Abstract
Poor maternal nutrition during gestation can have immediate and life-long negative effects on offspring growth and health. In livestock, this leads to reduced product quality and increased costs of production. Based on previous evidence that both restricted- and overfeeding during gestation decrease offspring muscle growth and alter metabolism postnatally, we hypothesized that poor maternal nutrition during gestation would reduce the growth and development of offspring muscle prenatally, reduce the number of myogenic progenitor cells, and result in changes in the global expression of genes involved in prenatal muscle development and function. Ewes were fed a control (100% NRC)-, restricted (60% NRC)-, or overfed (140% NRC) diet beginning on day 30 of gestation until days 45, 90, and 135 of gestation or until parturition. At each time point fetuses and offspring (referred to as CON, RES, and OVER) were euthanized and longissimus dorsi (LM), semitendinosus (STN), and triceps brachii (TB) were collected at each time point for histological and RNA-Seq analysis. In fetuses and offspring, we did not observe an effect of diet on cross-sectional area (CSA), but CSA increased over time (P < 0.05). At day 90, RES and OVER had reduced secondary:primary muscle fiber ratios in LM (P < 0.05), but not in STN and TB. However, in STN and TB percent PAX7-positive cells were decreased compared with CON (P < 0.05). Maternal diet altered LM mRNA expression of 20 genes (7 genes downregulated in OVER and 2 downregulated in RES compared with CON; 5 downregulated in OVER compared with RES; false discovery rate (FDR)-adj. P < 0.05). A diet by time interaction was not observed for any genes in the RNA-Seq analysis; however, 2,205 genes were differentially expressed over time between days 90 and 135 and birth (FDR-adj. P < 0.05). Specifically, consistent with increased protein accretion, changes in muscle function, and increased metabolic activity during myogenesis, changes in genes involved in cell cycle, metabolic processes, and protein synthesis were observed during fetal myogenesis. In conclusion, poor maternal nutrition during gestation contributes to altered offspring muscle growth during early fetal development which persists throughout the fetal stage. Based on muscle-type-specific effects of maternal diet, it is important to evaluate more than one type of muscle to fully elucidate the effects of maternal diet on offspring muscle development.
Keywords: fetal programming, muscle, myogenesis, nutrition, sheep
Introduction
With the estimated increase in the human population to 9.8 billion by 2050 (United Nations, 2017), it is imperative that we identify methods to improve efficiency of production that provide adequate, affordable, and high-quality animal protein to consumers. Genetics, diet, management, and health all have important roles in livestock production and efficiency of growth. In particular, there is considerable evidence demonstrating that the quality of maternal nutrition during pregnancy has a major role in offspring development and growth (Hales and Barker, 2001; Armitage et al., 2004; Du et al., 2010a; Ford and Long, 2011; Wu et al., 2012; Hoffman et al., 2017). Specifically, poor maternal nutrition, which can be defined as either restricted or excessive macro- or micro-nutrient intake during gestation, has been found to negatively affect a variety of tissues, organs, and cell lineages in the offspring both pre- and postnatally (Wu et al., 2006; Zhu et al., 2006; Nesterenko and Aly, 2009; Reed et al., 2014; Hoffman et al., 2016b; Long et al., 2015; Pillai et al., 2017). Exposure to adverse conditions during gestation, including poor maternal nutrition, predispose offspring to altered metabolic and endocrine regulation of growth and maintenance later in life as a long-term consequence of fetal programming (Barker, 1995; Wu et al., 2006). Despite accumulating phenotypic evidence (e.g., reduced muscle, increased adipose, and altered metabolism) of the effects of poor maternal nutrition on offspring development, the mechanisms contributing to these changes are not completely understood.
Skeletal muscle is an important protein source and has a key role in glucose homeostasis and oxidative metabolism. Poor maternal nutrition is known to reduce postnatal muscle mass, thereby reducing insulin sensitivity and altering insulin-mediated metabolic pathways in the offspring (Morrison et al., 2010). Given that net muscle fiber number is established prenatally, adequate postnatal muscle development is dependent upon prenatal myogenesis and muscle development (Rehfeldt et al., 2004; Zhu et al., 2006; Reed et al., 2014). Indeed, we and others have found that both maternal nutrient restriction and overnutrition during gestation reduce muscle mass and muscle fiber cross-sectional area (CSA), reduce muscle fiber number, and increase adiposity in offspring during postnatal growth (Huang et al., 2010; Yan et al., 2012; Reed et al., 2014; Hoffman et al., 2016a). These findings demonstrate that prenatal muscle development is impaired by poor maternal nutrition, both restricted- and overfeeding, with consequences that persist into postnatal growth where long-term development and growth can be compromised. Therefore, to elucidate the mechanisms contributing to these changes, evaluating both diets in the same experiment is necessary.
The development of skeletal muscle is reliant on the proper differentiation of myogenic progenitor cells. Myogenic progenitor cells are regulated by the temporal expression of the myogenic regulatory factor (MRF) family and disruption in the pattern or timing of their expression can adversely affect prenatal myogenesis, and therefore postnatal muscle structure and composition (Yablonka-Reuveni and Rivera, 1994; Du et al., 2010a). We previously found that maternal nutrient restriction alters the temporal expression of MRF, MyoD, and myogenin, in satellite cells isolated from offspring at both birth and 3 months of age (Raja et al., 2016); however, the effects of maternal diet on the temporal expression of these MRFs in vivo during fetal development have been less studied. Proper temporal expression of MRFs results in the differentiation of myogenic progenitor cells and fusion into primary and secondary fibers during early and mid-gestation. These fibers then continue to mature and hypertrophy during late gestation. Thus, it is important to assess time points reflecting the three major steps in myogenesis: primary myogenesis, secondary myogenesis, and hypertrophy.
Based on our previous work, we hypothesized that poor maternal nutrition during gestation would reduce the growth and development of offspring muscle prenatally as well as affect the number of myogenic progenitor cells. Additionally, we hypothesized that maternal diet would alter the global gene expression of factors involved in prenatal muscle development and function. To test these hypotheses, we combined transcriptomic and immunohistochemical approaches to determine the effects of both maternal nutrient restriction and overnutrition on offspring prenatal muscle development, as well as temporal expression of genes during fetal development.
Materials and Methods
Animals
All animal experiments were reviewed and approved by the University of Connecticut Institutional Animal Care and Use Committee (A13-059).
As previously described (Pillai et al., 2017), 82 multiparous Western White-faced ewes were estrus synchronized using a progesterone controlled intravaginal drug release device (Easi-Breed CIDR Sheep Insert, Zoetis Inc., Parsippany, NJ), after which a single intramuscular injection of prostaglandin F2α was administered (Lutalyse, Zoetis Inc.). Ewes were then housed with one of four related Dorset breeding rams and breeding date was determined via rump markings on ewes. After confirmation of pregnancy using ultrasound at day 28 ± 0.5 of gestation (Jones et al., 2016), animals were individually housed and assigned to one of three diets; control-fed (100%; n = 27), restricted-fed (60%; n = 28) or overfed (140%; n = 27) based on the NRC requirements for a ewe carrying twins (National Research, 2007) starting at day 30.2 ± 0.2 of gestation. Diets were adjusted weekly based on individual ewe body weight.
Ewes remained on their respective diets until day 45, 90, or 135 of gestation. At each of these time points, a subset of ewes (n = 5 to 7 per dietary treatment) was euthanized with an i.v. injection of Beuthansia-D Special (390 mg/mL sodium pentobarbital and 50 mg/mL phenytoin; Merck Animal Health, Summit, NJ) based on BW, followed by exsanguination. A hysterectomy was performed to remove the uterus and all fetuses for fetal sample collection. Another subset of ewes (birth; n = 5 to 7 per dietary treatment) was allowed to undergo natural parturition and lambs nursed for up to 24 h, after which they were euthanized as described above to collect samples. Offspring born to control-, restricted-, and overfed ewes are referred to as CON, RES, and OVER, respectively. A complete description of litter size and gender distribution was previously described (Pillai et al., 2017).
Sample collection and processing
Muscle samples for histological analysis were collected from the mid-belly of the longissimus dorsi (LM), semitendinosus (STN), and triceps brachii (TB) at each time point from each fetus (n = 10 to 15 fetuses or lambs per dietary treatment per time point). At day 45, the entire muscle was collected. At days 90, 135, and within 24 h of birth, an ~1 cm3 core sample was collected from each muscle. We performed these analyses during early- mid-, and late-gestation to coincide with developmental periods of myogenesis, and included an early postnatal time point (Du et al., 2015). Muscle samples were embedded in Tissue-Tek optimal cutting temperature medium (Fisher Scientific, Pittsburg, PA) and frozen in liquid nitrogen cooled isopentane. Samples for RNA extraction were taken from the LM at days 90, 135, and birth and immediately snap frozen in liquid nitrogen. Samples were stored at −80 °C until further use.
Immunohistochemistry
Muscle fiber CSA, number of primary and secondary fibers, and percentage of paired box (PAX)7-positive [PAX7(+)] progenitor cells were visualized using immunohistological procedures [Figure 1; (Town et al., 2004; Reed et al., 2014)]. Muscle samples were sectioned at 10 µm using a Microm HM 525 cryostat (Thermo Scientific, Waltham, MA) and fixed in 4% paraformaldehyde for 20 min followed by three 5 min washes with PBS. Sections were blocked with 5% horse serum, 0.2% Triton X-100 in PBS for 20 min. To determine the number of primary and secondary fibers and the corresponding CSA, muscle sections from day 90 samples were incubated with primary antibody raised against myosin heavy chain 1 β (BA-D5 concentrated antibody; 1:1,000; Developmental Studies Hybridoma Bank, Iowa City, IA; (Schiaffino et al., 1989; Yates et al., 2012) and wheat germ agglutinin (WGA; Alexa Fluor 594; 1:50; Invitrogen, Carlsbad, CA) overnight at 4 °C in a humidified chamber. Sections were then rinsed with three 5 min PBS washes and incubated with secondary antibody (goat anti-mouse Alexa Fluor 488; 1:250; Invitrogen) for 1 h, followed by additional washes with PBS. Slides were coated with 9:1 Glycerol:PBS solution and cover-slip applied.
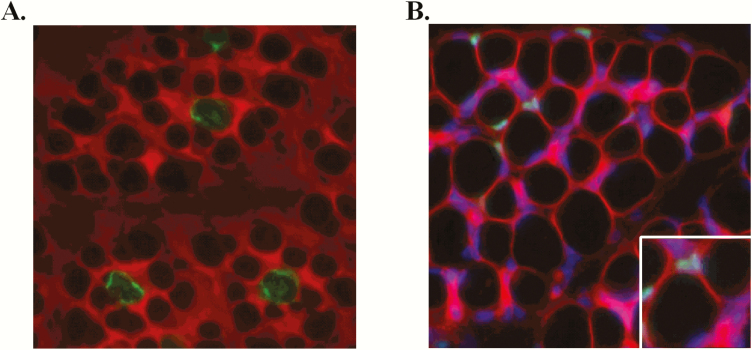
Figure 1.
Representative immunohistochemistry. (A) Muscle sections were immunostained against myosin heavy chain 1 (BA-D5) to identify primary fibers (green) at day 90 of gestation. Wheat germ agglutinin was used to visualize the cell membranes (red). (B) Muscle sections were immunostained against PAX7 (green) to identify satellite cells. Wheat germ agglutinin was used to visualize the cell membranes (red) and Hoescht 33342 was used to visualize nuclei (blue). Inset shows magnified view of two PAX7-positive cells adjacent to muscle fibers.
To determine the percentage of PAX7(+) cells, sections were incubated with an antibody raised against PAX7 (PAX7; 1:1,000; Developmental Studies Hybridoma Bank) overnight in a 4 °C humidified chamber. Sections were rinsed and incubated with secondary antibody (Goat Anti-Mouse Alexa Fluor 488; 1:250; Invitrogen) to visualize PAX7(+) cells. Hoescht 33342 (1:2,000; Invitrogen) was used to visualize nuclei. Alexa Fluor 568 conjugated WGA (1:50; Invitrogen) was used to visualize the sarcolemma membrane.
All images for immunohistochemistry analyses were captured with an AxioCam camera (Zeiss, Inc., Jena, Germany) mounted to an AxioObserver microscope (Zeiss, Inc.) with 5 to 10 images taken from 3 to 4 independent sections per muscle (taken at least 50 µm apart). Images were false colored and merged using ImageJ (National Institutes of Health, Bethesda, MD). Primary and secondary fibers were quantified manually, where fibers which immunostained positive with BA-D5 represented primary fibers and unstained fibers represented secondary fibers (Town et al., 2004). A minimum of 75 primary fibers and the related secondary fibers were counted per animal. Muscle fiber CSA was quantified using the area tool in ImageJ as previously described (Reed et al., 2011, 2014). The percentage of PAX7(+) cells was determined by dividing the number of PAX7(+) nuclei by the total number of nuclei per 20× image. A minimum of 250 fibers were analyzed at day 45 of gestation, a minimum of 300 fibers were analyzed at day 90 of gestation, and a minimum of 500 fibers were analyzed at day 135 and birth.
RNA-Seq and data analysis
Longissimus dorsi tissue samples were collected from a subset of male fetuses at day 90 (n = 11, CON = 4, OVER = 3, RES = 4), day 135 (n = 10, CON = 3, OVER = 3, RES = 4), and lambs at birth (n = 11, CON = 4, OVER = 4, RES = 3) and stored at −80 °C until RNA-Seq analyses. RNA was isolated as described by Reed et al. (2014). Briefly, tissue was homogenized in 1 mL Qiazol using a Qiagen Tissuelyser (Qiagen, Valencia, CA) and RNA was extracted using a Qiagen Mini Kit according to the manufacturer protocol (Qiagen). A Turbo DNA Free kit (Ambion, Foster City, CA) was used to remove genomic DNA. RNA quality was determined using a Bioanalyzer analysis system (Agilent Technologies, Santa Clara, CA).
A total of 50 ng of RNA (RNA integrity number of 8 or above) from each animal was used to prepare sequencing libraries following Illumina’s TruSeq stranded mRNA Sample Preparation Guide for library preparation protocol. Libraries were sequenced using an Illumina NextSeq500 (San Diego, CA) at the University of Connecticut Center for Genomic Innovations. Additionally, 2% PhiX control was added to the sequencing reaction as an endogenous control.
Quality control (QC) was performed using Sickle to eliminate sequences that were ≤35 bp in length and had a Phred score ≤35. Paired ends reads of 150 base pairs and a Q score of ≥30 were used for analysis. Sequences were mapped to the Ovis aries (Oar_V.4.0) genome reference annotation using STAR aligner (Dobin et al., 2013) and splice junctions were defined using NCBI GFF files for O. aries (Oar_V.4.0). The average number of raw reads were 26,731,710 for CON, 25,392,547 for RES, and 26,072,272 for OVER. The average number of short reads used for analysis after QC and trimming were 22,536,680 for CON, 21,693,193 for RES, and 22,056,024 for OVER. Of the post-QC reads, 84%, 85%, and 85% mapped to the O. aries reference for CON, RES, and OVER, respectively. Refseq was used for analysis and non-uniquely mapped reads were omitted and PCR duplicates were removed using samtools in STAR. The Htseq counts package was used to count the abundance of aligned reads (Anders et al., 2015) providing Htseq-count data for 27,721 genomic regions. A negative binomial model was fit to the counts for each gene, accounting for diet and time main effects as well as a diet × time interaction. Only genes who had read counts of at least 10 in at least 20% of the samples (and had greater overall coefficients of variability) were tested for differential expression [14,957 of 27,721 genes (54%) passed this filter]. Differential gene expression in a 3 × 3 factorial design (with factors diet and time as main effects, and their interaction) was determined [for genes passing a filter of greater variability in expression (Hackstadt and Hess, 2009)] using the DESeq2 package (Love et al., 2014) in R (Team, 2013). Genes were considered differentially expressed when the false discovery rate-corrected P-value (q-value) was ≤ 0.05. To rule out systematic differences in read frequency distributions (between diet and time levels), box plots of the primary data (log of read count + 1) were considered, with samples ordered by diet and time separately; no systematic differences in overall distributions were observed.
Gene ontology (GO) terms (biological processes, molecular functions, and cellular components) are of frequent interest, and therefore the focus of the GO analysis. Each term annotated in the GO database has an associated set of genes whose gene products are known to contribute to the biological process. The expression of those genes can be considered a proxy measure of the activity of the biological process. Because the current application includes simultaneous interest in several comparisons (restricted vs. control, overfed vs. control, and overfed vs. restricted; day 135 vs. day 90, birth vs. day 90, and birth vs. day 135), the R package mvGST (for multivariate gene set testing) was employed (Stevens, 2012; Mecham, 2014; Banks, 2015). Briefly, the mvGST approach takes the raw P-values for each gene in a comparison of interest (such as from the contrast for day 90 vs. day 135 in the DESeq2 negative binomial model), and uses meta-analytic methods to systematically combine P-values within each gene set (e.g., corresponding to a biological process), and the gene set (or GO term, such as a specific biological pathway) is classified as significantly more active, less active, or not differentially active (e.g., in day 135 than day 90). Across all gene sets considered, the false discovery rate is controlled (defined as 0.01 in the current report). One of the advantages of this approach is that it allows a gene set (or its associated biological process) to be identified as differentially active even if none of the genes in the set are called differentially expressed as long as the genes show a consensus of differential direction. Data are accessible through GEO series accession number (GSE124327).
Available gene annotations for O. aries were used to identify gene sets for GO biological processes. Specifically, the biomaRt package (Durinck et al., 2009) in R was used to access the October 2016 build of the ENSEMBL database for O. aries. The RNA-Seq platform (and gene naming convention) used in this study had expression data for about 85% of the genes annotated in the ENSEMBL database; about 45% of the genes in this study had at least some annotation information in the ENSEMBL database. To avoid testing overly-specific and overly-vague GO terms, only the GO terms with between 10 and 200 genes annotated thereto were tested for differential activity. Significant terms were not identified for treatment comparisons, therefore only terms for time comparisons are included.
Statistical analysis
Muscle fiber CSA and percentage of PAX7(+) cell data were analyzed using the mixed procedure in SAS (Cary, NC). Data were analyzed as a 3 × 4 factorial (diet by day of gestation as main effects). The number of primary fibers, secondary fibers, and secondary:primary fiber ratio were analyzed using the mixed procedure in SAS with diet as the main effect. Significance was considered at P ≤ 0.05.
Results
Muscle morphology
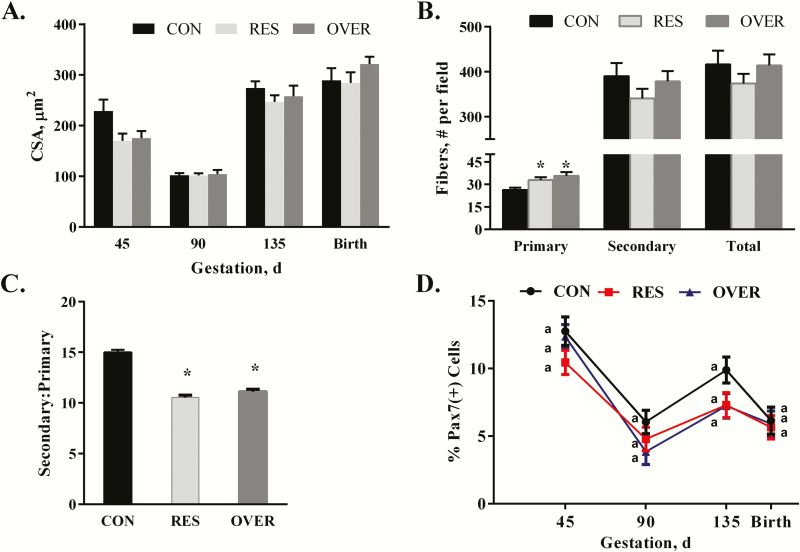
There was no significant effect of diet across time on muscle fiber CSA in the LM (P = 0.28; Figure 2A). However, there was a significant main effect of time on fiber CSA (P ≤ 0.0001). Average fiber CSA decreased from days 45 to 90, and then increased from days 90 to 135 to birth (P ≤ 0.004). In the LM, there was a 21% and 27% increase (Figure 2B) in the number of primary fibers per field in both the RES (P ≤ 0.03) and OVER (P ≤ 0.003), respectively compared with CON at day 90 of gestation. An effect of diet was not observed for the secondary or total fiber number per field (P ≥ 0.33; Supplement Figure S1). This resulted in a decreased secondary:primary fiber ratio (Figure 2C) in RES (P ≤ 0.0007) and OVER (P ≤ 0.003) offspring compared with CON. There was no effect of diet on the CSA of primary or secondary muscle fibers (P ≥ 0.51; data not shown) or the percentage of PAX7(+) cells within the LM of offspring (P = 0.57; Figure 2D).
Figure 2.
Poor maternal nutrition alters the number of primary fibers and the secondary to primary fiber ratio in the LM of offspring. Offspring born to control-, restricted- and overfed ewes are referred to as CON, RES, and OVER, respectively. (A) Muscle fiber CSA in CON, RES, and OVER offspring was determined at days 45, 90, and 135 of gestation and within 24 h of birth (145). (B) Average number of primary, secondary, and total fibers and (C) secondary:primary ratio was determined in CON, RES, and OVER offspring at day 90 of gestation. (D) Percentage of PAX7-positive cells was determined in CON, RES, and OVER offspring at days 45, 90, and 135 of gestation and within 24 h of birth (day 145). *P ≤ 0.05 compared with CON, different letters indicate P ≤ 0.05 compared with CON.
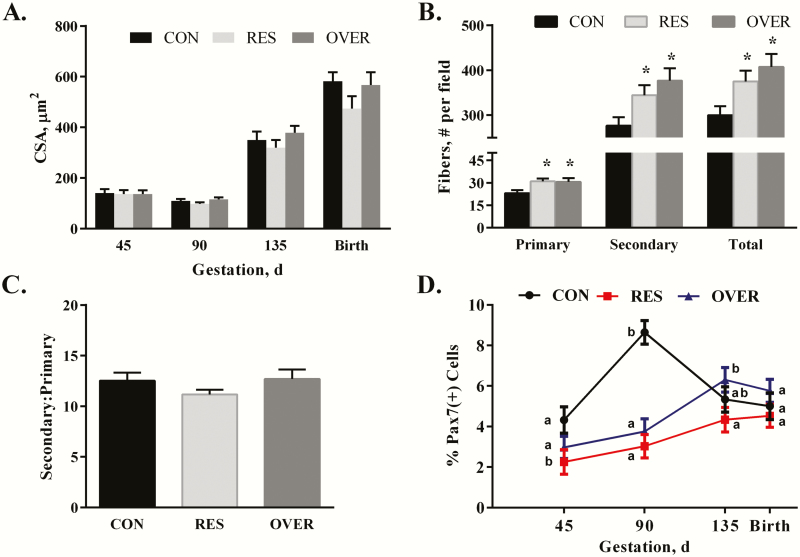
There was no effect of diet across time on muscle fiber CSA in the STN (P = 0.61S; Figure 3A). There was a main effect of time on fiber CSA (P ≤ 0.0001) where muscle fiber CSA increased over time from days 90 to 135 to birth (P ≤ 0.01). The number of primary fibers per field was increased in RES and OVER offspring by 26% and 25%, respectively (P ≤ 0.02; Figure 3B; Supplement Figure S1), compared with CON at day 90. Additionally, the number of secondary fibers per field was increased by 20% and 27% in RES (P ≤ 0.04) and OVER (P ≤ 0.004) offspring, respectively, compared with CON at day 90. The total number of fibers per field was increased in RES (P ≤ 0.03) and OVER (P ≤ 0.004) offspring by 20% and 26%, respectively, compared with CON. No differences were observed in the secondary:primary fiber ratio between diets (P ≥ 0.28; Figure 3C). There were no differences observed in the muscle fiber CSA of primary or secondary fibers due to maternal diet (P ≥ 0.12; data not shown). The percentage of PAX7(+) cells (Figure 3D) was reduced by 48% in RES (P ≤ 0.02) offspring compared with CON at day 45 of gestation. At day 90, the percentage of PAX7(+) cells was reduced by 65% in RES (P ˂ 0.0001) and 57% in OVER (P ˂ 0.0001) offspring compared with CON. Furthermore, at day 135 of gestation, the percentage of PAX7(+) cells was reduced by 31% in RES (P ≤ 0.026) compared with OVER. There were no differences in percentage of PAX7(+) cells observed between RES, OVER, and CON at birth (P ≥ 0.13).
Figure 3.
Poor maternal nutrition alters the number of primary and secondary fibers and reduces number of PAX7-positive PAX7(+) cells in the STN of offspring. Offspring born to control-, restricted-, and overfed ewes are referred to as CON, RES, and OVER, respectively. (A) Muscle fiber cross-sectional area in CON, RES, and OVER offspring was determined at days 45, 90, and 135 of gestation and within 24 h of birth (day 145). (B) Average number of primary and secondary fibers and (C) secondary:primary ratio was determined in CON, RES, and OVER offspring at day 90 of gestation. (D) The percentage of PAX7(+) cells was determined in CON, RES, and OVER offspring at days 45, 90, and 135 of gestation and within 24 h of birth (day 145). *P ≤ 0.05 compared with CON, a,bdifferent letters indicate P ≤ 0.05 compared with CON.
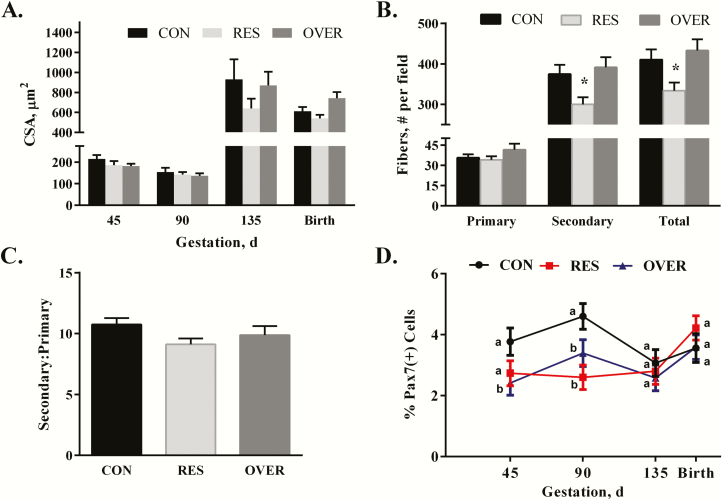
There was no effect of diet by time on muscle fiber CSA within the TB (P = 0.57; Figure 4A). However, there was a significant main effect of time on fiber CSA (P ≤ 0.0001). No difference was observed between days 45 and 90; however, muscle fiber CSA increased from days 90 to 135 and at birth (P ≤ 0.01). Secondary and total fiber numbers per field were reduced by 20% and 18%, respectively, in RES offspring (P ≤ 0.02; Figure 4B; Supplement Figure S1) compared with CON at day 90. However, there were no effects of diet on secondary:primary ratio (P = 0.14), or primary or secondary muscle fiber CSA (P ≥ 0.4; data not shown) at day 90. There was a significant interaction of diet by time observed for the percentage of PAX7(+) cells in the TB (P = 0.02; Figure 4D). The percentage of PAX7(+) cells was reduced by 36% in OVER (P ≤ 0.02) compared with CON at day 45 of gestation. At day 90, the percentage of PAX7(+) cells was reduced by 43% in the RES (P ≤ 0.001) and 26% in OVER (P ≤ 0.05) compared with CON. There were no differences in PAX7(+) cell percentage between diets at day 135 or birth (P ≥ 0.27).
Figure 4.
Poor maternal nutrition alters the number of secondary fibers and reduces number of PAX7(+) cells in the TB of offspring. Offspring born to control-, restricted-, and overfed ewes are referred to as CON, RES, and OVER, respectively. (A) Muscle fiber cross-sectional area in CON, RES, and OVER offspring was determined at days 45, 90, and 135 of gestation and within 24 h of birth (day 145). (B) The average number of primary and secondary fibers and (C) secondary:primary ratio was determined in CON, RES, and OVER offspring. (D) The percentage of PAX7(+) cells was determined in CON, RES, and OVER offspring at days 45, 90, and 135 of gestation and within 24 h of birth (145). *P ≤ 0.05 compared with CON, a,bdifferent letters indicate P ≤ 0.05 compared with CON.
Muscle RNA-Seq analysis
No significant interactions of diet by time were observed for differential gene expression analysis (q ≥ 0.22). Main effects of diet and time were observed for 20 and 2,205 genes, respectively (q ≤ 0.05; Supplement Tables S1 and S2). Specifically, in OVER vs. CON, 7 genes were downregulated (tripartite motif containing 9, GRAM domain containing 2, transmembrane channel like 2, transcription factor 7, transmembrane channel like 8, histone H1.3, and histone deacetylase 10; Table 1). In RES vs. CON, 2 genes were downregulated (tripartite motif containing 9 and kinesin family member 20A). In OVER vs. RES, 5 genes were downregulated (sterile alpha motif domain containing 14, adenosine A1 receptor, uroplakin 1B, protein tyrosine phosphatase, receptor type Z1, and fibulin 7). Six genes were identified that did not have available RefSeqID (OVER vs. CON: C3H9orf9 and LOC106991095 decreased 5-fold and LOC101104727 increased 19-fold; RES vs. CON: LOC101103869 and LOC101104727 decreased 6- and 25-fold, respectively; OVER vs. RES: LOC101104727 increased 43-fold).
Table 1.
Genes differentially expressed between CON, RES, and OVER offspring during gestation1
| Gene symbol | Gene name | Fold change | FDR-adj. P-value |
|---|---|---|---|
| OVER-CON | |||
| TRIM9 | Tripartite motif containing 9 | −7.28 | 0.0118 |
| GRAMD2 | GRAM domain containing 2 | −6.27 | 0.0326 |
| TMC2 | Transmembrane channel like 2 | −6.19 | 0.0283 |
| TCF7 | Transcription factor 7 (T-cell specific, HMG-box) | −6.03 | 0.0405 |
| TMC8 | Transmembrane channel like 8 | −5.41 | 0.0283 |
| LOC101109397 | Histone H1.3 | −4.15 | 0.0118 |
| HDAC10 | Histone deacetylase 10 | −3.52 | 0.0283 |
| RES-CON | |||
| TRIM9 | Tripartite motif containing 9 | −8.60 | 0.0087 |
| KIF20A | Kinesin family member 20A | −6.91 | 0.0097 |
| OVER-RES | |||
| SAMD14 | Sterile α motif domain containing 14 | −6.29 | 0.0090 |
| ADORA1 | Adenosine A1 receptor | −6.12 | 0.0450 |
| UPK1B | Uroplakin 1B | −5.99 | 0.0446 |
| PTPRZ1 | Protein tyrosine phosphatase, receptor type Z1 | −5.33 | 0.0450 |
| FBLN7 | Fibulin 7 | −4.88 | 0.0446 |
1Differentially expressed genes between treatments determined by RNA-Seq analysis for longissimus dorsi muscle from offspring, after accounting for the effect of time. Offspring born to control-, restricted-, and overfed ewes are referred to as CON, RES, and OVER, respectively. Fold change, log2 fold change; FDR-adj, false discovery rate adjusted P-value.
A main effect of time was observed for many genes (Supplement Table S2). Between day 90 vs. day 135, 36 genes were downregulated and 51 genes were upregulated. Between day 135 vs. birth, 48 genes were downregulated, and 16 genes were upregulated. Between day 90 vs. birth, 1,361 genes were downregulated and 338 genes were upregulated. To present a manageable number of genes, the top 20 down and upregulated genes with false discovery rate (FDR)-adjusted P-value of <0.02 are presented for each comparison (Tables 2, 3 and 4). Consistent with our objective to better understand the regulation of sheep fetal myogenesis, genes of various functions were identified to change over time. To further validate our methods and model, we manually curated a list of genes known to be involved in fetal myogenesis (e.g., growth hormone/insulin-like growth factor axis, Wnt signaling, PAX7, and MRF) and searched our entire list of differentially expressed genes to determine whether they were present. As expected several genes involved in myogenesis were found to be differentially expressed (Table 5) including insulin-like growth factor-I, insulin-like growth factor-binding protein, PAX7, MYH, and creatinine kinase.
Table 2.
Selected genes differentially expressed between day 90 and day 135 of gestation. 1
| Gene symbol | Gene name | Fold change | FDR-adj. P-value |
|---|---|---|---|
| SLC15A2 | Solute carrier family 15 member 2 | −6.15 | 0.0027 |
| ZCCHC12 | Zinc finger CCHC-type containing 12 | −5.81 | 0.0002 |
| NOTUM | NOTUM, palmitoleoyl-protein carboxylesterase | −5.71 | 0.0009 |
| PRR32 | Proline rich 32 | −5.23 | 0.0008 |
| BCAT1 | Branched chain amino acid transaminase 1 | −5.09 | 0.0138 |
| GABRG1 | Gamma-aminobutyric acid type A receptor gamma1 subunit | −5.02 | 0.0140 |
| CBLN2 | Cerebellin 2 precursor | −4.99 | 0.0116 |
| KCNH6 | Potassium voltage-gated channel subfamily H member 6 | −4.94 | 0.0007 |
| IL17B | Interleukin 17B | −4.80 | 0.0002 |
| NRCAM | Neuronal cell adhesion molecule | −4.48 | 0.0134 |
| NKAIN2 | Na+/K+ transporting ATPase interacting 2 | −4.43 | 0.0002 |
| NEFM | Neurofilament, medium polypeptide | −4.38 | 0.0043 |
| NKD1 | Naked cuticle homolog 1 | −4.23 | 0.0043 |
| CDH2 | Cadherin 2 | −4.00 | 0.0048 |
| AGTR2 | Angiotensin II receptor type 2 | −3.86 | 0.0113 |
| TNC | Tenascin C | −3.84 | 0.0185 |
| SCN5A | Sodium voltage-gated channel α subunit 5 | −3.78 | 0.0072 |
| PRSS35 | Protease, serine 35 | −3.78 | 0.0027 |
| CRMP1 | Collapsin response mediator protein 1 | −3.71 | 0.0009 |
| ERBB3 | Erb-B2 receptor tyrosine kinase 3 | −3.29 | 0.0150 |
| CRISPLD2 | Cysteine rich secretory protein LCCL domain containing 2 | 3.61 | 0.0102 |
| STEAP3 | STEAP3 metalloreductase | 3.74 | 0.0018 |
| PGFS | Prostaglandin F synthase 1 | 3.98 | 0.0116 |
| TUBA8 | Tubulin alpha 8 | 4.08 | 0.0195 |
| NT5C1A | 5’-Nucleotidase, cytosolic IA | 4.13 | 0.0061 |
| KLF9 | Kruppel-like factor 9 | 4.15 | 0.0000 |
| MYBPC2 | Myosin-binding protein C, fast type | 4.16 | 0.0004 |
| PADI2 | Peptidyl arginine deiminase 2 | 4.19 | 0.0059 |
| DUOX1 | Dual oxidase 1 | 4.23 | 0.0119 |
| TNFSF10 | Tumor necrosis factor superfamily member 10 | 4.29 | 0.0002 |
| ETNPPL | Ethanolamine-phosphate phospholyase | 4.48 | 0.0019 |
| ZBTB16 | Zinc finger and BTB domain containing 16 | 4.59 | 0.0004 |
| ACTN3 | Actinin α 3 | 4.77 | 0.0000 |
| FBP2 | Fructose-bisphosphatase 2 | 4.85 | 0.0151 |
| EEF1A2 | Eukaryotic translation elongation factor 1 α 2 | 5.35 | 0.0000 |
| CRHR2 | Corticotropin releasing hormone receptor 2 | 5.37 | 0.0080 |
| ACSM5 | Acyl-coa synthetase medium-chain family member 5 | 5.86 | 0.0004 |
| IGFN1 | Immunoglobulin-like and fibronectin type III domain containing 1 | 5.95 | 0.0000 |
| TMEM52 | Transmembrane protein 52 | 6.50 | 0.0000 |
| ALDH1L1 | Aldehyde dehydrogenase 1 family member L1 | 7.47 | 0.0011 |
1Differentially expressed genes between time points determined by RNA-Seq analysis for longissimus dorsi muscle from offspring at days 90 and 135 of gestation. The top 20 genes with the greatest significance and the greatest fold change (increased and decreased) are presented. Fold change, log2 fold change; FDR-adj, false discovery rate adjusted P-value.
Table 3.
Selected genes differentially expressed between day 135 of gestation and birth1
| Gene symbol | Gene name | Fold change | FDR-adj. P-value |
|---|---|---|---|
| MKI67 | Marker of proliferation Ki-67 | −6.64 | 0.0000 |
| C18H14orf132 | Chromosome 18 open reading frame, human C14orf132 | −5.59 | 0.0051 |
| TOP2A | Topoisomerase (DNA) II alpha | −5.32 | 0.0130 |
| KIF14 | Kinesin family member 14 | −4.87 | 0.0082 |
| CHRDL2 | Chordin like 2 | −4.83 | 0.0111 |
| ST6GALNAC5 | ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 | −4.78 | 0.0069 |
| FOXM1 | Forkhead box M1 | −4.72 | 0.0068 |
| HJURP | Holliday junction recognition protein | −4.66 | 0.0051 |
| GRIA3 | Glutamate ionotropic receptor ampa type subunit 3 | −4.26 | 0.0124 |
| PTX3 | Pentraxin 3 | −4.19 | 0.0002 |
| PAPLN | Papilin, proteoglycan like sulfated glycoprotein | −4.14 | 0.0080 |
| CCDC80 | Coiled-coil domain containing 80 | −4.13 | 0.0007 |
| NCKAP5 | NCK associated protein 5 | −4.05 | 0.0130 |
| SLITRK2 | SLIT and NTRK like family member 2 | −4.01 | 0.0082 |
| TNNT2 | Troponin T2, cardiac type | −3.90 | 0.0130 |
| MDK | Midkine (neurite growth-promoting factor 2) | −3.63 | 0.0051 |
| IGF2 | Insulin like growth factor 2 | −3.30 | 0.0051 |
| FBN3 | Fibrillin 3 | −3.09 | 0.0078 |
| NCAM1 | Neural cell adhesion molecule 1 | −3.08 | 0.0130 |
| PLXND1 | Plexin D1 | −3.07 | 0.0031 |
| PES1 | Pescadillo ribosomal biogenesis factor 1 | 2.16 | 0.0051 |
| MRPL55 | Mitochondrial ribosomal protein L55 | 2.50 | 0.0150 |
| DUSP15 | Dual specificity phosphatase 15 | 2.73 | 0.0130 |
| SESN1 | Sestrin 1 | 3.01 | 0.0169 |
| MLYCD | Malonyl-CoA decarboxylase | 3.87 | 0.0018 |
| AMPD3 | Adenosine monophosphate deaminase 3 | 4.51 | 0.0033 |
| ARMC12 | Armadillo repeat containing 12 | 5.78 | 0.0007 |
| CEP295NL | CEP295 N-terminal like | 5.89 | 0.0124 |
| PDK4 | Pyruvate dehydrogenase kinase 4 | 7.36 | 0.0000 |
1Differentially expressed genes between time points determined by RNA-Seq analysis for longissimus dorsi muscle from offspring at day 135 and birth. The top 20 genes with the greatest significance and the greatest fold change (increased and decreased) are presented. Fold change, log2 fold change; FDR-adj, false discovery rate adjusted P-value.
Table 4.
Selected genes differentially expressed between day 90 of gestation and birth1
| Gene symbol | Gene name | Fold change | FDR-adj. P-value |
|---|---|---|---|
| POSTN | Periostin | −10.12 | 0.0000 |
| MYL4 | Myosin light chain 4 | −8.57 | 0.0000 |
| MKI67 | Marker of proliferation Ki-67 | −8.42 | 0.0000 |
| PRR32 | Proline rich 32 | −7.79 | 0.0000 |
| TOP2A | Topoisomerase (DNA) II alpha | −7.67 | 0.0000 |
| FREM1 | FRAS1 related extracellular matrix 1 | −7.39 | 0.0000 |
| PEG10 | Paternally expressed 10 | −7.37 | 0.0000 |
| RRM2 | Ribonucleotide reductase regulatory subunit M2 | −7.10 | 0.0000 |
| BCAT1 | Breast carcinoma amplified sequence 4 | −7.07 | 0.0000 |
| KIF14 | Kinesin family member 14 | −7.05 | 0.0000 |
| SLC15A2 | Solute carrier family 15 member 2 | −7.03 | 0.0000 |
| CDCA8 | CUB domain containing protein 1 | −6.87 | 0.0000 |
| SHCBP1 | SHC binding and spindle associated 1 | −6.85 | 0.0000 |
| COL21A1 | Collagen type XXI α 1 chain | −6.82 | 0.0000 |
| CENPA | Centromere protein A | −6.81 | 0.0000 |
| HJURP | Holliday junction recognition protein | −6.61 | 0.0000 |
| FOXM1 | Forkhead box M1 | −6.60 | 0.0000 |
| SCN9A | Sodium voltage-gated channel α subunit 9 | −6.56 | 0.0000 |
| DPYSL5 | Dihydropyrimidinase like 5 | −6.52 | 0.0000 |
| KCNB2 | Potassium voltage-gated channel subfamily B member 2 | −6.52 | 0.0005 |
| CEP295NL | CEP295 N-terminal like | 5.94 | 0.0001 |
| PTRH2 | Peptidyl-tRNA hydrolase 2 | 5.95 | 0.0041 |
| KLHL34 | Kelch like family member 34 | 5.96 | 0.0000 |
| XDH | Xanthine dehydrogenase | 6.18 | 0.0001 |
| CKMT2 | Creatine kinase, mitochondrial 2 | 6.23 | 0.0000 |
| FBP2 | Fructose-bisphosphatase 2 | 6.24 | 0.0000 |
| ALDH1L1 | Aldehyde dehydrogenase 1 family member L1 | 6.39 | 0.0006 |
| EEF1A2 | Eukaryotic translation elongation factor 1 α 2 | 6.39 | 0.0000 |
| ARRDC2 | Arrestin domain containing 2 | 6.43 | 0.0000 |
| TUBA8 | Tubulin α 8 | 6.47 | 0.0000 |
| ANKRD2 | Ankyrin repeat domain 2 | 6.53 | 0.0000 |
| FOSL1 | FOS like 1, AP-1 transcription factor subunit | 6.60 | 0.0059 |
| FAM71E1 | Family with sequence similarity 71 member E1 | 6.63 | 0.0027 |
| IGFN1 | Immunoglobulin-like and fibronectin type III domain containing 1 | 6.91 | 0.0000 |
| ARMC12 | Armadillo repeat containing 12 | 7.79 | 0.0000 |
| FOXN4 | Forkhead box N4 | 7.83 | 0.0008 |
| ZG16 | Zymogen granule protein 16 | 7.91 | 0.0065 |
| SLC7A8 | Solute carrier family 7 member 8 | 7.99 | 0.0000 |
| PDK4 | Pyruvate dehydrogenase kinase 4 | 8.39 | 0.0000 |
| CALML6 | Calmodulin like 6 | 8.84 | 0.0158 |
1Differentially expressed genes between time points determined by RNA-Seq analysis for longissimus dorsi muscle from offspring at day 90 and birth. The top 20 genes with the greatest significance and the greatest fold change (increased and decreased) are presented. Fold change, log2 fold change; FDR-adj, false discovery rate adjusted P-value.
Table 5.
Genes known to be involved in myogenesis that are differentially expressed over time1
| Gene symbol | Gene name | Fold change | FDR-adj. P-value |
|---|---|---|---|
| Day 90 to birth | |||
| IGFBP2 | Insulin like growth factor binding protein 2 | −5.33 | 0.0000 |
| IGFBP2 | Insulin like growth factor binding protein 2 | −5.33 | 0.0000 |
| MDFI | MyoD family inhibitor | −4.89 | 0.0000 |
| IGF2BP1 | Insulin like growth factor 2 mRNA binding protein 1 | −4.09 | 0.0069 |
| MYF5 | Myogenic factor 5 | −3.96 | 0.0013 |
| IGF1 | Insulin like growth factor 1 | −3.94 | 0.0043 |
| WNT16 | Wnt family member 16 | −3.87 | 0.0121 |
| PAX7 | Paired box 7 | −3.79 | 0.0000 |
| MSTN | Myostatin | −3.74 | 0.0093 |
| DKK3 | Dickkopf WNT signaling pathway inhibitor 3) | −3.64 | 0.0022 |
| IGF2BP3 | Insulin like growth factor 2 mRNA-binding protein 3 | −3.59 | 0.0001 |
| IGF2 | Insulin like growth factor 2 | −3.05 | 0.0002 |
| IGFALS | Insulin like growth factor-binding protein acid labile subunit | −2.69 | 0.0258 |
| MYH10 | Myosin heavy chain 10 | −2.50 | 0.0188 |
| WNT9A | Wnt family member 9A | −2.09 | 0.0103 |
| MYH14 | Myosin heavy chain 14 | 1.32 | 0.0042 |
| MYH7 | Myosin heavy chain 7 | 2.59 | 0.0049 |
| CKM | Creatine kinase, M-type | 3.27 | 0.0003 |
| CKMT2 | Creatine kinase, mitochondrial 2 | 6.23 | 0.0000 |
| Day 135 to birth | |||
| IGF2 | Insulin like growth factor 2 | −3.30 | 0.0051 |
| Day 90 to day 135 | |||
| CKM | Creatine kinase, M-type | 2.84 | 0.0433 |
1Genes known to be involved in myogenesis were selected from the filtered gene list to determine changes in mRNA expression of genes involved in myogenesis over time. Fold change, log2 fold change; FDR-adj, false discovery rate adjusted P-value.
To gain a better understanding of the key processes involved in fetal myogenesis, for all genes in our analysis, GO classes biological processes, cellular component, and molecular function were classified based on the potential contribution to the process of each gene. As would be expected, numerous GO terms within each class were identified as playing a positive (upregulated) or negative (downregulated) role in fetal muscle development. The entire set of terms for all three comparisons (day 90 vs. day 135; day 135 vs. birth; day 90 vs. birth) is presented in Supplement Table S3. To present an overview of key pathways involved in fetal myogenesis we selected the top 10 terms with the smallest P-value for both up- and downregulated genes in each class at each time comparison (Tables 6, 7, and 8).
Table 6.
Selected GO descriptions for days 90 to 135 of gestation1
| Class | GO description and ID | Size | Size in data | FDR-adj. P-value |
|---|---|---|---|---|
| Downregulated | ||||
| Biological process | Centromere complex assembly (GO:0034508) | 17 | 17 | 0.0000 |
| Centrosome cycle (GO:0007098) | 60 | 58 | 0.0000 | |
| Regulation of ubiquitin protein ligase activity (GO:1904666) | 12 | 12 | 0.0001 | |
| Microtubule organizing center organization (GO:0031023) | 69 | 65 | 0.0001 | |
| Cilium organization (GO:0044782) | 118 | 115 | 0.0001 | |
| Centrosome duplication (GO:0051298) | 47 | 45 | 0.0001 | |
| Cartilage condensation (GO:0001502) | 16 | 15 | 0.0002 | |
| Cell aggregation (GO:0098743) | 16 | 15 | 0.0002 | |
| Chondrocyte development (GO:0002063) | 24 | 23 | 0.0002 | |
| Cilium assembly (GO:0060271) | 104 | 102 | 0.0002 | |
| Molecular function | Microtubule binding (GO:0008017) | 150 | 146 | 0.0010 |
| Voltage-gated sodium channel activity (GO:0005248) | 18 | 18 | 0.0014 | |
| Tubulin binding (GO:0015631) | 198 | 193 | 0.0014 | |
| Single-stranded DNA-dependent atpase activity (GO:0043142) | 11 | 11 | 0.0014 | |
| Voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential (GO:1905030) | 18 | 18 | 0.0014 | |
| Upregulated | ||||
| Biological process | Skeletal muscle contraction (GO:0003009) | 23 | 20 | 0.0000 |
| Tricarboxylic acid cycle (GO:0006099) | 15 | 15 | 0.0010 | |
| Aerobic respiration (GO:0009060) | 29 | 25 | 0.0014 | |
| Multicellular organismal movement (GO:0050879) | 33 | 29 | 0.0017 | |
| Musculoskeletal movement (GO:0050881) | 33 | 29 | 0.0017 | |
| Skeletal muscle adaptation (GO:0043501) | 12 | 12 | 0.0037 | |
| Citrate metabolic process (GO:0006101) | 18 | 18 | 0.0064 | |
| Tricarboxylic acid metabolic process (GO:0072350) | 22 | 22 | 0.0091 | |
| Molecular function | NADH dehydrogenase (ubiquinone) activity (GO:0008137) | 18 | 11 | 0.0014 |
| NADH dehydrogenase (quinone) activity (GO:0050136) | 18 | 11 | 0.0014 |
1GO terms were identified for all genes present in the current RNA-Seq analysis data set using multivariate gene set testing, thus providing us with a robust method to identify key pathways or processes involved in myogenesis between the three time points. Size in data, number of genes from our data set present in the GO term; FDR-adj, false discovery rate adjusted P-value.
Table 7.
Selected GO descriptions for day 135 of gestation to birth1
| Class | GO description and ID | Size | Size in data | FDR-adj. P-value |
|---|---|---|---|---|
| Downregulated | ||||
| Biological process | Extracellular matrix organization (GO:0030198) | 140 | 134 | 0.0000 |
| Extracellular structure organization (GO:0043062) | 140 | 134 | 0.0000 | |
| Collagen fibril organization (GO:0030199) | 24 | 23 | 0.0000 | |
| Gland morphogenesis (GO:0022612) | 102 | 98 | 0.0000 | |
| Endothelial cell migration (GO:0043542) | 112 | 108 | 0.0000 | |
| Epithelial cell migration (GO:0010631) | 167 | 161 | 0.0000 | |
| Cell junction assembly (GO:0034329) | 117 | 113 | 0.0000 | |
| Epithelium migration (GO:0090132) | 169 | 163 | 0.0000 | |
| Mesenchymal cell differentiation (GO:0048762) | 127 | 126 | 0.0000 | |
| Neuron projection guidance (GO:0097485) | 128 | 125 | 0.0000 | |
| Molecular function | Platelet-derived growth factor binding (GO:0048407) | 11 | 10 | 0.0000 |
| Extracellular matrix structural constituent (GO:0005201) | 31 | 30 | 0.0000 | |
| Semaphorin receptor activity (GO:0017154) | 11 | 11 | 0.0000 | |
| Ephrin receptor binding (GO:0046875) | 19 | 19 | 0.0000 | |
| Actin filament binding (GO:0051015) | 80 | 77 | 0.0000 | |
| Motor activity (GO:0003774) | 98 | 88 | 0.0000 | |
| Rho gtpase binding (GO:0017048) | 115 | 110 | 0.0000 | |
| Wnt-protein binding (GO:0017147) | 21 | 21 | 0.0000 | |
| Fibronectin binding (GO:0001968) | 14 | 13 | 0.0000 | |
| Integrin binding (GO:0005178) | 62 | 61 | 0.0000 | |
| Upregulated | ||||
| Biological process | Maturation of 5.8S rRNA (GO:0000460) | 10 | 10 | 0.0000 |
| Ribosomal large subunit biogenesis (GO:0042273) | 13 | 13 | 0.0023 | |
| rRNA processing (GO:0006364) | 54 | 50 | 0.0041 |
1GO terms were identified for all genes present in the current RNA-Seq analysis data set using multivariate gene set testing, thus providing us with a robust method to identify key pathways or processes involved in myogenesis between the three time points. Size in data, number of genes from our data set present in the GO term; FDR-adj, false discovery rate adjusted P-value.
Table 8.
Selected GO descriptions for day 90 of gestation to birth1
| Class | GO description and ID | Size | Size in data | FDR-adj. P-value |
| Downregulated | ||||
| Biological process | Mitotic sister chromatid segregation (GO:0000070) | 90 | 86 | 0.0000 |
| Cell cycle checkpoint (GO:0000075) | 118 | 113 | 0.0000 | |
| Mitotic cytokinesis (GO:0000281) | 28 | 28 | 0.0000 | |
| Sister chromatid segregation (GO:0000819) | 110 | 104 | 0.0000 | |
| Cytokinesis (GO:0000910) | 89 | 88 | 0.0000 | |
| Cartilage condensation (GO:0001502) | 16 | 15 | 0.0000 | |
| Osteoblast differentiation (GO:0001649) | 151 | 145 | 0.0000 | |
| Ureteric bud development (GO:0001657) | 78 | 77 | 0.0000 | |
| Formation of primary germ layer (GO:0001704) | 91 | 90 | 0.0000 | |
| Endoderm formation (GO:0001706) | 42 | 41 | 0.0000 | |
| Molecular function | Motor activity (GO:0003774) | 98 | 88 | 0.0000 |
| Microtubule motor activity (GO:0003777) | 58 | 56 | 0.0000 | |
| Protein tyrosine kinase activity (GO:0004713) | 95 | 94 | 0.0000 | |
| Transmembrane receptor protein tyrosine kinase activity (GO:0004714) | 44 | 44 | 0.0000 | |
| Ephrin receptor activity (GO:0005003) | 11 | 11 | 0.0000 | |
| Extracellular matrix structural constituent (GO:0005201) | 31 | 30 | 0.0000 | |
| Voltage-gated sodium channel activity (GO:0005248) | 18 | 18 | 0.0000 | |
| Microtubule binding (GO:0008017) | 150 | 146 | 0.0000 | |
| Tubulin binding (GO:0015631) | 198 | 193 | 0.0000 | |
| Rho GTPase binding (GO:0017048) | 115 | 110 | 0.0000 | |
| Upregulated | ||||
| Biological process | Tricarboxylic acid cycle (GO:0006099) | 15 | 15 | 0.0000 |
| Citrate metabolic process (GO:0006101) | 18 | 18 | 0.0000 | |
| Tricarboxylic acid metabolic process (GO:0072350) | 22 | 22 | 0.0000 | |
| Cellular respiration (GO:0045333) | 74 | 60 | 0.0000 | |
| Aerobic respiration (GO:0009060) | 29 | 25 | 0.0000 | |
| Regulation of oxidative phosphorylation (GO:0002082) | 12 | 10 | 0.0000 | |
| Oxidative phosphorylation (GO:0006119) | 36 | 25 | 0.0000 | |
| Skeletal muscle contraction (GO:0003009) | 23 | 20 | 0.0000 | |
| Maturation of 5.8S rRNA (GO:0000460) | 10 | 10 | 0.0000 | |
| Energy coupled proton transport, down electrochemical gradient (GO:0015985) | 13 | 11 | 0.0000 | |
| Molecular function | NADH dehydrogenase (ubiquinone) activity (GO:0008137) | 18 | 11 | 0.0000 |
| NADH dehydrogenase (quinone) activity (GO:0050136) | 18 | 11 | 0.0000 | |
| NADH dehydrogenase activity (GO:0003954) | 19 | 12 | 0.0000 | |
| Oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor (GO:0016655) | 22 | 15 | 0.0000 | |
| Structural constituent of ribosome (GO:0003735) | 87 | 84 | 0.0000 | |
| Electron transfer activity (GO:0009055) | 37 | 19 | 0.0000 | |
| Triglyceride lipase activity (GO:0004806) | 12 | 11 | 0.0000 | |
| Oxidoreductase activity, acting on NAD(P)H (GO:0016651) | 50 | 41 | 0.0000 | |
| Iron-sulfur cluster binding (GO:0051536) | 32 | 31 | 0.0000 | |
| Metal cluster binding (GO:0051540) | 32 | 31 | 0.0000 |
1GO terms were identified for all genes present in the current RNA-Seq analysis data set using multivariate gene set testing, thus providing us with a robust method to identify key pathways or processes involved in myogenesis between the three time points. Size in data, number of genes from our data set present in the GO term. FDR-adj, false discovery rate adjusted P-value.
For pathways that were downregulated, biological processes included cell division, cilium development, extracellular organization, cell migration and cycle, and germ layer formation. For pathways that were downregulated, cellular components included chromosome formation, and extracellular matrix formation and function. For pathways that were downregulated, molecular function included membrane function and trafficking, protein binding, and protein tyrosine kinase action (Tables 7 and 8).
For pathways that were upregulated, biological processes included metabolism, movement, and contraction. For pathways that were upregulated, cellular component included proteasome activity and mitochondrial respiration. For pathways that were upregulated, molecular function included redox activity, voltage channel function, proteasome complex, and metabolism (Tables 6, 7, and 8).
Discussion
The immunohistochemical approach used in the current experiment revealed a negative effect of both restricted- and overfeeding on offspring myogenic progenitor cells (PAX7+); however, this was muscle specific. The transcriptomic approach revealed that gestation was a greater driver of changes in gene expression in the offspring LM than maternal diet. The 20 genes altered by maternal diet were broadly involved in innervation and epigenetic regulation, whereas a much larger number of genes increased or decreased across gestation which is consistent with the temporal regulation of myogenesis and the transition to neonatal muscle growth. Consistent with our previous work (Hoffman et al., 2016b), these data demonstrate that although the muscle phenotype of offspring born to nutrient restricted and overnourished mothers is similar, changes at the transcriptomic level are different based on maternal diet.
To understand the impact of maternal restricted- and overfeeding during gestation on offspring muscle development and growth, we investigated the effects of poor maternal diets on skeletal muscle fiber CSA and the myogenic progenitor cell population in fetal and early neonatal muscle. Overfeeding ewes can result in reduced muscle fiber CSA at day 75 of gestation in offspring (Tong et al., 2009) and nutrient restriction can decrease myofiber number (Zhu et al., 2006). While we did not observe an effect of maternal restricted- or overfeeding on CSA at any time point in the present study, we observed that restricted- and overfeeding during gestation altered the number of primary and/or secondary fibers per field of view in RES and OVER offspring in a muscle-dependent manner. Consistent with Zhu et al. (2004), we observed a decreased ratio of secondary to primary fibers in the LM in response to nutrient restriction (Zhu et al., 2004). However, Zhu et al. (2004) did not report the number of primary and secondary fibers per field. Based on the observation of a greater number of primary fibers and a smaller number of secondary fibers per field of view, our data suggest that maternal diet delays or reduces secondary myogenesis in the LM. Varying feed intake during the peri-conception period (18 days before to 6 days after ovulation) reduced secondary:primary fiber ratio in the STN of offspring (Quigley et al., 2005). The differences between our study and others may be due to the timing of the dietary treatment [pre-conception to day 75 by Zhu et al. (2006); days 30 to 90 of gestation in the current work]. To our knowledge, there have been no other reports of primary and secondary muscle fiber quantification in the TB in sheep during gestation. The differences in response between muscles may be due to the temporal differences in muscle development, which occur cranially to caudally. However, this requires further investigation. The number of muscle fibers formed during primary and secondary myogenesis is critical, given that no additional fibers are formed following birth (Pearson, 1990). Thus, the number of muscle fibers formed during gestation prepares the animal for optimal postnatal muscle growth.
Paired box 7 is a key marker of both myogenic progenitor cells in prenatal development and satellite cells during postnatal development (Seale et al., 2000; Zammit et al., 2006). A subset of proliferating PAX7(+) muscle progenitor cells persist into late fetal development and ultimately become enveloped beneath the basal lamina of developing myofibers, where they adopt a satellite cell position and establish the satellite cell pool (Gros et al., 2005; Relaix et al., 2006). The reduction in PAX7(+) cell numbers in STN and TB of RES and OVER during early- and mid-gestation are consistent with results reported in of Gonzalez et al. (2013) who demonstrated a transient decrease in PAX7(+) cell number in offspring infraspinatus muscle as a result of nutrient restriction during early- and mid-gestation in cattle. Moreover, these findings support previous reports from our lab that these two extreme diets have similar negative phenotypic effects on myogenesis (Hoffman et al., 2016b). These changes are time, diet, and muscle specific, and in our data, do not translate into increased muscle growth at birth, as evidenced by a lack of change in muscle fiber CSA. In a previous study, we detected reduced postnatal muscle growth in myofibers of offspring born to restricted- and overfed ewes, which suggests that negative effects on hypertrophy may not be apparent until later postnatal time points, when satellite cells are recruited to support postnatal hypertrophy (Yin et al., 2013; Reed et al., 2014). The muscle-specific effects of maternal diet, reduced myogenic progenitor cells (PAX7+) at early- and mid-gestation in the TB and STN and reduced the ratio of secondary:primary fibers only in the offspring LM, emphasize the need to evaluate more than one muscle type when evaluating muscle development over time and in response to diet.
RNA-Seq analysis was performed to identify the effects of maternal diet on gene expression in offspring LM; however, only a few genes were differentially expressed. Although this could be due to smaller sample size, it is consistent with the minimal effect of maternal diet on LM morphology. Of the genes identified, the majority were specific to maternal nutrient restriction or overfeeding. This is similar to our previous finding that maternal diet (restricted and over) negatively impact muscle growth, but likely through different mechanisms based on gene expression (Hoffman et al., 2016b). One gene, tripartite motif-containing protein 9 (TRIM 9), which is an ubiquitin ligase involved in neuronal function and macrophage migration (Tanji et al., 2010), was downregulated in both treatments demonstrating potential effect of poor maternal nutrition on neuronal development and/or innervation in the muscle of offspring, as well as altered migration of myoblasts. Transmembrane channel-like (TMC) 2 is involved in membrane excitability (Yue et al., 2018) and disruption in mice impairs mechano-electrical transduction currents (Kawashima et al., 2011). Thus, the decreased expression of both TRIM9 and TMC2 warrants further investigation into the potential changes to muscle contraction in offspring born from maternal nutrient restriction and overnutrition. Transcription factor 7 (TCF7) binds β-catenin and activates downstream transcription in the Wnt-signaling pathway, a key pathway in myogenesis. Decreased expression of TCF7 in offspring of restricted-fed mothers may be one mechanism contributing to early effects on muscle growth. Changes in TCF7 expression and methylation are associated with increased adiposity and decreased insulin resistance (Martinez et al., 2009; Columbus et al., 2010). These are both phenotypes often observed in offspring of poorly fed mothers. Similarly, another gene involved in glucose homeostasis and insulin sensitivity, adenosine A1 receptor (Farhy and McCall, 2015), was downregulated, demonstrating a potential for alterations in genes expression during fetal development that may contribute to altered metabolism in muscle later in life.
Histone deacetylases (HDACs) have been implicated in the epigenetic regulation of key MRF (Feeney et al., 2014), and HDAC10 is a repressor of microRNA let-7g expression, which is important for adipogenesis (Sun et al., 2009; Li et al., 2015). Consistent with previous reports that microRNA let-7g is altered in skeletal muscle tissues of fetal offspring from obese mothers (Yan et al., 2012), HDAC10 expression decreased in OVER, suggesting epigenetic regulation of offspring muscle growth and/or composition as a result of maternal overnutrition. Decreased expression of HDAC10 and Histone H1.3, genes important for condensation of nucleosome chains and maintenance of chromatin structure are consistent with previous reports of changes in expression of genes involved in epigenetic modifications in muscle of offspring of restricted- and overfeeding ewes at birth (Hoffman et al., 2016b). Further, metabolomics data generated using these samples identified changes in the abundance of several metabolites involved in epigenetic changes in offspring of overfed ewes (Martin et al., 2019). Additional studies are needed to determine specific epigenetic modifications and their contributions to these phenotypic changes. Several other novel genes were identified, however their role in muscle, and in response to nutritional status are not known, demonstrating the need for further analysis of these genes, their proteins, and mechanisms contributing to altered muscle growth and metabolism in offspring.
Temporal changes in muscle morphometrics and transcriptome
Myogenesis is a highly coordinated process with critical time points throughout fetal development. Our experimental design, to evaluate effects of maternal diet at multiple time points during myogenesis (key developmental time points including primary and secondary myogenesis as well as late-term hypertrophy), provided the opportunity to evaluate temporal changes in normal myogenesis through morphometric and mRNA expression changes. We observed alterations to myofiber CSA over time independent of maternal diet. The smaller CSA of muscle fibers observed at day 90 was due to the development of secondary myofibers during mid-gestation (Zhu et al., 2004, 2008; Tong et al., 2009; Du et al., 2011). The increase in the myofiber CSA from days 90 to 135 and birth in all treatment groups can be attributed to late gestation myofiber hypertrophy observed in eutherian mammals (Du et al., 2010b, 2011, 2015).
Changes in CSA over time were similar across muscles; however, changes in the percent-positive PAX7 cells were tissue specific. In the LM, the percent PAX7(+) cells decreased with time, which is consistent with decreased gene expression of PAX7 over time. Consistent with the known role of GH/IGF axis and Wnt in myogenesis and reduced hyperplasia during late gestation, expression of genes involved in these signaling pathways were downregulated over time. Consistent with the increased hypertrophy in late gestation, expression of genes involved in protein accretion (myosin heavy chain and creatinine kinase) increased. These data demonstrate the effectiveness of RNA-Seq analysis to identify known regulators of myogenesis in our model. Using this model, we were able to provide a more global analysis of temporal changes in genes expression during myogenesis in LM.
Since thousands of genes were differentially expressed between day 90 of gestation and birth, we examined gene expression more globally to identify processes potentially important to distinct points of development. From day 90 to birth, biological processes and molecular functions were identified that are consistent with the complex regulation of myogenesis. As expected, several of the biological processes and molecular functions identified between day 90 and birth were also identified between days 90 and 135 of gestation, demonstrating the need for these changes to occur prior to birth. Specifically, GO pathways associated with cell aggregation, cell migration, centromere, and germ layer formation were downregulated over time. Specifically, expression of solute carrier family 15 member 2 and cadherin 2 and other key genes involved in early developmental processes may not be required during later development. Upregulation of genes involved in GO pathways associated with skeletal muscle contraction and movement (myosin-binding protein C 2, eukaryotic translation elongation factor 1 α 2 and creatinine kinase mitochodrina 2), metabolic processes (pyruvate dehydrogenase kinase 4), and cell respiration and oxidative phosphorylation is consistent with the role of muscle in metabolic activity, and nutrient storage and availability, and the developmental shifts that occur from mid-gestation to late-gestation (McCoard et al., 2001; Zhu et al., 2008; Du et al., 2011; Bentzinger et al., 2012; Yates et al., 2012). Further, these represent the perinatal metabolic shift from glucose to fatty acid oxidation (Makinde et al., 1998), which is further supported by the increase in TCA cycle processes and NADH dehydrogenase activity observed between days 90 and 135 of gestation in our model.
In summary, poor maternal nutrition through both restricted- and overfeeding during gestation alters the population of myogenic cells expressing PAX7 in a muscle-specific manner in the offspring. Thus, one mechanism by which maternal diet impairs postnatal offspring muscle growth may be through a limited progenitor cell population. Additionally, the gene expression of several biological processes and molecular functions such as cell cycle, metabolic processes, and protein synthesis, are altered during fetal myogenesis in support of increasing protein accretion, muscle function, and increased metabolic activity.
Conflict of interest statement None declared.
Supplementary Material
Acknowledgments
This work was supported by the USDA-AFRI National Institute of Food and Agriculture award number 2013-01919 and the Storrs and Utah Agricultural Experiment Stations. The authors thank the UConn Beef/Sheep unit and staff for assistance with animal care and Brandon Smith (UConn) for technical assistance with gene expression analysis.
Literature Cited
- Anders S., Pyl P. T., and Huber W.. . 2015. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage J. A., Khan I. Y., Taylor P. D., Nathanielsz P. W., and Poston L.. . 2004. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J. Physiol. 561(Pt 2):355–377. doi: 10.1113/jphysiol.2004.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks R. K. 2015. Annotation tools for multivariate gene set testing of non-model organisms [MS Thesis]. Logan (UT): Utah State University. [Google Scholar]
- Barker D. J. 1995. Intrauterine programming of adult disease. Mol. Med. Today 1:418–423. doi: 10.1016/s1357-4310(95)90793-9. [DOI] [PubMed] [Google Scholar]
- Bentzinger C. F., Wang Y. X., and Rudnicki M. A.. . 2012. Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4(2):1–16. doi: 10.1101/cshperspect.a008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbus J., Chiang Y., Shao W., Zhang N., Wang D., Gaisano H. Y., Wang Q., Irwin D. M., and Jin T.. . 2010. Insulin treatment and high-fat diet feeding reduces the expression of three Tcf genes in rodent pancreas. J. Endocrinol. 207:77–86. doi: 10.1677/JOE-10-0044 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R.. . 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., Tong J., Zhao J., Underwood K. R., Zhu M., Ford S. P., and Nathanielsz P. W.. . 2010a. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 88(13 Suppl.):E51–60. doi: 10.2527/jas.2009-2311 [DOI] [PubMed] [Google Scholar]
- Du M., Wang B., Fu X., Yang Q., and Zhu M. J.. . 2015. Fetal programming in meat production. Meat Sci. 109:40–47. doi: 10.1016/j.meatsci.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Du M., Yin J., and Zhu M. J.. . 2010b. Cellular signaling pathways regulating the initial stage of adipogenesis and marbling of skeletal muscle. Meat Sci. 86:103–109. doi: 10.1016/j.meatsci.2010.04.027 [DOI] [PubMed] [Google Scholar]
- Du M., Zhao J. X., Yan X., Huang Y., Nicodemus L. V., Yue W., McCormick R. J., and Zhu M. J.. . 2011. Fetal muscle development, mesenchymal multipotent cell differentiation, and associated signaling pathways. J. Anim. Sci. 89:583–590. doi: 10.2527/jas.2010-3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., and Huber W.. . 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4:1184–1191. doi: 10.1038/nprot.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy L. S., and McCall A. L.. . 2015. Glucagon - the new ‘insulin’ in the pathophysiology of diabetes. Curr. Opin. Clin. Nutr. Metab. Care 18:407–414. doi: 10.1097/MCO.0000000000000192 [DOI] [PubMed] [Google Scholar]
- Feeney A., Nilsson E., and Skinner M. K.. . 2014. Epigenetics and transgenerational inheritance in domesticated farm animals. J. Anim. Sci. Biotechnol. 5:48. doi: 10.1186/2049-1891-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S. P., and Long N. M.. . 2011. Evidence for similar changes in offspring phenotype following either maternal undernutrition or overnutrition: potential impact on fetal epigenetic mechanisms. Reprod. Fertil. Dev. 24:105–111. doi: 10.1071/RD11911 [DOI] [PubMed] [Google Scholar]
- Gonzalez J. M., Camacho L. E., Ebarb S. M., Swanson K. C., Vonnahme K. A., Stelzleni A. M., and Johnson S. E.. . 2013. Realimentation of nutrient restricted pregnant beef cows supports compensatory fetal muscle growth. J. Anim. Sci. 91:4797–4806. doi: 10.2527/jas.2013-6704 [DOI] [PubMed] [Google Scholar]
- Gros J., Manceau M., Thomé V., and Marcelle C.. . 2005. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435:954–958. doi: 10.1038/nature03572 [DOI] [PubMed] [Google Scholar]
- Hackstadt A. J., and Hess A. M.. . 2009. Filtering for increased power for microarray data analysis. BMC Bioinformatics 10:11. doi: 10.1186/1471-2105-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales C. N., and Barker D. J.. . 2001. The thrifty phenotype hypothesis. Br. Med. Bull. 60:5–20. doi: 10.1093/bmb/60.1.5 [DOI] [PubMed] [Google Scholar]
- Hoffman M. L., Peck K. N., Forella M. E., Fox A. R., Govoni K. E., and Zinn S. A.. . 2016a. The effects of poor maternal nutrition during gestation on postnatal growth and development of lambs. J. Anim. Sci. 94(2):789–799. doi: 10.2527/jas.2015-9933 [DOI] [PubMed] [Google Scholar]
- Hoffman M. L., Peck K. N., Wegrzyn J. L., Reed S. A., Zinn S. A., and Govoni K. E.. . 2016b. Poor maternal nutrition during gestation alters the expression of genes involved in muscle development and metabolism in lambs. J. Anim. Sci. 94:3093–3099. doi: 10.2527/jas.2016-0570 [DOI] [PubMed] [Google Scholar]
- Hoffman M. L., Reed S. A., Pillai S. M., Jones A. K., McFadden K. K., Zinn S. A., and Govoni K. E.. . 2017. The effects of poor maternal nutrition during gestation on offspring postnatal growth and metabolism. J. Anim. Sci. 95:2222–2232. doi: 10.2527/jas.2016.1229 [DOI] [PubMed] [Google Scholar]
- Huang Y., Yan X., Zhao J. X., Zhu M. J., McCormick R. J., Ford S. P., Nathanielsz P. W., Ren J., and Du M.. . 2010. Maternal obesity induces fibrosis in fetal myocardium of sheep. Am. J. Physiol. Endocrinol. Metab. 299:E968–E975. doi: 10.1152/ajpendo.00434.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. K., Gately R. E., McFadden K. K., Zinn S. A., Govoni K. E., and Reed S. A.. . 2016. Transabdominal ultrasound for detection of pregnancy, fetal and placental landmarks, and fetal age before Day 45 of gestation in the sheep. Theriogenology 85:939–945.e1. doi: 10.1016/j.theriogenology.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Kawashima Y., Géléoc G. S., Kurima K., Labay V., Lelli A., Asai Y., Makishima T., Wu D. K., Della Santina C. C., Holt J. R., . et al. 2011. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 121:4796–4809. doi: 10.1172/JCI60405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Peng L., and Seto E.. . 2015. Histone Deacetylase 10 Regulates the Cell Cycle G2/M Phase Transition via a Novel Let-7-HMGA2-Cyclin A2 Pathway. Mol. Cell. Biol. 35:3547–3565. doi: 10.1128/MCB.00400-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long N. M., Rule D. C., Tuersunjiang N., Nathanielsz P. W., and Ford S. P.. . 2015. Maternal obesity in sheep increases fatty acid synthesis, upregulates nutrient transporters, and increases adiposity in adult male offspring after a feeding challenge. PLoS One 10:e0122152. doi: 10.1371/journal.pone.0122152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S.. . 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinde A., Kantor P. F., and Lopaschuk G. D.. . 1998. Maturation of fatty acid and carbohydrate metabolism in the newborn heart. Mol. Cell. Biochem. 188:49–56. [PubMed] [Google Scholar]
- Martin D. E., Jones A. K., Pillai S. M., Hoffman M. L., McFadden K. K., Zinn S. A., Govoni K. E., and Reed S. A.. . 2019. Maternal Restricted- and Over-Feeding During Gestation Result in Distinct Lipid and Amino Acid Metabolite Profiles in the Longissimus Muscle of the Offspring. Front. Physiol. 10:515. doi: 10.3389/fphys.2019.00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G., Wijesinghe M., Turner K., Abud H. E., Taketo M. M., Noda T., Robinson M. L., and de Iongh R. U.. . 2009. Conditional mutations of beta-catenin and APC reveal roles for canonical Wnt signaling in lens differentiation. Invest. Ophthalmol. Vis. Sci. 50:4794–4806. doi: 10.1167/iovs.09-3567 [DOI] [PubMed] [Google Scholar]
- McCoard S. A., McNabb W. C., Birtles M. J., Harris P. M., McCutcheon S. N., and Peterson S. W.. . 2001. Immunohistochemical detection of myogenic cells in muscles of fetal and neonatal lambs. Cells. Tissues. Organs 169:21–33. doi: 10.1159/000047857 [DOI] [PubMed] [Google Scholar]
- Mecham D. S. 2014. mvGST: tools for multivariate and directional gene set testing. M.S. Thesis, Logan (UT): Utah State University. [Google Scholar]
- Morrison J. L., Duffield J. A., Muhlhausler B. S., Gentili S., and McMillen I. C.. . 2010. Fetal growth restriction, catch-up growth and the early origins of insulin resistance and visceral obesity. Pediatr. Nephrol. 25:669–677. doi: 10.1007/s00467-009-1407-3 [DOI] [PubMed] [Google Scholar]
- National Research Council. 2007. Nutrient Requirements of Small Ruminants: Sheep, Goats, Cervids, and New World Camelids. Washington (DC): The National Academies Press. [Google Scholar]
- Nesterenko T. H., and Aly H.. . 2009. Fetal and neonatal programming: evidence and clinical implications. Am. J. Perinatol. 26:191–198. doi: 10.1055/s-0028-1103027 [DOI] [PubMed] [Google Scholar]
- Pearson A. M. 1990. Muscle growth and exercise. Crit. Rev. Food Sci. Nutr. 29:167–196. doi: 10.1080/10408399009527522 [DOI] [PubMed] [Google Scholar]
- Pillai M. P., Jones A. E., Hoffman M. L., McFadden K. K., Reed S. A., Zinn S. A., and Govoni K. E.. . 2017. Fetal and organ development at gestational days 45, 90, 135 and at birth of lambs exposed to under- or over-nutrition during gestation. Trans. Anim. Sci. 1(1):77–89. doi: 10.2527/jas2016.0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley S. P., Kleemann D. O., Kakar M. A., Owens J. A., Nattrass G. S., Maddocks S., and Walker S. K.. . 2005. Myogenesis in sheep is altered by maternal feed intake during the peri-conception period. Anim. Reprod. Sci. 87:241–251. doi: 10.1016/j.anireprosci.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Raja J. S., Hoffman M. L., Govoni K. E., Zinn S. A., and Reed S. A.. . 2016. Restricted maternal nutrition alters myogenic regulatory factor expression in satellite cells of ovine offspring. Animal 10:1200–1203. doi: 10.1017/S1751731116000070 [DOI] [PubMed] [Google Scholar]
- Reed S. A., Raja J. S., Hoffman M. L., Zinn S. A., and Govoni K. E.. . 2014. Poor maternal nutrition inhibits muscle development in ovine offspring. J. Anim. Sci. Biotechnol. 5:43. doi: 10.1186/2049-1891-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. A., Senf S. M., Cornwell E. W., Kandarian S. C., and Judge A. R.. . 2011. Inhibition of IkappaB kinase alpha (IKKα) or IKKbeta (IKKβ) plus forkhead box O (Foxo) abolishes skeletal muscle atrophy. Biochem. Biophys. Res. Commun. 405:491–496. doi: 10.1016/j.bbrc.2011.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt C., Nissen P. M., Kuhn G., Vestergaard M., Ender K., and Oksbjerg N.. . 2004. Effects of maternal nutrition and porcine growth hormone (pGH) treatment during gestation on endocrine and metabolic factors in sows, fetuses and pigs, skeletal muscle development, and postnatal growth. Domest. Anim. Endocrinol. 27:267–285. doi: 10.1016/j.domaniend.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Relaix F., Montarras D., Zaffran S., Gayraud-Morel B., Rocancourt D., Tajbakhsh S., Mansouri A., Cumano A., and Buckingham M.. . 2006. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 172:91–102. doi: 10.1083/jcb.200508044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Gorza L., Sartore S., Saggin L., Ausoni S., Vianello M., Gundersen K., and Lømo T.. . 1989. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J. Muscle Res. Cell Motil. 10:197–205. doi: 10.1007/bf01739810 [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., and Rudnicki M. A.. . 2000. Pax7 is required for the specification of myogenic satellite cells. Cell 102:777–786. doi: 10.1016/s0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Stevens J. R., and Isom S. C.. . 2012. Gene set testing to characterize multivariate differentially expressed genes. Proceedings of Conference on Applied Statistics in Agriculture:125–137. [Google Scholar]
- Sun T., Fu M., Bookout A. L., Kliewer S. A., and Mangelsdorf D. J.. . 2009. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol. 23:925–931. doi: 10.1210/me.2008-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji K., Kamitani T., Mori F., Kakita A., Takahashi H., and Wakabayashi K.. . 2010. TRIM9, a novel brain-specific E3 ubiquitin ligase, is repressed in the brain of Parkinson’s disease and dementia with Lewy bodies. Neurobiol. Dis. 38:210–218. doi: 10.1016/j.nbd.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R. C 2013. A language and environment for statistical computing. Vienna, Austria:R Foundation for Statistical Computing. [Google Scholar]
- Tong J. F., Yan X., Zhu M. J., Ford S. P., Nathanielsz P. W., and Du M.. . 2009. Maternal obesity downregulates myogenesis and beta-catenin signaling in fetal skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 296:E917–E924. doi: 10.1152/ajpendo.90924.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town S. C., Putman C. T., Turchinsky N. J., Dixon W. T., and Foxcroft G. R.. . 2004. Number of conceptuses in utero affects porcine fetal muscle development. Reproduction 128:443–454. doi: 10.1530/rep.1.00069 [DOI] [PubMed] [Google Scholar]
- United Nations World Population Prospects: The 2017 Revision; 2017. https://www.un.org/development/desa/publications/world-population-prospects-the-2017-revision.html. [Google Scholar]
- Wu G., Bazer F. W., Wallace J. M., and Spencer T. E.. . 2006. Board-invited review: intrauterine growth retardation: implications for the animal sciences. J. Anim. Sci. 84:2316–2337. doi: 10.2527/jas.2006-156 [DOI] [PubMed] [Google Scholar]
- Wu G., Imhoff-Kunsch B., and Girard A. W.. . 2012. Biological mechanisms for nutritional regulation of maternal health and fetal development. Paediatr. Perinat. Epidemiol. 26(Suppl. 1):4–26. doi: 10.1111/j.1365-3016.2012.01291.x [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z., and Rivera A. J.. . 1994. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Huang Y., Zhao J. X., Rogers C. J., Zhu M. J., Ford S. P., Nathanielsz P. W., and Du M.. . 2012. Maternal obesity downregulates microRNA let-7g expression, a possible mechanism for enhanced adipogenesis during ovine fetal skeletal muscle development. International Journal of Obesity. 37:568–575. doi: 10.1038/ijo.2012.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates D. T., Macko A. R., Nearing M., Chen X., Rhoads R. P., and Limesand S. W.. . 2012. Developmental programming in response to intrauterine growth restriction impairs myoblast function and skeletal muscle metabolism. J. Pregnancy 2012:631038. doi: 10.1155/2012/631038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Price F., and Rudnicki M. A.. . 2013. Satellite cells and the muscle stem cell niche. Physiol. Rev. 93:23–67. doi: 10.1152/physrev.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X., Zhao J., Li X., Fan Y., Duan D., Zhang X., Zou W., Sheng Y., Zhang T., Yang Q., . et al. 2018. TMC Proteins Modulate Egg Laying and Membrane Excitability through a Background Leak Conductance in C. elegans. Neuron 97:571–585.e5. doi: 10.1016/j.neuron.2017.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit P. S., Relaix F., Nagata Y., Ruiz A. P., Collins C. A., Partridge T. A., and Beauchamp J. R.. . 2006. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 119(Pt 9):1824–1832. doi: 10.1242/jcs.02908 [DOI] [PubMed] [Google Scholar]
- Zhu M. J., Han B., Tong J., Ma C., Kimzey J. M., Underwood K. R., Xiao Y., Hess B. W., Ford S. P., Nathanielsz P. W., . et al. 2008. AMP-activated protein kinase signalling pathways are down regulated and skeletal muscle development impaired in fetuses of obese, over-nourished sheep. J. Physiol. 586:2651–2664. doi: 10.1113/jphysiol.2007.149633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. J., Ford S. P., Means W. J., Hess B. W., Nathanielsz P. W., and Du M.. . 2006. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J. Physiol. 575(Pt 1):241–250. doi: 10.1113/jphysiol.2006.112110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. J., Ford S. P., Nathanielsz P. W., and Du M.. . 2004. Effect of maternal nutrient restriction in sheep on the development of fetal skeletal muscle. Biol. Reprod. 71:1968–1973. doi: 10.1095/biolreprod.104.034561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.