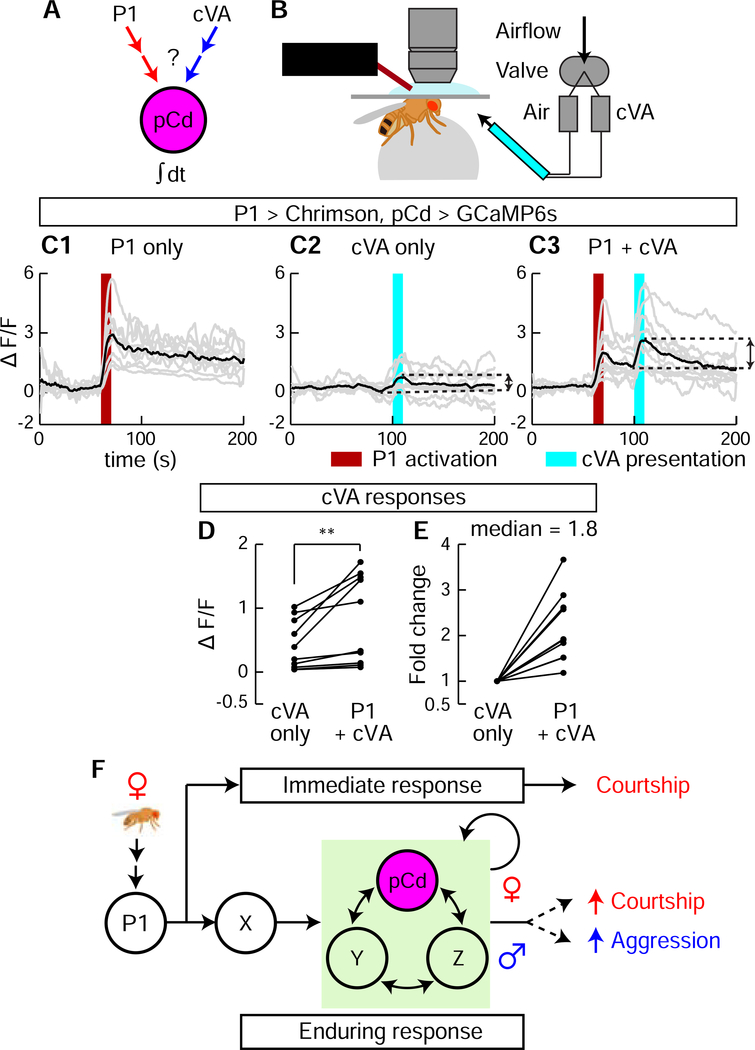
Individual pCd neurons respond to both P1 stimulation and the aggression-promoting pheromone cVA
The foregoing experiments indicated that when males are removed from the presence of females and confronted with another male, their behavior switches from courtship to aggression. To investigate whether pCd neurons themselves might also play a role in the detection of male cues that trigger this behavioral switch, we investigated whether they can respond to 11-cis Vaccenyl Acetate (cVA), a male-specific pheromone that has been shown to promote aggression (Wang and Anderson, 2010) (Figure 7A). Notably, cVA has already been shown to activate pCd cells in females (Zhou et al., 2014), where the pheromone promotes sexual receptivity. Although other pheromones have been shown to promote male aggression in Drosophila, such as 7-tricosene (Wang et al., 2011), the non-volatility of that compound made it difficult to deliver in a controlled manner to walking flies in our imaging preparation (Figure 7B) without physically disturbing them.
Figure 7. P1 activation of pCd neurons enhanced their responsiveness to cVA.
(A) Schematic illustrating experimental design. (B) In vivo GCaMP imaging. P1 neurons were optogenetically activated (660 nm LED), and cVA (or air) was delivered using an olfactometer synchronized and controlled by the imaging acquisition software. (C) GCaMP responses (ΔF/F ) to cVA of pCd neurons exhibiting persistent responses to P1 photostimulation (C1, dark red bar, 10 s, 10 Hz, 10 ms pulse-width). cVA alone (C2, cyan bar) or 30 s after a second (10 s) P1 stimulation (C3) were delivered 3 min apart in random order (Methods). Gray lines depict trial-averaged individual pCd cell responses (2–3 trials/cell, n=10 cells from 7 flies) and black lines show the mean for all cells. Double-headed arrows in (C2 and C3) indicate intervals for cVA responses calculated in (D, E). (D) Individual pCd cell responses (ΔF/F ) to cVA presented alone (“cVA only”) or 30 s after a 10 s P1 stimulation (“P1+cVA”). Statistical test used was a Wilcoxon signed-rank test. ** P < 0.01. (E) Fold change of pCd responses to cVA presentation after P1 stimulation, compared to cVA delivered alone. Data normalized to ΔF/F without P1 stimulation. (F) Models for how P1 and pCd neurons regulate immediate and enduring social behaviors.
To do this, we imaged pCd activity using GCaMP6s in flies exposed to the following stimuli at 5 minute intervals: 10 s of P1 activation; cVA vapour presentation; or P1 stimulation (10 s) followed 30 s later by cVA (Figure 7C). Among pCd neurons persistently activated by P1 stimulation (Figure 7C1), only half responded to cVA alone (defined as >2σ above baseline; Figure 7C2). However, delivery of cVA 30 s after P1 stimulation (i.e., during the persistent phase of the response) yielded cVA responses (>2σ above post-P1 activity) in 90% of the pCd cells (Figure 7C3). Moreover, peak cVA responses were significantly greater following P1 activation, than in flies exposed to the pheromone on its own (median increase 1.8-fold; Fig. 7D, E). Thus, individual pCd neurons that are activated by P1 stimulation in males can also respond to cVA (Figure 7A), and this response is enhanced during the persistent phase of the P1 response.
DISCUSSION
Optogenetic activation of P1 neurons evokes both courtship song, in a reflexive manner (Bath et al., 2014; Inagaki et al., 2014), and a persistent internal state of social arousal or drive (Anderson, 2016) that promotes aggression in the presence of a conspecific male (Hoopfer et al., 2015; Watanabe et al., 2017). Here we have identified a population of indirect persistent P1 follower cells, called pCd neurons (Zhou et al., 2014), whose activity is necessary for P1-triggered persistent aggression. pCd neurons are also necessary for persistent UWEs triggered by P1 activation, on a time scale outlasting P1 activity (as measured in separate imaging experiments). An earlier study (Zhang et al., 2018a) reported that P1 activity is continuously required during male courtship following initial female contact, but did not distinguish whether this requirement reflected continuous stimulation of P1 cells by non-contact-dependent female-derived cues (e.g., motion cues (Kohatsu and Yamamoto, 2015; Auer and Benton, 2016)), or a true fly-intrinsic persistent response. In contrast, the use of transient optogenetic stimulation here clearly demonstrates persistent fly-intrinsic responses. Nevertheless, we cannot exclude that persistent P1 activity may occur during natural courtship bouts (Zhang et al., 2018a). Importantly, however, we show that pCd but not P1 neurons are required for a persistent increase in aggressive state induced by transient female pre-exposure (Figure 6D). Together, these data suggest that pCd neurons participate in a network that may encode a persistent memory of a female, which can be combined with the detection of an opponent male at a later time to elicit aggression (Hoopfer et al., 2015; Anderson, 2016). The observation that P1 neuron activation enhances pCd responses to cVA, an aggression-promoting pheromone (Wang and Anderson, 2010), is consistent with this idea.
The effect of females to promote inter-male aggression are well-known and widespread throughout the animal kingdom (Homer, 850 B.C.; Lorenz, 1966). This effect is typically attributed to increases in circulating steroid hormones, such as testosterone or estrogen (Wingfield et al., 1990; Archer, 2006; Sobolewski et al., 2013), or to the effects of neuromodulators such as neuropeptides or biogenic amines (Gobrogge et al., 2007). Our data provide evidence that neural circuit dynamics involving persistent activity may also play a role in the effect of social experience with females to enhance male aggressiveness, in Drosophila. Whether such mechanisms also operate in mammalian systems where female exposure promotes aggressiveness (Remedios et al., 2017), remains to be determined.
Our physiological data suggest that pCd neurons are part of a circuit that temporally integrates P1 input to yield a slow response that decays over minutes (Figure 1E). The fact that transiently silencing pCd neurons using GtACR irreversibly interrupts this slow response argues that it indeed reflects persistent pCd activity, and not simply persistence of GCaMP6s fluorescence. It is likely that this integrator circuit comprises additional neurons, including non-Fru-expressing neurons. Evidently, P1 neurons activate this circuit in parallel with a “command” network, including pIP10 descending interneurons (von Philipsborn et al., 2011; Ding et al., 2019), that triggers rapid-onset courtship behavior. These results illustrate how acute and enduring responses to sensory cues may be segregated into parallel neural pathways, allowing behavioral control on different time scales, with different degrees of flexibility (Figure 7F). The incorporation of parallel neural pathways that allow behavioral responses to stimuli to be processed on multiple timescales may represent an important step in the evolution of behavior, from simple stimulus-response reflexes to more integrative, malleable responses (Anderson and Adolphs, 2014; Gibson et al., 2015; Bach and Dayan, 2017).
Recently, Zhang et al reported a role for pCd neurons in a recurrent circuit with NPF neurons that accumulates mating drive under conditions of extended sexual deprivation in Drosophila males (Zhang et al., 2019). In agreement with our results, Zhang et al found that constitutive silencing of pCd neurons partially reduces mating behavior. However, they also reported that acute activation of these neurons has no effect on courtship, and that behavioral effects are only observed following 12 hrs of continuous thermogenetic stimulation of these cells (Zhang et al., 2019). In contrast, we observed a clear effect of acute (30s) pCd stimulation to promote courtship, but only when males are provided with a source of female cues (Fig. 3D–F). Our data demonstrating a requirement for pCd neurons in aggression enhanced by 5 min of female pre-exposure (Fig. 6D) indicate that these cells regulate persistent internal states triggered by exposure to ecologically relevant stimuli, on a time-scale orders of magnitude shorter than those required for homeostatic influences on mating (Zhang et al., 2019). Whether NPF neurons are involved in this aggression-promoting function of pCd neurons remains to be determined; we previously reported a weak effect of NPF neuron stimulation to enhance aggression (Asahina et al., 2014), while another group reported that silencing of NPF neurons increased aggression (Dierick and Greenspan, 2007).
Our observations raise several new and interesting questions for future investigation. First, what cells provide direct synaptic inputs to pCd neurons, and what is the connectional relationship of these cells to P1 neurons? Second, the fact that pCd activity is necessary but not sufficient to trigger persistence suggests that other cells likely contribute to the integrator circuit; what are these cells (Figure 7F, Y, Z)? Finally, how is persistence encoded, and what is the role of pCd neurons in determining its duration? The data presented here provide insight into the complex networks that underlie behavioral temporal dynamics (Crickmore and Vosshall, 2013; Zhang et al., 2018a) in Drosophila, and offer a useful point-of-entry to this fascinating problem.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David J. Anderson (wuwei@caltech.edu). Fly lines generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rearing conditions
Flies were reared under standard conditions at 25°C and 55% humidity, on a 12 h light/12 h dark cycle. 2–5 days old virgin females were used to cross with different male stocks. The density of experimental flies (−5 pupae/cm2) was controlled by limiting the number of parents; crosses with too high or too low a density of progeny were discarded. Male flies were collected 0–2 days after eclosion and reared either individually (single-housed) or at 18 flies (group-housed) per vial for 5–6 days before the behavioral assays. Newly eclosed males were excluded from collection. For optogenetic experiments, eclosed males were reared in the dark with food containing 0.4 mM all-trans-retinal (Sigma-Aldrich, St. Louis, MO). For two-color optogenetic experiments, flies were reared in the dark from larval stage. Virgin females provided during behavioral tests were reared at high density (30 flies per vial) for 2–3 days. Flies carrying Gal4 and UAS-opsin transgenes were maintained in the dark to prevent uncontrolled activation of the opsins.
METHODS DETAILS
Generation of transgenic fly lines
The following lines were generated in this study. R41A01-LexA (vk00027 and attp2), R41A01-AD (attp40), and R41A01-DBD (attp2) were constructed based on the methods described in ref (Pfeiffer et al., 2008). R41A01 enhancer fragment was amplified from genomic DNA based on sequences in ref. (Adams et al., 2000). The primers used for amplification were designed based on recommendations in the Janelia FlyLight project and Bloominton Drosophila Stock Center (https://bdsc.indiana.edu/stocks/gal4/gal4_janelia.html). For making LexAop2-NLS-GCaMP6s (su(Hw)attp5), two nuclear localization signal (NLS) peptides, one from SV40 and the other from the Drosophila gene scalloped, were used. SV40-NLS (ccaaagaagaaaaggaaggta) was fused to the 5’ end, and the scalloped-NLS (agaaccaggaagcaagtcagttcgcacatccaagtgctggctcgccgtaaactccgcgagatc) was fused to the 3’ end of the codon-optimized GCaMP6s. A DNA fragment containing syn21-SV40-NLS-GCaMP6s-scalloped-NLS was ligated into pJFRC19–13LexAop2-IVS-myr::GFP-sv40 (Addgene plasmid # 26224) via XhoI and Xbal restriction enzyme sites. The sv40 terminator in the pJFRC19 was replaced with p10 terminator via Xbal and Fsel sites. To generate LexAop2-GtACR1 flies, the GtACR1 Drosophila-codon-optimized sequence (Mohammad et al., 2017) was subcloned into pJFRC19–13LexAop2-IVS-myr::GFP-sv40 (Addgene plasmid # 26224) plasmid. The GtACR1::eYFP fragment was swapped with the myr::GFP fragment using XhoI and Xba1.
Two-photon GCaMP imaging
Calcium imaging was performed using a custom-modified Ultima two-photon laser scanning microscope (Bruker). The primary beam path was equipped with galvanometers driving a Chameleon Ultra II Ti:Sapphire laser (Coherent) and used for GCaMP imaging (920 nm). The secondary beam path was equipped with separate set of galvanometers driving a Fidelity-2 Fiber Oscillator laser (Coherent) for GtACR1 actuation (1070 nm). The two lasers were combined using 1030 nm short-pass filter (Bruker). GCaMP emission was detected with photomultiplier-tube (Hamamatsu). Images were acquired with an Olympus 40x, 0.8 numerical aperture objective (LUMPLFLN) equipped with high-speed piezo-z (Bruker). All images acquisition was performed using PrairieView Software (Version 5.3). For fast volume imaging (Figure 1A, B and Figure S1), three 4-pm optical sections were collected at 180 × 180 pixel resolution with a frame rate ~0.83 Hz. All of the other images were acquired at 256 × 256 pixel resolution with a frame rate 1 Hz. Saline (108 mM NaCl, 5 mM KCl, 4 mM NaHCO3, 1 mM NaH2PO4, 5 mM trehalose, 10 mM sucrose, 5 mM HEPES, 0.5 mM CaCl2, 2 mM MgCl2, pH=7.5) was used to bathe the brain during functional imaging. Saline containing 90 mM KCl was added for high-resolution z stack after functional imaging to verify cell identity in Figure S1.
To prepare flies in vivo imaging, 6–8 days old flies were anesthetized on a cold plate and mounted on a thin plastic plate with wax. The wings, all legs, antenna, and arista were kept intact, wax-free, and free to move. Saline was added on the top side of the plate to submerge the fly head. A hole in the posterior-dorsal side of the head was opened using sharp forceps. Animals were then placed beneath the objective, and a plastic ball supported with air was positioned under the fly. The conditions inside of the imaging setup were maintained similar to the rearing conditions (25°C and 55% humidity). The flies were habituated for 30 min, and their behaviors were observed from the side using Point Grey Flea3 camera mounted with 0.5x-at-94 mm Infinistix lens fitted with a bandpass IR filter (830 nm, Edmund Optics) to block the two photon imaging laser and optogenetic stimulation lights. Animals that exhibited no movement, strenuous movement, and prolonged abdomen bending during and after habituation were discarded.
Chrimson activation during calcium imaging was performed as described in ref(Inagaki et al., 2014). A deep red (660 nm) fiber-coupled LED (Thorlab) with band-pass filter (660 nm, Edmund Optics) was used for light source to activate Chrimson. A 200 μm core multimode optic fiber placed 200 μm away from the brain was used to deliver 10 Hz, 10 ms pulse-width light. The light intensity at the tip of optic fiber was set to be 39.2 μW. For two photon GtACR1 actuation, 1070 nm laser (Fidelity-2, Coherent) was delivered by galvanometers to a circular area with diameter =~15 μm containing 1–3 cell bodies in focus for −10 s by spiral scanning (10 μm/pixel, 45.24 ms/repeat, 220 repeats). Galvanometers were re-calibrated weekly using a slide glass coated with thin layer of fluorescent dye. Field of view was adjusted in order to keep the spiral scanning area near the center of the imaging field. cVA was presented by directing a continuous airstream (80 mL/min) through a 4 mm diameter Teflon tube directed at the fly’s antennae. A custom-designed solenoid valve controller system was used to redirect the airstream between a blank cartridge and one containing cVA or Ethanol (solvent control). To make odour cartridges, 10 μL of undiluted cVA (Cayman Chemicals, 20 mg/mL) or Ethanol were placed on filter papers, and dried for 3 min to remove solvent before inserted into 15 mL pre-cleaned vials (Sigma-Aldrich).
Labeling neurons with Photoactivation after GCaMP imaging
Photoactivation experiments were performed in vivo using spiral scanning as described above. To perform GCaMP imaging and PA-GFP activation simultaneously, two Chameleon Ultra II Ti:Sapphire lasers (Coherent), one set at 920 nm and the other at 710 nm, are combined using 760 nm long pass filter (Bruker). Cell bodies of pCd neurons were identified by functional imaging using NLS-GCaMP6s, and a three-dimensional region of photoactivation was defined. The defined region of photoactivation was photoactivated by two cycles of spiral scanning (diameter =~7.5 μm, 45.24 ms/repeat, 20 repeats, 150 ms inter-repeat-intervals) separated by 20 min interval to allow diffusion of photoactivated PA-GFP molecules to the projections. 20 min after second cycle of the spiral scanning, 3-dimensional images were acquired at 1024 × 1024 pixel resolution. To reduce the fly’s movement and residual GCaMP signal, cold saline containing 1mM EDTA was perfused until the end of image acquisition. tdTomato signals and photoactivated PA-GFP signals were imaged simultaneously at 940 nm. Non-PPF1 PA-GFP and NLS-GCaMP basal fluorescence have been masked for clarity and z stack were created (Figure 1C3 and C4) using Fluorender (Wan et al., 2009) and Fiji (Schindelin et al., 2012; Schneider et al., 2012) software.
Immunohistochemistry
Brains from 7-to-10-day-old adult files were dissected and stained as previously described(Watanabe et al., 2017). The primary antibody mixture consisted of 1:1000 rabbit anti-GFP (Thermo Fisher Scientific, Cat#A11122), 1:1000 chicken anti-GFP (Aves Lab, Cat#GFP-1010), 1:100 mono-clonal (for GRASP experiment, Figure S4J–R) mouse anti-GFP (Sigma-Aldrich, Cat#G6539), 1:1000 rabbit anti-DsRed (Takara Bio, Cat#632496), 1:50 mouse anti-Brochpilot nc82 (Developmental Studies Hybridoma Bank), and 10% normal goat serum (Sigma-Aldrich) in PBST. Secondary antibodies used were 1:1000 goat anti-rabbit-Alexa488 (Thermo Fisher Scientific, Cat#A11008), 1:1000 goat anti-chicken-Alexa488 (Thermo Fisher Scientific, Cat#A11039), 1:1000 goat anti-mouse-Alexa488 (Thermo Fisher Scientific, Cat#A11001), 1:1000 goat anti-rabbit-Alexa568 (Thermo Fisher Scientific, Cat#A11011), and 1:1000 goat anti-mouse-Alexa633 (Thermo Fisher Scientific, Cat#A21050).
Confocal stacks were obtained with Fluoview FV1000 or FV3000 (Olympus). Fiji (Schindelin et al., 2012; Schneider et al., 2012) and Fluorender (Wan et al., 2009) software was used to create z stack images. For brain registration (Figure S4G–I), the two images shown in Figure S4B and D are registered to T1 template brain (Yu et al., 2010) using CMTK registration tools (Jefferis et al., 2007).
Behavioral assay
Temperature and humidity of the room for behavioral assay was set to 25°C and 55%, respectively. All naturally occurring behavior assays were performed between 2:00pm to 7:00pm. Optogenetically-induced behaviors were not performed at specific times. All the behavior assays except mating assay (Figure 4A, B) were performed in 8-well acrylic chamber (16 mm diameter × 10 mm height, modified from ref. (Inagaki et al., 2014), and side of the each well was coated with alnsect-a-Slip (Bioquip Products). Temperature probe (Vktech) was inserted into one side of the chamber to accurately monitor the chamber temperature. The clear top plates were coated with Sigmacote (Sigma-Aldrich), and the floor of the arenas was composed of clear acrylic covered with food (2.5% (w/v) sucrose and 2.25% (w/v) agarose in apple juice). Flies were introduced into the chambers by gentle aspiration using a mouth pipette, and the chambers were placed under the behavioral setup. Flies were allowed to acclimate to the chamber under the camera without disturbance for 90 s before the recording. Fly behaviors were recorded at 30 Hz using Point Grey Flea3 camera mounted with Fujinon lens (HF35HA-1B) fitted with a long pass IR filter (780 nm, Midwest Optical Systems). Camera was located ~0.5 m above the chamber, and IR backlighting (855 nm, SmartVision Lights) was used for illumination from beneath the arena.
Optogenetic activation was performed as described previously (Inagaki et al., 2014). Briefly, a 655 nm 10 mm Square LED (Luxeon Star) was used to deliver 0.48 mW/mm2 light for 30 seconds. For dead female presentation (Figure 3D–F and J–L, Figure 4C, and Figure S7), 2–5 day old wild-type Canton S virgin females were freeze-killed, and affixed in the middle of the arena with UV curable glue. The ventral end of the female abdomen was glued to prevent copulation.
For the female induced aggression assay (Figure 6A–D), single-housed male flies were transferred individually into empty vials containing a virgin female, and allowed to freely interact with the female for ~5 min. After this pre-exposure period, the male flies were gently transferred to the behavior arena covered with 2.25% (w/v) agarose in dH2O, instead of fly food. For GtACR1 stimulation (Figure 6C, D), a 530 nm 10 mm Square LED (Luxeon Star) was used to deliver 117 μW/mm2 light for 10 seconds. Male flies that initiated copulation during the 5 min pre-exposure period were not tested.
For the mating assay (Figure 4A, B),12-well two-layer chambers in which the layers were separated by a removable aluminum film. 2–5 day old wild-type Canton S virgin females were introduced into the lower layers, and males of a particular genotype were introduced in the upper layers. Flies were allowed to acclimate to the chamber for 90 s as described above before removing film. Behavior recording started right after film was removed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Imaging data analysis
All data analysis was performed in MATLAB (MathWorks). ROIs (region of interest) corresponding to individual cell bodies were manually selected and fluorescence signal from the ROIs were smoothed with a moving average (window =5 frames). For volume imaging (Figure 1A, B and Figure S1), a single focal plane in which we observed the highest ΔF/F was used for each cell. Normalized ΔF/F values for each trials were calculated by dividing ΔF/F by the maximum ΔF/F. The average signal before photostimulation was used as F0 to calculate the ΔF/F, and cells with peak ΔF/F responses < 4σ above baseline more than 1/3 trials were excluded. Decay constants (tau) were fit to minimize mean-squared error between observed ΔF/F traces and a five-parameter model of cell responses to optogenetic stimulation. Specifically, the ΔF/F trace evoked by three consecutive pulses of optogenetic stimulation was fit with a weighted sum of three impulse responses sharing a characteristic rise time tau_R and decay time tau: fit values of tau_R and tau were the same for all three evoked responses, while response amplitudes were fit independently. Fit impulse responses in the model were set to be 30 s apart, following experimental stimulation conditions. The best-fit 80% of cells (MSE<2.06) were used to generate plots of population-average responses. “Percent of peak” in Figure 5H and Figure S6E were calculated from mean normalized ΔF/F values between 10–30 s after GtACR1 actuation. cVA responses for Figure 7D were calculated by subtracting mean GCaMP signal 10 s before cVA presentation from those obtained during cVA presentation (10 s). cVA responses from each cell delivered 30s after P1 stimulation were divided by cVA responses without concurrent P1 stimulation (cVA only), to calculate fold change (Figure 7E). cVA alone or P1+cVA stimulation were delivered in random order following initial selection for P1-responsive pCd neurons. Individual cell responses used in Figure 7C–E were the average of 2–3 trials per cell.
Behavioral data analysis
Analysis of lunging and unilateral wing extension was performed as described in ref (Hoopfer et al., 2015). Briefly, fly posture was tracked from recorded videos using Caltech FlyTracker software, which is available for download at http://www.vision.caltech.edu/Tools/FlyTracker/, and bouts of behaviors were automatically annotated using the Janelia Automatic Animal Behavior Annotator (JAABA) (Kabra et al., 2013). All annotations were manually validated to remove false positives. Behavioral assays with dead females (Figure 3D–F and J–K) were manually scored without using JAABA due to inaccuracy. Data shown in Figure 3A–C and G–I were also manually scored for consistency. Copulation latency for Figure 4A and B was manually scored, and the total number of males that had engaged in copulation was summed across the 30-min period and plotted as a percentage of total flies for each time point. Courtship bouts shown in Figure S7 were manually annotated following the definition of courtship bouts described previously (Zhang et al., 2018a). Statistical analyses were performed using Matlab and Prism6 (GraphPad Software). All data were analyzed with nonparametric tests. The cutoff for significance was set as an α<0.05. Each experiment was repeated at least twice on independent group of flies. Outliers were defined as data points falling outside 1.5x the interquartile range of the data, and were excluded from plots for clarity, but not from statistical analyses.
Curve Fitting for Leaky bucket model
Rasters of courtship and lunging behavior in a 15-minute window were averaged across flies and binned in 10-second (for courtship) or 20-second (for lunging) time windows to produce a time-evolving population average behavior rate. Behavior rates for courtship and lunging were each fit with a three-parameter leaky integrator model with dynamics , which has analytical solution , where r is the behavior rate as a function of time t (in minutes), I is a constant sensory input, τ is the time constant of integration, and r0 is the initial behavior rate at the start of recording. Parameters I, τ, and r0 were fit to minimize the mean squared error between model and data, for courtship and for lunging. Parameter values were jointly fit across the two behaviors (courtship and lunging) and across the four experimental conditions: pCd > Kir2.1 (manipulation), pCd > GFP, BPD > Kir2.1, and BPD > GFP (controls). To reduce the number of free parameters, the sensory input I was constrained to take the same value for all groups and conditions, while r0 was fit separately for courtship and for aggression; only τ was fit independently for each group and each behavior.
DATA AND CODE AVAILABILITY
Source data and analysis code supporting the current study have not been deposited in a public repository, but are available from the corresponding author on request.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP (rabbit polyclonal) | Thermo Fisher Scientific | Cat# A11122 RRID: AB_221569 |
| Anti-GFP (chicken polyclonal, IgY Fraction) | Aves Labs | Cat# GFP-1010 RRID: AB_2307313 |
| Anti-GFP (mouse monoclonal) | Sigma-Aldrich | Cat# G6539 RRID: AB_259941 |
| Anti-DsRed (rabbit polyclonal) | Takara Bio | Cat# 632496 RRID: AB_10013483 |
| nc82 (mouse) | Developmental Studies Hybridoma Bank | Cat# nc82 RRID: AB_2314866 |
| Goat anti-rabbit-Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A11008 RRID: AB_143165 |
| Goat anti-chicken-Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A11039 RRID: AB_142924 |
| Goat anti-mouse-Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A11001 RRID: AB_2534069 |
| Goat anti-rabbit-Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A11011 RRID: AB_143157 |
| Goat anti-mouse-Alexa Fluor 633 | Thermo Fisher Scientific | Cat# A21050 RRID: AB_141431 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| All trans-retinal | Sigma-Aldrich | Cat# R2500 |
| Insect-A-Slip | BioQuip Products | Cat# 2871B |
| Sigmacote | Sigma-Aldrich | Cat# SL2 |
| Experimental Models: Organisms/Strains | ||
| Drosophila: R41A01-LexA (vk00027) | This study (Anderson Lab) | N/A |
| Drosophila: R41A01-LexA (attp2) | This study (Anderson Lab) | N/A |
| Drosophila: R41A01-AD (attp40) | This study (Anderson Lab) | N/A |
| Drosophila: R41A01-DBD (attp2) | This study (Anderson Lab) | N/A |
| Drosophila: 13xLexAop2-NLS-GCaMP6s (su(Hw)attp5) | This study (Anderson Lab) | N/A |
| Drosophila: 13xLexAop2-GtACR1 (attp40) | This study (Claridge-Chang Lab) | N/A |
| Drosophila: R15A01-LexA (attp2) | Gerald M. Rubin Lab | N/A |
| Drosophila: BDP-AD (attp40) | Gerald M. Rubin Lab | N/A |
| Drosophila: BDP-DBD (attp2) | Gerald M. Rubin Lab | N/A |
| Drosophila: 10xUAS-NLS-tdTomato (vk00022) | Gerald M. Rubin Lab | N/A |
| Drosophila: 13xLexAop-NLS-GFP (vk00040) | Gerald M. Rubin Lab | N/A |
| Drosophila: 10xUAS-Chrimson::tdTomato (su(Hw)attp1) | Gerald M. Rubin Lab | N/A |
| Drosophila: 10xUAS-Chrimson::tdTomato (su(Hw)attp18) | Gerald M. Rubin Lab | N/A |
| Drosophila: 20xUAS-Chrimson::tdTomato (su(Hw)attp5) | Gerald M. Rubin Lab | N/A |
| Drosophila: 13xLexAop2-myr::tdTomato (attp18) | Gerald M. Rubin Lab | N/A |
| Drosophila: 13xLexAop2-OpGCaMP6s (su(Hw)attp8) | Gerald M. Rubin Lab | N/A |
| Drosophila: 20xUAS-OpGCaMP6s (su(Hw)attp5) | Gerald M. Rubin Lab | N/A |
| Drosophila: 13xLexAop2-mPA-GFP (su(Hw)attp8) | Gerald M. Rubin Lab | N/A |
| Drosophila: 13xLexAop2-Kir2.1::eGFP (vk00027) | Gerald M. Rubin Lab | N/A |
| Drosophila: 10xUAS-Kir2.1::eGFP (attp2) | Gerald M. Rubin Lab | N/A |
| Drosophila: 10xUAS-GFP (attp2) | Gerald M. Rubin Lab | N/A |
| Drosophila: R21D06-LexA (attp2) | Gerald M. Rubin Lab | N/A |
| Drosophila: dsx-DBD | (Pavlou et al., 2016) | N/A |
| Drosophila: Fru-LexA | (Mellert et al., 2010) | N/A |
| Drosophila: Orco-LexA | (Lai and Lee, 2006) | N/A |
| Drosophila: UAS-CD4::spGFP1-10 | (Gordon and Scott, 2009) | N/A |
| Drosophila: LexAop-CD4::spGFP11 | (Gordon and Scott, 2009) | N/A |
| Drosophila: 20xUAS-GtACR1::eYFP (attp2) | (Mohammad et al., 2017) | N/A |
| Drosophila: Canton S | (Hoyer et al., 2008) | N/A |
| Drosophila: BDP-LexA (attp40) | Bloomington Drosophila Stock Center | RRID: BDSC_77691 |
| Drosophila: R71G01-Gal4 (attp2) | Bloomington Drosophila Stock Center | RRID: BDSC_39599 |
| Drosophila: R71G01-DBD (attp2) | Bloomington Drosophila Stock Center | RRID: BDSC_69507 |
| Drosophila: R15A01-Gal4 (attp2) | Bloomington Drosophila Stock Center | RRID: BDSC_48670 |
| Drosophila: R15A01-AD (attp40) | Bloomington Drosophila Stock Center | RRID: BDSC_68837 |
| Drosophila: R41A01-Gal4 (attp2) | Bloomington Drosophila Stock Center | RRID: BDSC_39425 |
| Drosophila: R41A01-LexA (attp40) | Bloomington Drosophila Stock Center | RRID: BDSC_54787 |
| Drosophila: R21D06-DBD (attp2) | Bloomington Drosophila Stock Center | RRID: BDSC_69873 |
| Drosophila: ChAT-DBD | Bloomington Drosophila Stock Center | RRID: BDSC_60318 |
| Drosophila: VGlut-DBD | Bloomington Drosophila Stock Center | RRID: BDSC_60313 |
| Drosophila: Gad1-p65AD | Bloomington Drosophila Stock Center | RRID: BDSC_60322 |
| Drosophila: UAS-Denmark; UAS-Syt-eGFP | Bloomington Drosophila Stock Center | RRID: BDSC_33064 |
| Drosophila: GH146-Gal4 | Bloomington Drosophila Stock Center | RRID: BDSC_30026 |
| Drosophila: 13xLexAop2-CsChrimson::mVenus (attp40) | Bloomington Drosophila Stock Center | RRID: BDSC_55138 |
| Drosophila: 10xUAS-myr::GFP (su(Hw)attp8) | Bloomington Drosophila Stock Center | RRID: BDSC_32196 |
| Drosophila: 10xUAS-myr::GFP (attp2) | Bloomington Drosophila Stock Center | RRID: BDSC_32197 |
| Drosophila: UAS-Kir2.1::eGFP | Bloomington Drosophila Stock Center | RRID: BDSC_6595 |
| Software and Algorithms | ||
| Caltech FlyTracker | Pietro Perona Lab, Caltech http://www.vision.caltech.edu/Tools/FlyTracker/index.html | N/A |
| Janelia Automatic Animal Behavior Annotator (JAABA) | Kristin Branson, Janelia Research Campus http://jaaba.sourceforge.net/ | N/A |
| Prism6 | GraphPad Sotware | N/A |
| Matlab R2015a | Matworks | RRID: SCR_001622 |
| FluoRender | http://www.sci.utah.edu/software/fluorender.html | RRID: SCR_014303 |
| Fiji | https://fiji.sc/ | RRID: SCR_002285 |
Highlights.
Repeated P1 neuron activation triggers pCd activity, which outlasts that of P1 cells
pCd neurons enhance and prolong courtship and aggression promoted by female cues
pCd neurons are necessary for, but not sufficient to trigger, physiologic persistence
Persistent enhancement of aggression by brief female exposure requires pCd activity
Acknowledgements
We thank H. Inagaki for comments on the manuscript, B. Pfeiffer and G. M. Rubin for fly strains, A. M. Wong for help with initial development of all-optical stimulation and imaging, A. Sanchez for maintaining fly stocks, C. Chiu and X. Da for lab management and G. Mancuso for administrative assistance. This work was supported in part by NIH grant R01 DA031389. D.J.A. is an Investigator of the Howard Hughes Medical Institute. The authors declare no conflicts-of-interest.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi. [DOI] [PubMed] [Google Scholar]
- Anderson DJ (2016). Circuit modules linking internal states and social behaviour in flies and mice. Nature reviews Neuroscience 17, 692–704. 10.1038/nrn.2016.125. [DOI] [PubMed] [Google Scholar]
- Anderson David J., and Adolphs R (2014). A Framework for Studying Emotions across Species. Cell 157, 187–200. 10.1016/j.cell.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J (2006). Testosterone and human aggression: an evaluation of the challenge hypothesis. Neuroscience & Biobehavioral Reviews 30, 319–345. doi. [DOI] [PubMed] [Google Scholar]
- Asahina K, Watanabe K, Duistermars BJ, et al. (2014). Tachykinin-Expressing Neurons Control Male-Specific Aggressive Arousal in Drosophila. Cell 156, 221–235. 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, and Benton R (2016). Sexual circuitry in Drosophila. Curr Opin Neurobiol 38, 18–26. 10.1016/j.conb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Bach DR, and Dayan P (2017). Algorithms for survival: a comparative perspective on emotions. In Nature reviews Neuroscience, pp. 311–319. [DOI] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, et al. (2001). Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci 21, 1523–1531. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI (2012). Beyond the connectome: How neuromodulators shape neural circuits. Bioessays 34, 458–465. doi. [DOI] [PubMed] [Google Scholar]
- Bath DE, Stowers JR, Hörmann D, et al. (2014). FlyMAD: rapid thermogenetic control of neuronal activity in freely walking Drosophila. In Nat Methods, pp. 756–762. [DOI] [PubMed] [Google Scholar]
- Cachero S, Ostrovsky AD, Yu JY, et al. (2010). Sexual dimorphism in the fly brain. Curr Biol 20, 1589–1601. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, and Fiete I (2016). Computational principles of memory. Nature neuroscience 19, 394–403. 10.1038/nn.4237. [DOI] [PubMed] [Google Scholar]
- Chen D, Sitaraman D, Chen N, et al. (2017). Genetic and neuronal mechanisms governing the sex-specific interaction between sleep and sexual behaviors in Drosophila. Nat Commun 8, 154 10.1038/s41467-017-00087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Essner RA, Kosar S, et al. (2019). Sustained NPY signaling enables AgRP neurons to drive feeding. eLife 8 10.7554/eLife.46348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin YC, Zimmerman CA, et al. (2016). Hunger neurons drive feeding through a sustained, positive reinforcement signal. eLife 5 10.7554/eLife.18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, et al. (2011). Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol 21, 1–11. 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, Iguchi S, Bussell JJ, et al. (2015). Multimodal Chemosensory Circuits Controlling Male Courtship in Drosophila. Neuron. 10.1016/j.neuron.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore MA, and Vosshall LB (2013). Opposing dopaminergic and GABAergic neurons control the duration and persistence of copulation in Drosophila. Cell 155, 881–893. 10.1016/j.cell.2013.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, et al. (2008). The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 452, 473–477. 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- Diao F, Ironfield H, Luan H, et al. (2015). Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell reports 10, 1410–1421. 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierick HA, and Greenspan RJ (2007). Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet 39, 678–682. 10.1038/ng2029. [DOI] [PubMed] [Google Scholar]
- Ding Y, Lillvis JL, Cande J, et al. (2019). Neural Evolution of Context-Dependent Fly Song. Curr Biol 29, 1089–1099 e1087 10.1016/j.cub.2019.02.019. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, et al. (2008). GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Gibson William T., Gonzalez Carlos R., Fernandez C, et al. (2015). Behavioral Responses to a Repetitive Visual Threat Stimulus Express a Persistent State of Defensive Arousal in Drosophila. Current Biology 25, 1401–1415. 10.1016/j.cub.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobrogge KL, Liu Y, Jia X, et al. (2007). Anterior hypothalamic neural activation and neurochemical associations with aggression in pair-bonded male prairie voles. In J Comp Neurol (Wiley Subscription Services, Inc., A Wiley Company), pp. 1109–1122. [DOI] [PubMed] [Google Scholar]
- Gordon MD, and Scott K (2009). Motor control in a Drosophila taste circuit. Neuron 61, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZV, Inagaki HK, Daie K, et al. (2017). Maintenance of persistent activity in a frontal thalamocortical loop. Nature 545, 181–186. 10.1038/nature22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer (850 B.C.). The Iliad. Translated by R. Lattimore. (Chicago, IL, University of Chicago Press; ). [Google Scholar]
- Hoopfer ED (2016). Neural control of aggression in Drosophila. Curr Opin Neurobiol 38, 109–118. 10.1016/j.conb.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Hoopfer ED, Jung Y, Inagaki HK, et al. (2015). P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. eLife 4 10.7554/eLife.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer SC, et al. (2008). Octopamine in male aggression of Drosophila. Current biology : CB 18, 159–167. [DOI] [PubMed] [Google Scholar]
- Inagaki HK, Fontolan L, Romani S, et al. (2019). Discrete attractor dynamics underlies persistent activity in the frontal cortex. Nature 566, 212–217. 10.1038/s41586-019-0919-7. [DOI] [PubMed] [Google Scholar]
- Inagaki HK, Jung Y, Hoopfer ED, et al. (2014). Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat Methods, −. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, et al. (2007). Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell 128, 1187–1203. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo T-TB, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell reports 2, 991–1001. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabra M, Robie AA, Rivera-Alba M, et al. (2013). JAABA: interactive machine learning for automatic annotation of animal behavior. In Nat Methods, pp. 64–67. [DOI] [PubMed] [Google Scholar]
- Kallman BR, Kim H, and Scott K (2015). Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. eLife 4, 2027 10.7554/eLife.11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K-I, Hachiya T, Koganezawa M, et al. (2008). Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. In Neuron, pp. 759–769. [DOI] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, et al. (2014). Independent optical excitation of distinct neural populations In Nat Methods (Nature Publishing Group; ), pp. 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohatsu S, Koganezawa M, and Yamamoto D (2011). Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron 69, 498–508. 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Kohatsu S, and Yamamoto D (2015). Visually induced initiation of Drosophila innate courtship-like following pursuit is mediated by central excitatory state. Nature Communications 6 10.1038/ncomms7457. [DOI] [PubMed] [Google Scholar]
- Kunwar PS, Zelikowsky M, Remedios R, et al. (2015). Ventromedial hypothalamic neurons control a defensive emotion state. eLife 4 10.7554/eLife.06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, and Lee T (2006). Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature neuroscience 9, 703–709. [DOI] [PubMed] [Google Scholar]
- Lim RS (2014). PhD Thesis In Division of Biology (California Institute of Technology). [Google Scholar]
- Lim RS, Eyjolfsdottir E, Shin E, et al. (2014). How food controls aggression in Drosophila. PloS one 9, e105626 10.1371/journal.pone.0105626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K (1966). On Aggression (New York, Harcourt, Brace & World; ). [Google Scholar]
- Lorenz K, and Leyhausen P (1973). Motivation of human and animal behavior; an ethological view, Vol xix (New York, Van Nostrand-Reinhold; ). [Google Scholar]
- Manoli DS, Foss M, Villella A, et al. (2005). Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436, 395–400. 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- Mellert DJ, Knapp JM, Manoli DS, et al. (2010). Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development 137, 323–332. 10.1242/dev.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Stewart JC, Ott S, et al. (2017). Optogenetic inhibition of behavior with anion channelrhodopsins. Nat Methods. doi. [DOI] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, et al. (2010). Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci USA 107, 20553–20558. 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Robinett CC, and Baker BS (2011). Turning males on: activation of male courtship behavior in Drosophila melanogaster. PloS one 6, e21144 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou HJ, et al. (2016). Neural circuitry coordinating male copulation. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. In Proc Natl Acad Sci USA (National Acad Sciences), pp. 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remedios R, Kennedy A, Zelikowsky M, et al. (2017). Social behaviour shapes hypothalamic neural ensemble representations of conspecific sex. Nature 550, 388–392. 10.1038/nature23885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, and Tank DW (2009). Two-photon excitation of channelrhodopsin-2 at saturation. Proceedings of the National Academy of Sciences of the United States of America 106, 15025–15030. 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Billeter J-C, and Goodwin SF (2007). The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Current biology : CB 17, 1473–1478. 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolewski ME, Brown JL, and Mitani JC (2013). Female parity, male aggression, and the Challenge Hypothesis in wild chimpanzees. Primates 54, 81–88. doi. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, et al. (2005). Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807. 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Tauber E, and Eberl DF (2003). Acoustic communication in Drosophila. In Behav Processes, pp. 197–210. [Google Scholar]
- Tinbergen N (1951). The study of instinct (Oxford Eng., Clarendon Press; ). [Google Scholar]
- von Philipsborn AC, Liu T, Yu JY, et al. (2011). Neuronal control of Drosophila courtship song. Neuron 69, 509–522. doi. [DOI] [PubMed] [Google Scholar]
- Wan Y, Otsuna H, Chien CB, et al. (2009). An interactive visualization tool for multi-channel confocal microscopy data in neurobiology research. IEEE transactions on visualization and computer graphics 15, 1489–1496. 10.1109/TVCG.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, and Anderson DJ (2010). Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature 463, 227–231. doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dankert H, Perona P, et al. (2008). A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci USA 105, 5657–5663. doi PMC2311352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Han X, Mehren J, et al. (2011). Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nature neuroscience 14, 757–762. 10.1038/nn.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Chiu H, Pfeiffer BD, et al. (2017). A Circuit Node that Integrates Convergent Input from Neuromodulatory and Social Behavior-Promoting Neurons to Control Aggression in Drosophila. Neuron 95, 1112–1128 e1117 10.1016/j.neuron.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JC, Hegner RE, Dufty AM, et al. (1990). The challenge hypothesis--theoretic implications for patterns of testosterone secretion, mating systems and breeding strategies. Am Nat 136, 829–846. doi. [Google Scholar]
- Yamamoto D, and Koganezawa M (2013). Genes and circuits of courtship behaviour in Drosophila males. Nature Rev Neurosci 14, 681–692. 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- Yu JY, Kanai MI, Demir E, et al. (2010). Cellular organization of the neural circuit that drives Drosophila courtship behavior. Current biology : CB 20, 1602–1614. 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Zhang SX, Miner LE, Boutros CL, et al. (2018a). Motivation, Perception, and Chance Converge to Make a Binary Decision. Neuron 99, 376–388 e376 10.1016/j.neuron.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SX, Rogulja D, and Crickmore MA (2016). Dopaminergic Circuitry Underlying Mating Drive. Neuron 91, 168–181. 10.1016/j.neuron.2016.05.020. [DOI] [PubMed] [Google Scholar]
- Zhang SX, Rogulja D, and Crickmore MA (2019). Recurrent Circuitry Sustains Drosophila Courtship Drive While Priming Itself for Satiety. Curr Biol. 10.1016/j.cub.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Guo C, Chen D, et al. (2018b). Hierarchical Control of Drosophila Sleep, Courtship, and Feeding Behaviors by Male-Specific P1 Neurons. Neuroscience bulletin. 10.1007/s12264-018-0281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Rodesch CK, and Broadie K (2002). Living synaptic vesicle marker: synaptotagmin-GFP. Genesis 34, 142–145. 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- Zhou C, Pan Y, Robinett CC, et al. (2014). Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron 83, 149–163. 10.1016/j.neuron.2014.05.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data and analysis code supporting the current study have not been deposited in a public repository, but are available from the corresponding author on request.