Abstract
Aim: Musashi 2 (MSI2), which is an RNA-binding protein, plays a fundamental role in the oncogenesis of several cancers. The aim of this study is to investigate the expression of MSI2 in Oral Squamous Cell Carcinoma (OSCC) and evaluate its correlation to clinic-pathological variables and prognosis. Materials and Methods: A bioinformatic analysis was performed on data downloaded from The Cancer Genome Atlas (TCGA) database. The MSI2 expression data were analysed for their correlation with clinic-pathological and prognostic features. In addition, an immmunohistochemical evaluation of MSI2 expression on 108 OSCC samples included in a tissue microarray and 13 healthy mucosae samples was performed. Results: 241 patients’ data from TCGA were included in the final analysis. No DNA mutations were detected for the MSI2 gene, but a hyper methylated condition of the gene emerged. MSI2 mRNA expression correlated with Grading (p = 0.009) and overall survival (p = 0.045), but not with disease free survival (p = 0.549). Males presented a higher MSI2 mRNA expression than females. The immunohistochemical evaluation revealed a weak expression of MSI2 in both OSCC samples and in healthy oral mucosae. In addition, MSI2 expression directly correlated with Cyclin-D1 expression (p = 0.022). However, no correlation has been detected with prognostic outcomes (overall and disease free survival). Conclusions: The role of MSI2 expression in OSCC seems to be not so closely correlated with prognosis, as in other human neoplasms. The correlation with Cyclin-D1 expression suggests an indirect role that MSI2 might have in the proliferation of OSCC cells, but further studies are needed to confirm such results.
Keywords: MSI2, OSCC, oral cancer, musashi 2, prognosis
1. Introduction
Oral cancer (OC) belongs to the wider family of Head and Neck Cancers (HNCs). OC is a highly relevant problem for global public health, with a clinical impact in terms of incidence, prevalence, and mortality rates that do not tend to improve. It is reported to be the 11th most common malignancy worldwide [1]. Around 90% of OCs are histologically classified as squamous cell carcinoma (OSCC), involving the mucosal surface of the oral cavity and tongue [2]. Oral carcinogenesis encompasses multistep processes that drive the progression from normal mucosa to OSCC [3]. Changes in the DNA sequence, accumulation of somatic mutations and epigenetic events are the main mechanisms that are involved in tumor progression. In particular, epigenetic and post-transcriptional events, gained an important role in cancer [4]. Key-regulators of these mechanisms are the RNA-Binding Proteins (RBPs) that cause variations in protein expression, due to their involvement in splicing, mRNA-polyadenylation, editing, and r-tRNA stabilization [5]. Musashi-2 (MSI2) is one of the most studied RBPs. In particular, different studies evaluated its role in cancer. For example, MSI2 overexpression was linked to an increase of invasion and metastasis in non-small cell lung carcinoma, whereas its depletion showed a decrease of epithelia-mesenchymal transition [6]. In bladder cancer, the differentiation antagonizing non-protein noding RNA (DANCR) long non-coding RNA (lncRNA) acts by sponging miR-149 increasing the expression of MSI2, getting worse a malignant phenotype [7]. In addition, MSI2 seems to be involved in patients’ prognosis, resulting as prognostic factor in gastric [8] and cervical cancer [9]; in lung cancer, MSI2 emerged as a novel therapeutic target [10]. Several other factors that are responsible of the regulation of cell proliferation and cell cycle control have been proposed as diagnostic, prognostic, and therapeutic markers for certain malignancies. Among these, the cyclin D1 has been deeply investigated and were shown to be essential for the tumorigenesis of melanoma, breast cancer, and colon and oral squamous cell carcinoma (OSCC) [11]. Cyclin D1 belongs to the family of Cyclins and it is essential in the regulation of cell proliferation, DNA repair, and cell migration control [12]. The aim of this study was to investigate the expression of MSI2 in OSCC samples, through a histologic and bioinformatics analysis in order to evaluate its correlation to clinic-pathological variables and prognosis. Furthermore, a staining for Cyclin-D1 has been performed on OSCC tissue microarray (TMA) and the correlation of Cyclin D1 expression with MSI2 was investigated.
2. Results
2.1. Analysis of MSI2 Mutations, Gene Methylation and mRNA Expression in TCGA Database
A total 241 patients’ records were included in this analysis after extracting and matching clinic-pathological data from the TCGA database. Table 1 summarizes the main clinical-pathological characteristics of the included patients. DNA mutations and copy number alterations were not detected for the MSI2 gene in patients with OSCC included in the TCGA database (0/241, 0%). The expression of MSI2 mRNA (log2 (fpkm+1)) was relatively low ranging from 0.2785 to 2.7117 with a mean of 1.270734 (S.E. 0.030) and a median of 1,40673. According to the median value, patients were divided in low (≤1.40673) and high (>1.40673) MSI2 mRNA expression. Methylation status, measured in beta unit, showed a hyper methylated condition of the gene in all of the patients analyzed, the values of gene methylation ranged from 0.6802 to 0.9910 with a mean of 0.974483 (S.E. 0.0018993). The Spearman rank correlation test did not show a significant correlation between mRNA expression and methylation status of the gene (ρ = −0.44; p = 0.498); however, a higher methylation status was detected for the low-expression group with results that were close to the statistical significance (Mann–Whitney p = 0.095). MSI2 mRNA expression correlated with Grading (ρ = 0.169; p = 0.009) and showed a differential expression according to the gender (Mann–Whitney p = 0.001) with males’ samples showing a higher expression, while MSI2 methylation profile correlated to the age of patients (ρ = 0.140; p = 0.03) (Table 2). Univariate and multivariate analyses were performed, aiming to investigate whether MSI2 mRNA expression in the TCGA database was able to predict prognosis. The results of the univariate analysis were promising, showing a significant association between MSI2 mRNA expression (High vs low) and overall survival (Hazard Ratio, HR = 1.488; 95% C.I. 1.013–2.185; p = 0.045); furthermore, the results of the multivariate analysis (HR = 1.437; 95% C.I. 0.952–1.970; p = 0.084) were close to the threshold of statistical significance. Conversely MSI2 mRNA expression did not correlate with disease free survival (HR = 0.827; 95% C.I. 0.443–1.542; p = 0.549) in OSCC patients.
Table 1.
Clinical-pathological characteristics of patients included in The Cancer Genome Atlas (TCGA) analysis.
| Clinic-Pathological Information | Groups | Number of Patients |
|---|---|---|
| Age | ≤ 65 years old | 144/241 |
| > 65 years old | 97/241 | |
| Gender | Male | 158/241 |
| Female | 83/241 | |
| Grade | 1 | 43/241 |
| 2 3 |
147/241 51/241 |
|
| Stage | 1–2 | 76/241 |
| 3–4 | 165/241 | |
| Subsite | Tongue | 103/241 |
| Gingivo-buccal | 30/241 | |
| Floor of the mouth | 46/241 | |
| Others | 62/241 |
Table 2.
Spearman rank correlation for the 241 Oral Squamous Cell Carcinoma (OSCC) patients included in the TCGA database. * p < 0.05; ** p < 0.001.
| Variable | Age | Grade | Stage | Gender | Perineural Invasion | MSI2 Methylation | MSI2 mRNA Expression | Ki-67 mRNA Expression | Cyclin-D mRNA Expression |
|---|---|---|---|---|---|---|---|---|---|
| Age | ρ = 1 | 0.088 | −0.084 | 0.243 | 0.060 | 0.140 | −0.121 | −0.069 | 0.033 |
| p-value = 1 | 0.175 | 0.197 | 0.001 ** | 0.414 | 0.03 * | 0.06 | 0.287 | 0.607 | |
| Grade | ρ = 1 | −0.003 | −0.066 | 0.100 | −0.094 | 0.169 | −0.105 | 0.053 | |
| p-value = 1 | 0.959 | 0.307 | 0.176 | 0.146 | 0.009 ** | 0.105 | 0.418 | ||
| Stage | ρ = 1 | −0.026 | 0.199 | −0.031 | −0.035 | 0.038 | −0.062 | ||
| p-value = 1 | 0.686 | 0.006 ** | 0.632 | 0.587 | 0.561 | 0.340 | |||
| Gender | ρ = 1 | −0.090 | −0.045 | −0.220 | 0.042 | 0.043 | |||
| p-value = 1 | 0.907 | 0.485 | 0.001 ** | 0.520 | 0.505 | ||||
| Perineural Invasion | ρ = 1 | −0.178 | 0.033 | −0.132 | −0.038 | ||||
| p-value = 1 | 0.014 * | 0.650 | 0.071 | 0.606 | |||||
| MSI2 Methylation | ρ = 1 | −0.044 | 0.094 | 0.027 | |||||
| p-value = 1 | 0.498 | 0.144 | 0.673 | ||||||
| MSI2 mRNA expression | ρ = 1 | −0.093 | 0.003 | ||||||
| p-value = 1 | 0.150 | 0.963 | |||||||
| Ki-67 mRNA expression | ρ = 1 | 0.253 | |||||||
| p-value = 1 | 0.000 ** | ||||||||
| Cyclin-D mRNA expression | ρ = 1 | ||||||||
| p-value = 1 |
2.2. Immunohistochemical Analysis of MSI2 Expression on TMA
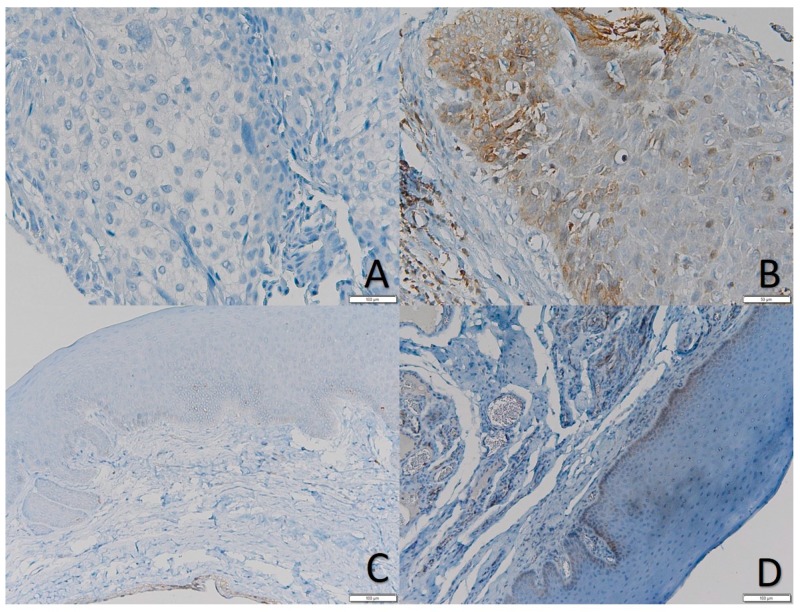
The IHC analysis of MSI2 protein expression was performed on a total of 108 patient’ samples included in the TMA; such patients had been treated at the National Cancer Institute “Giovanni Pascale” between 1997 and 2012. Table 3 reports the clinical pathological information of patients included in the cohort. An analysis of protein expression in the TMA samples revealed that MSI2 is not frequently expressed in OSCC, in fact 58.3% (63/108) cases analyzed resulted in being negative for MSI2 expression. Of the remaining 41.7% (45/108) samples, only 5.6% (6/108) showed higher level of MSI2 expression. The presence of MSI2 expression directly correlated with Cyclin-D1 expression (ρ = 0.279; Chi-Squared p-value = 0.022) (Table 4), this last one resulted to be higher expressed in males than in females (Mann–Whitney p-value = 0.024). The presence of MSI2 expression in the TMA cohort did not correlate with overall survival (HR = 0.575; 95% C.I. 0.278–1.190; p = 0.136) (Table 5). In the 13 oral healthy mucosae analyzed the expression of MSI2 was faint and mainly confined to the basal layer with a percentage of expression lower than 5% of the whole number of epithelial cells (Figure 1).
Table 3.
Clinical-pathological characteristics of patients included in the immunoistochemical analysis.
| Clinic-Pathological Information | Groups | Number of Patients |
|---|---|---|
| Age | ≤ 65 years old | 45/108 |
| > 65 years old | 63/108 | |
| Gender | Male | 79/108 |
| Female | 29/108 | |
| Grade | 1 2 |
22/108 50/108 |
| 3 | 36/108 | |
| Stage | 1–2 | 35/108 |
| 3–4 | 73/108 | |
| Subsite | Tongue | 67/108 |
| Gingivo-buccal | 23/108 | |
| Floor of the mouth | 13/108 | |
| Others | 5/108 |
Table 4.
Spearman rank correlation for patients included in the TMA. Age. Cyclin-D1 and Ki-67 were included as continuous variables. while Grade. Stage. Gender. and MSI2 as categorical variables. * p < 0.05; ** p < 0.001.
| Variable | Age | Grade | Stage | Gender | MSI2 Expression (Neg/Pos) | Cyclin-D1 Expression | Ki-67 Expression |
|---|---|---|---|---|---|---|---|
| Age | ρ = 1 | 0.049 | −0.151 | −0.062 | −0.006 | 0.021 | 0.148 |
| p-value = 1 | 0.617 | 0.122 | 0.528 | 0.955 | 0.865 | 0.226 | |
| Grade | ρ = 1 | 0.098 | 0.034 | 0.223 | 0.060 | 0.398 | |
| p-value = 1 | 0.313 | 0.726 | −0.066 | −0.044 | 0.001 * | ||
| Stage | ρ = 1 | −0.008 | 0.497 | 0.721 | −0.004 | ||
| p-value = 1 | 0.993 | 0.676 | 0.468 | 0.977 | |||
| Gender | ρ = 1 | −0.088 | −0.277 | −0.285 | |||
| p-value = 1 | 0.364 | 0.023 * | 0.031 * | ||||
| MSI2 expression (Neg/Pos) | ρ = 1 | 0.279 | 0.122 | ||||
| p-value = 1 | 0.022 * | 0.315 | |||||
| Cyclin-D1 expression | ρ = 1 | 0.485 | |||||
| p-value = 1 | 0.000 ** | ||||||
| Ki-67 expression | ρ = 1 | ||||||
| p-value = 1 |
Table 5.
Multivariate Cox regression analysis for MSI2 adjusted for other clinic-pathological parameters. * p < 0.05.
| Overall Survival | |||
|---|---|---|---|
| Variables | Hazard Ratio | 95.0% C.I. | p-Value |
| Gender | 1.904 | 0.964–3.759 | 0.064 |
| Grade | 1.162 | 0.706–1.911 | 0.555 |
| Age | 1.018 | 0.986–1.052 | 0.274 |
| Stage | 2.968 | 1.844–4.779 | 0.000 * |
| MSI2 (Pos/Neg) | 0.621 | 0.299–1.288 | 0.201 |
Figure 1.
(A) Negative OSCC for MSI2; (B) Basal MSI2 positivity intensity +2; (C) 40% Normal mucosa negative for MSI2 with mild positivity in a ductal epithelium; and, (D) normal mucosa mild positive for MSI2 in the basal layer.
3. Discussion
OSCC represents more than 90% of oral cancers and it is one of the most aggressive cancers, being characterized by a mortality rate reaching 50% of patients on average [13]. Continuous efforts are made to better understand the processes that lead to its onset and progression, as well as to the discovering of potential targets for its therapy. Similarly to other solid tumors, the onset of OSCC results from the accumulation of a certain number of genetic or epigenetic alterations into the cells, which cause cell cycle dysregulation and uncontrolled cell proliferation [14] Recently, the new “omic” sciences provided a great amount of data that characterize the tumors at the molecular level and that can lead to discovering specific biomarkers that could make the tumor treatment more efficient, precise, and predictable [15]. A great contribution has been obtained from bioinformatics that allowed for analyzing this enormous amount of data, giving them a clinical significance [16].
Musashi RNA-binding protein 2 (MSI-2) has been demonstrated to be involved in several solid and blood cancers, where its expression emerged to be higher than in normal tissues and correlate with the prognosis [17]. Its role seems to be explicated in different processes, among which: epithelial-mesenchymal transition, migration, invasion, cell proliferation, and drug resistance [17]. While for many tumors, such as those arising from breast [18], cervical [9], colon [19], lung [6], etc. the role of MSI-2 proteins has been extensively studied and some target therapies proposed, no results regarding the role of MSI-2 in OSCC are reported in the literature. In this work, we combined both a bioinformatics analysis of data that were extracted from electronic TCGA database and an immunohistochemical evaluation of our samples to better understand the role of MSI-2 in oral cancer. The first interesting result emerged from the analysis of genomic data revealed that no DNA mutations and copy number alterations are detected for the MSI2 gene in patients with OSCC. A hyper methylated condition of the gene emerged in all the patients from the investigation of epigenetic modifications. The analysis of transcriptomic data showed a relatively low expression of MSI2 mRNA in the OSCC samples. However, the mRNA expression did not result to be significantly correlated to the methylation status of the gene.
The associations between its mRNA expression and some clinicopathological characteristics of patients have been analyzed to explore the clinical value of MSI2. The analysis revealed that MSI2 mRNA expression correlates with tumor grading and males show a higher level of MSI2 mRNA expression when compared to female patients. No other clinical features seemed to significantly correlate with MSI2 mRNA expression.
As to our knowledge, no difference between sexes has been highlighted in the literature until now regarding the expression of these regulating molecules. Nevertheless, in our study Males showed significantly higher expression levels of both MSI2 and Cyclin-D1. Such results emerged from the analysis of TCGA database and they have been confirmed by our samples. This could be the decisive contribution that big data can give us in the path towards increasingly personalized medicine [16].
Another question of this work aimed to investigate whether the expression of MSI2 in OSCC patients could predict the prognosis. The results emerged from correlation analysis lay for a significant association between high MSI2 mRNA expression and poor overall survival rate; meanwhile, the disease free survival seems not to be correlated with the MSI2 mRNA expression level.
We performed the immunohistochemical evaluation of healthy mucosae and OSCC samples to determine the expression levels of MSI2, in parallel with this bioinformatic analysis. It was of great interest to discover that the protein expression of MSI2 was low or even absent both in healthy samples and OSCC TMA and it did not correlate with any prognostic behavior. These data, combined with those deriving from TCGA analysis, lead us to affirm that, unlike many other cancers [8,20,21], for OSCC the expression of MSI2 appears to be a poor prognostic biomarker.
In addition to these results, some information regarding the potential biological functions of MSI2 in oral cancer emerged from the evaluation of the TMAs. In particular, the presence of MSI2 expression directly correlated with Cyclin-D1 expression. Cyclin-D1 is a protein that plays a crucial role in cell cycle regulation, including cell proliferation and growth, as well as DNA repair and cell migration control [22]. Its key role in tumorigenesis of several tumors, among which oral cancer, has been proposed [12], and a poor prognosis correlated to its overexpression [23]. Several mechanisms of cyclin D1 overexpression in OSCC have been identified. They range from amplification to polymorphisms and mutational events involving the oncogene CCND1, but a fundamental role of some signaling pathway intermediaries has been also suggested [24]. Zhang et al. [25] demonstrated in their study how MSI2 silencing inhibited leukemic cell growth and caused a decreasing of Cyclin D1 expression. Han et al. [26] drew the same conclusions, showing how the MSI2 silencing induced cell cycle arrest in G0/G1 phase, with decreased Cyclin D1 and increased p21 expression. In the same way, a study investigating the role of MSI2 in Hematopoietic stem cell activity discovered a close correlation between the expression profile of MSI2 and that of Cyclin D1 [27]. Given the results of our study, in a similar manner, MSI2 could affect the Cyclin D1 expression in the cells of OSCC, but further studies are needed to affirm this.
4. Methods and Materials
4.1. Analysis of MSI2 Expression and Methylation in The Cancer Genome Atlas (TCGA)
The gene expression RNAseq data HTSq-Fragments Per Kilobase Million (FPKM) were downloaded from UCSC Xena Browser (https://xena.ucsc.edu/) [28]. Data were downloaded for MSI2, MKI67, and CCND1 mRNA expression. This platform was also used to access and download the methylation profile quantification for MSI2 (https://gdc.xenahubs.net/download/TCGA-HNSC/Xena_Matrices/TCGA-HNSC.methylation450.tsv.gz; Full metadata—Illumina Human Methylation 450 expressed as beta unit). Data were organized in Microsoft Excel sheet and then pasted in cBioPortal for Cancer genomics in order to display visually the patients’ profile (http://www.cbioportal.org) [29]. Clinic-pathological and follow-up information were downloaded from Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov/) [30].
4.2. Immunohistochemistry of MSI2 Expression in OSCC Tissue Microarray
The reporting recommendations for tumor marker prognostic studies (REMARK guidelines) [31] were taken as a reference for carrying out this study. All of the patients filled written informed consent for the use of their samples, according to the institutional regulations and the ethics committee of the National Cancer Institute “Giovanni Pascale”, as “Bio-Banca Istituzionale BBI” Deliberation NO. 15 del 20 Jan. 2016, approved and registered the study. We decided to exclude patients with HPV-positive tumors and those arising from the base of the tongue, tonsils, oropharynx, and lips. Patients with a follow-up lower than eight months were also excluded. A total of 122 patients were included in this study. Of them, 103 reported follow-up information (from 8 to 150 months—mean of 47.34 S.D. 34.609), meanwhile the microarray tissue was not evaluable for 14 patients. None of the patients had undergone treatments prior to tissue collection. The patients were diagnosed of OSCC and 7th American Joint Committee on Cancer (AJCC) staging system was applied. An evaluation of 13 mucosae samples from healthy subjects has been also performed. The paraffin blocks were cored in a 0.6 mm support (area of 0.28 mm2) and then transferred to the recipient master block while using Galileo TMA CK 3500® Tissue Microarrayer. As the control, we used an H&E staining of a 4-μm TMA section. Immuno-histochemical staining was performed by using a mouse monoclonal antibody (Ab), which was supplied by abcam (Mouse monoclonal Anti-MSI2 antibody [OTI2F10]—ab156770), in addition staining for Cyclin-D1 (Ventana-Roche, SP4-R) and Ki-67 (Ventana-Roche) was also performed. We used an automated staining device (Ventana-Roche), with a streptavidin-biotin horseradish peroxidase technique (LSABHRP), in order to uncover the primary Abs. An optical microscope (OLYMPUS BX53, at ×200) detected immune-stained spots in four high power fields (HPFs) and they were analyzed by ISE TMA Software (Integrated System Engineering, Milan, Italy). Two of the authors (GP and GT) performed the observational quantification analysis in a joint session. Detre S. et al. method was applied to assess the scoring of immunostaining [32]. The intensity (I) of expression was scored from 0 to 3 (0 = no staining; 1 = yellow; 2 = light brown; and, 3 = black brown/black). The relative number of the positive stained cells (%) was scored from 0 to 4 (0 = 0%; 1 < 10%; 2 = 10-50%; 3 = 51-80%; 4 > 80%). All of the samples resulted in only being stained in the cytoplasm. We decided to categorize patients in negative/positive tumors because of the relative low expression of MSI2 in OSCC.
4.3. Statistical Analysis
SPSS statistical software 21.0 was used to perform all of the statistical analyses. Spearman rank correlation analysis was performed to investigate the correlation of MSI2 expression to clinic-pathological variables. We decided to use non-parametric tests because of the non-normal distribution of the variable checked by means of the Shapiro–Wilk normality test. For such reason, the Mann–Whitney test was applied to explore the difference in expression between groups. Univariate survival analysis was performed with the Kaplan–Meier method to estimate both overall survival rates and disease-free survival, while comparing results between groups with the log-Rank test. A multivariate Cox Proportional Hazard Model was built in order to assess the prognostic significance of MSI2 expression after adjusting for covariates, including the clinic-pathological variables: age, grading, staging (8th AJCC edition), and gender, as covariates.
5. Conclusions
The role of MSI2 expression in OSCC seems to be not so closely correlated with prognosis, as in other solid and blood tumors. The MSI2 mRNA expression in oral cancer is higher in males and it is correlated with tumor grade. The protein expression of MSI2 in OSCC samples is relatively low, but significantly correlated with Cyclin D1 expression. Further studies are necessary to investigate its role in OSCC genesis, progression, and prognosis.
Author Contributions
G.T.: V.C.A.C., L.L.R. performed bioinformatic analysis, G.B., G.A., N.S.L., M.C.P., and G.P. collected, prepared and analyzed semples, K.Z., C.A., D.C., and F.M. contributed to write the manuscript, G.T., L.L.M., and G.P. conceived and supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
The research has been funded by authors’ own institutions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ghantous Y., Abu Elnaaj I. Global Incidence and Risk Factors of Oral Cancer. Harefuah. 2017;156:645–649. [PubMed] [Google Scholar]
- 2.Attar E., Dey S., Hablas A., Seifeldin I.A., Ramadan M., Rozek L.S., Soliman A.S. Head and neck cancer in a developing country: A population-based perspective across 8 years. Oral Oncol. 2010;46:591–596. doi: 10.1016/j.oraloncology.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingen M.W., Pinto A., Mendes R.A., Franchini R., Czerninski R., Tilakaratne W.M., Partridge M., Peterson D.E., Woo S.B. Genetics/epigenetics of oral premalignancy: Current status and future research. Oral Dis. 2011;17(Suppl. 1):7–22. doi: 10.1111/j.1601-0825.2011.01789.x. [DOI] [PubMed] [Google Scholar]
- 4.Fritzell K., Xu L.D., Lagergren J., Ohman M. ADARs and editing: The role of A-to-I RNA modification in cancer progression. Semin Cell Dev. Biol. 2018;79:123–130. doi: 10.1016/j.semcdb.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Soni S., Anand P., Padwad Y.S. MAPKAPK2: The master regulator of RNA-binding proteins modulates transcript stability and tumor progression. J. Exp. Clin. Cancer Res. 2019;38:121. doi: 10.1186/s13046-019-1115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudinov A.E., Deneka A., Nikonova A.S., Beck T.N., Ahn Y.H., Liu X., Martinez C.F., Schultz F.A., Reynolds S., Yang D.H., et al. Musashi-2 (MSI2) supports TGF-beta signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. USA. 2016;113:6955–6960. doi: 10.1073/pnas.1513616113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan Y., Chen Z., Li Y., He A., He S., Gong Y., Li X., Zhou L. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J. Exp. Clin. Cancer Res. 2018;37:273. doi: 10.1186/s13046-018-0921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z., Li J., Shi Y., Li L., Guo X. Increased musashi 2 expression indicates a poor prognosis and promotes malignant phenotypes in gastric cancer. Oncol. Lett. 2019;17:2599–2606. doi: 10.3892/ol.2019.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Fan Y., Wang X., Huang Z., Shi K., Zhou B. Musashi-2 is a prognostic marker for the survival of patients with cervical cancer. Oncol. Lett. 2018;15:5425–5432. doi: 10.3892/ol.2018.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M.R., Xi S., Shukla V., Hong J.A., Chen H., Xiong Y., Ripley R.T., Hoang C.D., Schrump D.S. The Pluripotency Factor Musashi-2 Is a Novel Target for Lung Cancer Therapy. Ann. Am. Thorac. Soc. 2018;15:S124. doi: 10.1513/AnnalsATS.201707-608MG. [DOI] [Google Scholar]
- 11.Santarius T., Shipley J., Brewer D., Stratton M.R., Cooper C.S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 12.Ramos-Garcia P., Gil-Montoya J.A., Scully C., Ayen A., Gonzalez-Ruiz L., Navarro-Trivino F.J., Gonzalez-Moles M.A. An update on the implications of cyclin D1 in oral carcinogenesis. Oral Dis. 2017;23:897–912. doi: 10.1111/odi.12620. [DOI] [PubMed] [Google Scholar]
- 13.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 14.Feller L.L., Khammissa R.R., Kramer B.B., Lemmer J.J. Oral squamous cell carcinoma in relation to field precancerisation: Pathobiology. Cancer Cell Int. 2013;13:31. doi: 10.1186/1475-2867-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonella L., Giannoccaro M., Alfieri S., Canevari S., De Cecco L. Gene Expression Signatures for Head and Neck Cancer Patient Stratification: Are Results Ready for Clinical Application? Curr. Treat. options in Oncol. 2017;18:32. doi: 10.1007/s11864-017-0472-2. [DOI] [PubMed] [Google Scholar]
- 16.Chin L., Andersen J.N., Futreal P.A. Cancer genomics: From discovery science to personalized medicine. Nat. Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 17.Kudinov A.E., Karanicolas J., Golemis E.A., Boumber Y. Musashi RNA-Binding Proteins as Cancer Drivers and Novel Therapeutic Targets. Clin. Cancer Res. 2017;23:2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M.H., Jeong K.J., Kim W.Y., Lee H.J., Gong G., Suh N., Gyorffy B., Kim S., Jeong S.Y., Mills G.B., et al. Musashi RNA-binding protein 2 regulates estrogen receptor 1 function in breast cancer. Oncogene. 2017;36:1745–1752. doi: 10.1038/onc.2016.327. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang S.W., Liu T.T., Liu X.S., Zhu F.X., Zhu F.M., Liu X.N., Peng Z.H. USP10 regulates Musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett. 2019;593:406–413. doi: 10.1002/1873-3468.13323. [DOI] [PubMed] [Google Scholar]
- 20.Zong Z., Zhou T., Rao L., Jiang Z., Li Y., Hou Z., Yang B., Han F., Chen S. Musashi2 as a novel predictive biomarker for liver metastasis and poor prognosis in colorectal cancer. Cancer Med. 2016;5:623–630. doi: 10.1002/cam4.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong P., Xiong Y., Hanley S.J.B., Yue J., Watari H. Musashi-2, a novel oncoprotein promoting cervical cancer cell growth and invasion, is negatively regulated by p53-induced miR-143 and miR-107 activation. J. Exp. Clin. Cancer Res. 2017;36:150. doi: 10.1186/s13046-017-0617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z., Wang C., Jiao X., Lu Y., Fu M., Quong A.A., Dye C., Yang J., Dai M., Ju X., et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol. Cell. Biol. 2006;26:4240–4256. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y., Yu D., Li H., Nie P., Zhu Y., Liu S., Zhu M., Fang B. Cyclin D1 overexpression is associated with poor clinicopathological outcome and survival in oral squamous cell carcinoma in Asian populations: Insights from a meta-analysis. PLoS ONE. 2014;9:e93210. doi: 10.1371/journal.pone.0093210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen K.E., Diehl J.A., Haiman C.A., Knudsen E.S. Cyclin D1: Polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Tan S., Wang J., Chen S., Quan J., Xian J., Zhang S., He J., Zhang L. Musashi2 modulates K562 leukemic cell proliferation and apoptosis involving the MAPK pathway. Exp. Cell Res. 2014;320:119–127. doi: 10.1016/j.yexcr.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Han Y., Ye A., Zhang Y., Cai Z., Wang W., Sun L., Jiang S., Wu J., Yu K., Zhang S. Musashi-2 Silencing Exerts Potent Activity against Acute Myeloid Leukemia and Enhances Chemosensitivity to Daunorubicin. PLoS ONE. 2015;10:e0136484. doi: 10.1371/journal.pone.0136484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hope K.J., Cellot S., Ting S.B., MacRae T., Mayotte N., Iscove N.N., Sauvageau G. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 2010;7:101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D., et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossman R.L., Heath A.P., Ferretti V., Varmus H.E., Lowy D.R., Kibbe W.A., Staudt L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McShane L.M., Altman D.G., Sauerbrei W., Taube S.E., Gion M., Clark G.M., Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat. Clin. Pract. Oncol. 2005;2:416–422. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 32.Detre S., Saclani Jotti G., Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]