Abstract
Utricularia belongs to Lentibulariaceae, a widespread family of carnivorous plants that possess ultra-small and highly dynamic nuclear genomes. It has been shown that the Lentibulariaceae genomes have been shaped by transposable elements expansion and loss, and multiple rounds of whole-genome duplications (WGD), making the family a platform for evolutionary and comparative genomics studies. To explore the evolution of Utricularia, we estimated the chromosome number and genome size, as well as sequenced the terrestrial bladderwort Utricularia reniformis (2n = 40, 1C = 317.1-Mpb). Here, we report a high quality 304 Mb draft genome, with a scaffold NG50 of 466-Kb, a BUSCO completeness of 87.8%, and 42,582 predicted genes. Compared to the smaller and aquatic U. gibba genome (101 Mb) that has a 32% repetitive sequence, the U. reniformis genome is highly repetitive (56%). The structural differences between the two genomes are the result of distinct fractionation and rearrangements after WGD, and massive proliferation of LTR-retrotransposons. Moreover, GO enrichment analyses suggest an ongoing gene birth–death–innovation process occurring among the tandem duplicated genes, shaping the evolution of carnivory-associated functions. We also identified unique patterns of developmentally related genes that support the terrestrial life-form and body plan of U. reniformis. Collectively, our results provided additional insights into the evolution of the plastic and specialized Lentibulariaceae genomes.
Keywords: evolution, genome fractionation, ABC transporters, transcription factors, transposable elements, whole-genome duplication
1. Introduction
The carnivorous plants from the Lentibulariaceae Rich. family exhibit different life-forms according to their habitats (terrestrial, aquatic, lithophytes, epiphytes, and rheophytes), showing an enormous diversity and worldwide distribution [1]. The family is composed of three genera: Genlisea A. St.-Hil. (corkscrew plants), Pinguicula L. (butterworts), and Utricularia L. (bladderworts). The sequencing of four Lentibulariaceae genomes: Genlisea aurea A. St.-Hil. [2], G. nigrocaulis Steyerm, G. hispidula Stapf [3], and Utricularia gibba L. [4,5], along with the estimation of genome size [6,7,8] and karyological analyses (e.g., [3,9,10,11,12]), revealed a wide distribution of genome size and chromosome numbers. In Utricularia, genome size ranges from ultra-small (~79 Mb) in U. purpurea Walter to large (~706 Mb) in U. caerulea L., with a variable number of chromosomes (e.g., 2n = 18, 28, 30, 36, 38, 40, 44, 48, and 56), supporting the occurrence of polyploidy and aneuploidy/dysploidy (for review see [8,12]).
Lentibulariaceae genome shrinkage has been attributed to a decrease in the number and length of introns and intergenic regions, associated with transposable elements (TEs) silencing. This phenomenon was primarily observed in U. gibba (101 Mb) and G. aurea (63–131 Mb according to [8,12]) genomes [2,13,14]. One of the leading hypotheses to explain the genome shrinkage is based on the high respiration rates caused by the carnivorous traps [15], which can increase reactive oxygen species (ROS) formation. Moreover, the U. gibba and the 86 Mb G. nigrocaulis genomes have suffered high rates of gene deletion in comparison to other eudicots, and in the former, a positive selection of tandemly repeated genes implicated in the adaptation to the carnivorous habit is observed [3,5,16].
In contrast, genome size expansion is generally attributed to TEs proliferation, whole-genome duplication (WGD) (represented by retained polyploid duplicates), and small-scale duplication (mainly represented by proximal and tandem copies) [17,18], which are the standard processes for plant genome evolution [19,20]. Together, TEs and WGDs are considered to be the key players in generating genomic novelties and genomic diversification [21,22,23]. Even among the minimal Lentibulariaceae genomes, the importance of these events it is clear, since the changes in size between the large, 1.5-Gb genome of G. hispidula and that of G. nigrocaulis (86 Mb) are consequences of WGDs, along with the LTR-retrotransposons expansion in the former, and the double-strand break repair-mediated deletions in the latter [3]. Additionally, the LTR-retrotransposons silencing and significant fractionation, which returned several genes to a single copy after multiple rounds of WGDs molded the U. gibba genome [4]. Therefore, the dynamic and specialized Lentibulariaceae genomes can provide a natural platform for the study of genome duplication, fractionation, and TEs expansion and silencing, and their impact on the evolution of the angiosperms [4,5,8].
Utricularia reniformis A.St.-Hil. is endemic to the Brazilian Atlantic Forest, growing as a terrestrial species in wet grasslands or as an epiphyte in moist habitats [1,24]. Some but limited information is available on the U. reniformis nuclear genome, such as its high levels of polymorphism [25], its genome size of 292 Mb (which is an intermediate size in comparison to other Utricularia) and a GC content of 38% [8]. We have also previously reported the sequence analyses of U. reniformis chloroplast (cpDNA) and mitochondrial (mtDNA) genomes [26,27]. In order to develop insights into the forces that have shaped the genomes of the Lentibulariaceae species, here we estimated the genome size and chromosome number, and deep-sequenced the terrestrial bladderwort U. reniformis genome and transcriptome. We also compared the U. reniformis draft genome against the aquatic species U. gibba and other angiosperms, revealing that the U. reniformis has a distinct genome structure, mostly due to lineage-specific WGDs and a TE expansion, reflecting a diversified repertoire of plant developmental and carnivory-associated genes and their underlying terrestrial adaptation.
2. Results
2.1. The Utricularia reniformis Genome
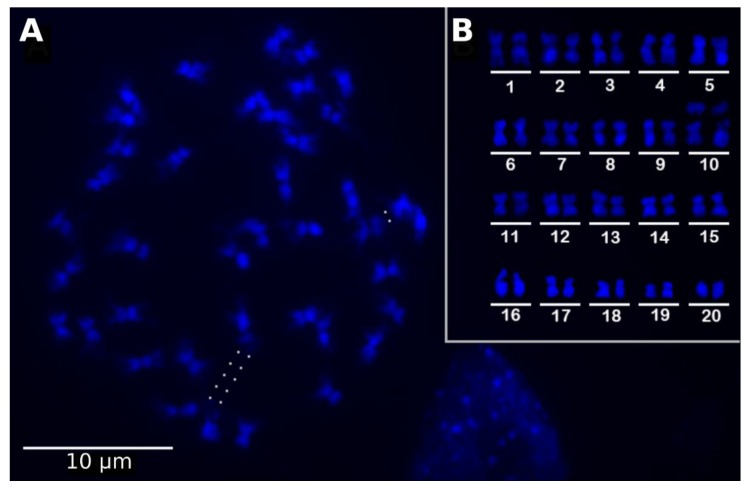
We first determined the chromosome number of U. reniformis as 2n = 40 (Figure 1A,B). The small chromosomes (~1.65 µm) varied between metacentric to submetacentric, with one pair holding a distended satellite, probably representing the ribosomal DNA 45S sites (see dots in Figure 1A and the chromosome pair 10 in Figure 1B). The genome size, estimated by the flow cytometry of the nuclei from leaves, was found to be 2C = 0.652 pg (SD = 1.48%), which represents a haploid genome size of 1C = 317.1 Mb (or 0.324 pg) (Table S1).
Figure 1.
Utricularia reniformis metaphases with 2n = 40. (A) Metaphase stained with DAPI. (B) Karyogram using the same metaphase presented in (A). Dots in (A) represent the distended and unstained region. The bar is equivalent to 10 µm.
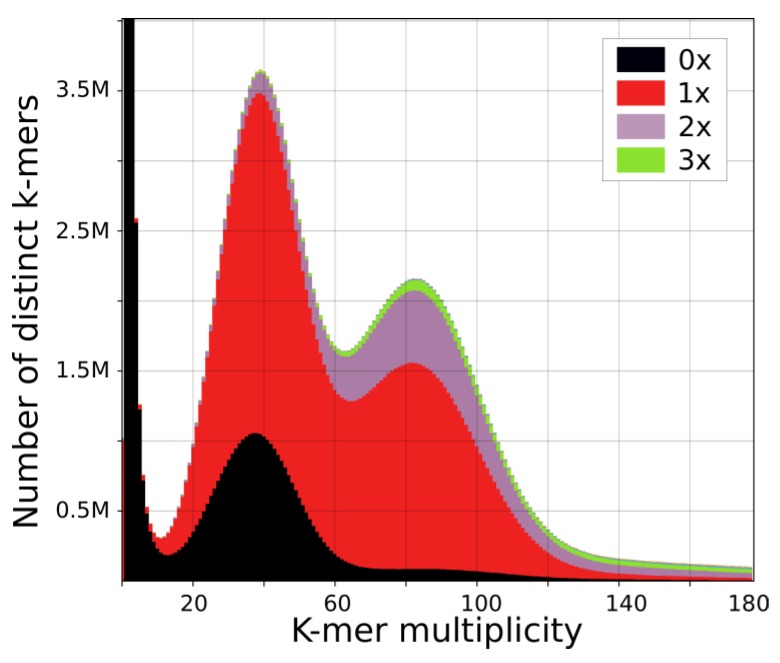
We generated 350 gigabytes of genomic and transcriptomic data for U. reniformis (Table 1). The k-mer frequency analysis demonstrated that U. reniformis exhibits a high heterozygosity rate of 1.8% (Figure 2 and Figure S1). In addition, the k-mer frequency plot was consistent with a tetraploid genome with the haploid at 149 Mb. Moreover, the predicted genome unique content and repeated content lengths are ~88 Mb (29%) and ~220 Mb (71%), respectively.
Table 1.
DNA and RNA read produced for the Utricularia reniformis genome assembly and annotation.
| Technology | Library | Read Length | Raw Reads | Trimmed Reads | |
|---|---|---|---|---|---|
| DNAseq | Illumina HiScan and MiSeq | Paired-end (~350 bp) | 100 bp | 285,403,944 | 187,288,003 |
| Paired-end (~450 bp) | 300 bp | 49,185,074 | 35,963,241 | ||
| Mate-paired (3–9 Kb) | 100 bp | 177,044,066 | 60,011,867 | ||
| RNAseq | Ion Proton | Single-end - Leaves - Stolon - Utricules (preys) |
~200 bp | 46,622,745 41,894,450 112,414,083 |
40,853,284 34,716,966 97,821,388 |
Figure 2.
Genome assembly analysis: Read k-mer frequency versus Utricularia reniformis assembly copy number stacked histograms generated by the KAT comp tool. Read content in black is absent from the assembly, red occurs once, purple twice, and green occurs three times, indicating a scenario of both heterozygous and polyploidy genome. The heterozygous content is represented by the first peak at x = 40 and the homozygous content in the second peak at x = 80. The hidden content (the black peak) represents the heterozygous content that is lost when the assembly bubbles are collapsed. The genome assembly contains most (but not all) of the heterozygous content and introduces duplications on both heterozygous and homozygous content.
The assembled draft genome spanned 304 Mb (96% of the flow cytometry-estimated genome size), with an average coverage of 145× (based on the number and length of aligned reads, divided by the assembled genome size), had an NG50 of 466-Kb and a BUSCO completeness of 87.8%, showing a duplication rate of ~26%, which might correspond with a partial genome duplication (Table S2) and 42,582 gene model predictions. The estimated number of error-free bases was 93% (283 of 304 Mb), which also indicated that multiple repeated genomic regions (e.g., non-autonomous LTR elements, centromeres, and telomeres) remained partial or non-assembled. However, this finding also supported that a large amount of the heterozygous content present in U. reniformis genome was represented in the assembled draft genome (Table 2 and Figure 2).
Table 2.
Utricularia reniformis genome assembly status.
| U. reniformis Genome | ||
|---|---|---|
| Total size of scaffolds (bp) | 304,550,249 | |
| Number of scaffolds | 1830 | |
| Number of contigs | 5452 | |
| Useful amount of scaffold sequences (≥25-Kb) | 297,419,257 | |
| % of assembled genome that is useful | 93.8% | |
| Longest scaffold (bp) | 1,862,935 | |
| Longest contig (bp) | 926,419 | |
| Scaffolds longer than 1-Kb | 1830 (100%) | |
| Scaffolds longer than 100-Kb | 688 (37.6%) | |
| Scaffolds longer than 1 Mb | 47 (2.6%) | |
| NG50 scaffold length (bp) | 466,988 | |
| LG50 scaffold count | 196 | |
| N50 contig length (bp) | 161,226 | |
| Percentage of assembly in scaffolded contigs | 91 | |
| Gaps number | 3677 | |
| Unknown bases (Ns) (bp) | 5,790,542 | |
| Average gap size (bp)/Longest gap (bp) | 1575 | 10,802 |
Moreover, several cpDNA- and mtDNA-derived regions were also identified, thus, providing support for the lateral transfer between the organelles and the nuclear genome, as previously reported [26,27]. We also detected simple sequence repeat (SSRs) markers already developed for U. reniformis [25], in our draft genome. Among all markers, for two SSR loci, we found corresponding regions in U. gibba and U. reniformis (Table S3). In both cases, we recovered two scaffolds in U. reniformis and only one syntenic genomic region in U. gibba (Figure S2). For all other markers, we were not able to detect loci shared in both genomes. In addition to the Arabidopsis-type telomeric repeats identified at the scaffolds ends, we also found Chlamydomonas- and Genlisea-type telomeric repeats in their vicinity, as previously observed for U. gibba [5]. Furthermore, U. reniformis exhibited a considerable amount of TE-derived sequences, when compared to U. gibba (56% vs. 32%).
2.2. Structural Comparative Analysis
In order to infer U. reniformis genome structure and WGD history, we compared the draft genome to U. gibba and other angiosperms. In spite of the low assembly contiguity, we were able to detect at least one WGD round since the core eudicot γ whole-genome triplication (WGT), according to the Ks values for estimates of individual genome duplication events (Figure S3A). Moreover, blockwise relationships between U. reniformis and U. gibba were about 4:2 (Figure S3B), supporting that U. reniformis is a tetraploid, as compared to U. gibba. This finding also suggested that the two Utricularia genomes responded distinctly after the WGD events.
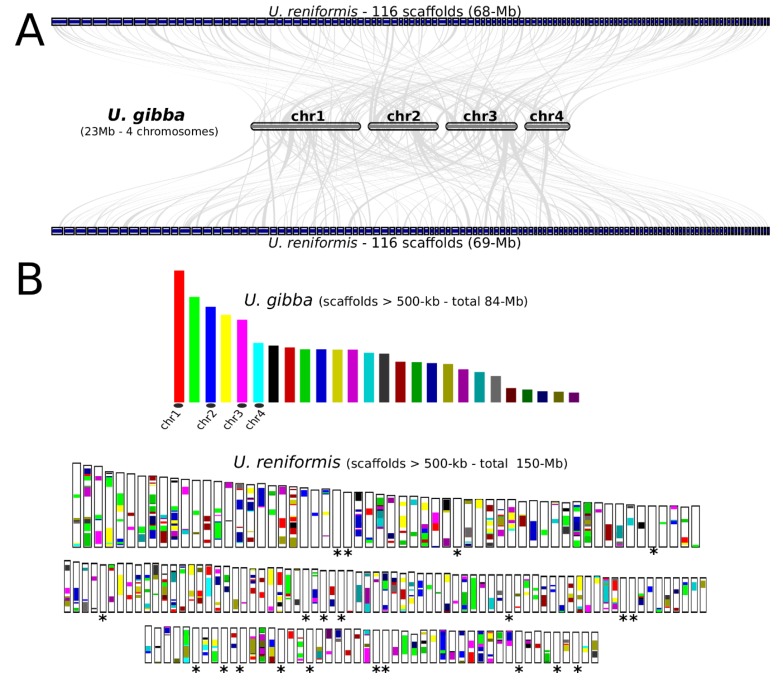
The U. reniformis and U. gibba structural genome differences become more obvious by comparative pairwise alignment. We observed 77% (SD:4%) average nucleotide identity (ANI) between the U. reniformis and U. gibba assemblies by reciprocal best blast (RBB). Additionally, we observed low global collinearity, which indicated that the U. reniformis genome presents a mosaic structure in comparison to U. gibba (Figure 3A,B). Approximately 114 Mb of the U. reniformis genome exhibited partial matches (average length of 84-Kb) to U. gibba (Table S4). The unaligned blocks corresponded to the segments exclusive to U. reniformis, which were mostly related to the TE-related regions (Figure 3B).
Figure 3.
Utricularia reniformis vs. U. gibba genome comparisons. (A) Macrosynteny karyotype visualization of U. gibba chromosome 1 (CM007989.1), 2 (CM007990.1), 3 (CM007991.1), and 4 (CM007992.1), against the corresponding U. reniformis scaffolds showing matches (generated with the MCscan tool with minspan = 10 option). (B) Dual synteny bar plot, created by the ‘dual-bar plotter’ from the MCScanX. The upper panel represents U. gibba chromosomes and scaffolds; the lower panel represents the U. reniformis scaffolds. The syntenic blocks are shown in colors. Most of the white blocks present in each U. reniformis scaffolds corresponds to exclusive regions and transposable elements. The asterisks correspond to the U. reniformis scaffolds showing no matches to U. gibba chromosomes and scaffolds.
Additionally, we mapped less than 37% of the U. reniformis trimmed paired-end reads consistently with the right distance and orientation onto the U. gibba assembly, which also supported the considerable structural differences between these species (Table S5). As a result, it was not possible to employ the reference-assisted assembly procedure to anchor the U. reniformis scaffolds into the U. gibba fully-assembled chromosomes to reconstruct potential pseudo-molecules.
Moreover, in comparison to other angiosperms, U. reniformis also showed a lower percentage of macrosyntenic blocks. For instance, U. reniformis retained at least 47 Mb (average length of 76-Kb and ANI of 82%), 35 Mb (average length of 46-Kb and ANI of 76%), and 53 Mb (average range of 63-Kbp and ANI of 77%) of shared blocks to A. thaliana, V. vinifera, and S. lycopersicum. In contrast, its closest relative, U. gibba, retained 71 Mb (average length of 44-Kb and ANI of 80%), 57 Mb (average length of 26-Kb and ANI of 75%), and 58 Mb (average length of 35-Kb and ANI of 77%) blocks, respectively (Table S4).
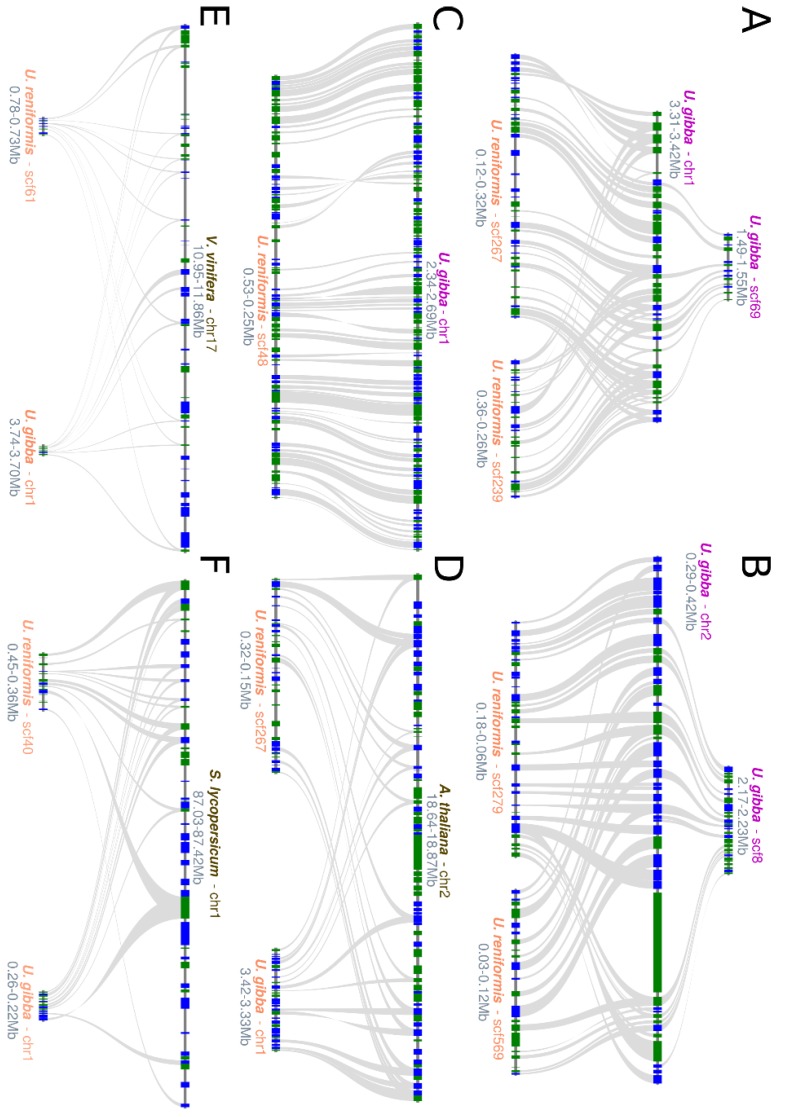
We further evaluated the microsyntenic level and verified that U. reniformis and U. gibba present distinct patterns of retention and alternative deletion of duplicated genes between the polyploid subgenomes, which is consistent with the distinct post-speciation evolutionary paths (Figure 4A–C). This feature is also observed among the Utricularia species and A. thaliana, yet with fewer microsynteny blocks (Figure 4D). When both Utricularia genomes are compared to the phylogenetically distant eudicotyledons, such as V. vinifera and S. lycopersicum, a significant fractionation is prominent (Figure 4E,F).
Figure 4.
Microsynteny analysis of Utricularia reniformis against U. gibba and other eudicotyledons: (A,B) Polyploid subgenomes of U. reniformis vs. U. gibba microsynteny showing an alternative deletion of the duplicated genes. (C) Highly conserved regions of U. reniformis vs. U. gibba microsynteny. (D) Arabidopsis thaliana vs. U. reniformis, with U. gibba showing moderately conserved regions of microsynteny. (E) Vitis vinifera vs. U. reniformis, with U. gibba showing a considerable fractionation. (F) Lycopersicon esculentum vs. U. reniformis, with U. gibba showing a moderate fractionation.
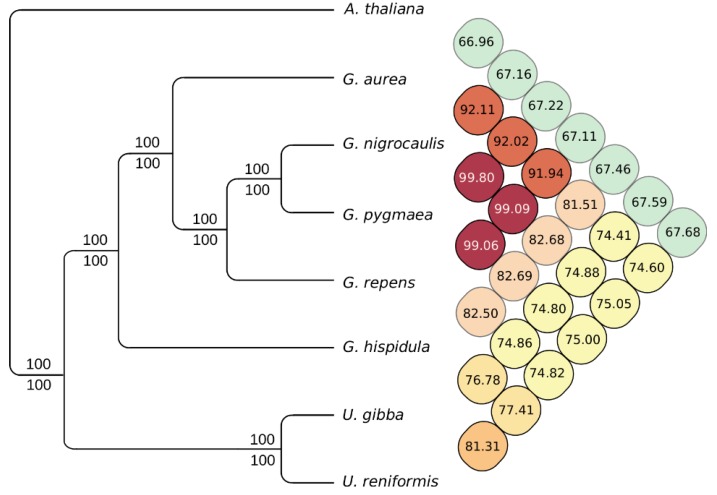
However, in general, the two Utricularia species were far more similar in gene order to each other than either were to A. thaliana, V. vinifera, and S. lycopersicum. The phylogenetic and ANI analyses based on 336 core eukaryotic genes corroborated our microsynteny analysis, supporting the same common ancestor for U. reniformis and U. gibba, and also for the Utricularia and Genlisea clades (Figure 5).
Figure 5.
Overall similarity heatmap and maximum likelihood and Bayesian inference phylogenetic tree of 336 core-eukaryotic genes shared among seven carnivorous plant genomes (Utricularia reniformis, U. gibba, Genlisea aurea, G. nigrocaulis, G. pygmaea, G. repens, and G. hispidula) and Arabidopsis thaliana as an out-group. Numbers above and below the lines indicate the maximum likelihood bootstrap values and the Bayesian posterior probabilities for each clade. The G. pygmaea and G. repens genomes used in this analysis are yet to be published. However, the concatenated core nucleotide gene fasta sequence used to construct this dataset is freely available at http://doi.org/10.5281/zenodo.3268745.
2.3. Utricularia reniformis Comparative Annotation
In general, U. reniformis and U. gibba show different arrays of gene duplication that might directly reflect support that they are retaining genes in a biased manner. This was exemplified by the more significant number of tandem, proximal, and dispersed gene pairs identified in U. reniformis, when compared to U. gibba (Table 3). In contrast, U. gibba displayed more singletons and segmental duplicates. However, the more significant number of segmental copies identified in U. gibba might be related to the assembled genome using long-reads, which permitted a better identification of large syntenic blocks, in comparison to the U. reniformis genome based on short-reads. Moreover, the considerable number of singletons in U. gibba might also represent retained duplicates that have reverted to a single copy after the WGD, which supported the genome size reduction observed in this species.
Table 3.
Utricularia reniformis and U. gibba comparative and structural annotation.
| U. reniformis | U. gibba | |
|---|---|---|
| Total number of identified genes | 42,582 | 25,509 |
| - Singletons | 3083 | 5120 |
| - Dispersed duplicates | 26,546 | 8831 |
| - Proximal duplicates | 1679 | 683 |
| - Tandem duplicates | 2994 | 999 |
| - Segmental duplicates | 8280 | 9876 |
| Annotation status | ||
| - Annotated genes † | 35,899 | 21,283 |
| - Genes with GOs | 27,751 | 17,760 |
| - Unknown and Hypothetical genes | 6683 | 4348 |
| Total gene length (bp) | 79,419,967 | 51,346,373 |
| Total exon length (bp) | 44,971,966 | 28,891,859 |
| Total intron length (bp) | 29,871,202 | 16,875,684 |
| Longest gene (bp) | 84,570 | 61,898 |
| Longest exon (bp) | 7309 | 6002 |
| Longest intron (bp) | 78,774 | 53,378 |
| Longest CDS (bp) | 15,201 | 15,801 |
| Mean gene length (bp) | 1872 | 2016 |
| Mean exons per gene | 5 | 5 |
| % of genome covered by genes | 26 | 51 |
| % of genome covered by CDS | 15 | 38 |
| % of genome covered by TEs-like regions | 56 | 32 |
† high-confidence genes models (showing a potential product, GO term, and hits to UniProt viridiplantae clade).
It is noteworthy that the gene space in U. reniformis represents ~26% of the genome, which also demonstrated that the intergenic regions are replete with TEs-related sequences constituting more than half of the genome size. Conversely, TEs covered ~32% of U. gibba genome, while the gene space represented ~51%.
2.4. Utricularia reniformis Shows a Massive Expansion of LTR from the Gypsy Superfamily
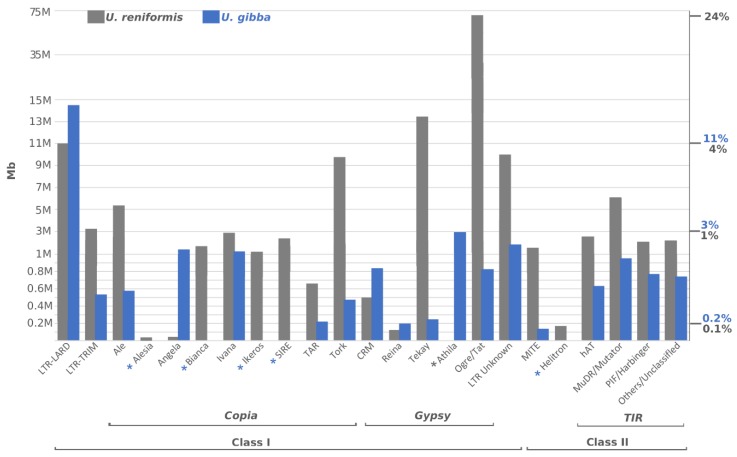
Utricularia reniformis presents a prominent LTR-retrotransposons expansion, representing up to 145 Mb (Figure 6 and Table S6). In general, a separate array of expansion and contraction of distinct evolutionary lineages from the Copia and Gypsy superfamilies stand as the main differences between U. reniformis and U. gibba. The most noticeable increase of the Gypsy superfamily was observed within the Ogre evolutionary lineage, accounting up to 72 Mb of U. reniformis genome. It is noteworthy that the LTR-LARDs, Angela, CRM, are more prevalent in U. gibba, including the Athila evolutionary lineage, which was exclusively found in U. gibba. In contrast, from the Copia superfamily, the evolutionary lineages Alesia, Bianca, Ikeros, and SIRE were found solely in U. reniformis. As observed in other plant genomes, the Class II elements were less prominent in both species, and except for the Helitron super-family which we were not able to identify in U. gibba, mostly known super-families and evolutionary lineages commonly found in plant genomes were present.
Figure 6.
Transposable elements discovery in Utricularia reniformis and U. gibba genomes: Length occupied (bp) of each super-family and evolutionary lineage. A detailed distribution of each element identified is presented in Table S6. The y-axis on the left indicated the total length occupied in the megabases of each evolutionary lineage, whereas the y-axis on the right meant the % of the corresponding genome. Blue and Grey asterisk correspond to the absent evolutionary lineages in U. gibba and U. reniformis, respectively.
2.5. Distinct Functional Enrichment Patterns Among Tandem and Dispersed Duplicate Genes
Self–self-genome comparisons among the singletons and duplicated genes (dispersed, proximal + tandem, and segmentals) revealed exciting patterns of enriched GO terms and KEGG enzymes (p < 0.05, Fisher’s exact test, Bonferroni corrected) between U. reniformis and U. gibba (Tables S7–S9). Both species retained a conserved arrangement of cellular components related to organelles and nucleus among the singletons, such as, chloroplast (GO:0009507), thylakoid (GO:0009579), mitochondrion (GO:0005739), nucleoplasm (GO:0005654), nuclear envelope (GO:0005635), endoplasmic reticulum (GO:0005783), and biological processes related to photosynthesis (GO:0015979) and DNA metabolic process (GO:0006259). Additionally, KEGG enzymes with associated functions that act on NADH or NADPH (EC:1.6.99.5), NADH dehydrogenase (EC:1.6.99.3), NADH—ubiquinone reductase (EC:7.1.1.2) were also augmented among the singletons. Together, these findings support a conserved organellar functional activity including the control of energy homeostasis functions among the singletons.
Among the dispersed duplicates, the nuclease activity (GO:0004518) was enriched in both species. However, the hydrolase activity (GO:0016787), transcription regulator activity (GO:0140110), DNA-binding transcription factor activity (GO:0003700), and tropism (GO:0009606) were significantly overrepresented in U. reniformis, while carbohydrate metabolic process (GO:0005975), transport (GO:0006810), kinase activity (GO:0016301), and lipid metabolic process (GO:0006629) were enriched in U. gibba.
It was previously observed in many angiosperm genomes that secondary metabolic function and transcriptional function are enriched among tandems and segmental duplicates, respectively [5,28,29,30]. Our results corroborate this observation for secondary metabolic function in the tandem repeats of both Utricularia, and the transcriptional functions only in U. gibba segmental duplicates. Interestingly, and opposite to what has been previously observed, the transcriptional features were augmented in U. reniformis dispersed duplicates genes. Since the dispersed copies could arise from TEs movements, and the U. reniformis genome had a high level of LTR-elements, this result might suggest a central role of LTR transposition in molding these genomic differences.
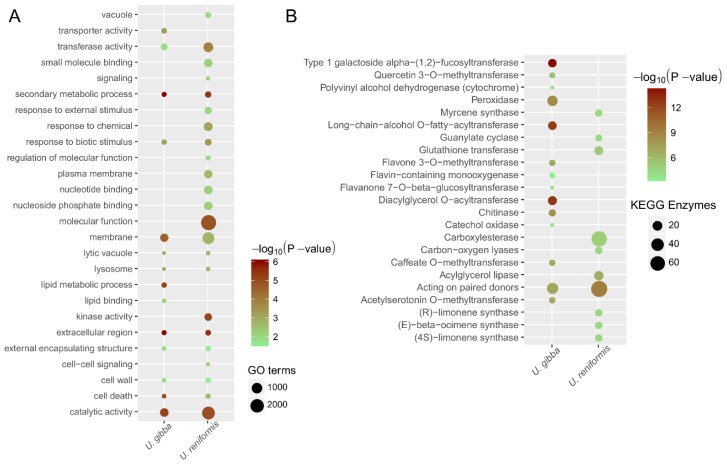
We also checked the potential role of tandem and proximal duplicates with the evolution of novel functions, such as those related to carnivory and the ROS metabolism, which revealed an enrichment of many GO terms and KEGG enzymes in both species (Figure 7). For instance, catalytic activity (GO:0003824), biotic stimulus (GO:0009607), transferase activity (GO:0016740), and the cellular components—lysosome (GO:0005764), lytic vacuole (GO:0000323), and the cell wall (GO:0005618) were overrepresented. The enrichment of the cell wall (GO:0005618) term, could be related to the trap movement during the prey capture, which demands profound and dynamic cell-wall changes [5], and the lysosome and lytic vacuole enrichment might be related to the digestion of preys. However, we also identified distinct GO and KEGG enrichment arrangement among species, strongly suggesting a functional divergence, subfunctionalization, or neofunctionalization among the tandem and proximal duplicates (Figure 7). Comparatively, U. reniformis showed an enrichment of response to chemicals (GO:0042221), a response to external stimulus (GO:0009605), and signaling (GO:0023052), whereas, as observed in the dispersed duplicates, the lipid metabolic process (GO:0006629) was also augmented in U. gibba tandem duplicates. Regarding the well-known carnivory [4,5] and ROS enzymatic functions, U. gibba presented chitinase (EC:3.2.1.14), cysteine protease (EC:3.4.22), and peroxidases (EC:1.11.1.7) enriched in tandem duplicates regions, as previously described [5], whereas U. reniformis exhibited glutathione transferases (EC:3.4.16), carboxylesterase (EC:2.5.1.18), acylglycerol lipase (EC:3.1.1.23), pectinesterase (EC:3.1.1.11), and enzymes that acted on the paired donors, with an incorporation or reduction of molecular oxygen (EC:1.14.13). Therefore, these findings indicated that both species presented distinct enrichment patterns of GO terms and KEGG enzymes, supporting an ongoing gene birth–death–innovation process that included metabolism and carnivory-associated functions, among the tandem and proximal duplicated genes.
Figure 7.
Functional enrichment comparative analysis of tandem duplicates among Utricularia reniformis and U. gibba: The enriched GO terms and KEGG enzymes with the corrected p-value < 0.05 are presented. The color of the circle represents the statistical significance of the enriched GO terms (A) and the KEGG Enzymes (B). The size of the circles represents the number of occurrences of a GO term (A) and the KEGG Enzyme (B).
Interestingly, among the segmental duplicates, similar patterns of functions related to the primary metabolism, and the plant developmental process were enriched in both species, for example the amide biosynthetic process (GO:0043604), anatomical structure development (GO:0048856), reproduction (GO:0000003), nitrogen compound metabolic process (GO:0006807), peptide metabolic process (GO:0006518), and the primary metabolic process (GO:0044238).
2.6. Utricularia reniformis Displays Unique Patterns of Carnivory-Associated, Land Adaptation, and Developmentally Related Genes
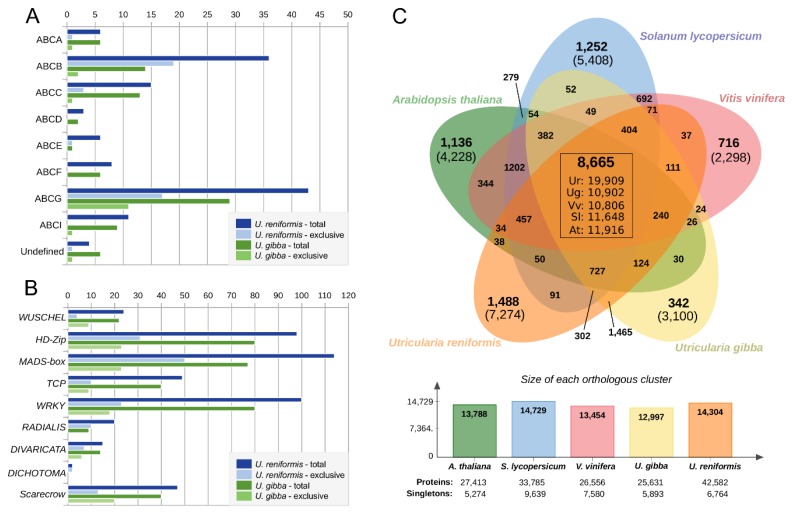
Previous studies, including literature review and comparative annotation approaches, accurately identified GO terms and KEGG enzymes strictly related to the carnivory-associated functions among the sequenced carnivorous plants available in public databases [31,32]. Based on this data, we investigated our functional annotation results and observed that U. reniformis had an extensive gene repertoire of carnivory-associated and ROS-related functions (Table 4). In addition, we also identified an extensive repertoire of plant development, reproduction, and life-form adaption GO terms in U. reniformis (Table 5), which might be related to the different life-forms in comparison to U. gibba. Moreover, distinct patterns of ABC transporters were observed between U. reniformis and U. gibba (Figure 8A,B). Terrestrial plants exhibited, on average, a repertoire of 128 ABC transporters, while the aquatic plants had fewer copies [33]. At least 132 and 86 ABC transporters were identified in U. reniformis and U. gibba, respectively (Figure 8A and Table S10), suggesting that U. reniformis had more copies due to its terrestrial adaptation. This was especially apparent from the expansion of the ABC transporter B (Figure 8A and Figure S4) and ABC transporter C families, which are commonly related to developmental processes and functions necessary for life on dry land, and the ABC G family (Figure 8A and Figure S5), which are related to response to biotic stress [33]. Furthermore, plant developmental-related transcription factor (TFs) containing the wuschel-like homeobox (WOX), homeobox-leucine zipper (HD-Zip), agamous-like (MADS-box), TCP, WRKY, RADIALIS, DIVARICATA, DICHOTOMA, and the scarecrow-like gene families present distinct patterns of expansion and contraction among both species (Figure 8B and Tables S11–S14), and thus, provide support for the two distinct life-forms between these species.
Table 4.
Carnivory-associated and reactive oxygen species (ROS) detoxification GO terms distribution. Utricularia reniformis (7200 unique genes: Singletons—3.5%, Dispersed—65.5%, Tandem and Proximal—10.5%, and Segmental—20.5%) and U. gibba (3918 unique genes: Singletons—10%, Dispersed—48%, Tandem and Proximal—6%, and Segmental—36%).
| Gene Ontology Term | GO Code | U. renif n# Genes | U. gibba n# Genes |
|---|---|---|---|
| amidase activity | GO:0004040 | 13 | 10 |
| actin filament | GO:0005884 | 20 | 11 |
| alpha-galactosidase activity | GO:0004557 | 15 | 13 |
| alternative oxidase activity | GO:0009916 | 6 | 4 |
| ammonium transmembrane transport | GO:0008519; 0072488 | 18 | 16 |
| aspartic-type endopeptidase activity | GO:0004190 | 289 | 94 |
| ATP:ADP antiporter activity | GO:0005471 | 9 | 9 |
| ATPase activity | GO:0016887 | 1831 | 792 |
| beta-galactosidase activity | GO:0004565 | 720 | 125 |
| catalase activity | GO:0004096 | 17 | 11 |
| cellulase activity | GO:0008810 | 62 | 34 |
| chitinase activity | GO:0004568 | 21 | 13 |
| cinnamyl-alcohol dehydrogenase activity | GO:0045551 | 34 | 15 |
| cyclic-nucleotide phosphodiesterase activity | GO:0004112 | 1 | 1 |
| cysteine-type peptidase activity | GO:0008234 | 384 | 185 |
| nuclease activity | GO:0004518 | 1955 | 1094 |
| fructose-bisphosphate aldolase activity | GO:0004332 | 12 | 10 |
| glutathione transferase activity | GO:0004364 | 54 | 21 |
| glutathione peroxidase activity | GO:0004602 | 14 | 9 |
| hydrolase activity, acting on ester bonds | GO:0016788 | 2767 | 1259 |
| heat shock protein activity | GO:0042026; 0006986; 0034620 | 129 | 72 |
| lipase activity | GO:0016298 | 231 | 140 |
| lipid transport | GO:0006869 | 170 | 113 |
| myosin heavy chain kinase activity | GO:0016905 | 36 | 23 |
| peroxidase activity | GO:0004601 | 192 | 140 |
| peptidase activity | GO:0008233 | 2650 | 1269 |
| phosphatase activity | GO:0016791 | 287 | 186 |
| phospholipase activity | GO:0004620 | 80 | 43 |
| polygalacturonase activity | GO:0004650 | 109 | 50 |
| polygalacturonase inhibitor activity | GO:0090353 | 5 | 1 |
| protein homodimerization activity | GO:0042803 | 809 | 525 |
| ribonuclease activity | GO:0004540 | 390 | 154 |
| serine-type carboxypeptidase activity | GO:0004185 | 82 | 38 |
| superoxide dismutase activity | GO:0004784 | 17 | 10 |
| symplast | GO:0055044 | 1060 | 679 |
| urease activity | GO:0009039 | 2 | 1 |
| water channel activity | GO:0015250 | 46 | 26 |
Table 5.
Distribution of GO terms related to the biological process related to plant development, reproduction, and life-form adaptation. (U. reniformis; 11,468 unique genes: Singletons—3%, Dispersed—62%, Tandem and Proximal—10%, and Segmental—23%. U. gibba; 7071 unique genes: Singletons—11%, Dispersed—41%, Tandem and Proximal—4%, and Segmental—43%).
| Gene Ontology Term | GO Code | U. renif n# Genes | U. gibba n# Genes |
|---|---|---|---|
| developmental process involved in reproduction | GO:0003006 | 1965 | 1237 |
| reproduction | GO:0000003 | 2546 | 1600 |
| reproductive process | GO:0022414 | 2223 | 1388 |
| multicellular organism development | GO:0007275 | 4749 | 2773 |
| embryo development | GO:0009790 | 744 | 465 |
| post-embryonic development | GO:0009791 | 2447 | 1578 |
| flower development | GO:0009908 | 915 | 601 |
| developmental maturation | GO:0021700 | 264 | 171 |
| developmental process | GO:0032502 | 4126 | 2704 |
| reproductive structure development | GO:0048608 | 1685 | 1065 |
| anatomical structure development | GO:0048856 | 4046 | 2639 |
| cellular developmental process | GO:0048869 | 1352 | 880 |
| reproductive shoot system development | GO:0090567 | 655 | 437 |
| pollination | GO:0009856 | 343 | 210 |
| tropism | GO:0009606 | 215 | 129 |
| circadian rhythm | GO:0007623 | 261 | 161 |
| response to stress | GO:0006950 | 4513 | 2869 |
| response to radiation and light stimulus | GO:0009314, GO:0009416 | 1012 | 711 |
| response to external stimulus | GO:0009605 | 2108 | 1352 |
| response to biotic stimulus | GO:0009607 | 1526 | 981 |
| response to abiotic stimulus | GO:0009628 | 3161 | 2041 |
| response to endogenous stimulus | GO:0009719 | 2665 | 1693 |
| response to chemical | GO:0042221 | 4590 | 2984 |
| response to stimulus | GO:0050896 | 6579 | 4277 |
Figure 8.
(A) Distribution of ABC transporters and (B) developmental-related transcription factors among Utricularia reniformis and U. gibba. Exclusive genes are species-specific genes for which no orthologs could be found in any of the other species compared. (C) Venn diagram showing the distribution of shared orthologous gene families among U. reniformis, U. gibba, Arabidopsis thaliana, Vitis vinifera, and Lycopersicon esculentum (using an MCL inflation factor of 1.5). The values present inside the box correspond to the number of core genes of each species. The number present inside the parentheses represent the total number of species-specific genes from each species cluster.
2.7. Comparative Annotation Among Other Angiosperms Genomes Reveals Species-Specific Genes Strictly Related to the Environment and Life-Form Adaptations
The comparative orthologous gene cluster analysis revealed that approximately 60% of U. reniformis and U. gibba genes are shared, indicating that they were inherited from the last common ancestor. Interestingly, U. reniformis presents an almost duplicated core gene set from that observed in U. gibba, V. vinifera, A. thaliana, and S. lycopersicum, and thus, supports a species-specific WGD event (Figure 8C and Table S15). In general, a significant fraction of the species-specific genes (genes for which no orthologs could be found in any of the other species compared) and single-copy gene clusters from U. reniformis and U. gibba encode uncharacterized or hypothetical proteins (63% for U. reniformis and 50% for U. gibba). Among the species-specific genes, we highlighted ABC transporters, and several developmental-related TF gene families (Figure 8A,B), supporting the role of the species-specific genes with plant development, reproduction, prey strategies, and life-from adaptations. GO enrichment analysis (p < 0.05, Fisher’s exact test, Bonferroni corrected) among the species-specific genes revealed an augmented functions related to the reproductive process (GO:0022414), hydrolase activity (GO:0016788), nuclease activity (GO:0004518), catalytic activity (GO:0003824), protein binding (GO:0005515), and organelles (GO:0043226) in both species. However, U. reniformis present a unique enrichment of kinase activity (GO:0016301), transferase activity (GO:0016740), carbohydrate metabolic process (GO:0005975), lytic vacuole (GO:0000323), and lysosome (GO:0005764), whereas, U. gibba exhibited cellular nitrogen compound metabolic process (GO:0034641), peptide metabolic process (GO:0006518), and signal transduction (GO:0007165). Taken together, these results indicate that the species-specific gene set might play an essential role in the diversification and adaptation to different life-forms.
3. Discussion
3.1. The Role of WGD in Utricularia reniformis Genome Evolution
Polyploid occurred multiple times during angiosperms evolution, and this is not an exception for Utricularia lineages. For instance, multi-way microsynteny analysis revealed three sequential WGD events in U. gibba, in addition to the γ WGT event shared by all core eudicots [4,5,34]. Additionally, out-crossing events might be broad among the Utricularia clade, and a comparative analyses revealed that the most recent U. gibba WGD was derived from an allopolyploidization event [5]. Allopolyploidization is recognized as a driving evolutionary innovation and is often associated with speciation when resulting in a duplicated genome structure [35,36,37]. Therefore, the current hypothesis is that not only was the U. gibba genome shaped by WGD events, but other Utricularia genomes and specifically the U. reniformis genome were shaped by WGD-induced variation and adaptation.
After a polyploidy event, each lineage undergoes distinct patterns of gene retention and fractionation, which generally leads to the genome returning to the diploid state [38]. Our results for U. reniformis and U. gibba support that each species has distinct patterns of deletions, duplicated genes, and rearrangements, suggesting that after speciation and WGD, they took distinct evolutionary paths that were consistent with their life-histories. This is exemplified by the significant number of tandem, proximal, and dispersed gene pairs identified in U. reniformis, when compared to U. gibba. Indeed, gene duplication is recognized as one of the major sources of evolutionary innovation [39]. For instance, tandem and proximal duplicated genes are often associated with the emergence of functional novelty, which commonly originated from isolated events in which an individual gene gets duplicated by unequal crossing-over between similar alleles [39]. Together with a large number of dispersed duplicates identified, the tandem duplicated genes might also arise by translocations mediated by TEs [39], generating single-gene transposition-duplication, and therefore, indicating for an essential role of TEs molding the genome structure and evolution. Additionally, WGD analysis confirmed that U. reniformis experienced at least one WGD round since the core eudicot WGT. However, it should be taken into account that the low assembly contiguity based on short-read technology hindered the identification of large syntenic blocks, and thus, complicated the evaluation of the complete history of U. reniformis WGD events.
3.2. LTR-Retrotransposons are Key Agents Governing Utricularia Genome Size Changes
One of the most common repetitive content of U. reniformis is related to TEs-related sequences, corresponding to at least 56% of the genome. The TEs drive the genome evolution, mainly by rearrangements, gene promoters repression, enhancers disruption, epigenetic regulation, and also by inflating the genome size by its copy-and-paste mechanism [40,41]. Apart from their deleterious effects, TEs also provide genome variation that can lead to the plant adapting more rapidly to new environmental conditions, and thus, leading to speciation [42,43]. The increased number of TEs in U. reniformis vs. U. gibba suggests that TEs might be playing a role in the genome innovation we are observing, but it could also just reflect a difference in the ability of the two genomes to purge mobile TEs.
The most prominent TE expansion in U. reniformis is related to LTR-retrotransposons elements from the Gypsy superfamily, which was also observed in the Solanaceae and Brassicaceae families [44,45]. In particular, elements from the Tat lineage were the most prominent, which are commonly broader in other plants, constituting up to 40% of the genome of some species [46].
Except for LTR-LARDs, all other TE groups are expanded in U. reniformis. The LTR-LARD and CRM are generally located in complex and highly repeated genomic loci, such as heterochromatic and pericentromeric regions [47,48]. These regions are often partially assembled using short-read technology, and we verified that most LTR-LARDs identified in U. reniformis are incompletely assembled, suggesting that they were partially determined and underestimated. Therefore, LTR-LARD and CRM elements might potentially represent a considerable fraction of the genome that remains unresolved in the U. reniformis assembly, and the results presented might be biased due to the assembly contiguity limits.
3.3. The Genomic Landmarks for Terrestrial Adaptation
Adaptation to the environment and life-form plasticity are hallmarks of the selective pressures that govern genome evolution in the Utricularia lineage. A common reproductive strategy for several Utricularia is to produce clones by stolon fragmentation, which is a usual asexual reproduction of aquatic species from the section Utricularia (the section in which U. gibba is nested), with some species being sterile and rarely flowering [49,50]. In contrast, U. reniformis presents a specialized sexual reproductive strategy that is entirely dependent on pollinators, and is specifically dependent on self-pollination [51]. Moreover, U. gibba seems to have a more severe degree of Fuzzy Arberian Morphology, such as no clear delimitation of distinct vegetative organs [52]. In contrast, U. reniformis presents a more traditional vegetative organ delimitation (as stems and leaves), similar to other angiosperms.
Consequently, the vegetative plasticity found in different Utricularia species might be essential strategies used to colonize the distinct habitats. Considering the different environments (water, soil, rock surface, etc.), the various trap designs [53,54,55] might be shaped as adaptation for different prey faunas [56,57,58,59]. Ultimately this might impact the absorption and assimilation of nutrients. At the genomic scale, both species display distinct patterns of development-related genes and TFs, such as WOX, HD-Zip, MADS-box, WRKY, TCP, RADIALIS, DIVARICATA, DICHOTOMA, and scarecrow-like gene families. These TFs play specialized roles in plant growth and development by regulating cell division and differentiation, in particular, the trap and floral meristem and trichome development, and might also develop an important role in the regulation of flowering time and the ontogeny of reproductive organs [16,60,61,62,63,64]. For instance, the WOX1 (3 vs. 5 species-specific genes) from the WOX family, is possibly associated with the trap development [16] SOC1 (11 vs. 6 species-specific genes) from the MADS-box family, which might develop a role with phosphorus scavenging from the trapped prey [4], and ATHB51 (5 vs. 6 species-specific genes) related to the leaf morphogenesis. These findings are suggestive to a different array of target genes, and the processes that these TFs control, and in addition to the distinct number of developmental genes identified in both species, might apply in different body plan adaptations and prey strategies.
Moreover, both species display a distinct repertoire of ABC transporters that are considered essential to terrestrial life-form adaption [33]. Some ABC transporters members are exclusively found in U. reniformis. For instance, the ABCB member 9 is related to plant hormone transport, regulation of growth, and in development [65]. Additionally, the mitochondrial ABCB family member 25 was directly involved with the molybdenum cofactor biosynthesis, and nitrogen assimilation from the soil [66]. Interestingly, aquatic plants mainly rely on ammonium as their primary nitrogen source, and the molybdenum cofactor biosynthesis deficiency is commonly associated with the use of ammonium as an alternative nitrogen source in habitats with increased relative humidity [66]. This suggests that the absence of ABCB members 25 in U. gibba might be directly related to their aquatic adaptation. Finally, the ABCG member 36, exclusively found in U. reniformis, is related to the export of related defence compounds and callose deposition at the site of pathogen contact to restrict pathogen development [67,68], supporting that U. reniformis needs extra protection for cellular damage, which is a common characteristic of terrestrial plants.
It is known that carnivorous plants experience different and variable levels of nutritional stress [69], and the emergence of the carnivory-associated function in each lineage was a result of multiple evolutionary paths [31]. Interestingly, U. reniformis presents a distinct and augmented set of well-known carnivory-associated functions. Among them, alpha-galactosidase, amidase, actin filament, aspartic-type endopeptidase, beta-galactosidase, cellulase, chitinase and cysteine-type peptidase, myosin heavy chain kinase, and polygalacturonases might be related to carbon and energy cycles maintenance with cellulose and its associated polymers-decomposing activity, suggesting a potential role in the breakdown of prey polysaccharides. Moreover, the enrichment of ATPase activities might be correlated with the trap acidity and molecular transport functions, such as the release of digestive enzymes and the absorption of digested material from the preys [5,31,70].
Conversely, in addition to carnivory-associated functions, the ROS-related enzymes are also augmented in U. reniformis. For instance, catalase, superoxide dismutase, peroxidase, and glutathione transferase might contribute to cellular homeostasis and impacting the control of the high respiration rates possibly originated through the carnivory process. In addition, U. reniformis exhibits much more dispersed, tandem, and proximal duplicated genes. The emergence of tandem duplicated genes might contribute to the evolution of novel functions and adaptation. Our results provide support for an ongoing gene birth–death–innovation, occurring mainly among tandem and dispersed duplicates, impacting a number of different GO terms and KEGG enzymes, including carnivory-associated functions across both Utricularia. This process might give a fine-tuning of carnivory-associated functions in each selective environment (aquatic and terrestrial). For instance, a high diversity of bacteria, fungi, algae, and protozoa compose the ecosystem inside the Utricularia trap and act synergistically to convert complicated organic matter into a nutrients source for the plants [71]. The trap ecosystem and prey biota can significantly vary between the terrestrial and aquatic habitat. The aquatic Utricularia species usually have a prey spectra dominated by copepodids, cladocerans, ostracods, rotifers, and aquatic insect larvae, while terrestrial species can also have acari, nematodes, and other terrestrial microorganisms [56,58,69,72,73]. Moreover, the environment and the capacity to capture preys can be determined by the trap and prey size [74]. Thus, the trap size difference between the two species (U. gibba possess traps c. 0.7–1.5 mm and U. reniformis c. 1–2.5 mm long; [1]) can also be the result of selection pressure for the different adaptation to aquatic (U. gibba) and terrestrial and epiphytic life-forms. Therefore, both plants might present a distinct trap ecosystem and an enzymatic digestive repertoire to uptake nutrients, and our results supported this hypothesis at a genomic scale. However, this hypothesis warrants further functional studies for confirmation.
4. Materials and Methods
4.1. Plant Material, Genome Size Estimation, and Cytogenetic Analysis
Utricularia reniformis samples were collected in the fall of 2015 in the Serra do Mar Atlantic Forest, in the Municipality of Salesópolis, SP, Brazil, and were deposited in the Herbarium JABU at the São Paulo State University (voucher—V.F.O. de Miranda et al., 1725). The plants were grown in a jar culture before procedures. These samples were recorded in the Brazilian National System of Management of Genetic Heritage and Associated Traditional Knowledge (SisGen) under the access number #A68D114, in accordance with the Brazilian Access and Benefit Sharing (ABS) legal framework introduced by the Federal Law No. 13,123 of 2015. No permission for collecting was necessary, as the sample was not collected in protected areas and U. reniformis is not a threatened species, according to the global IUCN (The IUCN Red List of Threatened Species: http://www.iucnredlist.org) and the Brazilian List of Threatened Plant Species.
For the genome size estimation, approximately 25 mg of leaf tissue was macerated in 1 mL of cold Ebihara buffer [75] supplied with 0.025 µg mL−1 RNAse, using a scalpel blade to release the nuclei into suspension with the same mass of the internal reference standard Raphanus sativus var. Saxa (2C = 1.11 pg; [76]). The nuclei suspensions were stained by adding 25 µL of a 1 mg mL−1 solution of propidium iodide (PI, Sigma). The analysis was performed using the FACSCanto II cytometer (Becton Dickinson, San Jose, CA, USA). The histograms were obtained through the FACSDiva software based on 5000 events, and the statistical evaluation was performed using the Flowing Software 2.5.1 (http://www.flowingsoftware.com/). Three individuals were analyzed in triplicates. The quality control of samples was based on the coefficient of variation (CV) of each measurement (which should be below 5%) and the standard deviation (SD) among the 2C-values (which should be below 3%). Such limits ensure that the variations observed inside and among measurements are due to technical factors and do not represent intraspecific variation among individuals [77].
For the cytogenetic analysis, stolon tips were pre-treated in 8-hydroxyquinoline (0.002 M) for 24 h, at 10 °C, fixed in ethanol:acetic acid (3:1, v/v) for 24 h, at room temperature, and were stored at −20 °C. The fixed stolon tips were washed in distilled water and digested in a 2% (w/v) cellulase (Onozuka)/20% (v/v) pectinase (Sigma-Aldrich, St. Louis, MO, USA)/1% macerozyme (Sigma-Aldrich, St. Louis, MO, USA) solution, at 37 °C for 15 min. The meristems were squashed in a drop of 45% acetic acid, and the coverslip was later removed in liquid nitrogen. Metaphases were DAPI (1 µg·mL−1) stained and photographed with an XM10 camera coupled to a BX 53 Olympus epifluorescence microscope.
4.2. Genome and Transcriptome Sequencing
One individual was sampled, producing the three different DNA libraries used in this study. DNA was extracted using the QIAGEN DNeasy Plant Maxi Kit extraction protocol (Qiagen, Hilden, Germany) to prepare libraries for Illumina TruSeq PCR-free and Nextera XT paired-end reads, with ~350 bp (2 × 100 bp) and ~450 bp (2 × 300 bp), insert sizes, and Illumina Nextera mate-pair gel free with insert sizes ranging from 3.5 to 10 Kb (2 × 100 bp). Illumina libraries were sequenced on HiScanSQ and MiSeq machines. The paired reads (2 × 100 bp and 2 × 300 bp) adapters were removed with Trimmomatic v0.27 [78], while the mate-pair reads (2 × 100 bp) internal adapters were trimmed with the NxTrim [79]. Sequences with phred value below 24 (<Q24) and spanning less than 50 bp were removed. For transcriptome sequencing, we used samples from leaves, stolon, and utricles. The RNA extraction and sequencing was performed according with the U. reniformis previous research [26,27]. Trimmed RNAseq reads from the leaves, stolon, and utricles were concatenated, and de novo assembled using Trinity v2.7.0 [80], and were exclusively used for gene model predictions (see below).
4.3. Genome Assembly and WGD Analysis
The trimmed reads were filtered against the cpDNA (NC_029719) and mtDNA (NC_034982) organellar genomes [26,27] with bowtie2 v2.26 [81]. The assembly was carried out with MaSuRCA v3.1.3 [82], SSPACE [83], and GapCloser [84]. REAPR v1.0.18 [85] was used to generate the corrected assembly and estimate the percentage of error-free bases (additional details are provided in Supplementary Materials). CEGMA v2.5 [86] and BUSCO v3 [87] using Eudicotyledons_odb10 from the OrthoDB database [88] were used to measure the genome completeness. We used GenomeScope v1 [89], the K-mer Analysis Toolkit (KAT) [90], and KmerSpectrumPlot.pl from ALLPATHS-LG [91] to estimate genome size and heterozygosity.
The predicted WGDs events that occurred in the U. reniformis evolutionary history were predicted on the basis of the distribution of synonymous substitutions per synonymous site (Ks) of gene pairs, using the SynMap2 tool from the Comparative Genomics Platform (CoGe) [92]. MCScanX [93] was employed to identify dispersed, proximal, and tandem (small-scale duplication) and segmental duplicates (WGDs). Tandem duplicates are defined as paralogs that are adjacent to each other, and the maximum distance to call a proximal copy was set to 10 genes. The anchor genes in collinear blocks inferred segmental duplicates. Structural and comparative analyses (pairwise alignment) were carried out with D-GENIES [94]. Macrosynteny and microsynteny analyses were carried out with VGSC2 and MCscan (Python version) (https://github.com/tanghaibao/jcvi.wiki.git).
4.4. Transposable Elements Identification, Classification, and Genome Annotation
The de novo detection and classification of TEs were carried out by the REPET v2.5 [95] with the “—struct” parameter. The identified elements from the REPET package were manually validated and characterized into super-families and evolutionary lineages with the usage of the Domain-based ANnotation of Transposable Elements (DANTE) with the Viridiplantae database v3.0 [96]. The TE masking was performed with the RepeatMasker Open-v4.0.7 (http://www.repeatmasker.org) using the parameter: “-s -cutoff 260”.
The gene prediction was performed using the BRAKER v2 pipeline [97], and the TE-related genes were discarded. PASA pipeline [98] was used to produce spliced alignment assemblies based on RNA-seq data. The gene predictions and transcript alignments were combined with the EVidence Modeler software [99]. Finally, two PASA pipeline iterations were used to update the EVidence Modeler consensus predictions, adding UTR annotations and models for the alternatively spliced isoforms. Functional annotation was performed using Blast2GO v5 [100], InterProScan 5 [101], and EggNOG-mapper v1.0.3 with the EggNOG 5.0 database [102]. For the protein assignment, the UniProtKB/TrEMBL [103] viridiplantae database was used. The final predicted gene models were scanned against the DANTE and RepeatMasker predictions results, and all TE-related genes were further discarded.
For consistency in the comparative analysis, Utricularia gibba transcriptomes read from NCBI Sequence Read Archive (SRA): SRX2368915, SRX247091, and SRX038704 were assembled, and the U. gibba genome (Genbank Accession Number: NEEC01000001-NEEC01000518) was re-annotated using the same pipeline.
4.5. Phylogenetic Analysis
The phylogenetic analyses were performed using maximum likelihood (ML), and Bayesian inference (BI) approaches. For the ML, RAxML v. 8.2.10 [104] was used with default parameters, and the clade support estimates were calculated using rapid bootstrapping of 1000 pseudoreplicates. The BI was performed with Mr. Bayes v.3.2.6 [105] using MCMCMC, with random starting trees, of about 5,000,000 generations, with sampling for each 100 trees, using two runs and four chains and the trees were considered only after a stationary position was reached, with 25% of initially elaborated trees discarded. The GTR + G + I was used as the best-of-fit model according to the AIC (Akaike Information Criterion) with the jModeltest v. 2.1.10 [106].
4.6. Comparative Analysis
OrthoVenn v2 [107] was used to determine the orthologous gene families among Utricularia reniformis, U. gibba, and other angiosperms (Arabidopsis thaliana, Vitis vinifera, and Solanum lycopersicum) retrieved from the Phytozome database [108]. Functional enrichment analyses of the GO terms and the KEGG enzymes were conducted using Fisher’s exact test and was corrected for multiple testing using two methods employed by the Blast2GO (GO terms and KEGG enzymes) and GOATOOLS (GO terms) [109]. The p-values were determined after the correction for multiple tests through False Discovery Rate (FDR) control, according to the Benjamini–Hochberg. The WOX, HD-Zip, MADS-Box Transcription Factors (TFs), and ABC transporters comparative analyses were based on previous [16,33] and manual annotation using the A. thaliana reviewed entries from UniProt Database as reference [110].
4.7. Availability of Supporting Data
The raw sequencing reads have been deposited into GenBank, BioProject PRJNA290588, linked directly to the SRA under the accession numbers SRR8289569, SRR8289570, and SRR8289571 for the Illumina datasets and SRR8289572 for Ion Torrent’s RNA-Seq data. This Whole Genome Shotgun project has been stored at DDBJ/ENA/GenBank under the accession RWGZ00000000. The version described in this paper is version RWGZ01000000. The gene models and genome browser of Utricularia reniformis are available at https://genomevolution.org/coge/GenomeInfo.pl?gid=54799. The Gene Ontology annotation (Blast2GO) and all the raw data generated for this study are freely available at the Zenodo platform at http://doi.org/10.5281/zenodo.3268745.
5. Conclusions
In this work, we generated a draft genome sequence and annotation for the terrestrial carnivorous plant U. reniformis and compared it against its sister, aquatic species U. gibba. Our results demonstrated that a massive proliferation and loss of lineage-specific LTR-retrotransposons, predominantly from the Gypsy super-family and WGDs are the main governing agents of the genome size changes and evolution between these species. Interestingly, our results strongly suggest that both genomes responded distinctly after the WGD events. This is mainly observed by specific patterns of gene fractionation and retention after the lineage split, which might be reflected in an ongoing gene birth–death–innovation process among the duplicated genes. Additionally, each species carries a diversified set of plant ontogeny and carnivory-associated gene repertoire, supporting that the reproductive isolation and terrestrial and aquatic life-form constraints (e.g., different nutrients repertoire and availability, prey diversity, and trap microbiome ecosystem) in an evolutionary time scale are associated with speciation.
Acknowledgments
We thank the Microbiology and Immunology Department of IBB-UNESP/Botucatu, Brazil that kindly made available the usage of the equipment FACSCanto II cytometer for the Genome size estimation through flow cytometry. We thank the Laboratory of Plant Systematics for all support, specially Yani Aranguren Díaz for help with the DNA extractions.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/1/3/s1.
Author Contributions
Conceptualization, S.R.S., A.M.V., and V.F.O.M.; methodology, A.M.V., T.P.M., V.F.O.M., S.R.S., and A.P.M.; software, A.M.V., T.P.M., S.R.S., and V.F.O.M.; validation, A.M.V., T.P.M., S.R.S., and V.F.O.M.; formal analysis, A.M.V., S.R.S., A.P.M., H.A.P., M.H.M.J., D.S.D., T.P.M., and V.F.O.M.; investigation, S.R.S., T.P.M., V.F.O.M., and A.M.V.; resources, A.P.M., V.F.O.M., and A.M.V.; data curation, A.M.V., T.P.M., and V.F.O.M.; writing—original draft preparation, A.M.V., S.R.S., and V.F.O.M.; writing—review and editing, A.M.V., S.R.S., V.F.O.M., H.A.P., A.P.M., M.H.M.J., D.S.D., and T.P.M.; visualization, S.R.S., T.P.M., V.F.O.M., and A.M.V.; supervision, A.M.V., T.P.M., and V.F.O.M.; project administration, V.F.O.M. and A.M.V.; funding acquisition, V.F.O.M. and A.M.V. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the São Paulo Research Foundation/Brazil, FAPESP [13/25164-6] to A.M.V. V.F.O.M. was funded by FAPESP [13/05144-0]. A.P.M. was funded by National Council for the Scientific and Technological Development—CNPq [460990/2014-4] and FAPESP Young Investigator Project [11/22215-3]. S.R.S. was funded by a post-doctoral research fellowship from FAPESP [18/02285-6], and H.A.P. was funded by the PNPD fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Taylor P. The Genus Utricularia-A Taxonomic Monograph. The Royal Botanic Gardens, Kew; London, UK: 1989. [Google Scholar]
- 2.Leushkin E.V., Sutormin R.A., Nabieva E.R., Penin A.A., Kondrashov A.S., Logacheva M.D. The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genom. 2013;14:476. doi: 10.1186/1471-2164-14-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vu G.T.H., Schmutzer T., Bull F., Cao H.X., Fuchs J., Tran T.D., Jovtchev G., Pistrick K., Stein N., Pecinka A., et al. Comparative genome analysis reveals divergent genome size evolution in a carnivorous plant genus. Plant Genome. 2015;8:1–14. doi: 10.3835/plantgenome2015.04.0021. [DOI] [PubMed] [Google Scholar]
- 4.Ibarra-Laclette E., Lyons E., Hernández-Guzmán G., Pérez-Torres C.A., Carretero-Paulet L., Chang T.H., Lan T., Welch A.J., Juárez M.J.A., Simpson J., et al. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan T., Renner T., Ibarra-Laclette E., Farr K.M., Chang T.H., Cervantes-Pérez S.A., Zheng C., Sankoff D., Tang H., Purbojati R.W., et al. Long-read sequencing uncovers the adaptive topography of a carnivorous plant genome. Proc. Natl. Acad. Sci. USA. 2017;114:E4435–E4441. doi: 10.1073/pnas.1702072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanson L., Mc Mahon K.A., Johnson M.A.T., Bennett M.D. First nuclear DNA C-values for another 25 angiosperm families. Ann. Bot. 2001;87:251–258. doi: 10.1006/anbo.2000.1325. [DOI] [PubMed] [Google Scholar]
- 7.Greilhuber J., Borsch T., Müller K., Worberg A., Porembski S., Barthlott W. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biol. 2006;8:770–777. doi: 10.1055/s-2006-924101. [DOI] [PubMed] [Google Scholar]
- 8.Veleba A., Bureš P., Adamec L., Šmarda P., Lipnerová I., Horová L. Genome size and genomic GC content evolution in the miniature genome-sized family Lentibulariaceae. New Phytol. 2014;203:22–28. doi: 10.1111/nph.12790. [DOI] [PubMed] [Google Scholar]
- 9.Kondo K. A Comparison of Variability in Utricularia cornuta and Utricularia juncea. Am. J. Bot. 1972;59:23. doi: 10.1002/j.1537-2197.1972.tb10059.x. [DOI] [Google Scholar]
- 10.Rahman D.M.O., Adamec L., Kondo K. Chromosome numbers of Utricularia bremii and Utricularia dimorphantha (Lentibulariaceae) Chromosom. Sci. 2001;5:105–108. [Google Scholar]
- 11.Casper S.J., Stimper R. Chromosome numbers in Pinguicula (Lentibulariaceae): Survey, atlas, and taxonomic conclusions. Plant Syst. Evol. 2009;277:21–60. doi: 10.1007/s00606-008-0097-9. [DOI] [Google Scholar]
- 12.Fleischmann A., Michael T.P., Rivadavia F., Sousa A., Wang W., Temsch E.M., Greilhuber J., Müller K.F., Heubl G. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Ann. Bot. 2014;114:1651–1663. doi: 10.1093/aob/mcu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibarra-Laclette E., Albert V.A., Herrera-Estrella A., Herrera-Estrella L. Is GC bias in the nuclear genome of the carnivorous plant Utricularia driven by ros-based mutation and biased gene conversion? Plant Signal. Behav. 2011;6:1631–1634. doi: 10.4161/psb.6.11.17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibarra-laclette E., Albert V.A., Pérez-torres C.A., Zamudio-Hernández F., de Ortega-estrada M.J., Herrera-Estrella A., Herrera-estrella L. Transcriptomics and molecular evolutionary rate analysis of the bladderwort (Utricularia), a carnivorous plant with a minimal genome. BMC Plant Biol. 2011;11:101. doi: 10.1186/1471-2229-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albert V.A., Jobson R.W., Michael T.P., Taylor D.J. The carnivorous bladderwort (Utricularia, Lentibulariaceae): A system inflates. J. Exp. Bot. 2010;61:5–9. doi: 10.1093/jxb/erp349. [DOI] [PubMed] [Google Scholar]
- 16.Carretero-Paulet L., Chang T.H., Librado P., Ibarra-Laclette E., Herrera-Estrella L., Rozas J., Albert V.A. Genome-wide analysis of adaptive molecular evolution in the carnivorous plant Utricularia gibba. Genome Biol. Evol. 2015;7:444–456. doi: 10.1093/gbe/evu288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grover C.E., Wendel J.F. Recent Insights into Mechanisms of Genome Size Change in Plants. J. Bot. 2010;2010:1–8. doi: 10.1155/2010/382732. [DOI] [Google Scholar]
- 18.Hakes L., Pinney J.W., Lovell S.C., Oliver S.G., Robertson D.L. All duplicates are not equal: The difference between small-scale and genome duplication. Genome Biol. 2007;8:R209. doi: 10.1186/gb-2007-8-10-r209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellicer J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes. 2018;9:88. doi: 10.3390/genes9020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schatz M.C., Witkowski J., McCombie W.R. Current challenges in de novo plant genome sequencing and assembly. Genome Biol. 2012;13:243. doi: 10.1186/gb-2012-13-4-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriyama Y., Koshiba-Takeuchi K. Significance of whole-genome duplications on the emergence of evolutionary novelties. Brief. Funct. Genom. 2018;17:329–338. doi: 10.1093/bfgp/ely007. [DOI] [PubMed] [Google Scholar]
- 22.Bennetzen J.L. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 2000;42:251–269. doi: 10.1023/A:1006344508454. [DOI] [PubMed] [Google Scholar]
- 23.Magallón S., Sánchez-Reyes L.L., Gómez-Acevedo S.L. Thirty clues to the exceptional diversification of flowering plants. Ann. Bot. 2018;123:491–503. doi: 10.1093/aob/mcy182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.BFG-The Brazil Flora Group Growing knowledge: An overview of Seed Plant diversity in Brazil. Rodriguésia. 2015;66:1085–1113. doi: 10.1590/2175-7860201566411. [DOI] [Google Scholar]
- 25.Clivati D., Gitzendanner M.A., Hilsdorf A.W.S., Araújo W.L., de Miranda V.F.O. Microsatellite markers developed for Utricularia reniformis (Lentibulariaceae) Am. J. Bot. 2012;99:375–378. doi: 10.3732/ajb.1200080. [DOI] [PubMed] [Google Scholar]
- 26.Silva S.R., Alvarenga D.O., Aranguren Y., Penha H.A., Fernandes C.C., Pinheiro D.G., Oliveira M.T., Michael T.P., Miranda V.F.O., Varani A.M. The mitochondrial genome of the terrestrial carnivorous plant Utricularia reniformis (Lentibulariaceae): Structure, comparative analysis and evolutionary landmarks. PLoS ONE. 2017;12:e0180484. doi: 10.1371/journal.pone.0180484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva S.R., Diaz Y.C.A., Penha H.A., Pinheiro D.G., Fernandes C.C., Miranda V.F.O., Michael T.P., Varani A.M. The chloroplast genome of Utricularia reniformis sheds light on the evolution of the ndh gene complex of terrestrial carnivorous plants from the Lentibulariaceae family. PLoS ONE. 2016;11:e0165176. doi: 10.1371/journal.pone.0165176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng F., Wu J., Cai X., Liang J., Freeling M., Wang X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants. 2018;4:258. doi: 10.1038/s41477-018-0136-7. [DOI] [PubMed] [Google Scholar]
- 29.Salojärvi J., Smolander O.-P., Nieminen K., Rajaraman S., Safronov O., Safdari P., Lamminmäki A., Immanen J., Lan T., Tanskanen J., et al. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 2017;49:904–912. doi: 10.1038/ng.3862. [DOI] [PubMed] [Google Scholar]
- 30.Rendón-Anaya M., Ibarra-Laclette E., Méndez-Bravo A., Lan T., Zheng C., Carretero-Paulet L., Perez-Torres C.A., Chacón-López A., Hernandez-Guzmán G., Chang T.-H., et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA. 2019;116:17081–17089. doi: 10.1073/pnas.1822129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler G.L., Carstens B.C. Evaluating the adaptive evolutionary convergence of carnivorous plant taxa through functional genomics. PeerJ. 2018;6:e4322. doi: 10.7717/peerj.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravee R., Mohd Salleh F.- ’Imadi; Goh, H.H. Discovery of digestive enzymes in carnivorous plants with focus on proteases. PeerJ. 2018;6:e26940. doi: 10.7717/peerj.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang J.-U., Song W.-Y., Hong D., Ko D., Yamaoka Y., Jang S., Yim S., Lee E., Khare D., Kim K., et al. Plant ABC Transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol. Plant. 2016;9:338–355. doi: 10.1016/j.molp.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Jiao Y., Leebens-Mack J.H., Ayyampalayam S., Bowers J.E., McKain M.R., McNeal J., Rolf M., Ruzicka D.R., Wafula E.K., Wickett N.J., et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldman M., Levy A.A. Allopolyploidy–A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005;109:250–258. doi: 10.1159/000082407. [DOI] [PubMed] [Google Scholar]
- 36.Alix K., Gérard P.R., Schwarzacher T., Heslop-Harrison J.S.P. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017;120:183–194. doi: 10.1093/aob/mcx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark J.W., Donoghue P.C.J. Whole-genome duplication and plant macroevolution. Trends Plant Sci. 2018;23:933–945. doi: 10.1016/j.tplants.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Bertioli D.J., Jenkins J., Clevenger J., Dudchenko O., Gao D., Seijo G., Leal-Bertioli S.C.M., Ren L., Farmer A.D., Pandey M.K., et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019;51:877–884. doi: 10.1038/s41588-019-0405-z. [DOI] [PubMed] [Google Scholar]
- 39.Panchy N., Lehti-Shiu M., Shiu S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedoroff N. Transposons and genome evolution in plants. Proc. Natl. Acad. Sci. USA. 2000;97:7002–7007. doi: 10.1073/pnas.97.13.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennetzen J.L., Wang H. The Contributions of Transposable Elements to the Structure, Function, and Evolution of Plant Genomes. Annu. Rev. Plant Biol. 2014;65:505–530. doi: 10.1146/annurev-arplant-050213-035811. [DOI] [PubMed] [Google Scholar]
- 42.Serrato-Capuchina A., Matute D.R. The role of transposable elements in speciation. Genes. 2018;9:254. doi: 10.3390/genes9050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubin M.J., Mittelsten Scheid O., Becker C. Transposons: A blessing curse. Curr. Opin. Plant Biol. 2018;42:23–29. doi: 10.1016/j.pbi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Kim K.M., Caetano-Anollés G. The proteomic complexity and rise of the primordial ancestor of diversified life. BMC Evolut. Biol. 2011;11:140. doi: 10.1186/1471-2148-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willing E.M., Rawat V., Mandáková T., Maumus F., James G.V., Nordström K.J.V., Becker C., Warthmann N., Chica C., Szarzynska B., et al. Genome expansion of Arabis alpi. Nat. Plants. 2015;1:1–7. doi: 10.1038/nplants.2014.23. [DOI] [PubMed] [Google Scholar]
- 46.Macas J., Neumann P. Ogre elements-A distinct group of plant Ty3/gypsy-like retrotransposons. Gene. 2007;390:108–116. doi: 10.1016/j.gene.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Neumann P., Navrátilová A., Koblí A., Kejnovský E., Eva H., Hobza R., Widmer A., Dole J. Plant centromeric retrotransposons: A structural and cytogenetic perspective. Mob. DNA. 2011:1–16. doi: 10.1186/1759-8753-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang N., Bao Z., Temnykh S., Cheng Z., Jiang J., Wing R.A., McCouch S.R., Wessier S.R. Dasheng: A recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics. 2002;161:1293–1305. doi: 10.1093/genetics/161.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kameyama Y., Toyama M., Ohara M. Hybrid origins and F1 dominance in the free-floating, sterile bladderwort, Utricularia australis F. australis (Lentibulariaceae) Am. J. Bot. 2005;92:469–476. doi: 10.3732/ajb.92.3.469. [DOI] [PubMed] [Google Scholar]
- 50.Astuti G., Peruzzi L. Are shoots of diagnostic value in Central European bladderworts (Utricularia L., Lentibulariaceae)? Plant Biosyst. 2018;152:1214–1226. doi: 10.1080/11263504.2018.1435573. [DOI] [Google Scholar]
- 51.Clivati D., Cordeiro G.D., Płachno B.J., de Miranda V.F.O. Reproductive biology and pollination of Utricularia reniformis A.St.-Hil. (Lentibulariaceae) Plant Biol. 2014;16:677–682. doi: 10.1111/plb.12091. [DOI] [PubMed] [Google Scholar]
- 52.Rutishauser R., Isler B. Developmental genetics and morphological evolution of flowering plants, especially bladderworts (Utricularia): Fuzzy Arberian Morphology complements Classical Morphology. Ann. Bot. 2001;88:1173–1202. doi: 10.1006/anbo.2001.1498. [DOI] [Google Scholar]
- 53.Fineran B.A. Glandular trichomes in Utricularia—A review of their structure and function. Isr. J. Bot. 1985;34:295–330. [Google Scholar]
- 54.Płachno B.J., Adamec L., Kamińska I. Relationship between trap anatomy and function in Australian carnivorous bladderworts (Utricularia) of the subgenus Polypompholyx. Aquat. Bot. 2015;120:290–296. doi: 10.1016/j.aquabot.2014.09.008. [DOI] [Google Scholar]
- 55.Płachno B.J., Stpiczyńska M., Krajewski Ł., Świątek P., Adamec L., Miranda V.F.O. Flower palate structure of the aquatic bladderworts Utricularia bremii Heer and U. minor L. from section Utricularia (Lentibulariaceae) Protoplasma. 2017;254:2007–2015. doi: 10.1007/s00709-017-1097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jobson R.W., Morris E.C. Feeding ecology of a carnivorous bladderwort (Utricularia uliginosa, Lentibulariaceae) Austral Ecol. 2001;26:680–691. doi: 10.1046/j.1442-9993.2001.01149.x. [DOI] [Google Scholar]
- 57.Reifenrath K., Theisen I., Schnitzler J., Porembski S., Barthlott W. Trap architecture in carnivorous Utricularia (Lentibulariaceae) Flora Morphol. Distrib. Funct. Ecol. Plants. 2006;201:597–605. doi: 10.1016/j.flora.2005.12.004. [DOI] [Google Scholar]
- 58.Sanabria-Aranda L., González-Bermúdez A., Torres N.N., Guisande C., Manjarrés-Hernández A., Valoyes-Valois V., Díaz-Olarte J., Andrade-Sossa C., Duque S.R. Predation by the tropical plant Utricularia foliosa. Freshw. Biol. 2006;51:1999–2008. doi: 10.1111/j.1365-2427.2006.01638.x. [DOI] [Google Scholar]
- 59.Rajasekar C., Rajendran A. Prey composition of Utricularia striatula Sm. (Lentibulariaceae): Lithophytic carnivore Southern Western Ghats, India. Int. J. Fish. Aquat. Stud. 2018;6:382–388. [Google Scholar]
- 60.Elhiti M., Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009;4:86. doi: 10.4161/psb.4.2.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lian G., Ding Z., Wang Q., Zhang D., Xu J. Origins and Evolution of WUSCHEL-Related Homeobox Protein Family in Plant Kingdom. Sci. World J. 2014;2014:1–12. doi: 10.1155/2014/534140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Messenguy F., Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/S0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 63.Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004;16:S1–S17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J., Ding C., Xu B., Chen C., Narsai R., Whelan J., Hu Z., Zhang M. A Casparian strip domain-like gene, CASPL, negatively alters growth and cold tolerance. Sci. Rep. 2015;5:14299. doi: 10.1038/srep14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho M., Cho H.-T. The function of ABCB transporters in auxin transport. Plant Signal. Behav. 2013;8:e22990. doi: 10.4161/psb.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teschner J., Lachmann N., Schulze J., Geisler M., Selbach K., Santamaria-Araujo J., Balk J., Mendel R.R., Bittner F. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell. 2010;22:468–480. doi: 10.1105/tpc.109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stein D.J., Chamberlain S.R., Fineberg N. An A-B-C Model of Habit Disorders: Hair-Pulling, Skin-Picking, and Other Stereotypic Conditions. CNS Spectr. 2006;11:824–827. doi: 10.1017/S1092852900014978. [DOI] [PubMed] [Google Scholar]
- 68.Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Juniper B.E., Robins R.J., Joel D.M. The Carnivorous Plants. Academic Press; London, UK: San Diego, CA, USA: 1989. [Google Scholar]
- 70.Brownlee C. Carnivorous plants: Trapping, digesting and absorbing all in one. Curr. Biol. 2013;23:R714–R716. doi: 10.1016/j.cub.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 71.Sirová D., Bárta J., Šimek K., Posch T., Pech J., Stone J., Borovec J., Adamec L., Vrba J. Hunters or farmers? Microbiome characteristics help elucidate the diet composition in an aquatic carnivorous plant. Microbiome. 2018;6:225. doi: 10.1186/s40168-018-0600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mette N., Wilbert N., Barthlott W. Food composition of Aquatic Bladderworts (Utricularia, Lentibulariaceae) in Various Habiats. Beiträge Biol. Pflanz. 2000;72:1–13. [Google Scholar]
- 73.Kurbatova S.A., Yershov I.Y. Crustaceans and Rotifers in the Predatory Feeding of Utricularia. Inland Water Biol. 2009;2:271–275. doi: 10.1134/S1995082909030122. [DOI] [Google Scholar]
- 74.Harms S. Prey selection in three species of the carnivorous aquatic plant Utricularia (bladderwort) Arch. Hydrobiol. 1999;146:449–470. doi: 10.1127/archiv-hydrobiol/146/1999/449. [DOI] [Google Scholar]
- 75.Ebihara A., Ishikawa H., Matsumoto S., Lin S.-J., Iwatsuki K., Takamiya M., Watano Y., Ito M. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am. J. Bot. 2005;92:1535–1547. doi: 10.3732/ajb.92.9.1535. [DOI] [PubMed] [Google Scholar]
- 76.Doležel J., Sgorbati S., Lucretti S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992;85:625–631. doi: 10.1111/j.1399-3054.1992.tb04764.x. [DOI] [Google Scholar]
- 77.Pellicer J., Leitch I.J. The application of flow cytometry for estimating genome size and ploidy level in plants. In: Bresse P., editor. Molecular Plant Taxonomy. Methods in Molecular Biology (Methods and Protocols) Humana Press; Totowa, NJ, USA: 2014. pp. 279–307. [DOI] [PubMed] [Google Scholar]
- 78.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Connell J., Schulz-Trieglaff O., Carlson E., Hims M.M., Gormley N.A., Cox A.J. NxTrim: Optimized trimming of Illumina mate pair reads. Bioinformatics. 2015;31:2035–2037. doi: 10.1093/bioinformatics/btv057. [DOI] [PubMed] [Google Scholar]
- 80.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zimin A.V., Marçais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boetzer M., Henkel C.V., Jansen H.J., Butler D., Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 84.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hunt M., Kikuchi T., Sanders M., Newbold C., Berriman M., Otto T.D. REAPR: A universal tool for genome assembly evaluation. Genome Biol. 2013;14:R47. doi: 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parra G., Bradnam K., Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 87.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 88.Kriventseva E.V., Kuznetsov D., Tegenfeldt F., Manni M., Dias R., Simão F.A., Zdobnov E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019;47:D807–D811. doi: 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vurture G.W., Sedlazeck F.J., Nattestad M., Underwood C.J., Fang H., Gurtowski J., Schatz M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mapleson D., Garcia Accinelli G., Kettleborough G., Wright J., Clavijo B.J. KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33:574–576. doi: 10.1093/bioinformatics/btw663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gnerre S., MacCallum I., Przybylski D., Ribeiro F.J., Burton J.N., Walker B.J., Sharpe T., Hall G., Shea T.P., Sykes S., et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haug-Baltzell A., Stephens S.A., Davey S., Scheidegger C.E., Lyons E. SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics. 2017;33:2197–2198. doi: 10.1093/bioinformatics/btx144. [DOI] [PubMed] [Google Scholar]
- 93.Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T., Jin H., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cabanettes F., Klopp C. D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ. 2018;6:e4958. doi: 10.7717/peerj.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flutre T., Duprat E., Feuillet C., Quesneville H. Considering Transposable Element Diversification in De Novo Annotation Approaches. PLoS ONE. 2011;6:e16526. doi: 10.1371/journal.pone.0016526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neumann P., Novák P., Ho N. Systematic survey of plant LTR- retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA. 2019;10:1–17. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoff K., Lomsadze A., Borodovsky M., Stanke M. Whole-Genome Annotation with BRAKER. In: Kollmar M., editor. Gene Prediction. Methods in Molecular Biology. Humana; New York, NY, USA: 2019. pp. 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K.S., Jr., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conesa A., Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., von Mering C., Bork P. Fast genome-wide functional annotation through orthology massignment by eggNOG-Mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pundir S., Magrane M., Martin M.J., O’Donovan C. Searching and navigating UniProt databases. Curr. Protoc. Bioinform. 2015;50:1–27. doi: 10.1002/0471250953.bi0127s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Darriba D., Taboada G.L., Doallo R., Posada D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu L., Dong Z., Fang L., Luo Y., Wei Z., Guo H., Zhang G., Gu Y.Q., Coleman-Derr D., Xia Q., et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klopfenstein D.V., Zhang L., Pedersen B.S., Ramírez F., Warwick Vesztrocy A., Naldi A., Mungall C.J., Yunes J.M., Botvinnik O., Weigel M., et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018;8:10872. doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Consortium T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.