Abstract
Colorectal cancer is a heterogeneous disease that develops via stepwise accumulation of well-characterized genetic and epigenetic alterations. We review the genetic changes associated with the development of pre-cancerous colorectal adenomas and their progression to tumors, as well as the effects of defective DNA repair, chromosome instability, microsatellite instability, and alterations in the serrated pathway and DNA methylation. We provide insights into the different molecular subgroups of colorectal tumors that develop via each of these different mechanisms and their associations with patient outcomes.
Keywords: CRC, genomic instability, DNA mismatch repair, serrated polyp
Since Fearon and Vogelstein proposed their multi-hit genetic model of colorectal carcinogenesis,1 accumulated insights have refined our understanding of the diverse genetic and epigenetic changes that underlie the initiation and progression of the adenoma-carcinoma sequence. A comprehensive analysis of mutations in 276 colorectal tumors by the Cancer Genome Atlas Network revealed that among all malignancies, colorectal tumors have one of the highest mutational burdens; dozens of somatic mutations have been identified in multiple colorectal tumors. Colorectal tumors can be broadly categorized as hypermutated (more than 12 mutations per 106 bases) or non-hypermutated (fewer than 8.24 mutations per 106 bases).2 Parallel efforts to subtype colorectal cancers based upon gene expression profiles resulted in a new classification system, comprising 4 consensus molecular subtypes (CMSs 1–4). Each CMS has unique histopathologic features and correlations with progression and clinical outcomes.3–6
Although colorectal tumors are heterogenous at a genetic level, they appear to develop via several distinct pathways. The pathways of chromosome instability (CIN), microsatellite instability (MSI), and serrated neoplasia provide some information about their mechanisms of pathogenesis, although and these pathways have some overlap. In addition, entirely new pathways continue to be recognized. Integrating data on gene expression signatures with tumor genotype has updated and refined these classifications.
We review the molecular changes associated with the transformation of normal colonic epithelium to histologically distinct precursor adenomatous lesions, and ultimately, malignant colorectal cancer (CRC). We discuss alterations in tumor suppressor genes, oncogenes, and DNA repair genes that contribute to colorectal carcinogenesis and review the correlations between these alterations and tumor progression.
Adenoma–Carcinoma Sequence
Most colorectal tumors arise from pre-cancerous polyps that are broadly categorized as either traditional tubular adenomas or serrated polyps. Adenomas develop when normal mechanisms that regulate DNA repair and cell proliferation are altered. Constant epithelial renewal is required due to the continued loss of surface cells from the intestinal mucosa; proliferation occurs only at the crypt base. As mutant cells advance towards the colonic lumen, the typically predictable process of terminal differentiation and eventual apoptosis is disrupted and discrete adenomas form. Over time, adenomatous polyps increase in size, develop increasingly dysplastic features, and can eventually acquire invasive potential.
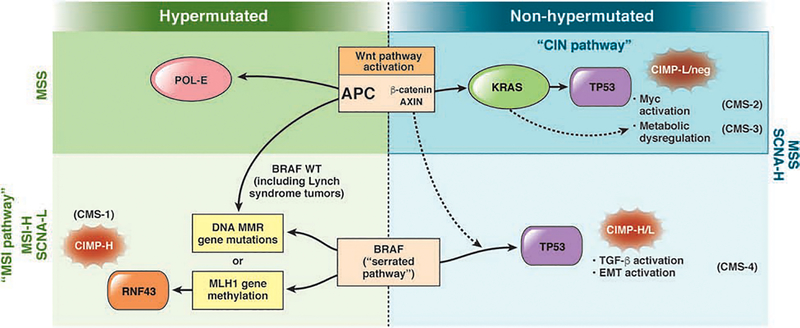
Sequential alterations in key growth regulatory genes mark the transition from normal to hyperproliferative epithelium. This stepwise progression that couples specific genetic alterations with advancing histologic features has served as a paradigm for solid tumorigenesis. Mutations in the adenomatous polyposis coli gene, (APC) which encodes a tumor suppressor,7 or the BRAF oncogene,8, 9 are initiating events that give rise to traditional adenomas or a serrated polyps, respectively. The subsequent events vary depending upon the specific pathway engaged (Fig. 1). Additional genetic alterations mark the progression from early to intermediate neoplasias, then to advanced polyps with high-grade dysplasia, and later, invasive tumors. However, not all adenomas advance to cancer—the accumulation of specific mutations in a particular order is essential for progression to malignancy. The timeline depends upon the specific pathway of tumorigenesis. For example, tumorigenesis via the CIN pathway can take 10 years or more, whereas tumor development via the comparatively accelerated MSI pathway can occur in a few years.
Figure 1. Pathways of Colorectal Carcinogenesis.
Activation of the Wnt pathway (primarily via APC mutation) or a mutation in BRAF can initiate colorectal tumorigenesis. BRAF mutations promote tumorigenesis via the serrated neoplasia pathway, leading to MSI with or without hypermutation (indicated in figure). Colorectal tumor classifications include CIN, MSI, and the serrated pathway (see CMS). L, low; H, high; EMT, epithelial to mesenchymal transition.
CIN pathway
The CIN pathway, observed in 65%–70% of sporadic colorectal tumors,10, 11 is characterized by chromosome changes that include somatic copy number alterations (SCNA) caused by aneuploidy, deletions, insertions, amplifications, or loss of heterozygosity. Tumors that develop through this pathway are considered non-hypermutated because of the relative paucity of base-pair mutations in coding sequences (Fig. 1). Mechanisms that lead to CIN are usually characterized by defects in chromosomal segregation, such as those that control sister chromatid separation; disordered cell senescence, induced by telomere shortening and culminating in genomic reorganization; dysfunctional DNA damage-response machinery; and loss of heterozygosity (LOH) at a tumor suppressor gene.
These karyotypic abnormalities are coupled with mutations in the tumor suppressor genes APC and TP53 and activating mutations in KRAS and phosphatidylinositol-4,5-bisphosphonate 3-kinase catalytic subunit alpha (PIK3CA). Mutation of APC appears to be the earliest genetic event in colorectal tumorigenesis1, 12. Loss of APC activity results in nuclear translocation of beta-catenin and activation of the Wnt signaling pathway.
In the absence of Wnt signaling, APC, axin, and glycogen synthase kinase 3 beta (GSK3B) complex with beta-catenin in the cytosol. This complex phosphorylates beta-catenin, marking it for degradation by the ubiquitin-mediated proteasomal pathway (in the presence of Wnt signaling, beta-catenin is stable). Mutant APC has decreased binding affinity for this multi-protein destruction complex, resulting in translocation of beta-catenin to the nucleus, where it heterodimerizes with the TCF and LEF family of transcription factors. This results in constitutive activation of genes that are regulated by Wnt signaling and are associated with tumorigenesis, including MYC, the cyclin D1 gene (CCND1), vascular endothelial growth factor (VEGF) genes, and the peroxisome proliferator activated receptor delta gene.13 Mutations in other genes regulated by the Wnt pathway, such as AXIN1, AXIN2, or CTNNB1, can amplify Wnt signaling in the absence of an APC mutation.14 The Wnt signaling pathway is therefore an important regulator of intestinal epithelial cell proliferation and provides an example for how the disruption of any component in specific pathway can affect transcription of multiple genes to promote tumorigenesis.15–17
The Wnt pathway is activated in nearly all CIN tumors, and APC mutations have identified in approximately 80% of these tumors.2, 3 In an effort to harmonize prior colorectal tumor functional profiles, which had only modest inter-study agreement,4, 6, 18, 19 the CRC Subtyping Consortium aggregated findings from more than 30 gene expression studies, performed with different platforms and sample preparation methods, into a single unified framework. Their efforts resulted in 4 classes of colorectal tumors based on robust, uniformly processed tumor gene expression profiles. Consensus molecular subtype 2 (CMS2) tumors are characterized by activation the Wnt and MYC signaling pathways and frequent SCNAs—features consistent with the CIN phenotype.3
MYC is a component of a heterodimeric basic helix-loop-helix transcription factor complex that regulates cell cycle progression, metabolism, and apoptosis. Mutations in MYC are not found in most colorectal tumors, but amplifications of MYC have been found in colorectal and other tumor types.20 Expression of MYC can be upregulated via activation of the Wnt signaling pathway.21 However, a meta-analysis found no clear association between tumor level of c-MYC protein and overall or disease-specific survival times.22
Autosomal dominant mutations in APC cause familial adenomatous polyposis (FAP), characterized by hundreds to thousands of adenomatous polyps.23 Biallelic germline variants in the mutY DNA glycosylase gene (MUTYH), which encodes a base excision repair protein, cause MUTYH-associated polyposis (MAP). Colon adenomas that develop in patients with MAP or FAP each have mutations in APC.24–26
Activating mutations in KRAS often arise after mutations in APC and are found in nearly 40% of colorectal tumors.27, 28 KRAS is a component of several growth factor signaling pathways, including the epidermal growth factor receptor (EGFR) pathway. In this pathway, activation of KRAS results in constitutive activation of the Raf-MEK-ERK pathway, phosphoinositide 3 kinase (PI3K) signaling via MTOR, and the transcription factor NF-kB. Proteins in the Raf family are serine/threonine kinases that activate MEK1 and MEK2, resulting in phosphorylation of ERK1 and ERK2 and then phosphorylation of enzymes that promote cell cycle progression.29 This pathway is activated in many tumors, including colorectal tumors, and might be targeted by therapeutic agents—especially metastatic colorectal tumors. Inhibitors of EGFR are used in treatment of metastatic tumors, but they are not effective in colorectal tumors with mutations in KRAS or BRAF, because these tumors maintain activity of this pathway.30–32
Analyses of gene expression patterns in colorectal tumors have identified a relationship between KRAS mutations and CMS3 (also called the metabolic phenotype). CMS3 tumors are characterized by CIN, but with fewer SCNAs than the CMS2 subtype. Gene set mRNA enrichment analysis of CMS3 tumors found evidence for dysregulation of metabolic pathways, including those that involve sugars (such as glucose and fructose), amino acids (such as glutamine), lysophospholipids, and fatty acids. These metabolic aberrations might support tumor growth3 and are consistent with reports that activation of KRAS affects glucose metabolism and hypoxia.33, 34
Many colorectal tumors with mutations in KRAS also contain mutations in genes encoding catalytic subunits of PI3K. PI3K is a heterodimeric lipid kinase that phosphorylates phosphatidylinositol, an cell signaling molecule. Increased activity of PI3K can increase prostaglandin synthesis to inhibit apoptosis in colorectal tumor cells.35 Activating mutations in PIK3CA arise late in the adenoma-carcinoma sequence and are found in 10%–20% of colorectal tumors, as well as in breast, brain, ovarian, liver, and lung tumors.36, 37 PIK3CA regulates cell proliferation and survival, inactivating proteins that promote apoptosis such as forkhead box protein O1 (FOXO1) and the Rac family of GTPases. Gain of function mutations in PIK3CA activate AKT signaling via MTOR to promote cell proliferation. Mutations in PIK3CA have been associated with tumors in female patients, with proximal location, and with MSI and mutations in KRAS.38–42 Interestingly, the ratio of mutational burden at exons 9 and 20 in tumors associate with their tissue of origin (colorectal, breast, gastric, or endometrial) and with survival times of patients with CRC.36, 43
Prostaglandin-endoperoxide synthase 2 (PTGS2, also called COX-2) is also involved in CRC development. COX-2 is an immediate to early response enzyme located on the luminal side of the endoplasmic reticulum and nuclear membrane.44 It can be activated by cytokines and other stimuli and is overexpressed in adenomas and colorectal tumors,45 although no somatic mutations in COX-2 have been found in colorectal tumors. During colorectal carcinogenesis, COX-2 might convert free arachidonic acid into prostanoids, including prostaglandins such as PGE2, which regulate proliferation of colorectal tumor cells.46 Mice with disruption of Ptgs2 have a decreased incidence of small- and large-bowel neoplasia.47, 48 COX-2 also regulates angiogenesis and promotes tumor vascularization.49, 50
COX-2 might contribute to invasive and migratory activities of colorectal cancer cells. During early stages of colorectal tumorigenesis, expression of COX-2 is increased in the stroma and connective tissue, rather than the colonic epithelium.48, 51, 52 These observations might account for the association between use of COX inhibitors such as non-steroidal anti-inflammatory drugs (including aspirin and selective COX-2 inhibitors) and reduced risk of lesions in adenoma-carcinoma sequence, including metastatic CRC.53–58 Supported by large-scale epidemiology studies, as well as data from primary prevention trials for cardiovascular diseases (CVD), the United States Preventative Services Task Force recommends low-dose aspirin therapy for primary prevention of CVD and CRC in adults 50–59 years old with an increased risk of CVD, no increased risk for bleeding complications, a life expectancy of at least 10 years, and a willingness to adhere to regular, long-term use.59
The TP53 gene, on human chromosome 17p, is the most commonly mutated gene in cancer. Its product, P53, regulates transcription of genes that regulate DNA repair and cell responses to oxidative stress. Loss of function alterations in P53 are detected more frequently in colorectal tumors than adenomas with microscopic foci of invasive cancer, and more frequently in these invasive adenomas than benign adenomas60. This association between the frequency of TP53 mutations and lesion stage indicates that mutant forms of P53 promote tumor development, at a late stage of tumorigenesis.61, 62 Approximately 60% of CIN tumors contain inactivating mutations in TP53.2 Patients with the Li-Fraumeni syndrome, characterized by germline mutations in TP53, have a modest increase in risk of CRC compared to the overall population. Analysis of data from patient registries found an association between germline alterations of TP53 and young-onset CRC (diagnosed in patients younger than 50 years).63 This observation is important because cases of of young-onset CRC have increased greatly in the United States and parts of Europe and Asia, in contrast to reported decreases in CRC incidence in other age groups. Inherited genetic factors might contribute to a subset of these cases.64–69
LOH at chromosome 18q is found in more than 70% of colorectal tumors (only of advanced stages).1 This observation indicates the presence of colorectal tumor suppressor genes at this region. Candidates include the DCC netrin 1 receptor gene (DCC) and genes that encode proteins in the transforming growth factor beta (TGFB) pathway SMAD2 and SMAD4.70–72 However, mutations in these genes are rare in human colorectal tumors and deletions of any of these genes in mice have not been associated with colorectal tumorigenesis. LOH at 18q has been associated with shorter survival times, although this result might be confounded by the association of 18q LOH with other negative prognostic factors, including high body mass index, higher tumor grade, and hypomethylation at long interspersed nucleotide element 1 elements of genomic DNA.73–75
MSI pathway
In contrast to the CIN pathway, characterized by a high frequency of genomic copy number alterations, colorectal tumors can also develop through hypermutable pathways, characterized by frequent somatic DNA base pair mutations. The MSI pathway is the primary mechanism for this hypermutable phenotype. Broadly, mutations in DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS2) or EPCAM (encodes a protein that regulates MSH2) cause instability within microsatellite regions. DNA microsatellites are repeated tandem sequences consisting of mono-, di-nucleotide, or even higher-order nucleotide repeats. These areas accumulate errors, as DNA polymerase struggles to efficiently bind these repeating genome sequences.
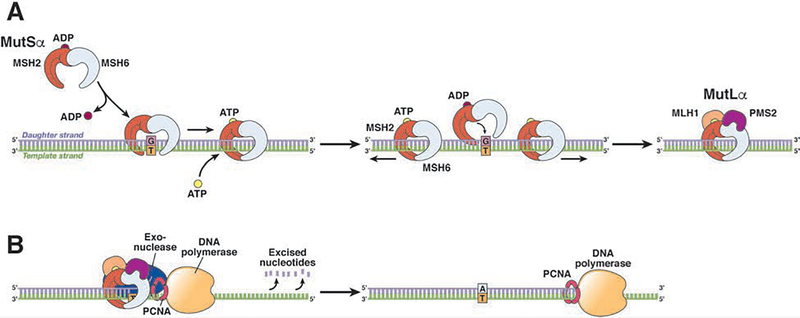
Failure to correctly assess the number of bases to insert into these invariable areas or even slippage during new strand synthesis (which generates a temporary insertion deletion loop) can produce a frameshift mutation that results in a truncated or non-functional protein. Ordinarily, in the absence of deleterious MMR gene mutations, the mismatch repair system recognizes these mistakes and performs DNA excision repair prior to daughter strand replication (see Fig. 2). Cells in tumors with the MSI phenotype do not properly detect and repair mismatched DNA, allowing them to maintain and replicate their mutations and acquire additional mutations (a hypermutation phenotype).
Figure 2. MMR of Single-pair DNA Mismatches and Insertion or Deletion Loops.
Upon detection of a DNA mismatch, MSH2 heterodimerizes with MSH6 or MSH3 to form MutSα (to correct single base-base or short insertion deletion loop mismatches) or MutSβ (principally to correct larger insertion deletion loop), respectively. These MutS complexes recognize the replication errors and form larger complexes, with MutL α, β, or γ (for example, MLH1 binds to PMS2, PMS1, or MLH3) to direct the action of exonuclease-1 and proliferating cell nuclear antigen (PCNA) and remove the misincorporated nucleotide(s). DNA polymerase and DNA ligase then reconstruct and reanneal the daughter strand to accurately reflect the parent strand.
MSI is observed in nearly 15% of sporadic colorectal tumors and nearly all colorectal tumors that develop in patients with Lynch syndrome, the most common hereditary colon cancer syndrome, caused by germline mutations in DNA MMR genes.76 However, most colorectal tumors with MSI are sporadic. The most common cause of the MSI phenotype is epigenetic silencing of the MLH1 gene, through promoter hypermethylation. Colorectal tumors of the MSI phenotype usually have high levels of methylation at regulatory regions throughout the genome, including the CpG island methylator phenotype (CIMP). MSI tumors also have an increased frequency of a mutation that encodes a V to E substitution in amino acid 600 of BRAF (in BRAFV600E), but a low frequency of APC and TP53 mutations, compared colorectal tumors that develop via the CIN pathway.8, 77, 78 In patients with Lynch syndrome, colorectal tumors that arise via a germline mutation in a DNA MMR gene also have MSI but do not usually have mutations in BRAF.79
MSI mutations include those in the TGFB receptor-2 gene (TGFBR2), which encodes a protein that prevents colon epithelial cell proliferation. TGFBR2 is mutated in more than 90% of MSI colorectal tumors.80 Mutations in TGFBR2 accumulate within a specific polyadenine tract to inactivate the receptor, so it can no longer signal to prevent proliferation. Other genes disrupted by MSI include those that encode proteins that regulate proliferation (GRB1, TCF4, WISP3, ACVR2, IGF2R, AXIN2, and CDX), cell cycle arrest or apoptosis ([CASP5, PRDM2, BCL10, PTEN, PA2G4, and FAS), and DNA repair (MBD4, BLM, CHK1, MLH3, RAD50, MSH3, and MSH6).81, 82
The events that initiate tumorigenesis via the MSI pathway vary. APC mutations are found in 35%–50% of MSI tumors,2, 3 so the initiating event of adenoma formation might be shared by MSI and CIN tumors. However, it is important to note that a distinct set of MSI tumors can develop via an initiating BRAF mutation (Fig. 1).8, 9 These tumors share traits of the MSI and serrated pathways. Once MSI occurs, colorectal tumors develop more rapidly than via the CIN pathway. Tumors with high levels of MSI (MSI-high) are associated with shorter times of progression, attributed to the hypermutable environment; CRC develops within 1–3 years as opposed to the decades required by CIN tumors.83
MSI status is determined using a standard panel of microsatellite markers defined by the NCI/Bethesda consensus guidelines. Tumors with frameshifts in 30% or more of marker genes are classified as MSI-high tumor. Tumors with frameshift mutations in fewer than 30% of marker genes are classified as MSI-low. Finally, those without instability are deemed microsatellite stable (MSS).84 PCR can reliably detect the presence of MSI, although immunohistochemical analysis of MMR proteins is the most common method. Tumor cells that lack MMR proteins, based on immunohistochemical analysis, are categorized as MSI-high. Genotypes of patients’ colorectal tumors are routinely analyzed, to identify factors that might be targeted therapeutically. MSI status can also be determined by DNA sequencing.85
Within the CMS framework, CMS1 tumors are characterized by hypermethylation of DNA, MSI-high, and infiltration by immune cells.3 Hypermethylation of promoter regions of the MLH1 gene, in particular, results in its silencing, accumulation of DNA mutations, and expression of many mutant forms of proteins. These are immunogenic, resulting in tumor infiltration by cytotoxic T cells and natural killer cells87–89, followed by expression of checkpoint molecules that inhibit T-cell activation and cytokine production, such as programmed cell death-ligand 1 (PD-L1). MSI-high tumors are therefore susceptible to treatment with PD-1 inhibitors,90, 91 resulting in a paradigm shift in treatment of colorectal cancer.
A high proportion of CMS1 tumors have the BRAF V600E mutation and CIMP-high status. However, patients with these tumors have a better prognosis than patients with other CRC subtypes. 86 These tumors have distinctive clinical and histopathologic features, in that they tend to arise in the proximal colon, contain mucin, and be poorly differentiated. There is growing consensus that adjuvant, single-agent, fluoropyrimidine-based chemotherapy is not effective, and might even be harmful, for patients with MSI-high colorectal tumors.92–96 Although these tumors have characteristics such as poor differentiation, the presence of MSI indicates a good prognosis, so determining MSI status is now routinely recommended for the staging of incident CRC.97 Among patients with localized tumors, those with MSI-high tumors had longer survival times than patients with MSS or MSI-low tumors of a similar stage.88, 96, 98–100
Serrated Neoplasia Pathway
Adenomas with intestinal type dysplasia are not the only precursor lesions to CRC. Advances in endoscopic technology have improved detection of serrated polyps—saw-toothed lesions with a varied spectrum of histopathologic correlates. Serrated polyps are believed to give rise to nearly 15% of CRCs, via the serrated neoplasia pathway.101 Serrated polyps are a heterogeneous group of lesions characterized by a stellate pattern of crypt in-folding and include benign hyperplastic polyps, pre-cancerous sessile serrated adenomas or polyps, or traditional serrated adenomas (TSAs).102 A large proportion of interval CRCs (those that develop within recommended surveillance periods, typically 3–5 years) are believed to arise via the serrated pathway.103–105
Hyperplastic polyps are the most frequently occurring subtype of serrated polyps, accounting for nearly two-thirds of all serrated lesions, and rarely undergo malignant transformation.106–108 Hyperplastic polyps have narrow crypt bases with serration confined to the upper portion and may be further subdivided by predominant mucin type: microvesicular, goblet cell rich, and mucin-poor hyperplastic polyps.
The second most common form of serrated lesions are sessile serrated adenomas and polyps, which are believed to account for one-third of all serrated lesions, although precise overall prevalence estimates have proven elusive.106, 107 Features that may make their identification challenging include a flat or sessile morphology that is easily missed by white-light imaging without chromoendoscopy and subtle distinguishing histopathologic traits that confound inter-observer reliability, even among expert gastrointestinal pathologists.109–111 Nuanced attributes such as abnormal proliferation, branching and dilated T- or L-shaped crypts, or subtle cellular dysplasia contribute to the heterogeneity of risk estimates.
TSAs account for a smaller proportion of serrated lesions. TSAs are histologically defined by protuberant growth patterns with villiform projections. These defining factors make distinctions between TSAs and tubulovillous adenomas more challenging. Cytologic dysplasia manifests as nuclear atypia with reduced differentiation and eosinophilic cytoplasm.112
The serrated pathway is a distinct mechanism of colorectal carcinogenesis. The pace of tumor progression is not fully characterized, but the subset of serrated tumors that acquire MSI are associated with a comparatively accelerated progression from precancerous lesion to carcinoma, like all MSI-high tumors. A distinguishing trait of the serrated pathway is the activating V600E mutation in BRAF, a component of the MAPK pathway.8 A large proportion of microvesicular hyperplastic polyps contain BRAF mutations, so this mutation is believed to occur early in the serrated pathway, causing constitutive activation of the MAPK—ERK pathway and uncontrolled cell division.8, 78, 113–117 BRAF is mutated in most sessile serrated adenomas, but rarely in conventional adenomas, supporting the concept that the serrated pathway is an alternative route to CRC.117
After mutation of BRAF, serrated tumors develop via 2 different routes (Fig. 1). One route converges with the MSI pathway—mutations in an MMR gene result in the MSI-high phenotype. These tumors typically develop from sessile serrated adenomas and share features with CMS1 tumors. Alternatively, tumors with mutations in BRAF can acquire TP53 mutations and activate several oncogenic pathways, including Wnt signaling, TGFB signaling, and the epithelial to mesenchymal transition; these do not result in MSI-high but rather MSS tumors (Fig. 1).3 These tumors typically develop through the traditional serrated adenoma as an intermediate lesion and have features of CMS4 (mesenchymal subtype) tumors. Unlike CMS1 tumors, CMS4 tumors have MSS with CIN, low levels of hypermutation, and high SCNA. CMS4 tumors have activation of pathways that facilitate an immunosuppressive microenvironment and permit stromal inflammation and tumor invasion, such as the angiogenic pathway These factors may contribute to the abilities of CMS4 tumors to evade the immune response, resulting in the lowest survival rates of the CMSs.118, 119 Compared with classic CIN tumors (MSS and no mutations in BRAF), patients with MSS and mutant BRAF tumors had shorter times of overall survival (hazard ratio, 2.16) and disease-specific survival (hazard ratio, 2.59). In contrast, patients with MSI and mutant BRAF tumors have longer times of overall survival (hazard ratio, 0.74) and disease-specific survival (hazard ratio, 0.74), respectively.119
Although BRAF mutations are the first detected event in the serrated pathway, it is not uncommon for the Wnt pathway to also become activated.120 In this scenario, the Wnt pathway is activated not by truncating mutations in APC, but rather by alternate routes including missense APC mutations or RNF43 mutations (Fig. 1).121, 122 RNF43 is an E3 ubiquitin ligase that can inhibit Wnt signaling through R-spondin. Up to 85% of MSI-high tumors with methylation at MLH1 have a somatic mutation in RNF43.122
Despite successes in other tumor types that carry BRAF mutations, such as melanoma, use of single agent BRAF inhibitors in BRAF-mutant colorectal cancers has been disappointing.123, 124 This has been attributed to the frequent development of resistance mutations in compensatory molecular pathways, such as upregulation of EGFR.125 Recently, a strategy that combines inhibition of BRAF with inhibitors of MEK and EGFR has demonstrated promising results in BRAF-mutant colon tumors, and this multi-drug approach is currently recommended by National Comprehensive Cancer Network (NCCN).126
Tumors that develop through either arm of the serrated neoplasia pathway also typically exhibit high levels of CpG island methylation. CpG islands are dense clusters of cytosine/guanine (CpG) dinucleotides linked by a phosphodiester bond that are particularly enriched in gene promoter regions.127 Hypermethylation of these promoter islands, particularly those upstream from tumor suppressor genes, abrogates their transcription, resulting in gene silencing and eventual tumor formation. The full spectrum of molecular mechanisms underlying the hypermethylation phenotype are not fully elucidated. However, one important mechanism depends upon the transcriptional repressor MAFG, which recruits BACH1, CHD8 and DNMT3B to bind and hypermethylate specific gene promoters including MLH1.128 Importantly, mutant BRAF upregulates levels of MAFG to enhance its binding to promoters.
The CpG island methylation phenotype (CIMP) has multiple definitions. Tumors were initially classified as CIMP-high (≥3) or CIMP-low (<2) based on the number of positive methylation markers found at locations of 5 genes (MINT1, MINT2, MINT31, CDKN2A, and hMLH1).129 Since then, multiple other panels have been proposed that include different genes (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1).130 In addition, assessment of CIMP can now be made through genome-wide methylation analyses.131 This lack of consistency has made it challenging to compare independent studies. Nevertheless, CIMP-high status is identified in approximately 20% of colorectal tumors, and this most often occurs in combination with a BRAF mutation and hypermethylation of MLH1, features that describe a large fraction of MSI-H tumors.130, 131 Although CIMP can be found in most sporadic MSI-H tumors, it is not restricted to MSI-H tumors. In fact, half of all tumors with CIMP do not have methylation of MLH1 or MSI.132, 133
CIMP can be first observed in early stages of tumorigenesis. For example, microvesicular hyperplastic polyps can be CIMP-high, and the frequency of CIMP increases compared with sessile serrated adenomas, TSAs, and other more advanced lesions.130, 134, 135 There is also an association between the CIMP in normal tissue and the occurrence of serrated polyps, indicating a field effect.136 Meta-analyses found that CIMP status correlated with mucinous or poorly differentiated tumors that contain BRAF mutations and MSI, often in the right colon, predominantly in older and female patients.137, 138 However, there does not appear to be a clear and reproducible prognostic role for the CIMP.
Other Pathways
Efforts to genotype large panels of colorectal tumors in an unbiased manner have revealed recurrent mutations in genes not previously associated with colorectal cancer pathogenesis. For example, The Cancer Genome Atlas project completed a comprehensive molecular analysis of 276 colorectal tumors, and in addition to expected mutations in APC, TP53, and KRAS, frequent mutations were identified in POLE, ARID1A, SOX9, and FAM123B.2 The identification of DNA polymerase protein mutations has specifically led to the characterization of a new molecular pathway. POLE, a catalytic subunit of DNA polymerase epsilon, and POLD1, its lagging-strand equivalent for DNA polymerase δ1, each provide proofreading capabilities to their respective enzyme complexes. Mutations in POLE generate a hypermutated phenotype with high frequency of single nucleotide variants in the absence of aneuploidy or MSI (Fig. 1).139 APC mutations appear to be the initiating event in this pathway, but the full spectrum of downstream mutations that develop in the context of this hypermutated environment is not yet described. Although these tumors do not have MSI, the hypermutated milieu nevertheless makes these tumors promising candidates for immunotherapy.140, 141
A new subgroup of colorectal tumors has been proposed; these tumors appear to have stable genomes, without significant levels of aneuploidy or hypermutation. These tumors also are initiated through APC, have modest levels of DNA hypermethylation (CIMP-low), and acquire mutations in KRAS, PIK3CA, SOX9, and PCBP1.139 The gene expression pattern of this subset of tumors most closely resembles the pattern of the CMS3 metabolic subtype, consistent with the frequent activation of KRAS. TP53 mutations were less frequently observed, which might account for the low rates of aneuploidy in these tumors. The outcomes of patients with this group of tumors have not been defined. More detailed characterizations of these subtypes are underway.
Implications for Diagnostics
Identification of molecular features of colorectal tumors could lead to new diagnostic tools. Multi-target stool-based tests have been developed to non-invasively screen for CRC and its precursors. These tests detect methylation patterns and genetic mutations in stool samples that are associated with the adenoma-carcinoma sequence.142 Heralded as the first commercialized CRC-based screening assay the Cologuard test detects mutant KRAS, aberrantly methylated promoter regions of BMP3 and NDRG4, and fecal hemoglobin through an immunological assay. Combined results from Cologuard and the fecal immunohistochemical test (FIT) identified patients with CRC with 92% sensitivity, compared to 74% sensitivity for the FIT alone, but with lower specificity (87% for the combined tests vs 95% for the FIT alone).
In 2016, the Food and Drug Administration approved a second-generation blood assay for a tumor-associated DNA marker for CRC screening. The septin 9 gene (SEPT9) encodes an eponymous GTP-binding protein involved in apoptosis, pseudopod protrusion, tumor cell migration, and invasion.143 SEPT9 is hypermethylated in CRC tissue compared with normal colon.144–146 Tests to detect methylated SEPT9 in plasma samples (Epi proColon 2.0) detect CRC with only 48% sensitivity and 92% specificity.147 Although the performance characteristics are inferior to those of the FIT, the SEPT-9 remains an option for patients unwilling to pursue other CRC screening strategies.
Quantification of circulating tumor cells or circulating cell-free DNA derived from tumor cells (circulating tumor DNA, or ctDNA) can be used to monitor for CRC recurrence and response to treatment, and is a promising area of investigation. Apart from the SEPT9 assay, the utility of so-called liquid biopsies has not been firmly established for CRC screening. Rather, the amount of CTCs or ctDNA detected in blood might be measured as a marker of disease recurrence after curative treatment.148 Serial, non-invasive surveillance of patients undergoing treatment for CRC might allow for more timely identification of resistant clonal evolution. There is evidence for the rapid emergence of cancer cells with mutations that mediate resistance to EGFR inhibitors that can be detected in circulating DNA, even before recurrence is detected by radiology.149–152
Future Directions
A wide variety of genetic and molecular changes mediate colorectal carcinogenesis. CRC is no longer considered to be a single disease entity. Advancement of care for patients with CRC will depend upon continued characterization of the different subtypes of colorectal tumors and development of therapeutic strategies based on these differences. In an effort to evaluate and summarize this progress, an expert panel represented by the American Society for Clinical Oncology, American Society for Clinical Pathology, Association for Molecular Pathology, and College of American Pathologists in 2017 offered 21 guideline statements for the use in analysis of colorectal tumors and care of patients.153
Identification of the colorectal tumor subtypes leads to important areas for future studies. For example, although the CIMP is well-described, we do not know how aberrations in DNA methylation occur or how reverse or overcome them. As we elucidate the effects of modifiable lifestyle and environmental factors on CRC risk, we will have to learn how features of the diet, obesity, and sedentary behavior contribute to colorectal tumor development. The association between Fusobacterium and other intestinal microbes with CRC risk will lead to many studies of the effects of the composition of the microbiome on intestinal epithelial cell transformation and tumorigenesis. There have been accomplishments, such as the discovery that immune checkpoint inhibitors are effective against MSI tumors, that non-steroidal anti-inflammatory and COX-2 inhibitors reduce risk of CRC, and that EGFR inhibitors are effective against tumors without mutations in KRAS. As we learn more about mechanisms of CRC pathogenesis and the different types of colorectal tumors, new therapeutic and diagnostic approaches will arise.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (Loan Repayment Program and T32CA009001 to LHN); the Crohn’s and Colitis Foundation (Research Fellowship Award to LHN). This work was also supported by CA72851, CA184792, CA202797, CA187956 and CA214254 grants from the National Cancer Institute to AG.
Footnotes
Conflicts of interest: The authors have no conflicts of interest to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 2.Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marisa L, de Reynies A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Sousa EMF, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013;19:614–8. [DOI] [PubMed] [Google Scholar]
- 6.Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 2013;19:619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparks AB, Morin PJ, Vogelstein B, et al. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res 1998;58:1130–4. [PubMed] [Google Scholar]
- 8.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 9.Rad R, Cadinanos J, Rad L, et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell 2013;24:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008;135:1079–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell SM, Zilz N, Beazer-Barclay Y, et al. APC mutations occur early during colorectal tumorigenesis. Nature 1992;359:235–7. [DOI] [PubMed] [Google Scholar]
- 13.Mann B, Gelos M, Siedow A, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci U S A 1999;96:1603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salahshor S, Woodgett JR. The links between axin and carcinogenesis. J Clin Pathol 2005;58:225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res 2005;306:357–63. [DOI] [PubMed] [Google Scholar]
- 16.Pinto D, Gregorieff A, Begthel H, et al. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 2003;17:1709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schepers A, Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol 2012;4:a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budinska E, Popovici V, Tejpar S, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol 2013;231:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlicker A, Beran G, Chresta CM, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics 2012;5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV. MYC on the path to cancer. Cell 2012;149:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science 1998;281:1509–12. [DOI] [PubMed] [Google Scholar]
- 22.He WL, Weng XT, Wang JL, et al. Association Between c-Myc and Colorectal Cancer Prognosis: A Meta-Analysis. Front Physiol 2018;9:1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996;87:159–70. [DOI] [PubMed] [Google Scholar]
- 24.Sieber OM, Lipton L, Crabtree M, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med 2003;348:791–9. [DOI] [PubMed] [Google Scholar]
- 25.Jones S, Emmerson P, Maynard J, et al. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-->T:A mutations. Hum Mol Genet 2002;11:2961–7. [DOI] [PubMed] [Google Scholar]
- 26.Venesio T, Molatore S, Cattaneo F, et al. High frequency of MYH gene mutations in a subset of patients with familial adenomatous polyposis. Gastroenterology 2004;126:1681–5. [DOI] [PubMed] [Google Scholar]
- 27.Santini D, Loupakis F, Vincenzi B, et al. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist 2008;13:1270–5. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchida N, Ryder T, Ohtsubo E. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science 1982;217:937–9. [DOI] [PubMed] [Google Scholar]
- 29.Pruitt K, Der CJ. Ras and Rho regulation of the cell cycle and oncogenesis. Cancer Lett 2001;171:1–10. [DOI] [PubMed] [Google Scholar]
- 30.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992–5. [DOI] [PubMed] [Google Scholar]
- 31.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 2009;27:5924–30. [DOI] [PubMed] [Google Scholar]
- 32.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008;26:374–9. [DOI] [PubMed] [Google Scholar]
- 33.Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149:656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald PC, Chafe SC, Brown WS, et al. Regulation of pH by Carbonic Anhydrase 9 Mediates Survival of Pancreatic Cancer Cells With Activated KRAS in Response to Hypoxia. Gastroenterology 2019. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Fu L, Sun H, et al. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology 2015;149:1884–1895 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res 2012;18:2257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [DOI] [PubMed] [Google Scholar]
- 38.Benvenuti S, Frattini M, Arena S, et al. PIK3CA cancer mutations display gender and tissue specificity patterns. Hum Mutat 2008;29:284–8. [DOI] [PubMed] [Google Scholar]
- 39.Taylor JG, DiSario JA, Buchi KN. Argon laser therapy for hemorrhagic radiation proctitis: long-term results. Gastrointest Endosc 1993;39:641–4. [DOI] [PubMed] [Google Scholar]
- 40.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia 2008;10:534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato S, Iida S, Higuchi T, et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer 2007;121:1771–8. [DOI] [PubMed] [Google Scholar]
- 42.Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 2010;16:790–9. [DOI] [PubMed] [Google Scholar]
- 43.Barbi S, Cataldo I, De Manzoni G, et al. The analysis of PIK3CA mutations in gastric carcinoma and metanalysis of literature suggest that exon-selectivity is a signature of cancer type. J Exp Clin Cancer Res 2010;29:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandrasekharan NV, Simmons DL. The cyclooxygenases. Genome Biol 2004;5:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eberhart CE, Coffey RJ, Radhika A, et al. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology 1994;107:1183–8. [DOI] [PubMed] [Google Scholar]
- 46.Greenhough A, Smartt HJ, Moore AE, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009;30:377–86. [DOI] [PubMed] [Google Scholar]
- 47.Oshima M, Murai N, Kargman S, et al. Chemoprevention of intestinal polyposis in the Apcdelta716 mouse by rofecoxib, a specific cyclooxygenase-2 inhibitor. Cancer Res 2001;61: 1733–40. [PubMed] [Google Scholar]
- 48.Oshima M, Dinchuk JE, Kargman SL, et al. Suppression of intestinal polyposis in Apc delta716 knockout mice by inhibition of cyclooxygenase 2 (COX-2). Cell 1996;87:803–9. [DOI] [PubMed] [Google Scholar]
- 49.Fukuda R, Kelly B, Semenza GL. Vascular endothelial growth factor gene expression in colon cancer cells exposed to prostaglandin E2 is mediated by hypoxia-inducible factor 1. Cancer Res 2003;63:2330–4. [PubMed] [Google Scholar]
- 50.Williams CS, Tsujii M, Reese J, et al. Host cyclooxygenase-2 modulates carcinoma growth. J Clin Invest 2000;105:1589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakagawa H, Liyanarachchi S, Davuluri RV, et al. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene 2004;23:7366–77. [DOI] [PubMed] [Google Scholar]
- 52.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature 2004;432:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dube C, Rostom A, Lewin G, et al. The use of aspirin for primary prevention of colorectal cancer: a systematic review prepared for the U.S. Preventive Services Task Force. Ann Intern Med 2007;146:365–75. [DOI] [PubMed] [Google Scholar]
- 54.Cole BF, Logan RF, Halabi S, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst 2009;101:256–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labayle D, Fischer D, Vielh P, et al. Sulindac causes regression of rectal polyps in familial adenomatous polyposis. Gastroenterology 1991;101:635–9. [DOI] [PubMed] [Google Scholar]
- 56.Giardiello FM, Hamilton SR, Krush AJ, et al. Treatment of colonic and rectal adenomas with sulindac in familial adenomatous polyposis. N Engl J Med 1993;328:1313–6. [DOI] [PubMed] [Google Scholar]
- 57.Higuchi T, Iwama T, Yoshinaga K, et al. A randomized, double-blind, placebo-controlled trial of the effects of rofecoxib, a selective cyclooxygenase-2 inhibitor, on rectal polyps in familial adenomatous polyposis patients. Clin Cancer Res 2003;9:4756–60. [PubMed] [Google Scholar]
- 58.Steinbach G, Lynch PM, Phillips RK, et al. The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N Engl J Med 2000;342:1946–52. [DOI] [PubMed] [Google Scholar]
- 59.Bibbins-Domingo K, Force USPST. Aspirin Use for the Primary Prevention of Cardiovascular Disease and Colorectal Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med 2016;164:836–45. [DOI] [PubMed] [Google Scholar]
- 60.Russo A, Bazan V, lacopetta B, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol 2005;23:7518–28. [DOI] [PubMed] [Google Scholar]
- 61.Leslie A, Carey FA, Pratt NR, et al. The colorectal adenoma-carcinoma sequence. Br J Surg 2002;89:845–60. [DOI] [PubMed] [Google Scholar]
- 62.Baker SJ, Preisinger AC, Jessup JM, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res 1990;50:7717–22. [PubMed] [Google Scholar]
- 63.Yurgelun MB, Masciari S, Joshi VA, et al. Germline TP53 Mutations in Patients With Early-Onset Colorectal Cancer in the Colon Cancer Family Registry. JAMA Oncol 2015;1:214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahnen DJ, Wade SW, Jones WF, et al. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc 2014;89:216–24. [DOI] [PubMed] [Google Scholar]
- 65.Siegel RL, Fedewa SA, Anderson WF, et al. Colorectal Cancer Incidence Patterns in the United States, 1974–2013. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siegel RL, Miller KD, Jemal A. Colorectal Cancer Mortality Rates in Adults Aged 20 to 54 Years in the United States, 1970–2014. JAMA 2017;318:572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey CE, Hu CY, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg 2015;150:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larsen IK, Bray F. Trends in colorectal cancer incidence in Norway 1962–2006: an interpretation of the temporal patterns by anatomic subsite. Int J Cancer 2010;126:721–32. [DOI] [PubMed] [Google Scholar]
- 69.Vuik FER, Nieuwenburg SAV, Bardou M, et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut 2019:gutjnl-2018–317592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehlen P, Fearon ER. Role of the dependence receptor DCC in colorectal cancer pathogenesis. J Clin Oncol 2004;22:3420–8. [DOI] [PubMed] [Google Scholar]
- 71.Takagi Y, Kohmura H, Futamura M, et al. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology 1996;111:1369–72. [DOI] [PubMed] [Google Scholar]
- 72.Takagi Y, Koumura H, Futamura M, et al. Somatic alterations of the SMAD-2 gene in human colorectal cancers. Br J Cancer 1998;78:1152–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheffer M, Bacolod MD, Zuk O, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009;106:7131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jen J, Kim H, Piantadosi S, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med 1994;331:213–21. [DOI] [PubMed] [Google Scholar]
- 75.Ogino S, Nosho K, Irahara N, et al. Prognostic significance and molecular associations of 18q loss of heterozygosity: a cohort study of microsatellite stable colorectal cancers. J Clin Oncol 2009;27:4591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–57. [PubMed] [Google Scholar]
- 77.Giannakis M, Hodis E, Jasmine Mu X, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014;46:1264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934. [DOI] [PubMed] [Google Scholar]
- 79.Bessa X, Balleste B, Andreu M, et al. A prospective, multicenter, population-based study of BRAF mutational analysis for Lynch syndrome screening. Clin Gastroenterol Hepatol 2008;6:206–14. [DOI] [PubMed] [Google Scholar]
- 80.Parsons R, Myeroff LL, Liu B, et al. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res 1995;55:5548–50. [PubMed] [Google Scholar]
- 81.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res 2002;62:2447–54. [PubMed] [Google Scholar]
- 82.Mori Y, Yin J, Rashid A, et al. Instabilotyping: comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res 2001;61:6046–9. [PubMed] [Google Scholar]
- 83.Umar A, Risinger JI, Hawk ET, et al. Testing guidelines for hereditary non-polyposis colorectal cancer. Nat Rev Cancer 2004;4:153–8. [DOI] [PubMed] [Google Scholar]
- 84.Dietmaier W, Wallinger S, Bocker T, et al. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res 1997;57:4749–56. [PubMed] [Google Scholar]
- 85.Latham A, Srinivasan P, Kernel Y, et al. Microsatellite Instability Is Associated With the Presence of Lynch Syndrome Pan-Cancer. J Clin Oncol 2019;37:286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fessler E, Drost J, van Hooff SR, et al. TGFbeta signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol Med 2016;8:745–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boland CR. The molecular biology of gastrointestinal cancer: implications for diagnosis and therapy. Gastrointest Endosc Clin N Am 2008;18:401–13, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993;260:816–9. [DOI] [PubMed] [Google Scholar]
- 89.Schwitalle Y, Kloor M, Eiermann S, et al. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology 2008;134:988–97. [DOI] [PubMed] [Google Scholar]
- 90.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003;349:247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Twelves C, Wong A, Nowacki MP, et al. Capecitabine as adjuvant treatment for stage III colon cancer. N Engl J Med 2005;352:2696–704. [DOI] [PubMed] [Google Scholar]
- 94.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609–18. [DOI] [PubMed] [Google Scholar]
- 95.Carethers JM, Smith EJ, Behling CA, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology 2004;126:394–401. [DOI] [PubMed] [Google Scholar]
- 96.Lanza G, Gafa R, Santini A, et al. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J Clin Oncol 2006;24:2359–67. [DOI] [PubMed] [Google Scholar]
- 97.Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin 2017;67:93–99. [DOI] [PubMed] [Google Scholar]
- 98.Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res 1993;53:5849–52. [PubMed] [Google Scholar]
- 99.Gryfe R, Kim H, Hsieh ET, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med 2000;342:69–77. [DOI] [PubMed] [Google Scholar]
- 100.Sinicrope FA, Mahoney MR, Smyrk TC, et al. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J Clin Oncol 2013;31:3664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bosman F, F C, Hruban RH, et al. WHO Classification of Tumours of the Digestive System, 2010.
- 103.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 2013;369:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burgess NG, Tutticci NJ, Pellise M, et al. Sessile serrated adenomas/polyps with cytologic dysplasia: a triple threat for interval cancer. Gastrointest Endosc 2014;80:307–10. [DOI] [PubMed] [Google Scholar]
- 105.Arain MA, Sawhney M, Sheikh S, et al. CIMP status of interval colon cancers: another piece to the puzzle. Am J Gastroenterol 2010;105:1189–95. [DOI] [PubMed] [Google Scholar]
- 106.Hazewinkel Y, de Wijkerslooth TR, Stoop EM, et al. Prevalence of serrated polyps and association with synchronous advanced neoplasia in screening colonoscopy. Endoscopy 2014;46:219–24. [DOI] [PubMed] [Google Scholar]
- 107.Carr NJ, Mahajan H, Tan KL, et al. Serrated and non-serrated polyps of the colorectum: their prevalence in an unselected case series and correlation of BRAF mutation analysis with the diagnosis of sessile serrated adenoma. J Clin Pathol 2009;62:516–8. [DOI] [PubMed] [Google Scholar]
- 108.Weston AP, Campbell DR. Diminutive colonic polyps: histopathology, spatial distribution, concomitant significant lesions, and treatment complications. Am J Gastroenterol 1995;90:24–8. [PubMed] [Google Scholar]
- 109.Higuchi T, Sugihara K, Jass JR. Demographic and pathological characteristics of serrated polyps of colorectum. Histopathology 2005;47:32–40. [DOI] [PubMed] [Google Scholar]
- 110.Aust DE, Baretton GB, Members of the Working Group GIPotGSoP. Serrated polyps of the colon and rectum (hyperplastic polyps, sessile serrated adenomas, traditional serrated adenomas, and mixed polyps)-proposal for diagnostic criteria. Virchows Arch 2010;457:291–7. [DOI] [PubMed] [Google Scholar]
- 111.Farris AB, Misdraji J, Srivastava A, et al. Sessile serrated adenoma: challenging discrimination from other serrated colonic polyps. Am J Surg Pathol 2008;32:30–5. [DOI] [PubMed] [Google Scholar]
- 112.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol 2011;42:1–10. [DOI] [PubMed] [Google Scholar]
- 113.O’Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 2006;30:1491–501. [DOI] [PubMed] [Google Scholar]
- 114.Spring KJ, Zhao ZZ, Karamatic R, et al. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology 2006;131:1400–7. [DOI] [PubMed] [Google Scholar]
- 115.Rosenberg DW, Yang S, Pleau DC, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res 2007;67:3551–4. [DOI] [PubMed] [Google Scholar]
- 116.Chan AO, Broaddus RR, Houlihan PS, et al. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol 2002;160:1823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kambara T, Simms LA, Whitehall VL, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004;53:1137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sinicrope FA, Shi Q, Allegra CJ, et al. Association of DNA Mismatch Repair and Mutations in BRAF and KRAS With Survival After Recurrence in Stage III Colon Cancers : A Secondary Analysis of 2 Randomized Clinical Trials. JAMA Oncol 2017;3:472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blaker H, Alwers E, Arnold A, et al. The Association Between Mutations in BRAF and Colorectal Cancer-Specific Survival Depends on Microsatellite Status and Tumor Stage. Clin Gastroenterol Hepatol 2019;17:455–462 e6. [DOI] [PubMed] [Google Scholar]
- 120.Borowsky J, Dumenil T, Bettington M, et al. The role of APC in WNT pathway activation in serrated neoplasia. Mod Pathol 2018;31:495–504. [DOI] [PubMed] [Google Scholar]
- 121.Bond CE, McKeone DM, Kalimutho M, et al. RNF43 and ZNRF3 are commonly altered in serrated pathway colorectal tumorigenesis. Oncotarget 2016;7:70589–70600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yan HHN, Lai JCW, Ho SL, et al. RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut 2017;66:1645–1656. [DOI] [PubMed] [Google Scholar]
- 123.Kopetz S, Desai J, Chan E, et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J Clin Oncol 2015;33:4032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
- 126.NCCN Clinical Practice Guidelines in Oncology: Colon Cancer. 2.2019 ed, 2019. [DOI] [PubMed]
- 127.Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986;321:209–13. [DOI] [PubMed] [Google Scholar]
- 128.Fang M, Ou J, Hutchinson L, et al. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG Island Methylator phenotype. Mol Cell 2014;55:904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan AO, Issa JP, Morris JS, et al. Concordant CpG island méthylation in hyperplastic polyposis. Am J Pathol 2002;160:529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787–93. [DOI] [PubMed] [Google Scholar]
- 131.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012;22:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005;129:837–45. [DOI] [PubMed] [Google Scholar]
- 133.Hawkins N, Norrie M, Cheong K, et al. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology 2002;122:1376–87. [DOI] [PubMed] [Google Scholar]
- 134.Fernando WC, Miranda MS, Worthley DL, et al. The CIMP Phenotype in BRAF Mutant Serrated Polyps from a Prospective Colonoscopy Patient Cohort. Gastroenterol Res Pract 2014;2014:374926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang S, Farraye FA, Mack C, et al. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol 2004;28:1452–9. [DOI] [PubMed] [Google Scholar]
- 136.Worthley DL, Whitehall VL, Buttenshaw RL, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene 2010;29:1653–62. [DOI] [PubMed] [Google Scholar]
- 137.Advani SM, Advani P, DeSantis SM, et al. Clinical, Pathological, and Molecular Characteristics of CpG Island Methylator Phenotype in Colorectal Cancer: A Systematic Review and Meta-analysis. Transl Oncol 2018;11:1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zong L, Abe M, Ji J, et al. Tracking the Correlation Between CpG Island Methylator Phenotype and Other Molecular Features and Clinicopathological Features in Human Colorectal Cancers: A Systematic Review and Meta-Analysis. Clin Transl Gastroenterol 2016;7:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu Y, Sethi NS, Hinoue T, et al. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell 2018;33:721–735 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Boland PM, Ma WW. Immunotherapy for Colorectal Cancer. Cancers (Basel) 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bourdais R, Rousseau B, Pujals A, et al. Polymerase proofreading domain mutations: New opportunities for immunotherapy in hypermutated colorectal cancer beyond MMR deficiency. Crit Rev Oncol Hematol 2017;113:242–248. [DOI] [PubMed] [Google Scholar]
- 142.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. [DOI] [PubMed] [Google Scholar]
- 143.Shankar J, Messenberg A, Chan J, et al. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res 2010;70:3780–90. [DOI] [PubMed] [Google Scholar]
- 144.Ahmed D, Danielsen SA, Aagesen TH, et al. A tissue-based comparative effectiveness analysis of biomarkers for early detection of colorectal tumors. Clin Transl Gastroenterol 2012;3:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wasserkort R, Kalmar A, Valcz G, et al. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 2013;13:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414–23. [DOI] [PubMed] [Google Scholar]
- 147.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yamada T, Matsuda A, Koizumi M, et al. Liquid Biopsy for the Management of Patients with Colorectal Cancer. Digestion 2019;99:39–45. [DOI] [PubMed] [Google Scholar]
- 149.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Diaz LA Jr., Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Misale S, Arena S, Lamba S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med 2014;6:224ra26. [DOI] [PubMed] [Google Scholar]
- 153.Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol 2017;35:1453–1486. [DOI] [PubMed] [Google Scholar]