We studied the dominant protists found in soils across the globe and their contributions to belowground food webs.
Abstract
Protists are ubiquitous in soil, where they are key contributors to nutrient cycling and energy transfer. However, protists have received far less attention than other components of the soil microbiome. We used amplicon sequencing of soils from 180 locations across six continents to investigate the ecological preferences of protists and their functional contributions to belowground systems. We complemented these analyses with shotgun metagenomic sequencing of 46 soils to validate the identities of the more abundant protist lineages. We found that most soils are dominated by consumers, although parasites and phototrophs are particularly abundant in tropical and arid ecosystems, respectively. The best predictors of protist composition (primarily annual precipitation) are fundamentally distinct from those shaping bacterial and archaeal communities (namely, soil pH). Some protists and bacteria co-occur globally, highlighting the potential importance of these largely undescribed belowground interactions. Together, this study allowed us to identify the most abundant and ubiquitous protists living in soil, with our work providing a cross-ecosystem perspective on the factors structuring soil protist communities and their likely contributions to soil functioning.
INTRODUCTION
Soil harbors a broad diversity of microorganisms that are well-recognized drivers of key ecosystem processes such as nutrient cycling and plant growth (1, 2). Just like aboveground plant and animal communities, the diversity and composition of soil microbial communities can vary markedly across the globe, with some of this variation predictable from environmental characteristics. Much of this previous research into the global-scale biogeographical patterns of soil microbes has focused on bacteria (3–5), fungi (5, 6), and archaea (7, 8). However, these are not the only microorganisms found in soil. Protists (nonfungal microbial eukaryotes) are ubiquitous and abundant in soil (9, 10), yet their global biogeography remains poorly resolved. The relative number of soil protist studies has actually declined over the past 15 years (11).
Protists are a key component of belowground ecosystems, both in density (a single gram of soil contains tens of thousands or more individuals) (9, 12) and in the ecosystem services they provide (13, 14). For example, as consumers, protists have an important role in controlling soil nutrient fluxes and plant nutrient uptake by stimulating carbon, nitrogen, phosphorus, and silica mineralization (9). Protists are also important primary producers, predators of other microeukaryotes, decomposers, and parasites or pathogens of most plants and animals (10, 15). Despite their substantial impacts on belowground food webs and soil processes, our understanding of the distributions and ecological preferences of soil protists across broad spatial scales (e.g., beyond local or regional scales) remains surprisingly limited (11). Previous studies of soil protists have focused on a few individual sites, or a relatively narrow set of environmental conditions (16), or a few specific lineages. Even for those protists known to have important effects on plant growth (e.g., pathogens and consumers), our ability to predict their spatial distributions in relation to soil, climate, and vegetation factors and the importance of the potential contributions of protists to soil processes and aboveground plant productivity remains limited. Identifying the environmental attributes that structure protistan communities was recently identified as a priority research direction, with such information deemed crucial to advancing our understanding of soil biology and associated ecosystem functions (11).
While some previous work has suggested that soil moisture availability is a key factor structuring protistan communities (12, 17), other studies have highlighted the importance of pH (18), total N (19), or plant species identity (20), and it remains unclear how predictable protistan communities are at larger spatial scales. The relative importance of climatic versus edaphic variables also remains unclear. In addition, protist distributions could be associated with bacterial distributions (21), with some protists even harboring specific bacterial taxa intracellularly (22). However, broad associations between soil protists and bacteria have not been well characterized. Moreover, there have been few direct comparisons of the biogeography of soil protists with the biogeography of the corresponding soil bacterial and archaeal communities (11).
Here, we address a set of questions fundamental to advancing our understanding of soil protists: (i) What are the dominant protists found in soils? (ii) What are the habitat preferences of these dominant protistan taxa and functional groups? (iii) How predictable is the composition of soil protistan communities across large spatial scales and environmental gradients? (iv) Are the biogeographical patterns observed for soil protists similar to those observed for soil bacteria and archaea? To address these questions, we analyzed the protistan communities found in 180 soils collected from six continents using an 18S ribosomal RNA (rRNA) gene sequencing–based approach to document the abundances and distributions of soil protists (fig. S1). We also obtained 18S rRNA gene sequences from shotgun metagenomic analyses of 46 soils to cross-validate the identities of the most dominant soil protists. We then used the protist distribution and corresponding environmental data to classify protistan taxa into groups based on their shared habitat preferences. Last, we acquired 16S rRNA amplicon data for 137 of 180 soils to directly compare the relative importance of environmental factors in shaping protistan versus bacterial and archaeal communities and to identify potential associations between co-occurring protists and prokaryotes in soil.
RESULTS AND DISCUSSION
What protistan lineages dominate soils globally?
Across the 180 soils, the most abundant protist lineages were Cercozoa and Ciliophora, which together comprised 73% of all reads (Fig. 1; see figs. S1 and S2 and table S1 for site information and soil properties). The glissomonads were the dominant lineage within Cercozoa and were composed mostly of Sandonidae, Allapsidae, and Cercomonadidae (Fig. 1). Our metagenomic comparisons confirmed the most abundant protist taxa, suggesting that protistan lineages detected by amplicon sequencing are generally representative of typical soil communities and not a product of the primer biases that can influence polymerase chain reaction (PCR)–based analyses (14, 23). We found concordance between the two datasets with regard to the identities of the most abundant protistan lineages (e.g., Cercozoa, Ciliophora, Apicomplexa, Chrysophyceae, and Amoebozoa; fig. S3). However, it is important to note some discrepancies between the approaches. For instance, some Oomycota were detected only in the amplicon 18S rRNA sequences. Conversely, Discosea-Flabellinia were only detected in the metagenomic data. Our assessment of the most abundant groups of soil protists aligns with previous studies, whereby the most abundant taxa were also abundant in other cultivation-independent surveys of soil protists (17, 24, 25). While our DNA-based study did not allow us to evaluate which protists are active, we note that the dominant taxa identified here also largely overlap with the protists that were found to be transcriptionally active in soils (14).
Fig. 1. Protistan communities, dominated by Cercozoa and Ciliophora, are composed of mostly consumers across the 180 global soils studied.
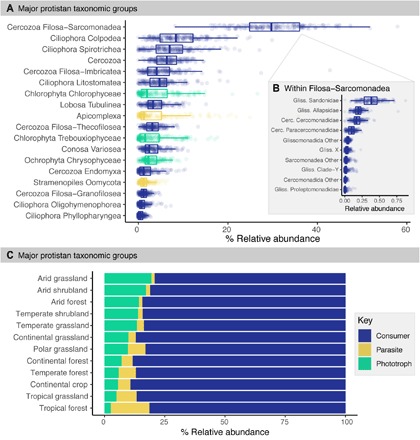
(A) Boxplots of the percent relative abundance (percentage of all protistan reads) of major soil protistan groups and (B) the percent relative abundance of lineages within the Filosea-Sarcomonadea (percentage of all Filosea-Sarcomonadea reads), and (C) the relative abundance of each general protist life history strategy (consumer, parasite, and phototroph), with samples averaged within each broad biome category.
What protistan life history strategies are most prevalent globally?
We investigated the dominant life history strategies (e.g., trophic functional groups) of soil protistan communities across ecotypes (Fig. 1). To do this, we assigned the major protistan lineages to their dominant mode of energy acquisition—either phototrophic, parasitic, or as consumers. Most soils were dominated by protistan consumers: On average, 85% of the overall protistan community was composed of consumers across the 180 soils. These consumers span a wide range of feeding strategies with diverse prey, including bacteria, fungi, algae, other protists, or even small metazoans (10, 26). Our finding that protist communities are dominated by consumers emphasizes the critical contribution of protists to turnover and energy transfer across trophic levels. Besides consumers, the protistan communities were composed of taxa putatively assigned as 10% phototrophs and 5% parasites (mean across all samples; Fig. 1). Phototrophic lineages were more abundant in arid soils, encompassing up to 40% of the total protist communities. This finding suggests that the contributions of protists to photosynthesis are particularly important in arid ecosystems. Algae outnumber cyanobacteria in some arid soils (27), and many of the phototrophs identified were algae known to be associated with soil surface biocrusts in arid environments (28). In contrast, parasitic protists were especially abundant in tropical soils, where they comprised up to 38% of the overall community.
To further investigate the relative distribution of parasites, we built environmental niche models for three predominately parasitic groups including the Apicomplexa (common parasites of soil invertebrates), the Oomycota (often plant pathogens), and the Ichthyosporea (which include many animal parasites) (15, 29). The distributions of these three parasite-dominated groups were all best predicted by climate, increasing in relative abundance with mean annual precipitation and temperature (Fig. 2). Apicomplexa exhibited particularly strong preferences for tropical soils (68.2% variation explained, P < 0.001). While previous sequence-based efforts have revealed the presence of parasites in nearly all soils (30), our results support a recent study showing increased proportional abundances of parasitic protists in the tropics (15), extending this finding with a direct comparison to colder and more arid systems. Our results suggest that parasitic protists likely have important roles in tropical ecosystems, where they may parasitize fauna, algae, or plants.
Fig. 2. Protistan parasite relative abundance is controlled by mean annual precipitation and temperature.

(A to C) Top: For the three parasite groups—Apicomplexa, Oomycota, and Ichthyosporea—the percent increase in mean squared error (MSE) of predictions when a given predictor is permuted randomly (e.g., a measure of variable importance) for all environmental predictors that were significant (P < 0.01) in the random forest models for explaining variation in parasite abundance across the 180 soils studied. For all parasite groups, mean annual temperature (MAT) and mean annual precipitation (MAP) explained the greatest percent of variation. Bottom: All three groups show a significant positive relationship with mean annual temperature and mean annual precipitation. Points represent the 180 soils, and for each group, the relative abundance of all amplicon sequence variants (ASVs) detected was summarized per site.
This evaluation of protistan communities by life history strategy has two important implications. First, given the relatively high abundance of consumers and parasites, protists may be important in controlling the abundances of other soil organisms. Second, protists are likely particularly important as primary producers in arid ecosystems. More broadly, our results demonstrate a strategy for linking the composition of protistan communities, determined via high-throughput sequencing, to the large body of literature on protist life history strategies to better understand the potential contributions of different protistan lineages in their respective ecosystems.
How predictable are protistan communities?
We found that the global-scale distribution of soil protistan communities was well explained by specific environmental parameters (fig. S4). The overall composition of protist communities was best predicted by mean annual precipitation and, to a lesser extent, mean annual temperature, soil pH, and percent total soil carbon (best model correlation with community dissimilarity = 0.77 and multiple regression analysis overall explanatory power: R2 = 61.5, P < 0.001). Although the geographic distance between sampling locations was significantly correlated with composition (Mantel ρ = 0.11, P = 0.007), this correlation was weak and did not improve the overall model fit. Our finding that precipitation is the most important factor in structuring protistan communities is consistent with previous studies that have investigated the relative importance of environmental factors (12, 17) and our understanding of the basic ecology of protists, given that most protistan lineages require water to move, feed, and reproduce (12, 31).
What are the habitat preferences of specific protistan lineages?
To assess which climatic and soil factors might be important for specific lineages, we built environmental niche models for 116 protistan lineages [Fig. 3, identified from grouping amplicon sequence variants (ASVs) into phylogenetic lineages]. For a complete list of the 116 lineages, associated habitat preferences, see table S2. We were able to determine the environmental preferences for 50 lineages (Fig. 3) from the soil and site characteristics. The top predictors were consistently climatic variables (namely, mean annual precipitation and temperature), although soil characteristics (e.g., pH and percent total C) were the best predictors for some lineages (fig. S5). We assigned the predictable lineages into six “ecological clusters” based on their shared habitat preferences (Fig. 3B), with each cluster composed of 2 to 13 protistan lineages (fig. S6). The ecological clusters were high pH (ρ = 0.81), low percent total C (ρ = −0.62), low precipitation and seasonality (ρ = −0.37), and dry (ρ = −0.82), cold (ρ = −0.74), and low pH (ρ = −0.39) environments (for all, P < 0.001).
Fig. 3. The phylogenetic distribution of protistan clades colored by their identified ecological preferences.
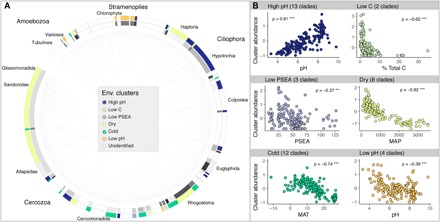
(A) Phylogenetic tree of protist clades (from 924 protist ASVs that were detected in >10% of the 180 soils). The inner ring shading indicates protist clade membership clustered at 96% patristic distance for clades with predictable ecological distributions, and the outer ring colors identify the environmental cluster memberships for those clades (see table S2 for details). PSEA, precipitation and seasonality. We note that this phylogeny is based on short read data and not intended to accurately represent broad supergroups. (B) Associations (Spearman’s ρ) between the standardized relative abundance of all clades within a particular environmental cluster (e.g., mean z scores) and the environmental predictor.
These clusters were not composed of phylogenetically concordant protistan lineages (Fig. 3A), as all clusters contained distantly related lineages, suggesting that habitat preferences are not necessarily shared across major lineages of soil protists. For example, cercozoan lineages spanned multiple ecological clusters, indicating that they can be found in a relatively wide range of habitats, a finding congruent with the observed ubiquity of Glissomonadida (32). The greatest number of protistan lineages was assigned to the “high pH” cluster (n = 13 clades; Fig. 3). This is likely because a majority of protists do not thrive in more acidic environments, although a few protists are known to withstand acidic conditions in aquatic environments (33). We recovered four lineages within the “low pH” cluster, suggesting that taxa within those lineages may also be suited to low pH soil environments (Fig. 3A and table S2).
How similar are the environmental factors that structure protistan versus prokaryotic communities?
We next investigated whether the environmental parameters that shape protistan communities were similar for bacterial and archaeal (i.e., prokaryotic) soil communities across the subset of the soil samples for which we obtained both 16S and 18S rRNA marker gene data (n = 137). Multiple regression analysis indicated that the best model for explaining variation in prokaryotic community composition included soil pH as the most important predictor, with a similar amount of overall explanatory power (R2 = 65.4, P < 0.001; fig. S4). This is consistent with the extensive previous research on the biogeography of soil prokaryotic communities (5, 8). Moreover, this finding confirms that the environmental parameters that shape protistan communities, which are most strongly associated with mean annual precipitation (fig. S4), are distinct from those that shape prokaryotic communities in soil.
Despite the contrasting environmental drivers, we still found a significant correlation between prokaryotic and protist community composition (Mantel ρ = 0.73, P = 0.001). This is likely to arise from shared habitats of particular bacterial and protistan lineages or, in some cases, potential interactions between some prokaryotic and protistan taxa in soil, such as bacterivorous protists selectively foraging on certain bacterial taxa (34). To further investigate the relationships between particular protistan and prokaryotic taxa, we visualized patterns of ASV co-occurrence (Fig. 4). Some of the observed protist-bacteria co-occurrences are likely due to shared environmental preferences, but some may be a product of direct interactions such as bacterial endosymbionts, grazing, or potential pathogenic relationships. For example, the amoebae Vermamoeba (formerly Hartmannella) is a well-documented host of pathogenic bacteria in water systems (35), and we found numerous correlations between Vermamoeba and particular bacterial taxa (including a number of actinobacteria). This either may be a product of predator-prey interactions for this common phagotroph or maybe because Vermamoeba hosts a range of intracellular bacterial taxa in soil systems. We also observed 40 correlations between Alphaproteobacteria and Colpodida. Some Alphaproteobacteria are readily consumed yet resistant to digestion by some ciliates (36), offering one potential explanation for their frequent co-occurrence. Previous work has also noted the potential relationship between some ciliate lineages and Proteobacteria (37). Although preliminary, our protist-bacteria analyses provide targets for future investigations into the myriad of ways by which bacteria and protists can interact in belowground ecosystems.
Fig. 4. Many protistan and bacterial taxa are highly correlated.
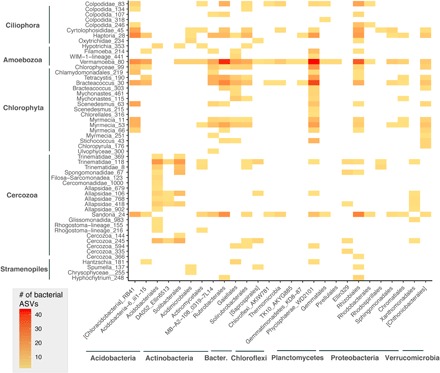
Pairwise correlation heatmap of co-occurrences between protistan and prokaryotic ASVs (those found at >10% of sites). The bacterial ASVs (x axis) are collapsed by order-level taxonomy for readability—the heatmap color indicates the number of bacterial ASVs that were correlated with a particular protistan ASV (y axis).
CAVEATS AND CONCLUSIONS
Our results, together with previous studies of soil protistan communities (12, 14, 15, 17), offer an emerging consensus of the taxa and functional groups that are most abundant in soil and how their distributions are related to soil and site characteristics. The soils included in our study span major environmental gradients representing a broad range in ecosystem types, edaphic characteristics, and climates. However, note that the geographic coverage of our collected soils was uneven, with some regions overrepresented in our sample set, including North America (n = 116 of 180 soils, with many from the United States). There are notable gaps in our sample set, as the geographic coverage of soils from Asia, Africa, and South America was limited. In addition, the distribution of samples among biomes is also uneven as we note in fig. S1. Future work studying soils from the more poorly sampled biomes and geographic regions will enable a better resolution of protist distributions and improve our understanding of which particular taxa may be restricted in their distributions.
We also note that it was not an objective of this study to quantify the total species-level diversity of protist communities found in individual soil samples or regions. Instead, our aim was to understand the distribution and ecological preferences of the more abundant and ubiquitous soil protist groups. While the total richness of protist communities across habitats and scales is an important topic, it remains difficult to accurately assess the alpha diversity of soil microbial communities using DNA sequencing–based approaches (1). This is due to well-recognized methodological challenges including potential primer biases against particular lineages and the potential overestimation of diversity arising from sequencing errors and the bioinformatic algorithms used to detect taxa (38, 39). In addition, accurately quantifying the total diversity of soil protists found in individual soils would require deeper sequencing depth (more 18S rRNA gene reads per sample) to improve the likelihood that we would capture rare taxa with our survey efforts (1).
In conclusion, our results indicate that soil protist community composition is best explained by differences in mean annual precipitation. This suggests that our knowledge of how belowground communities are structured, derived mostly from work on soil bacteria and archaea, is not generalizable to soil protists, which comprise a significant portion of the soil microbiome. By characterizing protists based on their life history strategies, we were able to quantify the relative functional contributions of protists to belowground ecosystems. Our finding that consumers dominate the protist communities in the soils studied here highlights the importance of protists as catalysts of nutrient turnover and energy transfer across trophic levels. A clear next step is to resolve the specific contributions of protistan taxa to belowground food webs beyond broad functional groups so we can begin to predict how shifts in belowground microbial communities will influence ecosystem processes. Our detection of particular soil protists and prokaryotes that co-occur globally highlights the potential importance of specific protist-bacterial interactions and their shared environmental preferences in structuring the soil microbiome. More generally, this work begins to integrate protists into the broader framework of soil microbiome research and establishes the ways in which protists differentially contribute to the functioning of belowground systems globally.
MATERIALS AND METHODS
Soil sample collection and characterization
We collected soils from 180 locations across six continents (fig. S1). The locations represent a broad range in climatic conditions (including arid, temperate, tropical, continental, and polar) and in dominant vegetation types (e.g., grasslands, forests, and shrublands). At each location, the coordinates were recorded with a Global Positioning System unit (table S1). We collected a composite mineral soil sample (top 10 cm) at each location. Immediately following field collection, each soil sample was sieved (2 mm) and separated into two subsamples. One subsample was air-dried for soil chemical analyses, and the other was frozen at −20°C for molecular analyses. For all soils, we measured pH, percent total organic carbon, percent total nitrogen, and total phosphorus concentrations using standard methods [table S1 and see (4)].
For the 180 sampling locations, we obtained climatic data from the WorldClim database based on latitude and longitude, including mean annual temperature, mean maximum temperature, mean minimum temperature, mean diurnal temperature range, precipitation and temperature seasonality, mean annual precipitation, and potential evapotranspiration. We used the normalized difference vegetation index as a proxy for net primary productivity (4). We calculated this value as a monthly average using all available data from 2000 to 2017.
Amplicon 18S rRNA gene sequencing to characterize protistan communities
To describe the protistan communities found in all 180 soils surveyed, we amplified and sequenced a portion of the 18S rRNA gene, using methods described previously (40). Briefly, we first PCR-amplified a 516–base pair (bp) portion of the V4-V5 region of 18S rRNA gene with the 616*F/1132R primer set (23), modified with Illumina adapters. This gene region provides high taxonomic resolution, as the targeted region is the most variable region in the 18S rRNA gene (23). Duplicate PCRs were completed for each of the 180 extracted samples with negative controls (both DNA extraction and no template controls) to identify any possible contamination. We cleaned the PCR products with the Qiagen UltraClean PCR Cleanup Kit, pooled the duplicate products, and performed a second round of PCR to ligate on the Nextera barcodes to enable multiplexed sequencing, as per the manufacturer’s instructions. We normalized sample concentrations with the SequalPrep Normalization Plate Kit (Invitrogen) and then sequenced samples with the 2 × 300 bp paired-end chemistry on the Illumina MiSeq platform at the University of Colorado Next Generation Sequencing Center.
Raw reads were demultiplexed and then merged and quality-filtered with UPARSE (38). We trimmed all merged reads to 400 bp to retain a larger portion of high-quality reads. We then identified ASVs with the UNOISE3 algorithm, as described previously (41). Sequences were quality-filtered to a maximum expected error threshold of 1.0, and then ASVs were identified using the UNOISE3 default settings. We chose to identify ASVs rather than clustering sequences based on an arbitrary sequence similarity threshold (e.g., 97%), as ASVs are more comparable across studies and permit more robust downstream analyses [as we are not basing our downstream analyses on a sequence representative of a cloud of similar sequences, see (42)]. In addition, a recent study on soil flagellates suggested that some diversity may be obscured when sequences are clustered at lower sequence similarity thresholds (43). However, we note that the ASVs identified here do not equate to species-level diversity (44), as some individual protistan genomes are known to contain distinct 18S rRNA genes due to intragenomic heterogeneity (45) and, conversely, some distinct protists may share identical sequences across the targeted gene region.
We used the Protist Ribosomal Reference database (PR2, v4.10) to assign taxonomic classifications to ASVs and filtered out those ASVs with less than an 80% identity match to any sequences in the PR2 database (46). We then removed any soil samples with ≥2500 protist reads per sample to remove those samples that failed to yield an adequate number of 18S rRNA gene reads for downstream analyses. We note that protistan richness and community composition were highly correlated whether the initial threshold was ≥2500 (n = 180 samples retained), 5000 (n = 170 samples retained), or 10,000 reads per sample (n = 132 samples retained) (Spearman ρ > 0.99 with P < 0.001 for all richness correlations and Spearman ρ > 0.99 for all Mantel correlations of dissimilarity in community composition). We therefore selected the lowest initial cutoff threshold of 2500 reads per sample to retain the greatest number of samples for downstream analyses. The 18S rRNA gene primer is conserved across eukaryotes, so we filtered out non-protistan ASVs, defined as those that were assigned to the Metazoa, Embryophyta, and Fungi, before downstream analyses. We also filtered out any ASVs represented by fewer than 10 reads across the dataset to be conservative in what ASVs we consider present and removed those ASVs that were not assigned to at least the class level of resolution in the PR2 database. We normalized the resulting ASV table via cumulative sum scaling (47).
Determination of dominant protistan lineages and community analyses
As some of the climatic and soil edaphic properties were well correlated across this sample set, we reduced collinearity by dropping a subset of variables that were highly correlated (Pearson’s r ≥ 0.8; fig. S2). Our final set of environmental predictors included measured soil properties (soil pH and total carbon) and associated climatic and geographic variables (latitude, longitude, mean annual temperature, maximum annual temperature, mean diurnal range, mean annual precipitation, potential evapotranspiration, and grassland or forest land type).
To estimate the percentages of different protistan life history strategies (e.g., functional groups) across ecotypes, we assigned the major protistan taxonomic groups to a broad life history strategy as either a consumer, parasite, or phototroph (48). We recognize that these broad categories do not necessarily capture all of the diversity within a particular lineage. For example, we assigned all photosynthetic lineages as “phototrophic,” although some lineages may not be exclusively phototrophic. Rather, we sought to represent the dominant life history strategy within each lineage [similar to (18)]. In addition, “consumers” include protists that feed on diverse prey from bacteria to algae, fungi, and even soil fauna such as nematodes and invertebrates and use diverse strategies for capturing prey (for example, via filter feeding, phagocytosis, or cytotrophy). However, for many lineages, too little is known of the specific feeding preferences to confidently categorize taxa by their preferences, so the risk of false-positive annotations would be too high. Thus, for a broad representation of the major life history strategies, we grouped all consumers together.
Dissimilarities in community composition were calculated with the Bray-Curtis metric on square root–transformed relative abundances (49) and visualized as ordinations with nonmetric multidimensional scaling (k = 2). We tested the correlation between pairwise distances in protist community composition to pairwise geographic distances by calculating geographic distances between sampling sites from latitude and longitude values (using geosphere package v. 1.5-7) and then using a Mantel test against Bray-Curtis community distances (Spearman’s rank correlations) with 999 permutations. To identify the best explanatory model associated with differences in overall protist community composition across soils, we first used BIOENV (49) to identify the subset of environmental variables that maximize that correlation. We then conducted multiple regression analysis on distance matrices to estimate the overall explanatory power of the model.
Shotgun metagenomic-based identification of dominant protistan taxa
The soils for which we obtained shotgun metagenomic data come from two sources: (i) a range of distinct ecosystem types found across U.S. Long Term Ecological Research Network sites [21 samples; the full field sampling methods are described in (50)] and (ii) the same sampling locations in Panama described above (25 samples; see table S3). We prepared the samples for shotgun metagenomic sequencing using an approach described previously (51), and the 46 libraries were sequenced on the Illumina NextSeq platform with 2 × 150 bp chemistry at the University of Colorado Next Generation Sequencing Facility. We filtered the raw reads with Sickle (52) using the parameters -q 50 and -I 20. Samples averaged ~11.6 million reads after quality filtering. We used Metaxa2 (53) to compare the detection of dominant protistan lineages from small subunit (SSU) rRNA gene sequences extracted from the metagenomic data to our amplicon-based data. We note that we were only able to detect the most abundant protists by analyzing the shotgun metagenomic data, as the 18S rRNA genes detected with Metaxa2 represented an extremely small percentage of the metagenomic reads on average (<0.0000015% of the ~366 million reads). To compare the two datasets, we summarized protist reads for metagenomic and amplicon samples separately at the fourth taxonomic level (e.g., class level, although PR2 taxonomic levels are unranked) and then computed the mean abundance of each major lineage across samples to compare detection of the top 50 protistan lineages (ranked by mean abundance) for both methods.
Identification of protistan lineages with shared ecological preferences
Our next aim was to identify the habitat preferences for particular protistan lineages that were relatively dominant across many of the soil sites. To do this, we focused on those ASVs that were found in >10% of the 180 sites for which we had amplicon 18S rRNA gene sequence data (e.g., detected in >18 sites) and then built a phylogeny to identify the dominant protist clades for modeling of ecological preferences (we modeled clades instead of individual ASVs because too few ASVs were shared across samples) (fig. S3). To build the phylogeny, we first aligned ASVs along with full-length nearest neighbors using the Silva Incremental Aligner (54). After alignment, we trimmed with a gap threshold of 0.2. We used FastTree with a GTR model of nucleotide evolution to obtain approximately maximum likelihood phylogenetic trees with the Crenarchaeota sequence (AQYM01000016) included as the outgroup (55). We clustered ASVs into dominant protist clades with RAMI (56) with a patristic distance of 96% and retained protist clades with at least two members (n = 116). We acknowledge that our approach will not allow for accurate phylogenetic inferences across the vast diversity of protistan life, which necessitates the use of multiple genes for accurate phylogenetic placement. However, our focus is on determining major differences across larger groups of protists without showing fine-scale phylogenetic differences across all groups.
We then used random forest analysis [as per (4)] to identify the environmental niche models for each of the ubiquitous protist clades. Our models included nine environmental predictors: soil pH, total C, mean annual precipitation, potential evapotranspiration, precipitation and seasonality, mean maximum temperature, mean diurnal temperature range, and dominant vegetation types (forest and grasslands). For random forest analyses, the importance of a predictor is assessed by determining the decrease in prediction accuracy (mean squared error) between observations and out-of-bag predictions (one-third of data are withheld and used to assess fit) when that predictor is permuted randomly (averaged across all trees). These analyses were done with the rfPermute package v.2.1 (57), with 1000 trees per model. We considered a protist clade predictable if the model explained at least 20% of the variation in the distribution of the clade. We then clustered the protist clades with predictable habitat preferences into ecological groups. To do this, we first used semipartial Spearman correlations to elucidate the directional response of protist lineages to environmental predictors and to quantify the uniquely predictable portion of variance for environmental predictors. We then used the correlation coefficients from significant semipartial correlations (P < 0.05) to cluster protist clades into ecological clusters with hierarchical cluster analysis (method = ward.D2). We also calculated the standardized relative abundances (z scores) of the protist clades within each cluster across samples and plotted these results by the dominant environmental preference for each ecological cluster.
We also separately investigated the habitat preferences of three major lineages of protists that are composed of predominately parasitic lineages (Apicomplexa, Oomycota, and Ichthyosporea). We chose to investigate these groups due to their ecological relevance as parasites (30, 58) and because many ASVs within these lineages were restricted to less than 10 locations. We first summarized the mean relative abundance of all ASVs for each parasite group across the 180 sites and then used random forest models to investigate the habitat preferences of these groups, as described above.
Identifying associations between soil protists and prokaryotes
We also acquired 16S rRNA gene sequence data (bacterial and archaeal) for 137 of 180 samples for which we have protist 18S rRNA gene data (4, 59). In brief, a portion of the V3-V4 region of the 16S rRNA gene was amplified and sequenced on the Illumina MiSeq. Raw reads were processed with the pipeline as described above, and all merged reads were trimmed to 410 bp. We then identified ASVs with UNOISE3 (41). Taxonomy was assigned with the Greengenes database (60). We set a minimum sequence threshold of 10,000 reads per sample and, as per our 18S rRNA gene sequencing dataset, normalized the taxa table with cumulative sum scaling. We compared the community structure of bacteria and archaea across the 137 soil samples, as we described above for protists. In brief, we assessed the factors that explain differences in overall community composition with BIOENV analysis to identify the best model and then multiple regression analysis on distance matrices to obtain an overall estimate for R2. To investigate which bacterial ASVs were closely associated with protistan ASVs, we computed pairwise Spearman correlations after filtering both datasets to only include those ASVs that were detected in greater than 10% of sites (site minimum = 14) and relatively abundant (filter threshold = 0.0001). We only considered two ASVs to be correlated if the Spearman’s ρ was greater than 0.6 with a P value of less than 1 × 10−9.
All statistical analyses were performed in R, and the raw amplicon and shotgun metagenomics sequence data have been deposited in the Sequence Read Archive (National Center for Biotechnology Information BioProject accession no. PRJNA554847), and the data used in this study are publicly available from Figshare (doi: 10.6084/m9.figshare.7845167).
Supplementary Material
Acknowledgments
We thank D. Agudo, M. Foley, M. Gebert, and J. Henley for assistance with field and laboratory work. Funding: A.M.O. acknowledges support from the NSF Graduate Research Fellowship and the CIRES Graduate Fellowship. S.G. acknowledges support from the NWO-VENI grant from the Netherlands Organisation for Scientific Research (016.Veni.181.078). M.D.-B. acknowledges the support from the Marie Skłodowska-Curie Actions of the Horizon 2020 Framework Programme H2020-MSCA-IF-2016 under REA grant agreement no. 702057 (CLIMIFUN). F.T.M. acknowledges support from European Research Council grant agreement no. 647038 (BIODESERT). Support to N.F. was provided by the Simons Foundation, the Smithsonian Tropical Research Institute, and the U.S. NSF (DEB 1556090). Author contributions: A.M.O., S.G., and N.F. designed the study. N.F., B.L.T., and F.T.M provided the soil. A.M.O. analyzed the data with input from S.G., M.D.-B., S.G., and N.F. A.M.O. and N.F. wrote the manuscript with contributions from all coauthors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Raw sequence data have been deposited to the SRA (BioProject accession no. PRJNA554847), and other data are publicly available from Figshare (doi: 10.6084/m9.figshare.7845167).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/4/eaax8787/DC1
Fig. S1. Site locations for the 180 unique soil samples for which we obtained 18S rRNA gene sequence data (green circles; 137 samples for which we also obtained 16S rRNA gene amplicon data) and 46 samples for which we obtained SSU rRNA gene data from shotgun metagenomic sequencing (blue circles).
Fig. S2. Pairwise correlation heatmap for the climatic and edaphic characteristics associated with the 180 soils analyzed.
Fig. S3. Detection of the top 50 dominant protistan lineages by 18S rRNA amplicon sequencing versus shotgun metagenomic sequencing.
Fig. S4. Factors shaping protistan community (namely, climatic variables) are distinct from those factors shaping prokaryotic communities (namely, pH).
Fig. S5. The top predictors of ubiquitous protist lineages (protistan ASVs were phylogenetically clustered into clades at 96% similarity).
Fig. S6. A heatmap of the correlation coefficients based on semipartial correlations between the relative abundance of protistan clades (one clade per row) that had significant semipartial correlations to environmental predictors (n = 43 of 50 protist clades that were predictable in distribution).
Table S1. Location and site information for the 180 soils for which we obtained 18S rRNA gene amplicon data.
Table S2. Information on the 116 abundant protistan lineages (phylogenetic clades) including taxonomic affiliation, the percent variance explained (var.expl), the top environmental predictor from random forest models (predictor1), the number of ASVs in the lineage (freq.), and the ecological cluster affiliation.
Table S3. Location and site information for the 46 soils for which we obtained shotgun metagenomic data.
REFERENCES AND NOTES
- 1.Fierer N., Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Bardgett R. D., van der Putten W. H., Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Fierer N., Jackson R. B., The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delgado-Baquerizo M., Oliverio A. M., Brewer T. E., Benavent-González A., Eldridge D. J., Bardgett R. D., Maestre F. T., Singh B. K., Fierer N., A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Bahram M., Hildebrand F., Forslund S. K., Anderson J. L., Soudzilovskaia N. A., Bodegom P. M., Bengtsson-Palme J., Anslan S., Coelho L. P., Harend H., Huerta-Cepas J., Medema M. H., Maltz M. R., Mundra S., Olsson P. A., Pent M., Põlme S., Sunagawa S., Ryberg M., Tedersoo L., Bork P., Structure and function of the global topsoil microbiome. Nature 560, 233–237 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Tedersoo L., Bahram M., Põlme S., Kõljalg U., Yorou N. S., Wijesundera R., Ruiz L. V., Vasco-Palacios A. M., Thu P. Q., Suija A., Smith M. E., Sharp C., Saluveer E., Saitta A., Rosas M., Riit T., Ratkowsky D., Pritsch K., Põldmaa K., Piepenbring M., Phosri C., Peterson M., Parts K., Pärtel K., Otsing E., Nouhra E., Njouonkou A. L., Nilsson R. H., Morgado L. N., Mayor J., May T. W., Majuakim L., Lodge D. J., Lee S. S., Larsson K.-H., Kohout P., Hosaka K., Hiiesalu I., Henkel T. W., Harend H., Guo L.-d., Greslebin A., Grelet G., Geml J., Gates G., Dunstan W., Dunk C., Drenkhan R., Dearnaley J., De Kesel A., Dang T., Chen X., Buegger F., Brearley F. Q., Bonito G., Anslan S., Abell S., Abarenkov K., Global diversity and geography of soil fungi. Science 346, 1256688 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Auguet J.-C., Barberan A., Casamayor E. O., Global ecological patterns in uncultured Archaea. ISME J. 4, 182–190 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Gubry-Rangin C., Hai B., Quince C., Engel M., Thomson B. C., James P., Schloter M., Griffiths R. I., Prosser J. I., Nicol G. W., Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U.S.A. 108, 21206–21211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adl M. S., Gupta V. S., Protists in soil ecology and forest nutrient cycling. Can. J. For. Res. 36, 1805–1817 (2006). [Google Scholar]
- 10.Geisen S., Mitchell E. A. D., Adl S., Bonkowski M., Dunthorn M., Ekelund F., Fernández L. D., Jousset A., Krashevska V., Singer D., Spiegel F. W., Walochnik J., Lara E., Soil protists: A fertile frontier in soil biology research. FEMS Microbiol. Rev. 42, 293–323 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Geisen S., Mitchell E. A. D., Wilkinson D. M., Adl S., Bonkowski M., Brown M. W., Fiore-Donno A. M., Heger T. J., Jassey V. E. J., Krashevska V., Lahr D. J. G., Marcisz K., Mulot M., Payne R., Singer D., Anderson O. R., Charman D. J., Ekelund F., Griffiths B. S., Rønn R., Smirnov A., Bass D., Belbahri L., Berney C., Blandenier Q., Chatzinotas A., Clarholm M., Dunthorn M., Feest A., Fernández L. D., Foissner W., Fournier B., Gentekaki E., Hájek M., Helder J., Jousset A., Koller R., Kumar S., La Terza A., Lamentowicz M., Mazei Y., Santos S. S., Seppey C. V. W., Spiegel F. W., Walochnik J., Winding A., Lara E., Soil protistology rebooted: 30 fundamental questions to start with. Soil Biol. Biochem. 111, 94–103 (2017). [Google Scholar]
- 12.Geisen S., Bandow C., Römbke J., Bonkowski M., Soil water availability strongly alters the community composition of soil protists. Pedobiologia 57, 205–213 (2014). [Google Scholar]
- 13.Bonkowski M., Protozoa and plant growth: The microbial loop in soil revisited. New Phytol. 162, 617–631 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Geisen S., Tveit A. T., Clark I. M., Richter A., Svenning M. M., Bonkowski M., Urich T., Metatranscriptomic census of active protists in soils. ISME J. 9, 2178–2190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahé F., de Vargas C., Bass D., Czech L., Stamatakis A., Lara E., Singer D., Mayor J., Bunge J., Sernaker S., Siemensmeyer T., Trautmann I., Romac S., Berney C., Kozlov A., Mitchell E. A. D., Seppey C. V. W., Egge E., Lentendu G., Wirth R., Trueba G., Dunthorn M., Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests. Nat. Ecol. Evol. 1, 0091 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Venter P. C., Nitsche F., Arndt H., The hidden diversity of flagellated protists in soil. Protist 169, 432–449 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Bates S. T., Clemente J. C., Flores G. E., Walters W. A., Parfrey L. W., Knight R., Fierer N., Global biogeography of highly diverse protistan communities in soil. ISME J. 7, 652–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont A. Ö. C., Griffiths R. I., Bell T., Bass D., Differences in soil micro-eukaryotic communities over soil pH gradients are strongly driven by parasites and saprotrophs. Environ. Microbiol. 18, 2010–2024 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Acosta-Mercado D., Lynn D. H., Soil ciliate species richness and abundance associated with the rhizosphere of different subtropical plant species. J. Eukaryot. Microbiol. 51, 582–588 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Turner T. R., Ramakrishnan K., Walshaw J., Heavens D., Alston M., Swarbreck D., Osbourn A., Grant A., Poole P. S., Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7, 2248–2258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saleem M., Fetzer I., Harms H., Chatzinotas A., Diversity of protists and bacteria determines predation performance and stability. ISME J. 7, 1912–1921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Görtz H.-D., Towards an understanding of the distribution, dynamics and ecological significance of bacterial symbioses in protists. Denisia 23, 307–311 (2008). [Google Scholar]
- 23.Hugerth L. W., Muller E. E. L., Hu Y. O. O., Lebrun L. A. M., Roume H., Lundin D., Wilmes P., Andersson A. F., Systematic design of 18S rRNA gene primers for determining eukaryotic diversity in microbial consortia. PLOS ONE 9, e95567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venter P. C., Nitsche F., Domonell A., Heger P., Arndt H., The protistan microbiome of grassland soil: Diversity in the mesoscale. Protist 168, 546–564 (2017). [DOI] [PubMed] [Google Scholar]
- 25.George P., Lallias D., Creer S., Seaton F. M., Kenny J. G., Eccles R. M., Griffiths R. I., Lebron I., Emmett B. A., Robinson D. A., Jones D. L., Divergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems. Nat. Commun. 10, 1107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seppey C. V. W., Singer D., Dumack K., Fournier B., Belbahri L., Mitchell E. A. D., Lara E., Distribution patterns of soil microbial eukaryotes suggests widespread algivory by phagotrophic protists as an alternative pathway for nutrient cycling. Soil Biol. Biochem. 112, 68–76 (2017). [Google Scholar]
- 27.Johansen J., Ashlel J., Rayburn W., Effects of rangefire on soil algal crusts in semiarid shrub-steppe of the lower Columbia Basin and their subsequent recovery. Gt. Basin Nat. 53, 73–88 (1993). [Google Scholar]
- 28.R. Rosentreter, J. Belnap, in Biological Soil Crusts: Structure, Function, and Management (Springer, 2001), pp. 31–50. [Google Scholar]
- 29.Sapkota R., Nicolaisen M., An improved high throughput sequencing method for studying oomycete communities. J. Microbiol. Methods 110, 33–39 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Geisen S., Laros I., Vizcaíno A., Bonkowski M., de Groot G. A., Not all are free-living: High-throughput DNA metabarcoding reveals a diverse community of protists parasitizing soil metazoa. Mol. Ecol. 24, 4556–4569 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Clarholm M., Protozoan grazing of bacteria in soil-impact and importance. Microb. Ecol. 7, 343–350 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Howe A. T., Bass D., Vickerman K., Chao E. E., Cavalier-Smith T., Phylogeny, taxonomy, and astounding genetic diversity of Glissomonadida ord. nov., the dominant gliding zooflagellates in soil (Protozoa: Cercozoa). Protist 160, 159–189 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Nixdorf B., Mischke U., Leßmann D., Chrysophytes and chlamydomonads: Pioneer colonists in extremely acidic mining lakes (pH < 3) in Lusatia (Germany). Hydrobiologia 369, 315–327 (1998). [Google Scholar]
- 34.Schulz-Bohm K., Geisen S., Wubs E. R. J., Song C., de Boer W., Garbeva P., The prey’s scent – Volatile organic compound mediated interactions between soil bacteria and their protist predators. ISME J. 11, 817–820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagnier I., Valles C., Raoult D., La Scola B., Isolation of Vermamoeba vermiformis and associated bacteria in hospital water. Microb. Pathog. 80, 14–20 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Gong J., Qing Y., Zou S., Fu R., Su L., Zhang X., Zhang Q., Protist-bacteria associations: Gammaproteobacteria and Alphaproteobacteria are prevalent as digestion-resistant bacteria in ciliated protozoa. Front. Microbiol. 7, 498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi A., Bellone A., Fokin S. I., Boscaro V., Vannini C., Detecting associations between ciliated protists and prokaryotes with culture-independent single-cell microbiomics: A proof-of-concept study. Microb. Ecol. 78, 232–242 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Edgar R. C., UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Xiong W., Li R., Gou S., Karlsson I., Jiao Z., Xun W., Kowalchuk G. A., Shen Q., Geisen S., Microbial amendments alter protist communities within the soil microbiome. Soil Biol. Biochem. 135, 379–382 (2019). [Google Scholar]
- 40.Oliverio A. M., Power J. F., Washburne A., Cary S. C., Stott M. B., Fierer N., The ecology and diversity of microbial eukaryotes in geothermal springs. ISME J. 12, 1918–1928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R. C., Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Callahan B. J., McMurdie P. J., Holmes S. P., Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venter P. C., Nitsche F., Scherwass A., Arndt H., Discrepancies between molecular and morphological databases of soil ciliates studied for temperate grasslands of central Europe. Protist 169, 521–538 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Caron D. A., Hu S. K., Are we overestimating protistan diversity in nature? Trends Microbiol. 27, 197–205 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Weber A. A.-T., Pawlowski J., Wide occurrence of SSU rDNA intragenomic polymorphism in Foraminifera and its implications for molecular species identification. Protist 165, 645–661 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L., Boutte C., Burgaud G., de Vargas C., Decelle J., del Campo J., Dolan J. R., Dunthorn M., Edvardsen B., Holzmann M., Kooistra W. H., Lara E., Le Bescot N., Logares R., Mahé F., Massana R., Montresor M., Morard R., Not F., Pawlowski J., Probert I., Sauvadet A.-L., Siano R., Stoeck T., Vaulot D., Zimmermann P., Christen R., The protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rrna sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. N. Paulson, metagenomeSeq: Statistical analysis for sparse high-throughput sequencing. Bioconductor Jp. (2014).
- 48.Adl S. M., Bass D., Lane C. E., Lukeš J., Schoch C. L., Smirnov A., Agatha S., Berney C., Brown M. W., Burki F., Cárdenas P., Čepička I., Chistyakova L., del Campo J., Dunthorn M., Edvardsen B., Eglit Y., Guillou L., Hampl V., Heiss A. A., Hoppenrath M., James T. Y., Karnkowska A., Karpov S., Kim E., Kolisko M., Kudryavtsev A., Lahr D. J. G., Lara E., Le Gall L., Lynn D. H., Mann D. G., Massana R., Mitchell E. A. D., Morrow C., Park J. S., Pawlowski J. W., Powell M. J., Richter D. J., Rueckert S., Shadwick L., Shimano S., Spiegel F. W., Torruella G., Youssef N., Zlatogursky V., Zhang Q., Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. Oksanen, F. G. Blanchet, R. Kindt, P. Legendre, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. Stevens, H. Wagner, Vegan: Community ecology package, R Package version 2.4-3 (2017); doi: 10.4135/9781412971874.n145. [DOI]
- 50.Crowther T. W., Maynard D. S., Leff J. W., Oldfield E. E., McCulley R. L., Fierer N., Bradford M. A., Predicting the responsiveness of soil biodiversity to deforestation: A cross-biome study. Glob. Chang. Biol. 20, 2983–2994 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Baym M., Kryazhimskiy S., Lieberman T. D., Chung H., Desai M. M., Kishony R., Inexpensive multiplexed library preparation for megabase-sized genomes. PLOS ONE 10, e0128036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.T. Y. James et al., Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. Bioinformatics (2011).
- 53.Bengtsson-Palme J., Hartmann M., Eriksson K. M., Pal C., Thorell K., Larsson D. G. J., Nilsson R. H., metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 15, 1403–1414 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Pruesse E., Peplies J., Glöckner F. O., SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price M. N., Dehal P. S., Arkin A. P., FastTree 2—Approximately maximum-likelihood trees for large alignments. PLOS ONE 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pommier T., Canbäck B., Lundberg P., Hagström Å., Tunlid A., RAMI: A tool for identification and characterization of phylogenetic clusters in microbial communities. Bioinformatics 25, 736–742 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E. Archer, rfPermute: Estimate permutation p-values for Random Forest importance metrics. R Package 2 (2016).
- 58.Siński E., Behnke J. M., Apicomplexan parasites: Environmental contamination and transmission. Pol. J. Microbiol. 53, 67–73 (2004). [PubMed] [Google Scholar]
- 59.C. Walsh, M. Gebert, M. Delgado-Baquerizo, F. T. Maestre, N. Fierer, A global survey of mycobacterial diversity in soil (2019); bioRxiv 562439 [Preprint]. 27 February 2019; 10.1101/562439. [DOI] [PMC free article] [PubMed]
- 60.McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., Hugenholtz P., An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/4/eaax8787/DC1
Fig. S1. Site locations for the 180 unique soil samples for which we obtained 18S rRNA gene sequence data (green circles; 137 samples for which we also obtained 16S rRNA gene amplicon data) and 46 samples for which we obtained SSU rRNA gene data from shotgun metagenomic sequencing (blue circles).
Fig. S2. Pairwise correlation heatmap for the climatic and edaphic characteristics associated with the 180 soils analyzed.
Fig. S3. Detection of the top 50 dominant protistan lineages by 18S rRNA amplicon sequencing versus shotgun metagenomic sequencing.
Fig. S4. Factors shaping protistan community (namely, climatic variables) are distinct from those factors shaping prokaryotic communities (namely, pH).
Fig. S5. The top predictors of ubiquitous protist lineages (protistan ASVs were phylogenetically clustered into clades at 96% similarity).
Fig. S6. A heatmap of the correlation coefficients based on semipartial correlations between the relative abundance of protistan clades (one clade per row) that had significant semipartial correlations to environmental predictors (n = 43 of 50 protist clades that were predictable in distribution).
Table S1. Location and site information for the 180 soils for which we obtained 18S rRNA gene amplicon data.
Table S2. Information on the 116 abundant protistan lineages (phylogenetic clades) including taxonomic affiliation, the percent variance explained (var.expl), the top environmental predictor from random forest models (predictor1), the number of ASVs in the lineage (freq.), and the ecological cluster affiliation.
Table S3. Location and site information for the 46 soils for which we obtained shotgun metagenomic data.


