Abstract
Introduction
Bioengineering an implantable artificial kidney (IAK) will require renal epithelial cells capable of reabsorption of salt and water. We used genome engineering to modify cells for improved Na+/H+ exchange and H2O reabsorption. The non-viral piggyBac transposon system enables genome engineering cells to stably overexpress one or more transgenes simultaneously.
Methods
We generated epitope-tagged human sodium hydrogen exchanger 3 (NHE3) and aquaporin-1 (AQP1) cDNA expressing piggyBac transposon vectors. Transgene expression was evaluated via western blot and immunofluorescence. Flow cytometry analysis was used to quantitate transporter expression in a library of genome engineered clones. Cell surface biotinylation was used evaluate surface protein localization. Blister formation assays were used to monitor cellular volumetric transport.
Results
piggyBac enabled stable transposon integration and overexpression of cumate-inducible NHE3 and/or constitutively expressing AQP1 in cultured renal (MDCK) epithelial cells. Cell surface delivery of NHE3 and AQP1 was confirmed using cell surface biotinylation assays. Flow cytometry of a library of MDCK clones revealed varying expression of AQP1 and NHE3. MDCK cells expressing AQP1 and cumate-inducible NHE3 demonstrated increased volumetric transport.
Conclusions
Our results demonstrate that renal epithelial cells an be genome engineered for enhanced volumetric transport that will be needed for an IAK device. Our results lay the foundation for future studies of genome engineering human kidney cells for renal tubule cell therapy.
Keywords: piggyBac, Transposon, Sodium hydrogen exchanger, Aquaporin, MDCK cells, Kidney
Introduction
Kidney disease affects one in seven Americans. Kidney transplant is limited by organ scarcity and dialysis offers an expensive and burdensome treatment that may delay death without high quality of life.16 The overarching goal of our group is to bioengineer a mass-produced universal donor kidney to eliminate the scarcity problem in kidney transplant.22 One requirement for our goal is functioning renal epithelial cells within the device capable of highly efficient salt and water reabsorption. In the native kidney, renal tubular epithelial cells serve to reabsorb salt and water filtered by the glomerulus while allowing uremic toxins to concentrate in the urine. However, renal tubular epithelial cells that are removed from their in vivo niche tend to lose their transport capacity as they dedifferentiate in cell culture in vitro.9,14
Genome engineering technology offers the capability to create cell lines with enhanced functionality. A promising non-viral technology for genome engineering is the piggyBac transposon system, which enables cut-and-paste integration of DNA cargo.2,7piggyBac can be used to deliver multiple genes simultaneously and can be adapted for inducible or constitutive transgene expression.5,11,23 Moreover, the use of a non-viral system may be safer for eventual patient use. piggyBac has already been used to successfully genome engineer human cells with the hope of eventual therapeutic application.27
Certain transporters play key roles in regulating salt and water reabsorption in the kidney. A key transporter for sodium reabsorption is the Na+/H+ exchanger NHE3. Normally expressed on the apical brush border of proximal tubule cells, NHE3 facilitates Na+ reabsorption in exchange for H+.1 A key transporter for water is the aquaporin AQP1. This transporter (AQP1) is expressed both apically and basolaterally facilitating reabsorption of water across renal tubular epithelial cells.18 Madin-Darby canine kidney (MDCK) cells are a well-studied polarized kidney epithelial cell line.8 We chose to genome engineer MDCK cells to overexpress NHE3 and AQP1 and evaluate the ability of the cells to transport volume as a step towards genome engineering human kidney epithelial cells for enhanced function in an IAK device.
Materials and Methods
Vectors
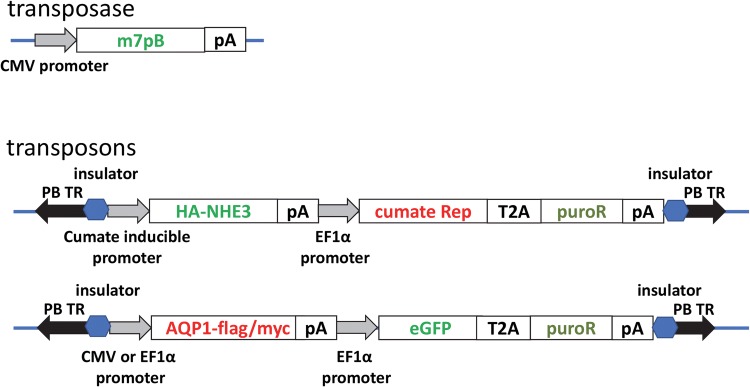
The piggyBac transposase plasmid pCMV-m7pB has been described previously.4SLC9A3 (NHE3) cDNA was PCR amplified from an NHE3 human ORF clone (Dharmacon, Lafayette, CO) and cloned into the pT-HS4-CuO-Puro plasmid vector (System Biosciences, Palo Alto, CA) using Gibson assembly and standard molecular biology techniques to create pT-HS4-CuO-NHE3-HA-Puro (CuNHE3). This vector is designed for cumate-inducible expression, and co-expresses the cumate repressor, CymR, and puromycin resistance from an EF1α promoter. A hemagglutinin epitope tag was incorporated into c-terminus of NHE3 using standard molecular biology techniques to facilitate detection of recombinant protein expression.
AQP1 cDNA was PCR amplified from an AQP1 human tagged ORF clone (OriGene, Rockville, MD) containing c-terminal myc and FLAG tags and cloned into the PB-CMV-MCS-EF1-GreenPuro (System Biosciences) plasmid vector using Gibson assembly and standard molecular biology techniques to create pCMV-AQP1-eGFP-puro (cAPQ1). This vector is designed for constitutive expression from a CMV promoter followed by co-expression of eGFP and puromycin resistance from an EF1α core promoter. FLAG and myc tags at the c-terminus of AQP1 facilitated detection of recombinant protein expression. For double transfection, the CMV promoter was excised by restriction digestion and replaced with a full length EF1α promoter to create p EF1α-AQP1-eGFP-puro (eAPQ1). Schematics of vectors are provided in Fig. 1. DNA sequencing was used to confirm the sequence of all DNA plasmids. All plasmids were prepped to be endotoxin free.
Figure 1.
Vector schematics. Expression of the hyperactive (m7pB) transposase is driven by the cytomegalovirus immediate early gene (CMV) promoter. Transposons are DNA segments flanked by inverted repeats (black arrows). Insulator elements flank the transgene expressing segments (blue hexagons). HA-tagged NHE3 is expressed via a cumate response system whereas AQP1 is constitutively expressed from the EF1a promoter. Both transposons harbor a puromycin resistance cassette for selection. T2A corresponds to a 2A-peptide cleavage sequence. pA, SV40 polyadenylation sequence.
Cell Culture and Transfection
MDCK cells (ATCC, Manassas, VA) were cultured at 37 °C with 5% CO2 in EMEM (Corning, Corning, NY) supplemented with 10% bovine growth serum (BGS, HyClone, Logan, UT), 2 mM l-glutamine, 100 U/mL penicillin and 100 mg/mL streptomycin (EMEM-CM). MDCK cells were co-transfected with CuNHE3 and m7pB plasmids or cAPQ1 and m7pB plasmids using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, Waltham, MA). Transfected cells were selected and maintained in EMEM-CM supplemented with 2–3 µg/mL puromycin (EMEM-puro). Clonal populations of CuNHE3 or cAQP1 were isolated with trypsin-infused cloning disks and expanded for further analysis. Double transfectants were created by re-transfecting a clonal population of CuNHE3 at passage 5 with eAQP1 and pCMV-m7pB using Lipofectamine LTX according to the manufacturer’s instructions (Invitrogen).
Immunoblot Analysis
Whole cell lysates were prepared in RIPA buffer (Sigma) supplemented with mammalian protease inhibitor mix (Sigma) and phosphatase inhibitor mix (Phos-Stop, Roche) and protein concentration determined by BCA. Lysates were normalized with RIPA buffer to equal concentrations and prepared for electrophoresis by adding 1X NuPAGE LDS Sample Buffer and 1X NuPAGE Reducing Agent (Invitrogen) and heating 70 °C for 10 min. Equal volumes were run on 4–12% NuPage Bis–Tris gels with MOPS buffer followed by transfer to nitrocellulose for immunoblotting. Blots were cut horizontally at ~ MW 60 and proteins in the top sections were incubated in rabbit anti-NHE3 (Santa Cruz) at 1:500 and rat anti-HA (clone 3F10, Roche) at 1:1000, and proteins in the bottom section were incubated in rabbit anti-AQP1 (Santa Cruz) at 1:500 and mouse anti-flag (clone M2, Sigma) at 1:1000, all at 4 °C overnight. IRD-labelled secondary antibodies (LICOR, Lincoln, NE) 680RD goat anti-rabbit, 800 goat anti-rat, and 800 donkey anti-mouse were each used at 1:15,000 for detection with an Odyssey Infrared Imaging System (LICOR). In some experiments, the bottom sections were reblotted with mouse α-β-actin (Novus, Centennial, CO) at 1:10,000 and secondary antibody 800 donkey anti-mouse as a loading control. TBS-based Odyssey blocking buffer and TBS with 0.1% Tween were used for all antibody dilutions and washing.
Immunofluorescence Analysis
Cells stably transfected with CuNHE3 at passage 5 were seeded on 12 mm-diameter polycarbonate Transwell membrane inserts (Corning, pore size: 0.4 µm) in 12 well culture plates at 2 × 105 cells/insert and grown to confluence in EMEM-puro with twice weekly media changes. After 2 weeks, cells were changed to fresh EMEM-puro ± 300 µg/mL cumate and incubated at 37 °C for an additional 6 days. Cells on membranes were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min and permeabilized with 0.1% Triton X-100 (Sigma, St. Louis, MO) in PBS for 10 min, then blocked with 5% normal goat serum (NGS) in PBS. Cells were incubated overnight at 4 °C with 500 ng/mL rat anti-HA (clone 3F10, Roche, Indianapolis, IN) and 2 µg/mL rabbit anti NHE3 (Santa Cruz, Santa Cruz, CA) in PBS containing 1% NGS added to both the upper and lower chambers of the Transwell. After washing with PBS, cells were incubated at room temperature in the dark for 2 h with 4 µg/mL Alexa Fluor 488 goat anti-rat (Invitrogen) and 4 µg/mL Alexa Fluor 594 goat anti-rabbit (Invitrogen) in PBS containing 0.1 µg/mL DAPI (Sigma), then washed with PBS. Membranes were cut from the Transwells and mounted cell side up on a slide with ProLong Gold Antifade Reagent without DAPI (Invitrogen). Images were collected with a 63X objective on an inverted LSM710 confocal microscope (Zeiss, Chester, VA). Untransfected cells or cells stably transfected with cAQP1 at passage 5 were seeded on coverslips in a 10 cm plate at 4 × 106 cells/plate. The next day, cells were fixed and permeabilized as described above, then blocked with 3% NGS and 3% BSA in PBS. Cells were incubated overnight at 4 °C with mouse anti-myc (clone 9E10, Vanderbilt Antibody and Protein Resource) 1:500 and rabbit anti-flag (Cell Signaling) 1:500 in blocking buffer. After washing with PBS, cells were incubated at room temperature in the dark for 3 h with 4 µg/mL Alexa Fluor 488 goat anti-mouse (Invitrogen) and 2 µg/mL Alexa Fluor 594 goat anti-rabbit in PBS containing 0.1 µg/mL DAPI, then washed with PBS. Coverslips were mounted with ProLong Gold Antifade Reagent without DAPI. Images were collected with a 40X objective on an AxioPlan 2 fluorescence microscope (Zeiss).
FACS Analysis of Double Transfectants
Transfected cells were expanded in EMEM-puro. All cells are resistant to puromycin, but only CuNHE3eAQP1 double-transfectants are eGFP positive. Stably transfected cells at passage 3 post-trasnfection were trypsinized, washed and resuspended at 5 × 106/mL in sorting medium (HBSS without phenol red supplemented with 5% BGS, 10 mM Hepes, 100 U/mL penicillin and 100 mg/mL streptomycin). GFP positive cells were isolated by fluorescence-activated cell sorting (FACS) on a FACSAria III cell sorter (BD Biosciences) into 24 well plates containing EMEM-puro supplemented with 10 mM Hepes at 10, 30, 100 or 300 cells/well. DAPI staining (1 µg/mL) was used to exclude dead cells from collection. Sorted cells were expanded and maintained in EMEM-puro. Out of 96 total wells, 89 independent clonal populations were established. Flow cytometry using intracellular staining for the FLAG tag on AQP1 and the HA tag on CuNHE3 identified the relative abundance and intensity of recombinant NHE3 and AQP1 expressed in these populations.
Cells expanded from each well of sorted double transfectants were cultured in EMEM-puro containing 300 µg/mL cumate (System Biosciences) for 72 h to induce NHE3 expression. Cells were then trypsinized, and 2 × 106 of each were transferred to a 96 well plate and fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS for 20 min at room temperature, followed by permeabilization in Intracellular Staining Permeabilization Wash Buffer (BioLegend, San Diego, CA). Cells were then stained with PE mouse α-HA (BioLegend) and APC mouse α-flag (Columbia Biosciences, Frederick MA) for 15–20 min at antibody concentrations determined by single-staining of known positive single transfectants with different dilutions of each antibody. Samples were analyzed on a LSRFortessa analytical cytometer equipped with High Throughput Sampler (BD Biosciences, San Jose, CA) for sample acquisition from microtiter plates. Unstained and single-stained controls along with untransfected cells and anti-mouse compensation beads (BD Biosciences) stained with PE mouse α-HA or APC mouse α-flag were used for instrument set-up. All Flow Cytometry experiments were performed in the VMC Flow Cytometry Shared Resource.
Biotinylation Assay
A biotinylation assay was used to determine whether recombinant proteins are expressed on the cell surface.26 Wild type MDCK and selected MDCK double transfectants at passage 5–7 were seeded at confluence on 75 mm Transwell polycarbonate membrane inserts (Corning, pore size: 0.4 µm) preincubated in culture medium for 30 min. The same cells were also seeded onto regular 10 cm tissue culture plates. Confluence and dome formation were monitored on the 10 cm plates, and media on all plates and Transwells was changed twice weekly. After one week, monolayer permeability in Transwells was assessed by adding 100 µg/mL FITC-Inulin (Sigma) in culture medium to the upper (apical) chamber of Transwells and fresh culture medium without FITC-inulin was added to the lower chamber (basolateral). Twenty-four hours later aliquots of medium were collected from the upper and lower chambers to a black 96-well plate. Fluorescence was measured on a FLUOstar Omega microplate reader (BMG Labtech, Cary, NC) and the concentration of FITC-inulin in each chamber was calculated by comparison to a standard curve. From these measurements, the % of FITC-inulin transferred from the apical to basal chamber in 24 h was determined. When the monolayers in Transwells exhibited 1.5% permeability or less in 24 h, all plates were changed to fresh medium ± 300 µg/mL cumate and cultured for an additional 3 days. Cell surface proteins were biotinylated and harvested using a Cell Surface Protein Isolation Kit (Thermo Fisher, Waltham, MA) and membrane proteins were isolated with a Mem-PER membrane protein extraction kit (Thermo Fisher) according to the manufacturers’ instructions with some modifications. Briefly, monolayers were washed with ice cold PBS and incubated at 4 °C for 30–45 min with gentle rocking with 0.5 mg/mL EZ-link Sulfo-NHS-SS-Biotin. After biotinylation of surface proteins, cells were washed with cold PBS and unbound biotin was quenched with Quenching Solution from the kit. Cells were scraped in Quenching Solution, centrifuged at 300×g 5 min at 4 °C, resuspended in Cell Wash Solution (from kit) and recentrifuged, washed, and respun. Cell pellets were resuspended in 5X pellet volumes Permeabilization Buffer (from Mem-PER kit) supplemented with mammalian protease inhibitor mix (Sigma) and phosphatase inhibitor mix (Phos-Stop, Roche), vortexed, and incubated 10 min at 4 °C with constant mixing at 2000 rpm. The resulting cell suspension was centrifuged at 4 °C, 16,000×g for 15 min. The supernatant containing cytosolic proteins was removed and the pellet washed with additional Permeabilization buffer to remove all traces of cytosol. The resulting membrane pellet was resuspended by pipetting in 5X pellet volumes Solubilization buffer (from Mem-PER kit) containing protease and phosphatase inhibitors and incubated 30 min at 4 °C with constant mixing at 2000 rpm. Solubilized proteins were centrifuged at 4 °C, 16,000×g for 15 min. The supernatant containing solubilized membrane and membrane-associated proteins (total membrane lysates) was transferred to a clean tube, snap-frozen and stored at − 80 °C for later isolation of biotinylated proteins. The protein concentration of total membrane lysates was determined by BCA assay (Pierce) and ~ 500 µg of each was normalized to the same volume (500 µL) with Solubilization buffer containing protease and phosphatase inhibitors and each added to a prewashed NeutrAvidin Agarose column (from biotinylation kit) to bind biotinylated proteins. After incubation for 1 h at room temperature with constant mixing, the flow through containing non-biotinylated proteins (FT) was collected by centrifugation at 1000×g for 1 min. The column was washed 3X with 500 µL Wash Buffer (from biotinylation kit) containing protease and phosphatase inhibitors, and biotinylated proteins were eluted by adding 330 µL of 50 mM DTT in 1X NuPage LDS Sample Buffer (Invitrogen) to column and heating 95–100 °C for 5 min. Isolated biotinylated proteins (IBP) were collected from the column by centrifugation 1000×g for 2 min. One tenth volume NuPage reducing agent (Invitrogen) was added to each FT and IBP and all were stored at − 20 °C for later analysis. Equal volumes of each FT and IBP lysate were analyzed by immunoblotting as described above.
Analysis of Blister Dome Formation
Images of wild type MDCK and selected MDCK double transfectants at passage 5–7 were seeded to 10 cm tissue culture plates for biotinylation were captured on a ZOE cell imager (Bio-Rad, Hercules, CA) after incubation ± 300 µg/mL cumate for 3 days as described above. For quantification, wild type MDCK and selected MDCK double transfectants were seeded in 6 well plates at 8 × 105/well, 1 plate per cell type. After 7 days, cells were changed to fresh medium ± 300 µg/mL cumate in triplicate wells for each cell type and cultured for an additional 3 days. On day 10, all cells were changed to fresh medium and blister domes > 100 µM were counted in 10 fields/well of all wells on a light microscope.
Results
We engineered piggyBac vectors capable of expression of human NHE3 and AQP1. We first attempted to express NHE3 driven by a CMV promoter; however, high NHE3 expression was toxic to the cells resulting in abnormally large cells (data not shown). Therefore, we chose to use an inducible expression system to provide user-controlled timing and dosage of NHE3 expression. We chose to use a cumate-inducible system as cumate is a non-toxic organic molecule capable of traversing cell membranes.17 With this system, the cumate repressor binds to the cumate operator sequences (CuO) with high affinity in the absence of cumate; therefore, addition of cumate activates transgene expression. We engineered a C-terminal HA tag for detection of NHE3 and the vector also harbored a puromycin resistance gene for selection. We next engineered a piggyBac transposon harboring AQP1 with FLAG and myc epitopes, eGFP for fluorescent detection, and puromycin resistance for selection. Schematics of the vectors utilized are shown in Fig. 1.
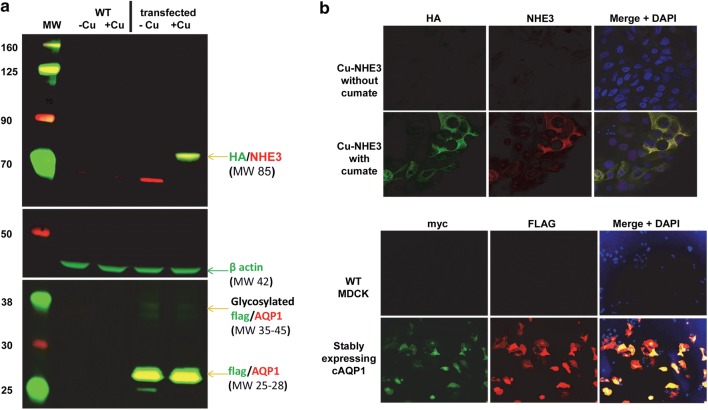
We used western blot analysis to confirm overexpression of NHE3 and AQP1. Our stably transfected CuNHE3 MDCK cells showed negligible leak of expression as there was no visible NHE3 without cumate. However, addition of 300 µg/mL cumate resulted in detectable NHE3 expression via blotting with anti-HA or anti-NHE3 antibodies (Fig. 2a). MDCK cells stably transfected with AQP1 demonstrated strong AQP1 expression with or without cumate as expected as AQP1 expression was driven by the EF1a promoter (Fig. 2a).
Figure 2.
Confirmed overexpression of NHE3 and AQP1. (a) western blot analysis confirming cumate-inducible NHE3 expression (detected by anti-HA and anti-NHE3 antibodies) and AQP1 expression (via anti-flag and anti-AQP1 antibodies). β-actin is provided as a loading control. Molecular weight (MW) markers are labeled to the left of the blots. (b) Immunofluorescent microscopy of transfected cells. Immunofluorescent microscopy of stably transfected MDCK cells expressing cumate-inducible NHE3 expression (detected by anti-HA and anti-NHE3 antibodies) and AQP1 expression (via anti-flag and anti-myc antibodies). Dapi indicates nuclear staining.
We next used immunofluorescent microscopy to evaluate overexpression of NHE3 and AQP1 in MDCK cells. Stably transfected CuNHE3 MDCK cells showed no detectable NHE3 expression using anti-HA or anti-NHE antibodies in the absence of cumate. However, addition of 300 µg/mL cumate resulted in overlapping HA/NHE3 immunofluorescence (Fig. 2b). We compared stably transfected cAQP1 MDCK cells to untransfected MDCK cells. cAQP1 cells demonstrated overlapping myc and FLAG immunofluorescent indicating expression of epitope-tagged AQP1 in those cells (Fig. 2c). These results demonstrated that we could genome engineer MDCK cells to overexpress AQP1 constitutively and NHE3 in a user-controlled manner using the piggyBac transposon system.
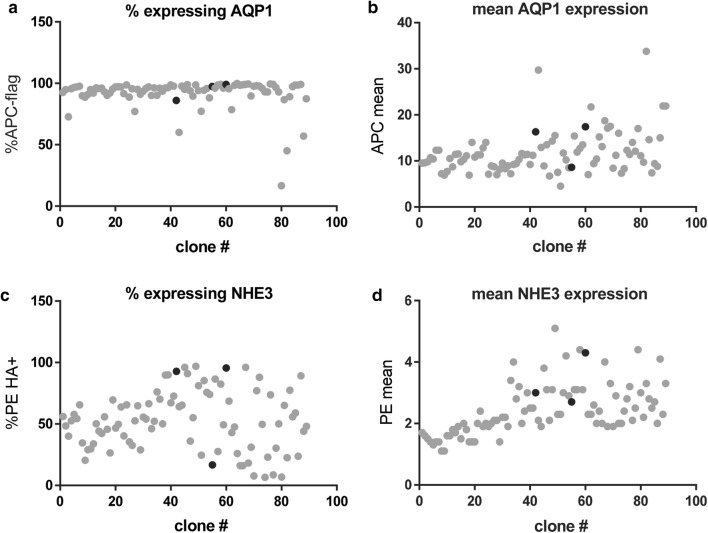
As it was not clear to us what ratio or amount of overexpression of NHE3 and AQP1 would lead to the best transport, we devised a flow cytometry analysis of double stably tranfsected NHE3 and AQP1 clones. Cells were first transfected with NHE3 vector and selected. This selected population of NHE3 expressing cells was re-transfected with AQP1 vector. Double transfectant clones were evaluated using flow cytometric quantification of both the number of expressing cells as well as the fluorescent intensity (level) of expression (Fig. 3). Almost all clones exhibited a high percentage of AQP1 expression, though at varying levels of intensity (Fig. 3a). The percentage of cumate-inducible NHE3 expressing cells was more variable (Fig. 3b). This is due to the fact that cumate-inducible NHE3 expressing cells were re-transfected with piggyBac and the AQP1 transposon. piggyBac can excise already integrated transposons,6,25 thereby potentially eliminating their ability to express NHE3. We subsequently selected three double stably-transfected clones for further evaluation (Fig. 3, black dots). These clones varied in their amount of AQP1 expression as well as amount and proportion of cells expressing NHE3 (Table 1).
Figure 3.
Flow cytometry analysis of NHE3/AQP1 expressing clones. (a) evaluation of % of AQP1 expressing cells as well as mean fluorescent intensity for each clone. (b) evaluation of % of cumate-inducible NHE3 expressing cells as well as mean fluorescent intensity for each clone. Black dots indicate clones 42, 55, and 60 on the corresponding graphs.
Table 1.
Flow cytometry analysis of clones.
| Clone | %APC flag+ | APC mean | %PE HA+ | PE mean | % flag and HA + |
|---|---|---|---|---|---|
| 42 | 86 | 16.3 | 92.8 | 3 | 81.7 |
| 55 | 97.4 | 8.6 | 16.7 | 2.7 | 20.3 |
| 60 | 99.1 | 17.4 | 95.5 | 4.3 | 96 |
Shown are the % expressing (APC or PE) as well as extent of overexpression (mean fluorescent intensity) of each clone
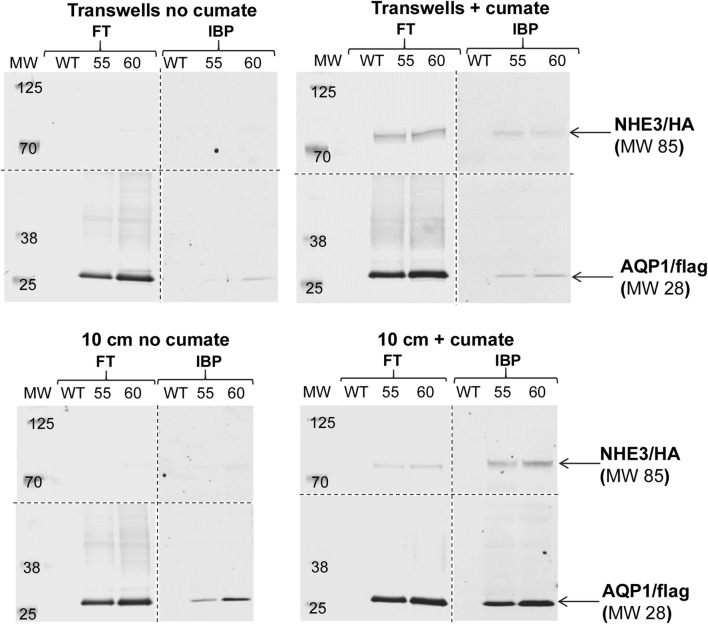
For NHE3 and AQP1 to function properly in MDCK cells, the transporter must achieve cell-surface localization. We subsequently used a cell surface biotinylation assay to evaluate for surface delivery of NHE3 and AQP1 in stably transfected MDCK cells. Membrane impermeant sulfo-reactive biotin was used to label surface proteins on WT and clonal MDCK cell lines (clones 55 and 60) grown on transwells or 10 cm dishes. Transwells provided an environment for cell polarization. As NHE3 is expected to be apically localized, and AQP1 both apical and basolateral, we biotinylated the apical surface of Transwells to evaluate for surface delivery of both proteins in Transwells. We performed western analysis of biotinylated and surface-delivered NHE3 and AQP1 (Fig. 4). We observed cumate-inducible surface expression of NHE3 in clones 55 and 60 as well as constitutive expression of AQP1 in those clones. These results confirm cell surface delivery of piggyBac-mediated expression of NHE3 and AQP1 in MDCK cells, which would be required for enhancing salt and water transport.
Figure 4.
Confirmation of cells surface delivery of NHE3 and AQP1. Cell surface biotinylation was used to confirm surface delivery of NHE3 and AQP in double stably-transfected clones. FT flow through, IBP isolated biotinylated proteins. Western detection was used to evaluate surface expression. Molecular weight (MW) marker weights are indicated on the blots. WT, wild type cells; 55, clone number 55; 60, clone number 60. Anti-HA antibodies were used for NHE3 detection and anti-flag antibodies were used for AQP1 detection.
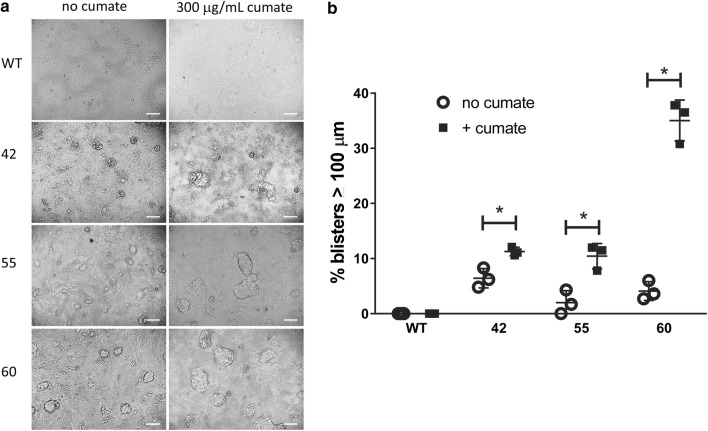
We next used a blister or dome formation assay to evaluate for transport of volume across double stably transfected clones (numbers 42, 55, and 60). The blister formation assay evaluates the formation of domes or blisters on cell culture plates that result from apical-to-basolateral transport of solute and water, and this assay has been used by others to evaluate pathways or proteins involved in transport of solutes and water in MDCK cells.3,13,24 Minimal blisters were formed in WT cells in the absence or presence of cumate (Fig. 5a). Clones 42, 55, and 60 demonstrated some blister formation in the absence of cumate, indicating AQP1 expression alone could enhance transport (Figs. 5a and 5b). However, addition of cumate to overexpress NHE3 in addition to constitutive AQP1 increased blister formation in all three clones analyzed, most notably in clone 60 (Figs. 5a and 5b). Clone 60 demonstrated the highest amount and percentage of cells expressing both AQP1 and cumate-inducible NHE3 between the three clones analyzed (Fig. 3; Table 1).
Figure 5.
NHE3/AQP1 overexpression enhance transport function in MDCK cells. (a) Phase contrast microscopy of WT and NHE3/AQP1 expressing clonal cell populations (clones 42, 55, and 60) in the absence or presence of cumate. The white bar in the lower right corner figure is a scale bar of 100 µm. (b) Quantitation of % blisters > 100 µM in the absence or presence of cumate (N = 3±SEM).
Discussion
An IAK has the potential to revolutionize treatment for patients with end-stage renal disease. However, such a device will require functional renal epithelial cells capable of reabsorbing salt and water in order to be feasible for patient use by minimizing urine output. Increasing the capability of renal epithelial cells to reabsorb salt and water would result in the use of a smaller device which would be more manageable for kidney disease patients.
Although there is a repertoire of genomic modifications that could be made to renal epithelial cells, increasing key proteins involved in salt and water transport seemed like a logical place to start. We chose to use the non-viral piggyBac transposon system to stably genome-modify cultured MDCK cells with NHE3 and AQP1 transgenes. We confirmed overexpression using standard techniques of western and immunofluorescence analysis. We developed a flow cytometry analysis that lead to isolation of multiple clonal populations of varying levels of expression of AQP1 and NHE3. We confirmed surface delivery of those proteins in MDCK cells and our analysis of three clones revealed that AQP1 and NHE3 expression resulted in enhanced transport as measured by blister/dome formation on cell culture plates. Other signaling pathways have been shown to increase blister/dome formation in MDCK cells including protein kinase A signaling and Stat3.12,24
In order for genome modified cells with enhanced volume transport to function in an IAK, the cells need to be permanently modified for stable expression. We have previously demonstrated piggyBac-mediated multiplexed stable genome modification of human cells with stable protein expression out to passage 38 in culture, engineering cell lines that were not possible to have previously been created.11 We have also genome modified adult mouse liver in vivo and observed stable transgene expression out to one year.23 Our cell-surface biotinylation and blister/dome formation experiments were performed on MDCK clonal cell lines at passage 5–7 after 8–10 weeks of culture indicating stable genome modification and protein expression. Therefore, our results, combined with our previous studies,11,23 indicate the ability of piggyBac to permanently genome modify renal epithelial cells for enhanced function in an eventual IAK.
piggyBac involves no viral components for achieving genome modification. Therefore, there is no possibility of cells producing virus or shedding viral proteins that might precipitate a host response. However, piggyBac transposition is untargeted though we and others have taken steps towards targeting piggyBac integrations.10,15 Nonetheless, untargeted integration is a potential safety concern. For our particular application, genome modified cells would be placed in an enclosed device potentially mitigating clonal expansion of modified cells that might affect a patient. We have previously evaluated piggyBac modification of human cells and found no evidence of clonal cell outgrowth or undesired genotoxic outcomes.20,21
Our studies lay the foundation for future studies directed at modifying human renal epithelial cells for increased transport function. These studies demonstrate that non-viral genome engineering technology can be used to bioengineer renal epithelial cells stably overexpressing one or more transporters with increased solute and water transport. In addition to transposon technology, other genome engineering technologies may be applied to human renal epithelial cells to upregulate endogenous genes in those cells.19
Future studies will be directed at bioengineering human renal epithelial cells for function within the IAK. Our vectors already contain human AQP1 and NHE3 transgenes. Subsequent studies will involve using our vector and analysis systems to optimize transport in human kidney epithelial cells for patient translation.
Acknowledgments
These studies were supported by DK093360 (NIH) and BX004285 (VA) to MHW, EB021214 to SR and WHF, and the Vanderbilt O’Brien Kidney Center Cell and Genome Engineering Core (DK114809). The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). Brittany K. Matlock in the VMC Flow Cytometry Shared Resource provided valuable assistance in developing the intracellular staining protocol for high-throughput sampling. Confocal immunofluorescence image collection and data analysis were performed in part through the use of the VUCell Imaging Shared Resource (supported by NIHgrants CA68485, DK20593, DK58404, DK59637 and EY08126) and NIH S10 Grant Number 1S10RR027396-01. Mouse anti-myc antibody was produced by the Vanderbilt Antibody and Protein Resource. The Vanderbilt Antibody and Protein Resource is supported by the Vanderbilt Institute of Chemical Biology and the Vanderbilt Ingram Cancer Center (P30 CA68485).
Conflict of interest
MHW, RAV, WL, and RCW declare no conflicts of interest. SR and WHF are founders of Silicon Kidney.
Ethical Standards
No human or animal studies were carried out by the authors for this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brant SR, Bernstein M, Wasmuth JJ, Taylor EW, McPherson JD, Li X, et al. Physical and genetic mapping of a human apical epithelial Na +/H + exchanger (NHE3) isoform to chromosome 5p15.3. Genomics. 1993;15(3):668–672. doi: 10.1006/geno.1993.1122. [DOI] [PubMed] [Google Scholar]
- 2.Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172(1):156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 3.Cattaneo I, Condorelli L, Terrinoni AR, Antiga L, Sangalli F, Remuzzi A. Shear stress reverses dome formation in confluent renal tubular cells. Cell. Physiol. Biochem. 2011;28(4):673–682. doi: 10.1159/000335813. [DOI] [PubMed] [Google Scholar]
- 4.Doherty JE, Huye LE, Yusa K, Zhou L, Craig NL, Wilson MH. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther. 2012;23(3):311–320. doi: 10.1089/hum.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty JE, Woodard LE, Bear AS, Foster AE, Wilson MH. An adaptable system for improving transposon-based gene expression in vivo via transient transgene repression. FASEB J. 2013;27(9):3753–3762. doi: 10.1096/fj.13-232090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elick TA, Bauser CA, Fraser MJ. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996;98(1):33–41. doi: 10.1007/BF00120216. [DOI] [PubMed] [Google Scholar]
- 7.Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect MolBiol. 1996;5(2):141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 8.Gaush CR, Hard WL, Smith TF. Characterization of an established line of canine kidney cells (MDCK) Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med (N.Y.). 1966;122(3):931–935. doi: 10.3181/00379727-122-31293. [DOI] [PubMed] [Google Scholar]
- 9.Hallman MA, Zhuang S, Schnellmann RG. Regulation of dedifferentiation and redifferentiation in renal proximal tubular cells by the epidermal growth factor receptor. J. Pharmacol. Exp. Ther. 2008;325(2):520–528. doi: 10.1124/jpet.107.134031. [DOI] [PubMed] [Google Scholar]
- 10.Hew BE, Sato R, Mauro D, Stoytchev I, Owens JB. RNA-guided piggyBac transposition in human cells. Synth Biol (Oxf). 2019;4(1):ysz018. doi: 10.1093/synbio/ysz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlig KM, Saridey SK, Kaja A, Daniels MA, George AL, Jr, Wilson MH. Multiplexed transposon-mediated stable gene transfer in human cells. Proc. Natl. Acad. Sci. USA. 2010;107(4):1343–1348. doi: 10.1073/pnas.0910383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klebe RJ, Grant A, Grant G, Ghosh P. Cyclic-AMP deficient MDCK cells form tubules. J. Cell. Biochem. 1995;59(4):453–462. doi: 10.1002/jcb.240590406. [DOI] [PubMed] [Google Scholar]
- 13.Lever JE. Regulation of dome formation in differentiated epithelial cell cultures. J. Supramol. Struct. 1979;12(2):259–272. doi: 10.1002/jss.400120210. [DOI] [PubMed] [Google Scholar]
- 14.Love HD, Ao M, Jorgensen S, Swearingen L, Ferrell N, Evans R, et al. Substrate elasticity governs differentiation of renal tubule cells in prolonged culture. Tissue Eng. Part A. 2019;25(13–14):1013–1022. doi: 10.1089/ten.tea.2018.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, Galvan DL, Woodard LE, Dorset D, Levy S, Wilson MH. Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res. 2017;45(14):8411–8422. doi: 10.1093/nar/gkx572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCullough KP, Morgenstern H, Saran R, Herman WH, Robinson BM. Projecting ESRD incidence and prevalence in the united states through 2030. J. Am. Soc. Nephrol. 2019;30(1):127–135. doi: 10.1681/ASN.2018050531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullick A, Xu Y, Warren R, Koutroumanis M, Guilbault C, Broussau S, et al. The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnol. 2006;6:43. doi: 10.1186/1472-6750-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen S, Smith BL, Christensen EI, Knepper MA, Agre P. CHIP28 water channels are localized in constitutively water-permeable segments of the nephron. J. Cell Biol. 1993;120(2):371–383. doi: 10.1083/jcb.120.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha S, Woodard LE, Charron EM, Welch RC, Rooney CM, Wilson MH. Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells. Nucleic Acids Res. 2015;43(3):1770–1782. doi: 10.1093/nar/gkv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito S, Nakazawa Y, Sueki A, Matsuda K, Tanaka M, Yanagisawa R, et al. Anti-leukemic potency of piggyBac-mediated CD19-specific T cells against refractory Philadelphia chromosome-positive acute lymphoblastic leukemia. Cytotherapy. 2014;16(9):1257–1269. doi: 10.1016/j.jcyt.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salani M, Roy S, Fissell WHT. Innovations in wearable and implantable artificial kidneys. Am J Kidney Dis. 2018;72(5):745–751. doi: 10.1053/j.ajkd.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Saridey SK, Liu L, Doherty JE, Kaja A, Galvan DL, Fletcher BS, et al. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol. Ther. 2009;17(12):2115–2120. doi: 10.1038/mt.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su HW, Yeh HH, Wang SW, Shen MR, Chen TL, Kiela PR, et al. Cell confluence-induced activation of signal transducer and activator of transcription-3 (Stat3) triggers epithelial dome formation via augmentation of sodium hydrogen exchanger-3 (NHE3) expression. J. Biol. Chem. 2007;282(13):9883–9894. doi: 10.1074/jbc.M606754200. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105(27):9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson M, Limbird L. Mechanisms regulating the cell surface residence time of the alpha(2A)-adrenergic receptor. Biochemistry. 2000;39(4):693–700. doi: 10.1021/bi9920275. [DOI] [PubMed] [Google Scholar]
- 27.Woodard LE, Wilson MH. piggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 2015;33(9):525–533. doi: 10.1016/j.tibtech.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]