Abstract
The engagement of programmed cell death protein 1 (PD-1; encoded by the PDCD1 gene) receptor expressed on activated T cells and its ligand programmed death-ligand 1 (PD-L1; encoded by the CD274 gene) is a major co-inhibitory checkpoint signaling that controls T-cell activities. Various types of cancers express high levels of PD-L1 and exploit the PD-L1/PD-1 signaling to evade T-cell immunity. Blocking the PD-L1/PD-1 pathway has consistently shown remarkable anti-tumor effects in patients with advanced cancers and is recognized as the gold standard for developing new immune checkpoint blockade (ICB) and combination therapies. However, the response rates of anti-PD-L1 have been limited in several solid tumors. Therefore, furthering our understanding of the regulatory mechanisms of PD-L1 can bring substantial benefits to patients with cancers by improving the efficacy of current PD-L1/PD-1 blockade or other ICBs. In this review article, we provide current knowledge of PD-L1 regulatory mechanisms at the transcriptional, posttranscriptional, post-translational, and extracellular level, and discuss the implications of these findings in cancer diagnosis and immunotherapy.
eTOC Blurb
Immune checkpoint programmed death-ligand 1 (PD-L1) plays a critical role in facilitating tumor immune evasion. Cha et al. discuss the mechanisms regulating PD-L1 expression and explore how targeting those mechanisms may lead to potential therapeutic strategies and biomarkers to improve response rates to immunotherapy.
Introduction
T-cell immunity is critical for maintaining our body’s homeostasis by selectively recognizing and eliminating pathogens and abnormal cells, including cancer cells. However, hyperactivation of uncontrolled T cells may also attack normal cells (Zhang and Bevan, 2011). To prevent such autoimmune reactions, co-inhibitory immune checkpoint proteins, such as cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed death-1 (PD-1; encoded by the PDCD1 gene), and programmed death-ligand (PD-L1; encoded by the CD274 gene), maintain an intricate regulation of T-cell activities in normal physiological conditions (Francisco et al., 2010).
In cancer cells, including renal cell carcinoma (RCC), and breast, colorectal, gastric, non-small cell lung (NSCLC), papillary thyroid, and testicular cancers (Thompson et al., 2004), high PD-L1 expression is detected and associated with poor prognosis (Ohaegbulam et al., 2015). Indeed, the binding between PD-L1 on cancer cells with PD-1 on tumor-infiltrating T cells (TILs) activates Src homology region 2 domain-containing phosphatases (SPH2), leading to suppression of the T-cell receptor (TCR) pathway and inhibition of T-cell activity. In cancer, interruption of immune surveillance promotes cancer cell survival by exploiting the PD-L1/PD-1 signaling (Schildberg et al., 2016).
In addition to cancer cells, multiple types of host cells in the tumor microenvironment (TME) and lymph nodes, including dendritic cells, macrophages, fibroblasts, and T cells, also express PD-L1 to reduce anti-tumor immunity (Curiel et al., 2003; Zou et al., 2016). Recently, Tang et al. reported that PD-L1 is upregulated by IFNγ on antigen-presenting cells (APCs) in the TME and lymph nodes to inhibit T-cell activation (Tang et al., 2018). Meanwhile, Lin et al. also reported that the efficacy of PD-1 antibody treatment alone or in combination with CTLA-4 antibody are correlated with the expression of PD-L1 on dendritic cells and macrophages in the tumor region and tumor-draining lymph nodes of patients with ovarian cancer or melanoma (Lin et al., 2018).
Based on the above findings, therapeutic antibodies against PD-L1 (e.g., atezolizumab, avelumab, and durvalumab) and PD-1 (e.g., nivolumab, pembrolizumab and cemiplimab) were developed and have demonstrated promising results in clinical trials for various types of cancer (Gong et al., 2018). Specifically, blocking the PD-L1/PD-1 signaling axis by antibody re-activates the exhausted immune cells in the TME and eliminates the cancer cells. This therapeutic strategy normalizes the imbalanced anti-tumor immunity and has achieved a 10–40 % response in the clinic (Zou et al., 2016). Currently, atezolizumab, nivolumab, and pembrolizumab are approved by the U.S. Food and Drug Administration (FDA) for the treatment of multiple cancer types, including melanoma, small cell lung cancer (SCLC), NSCLC, RCC, head and neck squamous cell carcinomas (HNSCC), classical Hodgkin lymphomas (cHL), and Merkel cell carcinoma.
Based on the promising therapeutic outcomes from anti-PD-1/PD-L1 therapy, PD-L1 has become a key protein in immuno-oncology, and its functions and regulatory mechanisms are being intensively studied. In addition, the expression of PD-L1 is intricately regulated by various processes, such as gene transcription, post-transcriptional and post-translational modifications, and exosomal transport. Therefore, it is important to broaden our understanding of the regulation of PD-L1 expression to improve the efficacy of current ICB and advance cancer immunotherapy.
1. Genomic alternations, and transcriptional and post-transcriptional mechanisms regulating PD-L1 expression
Aberrant signaling pathways and genomic mutations drive the formation of tumors. During cell transformation and tumorigenesis, upregulation of PD-L1 by these oncogenic pathways or gene mutations attenuates the activity of immune cells, allowing cancer cells to escape immunosurveillance and enhance their survival and metastatic potential. The TME provides a further enhanced niche for cancer immune escape by augmenting PD-L1 expression induced by pro-inflammatory cytokines, such as interferon γ, TNFα and IL-6 (Chan et al., 2019; Dong et al., 2002; Lim et al., 2016), and attenuating activation of immune cells, including TCR on T cells and the “don’t eat me” signaling on macrophages (Gordon et al., 2017; Zou et al., 2016). Hijacking these signals, which is induced by immune cell cytokines and enhanced expression of immune checkpoints, contributes to the adaptive resistance pathway (inducible expression) in tumor cells (Topalian et al., 2015) (Table 1). Below, we discuss the genomic alterations, and transcriptional and post-transcriptional mechanisms of PD-L1 in cancer cells and TME and their potential as biomarkers to improve and enhance the response rate of PD-L1/PD-1 therapy (Figure 1).
Table 1.
Regulatory mechanism of PD-L1 in the tumor microenvironment
| Stage of regulation | Key regulator/region | Regulatory mechanism | PD-L1 level | Potential/Proposed Therapeutic Strategies | References |
|---|---|---|---|---|---|
| Genomic alteration/rearrangements | 9p24.1 | PD-L1 amplifications and translocations in genome | Up | (Cancer Genome Atlas Research, 2014; Green et al., 2010; Ikeda et al., 2016; Roemer et al., 2016; Twa et al., 2014) | |
| Genomic alteration | DNA double-strand break (DSB) | DBS activates STAT signaling through ATM/ATR/Chk1 kinases, resulting in up-regulation of PD-L1 level | Up | Combining radiation therapy and PD-1/PD-L1 blockade | (Sato et al., 2017; Sun et al., 2018b). |
| Genomic alteration | 3’ -UTR of PD-L1 | Disruption of the 3’ - UTR of PD-L1 | Up | (Kataoka et al., 2016) | |
| Epigenetic regulation | Histone acetylation | Histone deacetylases inhibitors enhance histone acetylation and upregulate PD-L1 expression. Histone acetylation is critical for bromodomains and extraterminal (BET) proteins association to PD-L1 promoter. |
Up | Applying BET inhibitors to suppress PD-L1 and restore anti-cancer immunity | (Hogg et al., 2017; Woods et al., 2015; Zhu et al., 2016) |
| Epigenetic regulation | Methylation of H3K3me3 | Histone methyltransferase (HMTase) MLL1 catalyzes H3K4me3 and activate PD-L1 transcription in tumor cells | Up | (Lu et al., 2017) | |
| Transcriptional level | Dysregulation of oncogenic genes (intrinsic pathway) | Aberrant oncogenic pathway upregulated PD-L1 expression, including MYC, ALK, HIF1α, NF-κB pathway, MAPK pathway, PTEN/PI3K pathway, and EGFR pathway | Up | Combining kinase inhibitors and other immune checkpoint inhibitors | (Akbay et al., 2013; Barsoum et al., 2014; Casey et al., 2016; Jiang et al., 2013; Kataoka et al., 2016; Marzec et al., 2008; Xu et al., 2014) |
| Transcriptional level | Upregulated cytokines in inflammation condition (adaptive pathway) | Inflammatory cytokines in tumor region. Ex, IFN-α, β\ and γ, TNF-α, TLR3, TGF-β, IL-4/6/17/27. | Up | (Carbotti et al., 2015; Dong et al., 2002; Garcia-Diaz et al., 2017; M et al., 2016; Ni et al., 2012; Pulko et al., 2009; Quandt et al., 2014; Wang et al., 2017b; Zhang et al., 2016) | |
| Transcriptional level | Downregulated microRNA | Upregulating PD-L1 level via reducing microRNA expression in tumor cells. Ex, miR-200, miR-34a, miR-152, and miR-424 et al. | Up | (Chen et al., 2014; Cortez et al, 2016; Xie et al., 2017; Xu et al., 2016) | |
| Post-transcriptional level | Enhancing translational process | Upregulating Akt/mTOR/S6K1/eIF4E pathway | Up | Targeting Akt/mTOR pathway to reduce PD-L1 expression | (Parsa et al., 2007) |
| Post-transcriptional level | Enhancing RNA stability | RAS signaling can upregulate PD-L1 mRNA stability via modulation of the AU-rich element-binding protein tristetraprolin (TTP). | Up | (Coelho et al., 2017) | |
| Post-translational modification (Phosphorylation) | GSK3β | Phosphorylation of PD-L1 at T180 and SI84 recruits β-TrCP for degradation. PARP or c-Met inhibitors enhance PD-L1 expression by blocking GSK3β | Down | Blocking PRAP or c-Met to increase the therapeutic effect of anti-PD-1/PD-L1 antibody | (Jiao et al., 2017; Li et al., 2016; Li et al., 2019) |
| Post-translational modification (Phosphorylation) | AMPK | Abnormal ER mannose trimming by phosphorylating S195 induces PD-L1 degradation through ERAD pathway | Down | Combining metformin and anti-CTLA-4 | (Cha et al., 2018) |
| Post-translational modification (Phosphorylation) | JAK1 | Y112 tyrosine phosphorylation enhances STT3A mediated PD-L1 glycosylation | Up | Blocking IL-6/JAK1 pathway by IL-6 antibody and combining with other immune checkpoint inhibitors | (Chan et al., 2019) |
| Post-translational modification (Ubiquitination) | CSN5 | Deubiquitination of PD-L1 by proinflammatory cytokine enhance the stability of PD-L1 | Up | Combining curcumin and anti-CTLA-4 | (Lim et al., 2016) |
| Post-translational modification (Ubiquitination, lysosome-mediated degradation) | CMTM4/6 | CMTM4/6 block polyubiquitination of PD-L1 by STUB1 and co-localize with PD-L1 in recycling endosomes for preventing lysosome-mediated degradation | Up | (Burr et al., 2017; Mezzadra et al., 2017) | |
| Post-translational modification (Ubiquitination) | CDK4/6 | Destabilizing PD-L1 through proteasomal degradation mediated by Cullin 3-SPOP | Down | Blocking CDK4/6 by palbociclib or ribociclib to increase the therapeutic effect of anti-PD-1/PD-L1 antibody | (Zhang et al., 2018a) |
| Post-translational modification (Glycosylation) | B3GNT3 | LacNAc glycosylation on N192 and N200 is critical for interaction with PD-1 | Up | Inhibiting EGFR pathway to reduce B3GNT3 expression and PD-L1 glycosylation | (Li et al., 2018) |
| Post-translational modification (Glycosylation) | STT3A/B | Core glycan glycosylation of PD-L1 induced by TGF-β induce breast cancer EMT | Up | Inhibiting EGFR pathway to reduce B3GNT3 expression and PD-L1 glycosylation | (Hsu et al., 2018) |
| Post-translational modification (Glycosylation) | Sigma1 and FKBP51 | Sigma1 and FKBP51s cochaperone facilitate glycosylated PD-L1 folding and stability in the ER lumen | Up | Combining IPAG and SAFit inhibitor to reduce PD-L1 expression | (D’Arrigo et al., 2017; Maher et al., 2018) |
| Post-translational modification (Palmitoylation) | DHHC3/9 | C272 palmitoylation stabilizes PD-L1 for immunosuppression | Up | Inhibiting DHHC3/9 to reduce the stability of PD-L1 | (Yang et al., 2019; Yao et al., 2019) |
| Extracellular level (Exosome) | Exosomal machinery | Exosomal PD-L1 systemically inhibits the activity of T cells. | Up | (Chen et al., 2018; Poggio et al., 2019; Theodoraki et al., 2018; Yang et al., 2018) | |
| Extracellular level (Secretion) | RNA splicing | Alternative RNA splicing of transmembrane domain produces secreted PD-L1 variants. | Up | (Gong et al., 2019; Okuma et al., 2017; Okuma et al., 2018; Zhou et al., 2017) |
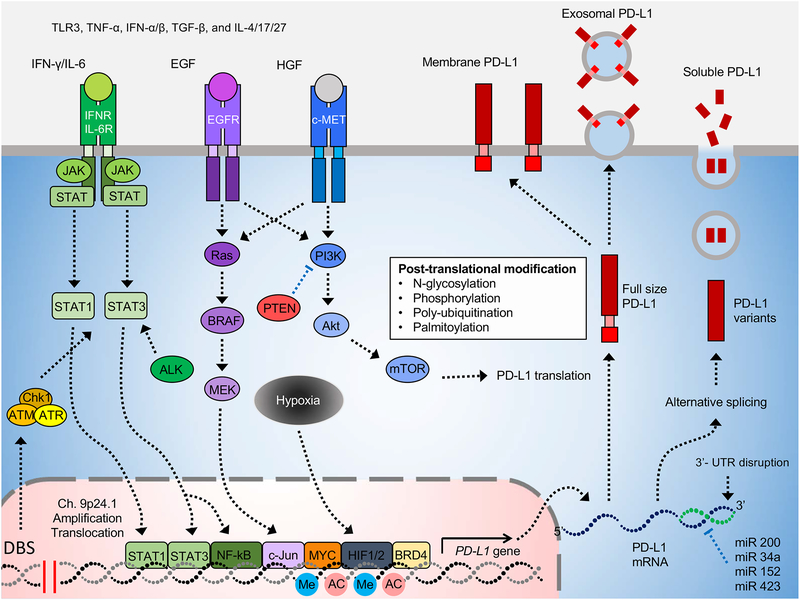
Figure 1. The regulatory mechanism of PD-L1 expression.
The precise and complex regulation of PD-L1 expression includes genomic alteration, transcriptional regulation, post-transcriptional and post-translational modifications, and exosomal transport. Me: methylation, AC: acetylation
1–1. Genomic alternation/rearrangements and epigenetic regulation of CD274 (PD-L1)
CD274 is located on chromosome 9p24.1, and genomic rearrangements in this region, including amplification and translocation, have been shown to upregulate its expression, leading to enhanced immune escape as reported for cHL, primary mediastinal large B-cell lymphoma (PMBCL), NSCLC, squamous cell carcinoma, and gastric adenocarcinoma (Cancer Genome Atlas Research, 2014; Green et al., 2010; Ikeda et al., 2016; Roemer et al., 2016; Twa et al., 2014). In addition, the rates of alterations in the CD274 or CD274 and PDCD1LG2 (encoding PD-L2) loci are significantly higher in cHL (29% in CD274 locus) and PMBCL (97% in CD274 and PDCD1LG2 loci) (Roemer et al., 2016; Twa et al., 2014). Interestingly, JAK2, which encodes Janus kinase 2, an upstream kinase that regulates PD-L1 expression, is also located on chromosome 9p with high alternation rates. It has been reported the amplification and mutation of the JAK family contribute to PD-L1 upregulation by increasing PD-L1 RNA expression The increased activities of the JAK2-signal transducers and activators of transcription (STAT) signaling pathway caused by this genomic alternation also increase PD-L1 expression (Green et al., 2010; Prestipino et al., 2018). Consistently, DNA double-strand breaks (DSB) activate STAT signaling through ataxia-telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR)/checkpoint kinase 1 (Chk1) kinases, resulting in upregulation of PD-L1 expression (Sato et al., 2017; Sun et al., 2018b). Moreover, structural variations in the 3′-untranslated region (UTR) of CD274 also increases its protein expression and enhances the cancer-immune evasion in multiple human cancers. The disruption of CD274 3′-UTR by CRISPR/Cas9 induces PD-L1 overexpression, which leads to immune evasion in the TME (Kataoka et al., 2016). Together, these findings suggested that genomic alterations play a critical role in the increased levels of PD-L1 in various cancer types and such alterations may be used as potential biomarkers to improve current anti-PD-1/PD-L1 therapy. (Sun et al., 2018a; Zhang et al., 2018b).
Epigenetic regulation, such as histone acetylation and methylation, are also involved in PD-L1 expression. Mechanistically, histone acetylation recruits bromodomains and extraterminal (BET) proteins, e.g., BRD4, to associate with the CD274 loci, which transcriptionally enhances PD-L1 mRNA production. Blocking the association of BET proteins on the CD274 locus by inhibitors reduces PD-L1 expression and increases immunosurveillance in the TME (Hogg et al., 2017; Zhu et al., 2016). Interestingly, another study also found that inhibiting histone deacetylase (HDAC) maintains histone acetylation of the CD274 locus and upregulates PD-L1 expression in tumor cells (Deng et al., 2019; Woods et al., 2015). These findings suggested that manipulating histone acetylation may be alternative strategies for future immunotherapy. Similarly, Lu and colleagues demonstrated that tri-methylation of histone H3 on lysine 4 (H3K4me3) also boosts PD-L1 expression in cancer cells. For example, mixed-lineage leukemia 1 (MML1) can bind directly to the CD274 promoter to catalyze H3K4me3 to upregulate its protein expression, and that targeting MML1 by inhibitors enhanced the efficacy of anti-PD-1/PD-L1 therapy (Lu et al., 2017). The expression of enhancer of zeste homolog 2 (EZH2), which catalyzes H3K4me3, is positively correlated with that of PD-L1 in lung adenocarcinoma (Toyokawa et al., 2019). Remarkably, the inhibitor of PARP1, a negative regulator of EZH2 upregulates PD-L1 level (Jiao et al., 2017; Yamaguchi et al., 2018), implying that EZH2 may be another epigenetic regulator of PD-L1. These studies provided evidence that PD-L1 expression is regulated epigenetically by various mechanisms.
1–2. Transcriptional upregulation of PD-L1 by aberrant oncogenic and inflammatory signaling pathways
The expression of immune checkpoint molecules is also regulated by intrinsic oncogenic and adaptive signaling pathways to facilitate cancer immunosurveillance escape in the TME. Several aberrant oncogenic pathways have been shown to contribute to the multiple hallmarks of tumorigenesis, including initiation of the intrinsic immune resistance to avoid detection and elimination by the immune system (Hanahan and Weinberg, 2011; Topalian et al., 2015). For instance, aberrant oncogenic pathways, which transcriptionally upregulate PD-L1 expression, directly reduce the anti-tumor immunity in the TME.
Elucidating the roles of oncogenic pathways driving PD-L1 expression not only identifies the functional mechanisms but also offers a rationale for future combination therapy consisting immune checkpoint blockade antibodies and inhibitors targeting those oncogenic signaling pathways (Sanmamed and Chen, 2018; Sun et al., 2018a). Specifically, overexpression of MYC, an oncogenic transcription factor, is observed in about 70% of tumorigenesis (Dang, 2012). Recent studies found that MYC binds to the PD-L1 promoter and enhances its expression in different cancer types. Moreover, genetic or pharmacological inactivation of MYC attenuates PD-L1 mRNA levels and re-establishes the anti-tumor immunity in the TME (Casey et al., 2016). Another driver of PD-L1 upregulation is anaplastic lymphoma kinase (ALK) in which hyperactivated ALK signaling pathway caused by the NPM-ALK gene fusion promotes PD-L1 expression via STAT3 (Marzec et al., 2008). Besides MYC and ALK, the HIF1/2α, NF-κB, MAPK, PTEN/PI3K and EGFR oncogenic pathways can also boost PD-L1 mRNA expression when they are mutated or hyperactivated (Akbay et al., 2013; Atefi et al., 2014; Barsoum et al., 2014; Jiang et al., 2013; Peng et al., 2015; Xu et al., 2014). Of note, numerous inhibitors that target these pathways have been approved by the FDA or are currently under investigation in the clinical trials. These findings suggested the feasibility of inhibiting these oncogenes in combination with anti-PD-1/PD-L1 therapy to achieve better therapeutic outcomes.
In the TME, cancer cells are threatened with immunosurveillance by both innate and adaptive immunity. Abundant inflammatory cytokines exist in this region and orchestrate the balance of anti-tumor immunity. However, cancer cells also hijack the inflammatory pathways (adaptive signaling pathway) to enhance PD-L1 expression and create favorable conditions for tumor progression by suppressing anti-tumor immunity (Chen and Han, 2015; Topalian et al., 2015). For instance, to avoid T-cell attack, cancer cells employ the IFN-γ/JAK/STAT1 pathway to increase PD-L1 mRNA expression (Dong et al., 2002; Garcia-Diaz et al., 2017). IFN-γ is a pro-inflammatory cytokine produced by T cells and NK cells, and enhances the major histocompatibility complex (MHC) expression to promote neoantigen presentation in tumor cells. By harnessing the IFN-γ/JAK/STAT1 pathway, PD-L1 expressed on cancer cells inactivate cytotoxic T cells and attenuate immunosurveillance in the TME. Similarly, several inflammatory cytokines also induce PD-L1 mRNA expression in tumor cells or tumor-associated stromal cells, e.g., TLR3, TNF-α, IFN-α/β, TGF-β, and IL-4/6/17/27 (Carbotti et al., 2015; Garcia-Diaz et al., 2017; M et al., 2016; Ni et al., 2012; Pulko et al., 2009; Quandt et al., 2014; Wang et al., 2017b; Zhang et al., 2016). Of note, detecting the expression of inflammatory cytokines in the plasma/serum samples has been reported to predict therapeutic outcome (Shao et al., 2017). Additional studies would be required to validate the correlation between inflammatory cytokines in the plasma/serum and PD-L1 expression in the TME as a non-invasion approach to predict the response of PD-L1/PD-1 blockade therapy.
1–3. Post-transcriptional and protein translational regulation of PD-L1
MicroRNAs (miRNAs) are 20–22 nucleotide RNAs that regulate genes expression by targeting the 3′-UTR and coding sequences to promote cleavage of mRNA transcripts. Current studies have demonstrated that dysregulated expression of miRNAs accelerates tumor metastasis and increases immune evasion in the TME (Zhang et al., 2014). The loss of certain miRNAs that reduce PD-L1 expression in tumor cells is one of the major mechanisms underlying cancer immune escape (Wang et al., 2017a). For instance, miR-200, a suppressor of epithelial-mesenchymal transition (EMT) and tumor metastasis, targets the 3′-UTR of CD274 directly to downregulate its expression. However, upregulation of zinc-finger E-box-binding homeobox 1 (ZEB1) in NSCLC inhibits the expression of miR-200, resulting in increased PD-L1 level and decreased cytotoxic T-cell activity in TME (Chen et al., 2014). In addition to miR-200, miR-34a, which also targets the CD274 3′-UTR, is downregulated in NSCLC and negatively correlated with PD-L1 expression. Restoring the expression of miR-34a by administrating microRNA-loaded liposomes improves the efficacy of radiotherapy and enhances T-cell immunity in the TME. Strikingly, both miR-200 and miR-34a are transcriptionally upregulated by p53 (Chang et al., 2011; Cortez et al., 2016). Thus, the loss of p53 function in tumor cells appears to be one of the underlying mechanisms contributing to cancer immune evasion due to dysregulated miRNA expression. Besides miR-200 and miR-34a, several other miRNAs, e.g., miR-152 and miR-424, have also been identified as suppressors of PD-L1 expression in different cancer types by targeting the 3′-UTR of CD274 (Wang et al., 2017a; Xie et al., 2017; Xu et al., 2016). Considering the structural variations in the 3′-UTR of CD274 in cancer cells, the PD-L1-suppressing miRNAs could miss the target sequence even under normal expression levels (Kataoka et al., 2016). Together, understanding the association of miRNA and PD-L1 mRNA may better clarify why tumor suppressor miRNAs lose their function in downregulating PD-L1 in tumor cells.
Interestingly, oncogenic pathways can also enhance PD-L1 RNA stability. For instance, tristetraprolin (TTP) destabilizes PD-L1 mRNA by binding to the AU-rich elements in CD274 3′-UTR. However, hyperactive MEK signaling pathway induced by mutated RAS phosphorylates and inhibits TTP via MK2 kinase, which in turn increases the half-life of PD-L1 mRNA. Higher RAS mutation rate and RAS pathway activation have been shown to correlate with elevated PD-L1 mRNA levels in both lung and colon adenocarcinomas (Coelho et al., 2017). Besides maintaining the stability of PD-L1 mRNA, PD-L1 expression can be upregulated by accelerating protein synthesis under the loss of PTEN in tumor cells, which leads to activation of the Akt/mTOR/S6K1 pathway and elevated PD-L1 protein translation rate as reported for glioma (Parsa et al., 2007). These findings provide additional insight into the potential mechanisms regulating PD-L1 expression in cancer cells. Nevertheless, the role of oncogenic signaling pathways in suppressing the anti-tumor immune response by upregulating PD-L1 expression after protein translation remains unclear.
2. Post-translational modification of PD-L1
Posttranslational modifications (PTM) of PD-L1 have emerged as important regulatory mechanisms that modulate immunosuppression in cancer. Following exposure to inflammatory cytokines, cancer cells and antigen-presenting cells, such as macrophages and dendritic cells, express PD-L1 to inhibit the activity of effector T cells through PD-1 engagement. PTM, e.g., glycosylation, phosphorylation, and ubiquitination, are known to play important roles in the regulation of protein stability, translocation, and protein-protein interactions of PD-L1. Recently, Yang et al. reported that palmitoylation also stabilizes PD-L1 (Yang et al., 2019; Yao et al., 2019). Furthermore, through unbiased approach, SUMOylation and acetylation have been suggested as potential PTMs of PD-L1 protein (Horita et al., 2017).
Aberrant alterations of PTM directly influence PD-L1-mediated immune resistance. On the basis of the newly identified regulatory signaling pathways of PD-L1 PTM, researchers have investigated the cancer therapeutic potential of natural food compounds, small-molecule inhibitors, and mAbs by targeting PD-L1 PTM. The results of these preclinical studies demonstrated that targeting PD-L1 PTM yields promising anti-tumor effects and that clinical translation of these therapeutic strategies is warranted (Figure 2).
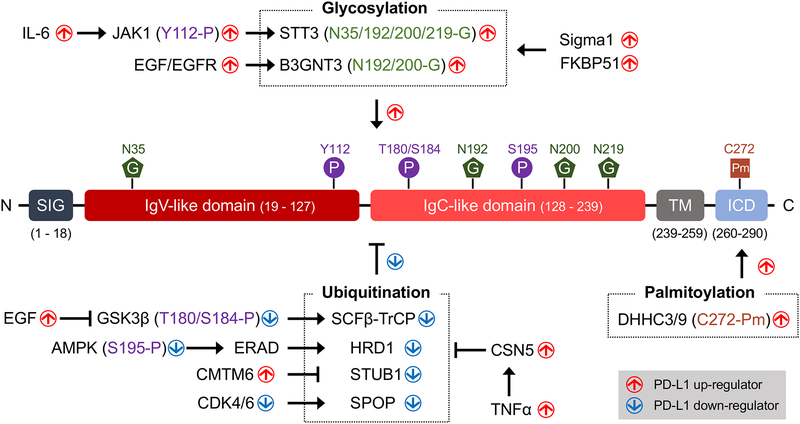
Figure 2. Post-translational modifications (PTM) of PD-L1.
After translation, the activity and stability of PD-L1 are regulated by various PTM. Glycosylation and palmitoylation are essential to maintain the stability of PD-L1. In addition, glycosylation affects the binding between PD-L1 and PD-1. In contrast, poly-ubiquitination is a negative regulator that induces PD-L1 degradation. Phosphorylation regulates PD-L1 level through cross-talk with the glycosylation and poly-ubiquitination.
2–1. N-linked glycosylation of PD-L1
Glycosylation is the most abundant PTM found in one-third of all proteins in mammals (Breitling and Aebi, 2013). N-linked glycosylation in which N-acetylglucosamine is linked to the amide side chain is regulated by the modification of glycosyltransferases and glycosidases (Schwarz and Aebi, 2011). During protein synthesis, oligosaccharyltransferase transfers a 14-sugar based core glycan from dolichol to an asparagine residue of an N-X-T/S motif (N: asparagine, X: any amino acid except proline, S: serine, and T: threonine) in the endoplasmic reticulum (ER) lumen (Xu and Ng, 2015). A recent study indicated that PD-L1 is heavily N-linked glycosylated and that inhibiting PD-L1 glycan synthesis by 2-DG activates T cells against triple-negative breast cancer (TNBC) (Li et al., 2016), suggesting the glycosylation is associated with TNBC malignancy (Shao et al., 2018). Further in-depth analysis revealed four N-X-T/S motifs spanning the extracellular domain of PD-L1 (N35, N192, N200, and N219) that are N-liked glycosylated. Ablation of PD-L1 glycosylation (4NQ mutant) enhances the anti-tumor immunity (Lim et al., 2016). These studies supported the oncogenic role of glycosylation on PD-L1.
Glycosylation is known to stabilize PD-L1, and although fully glycosylated PD-L1 has a half-life of about 12 hours, non-glycosylated PD-L1 undergoes rapid proteolysis with a half-life of about 4 hours. The massive glycan structure protects PD-L1 from GSK3β-mediated 26S proteasome machinery engagement and thus enhances its interaction with PD-1 on CD8+ T cells (Li et al., 2016). Sigma1 and FKBP51 co-chaperones facilitate glycosylated PD-L1 folding and stability in the ER lumen (D’Arrigo et al., 2017; Maher et al., 2018). Dysregulated PD-L1 glycosylation, on the other hand, undergoes ER-associated degradation (ERAD) (Cha et al., 2018). In addition, glycosylation enriches PD-L1 in cancer stemness. Specifically, the catalytic subunit of oligosaccharyltransferase STT3 transfers the core glycan structure to the PD-L1, resulting in epithelial-mesenchymal transition (Hsu et al., 2018). Another study also revealed that IL-6/JAK1 primes PD-L1 for STT3 interaction and PD-L1 glycosylation, suggesting a potential therapeutic combination for hepatocellular carcinoma treatment (Chan et al., 2019). Together, these findings support the important role of PD-L1 glycosylation in suppressing T-cell response against cancers.
The glycan structure of PD-L1 is also involved in the physical contact between PD-L1 and PD-1. Whereas fully glycosylated PD-L1 engages with PD-1, its non-glycosylated counterpart fails to do so in both in vitro and in vivo assays. In the study of EGF/EGFR signaling in TNBC, β−1, 3-N-acetylglucosaminyltransferase 3 (B3GNT3)-mediated poly-N-acetyllactosamine (poly-LacNAc) glycosylation on N192 and N200 of PD-L1 is required for PD-L1/PD-1 interaction (Li et al., 2018). Tomato lectin that specifically recognizes the poly-LacNAc moieties blocks the PD-L1 and PD-1 interaction. Indeed, 4T1 cells lacking B3GNT3 expression grew in SCID mice but not in immunocompetent BALB/c mice. These results suggested that the poly-LacNAc moieties on PD-L1 directly affect its interaction with PD-1.
2–2. Serine/Threonine and Tyrosine Phosphorylation of PD-L1
The extracellular domain of PD-L1 is phosphorylated by several kinases. Two GSK3β phosphorylation motifs (S/TXXXS/T, S: serine, T: threonine, and X: any amino acid) have been identified at T180 and S184 in PD-L1 (Li et al., 2016). Phosphorylation of PD-L1 by GSK3β induces its association with the E3 ligase β-TrCP which results in PD-L1 degradation in the cytoplasm. In addition, AMPK phosphorylates PD-L1 at S195 to induce abnormal PD-L1 glycosylation at all four N-glycosylation sites (N35, N192, N200, and N219). The accumulated PD-L1 is no longer transported to the Golgi and subsequently degraded via ER-associated protein degradation (ERAD) (Cha et al., 2018). Recently, Chan et al. reported that JAK1 phosphorylates PD-L1 at Y112, which is critical for STT3 complex formation in the ER. The ER-localized JAK1 primes PD-L1 phosphorylation at Y112, resulting in PD-L1 glycosylation and trafficking to the cell surface (Chan et al., 2019).
2–3. Polyubiquitination and Degradation of PD-L1
PD-L1 protein expression is extensively regulated by the ubiquitin-mediated proteasome degradation pathway (Burr et al., 2017; Li et al., 2016; Lim et al., 2016; Mezzadra et al., 2017; Zhang et al., 2018a). Studies on PD-L1 protein expression and stability have offered a strong molecular rationale to improve the efficacy of PD-1/PD-L1 blockade in clinics. Thus far, beta-transducin repeats-containing protein (β-TrCP) serves as the substrate recognition subunits for the SCFβ-TrCP E3 ubiquitin ligase and is known for degrading nonglycosylated PD-L1 (Li et al., 2016). The β-TrCP binding degron (DSG motif) at S176 catalyzes K48 ubiquitination of PD-L1. Deletion of the DSG motif in PD-L1 compromises ubiquitination-mediated PD-L1 degradation and increases PD-L1 expression, leading to reduced activity of tumor-infiltrating lymphocytes in mouse tumors. In contrast, overexpression of β-TrCP, but not β-TrCP F-box deletion mutant, reduced PD-L1 expression in TNBC cells. PARP1 inhibitor olaparib (Jiao et al., 2017), c-MET inhibitor (Li et al., 2019), tyrosine kinase inhibitor, and resveratrol (Li et al., 2016) all have been reported to directly or indirectly regulate GSK3β activity to alter PD-L1 and β-TrCP interaction. The HMG-CoA reductase degradation protein 1 (HRD1) also functions as an E3 ligase during ER-associated degradation (ERAD) targeting PD-L1 with abnormal glycan structures derived from S195 phosphorylation (Cha et al., 2018). In addition, speckle-type POZ protein (SPOP), an E3 ubiquitin ligase adaptor protein, stabilizes PD-L1 through cyclin D–cyclin-dependent kinase 4 in late G1 and S phases (Zhang et al., 2018a). PD-L1 is also a substrate of STIP1 homology and U-box containing protein 1 (STUB1/CHIP) which has been shown to polyubiquitinate and downregulate membrane-bound of PD-L1. Moreover, CKLF-like MARVEL transmembrane domain containing 6 (CMTM6) prevents PD-L1 from entering the recycling endosomes by blocking STUB1 and PD-L1 interaction (Burr et al., 2017; Mezzadra et al., 2017).
Another regulator of the ubiquitin conjugation pathway that mediates deubiquitination of SCF multisubunit complex (Skp1, Cullins, F-box proteins) E3 ligase is COP9 signalosome complex subunit 5 (CSN5). During chronic inflammation, deubiquitinase CSN5 catalyzes the removal of polyubiquitination from PD-L1 to suppress anti-tumor immune responses. Proinflammatory cytokine TNFα, secreted by M2 macrophages, induces CSN5 expression through IKKβ and NF-κB activation. Subsequently, CSN5-mediated deubiquitination and stabilization of PD-L1 enhance PD-L1/PD-1 interaction to escape from immune surveillance. In this regard, inhibition of NF-κB signaling by curcumin and aspirin, both of which were shown to inhibit CSN5, can reduce chronic inflammation-mediated PD-L1 expression (Lim et al., 2016).
2–4. Palmitoylation of PD-L1
Lipid modification of PD-L1 has emerged as a new PTM. Covalently linked palmitate to a cysteine residue on PD-L1 (C272) by zinc finger DHHC-type containing 3 (ZDHHC3, also known as DHHC9) palmitoyltransferase blocks PD-L1 ubiquitination to increase its stability (Yao et al., 2019). Competitive inhibition of palmitoyltransferase ZDHHC9 by cell-penetrating peptide sensitized tumor cells to T-cell killing and inhibited tumor growth (Yang et al., 2019). These two new reports open a direction for DHHC inhibitors to enhance the therapeutic efficacy of immune checkpoint therapy.
3. Extracellular PD-L1
PD-L1/PD-1 signaling is believed to restrict signaling locally through intercellular contacts as PD-L1 harbors a typical structure of membrane-bound ligand protein. Interestingly, however, recent studies suggested that PD-L1/PD-1 signaling can function as an expeditionary force dispatched from the mothership. These studies revealed that PD-L1 is secreted in the form of exosomes and/or soluble proteins and that exosomal PD-L1 has substantial bioactivity at the extracellular level, and provide insight into the resistance to ICB targeting PD-L1/PD-1 and the diagnosis to select patients eligible for ICB (Figure 1).
3–1. Exosomal PD-L1
Recently, the presence of exosomal PD-L1 was reported in various cancer types. For example, in a head and neck squamous cell carcinoma (HNSCC) model, PD-L1-positive exosomes purified from plasma of 40 HNSCC patients suppressed T-cell activity, and the levels of exosomal PD-L1 associated with HNSCC progression (Theodoraki et al., 2018). Yang et al. reported the presence of exosome with bioactive PD-L1 and demonstrated that administration of concentrated PD-L1-positive exosomes promoted tumor growth in an immunocompetent breast tumor mouse model. Consistently, blocking exosome secretion improved the therapeutic efficacy of anti-PD-L1 therapy was improved (Yang et al., 2018). Another study demonstrated enhanced secretion of PD-L1-positive exosomes mediated by IFN-γ signaling in a melanoma model. Importantly, it appears that the levels of exosomal PD-L1 varied based on the stage of anti-tumor immunity, suggesting that exosomal PD-L1 has potential to serve as a biomarker for patient selection of PD-L1/PD-1 therapy and indicator of clinical outcome (Chen et al., 2018). Recently, Poggio et al. found that circulating PD-L1-positive exosomes can systemically inhibit anti-tumor immunity. Consistent with the results in breast cancer models (Yang et al., 2018), genetic ablation of exosomal PD-L1 or blockage of exosome secretion suppressed tumor growth via antitumor immunity in prostate cancer models. Remarkably, blockage of exosome secretion attenuated exosomal PD-L1-mediated inhibition of T-cell activity in the lymph nodes, suggesting that circulating exosomal PD-L1 has systemic and substantial functions in adaptive immunity in cancer (Poggio et al., 2019).
3–2. Soluble PD-L1 (sPD-L1)
The existence of soluble PD-L1 (sPD-L1) has been reported by Okuma et al. Specifically, patients with advanced lung cancer had much higher levels of sPD-L1 in the plasma than did normal subjects, and such high levels of sPD-L1 are significantly related to poor prognosis (Okuma et al., 2017). Those findings provide some evidence to show that sPD-L1 in patients with NSCLC can have bioactivity to suppress T-cell immunity. In patients with low plasma sPD-L1 levels, 59% demonstrated response to anti-PD-1 therapy nivolumab whereas in patients with high plamsa sPD-L1, only 25% responded. Furthermore, the overall survival also exhibited a strong negative correlation with plasma sPD-L1 level (Okuma et al., 2018). Interestingly, in a melanoma model, Zhou et al. identified four PD-L1 splicing variants that lack the transmembrane domain and can be secreted. Moeover, the secretion of sPD-L1 was upregulated under IFNα, IFNγ, and TNFα treatment. The activities of both CD4+ and CD8+ T cells were suppresed by three of the four splicing variants in vitro. (Zhou et al., 2017). Consistently, five PD-L1 splicing variants were found to be secreted in NSCLC (Gong et al., 2019). Among them, two splicing variants (v229 and v242) were highly detected in patients with NSCLC who were resistant to anti-PD-L1 therapy, and effectively neutralized the activity of PD-L1 antibody through binding competition. (Gong et al., 2019). These results strongly supported the notion that sPD-L1 harbors systemic functions in T-cell immunity suppression in the bloodstream as well as tumor tissues. Further studies are warranted to validate those findings in vivo.
Perspectives and future directions
Inhibiting PD-L1 expression in tumor cells enhances immunosurveillance and reduces PD-L1-driven non-immune checkpoint function that attenuates DNA damage response and repair (Tu et al., 2019; Zou et al., 2016). Downregulating PD-L1 expression on the APCs and DCs also enhances the response rates to ICB combination therapy (Lin et al., 2018; Tang et al., 2018). The mechanisms discussed in this review provide many potential strategies to inhibit PD-L1 expression and its immune evasion function. However, the current clinical outcomes suggest that blockade of PD-L1/PD-1 pathway is not sufficient to restore anti-tumor immunity due to the presence of other immune checkpoints that also promote tumor immune escape in the TME or tumor-draining lymph nodes (Zou et al., 2016). Therefore, combined approaches comprised of oncogenic pathway inhibitors and immune checkpoint neutralizing antibodies (e.g., CTLA-4 and Tim-3) may increase the response rates. Interestingly, several different kinase-targeting therapies were reported to upregulate PD-L1 in the clinic and in mouse models by increasing the ability of tumor cells to evade immune response, leading to drug resistance (Li et al., 2019; Zhang et al., 2018a). Thus, neutralizing the upregulated PD-L1 by antibody has the potential to re-sensitize tumor cells to those kinase inhibitors.
In this regard, PD-L1-related inhibitors/activators have been largely defined in previous cancer studies, and these agents may be considered for use as adjuvants for current ICB. For instance, Li et al. showed that tyrosine kinase inhibitor (TKI) targeting EGFR (upstream signaling of PD-L1) improves the efficacy of PD-1 blockade in immunocompetent syngeneic mouse models (Li et al., 2016). In addition, Cha et al. proposed the use of metformin, which activates AMPK and subsequently induces ERAD of PD-L1, as an adjuvant to enhance the efficacy of CTLA-4 blockade (Cha et al., 2018). Another approach to consider is targeting interleukin-6 (IL-6), an upstream signaling of PD-L1, such as the anti-IL-6 and anti-TIM3 antibody combination reported by Chan et al (Chan et al., 2019). Another study demonstrated improved therapeutic efficacy when combining c-Met inhibitor with anti-PD-1 (Li et al., 2019). Moreover, the combined therapy of histone deacetylase (HDAC) inhibitors and PD-1 antibody decreases the tumor growth and increases the survival rate in a melanoma mouse model (Woods et al., 2015). On the other hand, PD-L1 glycosylation can also be targeted as reported by Hsu et al. in which etoposide inhibits the transcription level of STT3 (a glycosyltransferase of PD-L1) and sensitizes cancer cells to Tim-3 blockade (Hsu et al., 2018). Additionally, inhibition of CDK4/6 by palbociclib upregulates and CSN5 by curcumin downregulates the stability of PD-L1, leading to increased therapeutic effect of anti-PD-1 and anti-CTLA-4 therapy, respectively (Lim et al., 2016; Zhang et al., 2018a). Furthermore, analyzing potential candidates from CRISPR screens or PD-L1-interacting kinases may also lead to the discovery of novel mechanisms of PD-L1 regulation in tumor cells (Burr et al., 2017; Chan et al., 2019; Mezzadra et al., 2017).
Although extensive studies have evaluated various combination therapies with ICB, the benefits of those combination therapies have been limited, and in some cases, the side effects have increased (Kourie and Klastersky, 2016). One of the main reasons for the unsatisfactory outcomes is that the initial diagnosis may not be adequate to qualify patients for combination therapy. Cancers are known to accumulate abnormalities at multiple stages of progression involving regulators of PD-L1 expression. Furthermore, many factors which can affect PD-L1 expression levels are dynamically changed depending on the stage of anti-tumor immunity. Therefore, it is necessary to classify active regulatory pathways of PD-L1 in the TME and the stage of anti-tumor immunity to establish a diagnostic process that specifies the active pathways in individual patient specimens. In doing so, this could provide important information to determine the combination of adjuvant and ICB individually. These patient-tailored combination therapies are expected to significantly improve prognosis.
The activation of immune cells is tightly controlled by PD-1/PD-L1 interaction between immune cells, immune cells and tumor cells, or secreted PD-L1 and immune cells. The expression of PD-L1 and its PTM affect this association and subsequently the immune suppression signaling via PD-1. Therefore, accurate quantification of PD-L1 expression in the tumor or serum/plasma may be optimized for current anti-PD-1/PD-L1 cancer therapy. The tumor glyco-code is recognized as a novel signature for immunotherapy (RodrIguez et al., 2018), and specific glycan structure of PD-L1 (gPD-L1) can be targeted by antibody and detected by immunohistochemical (IHC) staining in TNBC samples (Li et al., 2018). On this basis of the results by Li et al., it would be interesting to further compare the sensitivity of current IHC method and detection via anti-gPD-L1 antibody in tumor samples. Given that the interaction of PD-1/PD-L1 relies on the glycan structure, the results from gPD-L1 staining is likely to more accurately represent the “levels of functional PD-L1” for immunosuppression in the TME. The levels of exosomal PD-L1 or sPD-L1 in serum are also reported to reflect the tumor malignancy in patients (Chen et al., 2018; Gong et al., 2019; Okuma et al., 2017; Theodoraki et al., 2018; Zhou et al., 2017). Importantly, Chen et al. demonstrated that exosomal PD-L1 levels differ based on the stage of anti-tumor immunity after ICB treatment, suggesting that exosomal PD-L1 level may be used not only as a biomarker to select patients receiving anti-PD-L1 / PD-1 therapy but also as an indicator for treatment response (Chen et al., 2018). Since the positive correlation between tumor PD-L1 and exosomal PD-L1 has not yet been validated and that sPD-L1 derived from the secretory pathway via alternative splicing is different from membrane PD-L1, current IHC diagnosis cannot rule out the secretion of exosomal PD-L1 and sPD-L1 into serum even if PD-L1 is negative in the tumor tissues. Therefore, the expression of PD-L1 in the tumor and serum/plasma should be collectively considered for diagnosis.
In addition, because the glycan structure is required to maintain the stability of PD-L1 and its engagement with PD-1 (Li et al., 2016), the exosomal PD-L1 or sPD-L1 is expected to also be highly glycosylated. Thus, determining the gPD-L1 level in serum from patients with cancers may provide a valuable biomarker for estimating the status of immunosuppression. Current studies suggest the possibility that both exosomal PD-L1 and sPD-L1 have bioactivity to suppress T-cell immunity systemically (Gong et al., 2019; Poggio et al., 2019; Zhou et al., 2017). Considering that systemic immunosuppression directly provides a niche for cancer metastasis, this approach may also can be used as a parameter to predict the risk of metastasis in patients with cancers.
Concluding remarks
In this review, we summarized the mechanisms that regulate PD-L1 through multiple processes. Overexpression of PD-L1 in different cell types, such as tumor cells, APCs, and macrophage, is recognized as a major player suppressing anti-tumor immunity in the TME and tumor-draining lymph nodes in a variety of cancer types, and high levels of PD-L1 are associated with increased response to ICB targeting PD-L1/PD-1. Therefore, furthering our understanding of the regulatory mechanisms of PD-L1 expression in different cell types in patients with cancers has the potential to improve the efficacy of ICB targeting PD-L1/PD-1 and/or overcome resistance to ICB. Moreover, activators and/or inhibitors of PD-L1 identified from mechanistic studies may have potential to increase the benefits in combination with inhibitors against other immune checkpoints.
Acknowledgments
This work was funded in part by the following: Cancer Prevention & Research Institute of Texas (MIRA grant RP160710); National Institutes of Health (R01 AI116722); National Breast Cancer Foundation, Inc.; Breast Cancer Research Foundation (BCRF-17-069); The University of Texas MD Anderson-China Medical University Sister Institution Fund and Inha University Institution Fund (to J.-H.C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, et al. (2013). Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 3, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atefi M, Avramis E, Lassen A, Wong DJ, Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et al. (2014). Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin Cancer Res 20, 3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum IB, Smallwood CA, Siemens DR, and Graham CH (2014). A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res 74, 665–674. [DOI] [PubMed] [Google Scholar]
- Breitling J, and Aebi M (2013). N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 5, a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, et al. (2017). CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbotti G, Barisione G, Airoldi I, Mezzanzanica D, Bagnoli M, Ferrero S, Petretto A, Fabbi M, and Ferrini S (2015). IL-27 induces the expression of IDO and PD-L1 in human cancer cells. Oncotarget 6, 43267–43280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, et al. (2016). MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH, Yang WH, Xia W, Wei Y, Chan LC, Lim SO, Li CW, Kim T, Chang SS, Lee HH, et al. (2018). Metformin Promotes Antitumor Immunity via Endoplasmic-Reticulum-Associated Degradation of PD-L1. Mol Cell 71, 606–620 e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, Wang HL, Yang WH, Yen EY, Chang WC, et al. (2019). IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest 130, 3324–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al. (2011). p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 13, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, et al. (2018). Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. (2014). Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 5, 5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, and Han X (2015). Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest 125, 3384–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, et al. (2017). Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity 47, 1083–1099 e1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, et al. (2016). PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, Krzysiek R, Knutson KL, Daniel B, Zimmermann MC, et al. (2003). Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med 9, 562–567. [DOI] [PubMed] [Google Scholar]
- D’Arrigo P, Russo M, Rea A, Tufano M, Guadagno E, Del Basso De Caro ML, Pacelli R, Hausch F, Staibano S, Ilardi G, et al. (2017). A regulatory role for the co-chaperone FKBP51s in PD-L1 expression in glioma. Oncotarget 8, 68291–68304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV (2012). MYC on the path to cancer. Cell 149, 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Hu Q, Zhang H, Yang F, Peng C, and Huang C (2019). HDAC3 Inhibition Upregulates PD-L1 Expression in B-Cell Lymphomas and Augments the Efficacy of Anti-PD-L1 Therapy. Mol Cancer Ther 18, 900–908. [DOI] [PubMed] [Google Scholar]
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al. (2002). Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8, 793–800. [DOI] [PubMed] [Google Scholar]
- Francisco LM, Sage PT, and Sharpe AH (2010). The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 236, 219–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, et al. (2017). Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 19, 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Kiyotani K, Sakata S, Nagano S, Kumehara S, Baba S, Besse B, Yanagitani N, Friboulet L, Nishio M, et al. (2019). Secreted PD-L1 variants mediate resistance to PD-L1 blockade therapy in non-small cell lung cancer. J Exp Med 216, 982–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Chehrazi-Raffle A, Reddi S, and Salgia R (2018). Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, Gupta R, Tsai JM, Sinha R, Corey D, et al. (2017). PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, et al. (2010). Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116, 3268–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, Beavis PA, Darcy PK, Martin BP, Spencer A, et al. (2017). BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep 18, 2162–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita H, Law A, Hong S, and Middleton K (2017). Identifying Regulatory Posttranslational Modifications of PD-L1: A Focus on Monoubiquitinaton. Neoplasia 19, 346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha JH, Chen CT, Liao HW, Kuo CW, Khoo KH, et al. (2018). STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun 9, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Okamoto T, Okano S, Umemoto Y, Tagawa T, Morodomi Y, Kohno M, Shimamatsu S, Kitahara H, Suzuki Y, et al. (2016). PD-L1 Is Upregulated by Simultaneous Amplification of the PD-L1 and JAK2 Genes in Non-Small Cell Lung Cancer. J Thorac Oncol 11, 62–71. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, and Hodi FS (2013). The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res 19, 598–609. [DOI] [PubMed] [Google Scholar]
- Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, et al. (2017). PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin Cancer Res 23, 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, et al. (2016). Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 534, 402–406. [DOI] [PubMed] [Google Scholar]
- Kourie HR, and Klastersky JA (2016). Side-effects of checkpoint inhibitor-based combination therapy. Curr Opin Oncol 28, 306–313. [DOI] [PubMed] [Google Scholar]
- Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, Cha JH, Xia W, Chan LC, Kim T, et al. (2018). Eradication of Triple-Negative Breast Cancer Cells by Targeting Glycosylated PD-L1. Cancer Cell 33, 187–201 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, et al. (2016). Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 7, 12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li CW, Li X, Ding Q, Guo L, Liu S, Liu C, Lai CC, Hsu JM, Dong Q, et al. (2019). MET Inhibitors Promote Liver Tumor Evasion of the Immune Response by Stabilizing PDL1. Gastroenterology 156, 1849–1861 e1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, et al. (2016). Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 30, 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, Szeliga W, Herbst R, Harms PW, Fecher LA, et al. (2018). Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest 128, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Paschall AV, Shi H, Savage N, Waller JL, Sabbatini ME, Oberlies NH, Pearce C, and Liu K (2017). The MLL1-H3K4me3 Axis-Mediated PD-L1 Expression and Pancreatic Cancer Immune Evasion. J Natl Cancer Inst 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M L, P PV, T K, M P, E S, J P, K VW, C L, F C, S D, et al. (2016). Essential role of HDAC6 in the regulation of PD-L1 in melanoma. Mol Oncol 10, 735–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CM, Thomas JD, Haas DA, Longen CG, Oyer HM, Tong JY, and Kim FJ (2018). Small-Molecule Sigma1 Modulator Induces Autophagic Degradation of PD-L1. Mol Cancer Res 16, 243–255. [DOI] [PubMed] [Google Scholar]
- Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, et al. (2008). Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci U S A 105, 20852–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, et al. (2017). Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni XY, Sui HX, Liu Y, Ke SZ, Wang YN, and Gao FG (2012). TGF-beta of lung cancer microenvironment upregulates B7H1 and GITRL expression in dendritic cells and is associated with regulatory T cell generation. Oncol Rep 28, 615–621. [DOI] [PubMed] [Google Scholar]
- Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, and Zang X (2015). Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol Med 21, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma Y, Hosomi Y, Nakahara Y, Watanabe K, Sagawa Y, and Homma S (2017). High plasma levels of soluble programmed cell death ligand 1 are prognostic for reduced survival in advanced lung cancer. Lung Cancer 104, 1–6. [DOI] [PubMed] [Google Scholar]
- Okuma Y, Wakui H, Utsumi H, Sagawa Y, Hosomi Y, Kuwano K, and Homma S (2018). Soluble Programmed Cell Death Ligand 1 as a Novel Biomarker for Nivolumab Therapy for Non-Small-cell Lung Cancer. Clin Lung Cancer 19, 410–417 e411. [DOI] [PubMed] [Google Scholar]
- Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et al. (2007). Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med 13, 84–88. [DOI] [PubMed] [Google Scholar]
- Peng J, Hamanishi J, Matsumura N, Abiko K, Murat K, Baba T, Yamaguchi K, Horikawa N, Hosoe Y, Murphy SK, et al. (2015). Chemotherapy Induces Programmed Cell Death-Ligand 1 Overexpression via the Nuclear Factor-kappaB to Foster an Immunosuppressive Tumor Microenvironment in Ovarian Cancer. Cancer Res 75, 5034–5045. [DOI] [PubMed] [Google Scholar]
- Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, et al. (2019). Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 177, 414–427 e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestipino A, Emhardt AJ, Aumann K, O’Sullivan D, Gorantla SP, Duquesne S, Melchinger W, Braun L, Vuckovic S, Boerries M, et al. (2018). Oncogenic JAK2(V617F) causes PD-L1 expression, mediating immune escape in myeloproliferative neoplasms. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulko V, Liu X, Krco CJ, Harris KJ, Frigola X, Kwon ED, and Dong H (2009). TLR3-stimulated dendritic cells up-regulate B7-H1 expression and influence the magnitude of CD8 T cell responses to tumor vaccination. J Immunol 183, 3634–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt D, Jasinski-Bergner S, Muller U, Schulze B, and Seliger B (2014). Synergistic effects of IL-4 and TNFalpha on the induction of B7-H1 in renal cell carcinoma cells inhibiting allogeneic T cell proliferation. J Transl Med 12, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RodrIguez E, Schetters STT, and van Kooyk Y (2018). The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat Rev Immunol. [DOI] [PubMed] [Google Scholar]
- Roemer MG, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, Connelly CF, Sun HH, Daadi SE, Freeman GJ, et al. (2016). PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol 34, 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmamed MF, and Chen L (2018). A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 175, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, Nuryadi E, Sekine R, Oike T, Kakoti S, et al. (2017). DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun 8, 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildberg FA, Klein SR, Freeman GJ, and Sharpe AH (2016). Coinhibitory Pathways in the B7-CD28 Ligand-Receptor Family. Immunity 44, 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, and Aebi M (2011). Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol 21, 576–582. [DOI] [PubMed] [Google Scholar]
- Shao B, Li CW, Lim SO, Sun L, Lai YJ, Hou J, Liu C, Chang CW, Qiu Y, Hsu JM, et al. (2018). Deglycosylation of PD-L1 by 2-deoxyglucose reverses PARP inhibitor-induced immunosuppression in triple-negative breast cancer. Am J Cancer Res 8, 1837–1846. [PMC free article] [PubMed] [Google Scholar]
- Shao YY, Lin H, Li YS, Lee YH, Chen HM, Cheng AL, and Hsu CH (2017). High plasma interleukin-6 levels associated with poor prognosis of patients with advanced hepatocellular carcinoma. Jpn J Clin Oncol 47, 949–953. [DOI] [PubMed] [Google Scholar]
- Sun C, Mezzadra R, and Schumacher TN (2018a). Regulation and Function of the PD-L1 Checkpoint. Immunity 48, 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LL, Yang RY, Li CW, Chen MK, Shao B, Hsu JM, Chan LC, Yang Y, Hsu JL, Lai YJ, et al. (2018b). Inhibition of ATR downregulates PD-L1 and sensitizes tumor cells to T cell-mediated killing. Am J Cancer Res 8, 1307–1316. [PMC free article] [PubMed] [Google Scholar]
- Tang H, Liang Y, Anders RA, Taube JM, Qiu X, Mulgaonkar A, Liu X, Harrington SM, Guo J, Xin Y, et al. (2018). PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest 128, 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, and Whiteside TL (2018). Clinical Significance of PD-L1(+) Exosomes in Plasma of Head and Neck Cancer Patients. Clin Cancer Res 24, 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RH, Gillett MD, Cheville JC, Lohse CM, Dong H, Webster WS, Krejci KG, Lobo JR, Sengupta S, Chen L, et al. (2004). Costimulatory B7-H1 in renal cell carcinoma patients: Indicator of tumor aggressiveness and potential therapeutic target. Proc Natl Acad Sci U S A 101, 17174–17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, and Pardoll DM (2015). Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokawa G, Takada K, Tagawa T, Hamamoto R, Yamada Y, Shimokawa M, Oda Y, and Maehara Y (2019). A Positive Correlation Between the EZH2 and PD-L1 Expression in Resected Lung Adenocarcinomas. Ann Thorac Surg 107, 393–400. [DOI] [PubMed] [Google Scholar]
- Tu X, Qin B, Zhang Y, Zhang C, Kahila M, Nowsheen S, Yin P, Yuan J, Pei H, Li H, et al. (2019). PD-L1 (B7-H1) Competes with the RNA Exosome to Regulate the DNA Damage Response and Can Be Targeted to Sensitize to Radiation or Chemotherapy. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twa DD, Chan FC, Ben-Neriah S, Woolcock BW, Mottok A, Tan KL, Slack GW, Gunawardana J, Lim RS, McPherson AW, et al. (2014). Genomic rearrangements involving programmed death ligands are recurrent in primary mediastinal large B-cell lymphoma. Blood 123, 2062–2065. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin W, Tang X, Li S, Guo L, Lin Y, and Kwok HF (2017a). The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang L, Huang F, Zhang Q, Liu S, Ma L, and You Z (2017b). Inflammatory cytokines IL-17 and TNF-alpha up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett 184, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DM, Sodre AL, Villagra A, Sarnaik A, Sotomayor EM, and Weber J (2015). HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol Res 3, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Li W, Li R, Wu K, Zhao E, Zhang Y, Zhang P, Shi L, Wang D, Yin Y, et al. (2017). Helicobacter Pylori Promote B7-H1 Expression by Suppressing miR-152 and miR-200b in Gastric Cancer Cells. PLoS One 12, e0168822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, Herter-Sprie GS, Akbay EA, Tchaicha JH, Altabef A, et al. (2014). Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 25, 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, and Ng DT (2015). Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol 16, 742–752. [DOI] [PubMed] [Google Scholar]
- Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, Song W, Chen Y, OuYang J, Chen J, et al. (2016). miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun 7, 11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Du Y, Nakai K, Ding M, Chang SS, Hsu JL, Yao J, Wei Y, Nie L, Jiao S, et al. (2018). EZH2 contributes to the response to PARP inhibitors through its PARP-mediated poly-ADP ribosylation in breast cancer. Oncogene 37, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu JL, Wei Y, Xia W, Hou J, Qiu Y, et al. (2019). Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res 29, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, Cha JH, Hou J, Hsu JL, Sun L, et al. (2018). Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res 28, 862–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, Lu H, Fang C, Zhang Y, Liang L, et al. (2019). Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng 3, 306–317. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, et al. (2018a). Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dang F, Ren J, and Wei W (2018b). Biochemical Aspects of PD-L1 Regulation in Cancer Immunotherapy. Trends Biochem Sci 43, 1014–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, and Bevan MJ (2011). CD8(+) T cells: foot soldiers of the immune system. Immunity 35, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu Z, and Huang JA (2016). The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int J Oncol 49, 1360–1368. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang P, and Wang XF (2014). Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol 24, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Mahoney KM, Giobbie-Hurder A, Zhao F, Lee S, Liao X, Rodig S, Li J, Wu X, Butterfield LH, et al. (2017). Soluble PD-L1 as a Biomarker in Malignant Melanoma Treated with Checkpoint Blockade. Cancer Immunol Res 5, 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, Yokoyama Y, Kossenkov AV, Bradner JE, Conejo-Garcia JR, et al. (2016). BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep 16, 2829–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Wolchok JD, and Chen L (2016). PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 8, 328rv324. [DOI] [PMC free article] [PubMed] [Google Scholar]