Abstract
Devosia are well known for their dominance in soil habitats contaminated with various toxins and are best characterized for their bioremediation potential. In this study, we compared the genomes of 27 strains of Devosia with aim to understand their metabolic abilities. The analysis revealed their adaptive gene repertoire which was bared from 52% unique pan-gene content. A striking feature of all genomes was the abundance of oligo- and di-peptide permeases (oppABCDF and dppABCDF) with each genome harboring an average of 60.7 ± 19.1 and 36.5 ± 10.6 operon associated genes respectively. Apart from their primary role in nutrition, these permeases may help Devosia to sense environmental signals and in chemotaxis at stressed habitats. Through sequence similarity network analyses, we identified 29 Opp and 19 Dpp sequences that shared very little homology with any other sequence suggesting an expansive short peptidic transport system within Devosia. The substrate determining components of these permeases viz. OppA and DppA further displayed a large diversity that separated into 12 and 9 homologous clusters respectively in addition to large number of isolated nodes. We also dissected the genome scale positive evolution and found genes associated with growth (exopolyphosphatase, HesB_IscA_SufA family protein), detoxification (moeB, nifU-like domain protein, alpha/beta hydrolase), chemotaxis (cheB, luxR) and stress response (phoQ, uspA, luxR, sufE) were positively selected. The study highlights the genomic plasticity of the Devosia spp. for conferring adaptation, bioremediation and the potential to utilize a wide range of substrates. The widespread toxin-antitoxin loci and ‘open’ state of the pangenome provided evidence of plastic genomes and a much larger genetic repertoire of the genus which is yet uncovered.
Subject terms: Microbiology, Genome informatics
Introduction
Devosia comprises a group of motile, gram-negative bacteria within the class Alphaproteobacteria and family Hyphomicrobiaceae1. The first recognized species of the genus was Pseudomonas riboflavina IFO13584 described by Foster in 19442 from riboflavin-rich soil which was reclassified into Devosia riboflavina in 19961. Since then, many members of this genus have been reported from diverse ecological niches. Although their distribution is ubiquitous including their presence in human cerebrospinal fluid3, nodules of legume plants4,5 and beach sediment6, members of this genus have been mainly reported from soils contaminated with hexachlorocyclohexane (HCH)7,8, mycotoxins (deoxynivalenol)9,10 and other hydrocarbon pesticides11.
In an effort to characterize the culturable diversity of soil contaminated with HCH12–18, we isolated and characterized four novel members of the genus Devosia viz. D. chinhatensis IPL187, D. crocina IPL208, D. albogilva IPL158 and D. lucknowensis L1519. Although isolated from HCH contaminated soil, these isolates were not able to degrade HCH isomers8. However, members of the genus are best studied for their potential to degrade several toxic compounds, establishing their promising candidature for bioremediation2,9. Previous studies have aimed to characterize their metabolic routes of detoxification20. In spite of their abundance in culture collections and public repositories, the genetic repertoire that enables them to survive in harsh environments have not been elucidated. Here, we report the first comparative genomic study of 27 members of genus Devosia, which provides valuable insights into their adaptations, the role of environment in shaping their genomes and the degree of genomic evolution in response to different environmental pressures.
Our study suggested the influx of new metabolic capabilities into the “open” pangenome of Devosia. Besides, the phylogenetic relationships of the group were fairly consistent. The study revealed that the genomes harbor a large diversity of transporters involved in uptake of di- and oligo-peptides from the environment. These peptide transport systems enable bacteria to take up short peptides of different amino-acid composition for satisfying nutritional demands and have been extensively studied in species of Lactococcus and Staphylococcus21,22. Besides increasing nutritional fitness, these permeases are also shown to be involved in signaling and virulence in Staphylococcus aureus, Borrelia burgdorferi and Bacillus thuringiensis23–26. Here, our analysis revealed the high diversity of these permeases encoded within genus Devosia for enabling efficient nutrient utilization and cell signaling required at such environments. Additionally, the large diversity of their substrate binding components reflected their wide range of substrates utilization. Positive evolution and selection of genes associated with growth and utilization of toxins highlights future applications in bioremediation. Further, the genomic repertoire adapted for utilization of organic sulfur, phosphorus and aromatic compounds are presumed to enable the members of the genus Devosia to survive in harsh sites. The presence of toxin-antitoxin (TA) loci within their genomes provided evidence of enhanced genome plasticity for maintaining a wide range of biological functions including stress response.
Results and Discussion
Genomic features
Genome analysis of twenty seven strains of the genus Devosia showed >96% completeness establishing the reliability of the datasets for comparative analyses. The overall genomic features of the strains are listed in Table 1. The genome size ranged from 3.5 to 5.8 Mbp with an average genome size of 4.3 ± 0.6 Mbp. Notably, the strains isolated from HCH contaminated sites namely, IPL-18, L15 and IPL-20 represented the three smallest genomes. It is difficult to explain the minimum genomic size of the organisms at such contaminated and nutrient depleted sites. However, in a previous study, where we isolated and described a Pseudomonas species that has the smallest genome with respect to its neighbours, this was attributed to the HCH isomer pressure shaping the genomic repertoire27. IPL18 and L15 also lacked genetic potential for utilization of organic phosphorus, rather found in other genomes. It is likely that the organisms lost several accessory gene clusters as part of adaptations to survive at HCH rich habitat. The two largest genomes of Root105 and Root413D1 harboured several hypothetical proteins in singletons along with the genes involved in drug resistance (daunorubicin and doxorubicin), serralysin and leukotoxin, type I secretion system, adhesion protein BmaC and polyamine synthesis proteins. These proteins are associated with protection, adhesion and biofilm formation and may facilitate the colonization of these strains in plant roots28,29. The %GC contents varied between 60.5–65.9% with an average of 63 ± 1.7%. Each genome, on an average consisted of 4,330 ± 620.4 protein coding genes. The number of predicted coding sequences correlated positively with the genome size (PMCC, r = 0.99). The large difference with respect to genome size and the coding potential among the species reflected towards the cadences in the genomic repertoire of the Devosia ecotypes in response to the different niches.
Table 1.
General attributes of the Devosia genomes analyzed in this study.
| Strain | Genome Size (bp) | No. of Contigs | GC Content (%) | CDS | rRNAs (5S, 16S, 23S) | tRNAs | CRISPRs | Source of Isolation | Accession Number | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Devosia insulae DS-56 | 5,750,119 | 410 | 65.3 | 5632 | 1,1,1 | 50 | — | Soil sample South Korea: Dokdo Island, East Sea of Korea | NZ_LAJE00000000.2 | 98 |
| D. limi DSM17137 | 4,297,227 | 25 | 62.7 | 4183 | 2,1,1 | 48 | — | Nitrifying inoculum of activated sludge in Gent, Belgium | NZ_FQVC00000000.1 | 98 |
| D. soli GH2-10 | 4,136,371 | 48 | 61 | 4183 | 3,1,1 | 48 | — | Greenhouse soil used to cultivate lettuce in Daejeon City, Korea | NZ_LAJG00000000.1 | 98 |
| D. epidermidihirudinis E84 | 3,859,784 | 47 | 61.1 | 3745 | 2,2,2 | 49 | — | Skin of medical leech Hirudo verbana, from Biebertal, Germany | NZ_LANJ00000000.1 | Unpublished data |
| D. riboflavina IFO13584 | 5,052,234 | 113 | 61.8 | 5042 | 1,1,1 | 52 | — | Riboflavin rich soil in Rahway, New Jersey | NZ_JQGC00000000.1 | 99 |
| D. chinhatensis IPL-18 | 3,497,719 | 98 | 62.3 | 3437 | 2,2,2 | 48 | — | Soil samples from an India Pesticide Limited plant at hexachlorocyclohexane (HCH) dump site, Lucknow, India. | NZ_JZEY00000000.1 | 91 |
| D. geojensis BD-c194 | 4,465,063 | 207 | 65.9 | 4432 | 1,1,1 | 49 | — | Diesel-contaminated soil in Geoje, Korea | NZ_JZEX00000000.1 | 100 |
| D. crocina IPL-20 | 3,723,990 | 7 | 61.3 | 3706 | 1,1,1 | 45 | 1 | Hexachlorocyclohexane (HCH)-contaminated site in Chinhat, Lucknow, India | NZ_FPCK00000000.1 | This study |
| D. psychrophila CGMCC 1.10210 | 4,328,275 | 85 | 61.2 | 4353 | 1,1,1 | 49 | — | Alpine glacier cryoconite, Tyrol, Austria | FOMB00000000.1 | Unpublished data |
| D. enhydra ATCC 23634 | 4,220,684 | 5 | 65.6 | 4107 | 2,1,2 | 48 | 1 | Freshwater from the Putah Creek overflow in Davis, Calif, California | NZ_FPKU00000000.1 | Unpublished data |
| D. lucknowensis L15 | 3,719,665 | 3 | 62.9 | 3722 | 1,1,1 | 46 | 1 | HCH contaminated pond soil in Ummari village, Lucknow, India | NZ_FXWK00000000.1 | This study |
| D. subaequoris HST3-14 | 4,123,118 | 20 | 60.9 | 4165 | 3,1,1 | 48 | — | Sediment sample from Hwasun Beach in Jeju, Republic of Korea | IMG Genome ID 2654587640 | Unpublished data |
| Devosia sp. LC5 | 4,202,858 | 47 | 62.3 | 4217 | 2,2,2 | 48 | — | Limestone Capitan Formation at −347 m in Lechuguilla Cave, New Mexico, U.S.A. | JNNO00000000.1 | 101 |
| Devosia sp. H5989 | 4,594,249 | 1 | 64.8 | 4574 | 2,2,2 | 51 | — | Human cerebrospinal fluid | NZ_CP011300.1 | 3 |
| Devosia sp. Root436 | 3,919,001 | 16 | 63.8 | 3890 | 1,1,1 | 46 | 1 | Root of Arabidopsis thaliana cultivated in greenhouse in Germany;Cologne | LMEM00000000.1 | 102 |
| Devosia sp. Root685 | 4,397,456 | 5 | 61.5 | 4228 | 1,1,1 | 48 | — | Root of Arabidopsis thaliana cultivated in greenhouse in Germany;Cologne | LMHK00000000.1 | 102 |
| Devosia sp. A16 | 5,032,994 | 1 | 65.8 | 4992 | 2,2,2 | 57 | — | Wheat field, China; Nanjing | NZ_CP012945.1 | 10 |
| Devosia sp. 17-2-E-8 | 4,684,238 | 124 | 64 | 4584 | 2,1,1 | 49 | — | Alfalfa soil sample that was enriched with F. graminearum-infested moldy corn for 6weeks, Canada;Ontario | JQGB00000000.1 | 99 |
| Devosia sp. Root105 | 5,850,117 | 21 | 65.4 | 5737 | 1,1,1 | 51 | — | Root of Arabidopsis thaliana cultivated in greenhouse in Germany;Cologne | LMCR00000000.1 | 102 |
| Devosia sp. Root413D1 | 5,851,361 | 14 | 65.4 | 5716 | 1,1,1 | 50 | — | Root of Arabidopsis thaliana cultivated in greenhouse in Germany;Cologne | LMEA00000000.1 | 102 |
| Devosia sp. Root635 | 3,816,628 | 24 | 64.1 | 3748 | 1,1,1 | 48 | 1 | Root of Arabidopsis thaliana cultivated in greenhouse in Germany;Cologne | LMGZ00000000.1 | 102 |
| Devosia nanyangense DDB001 | 4,669,456 | 95 | 64 | 4578 | 1,1,1 | 49 | — | Mycotoxin contaminated Wheat field soil in Nanyang, China | CCAO000000000.1 | 9 |
| Devosia sp. S37 | 3,878,148 | 151 | 64.1 | 3878 | 1,1,1 | 55 | — | Oil palm rhizospheric soil, Temerloh, Pahang, Malaysia | LVVY00000000.1 | Unpublished data |
| Devosia sp. Leaf64 | 4,244,488 | 24 | 60.5 | 4206 | 1,1,1 | 48 | — | Arabidopsis thaliana leaf natural site, Switzerland; Zurich | LMLO00000000.1 | 102 |
| Devosia sp. Leaf420 | 4,219,583 | 16 | 60.7 | 4128 | 1,1,1 | 50 | — | Arabidopsis thaliana leaf natural site, Switzerland; Zurich | LMQU00000000.1 | 102 |
| Devosia sp. YR412 | 3,831,215 | 11 | 62.5 | 3755 | 2,2,2 | 51 | — | Populus root and rhizosphere microbial communities from Tennessee, USA | FOFL00000000.1 | Unpublished data |
| Devosia sp. I507 | 4,005,916 | 1 | 61.9 | 4021 | 2,2,2 | 48 | — | Pit mud, Indian ocean | NZ_CP026747.1 | Unpublished data |
Phylogenomics analyses
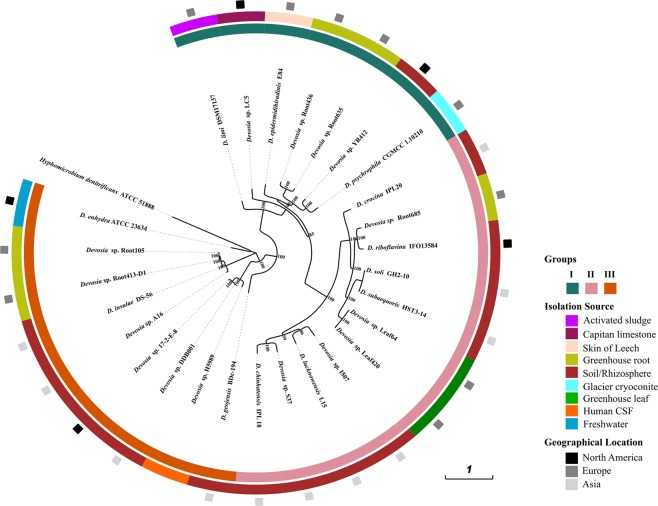
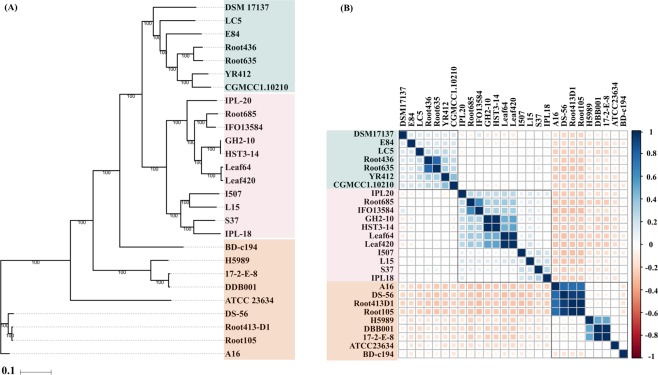
We deciphered the phylogenetic relationships of Devosia strains using marker genes, core genome and whole genome based average nucleotide identities. Maximum likelihood phylogeny based on the conserved set of 400 bacterial marker genes (Fig. 1)30 was reasonably consistent with that obtained from the concatenated alignments of 1,165 orthologous single copy core genes identified using OMCL algorithm (Fig. 2A). The phylogeny reconstructed from the whole genome wide ANIb also revealed identical topology (Fig. 2B). All the methods clearly resolved the genus into three different groups denoted as Group I, II and III with subclades (Figs. 1 and 2). Intriguingly, isolates from unrelated environments, for instance, CGMCC 1.10210 isolated from glacier cryoconite and YR412 isolated from rhizosphere clustered together while those from same habitats, such as isolates from Arabidopsis root appeared distantly in the phylogeny. This suggests that the role of environment in shaping bacterial genomes is still undefined.
Figure 1.
Phylogenomics analysis. The tree is based on the 400 conserved bacterial marker gene sequences constructed using maximum likelihood method with 1000 bootstrap replications. The innermost ring represents the three major groups of strains thus formed which are denoted as Group I, II and III. The colors in the middle ring represent the habitat of each strain and the outermost ring represents their geographic origin. The tree was constructed using iTOL (https://itol.embl.de/)84.
Figure 2.
Phylogenomics analyses. (A) Maximum likelihood tree based on the single copy core genetic content of the 27 analyzed members of the genus Devosia. Bootstrap values calculated from 100 bootstrap repetitions are denoted. (B) Correlation between the genomes on the basis of blast based average nucleotide identity (ANIb) values. The blue and pink squares denote high and low correlation values for a pair of genomes and the corresponding values of predicted Pearson correlation coefficients (-1 to 1.0) are shown in the adjacent bar.
We noticed high ANI values shared between the type strains D. soli GH2-10 and D. subaequoris HST3-14 (99.99%) with high percentage of conserved proteins (98.15%). However, as the percentage similarity shared between their submitted 16S rRNA gene sequences is less than 98.65%, it highlighted the need to redefine the boundaries for species demarcation due to low phylogenetic resolution of 16S rRNA marker gene31. Both the genomes were predicted to be 98.1% complete supporting the ANI based prediction. Similarly, we detected other pairs that might represent single species based on ANI values with the cutoff score of >95% defined for species demarcation that included the two DON degrading strains DDB001 and 17-2-E-8 (99%), Arabidopsis leaf isolates, Leaf64 and Leaf420 (96%), Arabidopsis root isolates, Root105 and Root413-D1 (98%). Moreover, the pairs also clustered together based on the comparative functional analysis while harboring the similar genetic repertoire. Further, the analysis showed that D. soli GH2-10 and D. subaequoris HST3-14 are likely the same species with a high ANIb value of 99.9% (Fig. 2B).
Pangenome analysis
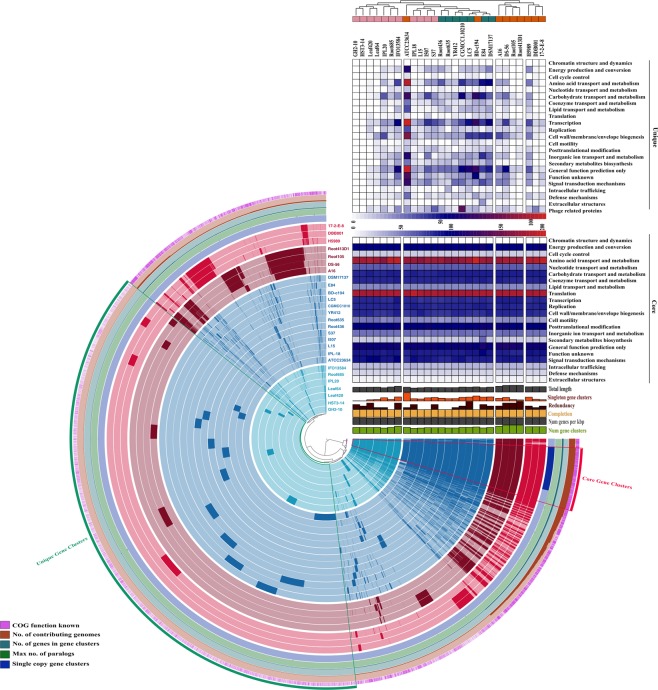
The pangenome of Devosia was analysed with aim to determine its genetic potential. The pangenome is defined as entire set of gene clusters present in a group and is constituted by the core and accessory genomes32. The core genome is formed of the conserved set of genomic functions found in all strains of the group. While accessory genome consists of the dispensable component which is present in a subset of genomes and the strain-specific content (singletons) that is unique to only one strain out of all the analysed genomes. The pangenome of Devosia was shown to be formed by 23,421 gene clusters (Distance: Euclidean; Linkage: Ward)33 that included 1,257 core, 10,383 dispensable and 11,781 strain-specific gene clusters. The small sized core (5.4%) and unique content (50.3%) forming more than half of the pangenome indicated that the species are highly diverged (Fig. 3). A robust analysis of the changes in pan- and core genome sizes upon sequential addition of genomes and their regression trends plotted as Tettelin best fit curve revealed indefinite increase in pangenome size up to the addition of the last genome. Therefore, the pangenome of Devosia may be classified as ‘open’ for expansion (Supplementary Fig. S1).
Figure 3.
Pangenome analysis. Clustering of genomes based on the presence/absence patterns of 23,421 pangenomic clusters. The genomes are organized in radial layers as core, unique and accessory gene clusters [Euclidean distance; Ward linkage] which are defined by the gene tree in the center. The clades are colored based on the shared gene clusters as shown in the tree in the right top above the heatmaps and the phylogenomic groups of the strains are denoted by the corresponding colors in the pangenome tree as in Fig. 1. Heat maps denote the functions enriched in the core- (below) and strain-specific (top) gene contents based on annotated clusters of orthologous groups (COG) categories. The core- and strain-specific gene clusters are highlighted to distinguish them from dispensable genome. The figure was constructed using Anvi’o pangenomics workflow (http://merenlab.org/software/anvio/)33.
The distribution of strains based on the pangenomic clusters was deviated from the phylogenomic clustering which was partly reflected in the differences in the accessory genomic contents of Group III strains (Fig. 3). The core and unique gene clusters were further annotated into COG classes. The core genome was mostly conserved in the following: amino acid transport and metabolism (11.12%), translation and ribosomal biogenesis (11.27%), post translational modifications (6.42%), energy production and conversion (6.36%) and signal transduction (5.65%). As largely the species are soil microbes that inhabit intoxicated environments, their genomes are thus enriched in genes for efficient uptake of the restrained nutrients, sense chemotactic stimuli and transduce the signals for colonization. The classes for carbohydrate metabolism (5.44%), transcription (5.25%), replication (5.32%) and cell envelope synthesis (5.18%) were moderately abundant while intracellular trafficking, secondary metabolite synthesis, defense mechanisms and extracellular structures were limited in the core (0.5–2%). About 70% of the average genome size of the genus was not conserved indicating a high degree of genomic diversity. Isolate ATCC23634 was found to harbor highest numbers of singletons which is expected as it is the only isolate from freshwater (Table 1). Besides, it also harbored a large CRISPR locus with 16 spacer sequences unveiling its adaptive immunity resulting from previous viral encounters.
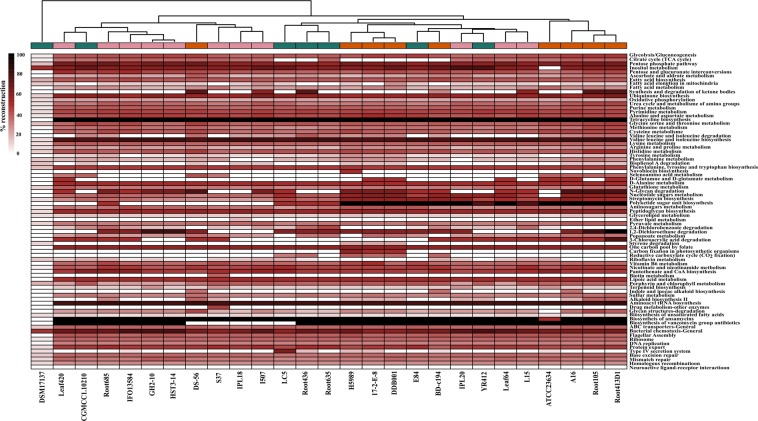
Comparative functional profiles
To gain more insights into specific functions, the top metabolic pathways of the genus were minimally reconstructed within individual genomes. Interestingly, the phylogenetically consistent groups of strains displayed different functional profiles and revealed an altogether different clustering pattern (Fig. 4). It suggests that their functional profiles might have been selected by the environment owing to evolutionary processes such as gene gain or loss and lateral gene transfer. The top metabolic pathways that were reconstructed within the genomes involved metabolism of sugars, fatty acids and amino acids, biosynthesis of antibiotics such as tetracycline, ansamycins and vancomycins, flagellar assembly and chemotaxis, ABC class transporters and degradation of chlorinated hydrocarbon compounds. These abundant functions are anticipated to provide survival benefits to the strains at the diverse niches that they inhabit. Strain DSM17137 was uncovered to be the most diverged strain within the genus with respect to its overall functional profile as the top metabolic pathways could not be reconstructed within its genome (Fig. 4). A major difference in the clades thus obtained was observed in the genes for synthesis of polyketide sugars that are important antimicrobial agents34 indicating that defence is not a primary function and hence not a part of the core genome. Concurrently, we noted that the clustering based on functional profiles was not strictly habitat-dependent. For instance, the strains isolated from plant leaves, Leaf64 and Leaf420 showed key differences in selenoamino acids utilization and polyketide sugar unit biosynthesis. This may be explained based on the fact that the process of gene gain or loss does not necessarily occur at the same rate in the isolates from similar habitats and hence the differences were observed. Similarly, the isolates from HCH contaminated soils showed different profiles for degradation of 1,2-dichloroethane and 3-chloroacrylic acid and for synthesis and degradation of ketone bodies. This suggests their dynamic genome repertoire and that the strains might be in the process of acquiring the genes for degradation of chlorinated hydrocarbons at this site.
Figure 4.
Comparative metabolic pathway analysis. The top metabolic pathways within each genome are compared based on their percentage reconstruction. A dendrogram constructed based on the metabolic profiles is shown at the top and the different phylogenetic groups are shown with corresponding colors. The heatmap was constructed using pheatmap92 in R (R Development Core Team, 2015).
Abundance of Oligo- and Di- peptide ABC transporters
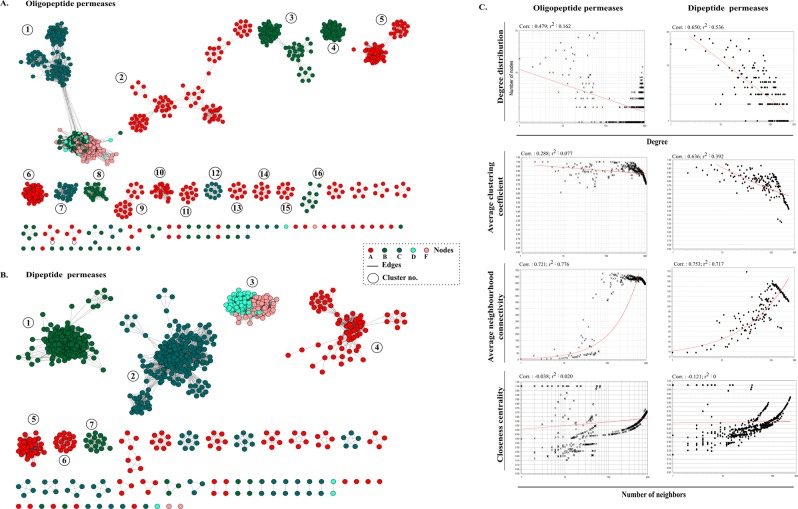
As amino acid transport and metabolism emerged as one of the most abundant functions of the genus, we studied the genes of this class for determining the important survival strategies of Devosia. More precisely, we found these genomes to be enriched in the oligo-peptide permeases (Opp) and di-peptide permeases (Dpp). Opp and Dpp permeases are present in the bacterial membranes as multi-subunit protein complexes and function primarily in the uptake of peptides from the environment to serve as a source of carbon and nitrogen. These transport systems have been widely studied in species of Lactococcus, Staphylococcus, Borrelia and Bacillus where they have been shown to be involved in growth, signalling and virulence21–26. The permease complex has a typical structure of an ABC class transporter: a substrate binding protein OppA/DppA, two transmembrane proteins OppB/DppB and OppC/DppC and two membrane bound cytoplasmic ATP-binding proteins OppD/DppD and OppF/DppF35. The copy number of each of these transporters within the analysed strains is given in Supplementary Fig. S2. Their large diversity and abundance in Devosia was further checked by comparing these permeases with those in representative genomes (n = 27) of other genera of family Hyphomicrobiaceae (Supplementary Table S1). A large diversity in the organization of genes within operons was observed and many individual genes were found segregated throughout the genomes. As the presence of each of the gene in the cluster is not a prerequisite for the operon to be functional, their abundance might be an adaptation for uptake of large variety of peptides for optimal nutrition36. The gene copy number varied from 21 to as high as 93 Opp operon associated genes with an average of 60.7 ± 19.1 copies within each genome. Also, the genomes were abundant in Dpp permeases with 17 to 54 copies of associated genes within a genome and each genome carried an average number of 36.5 ± 10.6 genes. Their genetic diversity across the genus was determined by eliminating ~6.8% of the redundant sequences in each case (sequence identity = 100%) from a total of predicted 1,640 Opp and 986 Dpp sequences indicating high diversity of these transporters. An empirical measure of the diversity among the permeases and comparison of pairwise relationships was determined through sequence similarity network (SSN) analysis. In SSN, each protein sequence is represented by a node and any two nodes are connected by edges if they share more than the defined threshold similarity. The similarity networks for all non-redundant Opp and Dpp sequences were visualized, using the threshold pairwise Blastp e-value of 1e-30 and 1e-25 respectively. Each node in the resulting networks could not be connected with all other nodes through a finite path (Fig. 5A,B). OppABCDF and DppABCDF partitioned into 65 and 55 connected components respectively that included both homologous and heterologous clusters and isolated nodes. Through network analysis, we identified 29 Opp and 19 Dpp sequences that did not share homology with any other sequence suggesting an expansive short peptidic transport system within Devosia. Average neighborhood connectivity within the networks was interpreted as an increasing function in k both in case of Opp (correlation = 0.72, r2 = 0.77) and Dpp (correlation = 0.75, r2 = 0.71) suggesting scarce edges between low connected and highly connected nodes and highlighting the diversity among the sequences (Fig. 5C). Furthermore, closeness centrality that measures the closeness of a node with all other nodes was negatively correlated with the number of neighbors in both Opp (-0.038, r2 = 0.020) and Dpp (−0.121, r2 = 0) (Fig. 5C). More specifically, we analysed the diversity of substrate binding components (SBCs): OppA and DppA within these complexes. OppA partitioned into 12/29 isolated nodes while DppA constituted 9/19 isolated nodes. The network parameters are noted in Table 2. Notably, all the isolated nodes of SBCs belonged to the species of the Group III strains that were most diverged in phylogeny (Fig. 1). Both the networks were very sparse and analysis of the networks revealed that a random Opp sequence was similar to only 20.5% of all the sequences which was even less 6.2% in case of OppA (n = 343) (Table 2). At the same time, any random Dpp sequence was similar to only 5.5% of the sequences while the similarity between any two DppA sequences (n = 192) was estimated to be 8.3%. Phylogenetic diversity of these SBCs was further compared with those predicted in the representative genomes (n = 27) from other genera of family Hyphomicrobiaceae by constructing a neighbour joining tree (Supplementary Fig. S3).
Figure 5.
Sequence similarity network analyses. Diversity of (A) Oligopeptide (Opp) and (B) Dipeptide (Dpp) permeases in analysed genomes. The nodes represent sequences connected through edges if the similarity exceeds the cutoff score. The networks are thresholded at e-value cutoff of 1e-30 and 1e-25 respectively. The ABCDF components of the permeases are represented by different colors. The clusters are ranked in order of decreasing number of nodes. Clusters with more than 10 nodes are numbered. (C) Topological properties of the similarity networks: degree distribution, average clustering coefficient, average neighborhood connectivity and closeness centrality are plotted against the number of neighbors. The power law fit curves are shown within each graph.
Table 2.
Parameters of the sequence similarity networks.
| Network Parameters | Opp | OppA | Dpp | DppA |
|---|---|---|---|---|
| No. of Nodes | 1529 | 343 | 919 | 192 |
| No. of Edges | 2,39,422 | — | 23,141 | — |
| Average degree | 313.17 | 21.24 | 50.36 | 15.94 |
| Connected components | 65 | — | 55 | — |
| Isolated nodes | 29 | 12 | 19 | 9 |
| Network Density | 0.20 | 0.06 | 0.05 | 0.08 |
| Characteristic path length | 1.92 | — | 2.02 | — |
| Shortest path | 38% | — | 18% | — |
| Network centralization | 0.298 | 0.096 | 0.19 | 0.12 |
| Clustering coefficient | 0.87 | 0.9 | 0.8 | 0.85 |
A relatively high diversity of the substrate binding proteins in Devosia unveiled the high nutritional demands and efficiency of the genus towards uptake of a wide range of structurally and chemically diverse amino acid side chains from environment. Apart from nutritional significance, the permeases are also gates to acquire natural and non-natural cargo molecules attached with amino acid side chains of peptides thereby acting as environmental sensors37,38. These signals drive the bacterial chemotaxis and form the basis of bacterial tolerance and bioremediation of environmental pollutants by bacteria39. Thus, the genus might as well have adopted this strategy for chemosensing and mediating signals to help them regulate their cellular processes for tolerating environmental stress.
Genome scale positive selection
For determining the genes under positive selection pressure, the orthologous gene clusters identified in all the genomes were filtered for eliminating clusters with low quality sequences. A total of 2000 valid clusters thus obtained were tested for presence of recombination and filtered based on FDR < 10% and dN/dS values were calculated. The dN/dS values compare the rate of substitutions at non-synonymous sites (dN) with the rate of substitutions at synonymous sites (dS) in protein orthologs. Values greater than 1 indicates positive selection while values less than one indicate that the protein is under purifying selection. The genes which were present in at least 25 genomes (1263 gene clusters) were considered to denote the positively selected genes of the genus. 24 genes were found to be under positive selection pressure with dN/dS values (ω) greater than 1 (Table 3).
Table 3.
List of genes identified to be under positive selection across the genus.
| Gene | Function | ω | p-value | q-value |
|---|---|---|---|---|
| Pyrroline-5-carboxylate | Proline synthesis and osmotic stress | 15.385168 | 0.000456 | 0.004044 |
| Alpha/beta hydrolase | Hydrolysis | 13.417266 | 0.00122 | 0.008221 |
| LamB | Lactam utilization | 12.54433 | 0.001888 | 0.012231 |
| Response regulator in two-component regulatory system with PhoQ | Response to divalent cation starvation; Resistance to antimicrobial peptides | 21.08824 | 0.000026 | 0.000738 |
| Translation initiation factor 3 | Translation | 14.490274 | 0.000714 | 0.005226 |
| Acetyl-coenzyme A carboxyl transferase alpha chain | Membrane lipid synthesis | 17.42453 | 0.000165 | 0.001848 |
| probable iron binding protein from the HesB_IscA_SufA family | Iron starvation | 20.783648 | 0.000031 | 0.000738 |
| Exopolyphosphatase (EC 3.6.1.11) | Inorganic polyphosphate utilization, adaptation to amino acid starvation | 17.073976 | 0.000196 | 0.002064 |
| NifU-like domain protein | Maturation of nitrogenase; scaffold for Fe-S cluster assembly | 11.071378 | 0.003943 | 0.02372 |
| DNA-directed RNA polymerase omega subunit (EC 2.7.7.6) | Transcription | 11.501884 | 0.00318 | 0.019835 |
| Molybdopterin biosynthesis protein MoeB | Cofactor for detoxifying enzymes | 9.045598 | 0.010859 | 0.053791 |
| Transcriptional regulator, LuxR family | Quorum sensing, motility | 19.678534 | 0.000053 | 0.000999 |
| Glutamate methylesterase CheB (EC 3.1.1.61) | Chemotaxis | 14.858994 | 0.000593 | 0.00476 |
| MutT/nudix family protein | Housekeeping enzyme | 10.824536 | 0.004462 | 0.025911 |
| hypothetical protein | — | 18.851394 | 0.000081 | 0.001132 |
| FtsZ (EC 3.4.24.-) | Cell division | 33.57748 | 0 | 0.000009 |
| SSU ribosomal protein S6p | Ribosomal protein | 17.630638 | 0.000148 | 0.001786 |
| Scaffold protein for [4Fe-4S] cluster assembly ApbC, MRP-like | Fe-S cluster assembly; Probable Iron binding protein | 24.659618 | 0.000004 | 0.000227 |
| PetP | HTH-type transcriptional regulator | 9.34578 | 0.009345 | 0.049185 |
| 3-isopropylmalate dehydratase small subunit (EC 4.2.1.33) | Biosynthesis of leucine and lysine | 9.057016 | 0.010797 | 0.053791 |
| Ribonuclease PH (EC 2.7.7.56) | tRNA processing | 18.16992 | 0.000113 | 0.001469 |
| Hypothetical protein | — | 23.900184 | 0.000006 | 0.000227 |
| Universal stress protein UspA and related nucleotide-binding proteins | Response to various stressors | 14.555118 | 0.000691 | 0.005226 |
| Sulfur acceptor protein SufE for iron-sulfur cluster assembly | Oxidative stress and iron starvation | 19.676042 | 0.000053 | 0.000999 |
The genes related to growth, osmotic stress response, inorganic polyphosphate utilization and amino acid and divalent cation starvation were under strong positive selection pressure. Apart from these, the gene responsible for cofactor molybdopterin synthesis was found to be under strong positive selection pressure. Molybdopterin acts as a cofactor for many enzymes responsible for detoxification such as sulphite oxidase, xanthine oxidase, aldehyde oxidase and formate dehydrogenase40. These molybdopterin dependent enzymes which were present in the genomes enable the optimal growth of strains by utilization of nitrate, inorganic sulfur and purines and pyrimidines as carbon and nitrogen sources. The genes involved in assembly of iron-sulfur (Fe-S) clusters were under positive selection pressure. Fe-S clusters are cofactors of proteins that perform a number of biological roles including electron transfer, redox and non-redox catalysis, and sensing for iron41. Besides, the universal stress protein (UspA) that gets activated in response to various stressors such as high temperature and salinity, antibiotics, nutrient starvation42 and LuxR family transcriptional regulator that plays a key role in quorum sensing, motility, and antibiotic synthesis43 were also positively selected. These positively selected genes signify the evolving environmental tolerance mechanisms among Devosia species.
Determination of positively evolving genes at HCH contaminated sites and differential osmotic stress response
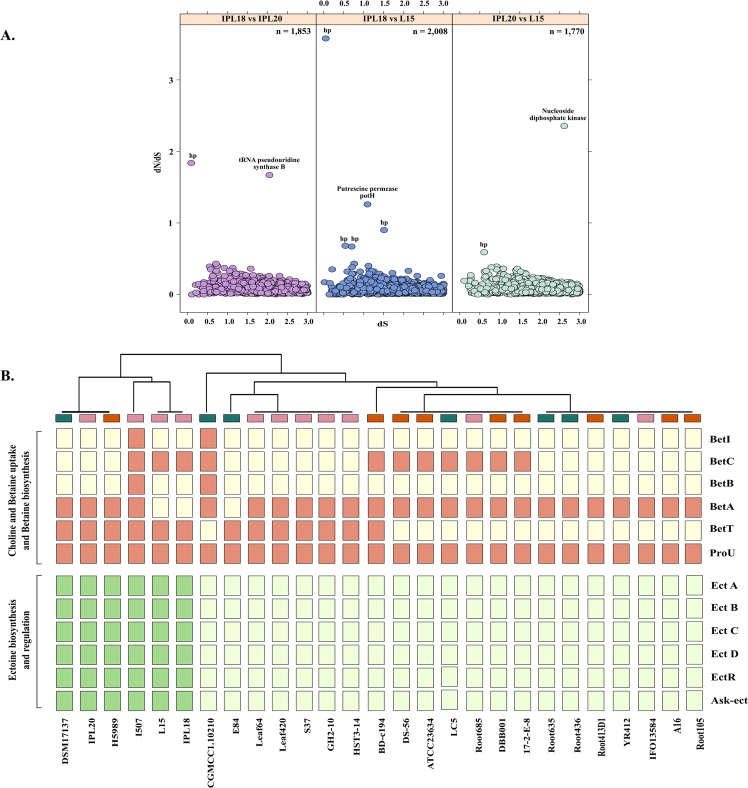
As the three strains IPL18, IPL20 and L15 isolated from HCH contaminated sites tolerate high levels of the chlorinated pollutant (450 mg/g of soil)44, we looked specifically at their genomic repertoire to uncover what enables them withstand high HCH stress. Through delineation of their orthologous proteins, we identified that their tolerance may be attributed to the abundance of two-component systems such as chemosensory phoB/phoR, cheA/cheW, cheB/cheR, cheD, cheY and methyl accepting chemotaxis protein I, might as well have been adopted to tolerate HCH stress as has been reported previously in a Pseudomonas genotypes27,45.
In order to determine the proteins encoded within their genomes that are under positive selection pressure to tolerate HCH stress, the orthologous proteins in independent pairs of three strains were subjected to positive selection detection. The majority of the proteins of all pairs were identified to be evolving under purifying selection with dN/dS values < 1 suggesting a conserved repertoire of genes is required for their survival (Fig. 6A). In IPL18 and IPL20, tRNA pseudouridine synthase subunit B was found to be under positive selection pressure (dN/dS = 1.7). Formation of pseudouridine is one of the important post-transcriptional modifications of the tRNAs. Most often these residues are confined to the functionally important part of tRNAs such that the genetic mutants lacking pseudouridine residues exhibit slow growth rates due to difficulties in translation and are not able to compete with wild type cells46. Therefore, the enzyme might confer selective advantage during competition at such a challenging niche47. In IPL18 and L15, putrescine transporter PotH was positively evolving (dN/dS = 1.25), which transports putrescine and is again involved in growth, as well as incorporation into the cell wall and biosynthesis of siderophore48. In IPL20 and L15, nucleoside diphosphate kinase showed dN/dS = 2.3. The enzyme facilitates bacterial cell growth and proliferation and mediates signal transduction49. Along with these, many hypothetical proteins were found to be under positive selection pressure (Fig. 6A). The hypothetical protein with the highest dN/dS of 3.58 belonged to GPCR family2-like protein with a query coverage of 76% using SmartBLAST (http://blast.ncbi.nlm.nih.gov/blast/smartblast/). In concordance with the previous results, all the positively selected proteins were related to growth or signalling mechanisms indicating the need to improve genetic fitness to cope high microbial competition at this nutrient depleted site.
Figure 6.
(A) Positively selected genes in genome pairs of strains isolated from hexachlorocyclohexane (HCH) contaminated sites. dN/dS values are plotted against dS values. The total number of predicted orthologs are for each pair that were subjected to the analysis are shown. The positively evolving poteins with dN/dS values > 1 are labelled. Hypothetical proteins are denoted as hp. (B) Presence absence pattern of the genes involved in the biosynthesis of osmolytes glycine betaine, ectoine and hydroxyectoine in response to osmotic sress response.
As the soils near the dumpsites are also reported to have high salinity levels44, we compared the profiles of osmotic stress response of these strains to determine any active gene transfers at this dumpsite and to gain insights on the plasticity of the genus Devosia. One of the strategies to cope osmotic stress is the uptake and synthesis of osmolytes such as glycine betaine, ectoine and hydroxyectoine50. Glycine betaine is synthesized from choline by betICBA operon where BetI is a sensory repressor and BetC converts choline-O-sulfate into choline. Choline uptake is mediated by BetT or ProU which is converted to glycine betaine by dehydrogenases BetA and BetB51.The tendency to synthesize the glycine betaine was restricted to I507 and CGMCC1.10210. However, the isolates from HCH dumpsite encoded complete clusters for synthesis of other two osmolytes ectoine and hydroxyectoine (Fig. 6B). Ectoine is synthesized from phosphorylation of aspartate to β-aspartyl phosphate by aspartokinase (Ask) which is then converted to a semialdehyde derivative. The derivative is successively converted to ectoine by ectABC gene cluster regulated by ectR52. Hydroxyectoine is produced from ectoine by a hydroxylase (EctD)53. The complete pathway for their synthesis was also determined in DSM17137, H5989 and I507 but was altogether absent in all other strains (Fig. 6B). The isolates from HCH and strain I507 appear to have acquired the potential for synthesis of ectoine and hydroxyectoine to overcome the osmotic stress posed by the high salinity in their respective niches.
Degradation of organic compounds
Utilization of phosphonates and sulphonates
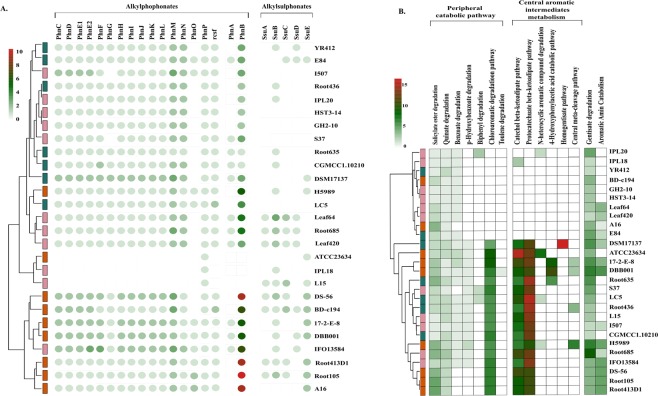
The sulphonates and phosphonates are added to environment through pesticides and are major source of sulfur and phosphorus in the soils54,55. Bacterial degradation of organic P and S play large role in global P and S cycling. As the Devosia are optimized for efficient utilization of nutrients, it evoked our interest in genus wide profiles for degradation of organic P and S.
Bacterial degradation of complex C-P bond in alkylphosphonates is catalyzed by C-P lyase encoded by a 14 gene cluster phnCDEFGHIJKLMNOP in which phnGHIJKLM code for the “core” components of the enzyme, PhnJ catalyzes the central reaction while phnNOP gene products play accessory roles56,57. phnCDE encode an ABC transporter and phnF a repressor protein. rcsF encodes a phosphoesterase analogous to phnP58. The degradation of aliphatic sulfonates is mediated by ssuEADCB gene cluster where SsuABC proteins constitute an ABC transport system while SsuD catalyzes the desulfonation of substrates and SsuE is an FMN reductase59. Our analysis revealed that the degradation of alkylphosphonates was widespread across Devosia while differential profiles for the degradation of alkylphosphonates were observed among the strains (Fig. 7A). Strains ATCC23634, IPL18 and L15 completely lacked potential to degrade alkylphosphonates (Fig. 7A). We argue that the strains IPL18 and L15 might have lost the catabolic ability in the process to tolerate the dominant pollutant i.e., HCH in their habitats. These functions are presumed to have been of environmental origin based on the clustering of genomes independent of their phylogeny. Overall, the analysis highlighted plasticity of Devosia genomes with potential for continued influx of novel functions and their evolution in response to environment.
Figure 7.
Biodegradation of organic compounds. Clustering of genomes based on the ability to degrade (A) alkylphosphonates and alkanesulphonates and (B) aromatic and xenobiotic compounds. The genomes are colored according to their original phylogenetic clustering at the tip of each branch in the tree.
Degradation of aromatic and xenobiotic compounds
The degradation of aromatic compounds by bacteria has immense environmental significance as they are the most prevalent class of natural carbon compounds which are also persistent pollutants60. So far, genera such as Pseudomonas, Acinetobacter, Geobacter, Dechloromonas and Novosphingobium have been extensively studied for their abilities of aromatic compounds degradation61–66. In this study, we examined the enzyme arsenal for remediation of aromatic compounds encoded wide the genus Devosia. The genomes were rich in the genes involved in both the branches of ß-ketoadipate utilization, one that converts catechol derived from various aromatic hydrocarbons, amino aromatics, and lignin monomers to beta-ketoadipate and another that converts protocatechuate, derived from phenolic compounds also to beta-ketoadipate for reduction through tricarboxylic acid cycle67. Among the peripheral catabolic pathways, the degradation of chloroaromatic compounds was most abundant among the strains (Fig. 7B). Again, the strains did not cluster in concordance with their phylogenetic distances. To note, strain DSM 17137 which showed maximum divergence with respect to overall functional profiles displayed maximum potential for homogentisate degradation pathway which were lacked by all other strains further confirming its functional divergence. In line with the previous observations, strain ATCC23634, the freshwater isolate was again the next most diverged among all analyzed genomes which displayed maximum potential for degradation of heterocyclic aromatic compounds (Fig. 7B). Overall, the profiles led us to consider that Devosia have acquired the potential of bioremediation during the course of evolution to adapt optimally to the environmental insults imposed on them. The conclusion was supported by the fact that the strains did not cluster based on their phylogeny but rather based on their abilities to degrade wide array of aromatic and xenobiotic compounds such as benzoate, p-hydroxybenzoate, biphenyl, catechol and chlorinated aromatic compounds.
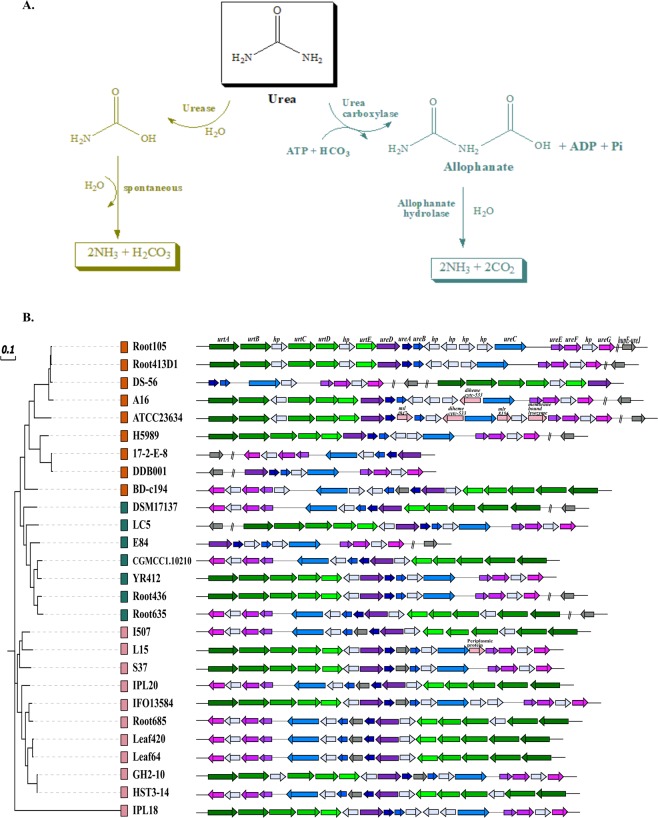
Metabolic versatility for decomposition of urea
Urea occurs as a source of organic nitrogen and its decomposition by bacteria is of immense significance for bacterial growth and nutrient cycling. Urea may be decomposed by either of the two different enzymatic pathways catalyzed by urease and urea amidolyase as illustrated in Fig. 8A 68. The second pathway catalyzed by urea amidolyase comprises activities of urea carboxylase and allophanate hydrolase69. This alternative pathway was only detected in few genomes (data not shown) and therefore, was not further inspected. Urease pathway was found to be the core pathway for urea decomposition as all the essential genes ureA, ureB, ureC encoding a functional urease and several accessory protein encoding genes ureDEFG, ureI or ureJ70 were present in all genomes (Fig. 8B). However, the genes for uptake of urea, urtABCDE were absent in DDB001, 17-2-E-8 and E84 that might have lost them or that might also harbor unique genes that still need to be characterized. Notably, the ureC gene coding for the α-subunit of urease was found to be evolving in strain DS-56 under strong positive selection pressure (dN/dS = 3.19). The ureC is the largest of the genes encoding urease functional subunits and is essential for a functional urease70,71. The strain DS-56 was isolated from the island soil near sea where urea acts as the dominant N source and thus the organism might be dependent upon its decomposition for building amino acids and hence proteins. We further tried to reconstruct the phylogeny in order to check the conservedness of the genes belonging to this pathway. The maximum-likelihood phylogeny was similar to phylogeny based upon conserved genes and marker proteins. This suggests that urea decomposition by urease is a conserved function of the genus. The conserved organization of the genes within operons also provided evidence of phylogenetic origin of this pathway.
Figure 8.
Metabolic versatility of urea decomposition. (A) The two different metabolic routes of decomposition of urea catalyzed by different enzymes namely urease and urea carboxylase. (B) A phylogram based on the genes involved in the urease pathway and their organization into operons within genomes. The phylogenetic clades are shown with the colored boxes in front of each genome name in the tree.
Determination of toxin-antitoxin (TA) systems
Bacterial toxin-antitoxin (TA) systems are key regulators of cellular processes that can respond to external stimuli and promote survival during periods of stress72. A TA locus is composed of two genes coding for a toxin and its cognate antitoxin73. Under favourable conditions, antitoxins typically inhibit their cognate toxins. While they are readily proteolysed upon stress encounters thereby unleashing the inhibitory effect of the toxin72. Widespread TA loci could be dissected within Devosia that all belonged to type II class in which both the toxin and anti-toxin are proteins73. Among the major TA systems within the genus were higB/higA and vapC/vapB but others such as parE/parD, yoeB/yefM, yafQ/dinJ and relB/relE were also present (Table 4). These small genetic modules are thought to epigenetically regulate bacterial survival controlling a wide range of biological functions including growth, persistence, programmed cell death, phage inhibition, biofilm formation and response to stress72,74. Besides, these loci are also known to stabilize the mobile genetic elements (MGEs) and enhance the genomic plasticity72. Therefore, the study could present a scenario that the environmental stress could have favored the accumulation of TA systems that confer selective advantage and competitiveness to the genus.
Table 4.
Various toxin-antitoxin (TA) systems identified within Devosia genomes.
| Genomes | Toxins and Antitoxins | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RelB/StbD | RelE/StbE | ParE | ParD | HigB | HigA | VapC | VapB | VapB1 | YoeB | YefM | YafQ | DinJ | |
| DDB001 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| 17-2-E-8 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| L15 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| GH2-10 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HST3-14 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| S37 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| DSM17137 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Root685 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| IPL20 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| BD-c194 | 0 | 0 | 1 | 1 | 3 | 2 | 4 | 5 | 1 | 1 | 1 | 0 | 0 |
| Root635 | 0 | 0 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| E84 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| A16 | 0 | 0 | 1 | 1 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Root105 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 1 |
| CGMCC 1.10210 | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| IFO13584 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| LC5 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| YR412 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| Root413-D1 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 1 |
| H5989 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Leaf420 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ATCC 23634 | 1 | 0 | 0 | 2 | 0 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 1 |
| Leaf64 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Root436 | 0 | 0 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| DS-56 | 1 | 1 | 3 | 2 | 0 | 0 | 5 | 2 | 0 | 1 | 1 | 1 | 0 |
Conclusions
In the present study, the genomes of 27 strains of the genus Devosia were analyzed which allowed the description of the open pangenome of the genus with half of the pangenome (50.32%) represented by the unique genes suggesting the role of their respective environments in shaping the genomic repertoire of the members. This was also indicated from the dissimilar phylogenetic pattern obtained based on conserved core genes and those obtained from the reconstruction of overall metabolic profiles. The phylogenetic relationships of the strains could be clearly resolved by the study. The clustering of the strains based on specific bioremediation linked functions and niche specific adaptations for example, the synthesis of osmolytes, utilization of sulphonates and phosphonates and degradation of aromatic and xenobiotic compounds revealed their plastic genomic repertoire subject to locally relevant environmental stressors. The uptake and utilization of nutrients for growth and survival was found to be the dominant function of the genus along with detoxification and degradation of organic pollutants. On this account, the genes associated with growth, motility, detoxification and nutrient starvation were found to be positively evolving. In concordance, the abundance of ABC class transporters for uptake of di- and oligo-peptides and potential of urea decomposition further revealed that the members have well adapted themselves for survival at hydrocarbons and organic compounds rich habitats by optimizing their genetic repertoire for optimal nutrient uptake and metabolism.
Materials and Methods
Genomic DNA extraction and sequencing
D. crocina IPL20 and D. lucknowensis L15 were isolated from soils contaminated with hexachlorocyclohexane (HCH) from dumpsites located at Chinhat and Ummari villages in Lucknow, India8,19. The strains were grown on Luria-Bertani (LB) agar incubated at 28 °C and genomic DNA was isolated by lysis with lysozyme and proteinase K followed by CTAB extraction using method described elsewhere75. Sequencing was performed on an Illumina HiSeq. 2500-1TB platform with Illumina regular fragment library of insert size 300 bp. A paired end library of read length 151 bp was generated for each genome. The sequencing and assembly was performed under the project ‘Genomic Encyclopedia of Type Strains, Phase III’ by the Joint Genome Institute (JGI) [Project ID: 1102317 (D. crocina IPL20) and 1102429 (D. lucknowensis L15)]. Whole genome sequences are available on NCBI under the accession numbers NZ_FPCK00000000.1 and NZ_FXWK00000000.1 respectively.
Selection and annotation of genomes
The whole genome sequences of all publicly available draft and complete genomes were retrieved from NCBI and JGI databases in March 2018 (n = 33). For all genomes, open reading frames (ORFs) were predicted using Prodigal76 and percentage completeness were estimated using 107 essential genes77 based on hidden Markov models (HMMs). Using the completeness criterion, we selected 27 strains (>96% complete) for comparative analyses (Table 1). Further, the putative protein-encoding genes were also predicted using GLIMMER-378 on RAST server v2.079. The rRNAs and tRNAs were predicted using RNAmmer v1.280 and ARAGORN81, respectively. The clustered regularly interspaced short palindromic repeat (CRISPR) elements were identified using CRISPR Finder82. Phage and prophage regions were determined using PHASTER83.
Phylogenomics analysis
The maximum likelihood phylogeny based on 400 ubiquitous and conserved marker proteins, was constructed using PhyloPhlan30 with 1000 bootstrap replications. iTOL v3 was used to visualize the tree84. In addition, phylogenetic analysis was also performed on the core genes identified in single copy within each genome. For this, amino acid alignments for each gene cluster were generated using KAlign v2.04 that employs Wu-Manber string-matching algorithm, to improve the accuracy of multiple sequence alignment85. The concatenated alignments were used to construct a maximum likelihood tree based on LG + F + R6 identified as the best fit model in IQ tree v1.686. The model generates a general amino acid replacement matrix87 using empirical amino acid frequencies and FreeRate model for calculating heterogeneity across sites. For genome-wide reconstruction of phylogeny, blast based pairwise Average Nucleotide Identity (ANIb) values computed using JSpecies web server88 were used to construct a Pearson correlation matrix and plotted in R (R Development Core Team, 2015).
Pan-gene clusters and identification of homologues
The pan-gene clusters were identified using microbial pangenomics workflow in anvi’o33 and the genomes were organized based on the distribution of gene clusters using MCL algorithm into core, dispensable and strain-specific contents (Distance: Euclidean; Linkage: Ward). The genes were annotated by BLASTp against the NCBI COG database. Heatmap based on the annotated COG functions of the core and singleton gene clusters were then plotted in R (R Development Core Team, 2015). The Tettelin best-fit curves32 of core and pangenomes were constructed using OMCL v1.4 implemented in GET_HOMOLOGUES pipeline89.
Comparative functional analyses
Functional annotation of genes was done on RAST v2.069 using the SEED subsystems approach. The ORFs were annotated by KAAS (KEGG Automatic Annotation Server)90 using Bi-directional Best Hit (BBH) algorithm. The top 50 metabolic pathways reconstructed within each genome using MinPath91 were plotted as heatmap using pheatmap package92 in R (R Development Core Team, 2015).
Sequence similarity network analysis
The di- and oligo-peptide permeases were identified within the genomes using Protein BLAST on NCBI database. The sequences were analysed by constructing similarity networks in which the relationships were read as independent pairwise alignments. The approach offers serious advantages over the phylogenetic trees in inferring relationships between large sequence data sets at defined cut-offs with ease. The sequences were filtered for the removal of 100% identical sequences using CD-HIT93. A pairwise BLAST of all non-redundant proteins was performed and sequence similarity networks (SSN) were constructed with a threshold alignment score of 50%. The threshold cutoff values of 1e-30 and 1e-25 were used for construction of opp and dpp sequence networks respectively upon analysing the trends of varying alignment length at different e-values. The networks were visualized in Cytoscape v3.6.1. The average numbers of neighbors or degree for a node or sequence was calculated as:
where K denotes the total number of edges and N denotes the total nodes. To estimate the diversity/similarity among sequences, the density of networks i.e. the fraction of all edges in the similarity networks was also calculated as:
Genome scale and pairwise positive selection detection
The orthologous gene clusters were determined using OrthoMCL v1.4. Orthologous groups with single copy genes were then filtered for determining orthologs under positive selection using POTION v1.1.394. Groups with evidence of recombination were removed from analysis using PhiPack95 that integrates three recombination tests: Phi, NSS and Max Chi2. For each group, multiple protein sequence alignments were generated using MUSCLE v 3.8.31 and trimmed using TrimAl v1.296. DNAML from phylip was used for phylogenetic tree reconstruction with 100 bootstraps. Later, groups were tested for positive selection using site-model analysis in codeml and a likelihood ratio test was conducted. The p-values were calculated as 2Δℓ (twice the difference in likelihood of the two nested models evaluated) based on the χ2 distribution with 2 ° of freedom followed by multiple hypothesis correction. Errors were minimised through False Discovery Rate (FDR) adjusted q-values (significance threshold cutoff of 10%).
To determine the evolutionary pressures at the HCH dumpsites, dN/dS values were calculated independently for the three HCH tolerating strains in a pairwise manner. The orthologous proteins were aligned using KAlign v2.04 and further converted to corresponding codon alignments using PAL2NAL script97. yn00 module in the PAML package was used to calculate dN/dS value for each orthologous pair.
Supplementary information
Acknowledgements
The sequence data were produced by the US Department of Energy Joint Genome Institute https://www.jgi.doe.gov/ in collaboration with the user community. This work was supported by funds from the Department of Biotechnology (DBT), National Bureau of Agriculturally Important Microorganisms (NBAIM) and DU-DST-PURSE grant, Government of India. C.T. and S.N. thank Council of Scientific and Industrial Research (CSIR) for providing doctoral fellowships.
Author contributions
C.T., S.N., R.L. and R.K.N. planned the study. C.T. and S.N. performed the analysis. C.T., S.N. and R.K. wrote the manuscript. R.L., R.K.N. and J.S. critically reviewed the manuscript and improved it. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chandni Talwar and Shekhar Nagar.
Supplementary information
is available for this paper at 10.1038/s41598-020-58163-8.
References
- 1.Nakagawa Y, Sakane T, Yokota A. Transfer of “Pseudomonas riboflavina” (Foster 1944), a gram-negative, motile rod with long-chain 3-hydroxy fatty acids, to Devosia riboflavina gen. nov., sp. nov., nom. rev. Int. J. Syst. Bacteriol. 1996;46:16–22. doi: 10.1099/00207713-46-1-16. [DOI] [PubMed] [Google Scholar]
- 2.Foster JW. Microbiological aspects of riboflavin. J. Bacteriol. 1944;47:27–41. doi: 10.1128/JB.47.1.27-41.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson AC, et al. Complete genome sequence of strain H5989 of a novel Devosia species. Genome Announc. 2015;3:e00934–15. doi: 10.1128/genomeA.00934-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivas R, et al. Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst. Appl. Microbiol. 2003;26:47–53. doi: 10.1078/072320203322337308. [DOI] [PubMed] [Google Scholar]
- 5.Bautista VV, Monsalud RG, Yokota A. Devosia yakushimensis sp. nov., isolated from root nodules of Pueraria lobata (Willd.) Ohwi. Int. J. Syst. Evol. Microbiol. 2010;60:627–32. doi: 10.1099/ijs.0.011254-0. [DOI] [PubMed] [Google Scholar]
- 6.Lee SD. Devosia subaequoris sp. nov., isolated from beach sediment. Int. J. Syst. Evol. Microbiol. 2007;57:2212–5. doi: 10.1099/ijs.0.65185-0. [DOI] [PubMed] [Google Scholar]
- 7.Kumar M, Verma M, Lal R. Devosia chinhatensis sp. nov., isolated from a hexachlorocyclohexane (HCH) dump site in India. Int. J. Syst. Evol. Microbiol. 2008;58:861–5. doi: 10.1099/ijs.0.65574-0. [DOI] [PubMed] [Google Scholar]
- 8.Verma M, Kumar M, Dadhwal M, Kaur J, Lal R. Devosia albogilva sp. nov. and Devosia crocina sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 2009;59:795–9. doi: 10.1099/ijs.0.005447-0. [DOI] [PubMed] [Google Scholar]
- 9.Onyango M, et al. First genome sequence of potential mycotoxin-degrading bacterium Devosia nanyangense DDB001. Genome Announc. 2014;2:e00922–14. doi: 10.1128/genomeA.00922-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin X, et al. Complete genome sequence of deoxynivalenol-degrading bacterium Devosia sp. strain A16. J. Biotechnol. 2016;218:21–22. doi: 10.1016/j.jbiotec.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Ryu SH, et al. Devosia geojensis sp. nov., isolated from diesel-contaminated soil in Korea. Int. J. Syst. Evol. Microbiol. 2008;58:633–636. doi: 10.1099/ijs.0.65481-0. [DOI] [PubMed] [Google Scholar]
- 12.Lal R, et al. Biochemistry of microbial degradation of hexachlorocyclohexane and prospects for bioremediation. Microbiol. Mol. Biol. Rev. 2010;74:58–80. doi: 10.1128/MMBR.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, et al. Parapedobacter indicus sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 2015;65:129–34. doi: 10.1099/ijs.0.069104-0. [DOI] [PubMed] [Google Scholar]
- 14.Mahato NK, Tripathi C, Nayyar N, Singh AK, Lal R. Pontibacter ummariensis sp. nov., isolated from a hexachlorocyclohexane contaminated soil. Int. J. Syst. Evol. Microbiol. 2016;66:1080–1087. doi: 10.1099/ijsem.0.000840. [DOI] [PubMed] [Google Scholar]
- 15.Rani P, Mukherjee U, Verma H, Kamra K, Lal R. Luteimonas tolerans sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 2016;66:1851–6. doi: 10.1099/ijsem.0.000956. [DOI] [PubMed] [Google Scholar]
- 16.Dwivedi V, Niharika N, Lal R. Pontibacter lucknowensis sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 2013;63:309–13. doi: 10.1099/ijsem.0.000956. [DOI] [PubMed] [Google Scholar]
- 17.Kaur J, et al. Sphingobium baderi sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 2013;63:673–8. doi: 10.1099/ijs.0.039834-0. [DOI] [PubMed] [Google Scholar]
- 18.Dadhwal M, Jit S, Kumari H, Lal R. Sphingobium chinhatense sp. nov., a hexachlorocyclohexane (HCH)-degrading bacterium isolated from an HCH dumpsite. Int. J. Syst. Evol. Microbiol. 2009;59:3140–3144. doi: 10.1099/ijs.0.005553-0. [DOI] [PubMed] [Google Scholar]
- 19.Dua A, Malhotra J, Saxena A, Khan F, Lal R. Devosia lucknowensis sp. nov., a bacterium isolated from hexachlorocyclohexane (HCH) contaminated pond soil. J. Microbiol. 2013;51:689–94. doi: 10.1007/s12275-013-2705-9. [DOI] [PubMed] [Google Scholar]
- 20.He JW, et al. Bacterial epimerization as a route for deoxynivalenol detoxification: the influence of growth and environmental conditions. Front. Microbiol. 2016;7:572. doi: 10.3389/fmicb.2016.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamarque M, et al. A multifunction ABC transporter (Opt) contributes to diversity of peptide uptake specificity within the genus. Lactococcus. J. Bacteriol. 2004;186:6492–500. doi: 10.1128/JB.186.19.6492-6500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, et al. Diversity and evolution of oligopeptide permease systems in staphylococcal species. Genomics. 2014;104:8–13. doi: 10.1016/j.ygeno.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Hiron A, Borezée-Durant E, Piard JC, Juillard V. Only one of four oligopeptide transport systems mediates nitrogen nutrition in. Staphylococcus aureus. J. Bacteriol. 2007;189:5119–5129. doi: 10.1128/JB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medrano MS, et al. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 2007;189:2653–9. doi: 10.1128/JB.01760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XG, Lin B, Kidder JM, Telford S, Hu LT. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J. Bacteriol. 2002;184:6198–6206. doi: 10.1128/jb.184.22.6198-6206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 2001;40:963–75. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- 27.Sharma A, et al. Pan-genome dynamics of Pseudomonas gene complements enriched across hexachlorocyclohexane dumpsite. BMC Genomics. 2015;16:313. doi: 10.1186/s12864-015-1488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael AJ. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018;293:18693–701. doi: 10.1074/jbc.TM118.005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogino PC, Oliva M, Sorroche FG, Giordano W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 2013;14:15838–59. doi: 10.3390/ijms140815838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segata N, Börnigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013;4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahato NK, et al. Microbial taxonomy in the era of OMICS: application of DNA sequences, computational tools and techniques. Antonie Van Leeuwenhoek. 2017;110:1357–1371. doi: 10.1007/s10482-017-0928-1. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome. Proc. Natl. Acad. Sci. USA. 2005;102:13950–55. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eren AM, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomes ES, Schuch V, de Macedo Lemos EG. Biotechnology of polyketides: new breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Braz. J. Microbiol. 2014;44:1007–34. doi: 10.1590/s1517-83822013000400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins CF. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 36.Green RM, Seth A, Connell ND. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000;68:429–436. doi: 10.1128/iai.68.2.429-436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuenzl T, et al. Mutant variants of the substrate-binding protein DppA from Escherichia coli enhance growth on nonstandard γ-glutamyl amide-containing peptides. Appl. Environ. Microbiol. 2018;84:e00340–18. doi: 10.1128/AEM.00340-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamarque M, et al. The peptide transport system Opt is involved in both nutrition and environmental sensing during growth of Lactococcus lactis in milk. Microbiology. 2011;157:1612–9. doi: 10.1099/mic.0.048173-0. [DOI] [PubMed] [Google Scholar]
- 39.Pandey G, Jain RK. Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl. Environ. Microbiol. 2002;68:5789–95. doi: 10.1128/aem.68.12.5789-5795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz G, Mendel RR. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 2006;57:623–47. doi: 10.1146/annurev.arplant.57.032905.105437. [DOI] [PubMed] [Google Scholar]
- 41.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science. 1997;277:653–9. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 42.Siegele DA. Universal stress proteins in Escherichia coli. J. Bacteriol. 2005;187:6253–54. doi: 10.1128/JB.187.18.6253-6254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Xie J. Role and regulation of bacterial LuxR-like regulators. J. Cell Biochem. 2011;112:2694–702. doi: 10.1002/jcb.23219. [DOI] [PubMed] [Google Scholar]
- 44.Sangwan N, et al. Comparative metagenomic analysis of soil microbial communities across three hexachlorocyclohexane contamination levels. PLoS ONE. 2012;7:e46219. doi: 10.1371/journal.pone.0046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zschiedrich CP, Keidel V, Szurmant H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. USA. 2003;100:12648–53. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutgsell N, et al. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA. 2000;6:1870–81. doi: 10.1017/s1355838200001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wortham BW, Patel CN, Oliveira MA. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 2007;603:106–15. doi: 10.1007/978-0-387-72124-8_9. [DOI] [PubMed] [Google Scholar]
- 49.Chakrabarty AM. Nucleoside diphosphate kinase: role in bacterial growth, virulence, cell signalling and polysaccharide synthesis. Mol. Microbiol. 1998;28:875–82. doi: 10.1046/j.1365-2958.1998.00846.x. [DOI] [PubMed] [Google Scholar]
- 50.Galinski EA, Pfeiffer HP, Trüper HG. 1,4,5,6,-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid, a novel cyclic acid from halophilic phototrophic bacteria of genus. Ectothiorodospira. Eur. J. Biochem. 1985;149:135–139. doi: 10.1111/j.1432-1033.1985.tb08903.x. [DOI] [PubMed] [Google Scholar]
- 51.Osterås M, Boncompagni E, Vincent N, Poggi MC, Le Rudulier D. Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. USA. 1998;95:11394–9. doi: 10.1073/pnas.95.19.11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters P, Galinski EA, Trüper HG. The biosynthesis of ectoine. FEMS Microbiol. Lett. 1990;71:157–162. doi: 10.1016/0378-1097(90)90049-V. [DOI] [Google Scholar]
- 53.Ingbar L, Labidot A. The structure and biosynthesis of new tetrahydropyrimidine derivatives in actinomycin D producer Streptomyces parvulus. Use of 13C- and 15N-labeled L-glutamate and 13C and 15N NMR spectroscopy. J. Biol. Chem. 1988;263:16014–22. [PubMed] [Google Scholar]
- 54.Autry AR, Fitzgerald JW. Sulfonate S: A major form of forest soil organic sulfur. Biol. Fertil. Soils. 1990;10:50–56. [Google Scholar]
- 55.McGrath JW, Chin JP, Quinn JP. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat. Rev. Microbiol. 2013;11:412–9. doi: 10.1038/nrmicro3011. [DOI] [PubMed] [Google Scholar]
- 56.Metcalf WW, Wanner BL. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene. 1993;129:27–32. doi: 10.1016/0378-1119(93)90692-v. [DOI] [PubMed] [Google Scholar]
- 57.Hove-Jensen B, Rosenkrantz TJ, Zechel DL, Willemoës M. Accumulation of intermediates of the carbon-phosphorus lyase pathway for phosphonate degradation in phn mutants of Escherichia coli. J. Bacteriol. 2010;192:370–4. doi: 10.1128/JB.01131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martínez A, Ventouras L. A, Wilson, S. T., Karl, D. M. & DeLong, E. F. Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front. Microbiol. 2013;4:340. doi: 10.3389/fmicb.2013.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Der Ploeg JR, Iwanicka-Nowicka R, Bykowski T, Hryniewicz MM, Leisinger T. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 1999;274:29358–65. doi: 10.1074/jbc.274.41.29358. [DOI] [PubMed] [Google Scholar]
- 60.Seo JS, Keum YS, Li QX. Bacterial degradation of aromatic compounds. Int. J. Environ. Res. Public Health. 2009;6:278–309. doi: 10.3390/ijerph6010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li D, et al. Genome-wide investigation and functional characterization of the β-ketoadipate pathway in the nitrogen-fixing and root-associated bacterium Pseudomonas stutzeri A1501. BMC Microbiology. 2010;10:36. doi: 10.1186/1471-2180-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbe V, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–79. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler JE, et al. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics. 2007;8:180. doi: 10.1186/1471-2164-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salinero KK, et al. Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics. 2009;10:351–10. doi: 10.1186/1471-2164-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, et al. Comparative genomics of degradative Novosphingobium strains with special reference to microcystin-degrading Novosphingobium sp. THN1. Front. Microbiol. 2018;9:2238. doi: 10.3389/fmicb.2018.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar R, et al. Comparative genomic analysis reveals habitat-specific genes and regulatory hubs within the genus Novosphingobium. mSystems. 2017;2:e00020–17. doi: 10.1128/mSystems.00020-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harwood CS, Parales RE. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996;50:553–90. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 68.Hausinger RP. Metabolic versatility of prokaryotes for urea decomposition. J. Bacteriol. 2004;186:2520–2. doi: 10.1128/jb.186.9.2520-2522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanamori T, Kanou N, Atomi H, Imanaka T. Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 2004;186:2532–9. doi: 10.1128/jb.186.9.2532-2539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mobley HLT, Hausinger RP. Microbial urease: significance, regulation, and molecular characterization. Microbiol. Rev. 1989;53:85e108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nolden L, et al. Urease of Corynebacterium glutamicum: organization of corresponding genes and investigation of activity. FEMS Microbiol. Lett. 2000;189:305–310. doi: 10.1111/j.1574-6968.2000.tb09248.x. [DOI] [PubMed] [Google Scholar]
- 72.Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 2013;340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- 73.Unterholzner SJ, Poppenberger B, Rozhon W. Toxin–antitoxin systems: Biology, identification, and application. Mob. Genet. Elements. 2013;3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen Y, Behiels E, Devreese B. Toxin–Antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 2014;70:240–249. doi: 10.1111/2049-632X.12145. [DOI] [PubMed] [Google Scholar]
- 75.Wilson K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001;2(2):4. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 76.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagesen K, et al. RNammer: consistent and rapid annotation of ribosomal rRNA genes. Nucleic Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lassmann T, Sonnhammer ELL. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 88.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ye Y, Doak TG. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 2009;5:e1000465. doi: 10.1371/journal.pcbi.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kolde, R. & Kolde, M.R. Package ‘pheatmap’. https://cran.r project.org/web/packages/pheatmap/pheatmap.pdf (2015).
- 93.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hongo JA, de Castro GM, Cintra LC, Zerlotini A, Lobo FP. POTION: an end-to-end pipeline for positive Darwinian selection detection in genome-scale data through phylogenetic comparison of protein-coding genes. BMC Genomics. 2015;16:567. doi: 10.1186/s12864-015-1765-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–81. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suyama M, Torrents M, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hassan YI, Lepp D, Zhou T. Genome assemblies of three soil-associated Devosia species: D. insulae, D. limi and D. soli. Genome Announc. 2015;3:e00514–15. doi: 10.1128/genomeA.00514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hassan YI, Lepp D, Zhou T. Draft genome sequences of Devosia sp. strain 17-2-E-8 and Devosia riboflavina strain IFO13584. Genome Announc. 2014;2:e00994–14. doi: 10.1128/genomeA.00994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hassan YI, Lepp D, Li XZ, Zhou T. Insights into the hydrocarbon tolerance of two Devosia isolates, D. chinhatensis strain IPL18T and D. geojensis strain BD-c194T, via whole-genome sequence analysis. Genome Announc. 2015;3:e00890–15. doi: 10.1128/genomeA.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gan HY, et al. Whole-genome sequences of five oligotrophic bacteria isolated from deep within Lechuguilla cave, New Mexico. Genome Announc. 2014;2:e01133–14. doi: 10.1128/genomeA.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bai Y, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.