Abstract
Photosynthetic organisms capture light energy to drive their energy metabolism, and employ the chemical reducing power to convert carbon dioxide (CO2) into organic molecules. Photorespiration, however, significantly reduces the photosynthetic yields. To survive under low CO2 concentrations, cyanobacteria evolved unique carbon-concentration mechanisms that enhance the efficiency of photosynthetic CO2 fixation, for which the molecular principles have remained unknown. We show here how modular adaptations enabled the cyanobacterial photosynthetic complex I to concentrate CO2 using a redox-driven proton-pumping machinery. Our cryo-electron microscopy structure at 3.2 Å resolution shows a catalytic carbonic anhydrase module that harbours a Zn2+ active site, with connectivity to proton-pumping subunits that are activated by electron transfer from photosystem I. Our findings illustrate molecular principles in the photosynthetic complex I machinery that enabled cyanobacteria to survive in drastically changing CO2 conditions.
Subject terms: Enzyme mechanisms, Cryoelectron microscopy, Biophysics
Cyanobacteria evolved carbon-concentration mechanisms to enhance the efficiency of photosynthetic CO2 fixation, but the molecular principles have remained unknown. Here authors use cryo-EM to reveal how modular adaptations enabled the photosynthetic complex I from the cyanobacterium Thermosynechococcus elongatus to concentrate CO2.
Introduction
Cyanobacteria evolved around 2.7 billion years ago with the ability to oxidise water into dioxygen (O2) using the energy captured from sunlight1. The released O2 powers respiratory chains in aerobic life, and the electrons extracted from the water oxidation are used for synthesis of organic molecules2. The light-driven water splitting catalysed by photosystem II (PSII), reduces plastoquinone (PQ) and establishes an electrochemical proton gradient across the thylakoid membrane that subsequently drives synthesis of adenosine triphosphate (ATP)3,4. During linear electron flow (LEF), the electrons are transferred to photosystem I (PSI), providing the reducing power for photosynthetic CO2 fixation that consumes nicotinamide adenine dinucleotide phosphate (NADPH) and drives the synthesis of complex organic compounds from inorganic carbon (Ci)2. Photosynthetic organisms also employ cyclic electron flow (CEF) around PSI to increase the ATP/NADPH ratio that powers the CO2 fixation.
CO2 concentrations drastically changed during the last 2.7 billion years, from >0.5% (5000 ppm) to today’s levels around 0.041% (410 ppm)5. To survive, cyanobacteria evolved carbon-concentrating mechanisms (CCMs) that enhance the efficiency of the photosynthetic CO2 fixation process6,7. In many organisms, hydration of the gaseous CO2 to the soluble bicarbonate (HCO3−) is catalysed by carbonic anhydrase (CA), one of the fastest known enzymes with a kcat ~ 106 s−1 8. However, cyanobacteria lack genes for canonical cytoplasmic CAs, and if artificially expressed in the cytoplasm, the bacteria do not survive in low CO2 concentrations < 20 ppm7. Cyanobacteria express instead the inducible NDH-1MS (NDH-13) and constitutive NDH-1MS’ (NDH-14) photosynthetic complex I isoforms that convert CO2 into HCO3− by kinetically shifting the reaction equilibrium towards bicarbonate, against high cytoplasmic HCO3− concentrations7,9. The HCO3− subsequently diffuses into the carboxysome micro-compartments, where it is converted by a carboxysomal CA to CO27, which further carboxylates ribulose-1,5-bisphosphate (RuBP) into carbohydrates by the action of RuBisCO (Ribulose-1,5-bisphosphate carboxylase/oxygenase)10. This CCM of the photosynthetic complex I, prevents CO2 to diffuse out of the cell by concentrating the Ci for RuBisCO, providing a basis for the efficient carbon fixation that is hampered during photorespiration7.
To determine the molecular architecture of NDH-1MS (NDH-13), we isolate the enzyme from the cyanobacterium Thermosynechococcus elongatus, solved its molecular structure at 3.2 Å resolution using cryo-EM (Fig. 1, Supplementary Fig. 1, Supplementary Table 1, and Supplementary Movie 1), and probe its molecular mechanism by classical and quantum mechanical simulations.
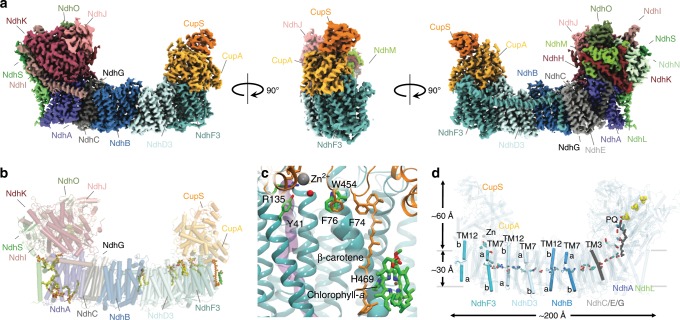
Fig. 1. Structure of the carbon-concentrating photosynthetic complex I.
a The electron density map of NDH-1MS (NDH-13) shown from a back view (left), side view (middle), and front view (right). b Experimentally refined lipid molecules and cofactors in the hydrophobic domain of NDH-1MS. c The structure of the Chl a/β-carotene motif of NdhF3 that connects to the active site of CupA. See Supplementary Fig. 3 for example densities. d Chain of charged elements transmitting the redox-signal into proton pumping and CO2 uptake. The PQ was computationally modelled into the experimental structure.
Results
Architecture of the photosynthetic complex I
The 0.5 MDa complex has an overall U-shape with 19 isolated subunits (Supplementary Fig. 2 and Supplementary Table 2). The structure of NdhV could not be resolved. The core structure is highly conserved across the complex I superfamily11–14, but in contrast to the respiratory enzyme, the photosynthetic complex lacks the N-module that accepts electrons from nicotinamide adenine dinucleotide (NADH) (Fig. 1a–d). Instead, the electrons directly enter a chain of three iron-sulphur (FeS) centres in the ferredoxin (Fd)-binding domain (Fig. 1d), similar to the recently characterised NDH-1L type photosynthetic complex I and membrane-bound hydrogenases15–18. The PQ-binding site is located ca. 20 Å above the membrane plane (Fig. 1d and Supplementary Fig. 3a–c, f), and the modelled PQ tail extends into the lipid membrane in the vicinity of the NdhL subunit, with experimentally resolved lipids stabilising the PQ entry gate (Supplementary Fig. 3b, f).
The modular membrane domain extends up to 200 Å away from the PQ reduction site, and it comprises the antiporter-like subunits NdhA, NdhB, NdhD3, and NdhF3, as well as the smaller transmembrane (TM) subunits NdhC/E/G/L. These subunits contain a chain of buried charged residues that establish central elements of the proton-pumping machinery11–17,19,20, with the isoform-specific NdhD3 and NdhF3 subunits at the terminal end of the enzyme (Fig. 1d, see below).
Structure and function of the CO2 concentrating module
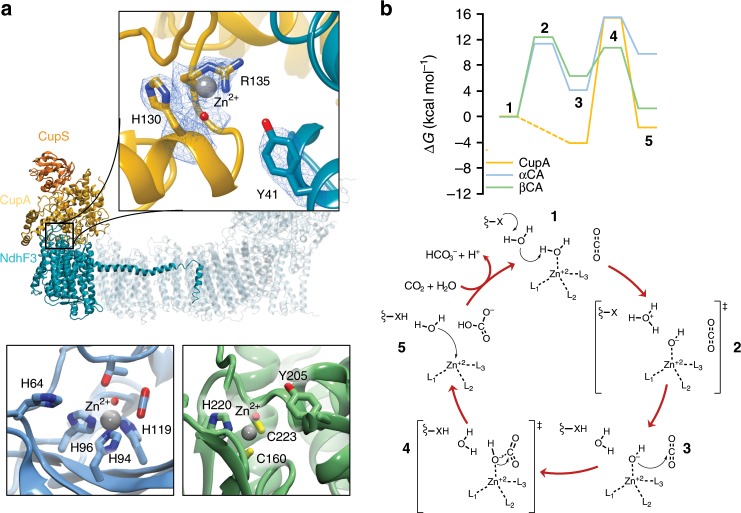
The CO2-concentrating CupA/S module (CO2 uptake, Cup/CO2 hydration protein, ChpY) is located on the cytoplasmic side of the membrane, extending up to 60 Å above the membrane plane (Figs. 1d and 2a and Supplementary Fig. 3d, e). The catalytic site responsible for the CO2 chemistry in the 50 kDa CupA protein electrostatically binds NdhF3 (Fig. 1 and Supplementary Fig. 4a). CupA has a fold, comprising α-helices, which drastically differs from known carbonic anhydrases (CAs) in which the catalytic zinc (Zn2+) ion binds to histidine (αCA family) or histidine/cysteine residues (βCA family) at the interface of β-sheets (Fig. 2a)8. CupA is capped by the 16 kDa α-helical CupS subunit that binds two helical lobes of the former, and could help in stabilising the CupA-NdhF3 interaction (Supplementary Fig. 4a). CupS undergoes a conformational change from its solution structure upon binding to CupA in which an α-helical lid (α2, α3) closes upon a β-sheet (Supplementary Fig. 4b)21. The active site of CupA shows a strong electron density that we assign to a Zn2+ ion (Fig. 2a), as also supported by inductively-coupled plasma optical emission spectrometry (ICP-OES), which indicates a protein-to-zinc ratio of ~0.7 in the NDH-1MS sample (Supplementary Fig. 2e). The Zn2+ ion is coordinated by His130, a putative water/hydroxide ligand or carbonate, and Arg135 (Fig. 2a). Our mass spectrometric (MS) measurements reveal no indication for citrullination of the arginine (Supplementary Fig. 2d, e), and our electrostatic calculations suggest that the residue is neutral with pKa << 7, due to its proximity to the Zn2+ ion (Fig. 2a and Supplementary Fig. 5). Such Zn2+-Arg coordination is rare, but similar structures are also found in a human genetic variant of carbonic anhydrase I (αCA type I)22.
Fig. 2. Structural comparison and mechanism of carbon concentration in CupA and canonical carbonic anhydrases.
a The active site structure of the CO2 concentrating CupA subunit, showing the density of Zn-coordinating residues (5.5 sigma value, contour level 2, top), α-carbonic anhydrase (αCA, PDB ID: 5YUI, bottom left), and β-carbonic anhydrase (βCA, PDB ID: 1EKJ, bottom right). The Zn-bound density has been modelled as a water ligand, although the character of the ligand cannot be unambiguously assigned based on the map. b Reaction mechanism and free energy profiles for the CO2 hydration process based on quantum chemical DFT models in CupA, αCA, and βCA with Tyr41, His64, and Tyr205 as proton acceptors, respectively. Free energies are reported at the B3LYP-D3/def2-TZVP/def2-SVP/ε = 4 theory level (see Methods). Ligands L1/2/3 = His130/Arg135/H2O, L4 = H2O/OH−, X = Tyr41 in CupA; L1/2/3 = His94/96/119, L4 = H2O/OH−, X = His64 in αCA; L1/2/3 = Cys160/223/His220, L4 = H2O/OH−, X = Tyr205 in βCA. The CupA reaction takes place via two water molecules, modelled based on MD simulations (Supplementary Fig. 5j). See Supplementary Fig. 5 for further details.
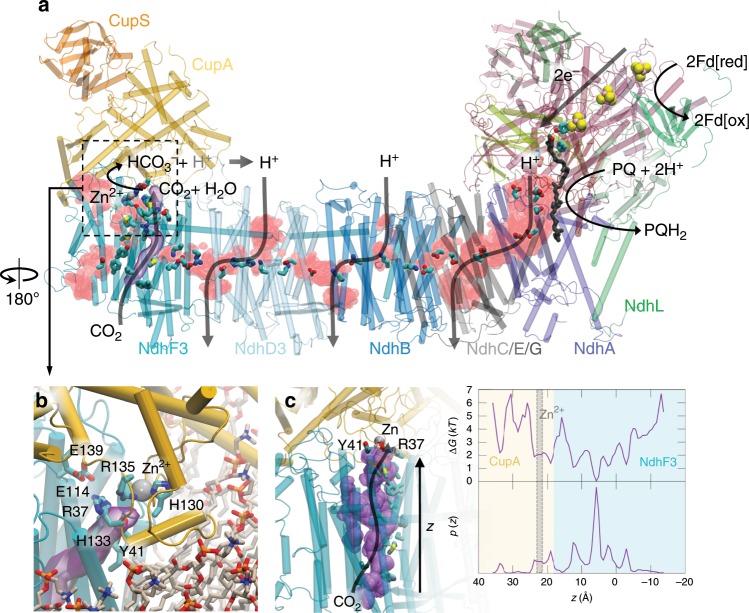
The Zn2+-site is located at the interface of two 12 Å helices, and CupA forms further contacts with NdhF3 and several lipid molecules (Fig. 3a, b and Supplementary Fig. 3d, e). The putative CO2 binding pocket is completed by residues from NdhF3: the Zn2+-H2O/OH− is stabilised by Arg37 that forms an ion-pair with Glu114 and Tyr41 of NdhF3. A non-polar tunnel starting from NdhF3 that leads to the Zn2+-site via this Arg37/Glu114/Tyr41 network could function as a CO2 conduction channel from the luminal side across the membrane (Fig. 3c). Remarkably, NdhF3 binds a chlorophyll a/β-carotene motif that connects via His469 of NdhF3 in a TM-helix to the CupA site (Fig. 1c and Supplementary Fig. 3d). Despite lacking experimental evidence of its functionality, we speculate that the motif could be involved in light-triggered regulation of the CA activity and/or to provide structural stability. A similar Chl a/β−carotene motif with unknown function is also found in cytochrome b6 f[ 23.
Fig. 3. Proton pathways and putative CO2 channel in the photosynthetic complex I.
a Location of proton pathways formed by water molecules (in red) during 250 ns atomistic molecular dynamics simulations. The figure shows the overall water density during MD. b Closeup of the active site of CupA at the NdhF3 interface (in cyan) and the lipid membrane from MD simulations. The putative CO2 channel (in purple), visualised based on CAVER analysis (see Methods), is surrounded by hydrophobic and bulky residues, and connects to the Zn2+-binding site by conserved Tyr41, Arg37, and Glu114 of the NdhF3 subunit (see Supplementary Fig. 6g for details). c Diffusion pathway of CO2 molecules from the luminal side to CupA during MD simulations. Left: Dynamics of CO2 in its putative gas channel in NdhF3. The average of the CO2 position sampled during dynamics is represented as a purple surface. Right: Probability distribution and resulting potential of mean force (pmf in units of kT), G(z) = −RT log(p(z)), of CO2 inside the channel along the membrane axis (z-axis) obtained from 200 ns MD simulation. Owing to sampling gaps, the pmf is subjected to large errors.
Mechanism of the CO2 hydration reaction
To probe the catalytic properties of the CupA module, we performed quantum chemical density functional theory (DFT) calculations on the CO2 hydration reaction and compared this to the reaction energetics in αCA and βCA (Fig. 2b and Supplementary Fig. 5, see Methods). The CO2 hydration is initiated by proton transfer (pT) from the Zn-bound water molecule followed by a nucleophilic attack of the hydroxide on CO2. In αCA (βCA), the rate-limiting reaction barrier of 11 (12) kcal mol−1 is connected with pT to His64 (Tyr205), which compares well with the experimentally-observed barriers of ca. 10–12 kcal mol−1 (Fig. 2b)8. In CupA, proton transfer from Zn-bound water to Tyr41 is slightly exergonic in our DFT models, whereas the nucleophilic attack of the Zn-bound OH− to the CO2 has a barrier of ca. 15 kcal mol−1, predicting that catalysis takes place in the millisecond timescale. The involvement of Tyr41, similarly to Tyr205 in βCA24, is also supported by QM/MM models (Supplementary Fig. 5e), as well as by MD simulations and pKa calculations (Supplementary Fig. 5f). These findings suggest that our resolved structure is catalytically efficient, with kinetic barriers similar to the canonical CAs, despite its unique molecular architecture. CupA becomes well-hydrated during the MD simulations, establishing protonic connectivity with the NdhD3 subunit (Fig. 3a and Supplementary Figs. 5d and 6c), and with global dynamics inferred from the local resolution of the cryo-EM map, closely resembling the motion extracted from the simulations (Supplementary Fig. 7). The proton channels are established across the membrane around charged residues in the broken helices TM7 and TM12 of the antiporter-like subunits NdhB, NdhD319,20, and also in NdhA/C/E/G (Figs. 1d and 3, and Supplementary Fig. 6a–d, f) at locations where conformational changes were recently observed in the mammalian enzyme (Supplementary Fig. 6e)12. The structural architecture thus supports that the long-range protonation signal could be triggered by dissociation of conserved ion-pairs in the antiporter-like subunits that leads to lateral proton transfer in the proton channels by coupled conformational and hydration changes (Fig. 4)16,19,20.
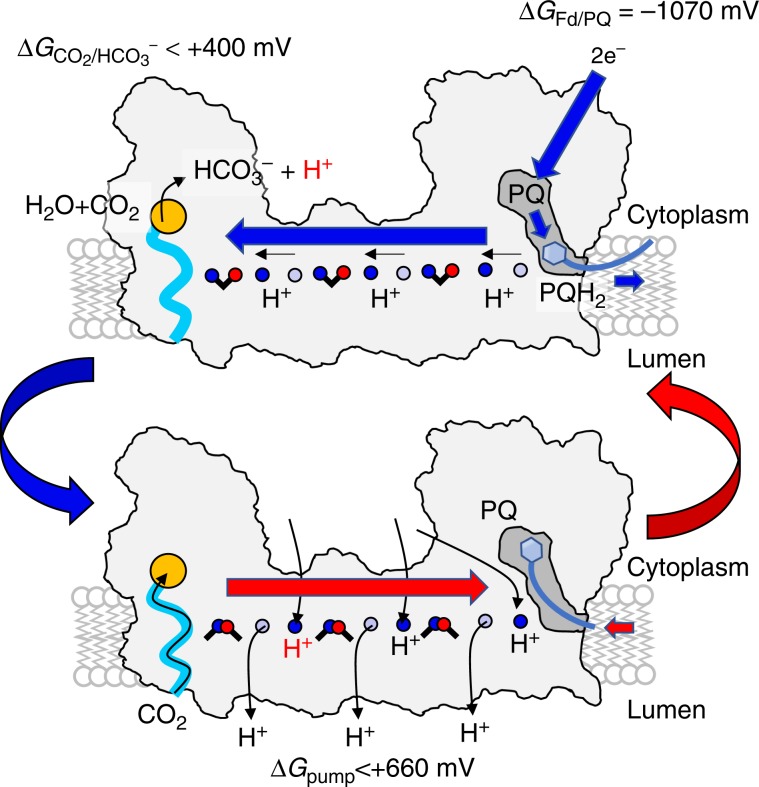
Fig. 4. Putative mechanism of redox-driven CO2 concentration in the photosynthetic complex I.
Top: PQ reduction to PQH2 leads to dissociation of the quinol to a membrane-bound PQ-binding site71 that triggers sequential opening of ion-pairs in NdhB, NdhD3, and NdhF3, by coupled hydration, conformational, and protonation changes, protonating terminal proton loading sites (blue forward arrow)19, 20. Bottom: Closing of ion-pairs in a sequential electrostatic backward pulse (red backward arrow) releases the protons to the luminal side of the membrane, and allows for re-protonation of the antiporter-like subunits by protons released from the CO2 hydration reaction in the active site of CupA (orange circle). Horizontal proton transfer reactions within each antiporter-like subunit are shown by small horizontal black arrows, and PQH2 (PQ) diffusion out (in) is indicated by small thick blue (red) arrows. CO2 is taken up by the putative gas channel (in light blue) that is expected to be open depending on the ion-pair conformation in NdhF3 (arrow along light blue channel).
Discussion
The NdhF3 comprises conserved ion-pairs at the interface to the NdhD3 subunit, similar as in the other antiporter-like subunits. However, charged elements in the lateral proton channel in NdhF3 are replaced by non-polar residues that could be employed to channel CO2 into CupA. Indeed, we observe that CO2 effectively diffuses along this channel to the active site of the CupA subunit in MD simulations (Fig. 3c and Supplementary Fig. 6g). Although the solubility of CO2 is similar in lipid membranes and water25, the gas tunnel could channel CO2 into the active site of the CupA subunit, and prevent its escape under low CO2 and high bicarbonate concentrations, arising from the substrate channelling to RuBisCO and bicarbonate pumps in the plasma membrane7,26. The thermodynamically unfavourable CO2 hydration reaction is thus kinetically gated in the bicarbonate direction when the protons released in the CO2 hydration process are pumped across the membrane by the antiporter-like subunits (Fig. 4). The driving force for this is achieved by the ca. 1070 mV redox span transduced from the PQ oxido-reduction chemistry (Em(Fd) ≈ −420 mV27; Em(PQ) ≈ + 117 mV28), that is transmitted to the terminal edge of the membrane domain (Fig. 4)19,20. The carbon concentration thus couples to generation of proton motive force (pmf) across the membrane, that in turn, is transduced in ATP synthesis and active transport3. With a high pmf of up to 220 mV29, the NDH-1MS is expected to thermodynamically drive the hydration of CO2 (at 410 ppm; [CO2] = 14 μM; [HCO3−] = 30 mM26; pH = 7), and couple this to pumping of at least three protons across the membrane, e.g., by NdhD3, NdhB, and NdhA/C/E/G11–14,19,20. However, the free energy available could allow the pump to operate also under considerably lower CO2 concentrations and pH. Under high pmf and a reduced quinone pool, the respiratory complex I operates in reverse direction by consuming the proton gradient to oxidize quinol30, and due to conserved functional elements, a similar operation mode is also expected for NDH-1MS. During such putative backward operation mode, coupled protonation and/or conformational changes at the NdhF3/NdhD3 interface could close the gas channel similar to conformational changes observed in the bacterial complex I19,31. Such changes might decouple the pump to avoid the back-reaction of HCO3− to CO2, and the diffusion of the latter out of the cell.
We have demonstrated here how unique molecular adaptation enabled the photosynthetic complex I to concentrate CO2 by redox-driven proton-pumping modules and powered by cyclic electron flow around photosystem I. Our findings show how combination of functional modules from different energy transduction machineries32 allowed primordial organisms to generate functionality and harness energy under changing environmental conditions.
Methods
Construction of the T. elongatus mutant and purification of the NDH-1MS complex
The C-terminal tagged CupS-TwinStrep-tag mutant of T. elongatus was generated by homologous recombination with plasmid DNA that contains the modified cupS gene and a kanamycin resistance cassette for selection of the mutant allele (see Supplementary Table 4). Cells of the fully segregated mutant were grown in liquid BG-11 medium and expression of the NDH-1MS complex was induced by low CO2 growth conditions. Preparation of the thylakoid membranes and isolation of the tagged complex were performed as described earlier15,33 with minor modifications. See SI Materials and Methods for details.
Cryo-EM grid preparation and data collection
To prepare cryo-EM grids, four microliter of NDH-1MS at 5 mg mL−1 were applied to R2/1 grids (Quantifoil) that were glow-discharged for 20 s immediately before use. The sample was incubated 30 s at 100% humidity and 4 °C before blotting for 3.5 s with blotforce 5 and then plunge-frozen into liquid ethane/propane mix cooled by liquid nitrogen using a Vitrobot Mark IV (FEI). Data was acquired on a Titan Krios electron microscope (ThermoFisher, FEI) operated at 300 kV, equipped with a K2 Summit direct electron detector (Gatan) and a GIF quantum energy filter (20 eV) (Gatan). Movies were recorded in counting mode at a pixel size of 1.35 Å per pixel using a cumulative dose of 40.20 e−/Å2 and 50 frames. Data acquisition was performed using SerialEM34 with four exposures per hole with a target defocus range of 0.5 to 3.0 μm.
Image processing
The dose-fractionated movies were gain normalised, aligned and dose-weighted using the motion correction algorithm (Motion Cor2)35 implemented in RELION 3.034,36. GCTF37 was used to estimate the defocus values, and particles were selected using Gautomatch. All subsequent processing steps were carried out in RELION 3.034. Following two-dimensional classification, an ab initio model was generated using stochastic gradient descent (SGD). The entire data set (526,925 particles) was subjected to three-dimensional (3D) classification into five classes using the previously determined 60 Å low-pass-filtered ab initio model. A single class consisting of the majority of particles (51.7%) resembled the complex structure. The particles were selected and subsequently sub-classified into 3 classes, using a finer angular sampling (3.75°). The best aligning class consisted of 170,151 particles (62.4%), which were subjected to 3D refinement, yielding an overall resolution of 3.9 Å. The particles were further polished using Bayesian-polishing and the CTF-values were refined on a per particle basis, improving the resolution of the reconstruction to 3.2 Å. The temperature factor (−89.9 Å2) and the resolution of the map were estimated by applying a soft mask around the protein density in the post-processing routine, using the gold standard Fourier shell correlation (FSC) = 0.143 criterion. Directional FSC curves and map anisotropy were assessed using the 3DFSC server38.
Model building and validation
The models for the conserved subunits were taken from the previously published NDH-1L complex (PDB ID: 6HUM)15. For the NdhD3 and NdhF3 paralog subunits homology models were generated with the Phyre2 server and fitted as rigid bodies with UCSF Chimera39. They were subsequently manually adjusted and rebuilt using Coot40. The carbonic anhydrase CupA and its binding partner CupS was built de novo using Coot. The model of NDH-1MS was refined against the cryo-EM map using the phenix.real_space_refine routine in the PHENIX software package41. The statistical quality of the final model was assessed using MOLPROBITY42 and EMRinger43. Figures were prepared using PyMOL44 or UCSF Chimera X45.
DFT calculations
DFT models of the CupA/NdhF3 active site were built based on the cryo-EM atomic coordinates of NDH-1MS. The model comprises the Zn2+ ion and its water/OH− ligands, Phe70, Lys78, Tyr79, His130, Ile131, His133, Arg135, Glu139, and Glu142 of CupA subunit, and Tyr41, Arg37, and Glu114 of the NdhF3 subunit, in addition to two water molecules and the CO2/HCO3− substrate. Water molecules between Zn and Tyr41 were modelled based on MD simulations (Supplementary Fig. 5j). The model comprises 185 atoms. DFT models of α-carbonic anhydrase were built based on type II human αCA (PDB ID: 5YUI46), comprising the Zn2+ ion, and the water/OH− group, His94 His96, His119, His64, Asn67, Gln92, Glu106, Thr199, Thr200, Val121, Val143, Trp209, and eight water molecules and a CO2 molecule solved in the crystal structure. The model comprises 155 atoms. DFT models of βCA were built based on the PDB ID: 1EKJ47. The model comprises the Zn2+ ion, Cys160, Cys223, His220, the water/OH− ligand, Gln151, Asp162, Arg164, Val165, Val184, Asn186, the backbone of Gly224, Tyr205, in addition to four water molecules and the CO2. The model comprises 132 atoms. Protein residues were cut at the Cα-Cβ bond and saturated with hydrogen atoms. The Cβ atoms were kept fixed during geometry optimisations that were performed at the B3LYP-D3/def2-SVP/def2-TZVP(Zn2+) level, and single point energies were evaluated at the B3LYP-D3/def2-TZVP level48–51. Solvation effects were treated using a polarisable dielectric medium with ε = 4 to model the protein environment52. Free energies were computed using the freeh module of TURBOMOLE based on electronic and zero-point energies (ZPE), and enthalpic (ΔH) and entropic (TΔS) effects, estimated at the B3LYP-D3/def2-SVP level by calculating the molecular Hessian. The free energy estimates do not consider dynamical sampling effects. Reaction pathways and transitions states were optimised using a chain-of-states method53,54. All QM calculations were performed with TURBOMOLE versions 6.6-7.355.
QM/MM calculations
Hybrid QM/MM calculations were performed based on our NDH-1MS structure. The models comprise the CupA and NdhF subunits, including the Zn-binding site, and water/lipid/ions in their 12-Å surroundings. The QM region comprises 135 atoms, including the Zn2+-OH− cofactor, Tyr79, His130, His133, Arg135, Glu139 of CupA subunit, and Arg37 and Tyr41 of the NdhF3 subunit. The QM/MM boundary was described by a link-atoms approach. The model was structure optimised at the QM/MM level using the Adopted Basis Newton-Raphson (ABNR) algorithm, and allowing a 10 Å-sphere within the QM region to relax, followed by QM/MM dynamics at T = 310 K using a 1 fs integration timestep. The QM region was described at the B3LYP-D3/def2-SVP(C,H,N,O)/def2-TZVP(Zn2+) level of theory, and the MM region was described by the CHARMM36 force field56. A minimum energy pathway scan between the Zn-H2O and Tyr41 was performed along a reaction coordinate modelled as linear combination of all proton donor and acceptor distances (R = r1 − r2 + r3 − r4, see Supplementary Fig. 5d). Our reported DFT models predict similar energetics for carbonic anhydrases as those reported in previous studies, cf.57 and refs. therein and further QM/MM models of these systems were therefore not considered. All QM/MM calculations were performed using an in-house version of the CHARMM/TURBOMOLE interface58.
Classical molecular dynamics simulations
Classical molecular dynamics (MD) simulations were performed based on the cryo-EM structure of NDH-1MS. Missing loops in the antiporter-like and NdhS subunits were modelled with MODELLER9.1959. The cryo-EM atomic coordinates were relaxed to the electron density map using molecular dynamics flexible fitting (MDFF)60. The MDFF relaxed model was further embedded in a POPC membrane and solvated with TIP3P water molecules and 100 mM NaCl concentration. Plastoquinone was modelled based on previous work16. The total system comprises ca. 580,000 atoms. MD simulations were performed at constant T = 310 K and p = 1 bar in an NPT ensemble. The CHARMM36 force field56 was used to model protein/lipid/water-ion atoms. Parameters for PQ, FeS centres, and the Zn-first coordination sphere were derived from DFT calculations. Long-range electrostatic interactions were described by the particle mesh Ewald (PME) approach. The system was gradually relaxed during the 2.5 ns with harmonic restraints, followed by 20 ns equilibration without restraints. An initial 1 fs integration timestep was used during the equilibration process, and 2 fs timestep during the remaining 250 ns production steps. MD simulations of the CO2 diffusion process were performed based on 20 independent trajectories each 10 ns with the CO2 molecule positioned along the putative cavity in the NdhF3 subunit, that was identified with CAVER61. All classical MD simulations were performed with NAMD262, and VMD63 and UCSF Chimera39 were used for analysis.
Poisson-Boltzmann continuum electrostatics calculations
pKa values of titratable residues in CupA/S/NdhF3 were estimated using the Adaptive Poisson-Boltzmann Solver (APBS)64 and by Monte Carlo (MC) sampling of 2N protonation states with Karlsberg+65,66. PBE/MC calculations can provide accurate estimation of pKa values in complex biochemical systems67,68, whereas constant pH-MD simulations that could further enhance the accuracy of the predictions69,70 are outside the scope of the present work due to the large size and complex surroundings of NDH-1MS. The protein was described using partial charges, embedded in an inhomogeneous dielectric medium with a dielectric constant of 4 inside the protein and 80 for water. The interface between the protein and solvent was calculated by the molecular surface routine, as implemented in APBS, using a solvent probe radius of 1.4 Å, and modelling an implicit ionic strength of 100 mM potassium chloride. The pKa values were computed as a difference of electrostatic free energy shifts between a model compound in water and the model compound in the protein for 80 structures obtained from the MDFF relaxation.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank H. Wulfhorst for construction of the CupS-TS mutant, M. Völkel for excellent technical assistance and Petra Düchting (Department MGPP, Ruhr University Bochum, Germany) for ICP-OES analysis. J.M.S. and S.K.S. would like to acknowledge E. Conti for unconditional support. J.M.S is grateful to B.D. Engel, J.M. Plitzko, and W. Baumeister for access to the cryo-EM infrastructure and early career support. This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme/grant agreement no. 715311 (to V.R.I.K.), the DFG research unit FOR2092 (836/3-2 to M.M.N.), the DFG priority programme 2002 (NO 836/4-1 to M.M.N.) and the DFG grant NO 836/1-1 (to M.M.N.). This work was also supported by Emergence of Life initiative (TRR235 to V.R.I.K.) and by the Knut and Alice Wallenberg Foundation (to V.R.I.K.). Computational resources were provided by SuperMuc/Leibniz-Rechenzentrum (LRZ, grant: pn34he) and PRACE (project nr: 2018194738), awarding us access to MareNostrum at the Barcelona Supercomputing Centre (BSC), Spain (project: pr1ejk).
Source data
Author contributions
J.T. and M.M.N. isolated and characterised the photosynthetic complex I; P.S., A.P.G.H. and V.R.I.K. performed molecular simulations; J.M.S. and S.K.S. prepared cryo-EM grids and collected data; J.M.S. and S.K.S. processed cryo-EM data; J.M.S., S.K.S. and V.R.I.K. built cryo-EM models; J.M.S., S.K.S. and V.R.I.K. analysed and interpreted the cryo-EM models; J.M.S., P.S., J.T., S.K.S., A.P.G.H., G.K., M.M.N. and V.R.I.K analysed the data; J.M.S., M.M.N. and V.R.I.K. directed the project; V.R.I.K. wrote the manuscript with input from all authors.
Data availability
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. The source data underlying Figs. 2b, 3c, 4c–e and Supplementary Figs. 2a, b, 3c, 5c, d are provided as a Source Data file. EMDB accession codes: EMD-10513, PDB ID: 6TJV.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Marcelo Martí, Thorsten Friedrich and Janet Vonck for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Patricia Saura, Jacqueline Thiemann
Contributor Information
Jan M. Schuller, Email: janschu@biochem.mpg.de
Marc M. Nowaczyk, Email: marc.m.nowaczyk@rub.de
Ville R. I. Kaila, Email: ville.kaila@dbb.su.se
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-14347-4.
References
- 1.Cardona T. Thinking twice about the evolution of photosynthesis. Open Biol. 2019;9:180246. doi: 10.1098/rsob.180246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson, N. & Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982 (2004). [DOI] [PubMed]
- 3.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida M, Muneyuki E, Hisabori T. ATP synthase: A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 5.Crowley TJ, Berner RA. CO2 and climate change. Science. 2001;292:870–872. doi: 10.1126/science.1061664. [DOI] [PubMed] [Google Scholar]
- 6.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 7.Price, G. D., Maeda, S., Omata, T. & Badger, M. R. Modes of active inorganic carbon uptake in the cyanobacterium, Synechococcus sp. PCC7942. Funct. Plant Biol. 29, 131–149 (2002). [DOI] [PubMed]
- 8.Silverman DN, Lindskog S. The catalytic mechanism of carbonic anhydrase: implications of a rate-limiting protolysis of water. Acc. Chem. Res. 1988;21:30–36. doi: 10.1021/ar00145a005. [DOI] [Google Scholar]
- 9.Peltier G, Aro E-M, Shikanai T. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu. Rev. Plant Biol. 2016;67:55–80. doi: 10.1146/annurev-arplant-043014-114752. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, et al. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature. 2019;566:131–135. doi: 10.1038/s41586-019-0880-5. [DOI] [PubMed] [Google Scholar]
- 11.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agip ANA, et al. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 2018;25:1–9. doi: 10.1038/s41594-018-0073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zickermann V, et al. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 15.Schuller JM, et al. Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science. 2019;363:257–260. doi: 10.1126/science.aau3613. [DOI] [PubMed] [Google Scholar]
- 16.Saura P, Kaila VRI. Molecular dynamics and structural models of the cyanobacterial NDH-1 complex. Biochim. Biophys. Acta—Bioenerg. 2019;1860:201–208. doi: 10.1016/j.bbabio.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laughlin TG, Bayne AN, Trempe JF, Savage DF, Davies KM. Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature. 2019;566:411–414. doi: 10.1038/s41586-019-0921-0. [DOI] [PubMed] [Google Scholar]
- 18.Yu H, et al. Structure of an ancient respiratory system. Cell. 2018;173:1636–1649.e16. doi: 10.1016/j.cell.2018.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaila VRI. Long-range proton-coupled electron transfer in biological energy conversion: Towards mechanistic understanding of respiratory complex I. J. R. Soc. Interface. 2018;15:20170916. doi: 10.1098/rsif.2017.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Luca A, Gamiz-Hernandez AP, Kaila VRI. Symmetry-related proton transfer pathways in respiratory complex I. Proc. Natl Acad. Sci. USA. 2017;114:E6314–E6321. doi: 10.1073/pnas.1706278114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korste A, Wulfhorst H, Ikegami T, Nowaczyk MM, Stoll R. Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus. Biochim. Biophys. Acta—Bioenerg. 2015;1847:1212–1219. doi: 10.1016/j.bbabio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Ferraroni M, et al. Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: Evidence for a second zinc binding site involving arginine coordination. Biochemistry. 2002;41:6237–6244. doi: 10.1021/bi0120446. [DOI] [PubMed] [Google Scholar]
- 23.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 24.Rowlett RS, et al. Kinetic characterization of wild-type and proton transfer-impaired variants of β-carbonic anhydrase from Arabidopsis thaliana. Arch. Biochem. Biophys. 2002;404:197–209. doi: 10.1016/S0003-9861(02)00243-6. [DOI] [PubMed] [Google Scholar]
- 25.Simon SA, Gutknecht J. Solubility of carbon dioxide in lipid bilayer membranes and organic solvents. Biochim. Biophys. Acta—Bioenerg. 1980;596:352–358. doi: 10.1016/0005-2736(80)90122-4. [DOI] [PubMed] [Google Scholar]
- 26.Mangan NM, Flamholz A, Hood RD, Milo R, Savage DF. pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc. Natl Acad. Sci. USA. 2016;113:E5354–E5362. doi: 10.1073/pnas.1525145113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tagawa K, Arnon DI. Oxidation-reduction potentials and stoichiometry of electron transfer in ferredoxins. Biochim. Biophys. Acta—Bioenerg. 1968;153:602–613. doi: 10.1016/0005-2728(68)90188-6. [DOI] [PubMed] [Google Scholar]
- 28.Golbeck JH, Kok B. Redox titration of electron acceptor Q and the plastoquinone pool in Photosystem II. Biochim. Biophys. Acta—Bioenerg. 1979;547:347–360. doi: 10.1016/0005-2728(79)90016-1. [DOI] [PubMed] [Google Scholar]
- 29.Ritchie J. Membrane potential and pH Control in The Cyanobacterium Synechococcus R-2 (Anacystis nidulans) PCC 7942. J. Plant Phys. 1991;137:409–418. doi: 10.1016/S0176-1617(11)80309-3. [DOI] [Google Scholar]
- 30.Lambert AJ, Brand MD. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. Biochem. J. 2004;382:511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Luca A, Mühlbauer ME, Saura P, Kaila VRI. How inter-subunit contacts in the membrane domain of complex I affect proton transfer energetics. BBA—Bioenerg. 2018;1859:734–741. doi: 10.1016/j.bbabio.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandt U. Adaptations of an ancient modular machine. Science. 2019;363:230–231. doi: 10.1126/science.aaw0493. [DOI] [PubMed] [Google Scholar]
- 33.Wulfhorst H, Franken LE, Wessinghage T, Boekema EJ, Nowaczyk MM. The 5kDa protein NdhP is essential for stable NDH-1L assembly in Thermosynechococcus elongatus. PLoS ONE. 2014;9:e103584. doi: 10.1371/journal.pone.0103584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zivanov J, Nakane T, Scheres SHW. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife7, pii: e42166 (2018). [DOI] [PMC free article] [PubMed]
- 37.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan YZ, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D. Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delano, W. L. The PyMOL molecular graphics system. Schrodingerhttp://www.pymol.sourceforge.net (2002).
- 45.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim CU, et al. Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals. Proc. Natl Acad. Sci. 2016;113:5257–5262. doi: 10.1073/pnas.1520786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimber MS. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 2000;19:1407–1418. doi: 10.1093/emboj/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 49.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 50.Schäfer A, Horn H, Ahlrichs R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992;97:2571–2577. doi: 10.1063/1.463096. [DOI] [Google Scholar]
- 51.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010;132:154104-1–19. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 52.Klamt A, Schüürmann G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 1993;2:799–805. doi: 10.1039/P29930000799. [DOI] [Google Scholar]
- 53.Plessow P. Reaction path optimization without NEB springs or interpolation algorithms. J. Chem. Theory Comput. 2013;9:1305–1310. doi: 10.1021/ct300951j. [DOI] [PubMed] [Google Scholar]
- 54.W E, Ren W, Vanden-Eijnden E. String method for the study of rare events. Phys. Rev. B –Condens. Matter Mater. Phys. 2002;66:523011–523014. [Google Scholar]
- 55.Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. Electronic structure calculations on workstation computers: The program system turbomole. Chem. Phys. Lett. 1989;162:165–169. doi: 10.1016/0009-2614(89)85118-8. [DOI] [Google Scholar]
- 56.Huang J, Mackerell AD. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riccardi D, Yang S, Cui Q. Proton transfer function of carbonic anhydrase: Insights from QM/MM simulations. Biochimica et. Biophysica Acta—Proteins Proteom. 2010;1804:342–351. doi: 10.1016/j.bbapap.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riahi S, Rowley CN. The CHARMM-TURBOMOLE interface for efficient and accurate QM/MM molecular dynamics, free energies, and excited state properties. J. Comput. Chem. 2014;35:2076–2086. doi: 10.1002/jcc.23716. [DOI] [PubMed] [Google Scholar]
- 59.Webb B, Sali A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinforma. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chovancova Eva, Pavelka Antonin, Benes Petr, Strnad Ondrej, Brezovsky Jan, Kozlikova Barbora, Gora Artur, Sustr Vilem, Klvana Martin, Medek Petr, Biedermannova Lada, Sochor Jiri, Damborsky Jiri. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Computational Biology. 2012;8(10):e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl Acad. Sci. 2002;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kieseritzky G, Knapp EW. Optimizing pKA computation in proteins with pH adapted conformations. Proteins Struct. Funct. Genet. 2008;71:1335–1348. doi: 10.1002/prot.21820. [DOI] [PubMed] [Google Scholar]
- 66.Rabenstein B, Knapp EW. Calculated pH-dependent population and protonation of carbon-monoxy myoglobin conformers. Biophys. J. 2001;80:1141–1150. doi: 10.1016/S0006-3495(01)76091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyer T, Knapp EW. pKa values in proteins determined by electrostatics applied to molecular dynamics trajectories. J. Chem. Theory Comput. 2015;11:2827–2840. doi: 10.1021/acs.jctc.5b00123. [DOI] [PubMed] [Google Scholar]
- 68.Kieseritzky G, Knapp EW. Improved pKa prediction: combining empirical and semimicroscopic methods. J. Comput. Chem. 2008;29:2575–2581. doi: 10.1002/jcc.20999. [DOI] [PubMed] [Google Scholar]
- 69.Baptista AM, Teixeira VH, Soares CMJ. Constant-pH molecular dynamics using stochastic titration. Chem. Phys. 2002;117:4184–4200. [Google Scholar]
- 70.Swails JM, York DM, Roitberg AE. Constant pH replica exchange molecular dynamics in explicit solvent using discrete protonation states: implementation, testing, and validation. J. Chem. Theory Comput. 2014;10:1341–1352. doi: 10.1021/ct401042b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warnau J, et al. Redox-coupled quinone dynamics in the respiratory complex I. Proc. Natl Acad. Sci. USA. 2018;115:E8413–E8420. doi: 10.1073/pnas.1805468115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. A reporting summary for this Article is available as a Supplementary Information file. The source data underlying Figs. 2b, 3c, 4c–e and Supplementary Figs. 2a, b, 3c, 5c, d are provided as a Source Data file. EMDB accession codes: EMD-10513, PDB ID: 6TJV.