Abstract
IL-10 is a critical immunoregulatory cytokine expressed in virtually all immune cell types. Maintaining a delicate balance between effective immune response and tolerance requires meticulous and dynamic control of IL-10 expression both epigenetically and transcriptionally. In this Review, we describe the epigenetic mechanisms controlling IL-10 expression, including chromatin remodeling, 3D chromatin loops, histone modification and DNA methylation. We discuss the role of transcription factors in directing chromatin modifications, with a special highlight on the emerging concept of pioneer transcription factors in setting up the chromatin landscape in T helper cells for Il10 induction. Besides summarizing the recent progress on transcriptional regulation in specialized IL-10 producers such as type 1 regulatory T cells, regulatory B cells and regulatory ILCs, we also discuss common transcriptional mechanisms for IL-10 regulation that are shared with other IL-10 producing cells.
Keywords: IL-10, chromatin remodeling, histone modification, DNA methylation, pioneer transcription factor, transcriptional regulation, T cell, Breg, ILC, myeloid cell
1. Introduction
Interleukin-10 (IL-10) is a pluripotent immune-regulatory cytokine indispensable for maintaining immune homeostasis and restricting autoimmunity[1]. It has broad-spectrum immunosuppressive activity on mainly the innate but also the adaptive arm of the immune system. IL-10 dampens activation of antigen presenting cells (APCs) by suppressing the expression of the major histocompatibility complex (MHC), co-stimulatory molecules and pro-inflammatory cytokines. It promotes the differentiation, survival and function of regulatory T cells, but can also directly inhibit the effector function of Th1, Th2 and Th17 cells. Under some circumstances, IL-10 can promote antibody responses of B cells and anti-tumor capabilities of CD8 T cells.
Corresponding to its widespread activity, IL-10 can be expressed by almost all types of immune cells. Type 1 regulatory T (Tr1) cells are specialized IL-10 producers in the T cell compartment that exert their major regulatory function through IL-10 [2–4], as do regulatory B (Breg) cells[5] in the B cell compartment and the recently discovered regulatory innate lymphoid cells (ILCs)[6]. Foxp3+ regulatory T (Treg) cells, all other T helper cells, CD8 T cells, as well as innate cells including dendritic cells, macrophages, mast cells, natural killer cells, eosinophils, and neutrophils, are capable of producing IL-10, which could then mitigate overexuberant immune responses.
Deficiency in IL-10 or IL-10 signaling leads to spontaneous colitis in mice and very early onset inflammatory bowel disease (VEO-IBD) in humans[7]. IL-10 deficient mice suffer from lethal immunopathology due to uncontrolled CD4 T cell responses in acute infections such as Toxoplasma gondii[8]. In addition, disease is exacerbated with IL-10 deficiency in animal models of autoimmune disease, including experimental autoimmune encephalomyelitis (EAE)[9], collagen-induced arthritis [10], and MRL/lpr lupus [11]. On the other hand, dysregulated enhancement in IL-10 expression can contribute to chronic infection[12]. Of note, the role of IL-10 under some circumstances can be dependent on the specific animal model that is examined. For example, unlike the MRL/lpr lupus model, neutralization of IL-10 in lupus-prone NZB/W F1 mice substantially delays the onset of lupus-like autoimmunity[13].
Highly dynamic yet precise mechanisms are required to regulate IL-10 transcription in order to maintain immune homeostasis in a wide range of cellular and environmental contexts while preserving the capability to mount effective immune responses. We now appreciate that regulation of IL-10 transcription involves multistep epigenetic reprogramming of chromatin structure and precise expression and recruitment of transcription factors. In this Review, we summarize the current state of knowledge regarding epigenetic mechanisms for IL-10 regulation and the role of pioneer and other transcription factors in orchestrating these epigenetic modifications. In addition to reviewing recent findings on specific transcription factors that regulate IL-10 in different immune cell types, we will also discuss how diverse signals in discrete cell types may converge to a similar transcriptional program to regulate IL-10 expression.
2. Epigenetic regulation of IL-10
2.1. Chromatin remodeling in Il10 locus by ATP-dependent chromatin-remodeling complexes
Genomic DNA of eukaryotic cells is tightly packaged to fit into the nucleus. Nucleosomes are the basic repeating unit of DNA packaging where 146 bp of DNA wrap around a histone octamer containing two each of the histones H2A, H2B, H3 and H4. In transcriptionally silent heterochromatic regions, nucleosomes are tightly spaced and further folded into higher order of chromatin condensation such as 30nm fibers. This chromatin organization occludes the target sequences for DNA binding proteins and creates a state of inaccessibility for the transcriptional machinery. The chromatin structure of tightly regulated genes such as IL-10 are dynamically regulated by a process called “chromatin remodeling” to create accessibility to the gene by transcription factors and RNA polymerases.
ATP-dependent chromatin-remodeling complexes provide a major means of chromatin remodeling by mediating nucleosome assembly, ejection and editing. In these processes, ATP hydrolysis is utilized to translocate DNA and reposition nucleosomes. The complexes are classified based on homology of their catalytic ATPase subunit: switch/sucrose non-fermentable (SWI/SNF) subfamily, imitation switch (ISWI) subfamily, chromodomain helicase DNA-binding (CHD) subfamily, and INO80 subfamily. Each subfamily is preferentially (but not solely) specialized for one of the following three functionalities: 1) ISWI and CHD subfamily complexes facilitate the assembly and maturation of nucleosomes, and space them at a relatively fixed distance apart. These processes can happen during the transcription process in which nucleosomes are dynamically ejected by the transcription machinery. 2) SWI/SNF subfamily complexes modify chromatin access by nucleosome sliding, eviction of nucleosome components or ejection of the whole nucleosome. 3) INO80 subfamily complexes have the unique ability to replace a specific histone with a canonical or variant histone and thus affect the recruitment and function of other factors[14,15]. These complexes target specific genes mainly through interaction with DNA-specific transcription factors and can contribute to both gene activation and suppression.
SWI/SNF subfamily complexes are major players in the regulation of chromatin accessibility and they orchestrate extremely diverse gene expression programs across a range of different tissues from embryonic stems cells[16] to postmitotic neurons[17]. Upon LPS stimulation in mouse macrophages, induction of primary and secondary response genes have a differential dependence on SWI/SNF[18,19]. SWI/SNF is required for chromatin remodeling during T cell development[20], Th1 and Th2 differentiation[21,22], as well as Treg function[23]. The functional specificity of SWI/SNF complexes is provided by its great diversity of composition. SWI/SNF complexes can contain more than eleven subunits, many of which have cell specific isoforms that are assembled in a combinatorial way to control gene expression in a cell- and context- dependent manner. The specific composition of SWI/SNF complexes that drive distinct gene programs in various immune cells is only beginning to be elucidated. In addition, cooperation with cell-type-specific transcription factors adds a second layer of specificity to SWI/SNF complexes.
Three versions of mammalian SWI/SNF (mSWI/SNF) complexes have been identified: BRG1/BRM-associated factor complexes (BAFs), polybromo-associated BAF complexes (PBAFs), and non-canonical BAFs (ncBAFs). BAFs use either BRG1 or BRM as ATPase, and contain subunit BAF250a or BAF250b; PBAFs use BRG1 but not BRM as ATPase and contain Protein polybromo-1 (PB1/BAF180). We currently have very limited knowledge regarding how IL-10 is regulated by specific chromatin remodeling complexes. Il10 stood out as the most differentially expressed gene in in-vitro differentiated Th2 cells that are genetically deficient in BAF180. BAF180 inhibits IL-10 production in Th2 cells without affecting their differentiation. BAF180 binds to the −29.8kb, −9kb and +6.2kb region of the Il10 locus and in the absence of BAF180, binding of BAF250 to Il10 is enhanced, indicating that PBAFs and BAFs might be playing opposite roles in IL-10 regulation. It is still not clear which isoforms of the other subunits are utilized in the IL-10 regulating PBAFs, and which transcription factors are directing the specificity of the target gene in this process. Besides Th2 cells, BRG1 also binds to the Il10 locus in Th1 and Th17 cells. As BRG1 can exist in both PBAF and BAF complexes, it remains to be addressed which complex is in charge of IL-10 regulation in these cell types[24].
2.2. Regulation of IL-10 by 3D chromatin loops
Cis-regulatory enhancers can be located 100bp to Mb away from the transcription starting site (TSS) of the gene they regulate. Distant enhancers are brought into close proximity to the promoters through 3D chromatin loops formed by DNA folding[25]. The formation of DNA loops between long-distance enhancers and promoters is best illustrated by two types of experimental evidence: 1) chromosome conformation capture (3C) technology, which chemically fixes and ligates DNA in close 3D proximity; 2) fluorescence in situ hybridization (FISH), which enables visualization of the in situ spatial proximity of enhancer and promoter[26]. Hi-C, a 3C-based technique that allows unbiased quantification of DNA-DNA interactions in the whole genome, revealed that the genome is compartmentalized into topologically associating domains (TADs) where intradomain interactions are strongly favored compared to interdomain interactions. Boundaries of TADs are demarcated by the zinc finger protein CTCF. The current working model holds that TADs are formed when a pair of cohesin rings slides in opposite directions along the chromatin and continuously extrudes a loop until a properly oriented CTCF site is reached on both sides[25]. Additionally, CTCF has binding sites within the TAD to facilitate promote-enhancer interactions[27]. Disruptions of CTCF-associated TADs cause rewiring of gene-enhancer interactions and can result in aberrant gene activation that leads to disease[28,29].
Precise control of 3D genome organization is critical for the development and function of immune cells. For example, commitment to the T cell lineage relies on changes in 3D genome organization[30], as does recombination of the TCR and BCR loci [31–33], and the germinal center response[34]. In addition to CTCF and cohesin, transcription factors are demonstrated to be important orchestrators in genome organization in immune cells: Bcl11b and Pax5 regulates genome organization during T and B cell development[30,35]; GATA3 and STAT6 orchestrate the chromatin interactions among Th2 genes including Il4, Il5 and Il13[36]; Musculin (MSC) strengthens a unidirectional Treg differentiation program by repressing Th2 differentiation, which involves antagonizing GATA3 and disruption of intrachromosomal interactions within the Th2 locus[37].
Disruption of TADs impairs IL-10 production in many contexts. For instance, genetic deletion of CTCF in T cells impairs IL-10 production in both Th1 and Th2 cells[38]. Similarly, conditional knockout of CTCF in macrophages impaired the production of IL-10 as well as other IL-10 family members such as IL-19, IL-20 and IL-24, which are located in the same locus[39]. It has to be pointed out that genetic deletion of CTCF disrupts all CTCF-associated TAD domains in the genome. Consequently, it’s difficult to interpret from these studies how the Il10 locus specifically is regulated by chromatin looping. It would be of interest to couple such genetic studies with chromosome conformation capture methods, or to mutate CTCF binding sites in or surrounding the Il10 locus rather than depleting CTCF globally.
As of now, the 3D chromatin organization at Il10 locus remains poorly characterized. Most cis-regulatory elements for Il10 were assigned to the Il10 simply because its TSS is the closest. The ENCODE Project and NIH Mouse Regulome Project are generating promoter-enhancer interactomes of many cell types including some immune cells, which will be very informational for discovery of distal enhancers of Il10. However, studies that focus on dynamic regulation of chromatin organization at the Il10 locus in relevant biological settings are still lacking.
2.3. Regulation of IL-10 by histone modifications
Covalent post-translational modification of histones not only marks distinct chromatin states associated with gene expression, but also critically regulates gene expression either by directly influencing chromatin structure or by recruiting “readers” with effector function. These modifications include methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation. The best studied and most prevalent modifications occur on lysines (Lys/K) in the N-terminal ‘tail’ of the histones, although they can also occur in the nucleosome core.
2.3.1.1. Histone acetylation
Histone acetylation facilitates gene expression by neutralizing the positive charge of lysine residues, thereby weakening the charge-dependent interactions between histone and nucleosomal DNA, and enhancing chromatin accessibility to transcription machinery. Acetylation can occur on numerous lysines in the histone tail and act in a cumulative way. For example, H3K27ac is highly associated with permissive chromatin and actively transcribed regions. Two groups of functionally opposing enzymes - histone acetyltransferases (HATs) and histone deacetylases (HDACs) act as “writers” and “erasers” of histone acetylation. HAT1 has been implicated in upregulation of IL-10 expression in breast cancer associated Treg cells that predominantly produce IL-10, but very little TGF-β. Treg cells cultured in the presence of supernatants from breast-cancer tissues showed an increase in IL-10 production accompanied by enhanced recruitment of HAT1 to the Il10 promoter. HAT1 predominantly acetylates histone H4 at K5 and K12 residues and to a lesser extent H2A at K5 residues in the Il10 locus. In HAT1-silenced tumor Tregs, histone acetylation at the IL-10 locus drops to a level that’s comparable to that of control Tregs, indicating that enhancement of histone acetylation in the IL-10 locus in tumor-associated Tregs is mainly mediated by HAT1[40]. In contrast, the deacetylase HDAC11 inhibits IL-10 production in macrophages in response to LPS. IL-12 expression, however, is not inhibited by HDAC11, illustrating that HDAC11 targets a specific gene program rather than mediating global suppression. HDAC11 physically interacts with a distal region (−807 to −1653bp) of the Il10 promoter, and interestingly, addition of this particular DNA element to the proximal Il12 promoter is sufficient to confer susceptibility to HDAC11-mediated inhibition. HDAC11 inhibits acetylation of both H3 and H4 proteins in the Il10 locus and hinders binding of the IL-10 promoting transcription factors including Sp and STAT3. In the meanwhile, it promotes the binding of transcriptional suppressor PU.1[41]. As HDACs don’t have a sequence specific DNA binding domain, it’s unclear how HDAC11 activity is targeted to a specific region in the Il10 locus. Additional HDACs likely play a role in regulation of IL-10 expression. The HDAC1–3 inhibitor MS-275 enhances IL-10 in macrophages and reduces cigarette smoke-induced airway inflammation in mice[42]; HDAC inhibitor TsA targeting HDAC 1, 3, 5, 6, 10 enhances IL-10 production in Th1 cells as well as B cells[43,44]. Unexpectedly, HDAC6 was found to promote IL10 in macrophages and dendritic cells (DCs) in response to LPS[41,45,46]. It’s unknown if the deacetylase activity is required in this process, but one study showed that HDAC6 can bind to STAT3 and promote its phosphorylation, which could potentially influence IL-10 production[46]. Another study found that HDAC6 promotes PD-L1 expression in melanoma cells through a similar mechanism, where PD-L1 downregulation due to HDAC6 deficiency can be rescued by overexpression of constitutively active STAT3[47].
The bromodomain and extra-terminal domain (BET) protein family use bromodomains as “readers” of histone acetylation that serve to bridge recognition of acetylated residues in chromatin with recruitment of chromatin remodelers (i.e. SWI/SNF), histone modifiers (i.e. HAT) and transcription regulators (i.e. PTEF-b), thereby changing chromatin structure and regulating gene expression[48]. This family comprises four members: BRD2, BRD3, BRD4 and BRDT. JQ1, a BET inhibitor, inhibits binding of BRD4 and NF-κB p65 to the proximal promoter of Il10 (−200bp) and downregulates IL-10 transcription in Breg cells in response to LPS [49].
2.3.1.2. Histone methylation
Unlike histone acetylation, histone methylation is electrically neutral. Depending on the chromatin factors recruited, histone methylation can be either stimulate or suppress gene expression. For instances, H3K9me3 and H3K27me3 are associated with heterochromatin and gene silencing; H3K4me3 is highly enriched at transcriptionally active promoters; Enhancers are characterized by high H3K4me1 and low H3K4me3 marks, and active enhancers can be further distinguished by H3K27ac marks written by HAT p300/CBP; H3K36me3 is located along actively transcribed gene bodies[50].
For the suppressive marks, H3K9 methylation can be catalyzed by multiple histone methyltransferases (HMTs), including G9a, yet H3K27 methylation is solely mediated by Ezh2, the catalytic subunit of polycomb repressive complex 2 (PRC2). H3K9me3 and H3K27me3 are recognized respectively by heterochromatin protein 1 (HP1) and polycomb repressive complex 1 (PRC1) to create compacted heterochromatin[51,52]. In addition, both HP1 and Ezh2 serve as a scaffold for DNA methyltransferases (DNMTs), therefore providing a direct link between histone methylation and DNA methylation at the repressed regions[53,54]. Deficiency in Ezh2 leads to upregulation of IL-10 transcription in Th0, Th2 and iTreg cells, but not Th1 cells[55]. Jarid2 is essential for recruiting PRC2 to their target sites in the genome. In Th17 cells, Jarid2 promotes H3K27me3 and suppresses the production of effector cytokines, including IL-17A, IL-17F, IL-22 as well as IL-10. Jarid2 mRNA is limited by miR-155 during Th17 differentiation to ensure proper cytokine production[56]. Bmi1 is a component of PRC1 that’s essential for the H2A ubiquitin E3 ligase activity and repressive function of PRC1 on gene expression. Activation of TLR4 in macrophages induces Bmi1 and it limits IL-10 expression[57]. In reverse, Jmjd3, a H3K27 demethylase that converts H3K27me3 to H3K27me1, controls M2 macrophage polarization[58] and is required for IL-10 induction in foamy macrophages during mycobacterial infection[59].
2.4. Regulation of IL-10 by DNA methylation
DNA methylation mediates chromatin inaccessibility and suppresses gene expression. The most widely studied type of DNA methylation 5-methylcytosine (5mC) is catalyzed and maintained by DNA methyltransferases (DNMTs), and TET cytosine dioxygenase family members are critical for DNA demethylation through oxidation of 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)[60,61]. The demethylation intermediate 5hmC marks active DNA demethylation and is associated with putative regulatory elements for signature genes that are turned on during T helper cell differentiation. Tet2 deficient T cells have defects in expression of signature cytokines in in-vitro differentiated Th1 and Th17 cells. In addition, Tet2 is required for DNA demethylation and expression of Il10 in both Th1 and Th17 cells. In Th17 cells, Tet2 is recruited to Il10 locus by STAT3. Tet2 conditional knockout mice driven by CD2-Cre develop more severe EAE due to the dominance of IL-10 deficiency. When IL-10 signaling is blocked with anti-IL-10R, Tet2 conditional knockout mice produce less of the Th1 and Th17 effector cytokines in vivo, and instead develop less severe EAE compared to IL-10R antibody treated control animals [62].
3. Transcription factors as the conductor of chromatin regulation
3.1. Pioneer transcription factors set up the chromatin landscape of Il10
The unique ability of transcription factors to bind specific DNA sites in a sequence-dependent manner with their DNA-binding domain puts them in control of epigenetic regulation, allowing them to directly recruit chromatin remodeling complexes and histone- modifying enzymes to their specific targets. A subset of transcription factors are capable of binding to target sequences occluded in nucleosomal DNA within closed chromatin and of initiating chromatin remodeling events that create nucleosome-free chromatin regions that allow further binding of additional regulatory factors. These transcription factors are termed “pioneer factors” as they are the first to engage chromatin during transitions in cell fate or cell status and to determine the chromatin landscape of accessible regulatory elements in a given cell type[63].
Naïve CD4 T cells lack open chromatin regions in the vicinity of the Il10 gene as measured by DNase I hypersensitivity assays[64]. However, in all differentiated T helper cells, the Il10 locus is in a transcriptionally competent state characterized by the presence of chromatin accessible regions by ATAC-seq ([65], and our data), enrichment of the active histone marker H3K4me3, and absence of the repressive histone marker H3K27me3[66,67]. Pioneer transcription factors are essential for chromatin remodeling that allows for poised or active IL10 transcription during the differentiation of various T helper cells.
The role of pioneer transcription factors in opening up the Il10 locus for transcription is best demonstrated in the context of Tr1 cells. Differentiation of naïve CD4 T cells into IL-10 producing Tr1 cells is induced by IL-27 signaling through STAT1 and STAT3[68–71]. IRF1 and Batf are induced by STAT1 and STAT3 respectively as early as 2 hours upon T cell activation in Tr1 cells, and act as pioneer factors for Tr1 cell differentiation. Deficiency of either IRF1 or Batf disables IL-10 production in Tr1 cells and impairs the suppressive function of Tr1 cells in autoimmunity in vivo. ATAC-seq analysis showed that Batf deficiency alters the accessibility of ~20,000 genomic loci in Tr1 cells, including the majority of loci that distinguish Tr1 cells from Th0 cells. IRF1, although having a more restricted effect, regulates accessibility of a distinct set of ~1200 Tr1 unique loci, most of which are not affected by Batf. In contrast, Ahr and cMaf, two other IL-10 regulators in Tr1 cells, don’t have much influence on chromatin accessibility. Furthermore, cMaf cannot induce IL-10 in the absence of IRF1 or Batf, nor can it bind to the Il10 locus, indicating that chromatin remodeling mediated by IRF1 and Batf is an indispensable step for IL-10 induction by other transcription factors in Tr1 cells[72]. Both IRF1 and Batf directly bind to the Il10 locus, but Batf lacks a transactivation domain[73] and only IRF1 can transactivate the Il10 promoter. Deficiency of either IRF1 and Batf limits binding of the other transcription factor to the Il10 locus, suggesting a cooperative binding mechanism [72]. It is, however, unclear how IRF1 and Batf contribute to regulation of the other unique loci in Tr1 cells, which may also directly or indirectly influence IL-10 production.
IL-10 production is characteristic of nonpathogenic immune-regulatory Th17 cells[74]. IL-6 and TGF-β synergistically induce IL-10 and generate nonpathogenic Th17 cells in a STAT3 dependent manner[68]. Cooperation between Batf and IRF4, another IRF family member, is required to set up the epigenetic landscape of Th17 cells. The master transcription factor of Th17 cells, RORγt, however, has a surprisingly small impact on Th17 related epigenetic features[67]. IRF4 itself binds DNA weakly due to a carboxy-terminal auto-inhibitory domain. Batf heterodimerizes with Jun proteins to form a complex that facilitates IRF4 binding to AP1–IRF composite elements (AICEs) in their target genes, including Il10[73,75]. Batf/Jun and IRF4 complexes cooperatively promote Il10 transcription in Th17 cells[76]. In contrast to IRF1/Batf interactions in Tr1 cells, IRF4 and Batf have a striking overlap in their genomic binding sites and deficiency in either affects chromatin accessibility of a largely shared sets of loci[67], indicating that the global effect of IRF4 is more dependent on Batf than IRF1. The specific signaling pathway that drives Batf/IRF4 expression in Th17 or Tr1 cells is not well addressed. Both Batf and IRF4 can be induced by TCR activation[77–79]. Besides Th17 cells, IRF4 regulates IL-10 in Th2[80,81] and Tregs cells[82], and Batf is also required for Th2 differentiation[83]. To what extent IRF4/Batf is involved in chromatin remodeling in these cell types is unknown.
A similar role for pioneering factors was defined in Th1 and Th2 cells in which STAT4 and STAT1 (for Th1 cells) and STAT6 (for Th2 cells) initiate lineage specification by establishing the chromatin landscape. Expression of STAT proteins in Th1 and Th2 cells is required for p300 binding to lineage specific enhancers and for characteristic chromatin markings, including H3K4me1. Overexpression of T-bet, the master transcription factor for Th1 cells, only restored 23% of active enhancers marked by histone acetyltransferase p300 that are lost in STAT4-deficient Th1 cells[84]. More specifically, STAT4 is required for IL-10 production in Th1 cells[85]. Although STAT6 and GATA3 overall follow a similar rule in Th2 cells, GATA3 has a greater capacity for remodeling the enhancer landscape than other lineage defining master transcription factors[84]. Overexpressing GATA3 in STAT6-deficient Th2 cells can rescue half of the active enhancer sites including those in the Il4-Il13 locus[66]. GATA3 can induce H3 and H4 acetylation and increase chromatin accessibility in the Il10 locus even in the absence of IL-4 signaling[86]. This is consistent with the notion of the GATA family as pioneer factors that can bind nucleosomal DNA and open chromatin[87]. GATA3 deficiency leads to reduced IL-10 in Th2 cells [88–90], but GATA3 doesn’t transactivate the Il10 promoter[66], even in the presence of multiple known enhancers (data not shown), indicating that GATA3 regulates IL-10 transcription mainly by setting the stage for recruitment of other transcription factors.
The above-mentioned studies all support hierarchical roles for different transcription factors to act in a precise temporal order for the epigenetic reprogramming and transcriptional activation during T helper cell differentiation and concurrent IL-10 induction. These studies underscore the profound effects of pioneer transcription factors in shaping the transcriptional program in T cell differentiation. Although the current data with genetic approaches support the role of Batf, IRF1, IRF4 and several STATs as pioneer factors, two key molecular properties must still be addressed: 1) Can these factors bind directly to nucleosomal DNA in heterochromatin? 2) Do they recruit chromatin modifiers and what are the chromatin modifiers they interact with? One preliminary study suggests that Batf can recruit CTCF to affect chromatin organization[91]. As both STAT4 and STAT6 have been proposed as pioneer factors, it would be reasonable to consider the possibility that STAT3 may have a similar function in Tr1 and Th17 cells. However, a comparison of the role of STAT3 in regulating the epigenetic landscape as compared to IRF1/Batf in Tr1 cells and IRF4/Batf in Th17 cells is still lacking.
3.2. Other transcription factors that modulate the chromatin status of Il10
Transcription factors other than pioneer factors can also modify the chromatin status of Il10 by recruiting various effector proteins using their protein-interaction domains. For example, Foxp3 is required for IL-10 production in tumor-associated Treg cells[40]. Foxp3 doesn’t appear to establish new enhancers as it is overwhelmingly bound to enhancers that pre-exist in precursor cells[92]. The Il10 promoter lacks a binding site for Foxp3, and Foxp3 decoy oligonucleotides (dODN) do not affect Il10 expression, indicating that regulation of IL-10 by Foxp3 is independent of direct DNA binding. Instead, Foxp3 promotes IL10 transcription by recruiting HAT1 complexes, which predominantly acetylate histone H4 at K5 and K12 residues at the STAT binding site of the Il10 promoter, making it more permissive to binding by phosphorylated STAT3[40]. Moreover, Nfil3, a transcription factor that contributes to IL-10 production in Th1, Th2, Treg as well as NKT cells, can promote H3 acetylation in the Il10 locus, although the HAT involved has not been identified[93]. A reverse example is that Ets1, a suppressor of IL-10 in Th1 and Th2 cells, promotes recruitment of HDAC1 to inhibit histone H3 acetylation at the Il10 promoter and enhancer regions, including at sites HSS+1.65kb and HSS+6.45kb, and thus reduces chromatin accessibility in the Il10 locus[43,94].
4. Recent progress on transcriptional regulation of IL-10
As the role of transcription factors in regulating IL-10 in various cell types has been extensively reviewed elsewhere[95–99], here we will focus on the more recent discoveries and their implications.
4.1. Transcriptional regulation of IL-10 in CD4 T cells
Given the potential clinical significance for Tr1 cells and IL-10, progress has been made in elucidating the detailed transcriptional circuits for IL-10 regulation in Tr1 cells. Egr2 was previously known to regulate T cell anergy and was recently demonstrated to be required for IL-10 production induced by IL-27. IL-27 induces Egr2 expression in a STAT3-dependent but STAT1-independent manner. Mechanistically, Egr2 doesn’t bind the Il10 promoter. It instead binds to and transactivates the promoter for Prdm1[100], which drives IL-10 production in response to IL-27 in both CD4 and CD8 T cells[101–103]. Eomes, on the other hand, acts downstream of Prdm1 and can promote IL-10 in mice through direct binding to the Il10 promoter [104]. Eomes can also induce IL-10 in human CD4 T cells, although a direct binding site of Eomes in the human Il10 locus has not been identified[105]. Aerobic glycolysis was found to be essential for Tr1 differentiation. At early time points, Hif1-α transcriptionally reprograms the metabolic status of the cell towards aerobic glycolysis to support Tr1 differentiation. However, it destabilizes Ahr and therefore inhibits IL-10 production during later time points[106].
Environmental cues can impact the differentiation and function of Tr1 cells, which may have clinical implications. A negative correlation between disease exacerbations and seasonal changes in melatonin levels has been observed in MS patients. Administration of melatonin to mice with EAE, a model of MS, ameliorated disease, at least in part by regulating the differentiation and function of Tr1 cells. Binding of melatonin to its membrane receptor MTNR1A activated p-Erk1/2, and melatonin binding to the nuclear receptor Ror-α promoted its binding to the Il10 promoter. Both p-Erk1/2 and Ror-α activation promoted the transcription of cMaf and Ahr, and Ror-α in turn synergized with Ahr and cMaf to transactivate the Il10 promoter[107].
In Th17 cells, the regulation of IL-10 and IL23R expression by multiple transcription factors was linked with Th17 cell pathogenicity. The master transcription factor Rorγt can exhibit a differential binding preference for the Il23r versus Il10 locus depending on availability of Rorγt ligands. Polyunsaturated fatty acids increased Rorγt binding to the Il10 locus, whereas saturated fatty acids decreased it. CD5L binds to fatty acid synthase and modulates the intracellular lipidome to promote Il10 production and the nonpathogenic phenotype in Th17 cells[108,109]. RBPJ is a key transcription factor that drives IL23r expression in Th17 cells, and contributes to the pathogenicity of Th17 cells by repressing IL-10 production induced by cMaf [110].
4.2. Transcriptional regulation of IL-10 in B cells
It is now recognized that B cell have indispensable regulatory roles in immune responses. IL-10-producing regulatory B (B10) cells are the most widely studied Breg subset, and their regulatory function is dependent upon IL-10. BCR signaling, CD40 ligation, Tim-1 ligation[111], and activation by TLR ligands have all been implicated in B10 development and IL-10 secretion from B cells. However, a transcriptional program that defines this population remains unknown[5,112]. Limited data link transcription factors to the regulation of IL-10 in B cells. For instance, conditional knockout of HIF-1α in B cells leads to a diminished IL-10-producing CD1dhiCD5+ Breg cell population. These mice display exacerbated collagen-induced arthritis (CIA) as well as EAE, which can be rescued by adoptive transfer of Hif1a-deficient CD1dhiCD5+ B cells with ectopic IL-10 expression. These results indicate that regulation of IL-10 by Hif1α is essential for the suppressive function of CD1dhiCD5+ Breg cells. HIF-1α binds to hypoxia-responsive element (HRE) sites in the Il10 locus and transactivates Il10. Additionally, HIF-1α physically interacts with p-STAT3 at the Il10 locus, although the functional consequence of this interaction is still unclear[113]. In addition, cMaf expression is associated with IL-10 production in B cells both in vitro and in vivo in the CIA model. Knocking-down cMaf expression downregulated IL- 10 expression in LPS- stimulated B cells in vitro, but the role of cMaf in regulating B10 cells in vivo remains to be investigated[114]. Conversely, mice with a conditional NFATc1 deficiency present with an increased number of IL-10 producing CD1dhiCD5+ Breg cells, develop milder EAE[115], and display attenuated inflammation in the imiquimod induced model of psoriasis[44]. NFATc1 binds to the Il10 gene and dampens IL-10 expression in association with HDAC1 upon imiquimod stimulation[44].
4.3. Transcriptional regulation of IL-10 in ILCs
ILCs are innate counterparts of adaptive CD4 T helper cells that are functionally defined discrete lineages (ILC1, ILC2, ILC3) mirroring Th1, Th2 and Th17 cells respectively. They share similar cytokine profiles with corresponding CD4 T cell subsets and are specified by the same master transcription factors[116]. A Foxp3 expressing ILC counterpart of Treg cells has not been identified. However, a newly discovered ILC population with regulatory properties (ILCreg) is characteristic of, and functionally dependent on production of IL-10[6]. Otherwise,IL-10 has only been reported in ILC2s, which are a major cellular source of IL-10 in the lung upon stimulation by IL-33 or chronic exposure to the allergen papain[117,118]. One distinctive feature of transcriptional regulation in ILCs is that the cis-regulatory elements that control signature cytokine expression are already poised or active in mature ILCs, whereas identical regions in naïve CD4 T cells only become accessible upon activation[119]. ILCregs are thought to develop from a distinct pathway from other ILC subsets that is dependent on Id3. It would be intriguing to investigate if the Il10 locus in ILCregs is already imprinted by Id3 or other transcription factors during development. Transcriptional mechanisms that control the dynamics of IL-10 production in ILCs is a new field that awaits exploration.
4.4. cMaf/Bhlhe40 orchestrate a shared mechanism for IL-10 regulation across cell types
Some aspects of transcriptional regulation of the IL-10 locus are shared among different cellular subsets. In T cells, TCR activation, CD28 co-stimulation and polarizing cytokine signaling all contribute to regulation of IL-10 transcription. Strong and chronic TCR stimulation favors IL-10 production via activation of p-Erk1/2 signaling[85,120], which then induces the expression of the IL-10 promoting transcription factor cMaf. p-Erk1/2 activation is required for IL-10 production in Th1, Th2 as well as Th17 cells. In addition, signaling of polarizing cytokine through various STATs (STAT4 in Th1 cells, STAT6 in Th2 cells, STAT3 in Th17 and Tr1 cells) can contribute to IL-10 production by generating a poised chromatin structure at the Il10 locus, and they can all induce cMaf as well[40]. cMaf is a common node for IL-10 regulation in all T helper cell subsets both in vitro and in vivo[121], including in Th1[85,101], Th2[122], Th17[74,123], Tr1[124,125] and Treg[126,127] cells. In contrast, a co-stimulatory signal via CD28 keeps IL-10 in check by inducing the transcription factor Bhlhe40. In the absence of Bhlhe40, CD4 T cells display a surge in IL-10 production and lose their effector function. Bhlhe40 deficient CD4 T cells are unable to induce EAE or clear infections such as Mycobacterium tuberculosis and Toxoplasma gondii, a phenotype that can be rescued by IL-10 blockade[128–130]. Suppression of IL-10 transcription by Bhlhe40 is also a shared mechanism in Th1, Th2, and Th17 cells[131]. Interestingly, Bhlhe40 and cMaf inversely regulate each other[121,132], suggesting that the balance between cMaf and Bhlhe40 serves as critical molecular switch for IL-10 production across all T helper cell subsets.
Beyond T cells, cMaf is required for IL-10 expression in B cells [114] and macrophages[133], and Bhlhe40 suppresses IL-10 production in myeloid cells[130]. It is therefore very likely that the cMaf/Bhlhe40 balance may regulate IL-10 regulation in other immune cell types, although we currently have a limited understanding about the signaling pathways that regulate cMaf and Bhlhe40 expression in contexts other than T cells.
5. Concluding remarks
As multiple immune cell types can produce IL-10, this raises the question of how IL-10 expression is regulated in each of these cellular contexts. A unique transcription factor that defines the lineage of IL-10 producing Tr1 or B10 cells has not been identified. As described above, shared mechanisms such as cMaf may contribute to IL-10 expression in these cell types. Additionally, unlike signature cytokines of T helper cell subsets that are predominantly controlled by one master transcription factor, as is the case for T-bet to IFN-γ, GATA3 for IL-4, and RORs for IL-17A, the regulation of IL-10 involves more complex interactions between different signaling pathways and transcription factors. Although cMaf by itself can transactivate Il10, cooperation with additional transcription factors such as Ahr and Ror-α is required for robust expression[107,125]. A number of additional activating and suppressing transcription factors work together with cMaf to integrate information from environmental and metabolic stimuli, and fine tune the expression of IL-10 in a context dependent manner. It’s likely that IL-10 expression represents more of a functional state rather than a discrete cell lineage, as stabilization of cell lineage often requires suppression of other cell fates mediated by master transcription factors. However, master transcription factors of T helper cell subsets, including T-bet[104,134], GATA3[86], and RORγt[108], do not inhibit but rather promote IL-10, and the transcriptional program for IL-10 regulation can co-exist with the effector cell program in the same cell type.
IL-10 expression is tightly regulated at multiple levels, including mechanisms involving chromatin structure, histone modifications, DNA methylation and active transcription factors, in a time- and context-restricted manner. More than 20 transcription factors have been shown to directly activate IL-10[95–99], including: Specificity factors (Sps), Signal Transducers and Activators of Transcription (STATs), Activator Proteins (AP-1), CCAAT/Enhancer Binding Proteins (C/EBPs), cAMP Response Element Binding Protein (CREB), Interferon Regulatory Factors (IRFs), Nuclear Factor-kappa B (NF-κB), cMaf, Prdm1, Hif1-α, Ahr, Nfil3, GATA3, ROR-α, RORγt, etc. The cMaf/Bhlhe40 axis is critical for IL-10 regulation in all T helper cells and potentially many other cell types including myeloid cells and B cells, yet it is not clear as to what extent the other transcription factors are playing a role among different cell types. It’s largely unknown how cMaf and other transcription factors interact with each other in the regulation of IL-10. Although a number of studies have revealed a role for epigenetic regulation of IL-10 expression, so far this has been shown only in a limited number of cellular contexts. Additional studies will be needed to connect specific chromatin remodeling complexes or histone modifiers with the specific transcription factors that target them to the Il10 locus in each specific cellular context.
It will be important to understand how the stability of IL-10 expression is influenced by epigenetic mechanisms in different cellular contexts. This question is of particular value for clinical applications of IL-10 producing regulatory cells. One study showed that repeatedly stimulated Th2 cells develop memory for IL-10, which is associated with H4 acetylation. In contrast, Th1 cells failed to obtain H4 acetylation during repeated stimulation and remained dependent on IL-12 for IL-10 expression[120]. The epigenetic mechanisms maintaining IL-10 expression in specialized IL-10 producing Tr1, B10 and ILCregs requires further investigation. A more comprehensive understanding of all the cis- and trans- regulatory elements for IL-10 will undoubtedly inform development of therapeutic strategies to modulate IL-10 production in inflammatory diseases.
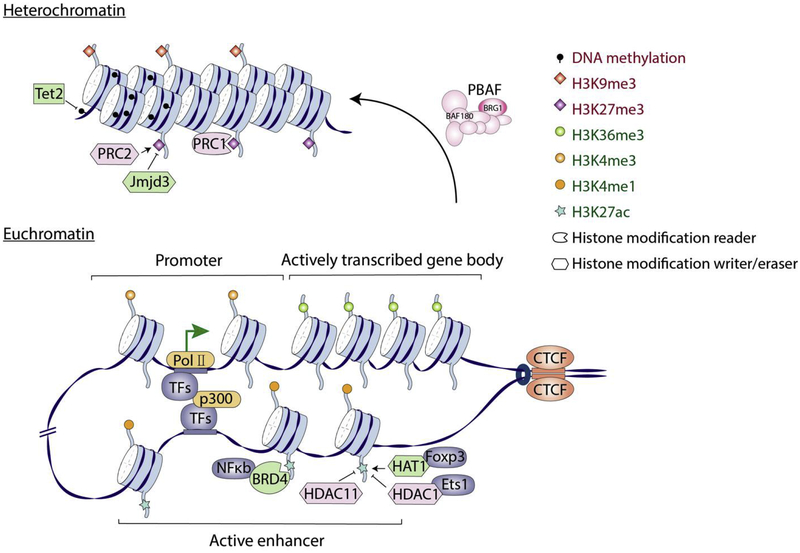
Figure 1. Epigenetic mechanisms for IL-10 regulation.
Heterochromatin and euchromatin are states of the chromatin that is respectively transcriptional silent or active. Epigenetic mechanisms regulate transcription of Il10 gene by influencing the chromatin status. 1) ATP-dependent chromatin remodelers regulate how tightly DNA is packaged around nucleosomes and accessibility of transcription machinery to the gene. Specifically, the PBAF chromatin remodeling complex containing the BAF180 subunit inhibits Il10. 2) Histone modifications affect chromatin structure. Histone acetylation enhances chromatin accessibility and promotes transcription. Foxp3 recruits acetylation “writer” HAT1 to promote Il10 transcription, while Ets1 recruits “eraser” HDAC1 to suppress Il10. HDAC11 also inhibits Il10 expression. The histone acetylation “reader” BRD4 recruits NFκb to promote Il10. In contrast, H3K9me3 and H3K27me3 are repressive histone markers. H3K27me3 is written by PRC2 complex and then recognized by PRC1 to close the chromatin and suppress Il10. Jmjd3 demethylates H3K27me3 and promotes Il10 transcription. 3) DNA methylation mediates gene inaccessibility and inhibit transcription. Tet2 is an DNA demethylation enzyme that promotes Il10 expression. 4) CTCF demarcates boundaries of topologically associating domains (TADs) and defines the scope of active promoter- enhancer interactions.
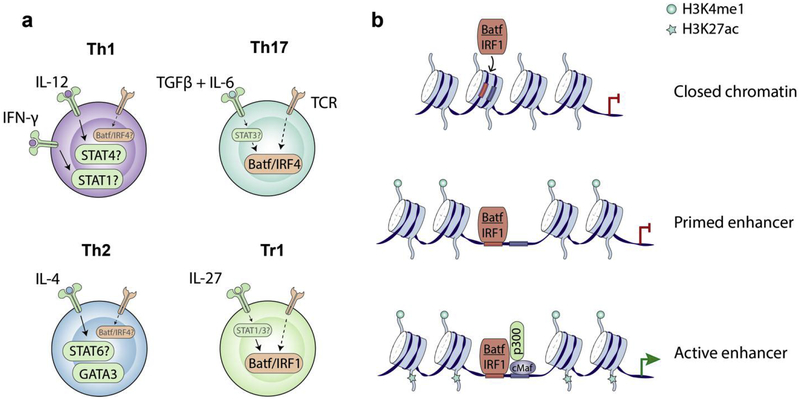
Figure 2. Pioneer transcription factors set up chromatin landscape for lineage specification and IL-10 expression in T helper cells.
a) Pioneer transcription factors are critical regulators of cell fate transition in that they possess unique capabilities to bind nucleosomal DNA and open the chromatin to allow binding of other lineage specific transcription factors. Il10 locus is closed in naïve T cells but all differentiated T helper cells can express IL-10. In Tr1 cells, IRF1 and Batf are induced by STAT1 and STAT3 signaling respectively and are required to set up the accessible chromatin landscape; whereas IRF4 instead of IRF1, together with Batf, are required in Th17 cells. It’s likely that Batf/IRF complexes play a similar role in Th1 and Th2 cells. STAT proteins induced by polarizing cytokines determine the deposition of p300 and H3K4me1 at cell lineage specific enhancers in Th1 and Th2 cells. Their roles in influencing chromatin accessibility remain to be investigated. Unlike other master transcription factors, GATA3 has the chromatin remodeling capability and can increase chromatin accessibility in the Il10 locus.
b) Batf/IRF1 complex opens up Il10 locus to allow binding of transcription factors such as cMaf, which further transactivates Il10 gene transcription in Tr1 cells.
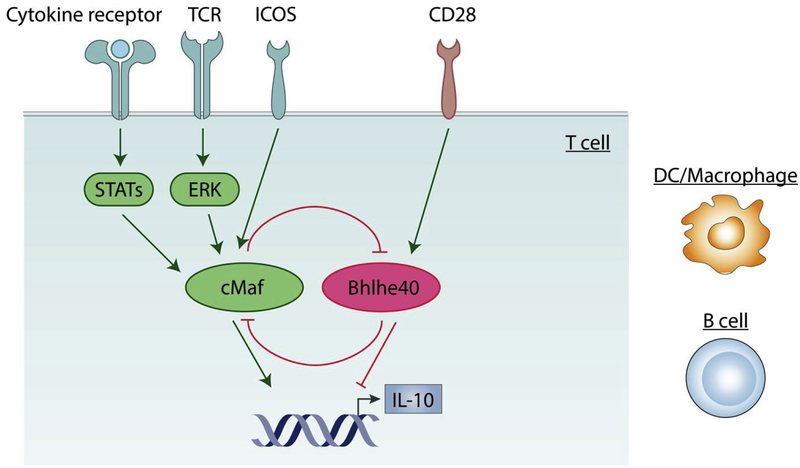
Figure 3. cMaf/Bhlhe40 as a shared mechanism for IL-10 regulation across cell types.
cMaf is required for IL-10 production in all T helper cells including Th1, Th2, Th17, Tr1 and Treg cells. Both TCR induced ERK activation and cytokine induced STAT signaling can induce cMaf expression in T cells. In addition, the co-stimulatory molecule ICOS promotes IL-10 in T helper cells via induction of cMaf [135–137]. Inversely, co-stimulation signal mediated by CD28 upregulates Bhlhe40, a potent inhibitor of IL-10 in Th1, Th2, and Th17 cells. cMaf and Bhlhe40 also antagonize the expression of each other. cMaf/Bhlhe40 axis may represent a core mechanism for IL-10 regulation that’s shared among different cell types because their function has also been implicated in myeloid cells and B cells.
Acknowledgement
We are most grateful to Mary Collins for advice and editing of the manuscript. The work presented in this review was funded by NIH grants (5P01AI073748-10, 5P01AI056299-15, 5P01AI039671-21, 1P01AI129880-01A, 5R01NS030843-27, 4R01NS045937-14)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Ouyang W, O’Garra A. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 2019;50:871–91. doi: 10.1016/j.immuni.2019.03.020. [DOI] [PubMed] [Google Scholar]
- [2].Roncarolo MG, Gregori S, Bacchetta R, Battaglia M, Gagliani N. The Biology of T Regulatory Type 1 Cells and Their Therapeutic Application in Immune-Mediated Diseases. Immunity 2018;49:1004–19. doi: 10.1016/j.immuni.2018.12.001. [DOI] [PubMed] [Google Scholar]
- [3].Pot C, Apetoh L, Awasthi A, Kuchroo VK. Induction of regulatory Tr1 cells and inhibition of T(H)17 cells by IL-27. Semin Immunol 2011;23:438–45. doi: 10.1016/j.smim.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pot C, Apetoh L, Kuchroo VK. Type 1 regulatory T cells (Tr1) in autoimmunity. Semin Immunol 2011;23:202–8. doi: 10.1016/j.smim.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rosser EC, Mauri C. Regulatory B cells: origin, phenotype, and function. Immunity 2015;42:607–12. doi: 10.1016/j.immuni.2015.04.005. [DOI] [PubMed] [Google Scholar]
- [6].Wang S, Xia P, Chen Y, Qu Y, Xiong Z, Ye B, et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell 2017;171:201–216.e18. doi: 10.1016/j.cell.2017.07.027. [DOI] [PubMed] [Google Scholar]
- [7].Zhu L, Shi T, Zhong C, Wang Y, Chang M, Liu X. IL-10 and IL-10 Receptor Mutations in Very Early Onset Inflammatory Bowel Disease. Gastroenterology Res 2017;10:65–9. doi: 10.14740/gr740w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gazzinelli RT, Wysocka M, Hieny S, Scharton-Kersten T, Cheever A, Kühn R, et al. In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J Immunol 1996;157:798–805. [PubMed] [Google Scholar]
- [9].Bettelli E, Das MP, Howard ED, Weiner HL, Sobel RA, Kuchroo VK. IL-10 is critical in the regulation of autoimmune encephalomyelitis as demonstrated by studies of IL-10- and IL-4-deficient and transgenic mice. J Immunol 1998;161:3299–306. [PubMed] [Google Scholar]
- [10].Finnegan A, Kaplan CD, Cao Y, Eibel H, Glant TT, Zhang J. Collagen-induced arthritis is exacerbated in IL-10-deficient mice. Arthritis Res Ther 2003;5:R18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yin Z, Bahtiyar G, Zhang N, Liu L, Zhu P, Robert ME, et al. IL-10 regulates murine lupus. J Immunol 2002;169:2148–55. doi: 10.4049/jimmunol.169.4.2148. [DOI] [PubMed] [Google Scholar]
- [12].Redford PS, Murray PJ, O’Garra A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol 2011;4:261–70. doi: 10.1038/mi.2011.7. [DOI] [PubMed] [Google Scholar]
- [13].Ishida H, Muchamuel T, Sakaguchi S, Andrade S, Menon S, Howard M. Continuous administration of anti-interleukin 10 antibodies delays onset of autoimmunity in NZB/W F1 mice. J Exp Med 1994;179:305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 2017;18:407–22. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 2013;5. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ho L, Crabtree GR. Chromatin remodelling during development. Nature 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol 2009;19:120–6. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev 2006;20:282–96. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009;138:129–45. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 2003;19:169–82. [DOI] [PubMed] [Google Scholar]
- [21].Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med 2006;203:1493–505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wurster AL, Pazin MJ. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol Cell Biol 2008;28:7274–85. doi: 10.1128/MCB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chaiyachati BH, Jani A, Wan Y, Huang H, Flavell R, Chi T. BRG1-mediated immune tolerance: facilitation of Treg activation and partial independence of chromatin remodelling. EMBO J 2013;32:395–408. doi: 10.1038/emboj.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wurster AL, Precht P, Becker KG, Wood WH, Zhang Y, Wang Z, et al. IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme. BMC Immunol 2012;13:9. doi: 10.1186/1471-2172-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Johanson TM, Chan WF, Keenan CR, Allan RS. Genome organization in immune cells: unique challenges. Nat Rev Immunol 2019. doi: 10.1038/s41577-019-0155-2. [DOI] [PubMed] [Google Scholar]
- [26].Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet 2013;14:288–95. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, et al. CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Mol Cell 2017;67:1049–1058.e6. doi: 10.1016/j.molcel.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016;529:110–4. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015;161:1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu G, Cui K, Fang D, Hirose S, Wang X, Wangsa D, et al. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity 2018;48:227–242.e8. doi: 10.1016/j.immuni.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng H-L, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature 2011;477:424–30. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol 2007;8:378–87. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- [33].Zhao L, Frock RL, Du Z, Hu J, Chen L, Krangel MS, et al. Orientation-specific RAG activity in chromosomal loop domains contributes to Tcrd V(D)J recombination during T cell development. J Exp Med 2016;213:1921–36. doi: 10.1084/jem.20160670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bunting KL, Soong TD, Singh R, Jiang Y, Béguelin W, Poloway DW, et al. Multi-tiered Reorganization of the Genome during B Cell Affinity Maturation Anchored by a Germinal Center-Specific Locus Control Region. Immunity 2016;45:497–512. doi: 10.1016/j.immuni.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Johanson TM, Lun ATL, Coughlan HD, Tan T, Smyth GK, Nutt SL, et al. Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat Immunol 2018;19:1257–64. doi: 10.1038/s41590-018-0234-8. [DOI] [PubMed] [Google Scholar]
- [36].Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- [37].Wu C, Chen Z, Dardalhon V, Xiao S, Thalhamer T, Liao M, et al. The transcription factor musculin promotes the unidirectional development of peripheral Treg cells by suppressing the TH2 transcriptional program. Nat Immunol 2017;18:344–53. doi: 10.1038/ni.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ribeiro de Almeida C, Heath H, Krpic S, Dingjan GM, van Hamburg JP, Bergen I, et al. Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J Immunol 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- [39].Nikolic T, Movita D, Lambers MEH, Ribeiro de Almeida C, Biesta P, Kreefft K, et al. The DNA-binding factor Ctcf critically controls gene expression in macrophages. Cell Mol Immunol 2014;11:58–70. doi: 10.1038/cmi.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hossain DMS, Panda AK, Manna A, Mohanty S, Bhattacharjee P, Bhattacharyya S, et al. FoxP3 acts as a cotranscription factor with STAT3 in tumor-induced regulatory T cells. Immunity 2013;39:1057–69. doi: 10.1016/j.immuni.2013.11.005. [DOI] [PubMed] [Google Scholar]
- [41].Villagra A, Cheng F, Wang H-W, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Leus NGJ, van den Bosch T, van der Wouden PE, Krist K, Ourailidou ME, Eleftheriadis N, et al. HDAC1–3 inhibitor MS-275 enhances IL10 expression in RAW264.7 macrophages and reduces cigarette smoke-induced airway inflammation in mice. Sci Rep 2017;7:45047. doi: 10.1038/srep45047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lee C-G, Kwon H-K, Sahoo A, Hwang W, So J-S, Hwang J-S, et al. Interaction of Ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J Immunol 2012;188:2244–53. doi: 10.4049/jimmunol.1101614. [DOI] [PubMed] [Google Scholar]
- [44].Alrefai H, Muhammad K, Rudolf R, Pham DAT, Klein-Hessling S, Patra AK, et al. NFATc1 supports imiquimod-induced skin inflammation by suppressing IL-10 synthesis in B cells. Nat Commun 2016;7:11724. doi: 10.1038/ncomms11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cheng F, Lienlaf M, Perez-Villarroel P, Wang H-W, Lee C, Woan K, et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol Immunol 2014;60:44–53. doi: 10.1016/j.molimm.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cheng F, Lienlaf M, Wang H-W, Perez-Villarroel P, Lee C, Woan K, et al. A novel role for histone deacetylase 6 in the regulation of the tolerogenic STAT3/IL-10 pathway in APCs. J Immunol 2014;193:2850–62. doi: 10.4049/jimmunol.1302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lienlaf M, Perez P. Essential role of HDAC6 in the regulation of PD- L1 in melanoma. Villarroel n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hsu SC, Blobel GA. The role of bromodomain and extraterminal motif (BET) proteins in chromatin structure. Cold Spring Harb Symp Quant Biol 2017;82:37–43. doi: 10.1101/sqb.2017.82.033829. [DOI] [PubMed] [Google Scholar]
- [49].Lee MB, Lee J-H, Hong SH, You JS, Nam ST, Kim HW, et al. JQ1, a BET inhibitor, controls TLR4-induced IL-10 production in regulatory B cells by BRD4-NF-κB axis. BMB Rep 2017;50:640–6. doi: 10.5483/bmbrep.2017.50.12.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gates LA, Foulds CE, O’Malley BW. Histone marks in the “driver’s seat”: functional roles in steering the transcription cycle. Trends Biochem Sci 2017;42:977–89. doi: 10.1016/j.tibs.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Laugesen A, Højfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med 2017;49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- [54].Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron P-F. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenetics 2018;10:17. doi: 10.1186/s13148-018-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang Y, Kinkel S, Maksimovic J, Bandala-Sanchez E, Tanzer MC, Naselli G, et al. The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood 2014;124:737–49. doi: 10.1182/blood-2013-12-544106. [DOI] [PubMed] [Google Scholar]
- [56].Escobar TM, Kanellopoulou C, Kugler DG, Kilaru G, Nguyen CK, Nagarajan V, et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity 2014;40:865–79. doi: 10.1016/j.immuni.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sienerth AR, Scheuermann C, Galmiche A, Rapp UR, Becker M. Polycomb group protein Bmi1 negatively regulates IL-10 expression in activated macrophages. Immunol Cell Biol 2011;89:812–6. doi: 10.1038/icb.2010.160. [DOI] [PubMed] [Google Scholar]
- [58].Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010;11:936–44. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- [59].Holla S, Prakhar P, Singh V, Karnam A, Mukherjee T, Mahadik K, et al. MUSASHI-Mediated Expression of JMJD3, a H3K27me3 Demethylase, Is Involved in Foamy Macrophage Generation during Mycobacterial Infection. PLoS Pathog 2016;12:e1005814. doi: 10.1371/journal.ppat.1005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lio C-WJ, Rao A. TET enzymes and 5hmc in adaptive and innate immune systems. Front Immunol 2019;10:210. doi: 10.3389/fimmu.2019.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Siegfried Z, Simon I. DNA methylation and gene expression. Wiley Interdiscip Rev Syst Biol Med 2010;2:362–71. doi: 10.1002/wsbm.64. [DOI] [PubMed] [Google Scholar]
- [62].Ichiyama K, Chen T, Wang X, Yan X, Kim B-S, Tanaka S, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 2015;42:613–26. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Im S-H, Hueber A, Monticelli S, Kang K-H, Rao A. Chromatin-level regulation of the IL10 gene in T cells. J Biol Chem 2004;279:46818–25. doi: 10.1074/jbc.M401722200. [DOI] [PubMed] [Google Scholar]
- [65].Miraldi ER, Pokrovskii M, Watters A, Castro DM, De Veaux N, Hall JA, et al. Leveraging chromatin accessibility for transcriptional regulatory network inference in T Helper 17 Cells. Genome Res 2019;29:449–63. doi: 10.1101/gr.238253.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wei L, Vahedi G, Sun H-W, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 2010;32:840–51. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol 2007;8:1363–71. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- [69].Awasthi A, Carrier Y, Peron JPS, Bettelli E, Kamanaka M, Flavell RA, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol 2007;8:1380–9. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- [70].Fitzgerald DC, Zhang G-X, El-Behi M, Fonseca-Kelly Z, Li H, Yu S, et al. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nat Immunol 2007;8:1372–9. doi: 10.1038/ni1540. [DOI] [PubMed] [Google Scholar]
- [71].Wang H, Meng R, Li Z, Yang B, Liu Y, Huang F, et al. IL-27 induces the differentiation of Tr1-like cells from human naive CD4+ T cells via the phosphorylation of STAT1 and STAT3. Immunol Lett 2011;136:21–8. doi: 10.1016/j.imlet.2010.11.007. [DOI] [PubMed] [Google Scholar]
- [72].Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, et al. Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat Immunol 2017;18:412–21. doi: 10.1038/ni.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- [74].Aschenbrenner D, Foglierini M, Jarrossay D, Hu D, Weiner HL, Kuchroo VK, et al. An immunoregulatory and tissue-residency program modulated by c-MAF in human TH17 cells. Nat Immunol 2018;19:1126–36. doi: 10.1038/s41590-018-0200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 2012;338:975–80. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 2012;490:543–6. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Iwata A, Durai V, Tussiwand R, Briseño CG, Wu X, Grajales-Reyes GE, et al. Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF-IRF4 transcription factor complex. Nat Immunol 2017;18:563–72. doi: 10.1038/ni.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol 2013;14:1155–65. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- [79].Allison KA, Sajti E, Collier JG, Gosselin D, Troutman TD, Stone EL, et al. Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. Elife 2016;5. doi: 10.7554/eLife.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lee C-G, Hwang W, Maeng K-E, Kwon H-K, So J-S, Sahoo A, et al. IRF4 regulates IL-10 gene expression in CD4(+) T cells through differential nuclear translocation. Cell Immunol 2011;268:97–104. doi: 10.1016/j.cellimm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- [81].Ahyi A-NN, Chang H-C, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol 2009;183:1598–606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol 2011;12:304–11. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- [83].Tussiwand R, Lee W-L, Murphy TL, Mashayekhi M, Kc W, Albring JC, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 2012;490:502–7. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Vahedi G, Takahashi H, Nakayamada S, Sun H-W, Sartorelli V, Kanno Y, et al. STATs shape the active enhancer landscape of T cell populations. Cell 2012;151:981–93. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O’Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity 2009;31:209–19. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Shoemaker J, Saraiva M, O’Garra A. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J Immunol 2006;176:3470–9. doi: 10.4049/jimmunol.176.6.3470. [DOI] [PubMed] [Google Scholar]
- [87].Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development 2018;145. doi: 10.1242/dev.164384. [DOI] [PubMed] [Google Scholar]
- [88].Pai S-Y, Truitt ML, Ho I-C. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci USA 2004;101:1993–8. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997;89:587–96. doi: 10.1016/S0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- [90].Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol 2004;5:1157–65. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- [91].Pham D, Moseley CE, Savic D, Myers R, Kee BL. Pioneer transcription factor BATF controls chromatin accessibility and CTCF-mediated chromatin architecture in CD4+ T cells 2018. [Google Scholar]
- [92].Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 2012;151:153–66. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Motomura Y, Kitamura H, Hijikata A, Matsunaga Y, Matsumoto K, Inoue H, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol 2011;12:450–9. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Grenningloh R, Kang BY, Ho I-C. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med 2005;201:615–26. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol 2010;10:170–81. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- [96].Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 2012;32:23–63. doi: 10.1615/CritRevImmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kubo M, Motomura Y. Transcriptional regulation of the anti-inflammatory cytokine IL-10 in acquired immune cells. Front Immunol 2012;3:275. doi: 10.3389/fimmu.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gabryšová L, Howes A, Saraiva M, O’Garra A. The regulation of IL-10 expression. Curr Top Microbiol Immunol 2014;380:157–90. doi: 10.1007/978-3-662-43492-5_8. [DOI] [PubMed] [Google Scholar]
- [99].Rutz S, Ouyang W. Regulation of Interleukin-10 Expression. Adv Exp Med Biol 2016;941:89–116. doi: 10.1007/978-94-024-0921-5_5. [DOI] [PubMed] [Google Scholar]
- [100].Iwasaki Y, Fujio K, Okamura T, Yanai A, Sumitomo S, Shoda H, et al. Egr-2 transcription factor is required for Blimp-1-mediated IL-10 production in IL-27-stimulated CD4+ T cells. Eur J Immunol 2013;43:1063–73. doi: 10.1002/eji.201242942. [DOI] [PubMed] [Google Scholar]
- [101].Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi M-F, Ahlers J, et al. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J Exp Med 2014;211:1807–19. doi: 10.1084/jem.20131548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat Immunol 2011;12:327–34. doi: 10.1038/ni.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Heinemann C, Heink S, Petermann F, Vasanthakumar A, Rothhammer V, Doorduijn E, et al. IL-27 and IL-12 oppose pro-inflammatory IL-23 in CD4+ T cells by inducing Blimp1. Nat Commun 2014;5:3770. doi: 10.1038/ncomms4770. [DOI] [PubMed] [Google Scholar]
- [104].Zhang P, Lee JS, Gartlan KH, Schuster IS, Comerford I, Varelias A, et al. Eomesodermin promotes the development of type 1 regulatory T (TR1) cells. Sci Immunol 2017;2. doi: 10.1126/sciimmunol.aah7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gruarin P, Maglie S, De Simone M. Eomesodermin controls a unique differentiation program in human IL- 10 and IFN- γ coproducing regulatory T cells. European Journal Of 2019. [DOI] [PubMed] [Google Scholar]
- [106].Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat Med 2015;21:638–46. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Farez MF, Mascanfroni ID, Méndez-Huergo SP, Yeste A, Murugaiyan G, Garo LP, et al. Melatonin contributes to the seasonality of multiple sclerosis relapses. Cell 2015;162:1338–52. doi: 10.1016/j.cell.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, et al. CD5L/AIM regulates lipid biosynthesis and restrains th17 cell pathogenicity. Cell 2015;163:1413–27. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, et al. Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity 2014;40:477–89. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Meyer Zu Horste G, Wu C, Wang C, Cong L, Pawlak M, Lee Y, et al. RBPJ Controls Development of Pathogenic Th17 Cells by Regulating IL-23 Receptor Expression. Cell Rep 2016;16:392–404. doi: 10.1016/j.celrep.2016.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Xiao S, Brooks CR, Sobel RA, Kuchroo VK. Tim-1 is essential for induction and maintenance of IL-10 in regulatory B cells and their regulation of tissue inflammation. J Immunol 2015;194:1602–8. doi: 10.4049/jimmunol.1402632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kalampokis I, Yoshizaki A, Tedder TF. IL-10-producing regulatory B cells (B10 cells) in autoimmune disease. Arthritis Res Ther 2013;15 Suppl 1:S1. doi: 10.1186/ar3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Meng X, Grötsch B, Luo Y, Knaup KX, Wiesener MS, Chen X-X, et al. Hypoxia-inducible factor-1α is a critical transcription factor for IL-10-producing B cells in autoimmune disease. Nat Commun 2018;9:251. doi: 10.1038/s41467-017-02683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Liu M, Zhao X, Ma Y, Zhou Y, Deng M, Ma Y. Transcription factor c-Maf is essential for IL-10 gene expression in B cells. Scand J Immunol 2018;88:e12701. doi: 10.1111/sji.12701. [DOI] [PubMed] [Google Scholar]
- [115].Bhattacharyya S, Deb J, Patra AK, Thuy Pham DA, Chen W, Vaeth M, et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin--NFAT signaling network. J Exp Med 2011;208:823–39. doi: 10.1084/jem.20100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell 2018;174:1054–66. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- [117].Seehus CR, Kadavallore A, Torre B de la, Yeckes AR, Wang Y, Tang J, et al. Alternative activation generates IL-10 producing type 2 innate lymphoid cells. Nat Commun 2017;8:1900. doi: 10.1038/s41467-017-02023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Kaye J, Seehus CR, Kadavallore A, de la Torre B. Characterization of an alternatively activated IL-10 producing innate lymphoid type-2 effector cell population 2017. [Google Scholar]
- [119].Shih H-Y, Sciumè G, Mikami Y, Guo L, Sun H-W, Brooks SR, et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell 2016;165:1120–33. doi: 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Chang H-D, Helbig C, Tykocinski L, Kreher S, Koeck J, Niesner U, et al. Expression of IL-10 in Th memory lymphocytes is conditional on IL-12 or IL-4, unless the IL-10 gene is imprinted by GATA-3. Eur J Immunol 2007;37:807–17. doi: 10.1002/eji.200636385. [DOI] [PubMed] [Google Scholar]
- [121].Gabryšová L, Alvarez-Martinez M, Luisier R, Cox LS, Sodenkamp J, Hosking C, et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells. Nat Immunol 2018;19:497–507. doi: 10.1038/s41590-018-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 1999;10:745–51. [DOI] [PubMed] [Google Scholar]
- [123].Xu J, Yang Y, Qiu G, Lal G, Wu Z, Levy DE, et al. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol 2009;182:6226–36. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Pot C, Jin H, Awasthi A, Liu SM, Lai C-Y, Madan R, et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol 2009;183:797–801. doi: 10.4049/jimmunol.0901233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 2010;11:854–61. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Neumann C, Blume J, Roy U, Teh PP, Vasanthakumar A, Beller A, et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat Immunol 2019;20:471–81. doi: 10.1038/s41590-019-0316-2. [DOI] [PubMed] [Google Scholar]
- [127].Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018;554:373–7. doi: 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Martínez-Llordella M, Esensten JH, Bailey-Bucktrout SL, Lipsky RH, Marini A, Chen J, et al. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J Exp Med 2013;210:1603–19. doi: 10.1084/jem.20122387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Yu F, Sharma S, Jankovic D, Gurram RK, Su P, Hu G, et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med 2018;215:1813–21. doi: 10.1084/jem.20170155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Huynh JP, Lin C-C, Kimmey JM, Jarjour NN, Schwarzkopf EA, Bradstreet TR, et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J Exp Med 2018;215:1823–38. doi: 10.1084/jem.20171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lin C-C, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, et al. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat Commun 2014;5:3551. doi: 10.1038/ncomms4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Jarjour NN, Schwarzkopf EA, Bradstreet TR, Shchukina I, Lin C-C, Huang SC-C, et al. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat Immunol 2019;20:687–700. doi: 10.1038/s41590-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol 2005;174:3484–92. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zhu C, Sakuishi K, Xiao S, Sun Z, Zaghouani S, Gu G, et al. An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat Commun 2015;6:6072. doi: 10.1038/ncomms7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Pot C, Jin H, Awasthi A, Liu SM, Lai CY. … induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing …. The Journal Of 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho I-C, Sharpe AH, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol 2009;10:167–75. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Nurieva RI, Duong J, Kishikawa H, Dianzani U, Rojo JM, Ho I cheng, et al. Transcriptional regulation of Th2 differentiation by inducible costimulator. Immunity 2003;18:801–11. [DOI] [PubMed] [Google Scholar]