Abstract
We designed, synthesized, and evaluated novel 2,6,9-trisubstituted purine derivatives for their prospective role as antitumor compounds. Using simple and efficient methodologies, 31 compounds were obtained. We tested these compounds in vitro to draw conclusions about their cell toxicity on seven cancer cells lines and one non-neoplastic cell line. Structural requirements for antitumor activity on two different cancer cell lines were analyzed with SAR and 3D-QSAR. The 3D-QSAR models showed that steric properties could better explain the cytotoxicity of compounds than electronic properties (70% and 30% of contribution, respectively). From this analysis, we concluded that an arylpiperazinyl system connected at position 6 of the purine ring is beneficial for cytotoxic activity, while the use of bulky systems at position C-2 of the purine is not favorable. Compound 7h was found to be an effective potential agent when compared with a currently marketed drug, cisplatin, in four out of the seven cancer cell lines tested. Compound 7h showed the highest potency, unprecedented selectivity, and complied with all the Lipinski rules. Finally, it was demonstrated that 7h induced apoptosis and caused cell cycle arrest at the S-phase on HL-60 cells. Our study suggests that substitution in the purine core by arylpiperidine moiety is essential to obtain derivatives with potential anticancer activity.
Keywords: cancer, purine derivatives, cytotoxicity, 3D-QSAR, apoptosis, cell cycle
1. Introduction
The generic term “cancer” refers to a large and complex group of diseases that can occur in virtually any part of the body. Due to abnormal growth of cells, these cells proliferate uncontrollably and, in some cases, metastasize [1]. As a consequence, there are more than 100 types of cancer [2], and despite the diversity and complexity of this disease, general principles governing the transformation of a normal cell into a malignant cell have been established [3]. The International Agency for Research on Cancer (IARC) estimates that 14.1 million new cancer cases occurred worldwide in 2012, and 8.2 million cancer deaths in the same year. Also, it is estimated that there are currently 32.4 million living cancer patients, despite being diagnosed with cancer the last five years. Lung and prostate cancer are the most common types for men, whereas for women, breast and colorectal cancer are the most frequent [4]. Today, chemotherapy is an important global research undertaking because of the urgent necessity to improve the longevity and the quality of life for those suffering with cancer. Although chemotherapy has become routine in most anticancer treatments, this therapeutic approach is currently limited by the ability of cancer cells to develop several resistance mechanisms to conventional drugs as an undesirable secondary effect. Therefore, new antitumor therapies aim at developing safer and more selective anticancer drugs that exhibit cytotoxic activity on malignant cells, without causing damage to healthy cells [5,6,7,8].
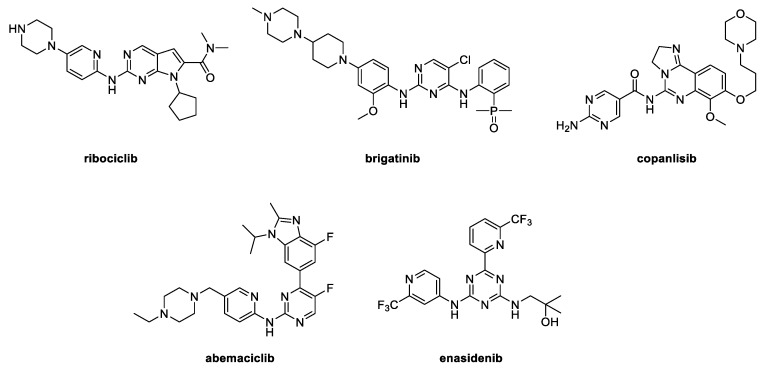
Chemically, most current synthetic anticancer drugs are heterocyclic compounds and nitrogen heterocycles [9,10,11,12]. This fact is observed by analyzing the chemical structures of some anticancer drugs approved by the FDA (Food and drug administration) in 2017, which share a pyrimidine scaffold as part of their structures (Figure 1). These compounds exert their antitumor activity through inhibition of protein kinases [13]. Therefore, this moiety is an interesting building block to consider in anticancer fragment-based drug discovery [14,15]. Interestingly this fragment is part of the purine core, which is considered a privileged scaffold in medicinal chemistry [16,17,18,19]. For this reason, a large number of purine derivatives have been synthetized and their antitumor activity has been reported [20,21,22,23]. Compounds I [24] and II [25] (Figure 2) are examples of di- and tri-substituted purine derivatives that have already been tested against cancer cells. Both of these derivatives show anticancer activity on some cancer cell lines with sub-micromolar IC50 values, and are therefore considered as potential drug candidates. In addition to the purine scaffold, compounds I and II contain a piperazinyl group and a small and rigid heterocyclic backbone. These characteristics amount to an attractive pharmacological scaffold used in other anticancer pharmaceuticals [26].
Figure 1.
Chemical structures of some FDA (Food and drug administration)-approved anticancer drugs in 2017.
Figure 2.
Chemical structures of 2,6,9-trisubstituted purines with biological properties (I–V) and proposed compounds of Series I–III.
In addition to the aforementioned, the purine nucleus has been used as a fragment in the development of ligands against a host of biological targets. Most of this work has been done on the design of purine derivatives such as kinase inhibitors, because the dysregulation of these proteins is implicated in several processes of carcinogenesis [17,27]. The substitution pattern of the purine ring has been explored, and therefore it is anticipated that introduction of substituents at the C-2, C-6, and N-9 positions will afford compounds with enhanced binding affinity and selectivity toward kinases [17,27]. Some purine-based compounds which target these proteins have been synthesized, and considering the essential role of some kinases in the regulation of the cell cycle or proliferation signaling, have transformed these compounds into new and potential anticancer agents [17,27,28]. Some examples of 2,6,9-trisubstituted purine with these biological properties are compounds III [29] and IV [30] (Figure 2). In this sense, III is a potent tyrosine kinase inhibitor that elicits inhibition on cyclin-dependent kinases (CDK), Src, and VEGFR2, and all of these targets are related to cancer therapy. Furthermore, IV is a potent and selective CDK2 inhibitor. In addition, the binding modes of IV with CDK1 and CDK2 were determined from their crystal structures, which have been useful to understand inhibition mechanism, as well as some features of the active site that are key for designing selective CDK inhibitors. On the other hand, compound V (Figure 2) was reported as an Hedgehog (Hh) signaling pathway inhibitor [31]. This aberrant regulated signaling pathway could lead to constitutive activation of a variety of cancers [32]. Compound V, a 2,6,9-trisubstituted purine derivative, was the most promising compound in this study, showing an IC50 value on a nanomolar scale into Gli-luciferase reporter assay.
Therefore, considering that the purine heterocycle scaffold is an attractive option for developing new antitumor compounds, we decided to design a new family of 2,6,9-trisubstituted purine derivatives (Series I–III). We analyzed the effectiveness of N-alkyl substitutions, as well as the substitutions at C-2 and C-6 by nitrogenated functions at the purine core, on antitumor activity. We then tested the synthesized compounds against eight cell lines in order to observe their cytotoxicity. These cell lines included MCR-5 cells and seven human-derived tumor cell lines. Our study also included 3D-QSAR studies using comparative molecular field analysis (CoMFA) [33], which allowed us to analyze the structure–activity relationships among them. Our results generated here will provide the basis for future design of new cytotoxic agents with better potency and selectivity.
2. Results
2.1. Synthesis
We synthesized new 2,6,9-trisubstituted purines 4a–k (Series I), 4l–u (Series II), and 7a–j (Series III), using short, simple, and efficient synthetic methods as described in Scheme 1 and Scheme 2 [22,34,35]. We obtained 21 compounds of Series I and II in three steps using 2-fluoro-6-chloropurine (1) as a starting material. Then, 10 compounds of Series III were achieved in two steps from 2,6-dichloropurine (5). In this series, the first step was the alkylation of 1 or 5 with the respective alkyl halides under basic conditions to give a mixture of N-9 and N-7-alkylated purines regioisomers 2a–e:2a’–e’ (or 6a:6a’), in a proportion 4:1 in the majority of cases [22]. For Series I and II, the second step was a regioselective Suzuki reaction at C-6 of the major regioisomers, 2a–e with trifluoromethoxyphenylboronic acid to give the arylpurine derivatives 3a–e. The third step was a nucleophilic substitution (SNAr) at position C-2 with several amines using n-butanol as solvent, N,N-diisopropylethylamine as base, at 110 °C for 12 h to obtain high yields of compounds 4a–u. For Series III, the second step was the regioselective SNAr at C-6 of 6a with different arylpiperazines or 4-phenylpiperidine, to yield purine derivatives 7a–j in moderate-to-high yields. We purified all compounds by column chromatography and established their structures based on their spectral properties (IR, MS, 1H NMR, and 13C NMR; see experimental section and Supplementary Information for HRMS, 1H NMR, and 13C NMR spectra).
Scheme 1.
Reagents and conditions: (1) Alkyl halides, K2CO3, DMF, rt, 12 h (34–60%); (2) 4-trifluoromethoxyphenylboronic acid, Pd(PPh3)2Cl2, K2CO3 2M, dioxane, reflux, 2 h (41–57%); (3) amine, DIPEA, n-BuOH, 110 °C, 12 h (54–96%).
Scheme 2.
Reagents and conditions: (1) Cyclopentyl bromide, K2CO3, DMF, rt, 12 h (55%); (2) arylpiperazines or phenylpiperidine, DIPEA, n-BuOH, 110 °C, 12 h (60–92%).
2.2. Crystallographic Studies
In addition, we determined molecular structures of two purine derivatives, 4f (CCDC 1475770) and 7e (CCDC 1475774), by X-Ray diffraction analyses (Figure 3). Derivative 4f showed a planar purine core (dihedral angle: 179.1(2)° N1–C2–C5–N3) with three different substituents. The n-propyl substituent was bonded to the N4 atom, and was outside of the purine plane with a dihedral angle of -93.1(2)° (C2–N4–C19–C20). The trifluoromethoxyphenyl group was attached to the C4 atom and was almost coplanar with the purine core and exhibited a dihedral angle of -14.1(3)° (N2–C4–C6–C11). Finally, the cyclohexylmethyl substituent bonded amine group exhibited a boat conformation, which was found outside of the amine plane with a dihedral angle of 77.0(6)° (C3–N5–C12–C13) and it was orientated in the same direction of the propyl substituent of the N4 atom. On the other hand, derivative 7e exhibited a planar purine core (dihedral angle: 179.0(1)° N1–C2–C5–N3) with two substituents. The piperidine moiety adopted a boat conformation and the phenyl group was almost orthogonal to the piperidine with a dihedral angle of -9.3(2)° (H8–C8–C11–C16). The phenylpiperidine fragment of the C4 was bent out of the purine plane with a dihedral angle of 132.1(6)° (C4–N5–C6–C7). The cyclopentyl substituent bonded to N4 was bent out of the purine plane (dihedral angle of -72.7(0)°, C2–N4–C17–C18), but in the opposite direction of the phenylpiperidine group.
Figure 3.
Molecular structure of selected purines 4f and 7e obtained by X-Ray diffraction analyses. Thermal ellipsoids are shown with 30% probability.
2.3. Cytotoxic Studies
In a first step for development of new potential antitumor drugs, it was crucial to determine in vitro antitumor action exhibited by these compounds by testing their cytotoxic effects on a panel of human cancer cell lines [36]. Likewise, it was important that a prospective antitumor drug must demonstrate low toxicity in untransformed cells; and therefore, a normal cell line must be used as a control. With this goal in mind, we fashioned a conventional colorimetric assay to determine IC50 values, which represent the necessary concentration of a compound required for 50% in vitro inhibition following 72 h of sustained exposure to such compound [22,37]. Serial dilutions (from 0.1 to 25, 50, or 100 µM, depending on solubility) for each compound were evaluated in triplicate, and cisplatin was used as a reference drug. In our study, the human cancer cell lines used were CFAPC- 1 (pancreatic adenocarcinoma); NCI-H460 (large cell lung); HL-60 (acute promyelocytic leukaemia); CACO2 (colorectal adenocarcinoma); HCT-116 (colorectal carcinoma); K562 (chronic myeloid leukaemia); MCF-7 (breast adenocarcinoma); and MRC-5 control cells (normal fibroblasts).
Table 1 gives IC50 values for 4a–u and 7a–j compounds’ cytotoxicity against the aforementioned cancer cell lines and the control group MRC-5 cells. Overall, the cytotoxicity of 2,6,9-trisubstituted purines was heterogeneous. This depended also on the type of cancer cell line, as well as for the series of the assayed compounds. For example, CACO2 cells appeared more resistant (all compounds tested with IC50 values >100 µM), except for 4r (IC50 = 27 µM); meanwhile, the most sensitive cell line was HL-60. Likewise, from a chemical point of view, compounds belonging to Series III were more active on the most cancer cell lines than those of Series I and II. The latter-described behavior was clearly observed for K562 and MCF-7 cell lines, in which all compounds from Series I and II were inactive (IC50 values >25 µM). Considering that a prospective antitumor compound must show low toxicity in mammalian host cells, we calculated the selective index (SI) values for the most active compounds (Table 2). However, after further cytotoxicity and selectivity analysis for each cancer cell line, we concluded that:
Table 1.
In vitro cytotoxicity of compounds 4a–u and 7a–j on cancer and MRC-5 cell lines.
| Entry | IC50 (µM) a | |||||||
|---|---|---|---|---|---|---|---|---|
| CFPAC-1 | NCI-H460 | HL-60 | CACO2 | HCT-116 | K562 | MCF-7 | MRC-5 | |
| 4a | >100 | >100 | 38 ± 3.0 | >100 | >50 | >25 | >25 | >100 |
| 4b | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4c | 67 ± 1.0 | 64 ± 2.0 | 25 ± 5.4 | >100 | >50 | >25 | >25 | 60 ± 1.0 |
| 4d | >100 | >100 | 21 ± 1.0 | >100 | >50 | >25 | >25 | >100 |
| 4e | >100 | >100 | 18 ± 4.0 | >100 | >50 | >25 | >25 | >100 |
| 4f | 69 ± 1.0 | >100 | 9.8 ± 1.5 | >100 | >50 | >25 | >25 | 68 ± 1.0 |
| 4g | 98 ± 5.0 | 72 ± 2.0 | 21 ± 5.0 | >100 | >50 | >25 | >25 | >100 |
| 4h | >100 | >100 | 14 ± 1.0 | >100 | >50 | >25 | >25 | >100 |
| 4i | >100 | >100 | 19 ± 3.9 | >100 | >50 | >25 | >25 | >100 |
| 4j | >100 | >100 | 6.0 ± 0.8 | >100 | >50 | >25 | >25 | >100 |
| 4k | >100 | >100 | 12 ± 0.6 | >100 | >50 | >25 | >25 | >100 |
| 4l | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4m | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4n | >100 | 42 ± 1.0 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4o | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4p | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4q | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4r | >100 | 32 ± 1.0 | 31 ± 8.8 | 27 ± 10 | >50 | >25 | >25 | 41 ± 1.0 |
| 4s | >100 | 91 ± 9.0 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4t | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 4u | >100 | >100 | >50 | >100 | >50 | >25 | >25 | >100 |
| 7a | >100 | 5.73 ± 0.37 | 2.1 ± 0.03 | >100 | 6.4 ± 1.3 | 0.87 ± 0.01 | 0.95 ± 0.14 | >100 |
| 7b | >100 | >100 | >50 | >100 | >50 | 3.06 ± 0.54 | 5.07 ± 0.90 | >100 |
| 7c | >100 | 4.79 ± 0.07 | 17 ± 2.3 | >100 | >50 | 0.39 ± 0.03 | 0.52 ± 0.05 | 56 ± 1.0 |
| 7d | >100 | 8.25 ± 0.11 | 4.6 ± 0.2 | >100 | >50 | 5.86 ± 1.06 | > 12.5 | 17 ± 1.0 |
| 7e | >100 | 11 ± 1.0 | 3.0 ± 0.6 | >100 | >50 | - | - | >100 |
| 7f | >100 | 6.71 ± 0.23 | 1.3 ± 0.5 | >100 | >50 | 4.22 ± 0.27 | 6.13 ± 0.25 | >100 |
| 7g | >100 | 2.21 ± 0.03 | 0.30 ± 0.06 | >100 | >50 | 0.71 ± 0.07 | 0.97 ± 0.07 | 2.54 ± 0.17 |
| 7h | >100 | 1.31 ± 0.07 | 0.40 ± 0.01 | >100 | 30 ± 7.1 | 1.02 ± 0.04 | 1.55 ± 0.34 | >100 |
| 7i | >100 | 8.54 ± 0.21 | 3.8 ± 1.5 | >100 | >50 | 9.96 ± 0.93 | 22 ± 1 | >100 |
| 7j | >100 | 7.08 ± 0.22 | 1.5 ± 0.07 | >100 | >50 | 3.38 ± 0.71 | 6.61 ± 0.67 | 30 ± 1.0 |
| Cisplatin | 3.16 ± 0.10 | 4.77 ± 0.13 | 6.2 ± 0.2 | 2.23 ± 0.10 | 3.89 ± 1.30 | 4.67 ± 0.38 | 10.9 ± 0.8 | 6.78 ± 0.18 |
From MTT assay after 72 h of treatment in the range 0.1 to 25, 50, or 100 µM, depending of the compound solubility. a IC50 values were determined in three independent experiments for triplicate and all of them are expressed as IC50 ± standard deviation (SD).
Table 2.
Selectivity Index (SI) a values for the most active purine derivatives.
| Compound | MRC-5/NCI-H460 | MRC-5/HL-60 | MRC-5/K562 | MRC-5/MCF-7 |
|---|---|---|---|---|
| 4j | - | 16.7 | - | - |
| 7a | 17.5 | 47.6 | 115 | 105 |
| 7b | - | - | 32.7 | 19.7 |
| 7c | 11.7 | 3.3 | 143.6 | 107.7 |
| 7d | 2.0 | 3.7 | 2.9 | - |
| 7e | 9.1 | 33.3 | - | - |
| 7f | 14.9 | 76.9 | 23.8 | 16.3 |
| 7g | 1.1 | 8.5 | 3.6 | 2.6 |
| 7h | 76.3 | 250 | 98 | 64.5 |
| 7i | 11.7 | 26.3 | 10 | 4.5 |
| 7j | 4.2 | 20.0 | 8.8 | 4.5 |
| Cisplatin | 1.4 | 1.1 | 1.5 | 0.6 |
a Selectivity index (SI) = IC50 of pure compound in an MRC-5/IC50 of the same pure compound in respective cancer cell lines.
(1) For CFPAC-1, CACO2 cells, and HCT-116 cells, all compounds were inactive compared with cisplatin. Except 4r for Caco2 cells or 7a, which elicited a weak activity on CACO2 and HCT-116 cancer cell lines, respectively.
(2) For NCI-H460 cells, compounds 7c, 7g, and 7h (IC50 values = 4.8, 2.2, and 1.3 μM, respectively) exhibited better or equal potency than cisplatin (IC50 = 4.8 μM). Nevertheless, 7a, 7d–f, 7i, and 7j showed moderate activity with IC50 values <10 µM. In relation to selectivity, all of the aforementioned compounds were more selective (SI values between 2–76) than cisplatin, except 7g (SI = 1.1). Interestingly, 7h gave the highest SI, being almost 70 times more selective to NCI-460 than MRC-5, which is a remarkable result in this study.
(3) Our best results were for HL-60 cells, in terms of potency and selectivity of the purine derivatives compared to cisplatin. Nine of these (4j, 7a, and 7d–j) elicited IC50 values of less than 6.0 μM, especially highlighting compounds 7g and 7h with IC50 values = 0.30 and 0.40 µM, respectively. In addition, these nine compounds not only showed high potency against this cancer cell line, but were also more selective (SI values higher than 3.0) than cisplatin (SI = 1.1). It is important to mention that 7f and 7h were almost 80–250 times more selective than cisplatin (SI values = 77 and 250) vs. MRC-5 cell line, being 7h again the most promising compound, showing a good potency and selectivity profile. Our group has not previously reported these results of selectivity [22,37].
(4) For K562 and MCF-7 cells, these purine derivatives had a similar behavior regarding potency and selectivity, although K562 cells were more sensitive than MCF-7 cells. Most compounds of Series III were active compared with cisplatin, where compounds 7a, 7c, and 7h showed the highest potency (IC50 values between 0.4–1.5 µM) and highest selectivity (SI values between 65–144) against both cell lines.
According to our analysis of potency (Table 1) and selectivity (Table 2), 7a, 7c, and 7f–h are interesting compounds for further biological studies, and we conclude that they are very good candidates for developing novel antitumor agents. This preliminary conclusion agrees with the National Cancer Institute (NCI) protocols, where compounds exhibiting IC50 values of <10 or 15 μM are considered active [36]. In addition to the aforementioned activity against the tested cancer cell lines and low toxicity towards normal human cells, the molecular properties of 7a, 7c, and 7f–h were predicted using the free online molecular calculation services provided by Molsoft (http//molsoft.com/mprop). These results are shown in Table 3. From the properties calculated from these compounds, all of them elicited acceptable molecular properties (the compounds agreed with Lipinski’s rules of five), which may be helpful to design more potent antitumor agents based on 2,6,9-trisubtititued purine scaffold.
Table 3.
Molecular properties of selected compounds. a
| Compound | MW | HBA | HBD | MolLogP | MolPSA(A2) | MV(A3) |
|---|---|---|---|---|---|---|
| Desirable value | <500 | <10 | <5 | <5 | <140 | - |
| 7a | 428.15 | 6 | 0 | 2.85 | 84.52 | 381.45 |
| 7c | 467.14 | 5 | 0 | 4.36 | 52.20 | 401.01 |
| 7f | 382.17 | 3 | 0 | 3.89 | 37.66 | 361.08 |
| 7g | 416.13 | 3 | 0 | 4.61 | 37.66 | 378.27 |
| 7h | 427.15 | 5 | 0 | 3.62 | 75.92 | 386.45 |
a MW: Molecular Weight; HBA: Number of hydrogen bond acceptors; HBD: Number of hydrogen bond donors; MolLogP: Log p-value predicted by Molsoft; MolPSA: Topological polar surface is; MV: Molecular volume.
2.4. Structure–Activity Relationship (SAR)
From the results shown in Table 1, it appears that an eventual structure relationship among these compounds depends on the cancer cell line, as well as the structural variations of compounds from Series I–III. In light of the IC50 values for the studied cancer cell lines, from a chemical standpoint, the following are interesting structural features found in our work:
(1) Substitution on C-2 of the purine core by a chlorine atom (Series III), instead of a nitrogenated fragment (Series I and II), could be responsible for an increase in cytotoxicity. This is the main structural difference among these purine derivatives.
(2) Interestingly, as mentioned in the introduction, arylpiperidinyl or arylpiperazinyl portions were fragments present in the most active compounds, but only if these groups were bonded at C-6 (Series III) instead of at C-2 (Series II).
(3) The substitution on N-9 seemingly is not determining of the cytotoxic effect. This observation is based on the consideration of 4g–k on HL-60 cells, where the potency was similar, independent of the large alkyl chain. However, 4j (on HL-60 cells), 4n (on NCI-H460 cells), and 4r (on NCI-H460, HL-60 and CACO2 cells) elicited major potency in their respective cell lines compared with their analogues, and all of them had a pentyl group on N-9.
(4) A comparison between compounds of Series I and II could indicate that for HL-60 cells, substitution by cycloalkyl or methylcyclohexyl at C-2 increases cytotoxicity (except for 4b) in contrast with arylpiperazine or arylpiperidine at C-2. Therefore, a reduction of the substituent volume at position C2 would be favorable for cytotoxic activity.
(5) For compounds from Series III, we could establish SAR analysis, given that there are more data from biological activities on NCI-H460, HL-60, K-562, and MCF-7 cells. Firstly, we observed a clear tendency in these four cancer cell line substitutions at para-position of the arylpiperidine moiety with electron-withdrawing groups (EWG = NO2, CF3, or Cl) to lead to an increment in antitumor activity. Secondly, for NCI-H460, K-562, and MCF-7 cells, the most active compounds had an EWG bond to the aryl fragment, which could be a benzene ring (7g or 7h) or a nitrogenated isoster ring (7a or 7c). In addition, compounds without an EWG (7d, 7e, 7f, or 7i) or with an electron donor group (EDG = OCH3, 7j) decreased antitumor activity on these three cancer cell lines. Thirdly, for HL-60 the arylpiperidine moiety required a benzene ring to arrive at the most cytotoxic compounds (7f–h and 7j), and this was the most important structural requirement if an EWG or an EDG was bonded at para-position. On the other hand, for HL-60 cells, less active compounds (7b–d and 7i) had a nitrogenated heterocyclic ring as part of the arylpiperidine moiety. Fourthly, for NCI-H460 and HL-60 cells, the phenylpiperidine moiety bonded to purine at C-6 led to a more cytotoxic compound (7f) than one that had a phenylpiperazine moiety (7e).
The aforementioned SAR analysis provided vital information about the effectiveness of chemical modification for these purine derivatives to increase cytotoxic activity in different cancer cell lines. Furthermore, 3D-QSAR analysis resulted in a precise conclusion about differences in cytotoxicity of the different compounds that we synthesized. Separate results and discussion about the 3D-QSAR assays are given in the following section.
2.5. 3D-QSAR
In order to obtain insight about the main structure–activity relationships of the compounds active in HL-60 and NCI-H460 cell lines, we performed a 3D-QSAR study through comparative molecular field analysis (CoMFA). This 3D-QSAR analysis was only possible for these cell lines considering the number of IC50 values available for consistency of calculations.
2.5.1. Statistical Results
Biological activities of the compounds in both cell lines were converted to pIC50 (= -logIC50, in molar concentration). The compounds were randomly divided into training sets (10 compounds = 71%, in the CoMFA NCI-H460 model; and 15 compounds = 75%, in the CoMFA HL60 model) and test sets (4 compounds = 29% in the CoMFA NCI-H460 model; and 5 compounds = 25% in the CoMFA HL60 model).
Statistical parameters of the models are listed in Table 4. Our obtained models presented high q2 (0.791 and 0.745), r2 (0.969 and 0.959), as well as a small standard error of estimation (SEE; 0.127 and 0.142), suggesting that they were both reliable and predictive. A good 3D-QSAR model must use a small number of components (< 33.3% of the studied compounds). As shown in Table 4, we built the models with a low number of components (N = 2 and 1) and they revealed high external predictive capability (r2pred = 0.968 and 0.976).
Table 4.
Statistical parameters of the CoMFA models a.
| Statistic Index | CoMFA NCI-H460 | CoMFA HL60 |
|---|---|---|
| q2 | 0.791 | 0.745 |
| N | 2 | 1 |
| SEP | 0.473 | 0.247 |
| SEE | 0.127 | 0.142 |
| r2 | 0.969 | 0.959 |
| F | 62.037 | 87.22 |
| S | 0.737 | 0.640 |
| E | 0.263 | 0.360 |
| SSD | 1.215 | 2.458 |
| PRESS | 0.039 | 0.060 |
| r2pred | 0.968 | 0.976 |
aq2 = the square of the LOO cross-validation (CV) coefficient; N = the optimum number of components; SEP = standard error of prediction; SEE = the standard error of estimation of non-CV analysis; r2 = the square of the non-CV coefficient; F = the F-test value; S and E = the steric and electrostatic contributions, respectively; r2pred = the predictive r2 for test set compounds; SSD = the sum of the squared deviation between the biological activity of molecules in the test set and the mean activity of the training set molecules; PRESS = the sum of the squared deviations between predicted and actual biological activity values for every molecule in the test set; r2pred, predictive r2 calculated from Equation (2).
Plots of the predicted pIC50 values versus the experimental ones for the CoMFA analysis are also shown in Figure 4 (in Tables S1 and S2, we report the experimental versus predicted activity for the compounds). In both models, most points were well distributed along the line Y = X, suggesting that the quality (robustness of internal and external predictability) of the 3D-QSAR models was good.
Figure 4.
Plots of experimental versus predicted pIC50 values for the training set and test set molecules in CoMFA studies.
2.5.2. CoMFA Contour Maps Analysis for NCI-H460
According to the CoMFA contour maps, the fact that the family of compounds 7a–j were the most active against this cell line could be explained by the following: As shown in Figure 5A, there is a large green polyhedron around the group at para-position of the aryl ring linked to the piperidine or piperazine core. This means that a bulky group in that position is highly favorable for activity. This can be corroborated by the fact that compounds without substituents at para-position were among the less active of the family (7d, 7e, and 7i) or the absence of a bulky group in compound 7b could have contributed to its inactivity, among other factors. On the other hand, the less active compounds among the 31, such as 4s (Figure 5C), did not have a piperazine or piperidine linker; therefore, they could not project a bulky group inside the green polyhedron. Likewise, in this steric contour map, a smaller yellow polyhedron is depicted near C-2 of purine core. However, the limitation to increase the size of the groups in this position seemed to be less restricted, which was also observed in HL-60 cells. Compounds 4c and 4g directed their cyclohexyl or cycloheptyl group inside the yellow polyhedron at C-2, and they were less active than compounds 4n and 4r; therefore, the use of short and bulky groups was worse than the use of the arylpiperazinyl fragment. On the other hand, at the end of the alkyl chain in compounds 4n, 4r, and 4s, there were two yellow polyhedrons (Figure 5C), which means that the use of long chains is detrimental for biological activity. Finally, the electrostatic contour map (Figure 5B) shows a blue polyhedral which intersects with the nitrogen atom of nitro group of compounds 7a and 7h, and a red polyhedral which intersects with Cl– and CF3– groups of compounds 7c and 7g, indicating the use of an EWG at C-4 of the aryl fragment was highly favorable for activity. In Figure 5D, it can be observed that the CF3– group of 4s did not reach the electronegative favorable region.
Figure 5.
CoMFA NCI-H460: Steric (A,C) and electrostatic (B,D) contour maps around the most active compound of the series, 7h (A,B), and the least active compound of the series, 4s (C,D).
2.5.3. CoMFA Contour Maps Analysis for HL-60
In the steric contour map (Figure 6A,C), a big yellow polyhedron can be observed around the substituents at position C-2 of the purine core. This means that the use of bulky groups at this position is not favorable for activity. In fact, this trend is corroborated due to compounds with bulky cycles like cyclohexyl or cycloheptyl (compounds 4c–k), or with arylpiperazinyl or arylpiperidinyl fragment bonds at C-2 (compounds 4l–u), are less potent like compounds that carry a chlorine atom in that position (compounds 7a–j, except 7b), which was consistent with the SAR analysis described in the previous section. Therefore, the use of halogens or smalls groups like methyl, ethyl, CF3–, or NO2–, among others, at position C-2 would be the best option to future drug designs. On the other hand, a restrictive yellow polyhedron surrounds the CF3– group attached at para-position of the benzene ring in compounds 4a–k and 4r (Figure 6C), suggesting that the use of small groups at this position would be favorable for activity. In the electrostatic contour map (Figure 6B,D), a blue polyhedron is near the CF3– groups (Figure 6D), which means that electropositive groups would be useful for the activity. Finally, there are two green polyhedrons around positions 3-, 4-, and 5- of the aryl moiety linked to the piperazine in compounds 7a–j (Figure 6A). This means that bulky groups at these positions is favorable. In fact, the most active compounds, 7h and 7i, have bulky groups like Cl– and NO2– at position 4. The change in position for these groups or the assay of halogens like Br– or I– would be interesting to explore in future studies.
Figure 6.
CoMFA HL-60: Steric (A,C) and electrostatic (B,D) contour maps around the most active compound of the series 7g (A,B) and the least active compound of the series 4a (C,D).
2.6. Effect of Purine Derivatives on Cell Death
Next, in order to determinate the cell death type induced by the most promising compounds, we performed a flow cytometry analysis of cell viability in cell line HL-60 treated with the most promising compounds (selected according to their IC50 values and SI). The HL-60 cancer cells were chosen due to their sensitivity to prepared compounds. The cells were incubated for 24 h in the presence of 50 μM of the 7a, 7d, 7e, 7g–j purine derivatives, and cisplatin (50 μM) was used as a positive control. As a preliminary apoptosis filter, propidium iodide (PI) staining was used through flow cytometry analysis which discriminated a population of viable cells impermeable to PI (PI negative) from necrotic cells PI-permeable (PI positive) and apoptotic cells (the difference of the previous two). The results are shown in Figure 7, illustrating that all purine derivatives tested induced apoptosis on HL-60 cells. Around 40% of apoptotic cells were detected in a culture treated with compounds 7a and 7i, 30% by 7e, 7g, and 7h and 70% by 7d and 7j. Meanwhile, cisplatin induced apoptosis in 66% of cells. Interestingly, necrosis was not observed as a cell death mechanism for the most of compounds studied in this cell line. (For dot plots, see Supplementary Information Figures S1–S3.)
Figure 7.
Viability of HL-60 cells. Cells were stimulated for 24 h with the different compounds (50 μM). Cells were then stained with propidium iodide (PI; 10 μg/mL), and cell viability and cell death by apoptosis or necrosis were evaluated by flow cytometry. (A) Graphical representation after quantification of the percentage of cells in each quadrant of the dot plot. (B) graphical representation of apoptosis percentage for the 7h compound at dose range of 1–50 µM. We obtained these results using the non-treated cells as control. The data represent the average ± SD of three independent experiments. We analyzed the data by t-test, *** p < 0.005, # p < 0.0001 compared to control (<1% DMSO).
To assess an unequivocal identification of apoptosis, annexin-V FITC is commonly combined with PI to understand if cells are viable, apoptotic, or necrotic through differences in plasma membrane integrity and permeability. To better understand the ability of compound 7h (the lowest IC50 values and high SI) to induce cell death, we performed the annexin-V FITC/PI assay. HL-60 cells were treated with 7h at the dose corresponding to 5 and 50 μM by 24 h, then harvested and stained with annexin-V FITC and PI (Figure 8). From these results, we could conclude that: 7h diminished cell viability with respect to control (from around 82–55% for 5 µM and up to 38% for 50 µM); and 7h led to an increase of late apoptosis (Q2) percentage when is compared to untreated control cells (36.5% for 5 µM and 37.7% for 50 µM). In addition, Figure 8 illustrates a discrete increase in the apoptosis (Q2 and Q4) from 5 to 50 µM, as well as an increase in necrosis at the concentration of 50 µM.
Figure 8.
Flow cytometry analysis of dead cell apoptosis assessed by Kit annexin-V FITC/PI Alexa Fluor 488. (A) Representative dot plot of HL-60 cells untreated (control) or treated for 24 h with 5.0 and 50 µM concentration for 7h. (B) Graphical representation after quantification of the percentage of cells in each quadrant of the dot plot with 5.0 and 50 µM concentrations. Results were obtained using the non-treated cells as control and average ± SD of two independent experiments. We analyzed annexin-V FITC/PI data by one-way ANOVA non-parametric Dunnett test. * p < 0.001.
2.7. Effect of 7h on Cell Cycle
Considering that apoptotic mechanisms are associated with the G1/S boundary cell cycle arrest [38] and the inhibition of cell cycle progression is an important factor to control cancer cell growth, we were interested in investigating whether 7h affects the cell cycle of HL-60 cells. We analyzed the cell cycle distribution of the cells stained with PI using flow cytometry. As shown in Figure 9, cells treated with 7h show an accumulation of cells in S cell cycle phase, accompanied by a notorious decrease of G2/M phase of the cell cycle, much like the reference compound cisplatin. This effect is in agreement with apoptotic mechanisms [38].
Figure 9.
Flow cytometry analysis of the DNA content of HL-60 cells at 48 h of treatment with 7h 25 µM. Quantification of the cells in different phases of cell cycle. Results are represented as average ± SD of two independent experiments. Data were analyzed by t-test, * p < 0.05.
Therefore, we demonstrated that 7h had no effect on p53 activation in the p53 reporter assays that we carried out (see Table S3 and Figure S6), which suggests that the apoptotic mechanism of 7h was independent of p53 activation. This mechanism has been widely reported for several compounds [39,40] and is interesting given that several cancer types are associated with mutant forms of p53 [41]. Future experiments are necessary to establish the ulterior apoptotic mechanism of 7h.
2.8. Search of Molecular Targets
Several other assays were performed in order to obtain more data, suggesting that some targets responsible for the cytotoxic effect were elicited, especially for the most promising compounds. For example, we performed a preliminary screening of compounds 7a–j towards several protein kinases in order to find possible targets. Due to the structural analogy to known CDK inhibitors [27,28,29,30], the screening included CDK2/cyclin E, CDK9/cyclin T, PKN3, and Abl kinases, but these compounds did not show any activity up to a dose of 10 µM (data not shown). In addition, we performed preliminary experiments with one of the most potent compounds also in the MCF7 cell line, which served as a model of solid cancer. Compound 7g (i.e., the compound with the highest potency in MCF7) slightly increased population of MCF7 cells in the G1-phase (Figure S5) and potently reduced the level of mitogenic transducer pERK1/2, albeit at a high concentration (10 µM; Figure 10). These results indicate that the antiproliferative activity of compound 7g (or other purines derivatives) is likely due to targeting other proteins and pathways. Therefore, further research is needed to elucidate the mechanism of cellular action responsible for the cytotoxicity and antiproliferative effects elicited by 7g and analogues.
Figure 10.
Compound 7g inhibits mitogenic transducer pERK1/2 in MCF-7 cells. MCF-7 cells were treated with vehicle or the indicated concentrations of 7g for 1 h.
3. Materials and Methods
3.1. Materials and Measurements
Melting points were determined on a Kofler Thermogerate apparatus and were uncorrected. Infrared spectra were recorded on a JASCO FT/IR-400 spectrophotometer. Nuclear magnetic resonance spectra were recorded, unless otherwise specified, on a Bruker AM-400 instrument using deuterated chloroform or dimethylsulfoxide solutions containing tetramethylsilane as an internal standard. Mass spectra were obtained on a HP 5988A mass spectrometer. HRMS–ESI-MS experiments were performed using a Thermo Scientific Exactive Plus Orbitrap spectrometer with a constant nebulizer temperature of 250 °C. The experiments were carried out in positive or negative ion mode, with a scan range of m/z 300.00–1510.40 with resolution 140,000. The samples were infused directly into the ESI source, via a syringe pump, at flow rates of 5 µL min−1, through the instrument’s injection valve. Thin layer chromatography (tlc) was performed using Merck GF-254 type 60 silica gel. Column chromatography was carried out using Merck type 9385 silica gel. The purity of the compounds was determined by tlc and high-resolution mass spectrometry (HRMS). For X-ray crystal structure analysis, data sets were collected with a STOE IPDS II two-circle-diffractometer using Mo Kα radiation (λ = 0.71073 Å). The intensities were corrected for absorption by an empirical correction with X-Area [42]. The structures were solved by direct methods (SHELXS) [43] and refined by full-matrix least-squares calculations on F2 (SHELXL-97). Anisotropic displacement parameters were refined for all non-hydrogen atoms.
3.2. General Procedure of the Synthesis for Compounds 2a–e and 6a
A mixture of 6-chloro-2-fluoropurine 1 or 2,6-dichloropurine 5 (1.0 mmol), the respective alkyl halide (1.5 mmol), and potassium carbonate (3.0 mmol) in DMF (5 mL) was stirred for 6 h, then the mixture was filtered and evaporated under vacuum. The products were separated by flash chromatography on silica gel eluting with EtOAc/CH2Cl2 (2:3).
6-Chloro-9-ethyl-2-fluoro-9H-purine 2a. White solid, yield 34%, m.p. 87–89 °C. 1H NMR (400 MHz, CDCl3) δ 8.15 (s, 1H), 4.33 (q, J. = 7.3 Hz, 2H), 1.59 (t, J. = 7.3 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.30–156.12 (d, 1JCF = 219.5 Hz), 153.60–153.43 (d, 3JCF = 17.5 Hz), 152.67–152.50 (d, 3JCF = 17.6 Hz), 145.39, 130.40–130.35 (d, 4JCF = 4.9 Hz), 39.72, 15.14.
6-Chloro-2-fluoro-9-propyl-9H-purine 2b. White solid, yield 38%, m.p. 81–82 °C. 1H NMR (400 MHz, DMSO) δ 8.70 (s, 1H), 4.19 (t, J. = 7.1 Hz, 2H), 1.92–1.79 (m, 2H), 0.86 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, DMSO) δ 157.63–155.51 (d, 1JCF = 213.7 Hz), 154.39–154.22 (d, 3JCF = 17.4 Hz), 150.72–150.54 (d, 3JCF = 18.2 Hz), 148.88–148.85 (d, 5JCF = 3.1 Hz) 130.36–130.31 (d, 4JCF = 4.9 Hz), 45.96, 22.72, 11.24.
9-Butyl-6-chloro-2-fluoro-9H-purine 2c. White solid, yield 36%, m.p. 76–78 °C. 1H NMR (200 MHz, CDCl3) δ 8.09 (s, 1H), 4.25 (t, J. = 7.2 Hz, 2H), 2.00–1.81 (m, 2H), 1.50–1.22 (m, 2H), 0.98 (t, J. = 7.3 Hz, 3H). 13C NMR (50 MHz, CDCl3) δ 159.44–155.08 (d, JCF = 219.6 Hz), 153.87–153.50 (d, JCF = 18.3 Hz), 152.82–152.47 (d, JCF = 17.6 Hz), 145.76–145.69 (d, JCF = 3.1 Hz), 130.32–130.22 (d, JCF = 5.0 Hz), 44.38, 31.65, 19.78, 13.38. 19F NMR (188 MHz, CDCl3) δ -49.69. IR (KBr, cm−1): 2959, 1605, 1579, 1511, 1408, 1332, 920. MS (ESI) for (C9H10ClFN4 [M + H]+): 229.1. Found: 229.1
6-Chloro-2-fluoro-9-pentyl-9H-purine 2d. Yellow solid, yield 47%, m.p. 92–93 °C. 1H NMR (400 MHz, CDCl3) δ 8.05 (s, 1H), 4.19 (t, J. = 7.3 Hz, 2H), 1.93–1.83 (m, 2H), 1.39–1.22 (m, 4H), 0.85 (t, J. = 6.9 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.35–156.17 (d, JCF = 219.6 Hz), 153.76–153.59 (d, JCF = 17.0 Hz), 152.71–152.54 (d, JCF = 17.6 Hz), 145.79–145.76 (d, JCF = 3.2 Hz), 130.31–130.26 (d, JCF = 5.0 Hz), 44.66, 29.37, 28.63, 22.04, 13.79. 19F NMR (376 MHz, CDCl3) δ -49.71. IR (KBr, cm−1): 2958, 1599, 1578, 1510, 1406, 1338, 920. MS (ESI) for (C10H13ClFN4 [M + H]+): 243.0. Found: 243.0
6-Chloro-2-fluoro-9-hexyl-9H-purine 2e. Yellow oil, yield 60%. 1H NMR (400 MHz, CDCl3) δ 8.08 (s, 1H), 4.21 (t, J. = 7.3 Hz, 2H), 1.95–1.85 (m, 2H), 1.30 (m, 6H), 0.85 (t, J. = 6.7 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.44–156.26 (d, JCF = 219.7 Hz), 153.87–153.70 (d, JCF = 16.9 Hz), 152.78–152.61 (d, JCF = 17.6 Hz), 145.93–145.90 (d, JCF = 3.1 Hz), 130.40–130.36 (d, JCF = 4.8 Hz), 44.78, 31.16, 29.74, 26.31, 22.48, 13.98. 19F NMR (376 MHz, CDCl3) δ -49.73 MS (ESI) for (C11H14ClFN4 [M + H]+): 257.0. Found: 257.0
2,6-Dichloro-9-cyclopentyl-9H-purine (6a). White solid, yield 55%, m.p. 106–108 °C. 1H NMR (400 MHz, CDCl3) δ 8.10 (s, 1H), 5.01–4.82 (m, 1H), 2.40–2.20 (m, 2H), 2.06–1.69 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 153.13, 152.60, 151.61, 144.13, 131.03, 56.86, 32.73 (2C), 23.85 (2C).
3.3. General Procedure of the Synthesis for Compounds 3a–e:
A stirred solution of 2a–e derivatives (1.0 mmol), (4-(trifluoromethoxy)phenyl)boronic acid (1.0 mmol), 2 M aqueous solution of potassium carbonate (1 mL), palladium(II)bis(triphenylphosphine) dichloride (0.1 mmol) in dioxane (5 mL) was heated to reflux during 2 h. Then, the solution was cooled to room temperature and extracted with ethyl acetate. The organic layers were dried with anhydrous sodium sulfate, filtered, and concentrated under vacuo. The crude product was purified on a silica gel column using dichloromethane as eluent.
9-Ethyl-2-fluoro-6-(4-(trifluoromethoxy)phenyl)-9H-purine (3a). White solid, yield 55%, m.p. 57–59 °C. 1H NMR (400 MHz, CDCl3) δ 8.87 (d, J. = 8.9 Hz, 2H), 8.11 (s, 1H), 7.37 (d, J. = 8.3 Hz, 2H), 4.31 (q, J. = 7.3 Hz, 2H), 1.58 (t, J. = 7.3 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 159.70–157.59 (d, 1JCF = 219.6 Hz), 155.52–155.37 (d, 2JCF = 15.3 Hz), 154.95–154.78 (d, 2JCF = 16.8 Hz), 151.68–151.67 (d, 4JCF = 1.6 Hz), 144.72, 144.69, 134.59–134.46 (t, 3JCF = 6.4 Hz), 132.98, 131.75, 129.72–129.67 (d, 3JCF = 4.3 Hz), 127.77–122.67 (d, 3JCF = 5.2 Hz), 121.68–119.12 (d, 1JCF = 258.6 Hz) 120.60 (d, 5JCF = 0.4 Hz), 116.18, 39.18, 15.16.
2-Fluoro-9-propyl-6-(4-(trifluoromethoxy)phenyl)-9H-purine (3b). White solid, yield 57%, m.p. 61–63 °C. 1H NMR (400 MHz, CDCl3) δ 8.89 (d, J. = 6.7 Hz, 2H), 8.09 (s, 1H), 7.38 (d, J. = 6.3 Hz, 2H), 4.23 (br, 2H), 1.98 (d, J. = 5.9 Hz, 2H), 1.00 (br, 3H). 13C NMR (101 MHz, CDCl3) δ 159.74–157.63 (d, 1JCF = 212.6 Hz), 155.52–155.37 (d, 2JCF = 15.2 Hz), 155.14–154.98 (d, 2JCF = 16.7 Hz), 151.68, 145.16–145.15 (d, 4JCF = 2.6 Hz), 132.99, 131.76 (2C), 129.65–129.61 (d, 3JCF = 3.8 Hz), 121.69–119.11 (d, 1JCF = 260.6 Hz), 120.60 (2C), 45.74, 23.11, 11.10.
9-Butyl-2-fluoro-6-(4-(trifluoromethoxy)phenyl)-9H-purine (3c). White solid, yield 41%, m.p. 67–69 °C. 1H NMR (400 MHz, CDCl3) δ 8.89 (d, J. = 8.9 Hz, 2H), 8.09 (s, 1H), 7.38 (d, J. = 8.3 Hz, 2H), 4.26 (t, J. = 7.3 Hz, 2H), 1.99–1.84 (m, 2H), 1.48–1.35 (m, 2H), 0.99 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 159.77–157.66 (d, 1JCF = 212.9 Hz), 155.58–155.43 (d, 2JCF = 15.3 Hz), 155.15–154.99 (d, 2JCF = 16.7 Hz), 151.71, 145.13–145.10 (d, 3JCF = 3.0 Hz), 133.02, 131.78 (2C), 129.66–129.62 (d, 3JCF = 4.3 Hz), 121.70, 120.64 (2C), 119.13, 43.92, 31.74, 19.86, 13.45. MS (ESI) for (C16H14F4N4O [M + H]+): 355.1. Found: 354.9.
2-Fluoro-9-pentyl-6-(4-(trifluoromethoxy)phenyl)-9H-purine (3d). White solid, yield 57%, m.p. 72–73 °C. 1H NMR (400 MHz, CDCl3) δ 8.90 (d, J. = 8.5 Hz, 1H), 8.09 (s, 1H), 7.39 (d, J. = 8.2 Hz, 1H), 4.26 (t, J. = 7.1 Hz, 1H), 2.03–1.90 (m, 1H), 1.37 (s, 2H), 0.91 (t, J. = 6.5 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 159.79–157.67 (d, 1JCF = 213.0 Hz), 155.61–155.46 (d, 2JCF = 14.7 Hz), 155.15–155.00 (d, 2JCF = 15.0 Hz), 151.71, 145.11, 133.02, 131.78 (2C), 131.66, 129.64, 120.65 (2C), 44.18, 29.45, 28.72, 22.10, 13.82. MS (ESI) for (C17H16F4N4O [M + H]+): 369.1. Found: 368.9.
2-Fluoro-9-hexyl-6-(4-(trifluoromethoxy) phenyl)-9H-purine (3e). White solid, yield 51%, m.p. 80–82 °C. 1H NMR (400 MHz, CDCl3) δ 8.73 (d, J. = 8.8 Hz, 2H), 7.92 (s, 1H), 7.22 (d, J. = 8.5 Hz, 2H), 4.09 (t, J. = 7.3 Hz, 2H), 1.83–1.71 (m, 2H), 1.26–1.07 (m, 6H), 0.72 (t, J. = 6.8 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 159.78–157.67 (d, 1JCF = 213.0 Hz), 155.60–155.45 (d, 2JCF = 15.4 Hz), 155.15–154.98 (d, 2JCF = 16.8 Hz), 151.70, 145.14–145.11 (d, 3JCF = 3.0 Hz), 133.03, 131.79 (2C), 129.67–129.63 (d, 3JCF = 4,2 Hz), 121.70–119.13 (d, 1JCF = 259.6 Hz), 120.65 (2C), 44.20, 31.14, 29.72, 26.29, 22.43, 13.90. MS (ESI) for (C18H18F4N4O [M + H]+): 382.1. Found: 382.8.
3.4. General Procedure of the Synthesis for Compounds 4a–u and 7a–j:
Compounds 3a–e or 6a (1.0 mmol), the respective amine (3.0 mmol), and DIPEA (3.0 mmol) were dissolved in n-BuOH (5 mL) and the mixture was heated to 110 °C for 12 h. Then, the reaction mixture was cooled and concentrated under vacuum. The crude product was purified on silica gel column using chloroform as mobile phase.
N-cyclopentyl-9-ethyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4a). White solid, yield 87%, m.p. 85–87 °C. 1H NMR (400 MHz, CDCl3) δ 8.75 (d, J. = 8.7 Hz, 2H), 7.76 (s, 1H), 7.34 (d, J. = 8.6 Hz, 2H), 5.20 (d, J. = 6.7 Hz, 1H), 4.46–4.34 (m, 1H), 4.16 (q, J. = 7.3 Hz, 2H), 2.12 (m, 2H), 1.82–1.60 (m, 4H), 1.54 (m, 5H). 13C NMR (101 MHz, CDCl3) δ 159.15, 154.49, 153.64, 150.72–150.70 (d, 4JCF = 1.7 Hz) 140.73, 134.98, 131.12 (2C), 124.92, 121.76–119.20 (d, 1JCF = 258.6 Hz), 120.52 (2C), 53.50, 38.29, 33.35 (2C), 23.83 (2C), 15.16. IR (KBr, cm−1): 3442, 2959, 1607, 1258, 1164. HRMS for (C19H20F3N5O [M + H]+). Calcd: 392.1693. Found: 392.1705.
N-cyclohexyl-9-ethyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4b). White solid, yield 92%, m.p. 82–84 °C. 1H NMR (400 MHz, CDCl3) δ 8.75 (d, J. = 8.8 Hz, 2H), 7.76 (s, 1H), 7.35 (d, J. = 8.2 Hz, 2H), 5.08 (d, J. = 7.7 Hz, 1H), 4.16 (q, J. = 7.3 Hz, 2H), 4.02–3.91 (m, 1H), 2.20–2.07 (m, 2H), 1.84–1.74 (m, 2H), 1.52 (t, J. = 7.3 Hz, 3H), 1.49–1.39 (m, 2H), 1.33–1.24 (m, 4H). 13C NMR (101 MHz, CDCl3) δ 158.76, 154.57, 153.71, 150.72–150.71 (4JCF = 1.6 Hz), 140.69, 134.99, 131.10 (2C), 124.94, 121.77–119.20 (1JCF = 259.6 Hz), 120.55 (2C), 50.23, 38.26, 33.22 (2C), 25.89, 24.98 (2C), 15.18. IR (KBr, cm−1): 3444, 2928, 1535, 1221, 1168. HRMS for (C20H22F3N5O [M + H]+). Calcd: 406.1849. Found: 406.1872.
N-(cyclohexylmethyl)-9-ethyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4c). White solid, yield 79%, m.p. 80–82 °C. 1H NMR (400 MHz, CDCl3) δ 8.75 (d, J. = 8.8 Hz, 2H), 7.76 (s, 1H), 7.34 (d, J. = 8.5 Hz, 2H), 5.29 (s, 1H), 4.15 (q, J. = 7.3 Hz, 2H), 3.39 (t, J. = 6.3 Hz, 2H), 1.83 (m, 2H), 1.79–1.70 (m, 2H), 1.71–1.57 (m, 1H), 1.51 (t, J. = 7.3 Hz, 3H), 1.33–1.14 (m, 4H), 1.02 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 159.69, 154.50, 153.66, 150.73–150.71 (4JCF = 1.6 Hz), 140.70, 134.96, 131.13 (2C), 124.90, 121.77–119.21 (1JCF = 258.6 Hz), 120.52 (2C), 116.25, 48.33, 38.26, 31.11 (2C), 26.53, 26.00 (2C), 15.13. IR (KBr, cm−1): 3364, 2926, 1608, 1267, 1160. HRMS for (C21H24F3N5O [M + H]+). Calcd: 420.2006. Found: 420.2023.
N-cycloheptyl-9-ethyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4d). White solid, yield 67%, m.p. 66–68 °C. 1H NMR (400 MHz, CDCl3) δ 8.63 (d, J. = 8.7 Hz, 2H), 7.63 (s, 1H), 7.22 (d, J. = 8.4 Hz, 2H), 5.03 (d, J. = 7.5 Hz, 1H), 4.03 (q, J. = 7.3 Hz, 3H), 2.00–1.94 (m, 2H), 1.65–1.32 (m, 13H). 13C NMR (101 MHz, CDCl3) δ 158.59, 154.55, 153.69, 150.73–150.71 (d, 4JCF = 1.7 Hz), 140.68, 134.99, 131.09 (2C), 124.87, 121.76–119.20 (d, 1JCF = 258.6 Hz), 120.53 (2C), 52.39, 38.28, 34.91 (2C), 28.37 (2C), 24.35 (2C), 15.13. IR (KBr, cm−1): 3279, 2929, 1604, 1255. HRMS for (C21H24F3N5O [M + H]+). Calcd: 420.2006. Found: 420.2029.
N-cyclopentyl-9-propyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4e). White solid, yield 73%, m.p. 78–80 °C. 1H NMR (400 MHz, CDCl3) δ 8.76 (d, J. = 8.7 Hz, 2H), 7.74 (s, 1H), 7.35 (d, J. = 8.6 Hz, 2H), 5.16 (d, J. = 6.7 Hz, 1H), 4.45–4.34 (m, 1H), 4.08 (t, J. = 7.1 Hz, 2H), 2.17–2.09 (m, 2H), 1.99–1.86 (m, 2H), 1.81–1.61 (m, 4H), 1.54 (m, 2H), 0.97 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 159.17, 154.72, 153.61, 150.73 (d, 4JCF = 1.7 Hz), 141.28, 135.00, 131.11 (2C), 124.89, 121.77–119.21 (d, 1JCF = 258.6 Hz), 120.54 (2C), 53.51, 44.96, 33.38 (2C), 23.87 (2C), 23.05, 11.25. IR (KBr, cm−1): 3286, 2964, 1606, 1260, 1165. HRMS for (C20H22F3N5O [M + H]+). Calcd: 406.1849. Found: 406.1860.
N-cyclohexyl-9-propyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4f). White solid, yield 81%, m.p. 73–75 °C. 1H NMR (400 MHz, CDCl3) δ 8.75 (d, J. = 8.7 Hz, 2H), 7.74 (s, 1H), 7.35 (d, J. = 8.4 Hz, 2H), 5.07 (d, J. = 7.5 Hz, 1H), 4.08 (t, J. = 7.1 Hz, 2H), 4.03–3.89 (m, 1H), 2.13 (d, J. = 9.3 Hz, 2H), 2.00–1.59 (m, 6H), 1.55–1.38 (m, 2H), 1.38–1.19 (m, 2H), 0.98 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.78, 154.77, 153.68, 150.71 (d, 4JCF = 1.7 Hz), 141.27, 135.00, 131.09 (2C), 124.86, 121.76–119.20 (d, 1JCF = 259.6 Hz), 120.55 (2C), 50.25, 44.95, 41.97, 33.21, 27.01, 25.90, 24.97, 23.05, 11.26. IR (KBr, cm−1): 3445, 2920, 1572, 1285, 1108. HRMS for (C21H24F3N5O [M + H]+). Calcd: 420.2006. Found: 420.1988.
N-(cyclohexylmethyl)-9-propyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4g). White solid, yield 54%, m.p. 65–67 °C. 1H NMR (400 MHz, CDCl3) δ 8.76 (d, J. = 8.8 Hz, 2H), 7.73 (s, 1H), 7.35 (d, J. = 8.3 Hz, 2H), 5.28 (d, J. = 1.7 Hz, 1H), 4.07 (t, J. = 7.1 Hz, 2H), 3.38 (t, J. = 6.3 Hz, 2H), 1.96–1.82 (m, 4H), 1.76–1.72 (m, 2H), 1.70–1.56 (m, 2H), 1.31–1.14 (m, 3H), 1.04 (m, 2H), 0.97 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 159.71, 154.71, 153.58, 150.71, 150.70 (d, 4JCF = 1.7 Hz), 141.25, 135.00, 131.12 (2C), 124.85, 121.77–119.21 (d, 1JCF = 258.6 Hz), 120.52 (2C), 48.35, 44.93, 38.24, 31.12 (2C), 26.54, 26.00, 23.03, 11.22. IR (KBr, cm−1): 3273, 2928, 1605, 1257. HRMS for (C22H26F3N5O [M + H]+). Calcd: 434.2162. Found: 434.2170.
N-cycloheptyl-9-propyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4h). Colorless oil, yield 82%. 1H NMR (400 MHz, CDCl3) δ 8.75 (d, J. = 8.9 Hz, 2H), 7.73 (s, 1H), 7.34 (d, J. = 8.2 Hz, 2H), 5.23 (d, J. = 6.5 Hz, 1H), 4.13 (t, J. = 7.1 Hz, 2H), 4.20–3.98 (m, 1H), 2.20–1.99 (m, 2H), 2.00–1.80 (m, 2H), 2.43–1.77 (m, 10H), 0.96 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.46, 154.78, 153.40, 150.74–150.61 (d, 4JCF = 1.7 Hz), 141.34, 134.75, 131.10 (2C), 124.71, 123.00–117.88 (d, 1JCF = 257.6 Hz), 120.51 (2C), 52.41, 44.95, 34.85 (2C), 28.33 (2C), 24.34 (2C), 23.00, 11.21. IR (KBr, cm−1): 3276, 2930, 1604, 1259, 1154. HRMS for (C22H26F3N5O [M + H]+). Calcd: 434.2162. Found: 434.2169.
9-Butyl-N-cycloheptyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4i). White solid, yield 60%, m.p. 72–74 °C. 1H NMR (400 MHz, CDCl3) δ 8.63 (d, J. = 8.7 Hz, 2H), 7.60 (s, 1H), 7.22 (d, J. = 8.4 Hz, 2H), 5.04 (d, J. = 7.4 Hz, 1H), 3.97 (t, J. = 7.1 Hz, 3H), 2.02–1.91 (m, 2H), 1.79–1.68 (m, 2H), 1.64–1.39 (m, 10H), 1.31–1.18 (m, 2H), 0.84 (t, J. = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.62, 154.76, 153.65, 150.72–150.70 (d, 4JCF = 1.7 Hz), 141.18, 135.02, 131.10 (2C), 124.77, 121.77–119.21 (d, 1JCF = 258.6 Hz), 120.53 (2C), 52.47, 42.97, 34.91 (2C), 31.73, 28.34 (2C), 24.39 (2C), 19.85, 13.49. IR (KBr, cm−1): 3375, 2930, 1606, 1260, 1161. HRMS for (C23H28F3N5O [M + H]+). Calcd: 448.2319. Found: 448.2321.
N-cycloheptyl-9-pentyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4j). White solid, yield 93%, m.p. 50–53 °C. 1H NMR (400 MHz, CDCl3) δ 8.63 (d, J. = 8.8 Hz, 2H), 7.61 (s, 1H), 7.22 (d, J. = 8.2 Hz, 2H), 5.10 (s, 1H), 3.97 (t, J. = 7.1 Hz, 2H), 4.21–3.96 (m, 1H), 1.97 (t, 2H), 1.86–1.69 (m, 2H), 1.67–1.39 (m, 11H), 1.32–1.14 (m, 5H), 0.77 (t, J. = 7.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.52, 154.79, 153.54, 150.76–150.74 (d, 4JCF = 1.6 Hz), 141.29, 134.84, 131.13 (2C), 124.74, 121.56–119.20 (d, 1JCF = 257.6 Hz), 120.53 (2C) 52.47, 43.28, 34.90 (2C), 29.37, 28.77, 28.34 (2C), 24.39 (2C), 22.12, 13.86. IR (KBr, cm−1): 3375, 2930, 1606, 1260, 1161. HRMS for (C24H30F3N5O [M + H]+). Calcd: 462.2475. Found: 462.2471.
N-cycloheptyl-9-Hexyl-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4k). White solid, yield 93%, m.p. 68–70 °C. 1H NMR (400 MHz, CDCl3) δ 8.63 (d, J. = 8.8 Hz, 2H), 7.61 (s, 1H), 7.22 (d, J. = 8.3 Hz, 2H), 5.09 (s, 1H), 3.97 (t, J. = 7.2 Hz, 3H), 3.95–4.3 (m, 1H) 2.03–1.89 (m, 2H), 1.87–1.67 (m, 2H), 1.65–1.36 (m, 11H), 1.26–1.06 (m, 7H), 0.75 (t, J. = 7.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.48, 154.74, 150.71–150.70 (d, 4JCF = 1.6 Hz), 141.24, 134.81, 131.08 (2C), 124.70, 121.72–119.16 (d, 1JCF = 257.7 Hz), 120.49 (2C), 52.41, 43.26, 34.86 (2C), 31.16, 29.59, 28.31 (2C), 26.27, 24.32 (2C), 22.41, 13.89. IR (KBr, cm−1): 3391, 2927, 1607, 1261, 1170. HRMS for (C25H32F3N5O [M + H]+). Calcd: 476.2632. Found: 476.2657.
9-Hexyl-2-(4-(5-nitropyridin-2-yl)piperazin-1-yl)-6- (4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4l). White solid, yield 84%, m.p. 159–161 °C. 1H NMR (400 MHz, CDCl3) δ 8.80 (d, J. = 8.8 Hz, 2H), 7.73 (s, 1H), 7.36–7.15 (m, 7H), 5.12 (d, J. = 13.2 Hz, 2H), 4.09 (t, J. = 7.1 Hz, 2H), 3.01 (td, J. = 12.9, 2.1 Hz, 2H), 2.79 (t, J. = 12.1 Hz, 1H), 2.07–1.66 (m, 6H), 1.43–1.16 (m, 7H), 0.85 (t, J. = 7.0 Hz, 3H). 13C NMR (50 MHz, CDCl3) δ 160.38, 158.34, 154.56, 153.00, 150.85–150.82 (d, 4JCF = 1.7 Hz), 146.40, 141.93, 135.06, 134.82, 132.98, 131.16 (2C), 124.58, 122.98–117.86 (d, 1JCF = 257.8 Hz), 120.51 (2C), 104.51, 44.60 (2C), 43.96 (2C), 43.32, 31.11, 29.56, 26.25, 22.40, 13.91. IR (KBr, cm−1): 2931, 1598, 1296, 1250. HRMS for (C27H29F3N8O3 [M + H]+). Calcd: 571.2387. Found: 571.2390.
9-Hexyl-2-(4-(pyridin-2-yl)piperazin-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4m). Yellow solid, yield 67%, m.p. 172–173 °C. 1H NMR (400 MHz, CDCl3) δ 8.68 (d, J. = 8.5 Hz, 2H), 8.09 (d, J. = 4.4 Hz, 1H), 7.64 (s, 1H), 7.38 (s, 1H), 7.22 (d, J. = 8.4 Hz, 2H), 6.65–6.45 (m, 2H), 4.07–3.89 (m, 6H), 3.65–3.52 (m, 4H), 1.75 (t, J. = 5.7 Hz, 2H), 1.29–1.08 (m, 6H), 0.74 (t, J. = 6.5 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.72, 155.06, 153.00, 150.75–150.76 (d, 4JCF = 1.2 Hz), 141.75, 137.73, 135.09, 131.16 (2C), 124.69, 121.74–119.18 (d, 1JCF = 257.6 Hz), 120.52 (2C), 113.41, 107.45, 45.22, 44.27 (2C), 43.23, 31.14, 29.60, 26.27, 22.43 (2C), 13.94. IR (KBr, cm−1): 2928, 1519, 1281, 1229. HRMS for (C27H30F3N7O [M + H]+). Calcd: 526.2537. Found: 526.2523.
9-Hexyl-2-(4-(5-(trifluoromethyl)pyridin-2-yl)piperazin-1-yl)-6- (4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4n). Yellow solid, yield 80%, m.p. 154–156 °C. 1H NMR (400 MHz, CDCl3) δ 8.74 (d, J. = 8.8 Hz, 2H), 8.36 (s, 1H), 7.71 (s, 1H), 7.58 (dd, J. = 9.0, 2.2 Hz, 1H), 7.28 (d, J. = 8.4 Hz, 2H), 6.61 (d, J. = 9.0 Hz, 1H), 4.31–3.92 (m, 6H), 3.86–3.67 (m, 3H), 1.94–1.66 (m, 2H), 1.43–1.10 (m, 7H), 0.80 (t, J. = 6.9 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 160.29, 158.59, 154.68, 153.05, 150.85–150.84 (d, 4JCF = 1.2 Hz), 145.67–145.63 (d, 3JCF = 4.2Hz), 141.88, 135.03, 134.65–134.62 (d, 3JCF = 3.0 Hz), 131.21 (2C), 125.92–123.23 (d, 1JCF = 270.4 Hz), 124.77, 121.78–119.21 (d, 1JCF = 257.8 Hz), 120.56 (2C), 115.58–115.16 (d, 2JCF = 33.1 Hz), 105.74, 44.61 (2C), 44.11 (2C), 43.30, 31.17, 29.62, 26.31, 22.46, 13.96. IR (KBr, cm−1): 2933, 1608, 1518, 1271, 1259. HRMS for (C28H29F6N7O [M + H]+). Calcd: 594.2411. Found: 594.2403.
9-Hexyl-2-(4-(pyrazin-2-yl)piperazin-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4o). White solid, yield 94%, m.p. 181–183 °C. 1H NMR (400 MHz, CDCl3) δ 8.77 (d, J. = 8.9 Hz, 2H), 8.17 (d, J. = 1.0 Hz, 1H), 8.12–7.98 (m, 1H), 7.84 (d, J. = 2.6 Hz, 1H), 7.76 (s, 5H), 7.32 (d, J. = 8.3 Hz, 2H), 4.24–4.00 (m, 6H), 3.83–3.65 (m, 4H), 1.97–1.79 (m, 2H), 1.64–1.44 (m, 1H), 1.26 (dt, J = 28.9, 12.7 Hz, 5H), 0.96–0.70 (m, 3H). 13C NMR (101 MHz, CDCl3) δ 158.55, 155.04, 154.63, 153.02, 150.78–150.77(d, 4JCF = 1.7 Hz), 141.84, 141.77, 134.96, 133.06, 131.15 (2C), 131.06, 124.75, 121.71–119.14 (d, 1JCF = 257.7 Hz), 120.50 (2C), 44.33 (2C), 44.04 (2C), 43.23, 31.11, 29.57, 26.25, 22.40, 13.91. IR (KBr, cm−1): 2928, 1604, 1283, 1229. HRMS for (C26H29F3N8O [M + H]+). Calcd: 527.2489. Found: 527.2488.
9-Hexyl-2-(4-phenylpiperidin-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4p). Yellow solid, yield 86%, m.p. 132–134 °C. 1H NMR (400 MHz, CDCl3) δ 8.80 (d, J. = 8.8 Hz, 2H), 7.73 (s, 1H), 7.26 (ddd, J. = 19.5, 17.8, 10.5 Hz, 7H), 5.12 (d, J. = 13.2 Hz, 2H), 4.09 (t, J. = 7.1 Hz, 2H), 3.01 (td, J. = 12.9, 2.1 Hz, 2H), 2.79 (ddd, J. = 12.1, 8.7, 3.5 Hz, 1H), 2.06–1.67 (m, 5H), 1.40–1.15 (m, 7H), 0.85 (t, J. = 7.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.80, 154.79, 152.94, 150.67–150.65 (d, 4JCF = 1.2 Hz), 146.19, 141.45, 135.31, 131.13 (2C), 128.48 (2C), 128.44 (2C), 126.82, 126.71, 126.21, 124.25, 121.74–119.18 (d, 1JCF = 257.6 Hz), 120.47 (2C), 45.29 (2C), 43.22, 43.12, 33.20 (2C), 31.14, 29.59, 26.26, 22.42, 13.92. IR (KBr, cm−1): 2931, 1598, 1296, 1250. HRMS for (C29H32F3N5O [M + H]+). Calcd: 524.2632. Found: 524.2632.
9-Pentyl-2-(4-(5-nitropiridin-2-yl)piperazinil-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purine (4q). Orange solid, yield 96%, m.p. 169–171 °C. 1H NMR (400 MHz, CDCl3) δ 9.06 (s, 1H), 8.78 (d, J. = 8.7 Hz, 2H), 8.23 (d, J. = 9.5 Hz, 1H), 7.80 (s, 1H), 7.34 (d, J. = 8.3 Hz, 2H), 6.61 (d, J. = 9.5 Hz, 1H), 4.24–4.01 (m, 6H), 4.01–3.74 (m, 4H), 1.98–1.83 (m, 2H), 1.43–1.27 (m, 4H), 0.89 (t, J. = 7.0 Hz, 3H). 13C NMR (50 MHz, CDCl3) δ 160.48, 158.44, 154.66, 153.10, 150.95–150.92 (d, 4JCF = 1.7 Hz), 146.50, 142.03, 135.16, 134.92, 133.08, 131.26 (2C), 124.68, 123.08–117.96 (d, 1JCF = 257.9 Hz), 120.61 (2C), 104.61, 44.70 (2C), 44.06 (2C), 43.42, 31.21, 26.35, 22.51, 14.01. IR (KBr, cm−1): 2924, 1637, 1542. HRMS for (C26H27F3N8O3 [M + H+]). Calcd: 555.2206. Found: 555.2780.
9-Pentyl-2-(4-(piridin-2-yl)piperazinil-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purine (4r). Yellow solid, yield 88%, m.p. 187–190 °C. 1H NMR (400 MHz, CDCl3) δ 8.82 (d, J. = 8.9 Hz, 2H), 8.23 (dd, J. = 4.9, 1.4 Hz, 1H), 7.77 (s, 1H), 7.50 (ddd, J. = 8.9, 7.2, 1.9 Hz, 1H), 7.36 (d, J. = 8.3 Hz, 2H), 6.70 (d, J. = 8.6 Hz, 1H), 6.64 (dd, J. = 7.0, 5.0 Hz, 1H), 4.24–3.90 (m, 6H), 3.86–3.62 (m, 4H), 2.02–1.81 (m, 2H), 1.47–1.24 (m, 4H), 0.90 (t, J. = 7.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 159.51, 158.71, 154.64, 152.93, 150.72–150.70 (d, 1JCF = 1.6 Hz), 147.94, 141.70, 137.48, 135.08, 131.13 (2C), 124.62, 121.71–119.15 (d, 4JCF = 257.7 Hz), 120.47 (2C), 113.40, 107.16, 45.11 (2C), 44.25 (2C), 43.16, 29.28, 28.69, 22.03, 13.81. IR (KBr, cm−1): 2929, 1595, 1482, 1229, 1144. HRMS for (C26H28F3N7O [M + H+]). Calcd: 512.2380. Found: 512.2375.
9-Pentyl-2-(4-(5-trifluoromethyl)pirerazin-2-yl)piperazin-1-yl)-6- (4-(trifluoromethoxy)phenyl)-9H-purine (4s). Yellow solid, yield 93%, m.p. 133–135 °C. 1H NMR (400 MHz, CDCl3) δ 8.67 (d, J. = 8.9 Hz, 2H), 8.30 (s, 1H), 7.65 (s, 1H), 7.52 (dd, J. = 8.9, 2.4 Hz, 1H), 7.22 (d, J. = 8.4 Hz, 2H), 6.55 (d, J. = 9.0 Hz, 1H), 4.11–3.83 (m, 6H), 3.80–3.59 (m, 4H), 1.86–1.69 (m, 2H), 1.21 (ddd, J. = 20.2, 19.2, 7.8 Hz, 4H), 0.77 (t, J. = 7.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 160.40, 158.58, 154.57, 153.00, 150.82–150.84 (d, 4JCF = 1.5 Hz), 145.80–145.76 (d, 3JCF = 4.2Hz), 141.86, 135.01, 134.54–134.51 (d, 3JCF = 3.0 Hz), 131.18 (2C), 124.78, 124.77, 121.74–119.18 (d, 1JCF = 257.7 Hz), 121.54 (2C), 115.46–115.13 (d, 2JCF = 32.1 Hz), 105.62, 44.54 (2C), 44.10 (2C), 43.35, 29.34, 28.74, 22.08, 13.86. IR (KBr, cm−1): 2932, 1654, 1273, 1243. HRMS for (C27H27F6N7O [M + H+]). Calcd: 580.2254. Found: 580.2249.
9-Pentyl-2-(4-phenylpiperidin-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purine (4t). White solid, yield 93%, m.p. 150–153 °C. 1H NMR (400 MHz, CDCl3) δ 8.81 (d, J. = 8.6 Hz, 2H), 7.73 (s, 1H), 7.47–7.07 (m, 7H), 5.12 (d, J. = 13.0 Hz, 2H), 4.09 (t, J. = 7.0 Hz, 2H), 3.01 (t, J. = 12.6 Hz, 2H), 2.79 (t, J. = 12.1 Hz, 1H), 2.05–1.63 (m, 6H), 1.45–1.22 (m, 4H), 0.88 (t, J. = 6.9 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.87, 154.87, 153.00, 150.74–150.72 (d, 4JCF = 1.6 Hz), 146.26, 141.52, 135.40, 131.21 (2C), 128.51 (2C), 126.89 (2C), 126.28, 124.34, 121.82–119.26 (d, 1JCF = 257.6 Hz), 120.54 (2C), 45.36 (2C), 43.30, 43.18, 33.28 (2C), 29.40, 28.80, 22.14, 13.91. IR (KBr, cm−1): 2930, 1263. HRMS for (C28H30F3N5O [M+3H+]). Calcd: 512.2621. Found: 512.2385
9-Pentyl-2-(4-(pyrazin-2-yl)piperazin-1-yl)-6-(4-(trifluoromethoxy)phenyl)-9H-purin-2-amine (4u). Yellow solid, yield 83%, m.p. 178–180 °C. 1H NMR (400 MHz, CDCl3) δ 8.67 (d, J. = 8.8 Hz, 2H), 8.07 (s, 1H), 7.96 (s, 1H), 7.74 (d, J. = 2.4 Hz, 1H), 7.66 (s, 1H), 7.22 (d, J. = 8.5 Hz, 2H), 4.09–3.85 (m, 6H), 3.67–3.54 (m, 4H), 1.83–1.65 (m, 2H), 1.31–1.08 (m, 4H), 0.77 (t, J. = 7.0 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 158.61, 155.10, 154.68, 153.09, 150.83, 141.91, 141.85, 135.01, 133.07, 131.20 (2C), 131.08, 124.78, 121.76–119.20 (1J = 257.9 Hz), 120.56 (2C), 44.38 (2C), 44.10 (2C), 43.28, 29.36, 28.76, 22.10, 13.89. IR (KBr, cm−1): 2955, 1608, 1426, 1277. HRMS for (C25H27F3N8O [M + H+]). Calcd: 513.2333. Found: 513.2350.
2-Chloro-9-cyclopentyl-6-(4-(5-nitropyridin-2-yl)piperazin-1-yl)-9H-purine (7a). Orange solid, yield 81%, m.p. 130–132 °C. 1H NMR (400 MHz, CDCl3) δ 9.00 (d, J. = 2.3 Hz, 1H), 8.18 (dd, J. = 9.5, 2.6 Hz, 1H), 7.74 (s, 1H), 6.57 (d, J. = 9.5 Hz, 1H), 4.96–4.80 (m, 1H), 4.39 (s, 4H), 4.00–3.78 (m, 4H), 2.35–2.12 (m, 2H), 1.97–1.65 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 160.25, 153.82, 153.52, 152.33, 146.27, 137.10, 135.36, 133.08, 119.05, 104.56, 55.60, 44.58 (2C), 32.82 (3C), 23.76 (3C). IR (KBr, cm−1): 1580, 1293, 1246, 969. HRMS for (C19H21ClN8O2 [M + H+]). Calcd: 429.1547. Found: 429.1547.
2-Chloro-9-cyclopentyl-6-(4-(pyridin-2-yl)piperazin-1-yl)-9H-purine (7b). White solid, yield 81%, m.p. 146–148 °C. 1H NMR (400 MHz, CDCl3) δ 7.96 (d, J. = 4.6 Hz, 1H), 7.51 (s, 1H), 7.33–7.16 (m, 1H), 6.41 (dd, J. = 10.2, 7.3 Hz, 2H), 4.76–4.57 (m, 1H), 4.16 (s, 4H), 3.52–3.32 (m, 4H), 2.12–1.93 (m, 2H), 1.74–1.44 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 159.10, 153.87, 153.51, 152.15, 147.90, 137.50, 136.66, 118.92, 113.66, 107.01, 55.43, 45.13 (2C), 32.75 (3C), 23.69 (3C). IR (KBr, cm−1): 1580, 1478, 1536, 1241, 773. HRMS for (C19H22ClN7 [M + H+]). Calcd: 384.1698. Found: 384.1695.
2-Chloro-9-cyclopentyl-6-(4-(5-(trifluoromethyl)pyridin-2-yl)piperazin-1-yl)-9H-purine (7c). Yellow solid, yield 60%, m.p. 160–162 °C. 1H NMR (400 MHz, CDCl3) δ 8.38 (s, 1H), 7.73 (s, 1H), 7.62 (d, J. = 8.9 Hz, 1H), 6.63 (d, J. = 9.0 Hz, 1H), 4.99–4.77 (m, 1H), 4.37 (s, 4H), 3.86–3.68 (m, 4H), 2.35–2.12 (m, 2H), 1.99–1.65 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 160.18, 153.96, 153.63, 152.34, 145.77 (q, 3JCF = 4.3 Hz), 136.95, 134.65 (d, 3JCF = 3.1 Hz), 124.50 (q, 1JCF = 270.4 Hz), 119.08, 115.68 (q, 2JCF = 33.4 Hz), 105.63, 55.61, 44.61 (2C), 32.88 (3C), 23.81 (3C). IR (KBr, cm−1): 1581, 1330, 1243, 1003, 637. HRMS for (C20H21ClF3N7 [M + H+]). Calcd: 452.1572. Found: 452.1589.
2-Chloro-9-cyclopentyl-6-(4-(pyrazin-2-yl)piperazin-1-yl)-9H-purine (7d). White solid, yield 74%, m.p. 144–147 °C. 1H NMR (400 MHz, CDCl3) δ 8.13 (s, 1H), 8.05 (d, J. = 1.4 Hz, 1H), 7.84 (d, J. = 2.5 Hz, 1H), 7.72 (s, 1H), 4.98–4.75 (m, 1H), 4.37 (s, 4H), 3.82–3.58 (m, 4H), 2.30–2.05 (m, 2H), 2.05–1.61 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 154.73, 153.87, 153.57, 152.28, 141.73, 136.87, 133.48, 130.93, 119.00, 55.52, 44.42 (2C), 32.82 (3C), 23.75 (3C). IR (KBr, cm−1): 1581, 1248, 1008. HRMS for (C18H21ClN8 [M + H+]). Calcd: 385.1650. Found: 385.1661.
2-Chloro-9-cyclopentyl-6-(4-phenylpiperidin-1-yl)-9H-purine (7e). White solid, yield 84%, m.p. 111–113 °C. 1H NMR (400 MHz, CDCl3) δ 7.76 (s, 1H), 7.38–7.14 (m, 5H), 5.63 (s, 2H), 5.03–4.85 (m, 1H), 3.15 (s, 2H), 2.96–2.75 (m, 1H), 2.41–2.19 (m, 2H), 2.12–1.69 (m, 10H). 13C NMR (101 MHz, CDCl3) δ 153.87, 153.69, 152.18, 145.35, 136.31, 128.48 (2C), 126.72 (2C), 126.37, 118.89, 55.40, 45.99 (2C), 42.83, 33.39 (2C), 32.85 (2C), 23.76 (2C). IR (KBr, cm−1): 1579, 1309, 1242. HRMS for (C21H24ClN5 [M + H+]). Calcd: 382.1793. Found: 382.1788.
2-Chloro-9-cyclopentyl-6-(4-phenylpiperazin-1-yl)-9H-purine (7f). White solid, yield 88%, m.p. 115–117 °C. 1H NMR (400 MHz, CDCl3) δ 7.71 (s, 1H), 7.23 (dd, J. = 10.0, 5.7 Hz, 2H), 6.98–6.69 (m, 3H), 4.96–4.76 (m, 1H), 4.39 (s, 4H), 3.34–3.13 (m, 4H), 2.33–2.09 (m, 2H), 1.93–1.62 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 153.71, 153.49, 152.14, 150.92, 136.61, 129.06 (2C), 120.19, 118.86, 116.37 (2C), 55.40, 49.39 (2C), 44.87 (2C), 32.70 (2C), 23.66 (2C). IR (KBr, cm−1): 1582, 1233, 1009, 759. HRMS for (C20H23ClN6 [M + H+]). Calcd: 383.1845. Found: 383.1847.
2-Chloro-6-(4-(4-chlorophenyl)piperazin-1-yl)-9-cyclopentyl-9H-purine (7g). White solid, yield 85%, m.p. 129–131 °C. 1H NMR (400 MHz, CDCl3) δ 7.79 (s, 1H), 7.24 (d, J. = 8.9 Hz, 2H), 6.89 (d, J. = 8.9 Hz, 2H), 5.02–4.86 (m, 1H), 4.46 (s, 4H), 3.36–3.16 (m, 4H), 2.39–2.17 (m, 2H), 2.03–1.67 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 153.73, 153.53, 152.22, 149.57, 136.74, 128.97 (2C), 125.12, 118.91, 117.61 (2C), 55.48, 49.43 (2C), 44.90 (2C), 32.77 (2C), 23.71 (2C). IR (KBr, cm−1): 1585, 1236, 1006, 808. HRMS for (C20H22Cl2N6 [M + H+]). Calcd: 417.1356. Found: 417.1349.
2-Chloro-9-cyclopentyl-6-(4-(4-nitrophenyl)piperazin-1-yl)-9H-purine (7h). Orange solid, yield 65%, m.p. 130–133 °C. 1H NMR (400 MHz, CDCl3) δ 8.10 (d, J. = 9.2 Hz, 2H), 7.74 (s, 1H), 6.82 (d, J. = 9.3 Hz, 2H), 4.99–4.79 (m, 1H), 4.42 (s, 4H), 3.62–3.44 (m, 4H), 2.34–2.14 (m, 2H), 1.95–1.70 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 154.52, 153.77, 153.56, 152.33, 138.88, 137.09, 125.90 (2C), 119.05, 112.81 (2C), 55.61, 46.92 (2C), 44.46 (2C), 32.83 (2C), 23.77 (2C). IR (KBr, cm−1): 1575, 1319, 1237. HRMS for (C20H22ClN7O2 [M + H+]). Calcd: 428.1596. Found: 428.1617.
2-Chloro-9-cyclopentyl-6-(4-(pyridin-4-yl)piperazin-1-yl)-9H-purine (7i). White solid, yield 68%, m.p. 155–157 °C. 1H NMR (400 MHz, CDCl3) δ 8.14 (d, J. = 6.2 Hz, 2H), 7.65 (s, 1H), 6.54 (d, J. = 6.3 Hz, 2H), 4.83–4.68 (m, 1H), 4.27 (s, 4H), 3.44–3.23 (m, 4H), 2.23–2.01 (m, 2H), 1.88–1.55 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 154.36, 153.44, 153.19, 151.98, 149.82 (2C), 136.80, 118.71, 108.10 (2C), 55.32, 45.54, 44.07 (2C), 32.48 (2C), 23.47 (2C). IR (KBr, cm−1): 1599, 1519, 1252, 997. HRMS for (C19H22ClN7 [M + H+]). Calcd: 384.1698. Found: 384.1697.
2-Chloro-9-cyclopentyl-6-(4-(4-methoxyphenyl)piperazin-1-yl)-9H-purine (7j). White solid, yield 92%, m.p. 122–124 °C. 1H NMR (400 MHz, CDCl3) δ 7.72 (s, 1H), 6.79–6.65 (m, 4H), 3.89–3.85 (m, 1H), 3.83–3.79 (m, 3H), 3.75–3.67 (m, 4H), 3.67–3.56 (m, 4H), 2.18–2.02 (m, 2H), 1.92–1.51 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 159.11, 153.40, 152.81, 148.20, 143.34, 141.69, 135.06, 116.20 (2C), 116.30 (2C), 57.38, 56.04, 49.48 (2C), 47.88 (2H), 34.32 (2C), 25.47 (2C). IR (KBr, cm−1): 1580, 1436, 1241. HRMS for (C21H25ClN6O [M + H+]). Calcd: 413.1851. Found: 413.1849.
3.5. Anticancer Activity: In Vitro Studies
3.5.1. Materials
Cell culture, fetal bovine, and penicillin–streptomycin were purchased from Biological industries. Cisplatin, 3-(4,5-dymethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), propidium iodide and DMSO were obtained from Sigma-Aldrich.
3.5.2. Cell Culture and Treatments
CFAPC-1, NCI-H460 H1975, HL-60, HCT-116, CACO2, K562, MCF-7, and MCR-5 were grown and maintained in RPMI 1640 and DMEM F12 as appropriate with 10% fetal bovine serum, 100 UI/mL penicillin–streptomycin, and cultured al 37 °C in 5% CO2 humidified atmosphere. Control cultures were treated with dimethyl sulfoxide (1%) alone at the same concentration as in the purine derivatives treatment. Cell viability was determined by trypan blue dye exclusion. Cell numbers were counted in hemocytometer by light microscopy.
3.5.3. Drugs
Purine derivatives and cisplatin were prepared as fresh solutions with dimethyl sulfoxide (DMSO) immediately prior to any experiment and stored at −20 °C in amber flask until new use.
3.5.4. Cytotoxicity Study
Cytotoxicity assays were performed by using the MTT reduction method as described in the literature [23,37]. Briefly, cancer cell lines were plated in a flat-bottom 96-wells plate at 1 × 104 cells per well density. Then, the cells were incubated with synthetized compounds at different concentrations (from 0.1 to 100 µM) in 200 µL of 10% fetal bovine serum-RPMI or EMEM culture medium at 37 °C for 72 h. Then, 10 µL of MTT was added at a final concentration of 0.5 mg/mL, incubated at 37 °C for 4 h, and then solubilized with 10% sodium dodecyl sulphate (SDS) in 0.1 mM HCl and incubated overnight at 37 °C. Formazan formation was measured at 570 nm in a multi-well reader (StatFax 4200).
3.5.5. Cell Viability Assessed by Propidium Iodide Assay
Cell viability was analyzed by flow cytometry as previously described [22,44]. In this assay, after setting the baseline to exclude cell debris, cells impermeable to propidium iodide (PI negative) are considered as viable. Two populations of PI-permeable dead cells are distinguished based on fluorescence intensity, corresponding to either hypodiploid apoptotic cells or necrotic cells with intact DNA. Here, HL60 cells were incubated with concentrations of 50 µM of each compound for 24 h at 37 °C. Cells were harvested and stained with 10 µg/mL of propidium iodide to determine cell viability. Samples containing roughly 1 × 104 cells were evaluated by flow cytometry (FACScanto II; Becton Dickinson, Mountain View, CA) and analyzed using FCS Express v6 software. Three independent biological replicates were performed.
3.5.6. Cell Viability Assessed by AnexinV/PI Assay
HL60 cells were seeded into a 12-well plate (5 × 105 per well) and incubated in RPMI 1640 with 50 µM of 7h for 24 h at 37 °C—then, replaced with fresh medium containing the tested compound 7h at indicated concentrations for another 24 h. The cells were collected and then used the Annexin-V/PI Alexa Fluor 488 apoptosis kit (Invitrogen, Carlsbad, CA, USA) according to the instruction of manufacturer to detect the apoptosis cells, which were analyzed by flow cytometry (BD; FACScanto II, Mountain View, CA) and data were processed using FCS Express v6 software. Two independent biological replicates were performed.
3.5.7. Cell Cycle Analysis
HL60 cells were plated in 6-well plates at a density of 2.0 × 105 cells/1.5 mL/well, and grown overnight at 37 °C in a 5% CO2 humidified atmosphere. Cells were treated with compounds 7h at 25 µM concentration or 1% DMSO (controls) for 48 h. Aliquots of cells were collected, pelleted, washed, and fixed with ethanol (70% v/v) for at least 30 min at 4 °C. Cells were twice resuspended in PBS, and after centrifugation and elimination of the supernatant, resuspended in a solution containing PBS, PI (50 mg/mL, Invitrogen), RNase A (20 mg/mL, Invitrogen). After a final incubation for 1 h in the dark at 37 °C; PI signal was measured using a FACS canto II flowcytometer (BD; Mountain View, CA) with a 488 nm excitation laser, captured, and FACS Diva was used as the acquisition software. The percentage of cells in each phase was analyzed using the FCS Express v6 software. Three independent biological replicates were performed.
3.5.8. Western Blotting
Cell lysates were prepared by harvesting cells in Laemmli sample buffer. Proteins were separated on SDS polyacrylamide gels and electroblotted onto nitrocellulose membranes. After blocking, the membranes were incubated with specific primary antibodies overnight, washed, and then incubated with peroxidase-conjugated secondary antibodies. Finally, peroxidase activity was detected with SuperSignal West Pico reagents (Thermo Scientific, Waltham, MA, USA) using a CCD camera LAS-4000 (Fujifilm). Specific antibodies were purchased from Sigma-Aldrich (anti-α-tubulin, clone DM1A; peroxidase-labeled secondary antibodies) and Cell Signaling (anti-PARP, clone 46D11; anti-ERK1/2; anti-phospho-ERK1/2 T202/Y204).
3.6. 3D-QSAR
3.6.1. Selection of Conformers and Molecular Alignment
CoMFA studies were performed with Sybyl X-1.2 software installed in a Windows 7 environment on a PC with an Intel core i7 CPU. In order to acquire the best conformers for each molecule, every compound was subjected to a preliminary geometry optimization of 1000 iterations using the Tripos force field implemented in Sybyl [45]. The convergence criterion of the energy gradient was set to 0.005 kcal/molÅ, and Gasteiger–Hückel charges were assigned to each atom [46], after which 10 cycles of simulated annealing dynamics were run, heating the molecules to 2000 K for 2000 fs, followed by the annealing of the compounds at 0 K for 10,000 fs. From this analysis, the conformers with minimal total energy for each compound were chosen for the definitive CoMFA studies. The minimized structures were superimposed by Distill rigid method, as is implemented in Sybyl.
3.6.2. CoMFA Field Calculation
To derive CoMFA descriptor fields, the aligned training set molecules were placed in a three-dimensional (3D) cubic lattice with grid spacing of 2 Å in x, y, and z directions, such that the entire set was included. CoMFA steric and electrostatic field energies were calculated using a sp3 carbon probe atom with a Van der Waals radius of 1.52 Å and a charge of +1.0. Cut-off values for both steric and electrostatic fields were set to 30.0 kcal/mol.
3.6.3. Internal Validation and Partial Least Squares (PLS) Analysis
PLS analysis was used to construct a linear correlation between the CoMFA descriptors (independent variables) and the activity values (dependent variables) [47]. To select the best model, the cross-validation analysis was performed by using the LOO method (and SAMPLS), which generates the square of the cross-validation coefficient (q2) and the optimum number of components (N). The non-cross-validation was performed with a column filter value of 2.0 in order to speed up the analysis and reduce the noise. The q2, which is a measure of the internal quality of the models, was obtained according to Equation (1):
| (1) |
where , , and are observed, mean, and predicted activity in the training, respectively.
3.6.4. External Validation of the CoMFA Model
The predictive power of the models was assessed by calculation of the predictive r2 (r2pred) [48,49]. r2pred measures the predictive performance of a PLS model and is defined according to Equation (2):
| (2) |
where SD is the sum of the squared deviations between the biological activities of the test set compounds and mean activity of the training set compounds, and PRESS is the sum of squared deviations between observed and predicted activities of the test set compounds.
4. Conclusions
We studied new 2,6,9-trisubstituted purines against seven cancer cell lines and MCR-5 cells, concluding that chemical modifications to the purine moiety increased cytotoxicity and selectivity. We found that some of these compounds were more potent than cisplatin and demonstrated unprecedented selectivity against four cancer cell lines when compared to MCR-5 cells, especially 7h. Our preliminary analysis upholds earlier conclusions that the purine core is a privilege scaffold. Structure–activity relationships indicated that steric properties were more relevant to explain the cytotoxicity of compounds than electronic properties. Likewise, an arylpiperazinyl system connected at position C-6 of the purine ring was pivotal for cytotoxic activity in the main cancer cell lines studied, and the alkylation at N-9 was not relevant for thus activity. Hence, the primary action detected for 7h in HL-60 cells was the induction of apoptosis, specifically late apoptosis. Similar to the already market drug cisplatin, 7h did not have an effect on proliferation on HL-60 cells and arrested the cell cycle in S-phase. These results showed that 7h is a promising lead, and several current efforts are aimed at elucidating the targets involved in antitumor activity.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/1/161/s1.
Author Contributions
Writing—original draft preparation, C.O.S., V.K., and C.E.-B.; writing—review and editing, C.O.S., R.A.T., and C.E.-B.; synthesis, purification, and characterization of the chemical compounds, A.M.Z.; 3D-QSAR studies, J.M.; crystallographic studies, I.B. and A.R.C.; cytotoxic studies, M.F., V.K., J.B., M.I.L., D.H., and R.J.; flow cytometry analysis and cell proliferation, A.M.Z.; search of molecular targets, V.K., D.H., and R.J. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was received from FONDECYT (Research Grant N° 1161816) and FONDEQUIP program CONICYT EQM 160042, Czech Science Foundation (19-09086S) and Palacky University (IGA_PrF_2019_013) and Xunta de Galicia (ED431C 2018/21) and European Regional Development Fund (Project ENOCH, N° CZ.02.1.01/0.0/0.0/16_019/0000868).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Hassanpour S.H., Dehghani M. Review of cancer from perspective of molecular. J. Cancer Res. Pract. 2017;4:127–129. doi: 10.1016/j.jcrpr.2017.07.001. [DOI] [Google Scholar]
- 3.Heng H.H., Stevens J.B., Bremer S.W., Ye K.J., Liu G., Ye C.J. The evolutionary mechanism of cancer. J. Cell. Biochem. 2010;109:1072–1084. doi: 10.1002/jcb.22497. [DOI] [PubMed] [Google Scholar]
- 4.Aghi M., Chou T.C., Suling K., Breakefield X.O., Chiocca E.A. Multimodal cancer treatment mediated by a replicating oncolytic virus that delivers the oxazaphosphorine/rat cytochrome P450 2B1 and ganciclovir/herpes simplex virus thymidine kinase gene therapies. Cancer Res. 1999;59:3861–3865. [PubMed] [Google Scholar]
- 5.Chan E.L., Chin C.H., Lui V.W. An update of ALK inhibitors in human clinical trials. Future Oncol. 2015;12:71–81. doi: 10.2217/fon.15.293. [DOI] [PubMed] [Google Scholar]
- 6.Rani R., Kumar V. Recent Update on Human Lactate Dehydrogenase Enzyme 5 (hLDH5) Inhibitors: A Promising Approach for Cancer Chemotherapy. J. Med. Chem. 2015;59:487–496. doi: 10.1021/acs.jmedchem.5b00168. [DOI] [PubMed] [Google Scholar]
- 7.Dörsam B., Fahrer J. The disulfide compound α-lipoic acid and its derivatives: A novel class of anticancer agents targeting mitochondria. Cancer Lett. 2016;371:12–19. doi: 10.1016/j.canlet.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Ni M., Esposito E., Raj V.P., Muzi L., Zunino F., Zuco V., Cominetti D., Penco S., Dal Pozzo A. New macrocyclic analogs of the natural histone deacetylase inhibitor FK228; design, synthesis and preliminary biological evaluation. Bioorg. Med. Chem. 2015;23:6785–6793. doi: 10.1016/j.bmc.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Sherer C., Snape T.J. Heterocyclic scaffolds as promising anticancer agents against tumours of the central nervous system: Exploring the scope of indole and carbazole derivatives. Eur. J. Med. Chem. 2015;97:552–560. doi: 10.1016/j.ejmech.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Dolezal M., Zitko J. Pyrazine derivatives: A patent review (June 2012—present) Expert Opin. Ther. Pat. 2015;25:33–47. doi: 10.1517/13543776.2014.982533. [DOI] [PubMed] [Google Scholar]
- 11.Shiro T., Fukaya T., Tobe M. The chemistry and biological activity of heterocycle-fused quinolinone derivatives: A review. Eur. J. Med. Chem. 2015;97:397–408. doi: 10.1016/j.ejmech.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Khan I., Ibrar A., Abbas N. Oxadiazoles as Privileged Motifs for Promising Anticancer Leads: Recent Advances and Future Prospects. Arch. Pharm. 2014;347:1–20. doi: 10.1002/ardp.201300231. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis L.M. The year in new drugs: FDA approvals hit a 20-year high in 2017, with cancer and rare-disease drugs dominating the list of new medicines. Chem. Eng. News. 2018;96:25–30. [Google Scholar]
- 14.Price A.J., Howard S., Cons B.D. Fragment-based drug discovery and its application to challenging drug targets. Essays Biochem. 2017;61:475–484. doi: 10.1042/EBC20170029. [DOI] [PubMed] [Google Scholar]
- 15.Romasanta A.K.S., van der Sijde P., Hellsten I., Hubbard R.E., Keseru G.M., van Muijlwijk-Koezen J., de Esch I.J.P. When fragments link: A bibliometric perspective on the development of fragment-based drug discovery. Drug Discov. Today. 2018;23:1596–1609. doi: 10.1016/j.drudis.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Welsch M.E., Snyder S.A., Stockwell B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010;14:347–361. doi: 10.1016/j.cbpa.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legraverend M., Grierson D.S. The purines: Potent and versatile small molecule inhibitors and modulators of key biological targets. Bioorg. Med. Chem. 2006;14:3987–4006. doi: 10.1016/j.bmc.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 18.Zhao H., Dietrich J. Privileged scaffolds in lead generation. Expert Opin. Drug Discov. 2015;10:781–790. doi: 10.1517/17460441.2015.1041496. [DOI] [PubMed] [Google Scholar]
- 19.Morales F., Ramirez A., Conejo-Garcia A., Morata C., Marchal J.A., Campos J.M. Anti-proliferative activity of 2,6-dichloro-9- or 7-(ethoxycarbonylmethyl)-9H- or 7H-purines against several human solid tumour cell lines. Eur. J. Med. Chem. 2014;76:118–124. doi: 10.1016/j.ejmech.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Yoon J.-s., Jarhad D.B., Kim G., Nayak A., Zhao L.X., Yu J., Kim H.-R., Lee J.Y., Mulamoottil V.A., Chandra G., et al. Design, synthesis and anticancer activity of fluorocyclopentenyl-purines and–pyrimidines. Eur. J. Med. Chem. 2018;155:406–417. doi: 10.1016/j.ejmech.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Malinkova V., Reznickova E., Jorda R., Gucky T., Krystof V. Trisubstituted purine inhibitors of PDGFRalpha and their antileukemic activity in the human eosinophilic cell line EOL-1. Bioorg. Med. Chem. 2017;25:6523–6535. doi: 10.1016/j.bmc.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 22.Calderon-Arancibia J., Espinosa-Bustos C., Canete-Molina A., Tapia R.A., Faundez M., Torres M.J., Aguirre A., Paulino M., Salas C.O. Synthesis and pharmacophore modelling of 2,6,9-trisubstituted purine derivatives and their potential role as apoptosis-inducing agents in cancer cell lines. Molecules. 2015;20:6808–6826. doi: 10.3390/molecules20046808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gucký T., Řezníčková E., Radošová Muchová T., Jorda R., Klejová Z., Malínková V., Berka K., Bazgier V., Ajani H., Lepšík M., et al. Discovery of N2-(4-Amino-cyclohexyl)-9-cyclopentyl-N6-(4-morpholin-4-ylmethyl-phenyl)-9H-purine-2,6-diamine as a Potent FLT3 Kinase Inhibitor for Acute Myeloid Leukemia with FLT3 Mutations. J. Med. Chem. 2018;61:3855–3869. doi: 10.1021/acs.jmedchem.7b01529. [DOI] [PubMed] [Google Scholar]
- 24.Demir Z., Guven E.B., Ozbey S., Kazak C., Atalay R.C., Tuncbilek M. Synthesis of novel substituted purine derivatives and identification of the cell death mechanism. Eur. J. Med. Chem. 2015;89:701–720. doi: 10.1016/j.ejmech.2014.10.080. [DOI] [PubMed] [Google Scholar]
- 25.Tuncbilek M., Kucukdumlu A., Guven E.B., Altiparmak D., Cetin-Atalay R. Synthesis of novel 6-substituted amino-9-(β-d-ribofuranosyl)purine analogs and their bioactivities on human epithelial cancer cells. Bioorg. Med. Chem. Lett. 2018;28:235–239. doi: 10.1016/j.bmcl.2017.12.070. [DOI] [PubMed] [Google Scholar]
- 26.Patel R.V., Park S.W. An evolving role of piperazine moieties in drug design and discovery. Mini Rev. Med. Chem. 2013;13:1579–1601. doi: 10.2174/13895575113139990073. [DOI] [PubMed] [Google Scholar]
- 27.Sharma S., Singh J., Ojha R., Singh H., Kaur M., Bedi P.M.S., Nepali K. Design strategies, structure activity relationship and mechanistic insights for purines as kinase inhibitors. Eur. J. Med. Chem. 2016;112:298–346. doi: 10.1016/j.ejmech.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Chang Y.T., Gray N.S., Rosania G.R., Sutherlin D.P., Kwon S., Norman T.C., Sarohia R., Leost M., Meijer L., Schultz P.G. Synthesis and application of functionally diverse 2,6,9-trisubstituted purine libraries as CDK inhibitors. Chem. Biol. 1999;6:361–375. doi: 10.1016/S1074-5521(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Metcalf C.A., Shakespeare W.C., Sundaramoorthi R., Keenan T.P., Bohacek R.S., van Schravendijk M.R., Violette S.M., Narula S.S., Dalgarno D.C., et al. Bone-Targeted 2,6,9-Trisubstituted purines: Novel inhibitors of Src tyrosine kinase for the treatment of bone diseases. Bioorg. Med. Chem. Lett. 2003;13:3067–3070. doi: 10.1016/S0960-894X(03)00648-6. [DOI] [PubMed] [Google Scholar]
- 30.Coxon C.R., Anscombe E., Harnor S.J., Martin M.P., Carbain B., Golding B.T., Hardcastle I.R., Harlow L.K., Korolchuk S., Matheson C.J., et al. Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines. J. Med. Chem. 2017;60:1746–1767. doi: 10.1021/acs.jmedchem.6b01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Xin M., Shen H., Wen J., Tang F., Tu C., Zhao X., Wei P. Five-membered heteroaromatic ring fused-pyrimidine derivatives: Design, synthesis, and hedgehog signaling pathway inhibition study. Bioorg. Med. Chem. Lett. 2014;24:3486–3492. doi: 10.1016/j.bmcl.2014.05.066. [DOI] [PubMed] [Google Scholar]
- 32.Espinosa-Bustos C., Mella J., Soto-Delgado J., Salas C.O. State of the art of Smo antagonists for cancer therapy: Advances in the target receptor and new ligand structures. Future Med. Chem. 2019;11:617–638. doi: 10.4155/fmc-2018-0497. [DOI] [PubMed] [Google Scholar]
- 33.Malathi K., Ramaiah S. Bioinformatics approaches for new drug discovery: A review. Biotechnol. Genet. Eng. Rev. 2018;34:243–260. doi: 10.1080/02648725.2018.1502984. [DOI] [PubMed] [Google Scholar]
- 34.Krajcovicova S., Soural M. Solid-Phase Synthetic Strategies for the Preparation of Purine Derivatives. ACS Comb. Sci. 2016;18:371–386. doi: 10.1021/acscombsci.6b00061. [DOI] [PubMed] [Google Scholar]
- 35.Fiorini M.T., Abell C. Solution-phase synthesis of 2,6,9-trisubstituted purines. Tetrahedron Lett. 1998;39:1827–1830. doi: 10.1016/S0040-4039(98)00098-7. [DOI] [Google Scholar]
- 36.NCI NCI/NIH Developmental Therapeutcs Program. [(accessed on 20 August 2014)]; Available online: http://dtp.nci.nih.gov/branches/btb/handlingprep.html.
- 37.Cañete-Molina Á., Espinosa-Bustos C., González-Castro M., Faúndez M., Mella J., Tapia R.A., Cabrera A.R., Brito I., Aguirre A., Salas C.O. Design, synthesis, cytotoxicity and 3D-QSAR analysis of new 3,6-disubstituted-1,2,4,5-tetrazine derivatives as potential antitumor agents. Arab. J. Chem. 2019;12:1092–1107. doi: 10.1016/j.arabjc.2017.04.002. [DOI] [Google Scholar]
- 38.Aubrey B.J., Kelly G.L., Janic A., Herold M.J., Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2017;25:104–113. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seung-Wook C. Structural insights into the transcription-independent apoptotic pathway of p53. BMB Rep. 2014;47:167–172. doi: 10.5483/BMBRep.2014.47.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanni J.S., Lowe S.W., Licitra E.J., Liu J.O., Jacks T. p53-independent apoptosis induced by paclitaxel through an indirect mechanism. Proc. Natl. Acad. Sci. USA. 1997;94:9679–9683. doi: 10.1073/pnas.94.18.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Orazi G., Cirone M. Mutant p53 and Cellular Stress Pathways: A Criminal Alliance That Promotes Cancer Progression. Cancers. 2019;11:614. doi: 10.3390/cancers11050614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoe & Cie GmbH; Darmstadt, Germany: 2000. Stoe, Cie, X-STEP32, Version 1.07b: crystallographic package. [Google Scholar]
- 43.Sheldrick G.M. A short history of SHELX. Acta Crystallogr. Sect. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 44.Cabrera A.R., Espinosa-Bustos C., Faundez M., Melendez J., Jaque P., Daniliuc C.G., Aguirre A., Rojas R.S., Salas C.O. New imidoyl-indazole platinum (II) complexes as potential anticancer agents: Synthesis, evaluation of cytotoxicity, cell death and experimental-theoretical DNA interaction studies. J. Inorg. Biochem. 2017;174:90–101. doi: 10.1016/j.jinorgbio.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Vinter J.G., Davis A., Saunders M.R. Strategic approaches to drug design. I. An integrated software framework for molecular modelling. J. Comput. Aided Mol. Des. 1987;1:31–51. doi: 10.1007/BF01680556. [DOI] [PubMed] [Google Scholar]
- 46.Gasteiger J., Marsili M. Iterative Partial Equalization of Orbital Electronegativity—A Rapid Access to Atomic Charges. Tetrahedron. 1980;36:3219–3228. doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- 47.Clark M., Cramer R.D., Van Opdenbosch N. Validation of the general purpose Tripos 5.2 force field. J. Comput. Chem. 1989;10:982–1012. doi: 10.1002/jcc.540100804. [DOI] [Google Scholar]
- 48.Oprea T.I., Waller C.L., Marshall G.R. Three-dimensional quantitative structure-activity relationship of human immunodeficiency virus (I) protease inhibitors. 2. Predictive power using limited exploration of alternate binding modes. J. Med. Chem. 1994;37:2206–2215. doi: 10.1021/jm00040a013. [DOI] [PubMed] [Google Scholar]
- 49.Waller C.L., Oprea T.I., Giolitti A., Marshall G.R. Three-dimensional QSAR of human immunodeficiency virus (I) protease inhibitors. 1. A CoMFA study employing experimentally-determined alignment rules. J. Med. Chem. 1993;36:4152–4160. doi: 10.1021/jm00078a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.