Abstract
Claudins are cell–cell adhesion proteins, which are expressed in tight junctions (TJs), the most common apical cell-cell adhesion. Claudin proteins help to regulate defense and barrier functions, as well as differentiation and polarity in epithelial and endothelial cells. A series of studies have now reported dysregulation of claudin proteins in cancers. However, the precise mechanisms are still not well understood. Nonetheless, studies have clearly demonstrated a causal role of multiple claudins in the regulation of epithelial to mesenchymal transition (EMT), a key feature in the acquisition of a cancer stem cell phenotype in cancer cells. In addition, claudin proteins are known to modulate therapy resistance in cancer cells, a feature associated with cancer stem cells. In this review, we have focused primarily on highlighting the causal link between claudins, cancer stem cells, and therapy resistance. We have also contemplated the significance of claudins as novel targets in improving the efficacy of cancer therapy. Overall, this review provides a much-needed understanding of the emerging role of claudin proteins in cancer malignancy and therapeutic management.
Keywords: claudins, cancer, stem cell, chemoresistance
1. Introduction
1.1. Tight Junctions
Tight junctions (TJs) are the sites where tissues interface directly with the external environment or internal compartments that are contiguous with the external environment and are lined by mucosal surfaces, where epithelial cells act insulation for the internal organ. These structures not only provide a protective layer but also act as a selective barrier between the body and the gut lumen that restricts free exchange across the paracellular space [1,2]. There are three main transport mechanisms across the epithelial layers, which include the trans-cellular pathway (passive diffusion), carrier dependent pathway (carrier or receptors), and the paracellular pathway (passage through spaces between cells). Among these transport mechanisms, the apical junctional complex, a crucial factor for the paracellular pathway, is composed of three junctions from apical to basal are known as the tight junction (zonula occludens), adherens junction (zonula adherens), and desmosome (macula adherens) [3]. The TJs are intercellular junctions, which act as permeability barriers in epithelial cells [1]. The tight junction proteins are diverse and include occludins (the first one to be found), claudins, tricellulin, cingulin, and junctional adhesion molecules (JAM). These proteins interact within themselves and with the cellular cytoskeleton to form a complex architecture [4,5,6,7,8]. Among these TJ proteins, claudins are key proteins, acting as both pores and barriers, aiding the paracellular pathway between epithelial cells [9,10].
1.2. Claudins
The functionalities of claudins are as follows: (1) Fence function, responsible for maintaining polarity by differentiating apical and basolateral cell domains; (2) Signaling molecule, involved in cell growth, survival, proliferation, and differentiation; (3) Barrier function, this gate function separates compartments with fluids to avoid intermixing [11]. Claudins were identified as a major integral membrane protein by Tsukita and his colleagues in 1998, before which the only known tight junction protein was occludin [12,13]. Studies conducted to overexpress claudins in fibroblasts, which do not have tight junctions, were able to reconstitute tight junction-like networks of strands, which shows the importance of claudins in tight junction assembly [14]. Several claudin isoforms have been identified in mammals. These have high sequence homology in the first and fourth transmembrane domains and extracellular loops. Further, the homologous classic claudins include claudins 1–10, 14, 15, 17, and 19, and non-classical claudins comprised of claudins 11–13, 16, 18, and 20–27, which are less homologous [15].
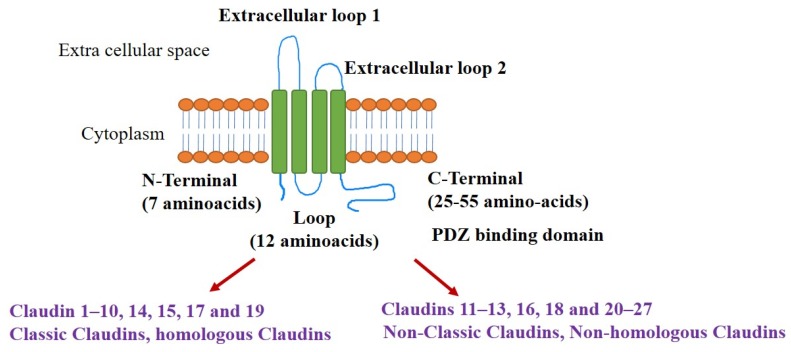
The structure of claudins is comprised of four transmembrane domains, the intracellular N and C termini, and the two extracellular loops (ECLs). The claudin structure encompasses N-termini (7 amino acids), C-termini (25–55 amino acids), and loops containing 25–55 aminoacids. The ECLs are involved in barrier and pore formations. There are two ECLs, ECL1 consists of ~50 amino acids with two conserved cysteines involved in the barrier function. Negative and positive charges in ECL1 contribute to pore formation. The schematic representation of the structure of claudins and its classification is depicted (Figure 1). The ECL2 is responsible for homo and heterotypic interactions and was recently shown to be involved in host cell binding and cytotoxicity for the Clostridium perfringens enterotoxin. The ECL2 usually has ~25 amino acids, but fewer in claudin-11 and more in claudin-18 [16]. Claudins interact with other TJ-associated proteins through carboxy-terminal tails, which contain a PDZ-domain binding motif [17].
Figure 1.
Structural organization of claudin proteins (monomer), and its classification based on homologous sequences between them. Colour code: Green- transmembrane domains; Orange: Bilipid layer, Blue–Extracellular loops/N and C termini.
2. Claudins as Oncogenic Signal Transducer
The expression of claudins varies among different tissue types [18]. As an important structure in regulating paracellular permeability, claudin overexpression influences trans-epithelial resistance (TER) and ion permeability [19,20,21,22]. Aberrant expressions of claudins have been reported in various cancers. Some of the claudins known to be frequently dysregulated in cancers are claudin-1, -3, -4, and -7 [23]. A large body of evidence highlights claudins as pro and anti-tumorigenic factors [24,25,26,27,28,29,30,31]. The potential of claudins to act as proto-oncogene or tumor promotor in various cancers are summarized in Table 1. In addition, several recent studies have also demonstrated the importance of claudins as tumor suppressors [24,25,26,27,28,29,30,31]. A recent study by Chang et al. in 2019 provided evidence for intestinal hyperplasia and adenomas in claudin-7 knockdown mice [32]. Consistent with this, claudin-7 was downregulated in colon cancer patient samples as compared to normal tissue [33]. These effects of claudin-7 were achieved by inhibiting phosphorylation and nuclear localization of Akt. Conversely, claudin-7 association with Epithelial cell adhesion molecule (EPCAM) supports proliferation, upregulation of anti-apoptotic proteins, and drug resistance [33]. Claudin-18 knockout mice spontaneously developed lung adenocarcinomas, and its mRNA expression was decreased in lung adenocarcinomas. Claudin-18 inhibits Akt signaling through modulation of yes-associated protein/Taz (Yap/Taz) and insulin-like growth factor (IGF-1R) signaling in lung cancer [34]. Further, the depletion of claudin-3 induced tumor burden by enhancing β-catenin activity through (IL)-6/STAT3 signaling in colon cancer [35]. Yet another study by Che et al. in 2018 [36] identified claudin-3 as a suppressor of lung squamous cell carcinoma cells, in which overexpression of claudin-3 inhibited invasion, migration, and EMT of lung squamous cell carcinoma. Similarly, claudin-4 accelerates cell migration and invasion in ovarian tumor cell lines, in support of this, peptide-mediated silencing of claudin-4 in ovarian cancer cells exhibited lower tumor burden [37]. Claudin-6 was shown to be a tumor suppressor through genetic manipulation studies in cervical carcinoma cells wherein loss of claudin-6 exacerbated cell proliferation and tumor growth [38,39]. An array of articles from Dhawan et al., have proved a significant role of claudin-1 as a tumor promoter in colon cancer [40,41]. In one of their reports, increased claudin-1 expression was causally associated with metastasis [40]. In contrast to claudin-1, claudin-7 has an inverse role on EMT, wherein it causes mesenchymal to epithelial transformation (MET) in Rab25 dependent manner to combat colon cancer [42]. Similarly, claudin-2 is upregulated in colon cancer and is involved in cancer progression. Claudin-2 suppression in colon cancer cells has led to decreased cell proliferation through the modulation of EGF signaling [43]. Opposite colon cancer, claudin-1 is frequently down-regulated in invasive human breast cancer. Recently, mutations of claudin-1 have been reported in breast cancer, which has led to claudin-1 transcript variants shorter than classical claudin-1 transcript [44]. Taken together, it appears that the deregulated claudin composition in any given epithelial cells sheet may modify the signaling and associated changes in protein partnering to modulate oncogenesis.
Table 1.
Claudins as tumour promotor/suppressor.
| Claudins Subtype | Cancer Type | Proto-Oncogene | Reference |
|---|---|---|---|
| Claudin-6 | Gastric cancer | Tumour promotor | [25] |
| Claudin-1 | Colon cancer | Tumour promotor | [40,45] |
| Claudin-3 | Ovarian cancer | Tumour promotor | [22] |
| Claudin-4 | Ovarian cancer | Tumour promotor | [22] |
| Claudin-6 | Breast cancer, Gastric cancer | Tumour promotor | [26,27] |
| Claudin-7 | Colon cancer | Tumour promotor | [46] |
| Claudin-2 | Lung cancer | Tumour promotor | [28] |
| Claudin-1 | Gastric cancer | Tumour suppressor | [47] |
| Claudin-1 | Lung cancer | Tumour suppressor | [29] |
| Claudin-3 | Ovarian cancer | Tumour suppressor | [31] |
| Claudin-4 | Ovarian cancer | Tumour suppressor | [31] |
| Claudin-7 | Lung cancer | Tumour suppressor | [30] |
| Claudin-11 | Gastric cancer | Tumour suppressor | [48] |
| Claudin-2 | Osteosarcoma | Tumour suppressor | [49] |
To glimpse how claudins can achieve its pro or anti-tumorigenic effect, understanding the regulation of claudins in normal and cancer cells is essential. Recently it has been demonstrated that claudins are not a static and rigid seal of the paracellular space; rather, they are dynamically capable of responding to various biochemical and mechanical stimuli through reshaping and remodeling [50,51]. Epigenetic regulation of claudins has recently gained significant importance. The claudin-3 promotor is known to possess low DNA methylation and high histone H3 acetylation for its expression in ovarian cancer cells [52]. DNA hypomethylation of the claudin-4 promotor is an important factor for its high expressions in gastric cancer [53]. Downregulation of claudin 1 via DNA promoter methylation is reported in estrogen receptor-positive breast cancer [54]. Claudins are also regulated at the transcriptional level by different transcription factors. A study has reported novel post-transcriptional regulation of claudin-1 in colon cancer cells [55], the authors documented the role of histone deacetylase (HDAC)-dependent histone acetylation as a key post-transcriptional regulation over claudin-1 expression, as found through HDAC inhibitor studies. Studies demonstrate the interaction of Slug and Snail (transcriptional factors) with the E-box element in the claudin-1 promoter causes inhibition of claudin-1. Snail is known to act as a transcription factor causing repression of E-cadherin (E-CAD) and has a potential role in promoting tumorigenesis. Slug is also a pivotal transcription factor involved in cell migration during embryogenesis and in tumor cell invasion and migration [56]. Yet another transcriptional factor known to be associated with claudin-1 is Runt-related transcription factor 3 (RUNX3), which is a gastric tumor suppressor [47]. Caudal homeobox proteins (Cdx1 & Cdx2) and GATA binding protein 4, GATA4) are known activators of claudin-1 promoters in colon cancer [57]. Sp1 is a transcriptional factor known to regulate claudin-3 and claudin-4 promoter activity in ovarian cancer [52,58]. Apart from these transcriptional regulations, claudins are also known to be regulated by post-translational modifications involved in their protein localization, interaction with other proteins, and overall turnover [59,60]. The post-translational modification of claudins includes palmitoylation, O-glycosylation, and phosphorylation [61,62]. Phosphorylation is one of the key regulatory modifications for the regulation of intracellular localization and degradation of claudins.
Claudins are phosphorylated by many different enzymes like protein-kinase A/C, protein phosphatase 2A and mitogen-activated protein kinase (MAPK) [63,64]. The localization or dissociation of claudins to TJs is regulated by phosphorylation. For phosphorylated claudin-1, -5, and -16 are localized in the TJs while in contrast, phosphorylated claudin-3 and -4 dissociate from TJs [64,65,66]. Furthermore, the rho family of small dimeric G proteins mediated phosphorylation of claudin-5 at T207 was recently reported [67]. The phosphorylation of claudin-1 at different serine sites (192, 205, 206, and T191) regulates its assembly at tight junctions [68]. The cAMP-dependent protein kinase (PKA) is known to phosphorylate of claudin-3 at amino acid 192 at the C terminus. Claudin-4 is phosphorylated by atypical PKC (aPKC) at serine195 [65]. Another important posttranslational modification playing a key role in claudin regulation is palmitoylation. Emerging articles have demonstrated the importance of palmitoylation in claudin localization into tight junctions. In claudin-5 self-assembly, palmitoylation restricts specific protein-protein conformations, as reported by Rajagopal et al. [61]. Claudin-7 interacts directly with EpCAM along the basal membrane. Palmitoylation regulates the ability of claudin-7 to interact with integrins, recruiting EpCAM, and concomitantly associate with the actin cytoskeleton [69].
3. Claudins and Stem Cells
Stem cells are crucial for the development and homeostasis of many different tissues. Stem cells are also involved in cell replacement therapies in the case of cell damage or degeneration [70]. Pluripotency of stem cells is defined as self-renewing and differentiating potential into all three germ layers. Human pluripotent stem cells are very promising in regenerative medicine. The stem cell further differentiates into a wide variety of cells under the influence of diverse signaling molecules, growth factors, and transcription factors [71]. Recent research is focused on understanding the signals, which maintain pluripotency or differentiation potential. Various intrinsic and extrinsic factors are involved in stem cell maintenance, self-renewal, and differentiation [71]. On the other hand, stem cells are also an important factor for many tumors. Dysregulated pluripotent stem cells in tumors are more aggressive and have the potential to reform the whole tumor [72]. Thus, it becomes important to selectively remove undifferentiated human pluripotent stem cells (hPSCs) from differentiated cultures. For achieving this, selective pluripotent-specific cell surface markers are needed, which can separate undifferentiating from the differentiated cells. While screening for a highly specific marker protein specific for the undifferentiated hPSCs, Uri Ben-David et al. [73] found claudin-6 to be highly specific for undifferentiated hPSCs. The expression of claudin-6 was 90-fold higher in undifferentiated hPSCs than in differentiated cells. The proof for the involvement of claudins in epithelial differentiation from embryonic stem cells was first reported by Sugimoto et al. [74], where the potential of claudin-6 to trigger epithelial morphogenesis in mouse stem cells was reported. Also, claudin-6 regulated other tight-junction and microvillus molecules claudin-7, occludin, Zonula occludens (ZO-1α+), and ezrin/radixin/moesin-binding phosphoprotein50, which strongly proved the role of claudins in epithelial differentiation [74]. This was further supported by other studies, which also showed the expression of claudin-6 is an early marker in embryonic stem cells [75,76]. Differentiation of Human Embryonic Stem Cells to Hepatocyte-Like Cells resulted in a decrease in stem cell markers Oct3/4 and Nanog as expected. Along with stem cell markers, claudin-1 declined eventually, whereas claudin-4 increased and was highest at the end stage of differentiation [77].
A growing body of evidence focuses on cancer stem cells in cancer biology. The drawbacks of cancer treatment failures and drug resistance are proved to originate from cancer stem cells, which are a small subpopulation in tumors. Recently the factors regulating cancer stem cells have gained significant importance and opened new avenues for targeted therapies and thus decrease the chance of recurrence of the disease [78]. Cancer stem cells (CSCs) represent a small group of cells in typically heterogeneous tumors, which possess tumor-initiating and self-renewal properties, giving rise to non-tumorigenic progeny. CSCs are enriched after chemotherapy and lead to therapy failure and thus recurrence of cancer. The role of CSCs in tumor relapse, metastasis, and therapeutic refractoriness is well described [79]. The role of claudins in cancer stem cell (CSC) biology is gaining much attention. The WNT pathway is well known to provide the key signals for achieving this particular phenotype. It is also established that the Wnt signal transduction pathway is important in normal and malignant stem cells [80]. Recent articles have highlighted the link between claudin and the Wnt/β-catenin signaling pathway and the role of CSC in this cross-talk. Claudin-1 and claudin-2 transcription is regulated by WNTt signaling, and they are known to regulate the β-Catenin- T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) signaling pathway to regulate CSC [81,82]. In contrast, other claudins negatively regulate WNTsignaling cascades, such as loss of claudin-3 inducing WNT/β-catenin activation, thus aiding in the promotion of colon cancer [35]. Darido et al. provided evidence for Tcf-4 and Sox-9 regulating the expression of claudin-7 [46]. In addition, studies by Prat et al. discovered a new claudin-low molecular subtype of breast cancer [83]. The key characteristics of this subtype are low expression of tight junction and junction adherens proteins (claudin-3, -4 and -7, and E-cadherin), and enriched in stem cell and EMT features. Patients having high-grade invasive ductal breast carcinoma in this subgroup had a poor prognosis, absence of luminal differentiation markers, enhanced EMT markers, expression of immune response genes, and most closely resembled mammary epithelial stem cells. This suggested that low claudin cells might emerge from more immature stem or progenitor cells and comprise cancer stem cells. Thus, identification of the low claudin subtype in breast cancer has shown the potential of claudins in regulating stem cells. In addition, claudin-3 is known to play an oncogenic role in non-small cell lung cancer (NSCLC). One of the major contributing factors for the role of claudin-3 is regulation of cancer stemness and chemoresistance in non-squamous NSCLC. The depletion of claudin-3 was able to combat the formation of spheres and tumor formation as well as increased sensitivity to cisplatin [84]. Further, claudin-3 inhibition by small-molecule inhibitors including withaferin A, estradiol and fulvestrant, suppressed cancer stemness and combated chemoresistance, giving strong evidence for the role of claudin-3 in inducing stemness. Another claudin playing an important role in stem cell regulation is claudin-18 in lung cancer [85], which has been reported to have a role in the aberrant proliferation of alveolar epithelial type II (AT2) cells, resulting in lung enlargement and parenchymal expansion by restrictions on stem/progenitor cell proliferation. Recently, claudin-2 was shown to be restricted in the stem/progenitor cell compartment of intestinal crypts. It enriches aldehyde dehydrogenase ALDHHigh cancer stem-like cells in heterogeneous colorectal cancer cell populations through the regulation of Yes-associated protein (YAP) activity and miR-222-3p expression [86]. Overall, these studies give an overview of the potential role of claudins in stem cell biology. The role of claudins in the regulation of stem cells is summarized in Table 2. The claudin mediated enrichment of stem cells provides a new axis-of-evil for a preferential therapeutic target, which has potential clinical consequences.
Table 2.
Claudins and stemness.
| Claudin Subtype | Stem Cell Related Functions | References |
|---|---|---|
| Claudin-6 | Early marker in embryonic stem cell.High expression in undifferentiated human pluripotent stem cells (hPSCs). Trigger epithelial morphogenesis in mouse stem cells. |
[73,74] |
| Claudin-1 and 2 | Known to regulate the β-Catenin-TCF/LEF signaling pathway to regulate CSC. | [81] |
| Claudin low subtype in breast cancer | Enriched in stem cells and more EMT. | [83] |
| Claudin-3 | Regulation on cancer stemness and chemoresistance in non-small cell lung cancer (NSCLC). | [84] |
| Claudin-18 | Triggers lung enlargement and parenchymal expansion by restrictions on stem/progenitor cell proliferation. | [85] |
| Claudin-2 | Enrich ALDHHigh cancer stem-like cells in heterogeneous colorectal cancer cell populations. | [86] |
4. Claudins in Chemoresistance
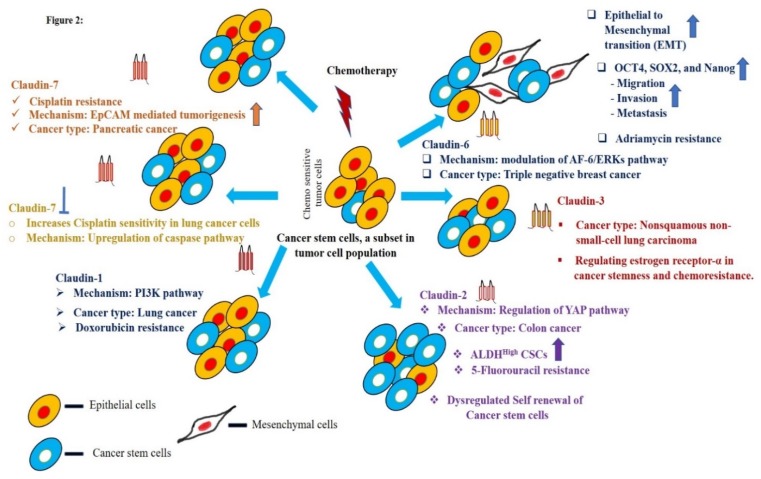
Most cancer patients initially respond to chemotherapy. Eventually, cancer relapses due to chemoresistance resulting in treatment failure causing death. The mechanisms of chemoresistance in cancers are still largely unknown [87]. Since the role of claudins in the regulation of cancer stem cells is well documented, their correlation with drug resistance and distant metastasis is inevitable and obvious [49,88,89]. In brief, claudin-3 and -6 are correlated with lymph node metastasis in squamous cell lung carcinomas [90,91]. Claudin-4 is highly expressed in primary and metastatic prostate cancer [92] and gastric cancer [93,94]. Claudin-1 and -7 have proved to have an inverse role in colon cancer, wherein claudin-1 elevates the metastasis of colon cancer cells. On the other hand, suppression of claudin-7 leads to liver metastasis [40,42]. Epithelial to mesenchymal transition (EMT) is a piece of vital machinery responsible for invasiveness and initiation of metastasis and chemoresistance of cancer cells. Claudins are known inducers of EMT in cancers. Claudin-1 is known to induce EMT in colon, liver, nasopharyngeal carcinoma, and breast cancers [40,95,96]. At the same time, claudin-7 is reported to be involved in establishing MET in colon cancer [36,42]. Claudin-3 suppresses EMT in lung cancer cells [36]. Overall, the potential role of claudins in EMT, Metastasis and CSC enrichment provides the rationale for exploring them as a key factor in establishing drug resistance. Claudins as chemo-resistance modulators is an emerging field of research. In a recent article, the potential of claudin-6 in enhancing chemoresistance to Adriamycin in triple-negative breast cancer (TNBC) was documented [97]. This effect of claudin-6 was mediated through its regulation over the AF-6/extracellular signal–regulated kinases (AF-6/ERK signaling pathway and up-regulation of cancer stem cells. Claudin-3 is also identified as a molecule to combat cisplatin chemoresistance in non-squamous lung carcinoma [84]. Here, claudin-3 overexpressing lung cancer cells were insensitive to cisplatin treatment compared to control cells. Adding to this, knockdown of claudin-3 or claudin-4 in ovarian cancer cells induced resistance to cisplatin by the regulation of Cu transporter CTR1 [98]. Another study by a Japanese group of researchers reported a high expression of claudin-4 in the ovarian cancer tissues of platinum-resistant patients [99]. In lung cancer, claudin-1 is a key deciding factor for metastasis and a responsible factor for drug resistance towards cisplatin through the up-regulation of Unc-51 Like Autophagy Activating Kinase 1 (ULK1) phosphorylation [100,101]. It is also known to enhance drug resistance in liver cancer cells by modulating autophagy to achieve drug resistance. The role of claudin-7 in drug resistance [102] has also been reported, wherein decreased drug resistance, increased apoptosis and diminished anti-apoptotic PI3K/Akt pathway was achieved by knocking down claudin-7, proving the potentiality in chemo-resistance [103]. It is well known that EpCAM associates with claudin-7 and is known to be involved in cancer metastasis. Florian et al. [69] have provided evidence for the increased migratory potential of pancreatic cancer cells upon EPCAM and claudin-7 association influencing cell-cell adhesion. Interestingly, the EPCAM and claudin-7 association seems to enhance drug resistance against cisplatin through enhancing MAPK and c-Jun N-terminal kinases (JNK) pathways. Altogether, these studies indicate the important role of claudins contributing to drug resistance in cancer cells. The pictorial representation of the role of claudins as a stem cell regulator and its impact in chemoresistance is shown in Figure 2.
Figure 2.
The central role of claudins in the regulation of epithelial to mesenchymal transition (EMT), cancer stem cells, and chemoresistance in various cancers.  - inhibition of claudin-7. The arrows indicate the upregulation and higher enrichment of the mentioned signaling molecules, colour is respective of each claudin.
- inhibition of claudin-7. The arrows indicate the upregulation and higher enrichment of the mentioned signaling molecules, colour is respective of each claudin.
5. Claudins in Prognosis
Emerging data defining mechanisms through which claudins augment cancer metastasis provides the rationale for exploring claudins as prognostic factors and therapeutic targets in cancer. The importance of claudins is established using cancer cell models, mouse models, and human patient samples. Target molecules for cancer surveillance in high-risk populations are desperately warranted. As a vital emerging modulator in molecular or cellular pathways related to cancers, claudins could be targeted or used as biomarkers for prognosis, diagnosis, and treatments. A number of recent studies have projected a role for claudins as key prognosis factors in cancers. In one of the study Lechpammer et al. [104] demonstrated the potential of claudins as a diagnostic and prognostic factor in renal cell carcinoma. Claudin-1, -3, -4, -7, and -8 were studied in human renal cell carcinomas and oncocytomas. The data from their research showed an inverse correlation between claudin-3 and -4 expression with overall survival in clear cell renal cell carcinomas, and these claudins could be considered for prognosis in renal cell carcinomas. Claudin-7 and 8 can be implied as useful markers in the identification of renal cell carcinomas from oncocytomas [105].
Claudin-6 was reported as a prognosis factor in NSCLC patients. In this report, the patients with low claudin-6 had a lower survival rate than the patients with high claudin-6. [91] reported low claudin-6 as an independent indicator of prognosis in NSCLC patients. In this study, they documented low claudin-6 in 61 of 123 NSCLC tissue samples, and patients with low claudin-6 expression correlated with lower survival rates than those with high claudin-6 expression. The influence of claudin-3, claudin-7, and claudin-18 in gastric cancer patients were also studied [106]. Claudin-3 and claudin-7 were expressed in 25.4% and 29.9% of the gastric cancer tissues, respectively. However, 51.5% of gastric cancer tissues exhibited reduced expression of claudin-18. Claudin-7 expression correlated with shorter overall survival in gastric cancer patients, while the overall survival was increased in patients with claudin-18 expression. Recently, claudin-3 and -7 are also considered as novel prognostic factors in triple-negative breast cancer (TNBC) through its aberrant immunohistochemical expressions [107]. Claudin-3 cytoplasmic expression is an indicator of poor survival in triple-negative breast cancer. In addition, epigenetic modifications of claudins are reported to be a promising prognosis marker of various cancers. Zhenzhen et al. [106] recently demonstrated that the methylation of claudin-3 is a prognostic factor in gastric adenocarcinoma.
Further, the serum levels of claudin-7 among patients with colorectal cancer (CRC) was significantly reduced and correlated with high tumor stage and high carcinoembryonic antigen levels [108]. Claudin-7 was found to be downregulated in CRC, as reported by Bhat et al. [42], and associated with diminished EMT and tumor progression. These studies give a strong rationale to consider claudin-7 as a biomarker for predicting the development, proliferation, and prognosis of CRC. A claudin-low molecular subtype of breast cancer has been described with a concomitant upregulation of several EMT markers and an enrichment in stem cell features [109]. In an interesting article by Danzinger et al., the importance of claudin-3 in triple-negative breast cancer (TNBC) was documented. It was reported that claudin-3 expression was correlated with a Breast cancer type 1 (BRCA1) mutation [107]. This could help in guiding the decision for BRCA testing for triple-negative breast cancer (TNBC). Also, the expression of claudin-11 has been suggested as a biomarker for advanced-stage cutaneous squamous carcinoma, and reflects the distinct stages of tumor development and differentiation [110]. The clinical significance of claudin-11 was addressed in Laryngeal Squamous Cell Carcinoma (LSCC) by Nissinen et al. [110]. In this study, elevated promoter methylation of claudin-11 in tumor tissues was observed. Patients with lymph node metastasis with an advanced clinical stage showed more methylation in the claudin-11 promoter, which associated with poor overall survival of LSCC patients. In TNBCs, claudin-1, -3, -4, and -7 higher expression rates are more frequent than in other subtypes [111]. Claudin-4 high/claudin-1 low, claudin-4 high/claudin-7 low, and claudin-4 high/claudin-1 low/claudin-7 low types were also significantly correlated with lymph node metastasis, and showed worse survival. Apart from this, a recent article from Upadhaya et al. documented the therapeutic potential of claudin-1 in oral epithelial dysplasia and oral squamous cell carcinoma [112]. Overall the differential expression pattern of claudins may reflect the distinct stages of tumor development and differentiation and have been implied as prognostic factors for early determination of the tumor state.
6. Claudins as Therapeutic Agents
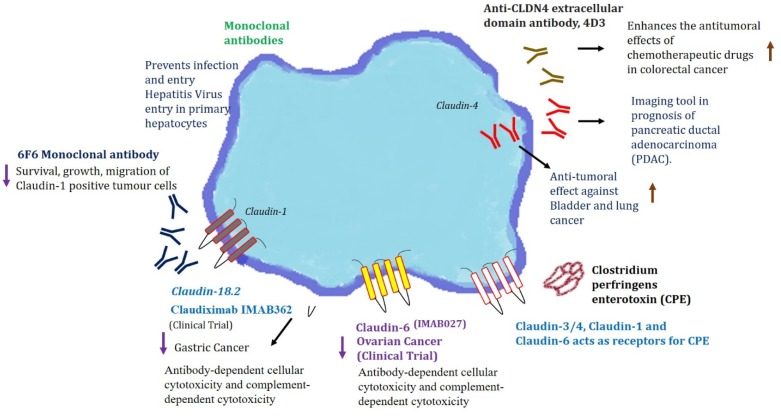
So far, over 100 monoclonal antibody (mAb) products are in clinical trials [113]. In an oncology setting, these monoclonal antibodies can mediate antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) against cancer cells [114]. There is a long-lasting history of antibody-mediated targeting of claudin-1 against hepatitis C virus (HCV) infections, and wherein many researchers have provided proof for the importance of claudins in HCV infections as viral entry point [115,116]. A study by Fofana et al. [117] designed monoclonal antibodies against claudin-1 to combat HCV entry. It was promising to see the antibodies raised against claudin-1 was able to block HCV entry. A recent study by Colpitts et al. has documented the humanization of a claudin-1-specific monoclonal antibody and was investigated in a large panel of primary human hepatocytes, and was found to be very promising for clinical HCV prevention and cure [118]. These studies hold significance because these antibodies could prevent HCV infection after liver transplantation, and virus spread in chronically infected patients. These antibodies are now being tested in cancer models. Claudins, as a potential target in antibody-based therapies for carcinomas, was investigated by Offner et al. [119]. In this study, the antibodies were raised against the extracellular domains of claudin-1, -3, and -4. Recently Romani et al. engineered a fully human anti-claudin-3 IgG1 antibody (IgGH6) [120], which is specific to claudin-3 and no cross-reactivity with other claudins was observed. Recent work by Cherradi et al. [121] investigated the importance of claudin-1 in different colorectal cancer (CRC) molecular subtypes. There is a differential expression pattern of claudin-1 based on the subtype. A murine monoclonal antibody against the extracellular part of human claudin-1 (6 F6 mAb) was designed and generated, which was specifically able to pick claudin-1 positive CRC cell lines, and no other cross-reactivity was observed. Furthermore, 6 F6 mAb was able to combat colony formation, xenograft growth and metastasis of claudin-1 positive CRC cells suggesting its utility as a therapeutic. Fujiwara et al. recently targeted claudin-4 in CRC using an anti-claudin-4 extracellular domain antibody [122]. The efficacy of the anti-claudin-4 antibody is promising and observed to enhance the anti-tumorigenic potential of 5-fluorouracil (FU) and anti-EGFR antibodies. These works demonstrate the proof of concept for exploiting claudins as targets for monoclonal antibodies in therapies.
Some of the monoclonal antibodies against claudins, including anti-claudin-18.2 (IMAB362-claudin-18.2) and the anti-claudin-6 (IMAB027-claudin-6), have also found their way into clinical trials [123]. Claudin-18.2 is expressed on the outer cell membrane of gastric cancer cells and binds to monoclonal antibodies. The IMAB362 was proven to be clinically safe as the patients were devoid of any side effects. Also, IMAB027 is in an ongoing clinical trial for recurrent advanced ovarian cancer (NCT02054351), and patients have not demonstrated any adverse effects [123]. Clinical trials for claudiximab (claudin-18 targeting) in advanced gastroesophageal cancer patients are also underway [124]. Recently, claudiximab was reported to be a first-in-class chimeric monoclonal antibody for the treatment of gastric cancer targeting claudin-18, which is an important factor in gastric cancer metastasis. This is just the beginning of an exciting journey and more research is warranted to revolutionize claudins targeted monoclonal antibodies in cancer therapy.
Another avenue to exploit Claudins as a therapeutics is their ability to behave as receptors for microbes. Clostridium perfringens enterotoxin (CPE) has the potential to bind with claudin receptors. CPE binds to the C-terminal CPE domain at both the first and second extracellular loops (ECL-1 and ECL-2) of claudins [125]. The affinity of CPE to claudins causes a pore leading to calcium influx responsible for host cell death. The claudin–CPE interaction is gaining significance in receptor decoy therapeutics for potential applications in gastrointestinal disease, cancer therapy/diagnoses, and drug delivery [125]. Claudin-3 and claudin-4 have been widely demonstrated to function as CPE receptors [126,127]. The binding ability of CPE to claudins, especially claudin-3 and claudin-4, has raised a great opportunity to target cancers with dysregulated claudin-3 and -4 cancers, especially breast, ovarian, and pancreatic cancers. The binding of CPE to claudin-3 and -4 was documented to induce dose-dependent cytolysis in breast cancer cells expressing claudin-3 and -4 [128]. Recent studies have exploited the CPE mediated targeting of claudin-3 and 4 cancers to target therapy-resistant ovarian cancer, pancreatic, and breast cancer xenografts possessing increased expressions of claudin-3 and -4. In one of the studies, the possibility of CPE binding claudin-3 as a visualization tool for identifying of micrometastatic chemotherapy-resistant ovarian cancer has been demonstrated [129]. The applicability of CPE, claudin-3, and -4 interactions is exploited in gene therapy against colon cancer. Recombinant (recCPE) and optimized CPE expressing vector (optCPE) were demonstrated to have a cytotoxic effect in claudin overexpressing colon cancer cells [130,131]. Further, the recent identification of the crystal structure of claudin-9 revealed that human claudin-9 has high-affinity for the CPE receptor and treatment with CPE caused cell death in human claudin-9 expressing cells [132]. In continuation of these studies, an interesting approach of nanoparticle-based targeting of cancer cells was documented by researchers, wherein the C-terminus of the CPE was conjugated to gold nanoparticles (AuNPs). This combination binds to claudin expressing tumor cells and kills the cells using gold nanoparticle-mediated laser perforation (GNOME-LP) technique [133,134]. Thus, the clinical relevance and functional importance of claudins in diverse cancers make them potential therapeutic targets.
7. Claudins as a Visualization Tool
The use of monoclonal antibodies against claudins have also been utilized in imaging modalities. Recently, claudin-4 was studied as an imaging tool for x-ray computed tomography (CT) in the prognosis of pancreatic ductal adenocarcinoma (PDAC) [135]. Claudin-4 is a known biomarker in PDAC detection. In this study, researchers reported a novel radiolabeled anti-claudin-4 monoclonal antibody in detecting PDAC using single-photon emission computed tomography (SPECT) imaging. The results showed promising uptake of anti-claudin-4 monoclonal antibody by PDAC tumors and were helpful in early detection and characterization of PDAC malignancy. Also, the researcher later targeted the extracellular domain of claudin-4 (4D3) with monoclonal antibodies (4D3) in combating bladder and lung cancer [136].
Colonoscopic aided screening and polyp and tumor removal have led to the reduced incidence and mortality of colorectal cancer (CRC). However, the lack of specificity is a major pitfall in these approaches and makes them less effective. It is especially difficult to detect the regions of flat dysplasia or serrated polyps, which also possess malignant potential. Thus, a targeted approach for advanced endoscopic techniques is a cornerstone requirement. A promising approach was recently demonstrated for the real-time endoscopic imaging of colonic adenomas [137]. In this study, the researchers exploited claudin-1 as a potential target in endoscopic imaging of colonic adenomas. As claudin-1 is highly expressed in the early development of CRC, endoscopic imaging might be useful for detecting either polypoid or flat precancerous lesions that are difficult to visualize [138]. Peptide (peptide sequence—RTSPSSR), specific to claudin-1, was developed against the extracellular loop of claudin-1. This peptide showed greater intensity for human adenomas, hyperplastic polyps and sessile serrated adenomas thus proposing the possibility of using claudin-1 peptide aided endoscopic imaging for the future clinical translation to detect precancerous lesions. Recently another study by our group demonstrated the significance of claudin-1 as a useful target for near-infrared antibody-based imaging for visualization of colorectal tumors [138]. When animals injected with colon cancer cells subcutaneously were imaged using claudin-1 antibody conjugated LI-COR IR800DyeCW through a LI-COR Pearl Trilogy Fluorescence Imaging System, the system was able to target tumors specifically. These studies pave the way for using claudins as a tool for fluorescence-guided surgery, which will help in more specific targeting of the tumors in a stage-specific manner. A comprehensive representation encompassing the role of claudins and the monoclonal antibodies against claudins as therapeutic and detection tools is given in Figure 3, and the role of claudins as a therapeutic, prognostic, and detection agents is tabulated in Table 3.
Figure 3.
Claudins as an employable platform for prognostic, diagnostic, and therapeutic targets. The upward arrow indicated upregulation and downwards arrow indicated downregulation of the mentioned signaling events.
Table 3.
Claudins as prognostic, therapeutic and detection agents.
| Claudins Subtype | Disease Type | Therapeutic Agent | Clinical Application | Reference |
|---|---|---|---|---|
| Claudin-1 | Hepatitis C virus infection | Residues within the first extracellular loop. | Hepatitis C virus co-receptor. | [139] |
| Humanization of a claudin-1-specific monoclonal antibody. | Clinical prevention and cure of Hepatitis C virus(HCV) infection. | [118] | ||
| Claudin-6 | Ovarian cancer | Clostridium perfringens enterotoxin (CPE) cytotoxicity. | CPE-mediated cytotoxicity in Ovarian cancer. | [127] |
| Claudin-3 | Ovarian cancer uterine carcinomas |
Human anti-claudin-3 IgG1 antibody. | Candidate for antibody-drug conjugate therapeutic applications. | [120,140] |
| Claudin-1 | Colon cancer | Human claudin-1 (6F6 mAb). |
Suppressed survival, growth, and migration of claudin-1 positive cells. Suppressed tumor growth and liver metastasis formation. |
[121] |
| Claudin-4 | Colorectal cancer | Anti-claudin-4 extracellular domain antibody. | Enhancer of anti-tumoral effects of chemotherapeutic agents. | [122] |
| Claudin-4 | Pancreatic Cancer (PDAC) | Indium-111 tagged anti-claudin-4 monoclonal antibody. | X-ray computed tomography sided detection of PDAC. | [135] |
| Claudin-18.2 | Gastric and gastroesophageal junction cancer | Chimeric monoclonal antibody that binds to claudin-18.2 (NCT03504397) | Cell death through antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity. | [123] |
| Claudin-4 | Pancreatic cancer | Claudin-4 binder C-CPE 194 | Enhances Tazeffects of anticancer agents via a MAPK pathway. | [141] |
| Claudin-3 and 4 | Prostate cancer | Claudin-3 and claudin-4 targeted Clostridium perfringens protoxin | Selectively cytotoxic to PSA-producing prostate cancer cells. | [126] |
| Claudin-1 | Colon cancer | Peptide RTSPSSR, specific to claudin-1 against the extracellular loop of claudin-1. | Specific to human adenomas, hyperplastic polyps, and sessile serrated adenomas. | [137] |
| Claudin-1 | Colon cancer | Claudin-1 antibody conjugated with LI-COR IR800DyeCW | Near-infrared antibody-based imaging for visualization of colorectal tumors. | [138] |
| Claudin-9 | Hepatitis C virus infection | Residues N38 and V45 in the first extracellular loop (EL1) of claudin-9 are responsible for HCV entry. Also found in PBMS (peripheral blood mononuclear cell) contributing to extrahepatic HCV infection. |
It can be implicated in the development of drugs to block HCV entry into the liver and peripheral blood mononuclear cell (PBMS). | [142] |
| Claudin-11 | Gastric Cancer | Hyper-methylation of claudin-11 promotor region leads to significant downregulation in gastric cancer. | Identification of the associated signaling cascades might lead to novel approaches in diagnosis and therapy for gastric cancer. | [48] |
| Claudin-7 | Non-small cell lung cancer (NSCLC) | Reduced expression—Poor outcome Claudin-7 low NSCLC—Poor survival. Claudin-7 high NSCLC—High Survival. |
Biomarker and a potential therapeutic target in patients with NSCLC. | [143] |
| Claudin-7 | Epithelial Ovarian cancer | Claudin-7 transcripts were significantly enhanced in epithelial ovarian carcinoma patients. Silencing claudin-7 displayed enhanced sensitivity to Cisplatin treatment. |
Independent prognostic factor and a key protein in regulating response to platinum-based chemotherapy in the treatment of epithelial ovarian cancer (EOC). | [144] |
| Claudin-2 | Irritable bowel disease (IBD) | Anti-claudin-2 mAb 1A2 | Prevent cis- and trans-interactions of claudin-2, attenuating the formation of leaky tight junction (TJ) seals. | [145] |
8. Future Perspectives
The quest for prognostic, diagnostic, and therapeutic markers for many cancers is of high importance. More reliable and earlier detection markers have implications for diagnostic and therapeutic targeting. As the role of claudins in the regulation and enrichment of cancer stem cells and chemo-resistance becomes obvious, targeting claudins for diminishing cancer stem cells, which are cancer-propagating subsets of malignant cells, would be very useful. The potential of the claudin–cancer stem cell axis provides great potential for combating invasive, metastatic, and drug resistance phenotypes of various cancers. Future studies focusing on the role of claudins in cancer stem cells will be warranted to specifically target these populations to curb down residual tumor cells left after standard therapies.
Claudins are gaining their importance as detection and therapeutic agents. Future engineering of more monoclonal antibodies against claudins will have potential applications in targeted therapy, and claudin assisted endoscopy, imaging of various tumors. Also, the antibody-based detections will provide ample opportunity for the early diagnosis of any inflammatory diseases before they reach cancer status. The ongoing clinical trials for monoclonal antibodies against claudins might lead to claudin directed immunotherapies. Recently, small molecules inhibitors have been gaining more attention in cancer biology, as they aid in targeted therapy. No known small molecule inhibitors are currently being researched for claudins. Thus, in the future, screening for more potent inhibitors against claudins is warranted. Overall, to strengthen the therapeutic window of claudins, a more translational view of claudins by researchers is warranted.
Author Contributions
Conceptualization, S.G., P.D., and A.B.S.; methodology, S.G. and P.D.; resources, S.G. and P.D.; writing—original draft preparation, S.G.; writing—review and editing, S.G., A.B.S., and P.D.; supervision, P.D.; project administration, P.D. and A.B.S.; funding acquisition, P.D. and A.B.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grant numbers BX002086 (VA merit) and CA216746 (NIH/NCI). Further funding came from a pilot project award from the Fred and Pamela Buffet Cancer Center, which was funded by a National Cancer Institute Cancer Center Support Grant, under award number P30 CA036727 to P.D., BX002761 (VA merit) to A.B.S., and Nebraska research initiative (NRI to P.D and A.B.S).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Farquhar M.G., Palade G.E. Junctional complexes in various epithelia. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneeberger E.E., Lynch R.D. The tight junction: a multifunctional complex. Am. J. Physiol.-Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 3.Niessen C.M. Tight junctions/adherens junctions: basic structure and function. J. Investig. Dermatol. 2007;127:2525–2532. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Mariscal L., Betanzos A., Nava P., Jaramillo B.E. Tight junction proteins. Prog. Biophys. Mol. Biol. 2003;81:1–44. doi: 10.1016/S0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson B.R., Siliciano J.D., Mooseker M.S., Goodenough D.A. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Citi S., Sabanay H., Jakes R., Geiger B., Kendrick-Jones J. Cingulin, a new peripheral component of tight junctions. Nature. 1988;333:272–276. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- 7.Furuse M., Itoh M., Hirase T., Nagafuchi A., Yonemura S., Tsukita S., Tsukita S. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J. Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes F.D., Lopez L.N., Lin H.W., Davies C., Azevedo R.B., Gow A., Kachar B. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J. Cell Sci. 2006;119:4819–4827. doi: 10.1242/jcs.03233. [DOI] [PubMed] [Google Scholar]
- 9.Gunzel D., Yu A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013;93:525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angelow S., Yu A.S. Claudins and paracellular transport: an update. Curr. Opin. Nephrol. Hypertens. 2007;16:459–464. doi: 10.1097/MNH.0b013e32820ac97d. [DOI] [PubMed] [Google Scholar]
- 11.Krause G., Protze J., Piontek J. Assembly and function of claudins: Structure-function relationships based on homology models and crystal structures. Semin. Cell Dev. Biol. 2015;42:3–12. doi: 10.1016/j.semcdb.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Tsukita S., Furuse M. Occludin and claudins in tight-junction strands: leading or supporting players? Trends Cell Biol. 1999;9:268–273. doi: 10.1016/S0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- 13.Tsukita S., Furuse M. Overcoming barriers in the study of tight junction functions: from occludin to claudin. Genes Cells. 1998;3:569–573. doi: 10.1046/j.1365-2443.1998.00212.x. [DOI] [PubMed] [Google Scholar]
- 14.Furuse M., Sasaki H., Fujimoto K., Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lal-Nag M., Morin P.J. The claudins. Genome Biol. 2009;10:235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelow S., Ahlstrom R., Yu A.S. Biology of claudins. Am. J. Physiol. Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunzel D., Fromm M. Claudins and other tight junction proteins. Compr. Physiol. 2012;2:1819–1852. doi: 10.1002/cphy.c110045. [DOI] [PubMed] [Google Scholar]
- 18.Soini Y. Expression of claudins 1, 2, 3, 4, 5 and 7 in various types of tumours. Histopathology. 2005;46:551–560. doi: 10.1111/j.1365-2559.2005.02127.x. [DOI] [PubMed] [Google Scholar]
- 19.Schlingmann B., Molina S.A., Koval M. Claudins: Gatekeepers of lung epithelial function. Semin. Cell Dev. Biol. 2015;42:47–57. doi: 10.1016/j.semcdb.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amasheh S., Meiri N., Gitter A.H., Schoneberg T., Mankertz J., Schulzke J.D., Fromm M. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J. Cell Sci. 2002;115:4969–4976. doi: 10.1242/jcs.00165. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Mumm J.B., Herbst R., Kolbeck R., Wang Y. IL-22 Increases Permeability of Intestinal Epithelial Tight Junctions by Enhancing Claudin-2 Expression. J. Immunol. 2017;199:3316–3325. doi: 10.4049/jimmunol.1700152. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal R., D’Souza T., Morin P.J. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 23.Morin P.J. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 24.Kage H., Flodby P., Zhou B., Borok Z. Dichotomous roles of claudins as tumor promoters or suppressors: Lessons from knockout mice. Cell. Mol. Life Sci. 2019;76:4663–4672. doi: 10.1007/s00018-019-03238-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohmoto T., Masuda K., Shoda K., Takahashi R., Ujiro S., Tange S., Ichikawa D., Otsuji E., Imoto I. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer. 2019:1–15. doi: 10.1007/s10120-019-01014-x. [DOI] [PubMed] [Google Scholar]
- 26.Yafang L., Qiong W., Yue R., Xiaoming X., Lina Y., Mingzi Z., Ting Z., Yulin L., Chengshi Q. Role of Estrogen Receptor-alpha in the Regulation of Claudin-6 Expression in Breast Cancer Cells. J. Breast Cancer. 2011;14:20–27. doi: 10.4048/jbc.2011.14.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rendon-Huerta E., Teresa F., Teresa G.M., Xochitl G.S., Georgina A.F., Veronica Z.Z., Montano L.F. Distribution and expression pattern of claudins 6, 7, and 9 in diffuse- and intestinal-type gastric adenocarcinomas. J. Gastrointest Cancer. 2010;41:52–59. doi: 10.1007/s12029-009-9110-y. [DOI] [PubMed] [Google Scholar]
- 28.Ikari A., Watanabe R., Sato T., Taga S., Shimobaba S., Yamaguchi M., Yamazaki Y., Endo S., Matsunaga T., Sugatani J. Nuclear distribution of claudin-2 increases cell proliferation in human lung adenocarcinoma cells. Biochim. Biophys. Acta. 2014;1843:2079–2088. doi: 10.1016/j.bbamcr.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 29.Chao Y.C., Pan S.H., Yang S.C., Yu S.L., Che T.F., Lin C.W., Tsai M.S., Chang G.C., Wu C.H., Wu Y.Y., et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 2009;179:123–133. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z., Ding L., Hong H., Hoggard J., Lu Q., Chen Y.H. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp. Cell Res. 2011;317:1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang X., Lin X., Alvarez E., Manorek G., Howell S.B. Tight junction proteins claudin-3 and claudin-4 control tumor growth and metastases. Neoplasia. 2012;14:974–985. doi: 10.1593/neo.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu C., Wang K., Ding Y.H., Li W.J., Ding L. Claudin-7 gene knockout causes destruction of intestinal structure and animal death in mice. World J. Gastroenterol. 2019;25:584–599. doi: 10.3748/wjg.v25.i5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nubel T., Preobraschenski J., Tuncay H., Weiss T., Kuhn S., Ladwein M., Langbein L., Zoller M. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol. Cancer Res. 2009;7:285–299. doi: 10.1158/1541-7786.MCR-08-0200. [DOI] [PubMed] [Google Scholar]
- 34.Shimobaba S., Taga S., Akizuki R., Hichino A., Endo S., Matsunaga T., Watanabe R., Yamaguchi M., Yamazaki Y., Sugatani J., et al. Claudin-18 inhibits cell proliferation and motility mediated by inhibition of phosphorylation of PDK1 and Akt in human lung adenocarcinoma A549 cells. Biochim. Biophys. Acta. 2016;1863:1170–1178. doi: 10.1016/j.bbamcr.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Ahmad R., Kumar B., Chen Z., Chen X., Muller D., Lele S.M., Washington M.K., Batra S.K., Dhawan P., Singh A.B. Loss of claudin-3 expression induces IL6/gp130/Stat3 signaling to promote colon cancer malignancy by hyperactivating Wnt/beta-catenin signaling. Oncogene. 2017;36:6592–6604. doi: 10.1038/onc.2017.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Che J., Yue D., Zhang B., Zhang H., Huo Y., Gao L., Zhen H., Yang Y., Cao B. Claudin-3 Inhibits Lung Squamous Cell Carcinoma Cell Epithelial-mesenchymal Transition and Invasion via Suppression of the Wnt/beta-catenin Signaling Pathway. Int. J. Med. Sci. 2018;15:339–351. doi: 10.7150/ijms.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hicks D.A., Galimanis C.E., Webb P.G., Spillman M.A., Behbakht K., Neville M.C., Baumgartner H.K. Claudin-4 activity in ovarian tumor cell apoptosis resistance and migration. BMC Cancer. 2016;16:788. doi: 10.1186/s12885-016-2799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Ruan Y., Li Y., Lin D., Quan C. Tight junction protein claudin-6 inhibits growth and induces the apoptosis of cervical carcinoma cells in vitro and in vivo. Med. Oncol. 2015;32:148. doi: 10.1007/s12032-015-0600-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Ruan Y., Li Y., Lin D., Liu Z., Quan C. Expression of apoptosis signal-regulating kinase 1 is associated with tight junction protein claudin-6 in cervical carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:5535–5541. [PMC free article] [PubMed] [Google Scholar]
- 40.Dhawan P., Singh A.B., Deane N.G., No Y., Shiou S.R., Schmidt C., Neff J., Washington M.K., Beauchamp R.D. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Investig. 2005;115:1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A.B., Sharma A., Smith J.J., Krishnan M., Chen X., Eschrich S., Washington M.K., Yeatman T.J., Beauchamp R.D., Dhawan P. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. 2011;141:2140–2153. doi: 10.1053/j.gastro.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat A.A., Pope J.L., Smith J.J., Ahmad R., Chen X., Washington M.K., Beauchamp R.D., Singh A.B., Dhawan P. Claudin-7 expression induces mesenchymal to epithelial transformation (MET) to inhibit colon tumorigenesis. Oncogene. 2015;34:4570–4580. doi: 10.1038/onc.2014.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhawan P., Ahmad R., Chaturvedi R., Smith J.J., Midha R., Mittal M.K., Krishnan M., Chen X., Eschrich S., Yeatman T.J., et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene. 2011;30:3234–3247. doi: 10.1038/onc.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanchard A.A., Zelinski T., Xie J., Cooper S., Penner C., Leygue E., Myal Y. Identification of Claudin 1 Transcript Variants in Human Invasive Breast Cancer. PLoS ONE. 2016;11:e0163387. doi: 10.1371/journal.pone.0163387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oku N., Sasabe E., Ueta E., Yamamoto T., Osaki T. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res. 2006;66:5251–5257. doi: 10.1158/0008-5472.CAN-05-4478. [DOI] [PubMed] [Google Scholar]
- 46.Darido C., Buchert M., Pannequin J., Bastide P., Zalzali H., Mantamadiotis T., Bourgaux J.F., Garambois V., Jay P., Blache P., et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008;68:4258–4268. doi: 10.1158/0008-5472.CAN-07-5805. [DOI] [PubMed] [Google Scholar]
- 47.Chang T.L., Ito K., Ko T.K., Liu Q., Salto-Tellez M., Yeoh K.G., Fukamachi H., Ito Y. Claudin-1 has tumor suppressive activity and is a direct target of RUNX3 in gastric epithelial cells. Gastroenterology. 2010;138:255–265 e251-253. doi: 10.1053/j.gastro.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 48.Agarwal R., Mori Y., Cheng Y., Jin Z., Olaru A.V., Hamilton J.P., David S., Selaru F.M., Yang J., Abraham J.M., et al. Silencing of claudin-11 is associated with increased invasiveness of gastric cancer cells. PLoS One. 2009;4:e8002. doi: 10.1371/journal.pone.0008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X., Wang H., Li Q., Li T. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int. 2018;18:160. doi: 10.1186/s12935-018-0662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki Y., Tokumasu R., Kimura H., Tsukita S. Role of claudin species-specific dynamics in reconstitution and remodeling of the zonula occludens. Mol. Biol. Cell. 2011;22:1495–1504. doi: 10.1091/mbc.e10-12-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuda M., Kubo A., Furuse M., Tsukita S. A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J. Cell Sci. 2004;117:1247–1257. doi: 10.1242/jcs.00972. [DOI] [PubMed] [Google Scholar]
- 52.Honda H., Pazin M.J., D’Souza T., Ji H., Morin P.J. Regulation of the CLDN3 gene in ovarian cancer cells. Cancer Biol. Ther. 2007;6:1733–1742. doi: 10.4161/cbt.6.11.4832. [DOI] [PubMed] [Google Scholar]
- 53.Kwon M.J., Kim S.H., Jeong H.M., Jung H.S., Kim S.S., Lee J.E., Gye M.C., Erkin O.C., Koh S.S., Choi Y.L., et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab. Investig. 2011;91:1652–1667. doi: 10.1038/labinvest.2011.117. [DOI] [PubMed] [Google Scholar]
- 54.Di Cello F., Cope L., Li H., Jeschke J., Wang W., Baylin S.B., Zahnow C.A. Methylation of the claudin 1 promoter is associated with loss of expression in estrogen receptor positive breast cancer. PLoS ONE. 2013;8:e68630. doi: 10.1371/journal.pone.0068630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishnan M., Singh A.B., Smith J.J., Sharma A., Chen X., Eschrich S., Yeatman T.J., Beauchamp R.D., Dhawan P. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene. 2010;29:305–312. doi: 10.1038/onc.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez-Estrada O.M., Culleres A., Soriano F.X., Peinado H., Bolos V., Martinez F.O., Reina M., Cano A., Fabre M., Vilaro S. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem. J. 2006;394:449–457. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhat A.A., Sharma A., Pope J., Krishnan M., Washington M.K., Singh A.B., Dhawan P. Caudal homeobox protein Cdx-2 cooperates with Wnt pathway to regulate claudin-1 expression in colon cancer cells. PLoS ONE. 2012;7:e37174. doi: 10.1371/journal.pone.0037174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Honda H., Pazin M.J., Ji H., Wernyj R.P., Morin P.J. Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J. Biol. Chem. 2006;281:21433–21444. doi: 10.1074/jbc.M603767200. [DOI] [PubMed] [Google Scholar]
- 59.Shigetomi K., Ikenouchi J. Regulation of the epithelial barrier by post-translational modifications of tight junction membrane proteins. J. Biochem. 2018;163:265–272. doi: 10.1093/jb/mvx077. [DOI] [PubMed] [Google Scholar]
- 60.Van Itallie C.M., Anderson J.M. Claudin interactions in and out of the tight junction. Tissue Barriers. 2013;1:e25247. doi: 10.4161/tisb.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajagopal N., Irudayanathan F.J., Nangia S. Palmitoylation of Claudin-5 Proteins Influences Their Lipid Domain Affinity and Tight Junction Assembly at the Blood-Brain Barrier Interface. J. Phys. Chem. B. 2019;123:983–993. doi: 10.1021/acs.jpcb.8b09535. [DOI] [PubMed] [Google Scholar]
- 62.Butt A.M., Khan I.B., Hussain M., Idress M., Lu J., Tong Y. Role of post translational modifications and novel crosstalk between phosphorylation and O-beta-GlcNAc modifications in human claudin-1, -3 and -4. Mol. Biol. Rep. 2012;39:1359–1369. doi: 10.1007/s11033-011-0870-7. [DOI] [PubMed] [Google Scholar]
- 63.French A.D., Fiori J.L., Camilli T.C., Leotlela P.D., O’Connell M.P., Frank B.P., Subaran S., Indig F.E., Taub D.D., Weeraratna A.T. PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int. J. Med. Sci. 2009;6:93–101. doi: 10.7150/ijms.6.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D’Souza T., Indig F.E., Morin P.J. Phosphorylation of claudin-4 by PKCepsilon regulates tight junction barrier function in ovarian cancer cells. Exp. Cell Res. 2007;313:3364–3375. doi: 10.1016/j.yexcr.2007.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D’Souza T., Agarwal R., Morin P.J. Phosphorylation of claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. J. Biol. Chem. 2005;280:26233–26240. doi: 10.1074/jbc.M502003200. [DOI] [PubMed] [Google Scholar]
- 66.Akizuki R., Shimobaba S., Matsunaga T., Endo S., Ikari A. Claudin-5, -7, and -18 suppress proliferation mediated by inhibition of phosphorylation of Akt in human lung squamous cell carcinoma. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:293–302. doi: 10.1016/j.bbamcr.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 67.Yamamoto M., Ramirez S.H., Sato S., Kiyota T., Cerny R.L., Kaibuchi K., Persidsky Y., Ikezu T. Phosphorylation of claudin-5 and occludin by rho kinase in brain endothelial cells. Am. J. Pathol. 2008;172:521–533. doi: 10.2353/ajpath.2008.070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmad W., Shabbiri K., Ijaz B., Asad S., Sarwar M.T., Gull S., Kausar H., Fouzia K., Shahid I., Hassan S. Claudin-1 required for HCV virus entry has high potential for phosphorylation and O-glycosylation. Virol. J. 2011;8:229. doi: 10.1186/1743-422X-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heiler S., Mu W., Zoller M., Thuma F. The importance of claudin-7 palmitoylation on membrane subdomain localization and metastasis-promoting activities. Cell Commun. Signal. 2015;13:29. doi: 10.1186/s12964-015-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biteau B., Hochmuth C.E., Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romito A., Cobellis G. Pluripotent Stem Cells: Current Understanding and Future Directions. Stem Cells Int. 2016;2016:9451492. doi: 10.1155/2016/9451492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duinsbergen D., Salvatori D., Eriksson M., Mikkers H. Tumors originating from induced pluripotent stem cells and methods for their prevention. Ann. N.Y. Acad. Sci. 2009;1176:197–204. doi: 10.1111/j.1749-6632.2009.04563.x. [DOI] [PubMed] [Google Scholar]
- 73.Ben-David U., Nudel N., Benvenisty N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat. Commun. 2013;4:1992. doi: 10.1038/ncomms2992. [DOI] [PubMed] [Google Scholar]
- 74.Sugimoto K., Ichikawa-Tomikawa N., Satohisa S., Akashi Y., Kanai R., Saito T., Sawada N., Chiba H. The tight-junction protein claudin-6 induces epithelial differentiation from mouse F9 and embryonic stem cells. PLoS ONE. 2013;8:e75106. doi: 10.1371/journal.pone.0075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L., Xue Y., Shen Y., Li W., Cheng Y., Yan X., Shi W., Wang J., Gong Z., Yang G., et al. Claudin 6: a novel surface marker for characterizing mouse pluripotent stem cells. Cell Res. 2012;22:1082–1085. doi: 10.1038/cr.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turksen K., Troy T.C. Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Dev. Dyn. 2001;222:292–300. doi: 10.1002/dvdy.1174. [DOI] [PubMed] [Google Scholar]
- 77.Erdelyi-Belle B., Torok G., Apati A., Sarkadi B., Schaff Z., Kiss A., Homolya L. Expression of Tight Junction Components in Hepatocyte-Like Cells Differentiated from Human Embryonic Stem Cells. Pathol. Oncol. Res. 2015;21:1059–1070. doi: 10.1007/s12253-015-9936-5. [DOI] [PubMed] [Google Scholar]
- 78.Abdullah L.N., Chow E.K. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013;2:3. doi: 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phi L.T.H., Sari I.N., Yang Y.G., Lee S.H., Jun N., Kim K.S., Lee Y.K., Kwon H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018;2018:5416923. doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 81.Miwa N., Furuse M., Tsukita S., Niikawa N., Nakamura Y., Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol. Res. 2001;12:469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 82.Gowrikumar S., Ahmad R., Uppada S.B., Washington M.K., Shi C., Singh A.B., Dhawan P. Upregulated claudin-1 expression promotes colitis-associated cancer by promoting beta-catenin phosphorylation and activation in Notch/p-AKT-dependent manner. Oncogene. 2019;38:5321–5337. doi: 10.1038/s41388-019-0795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma L., Yin W., Ma H., Elshoura I., Wang L. Targeting claudin-3 suppresses stem cell-like phenotype in nonsquamous non-small-cell lung carcinoma. Lung Cancer Manag. 2019;8:LMT04. doi: 10.2217/lmt-2018-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou B., Flodby P., Luo J., Castillo D.R., Liu Y., Yu F.X., McConnell A., Varghese B., Li G., Chimge N.O., et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J. Clin. Investig. 2018;128:970–984. doi: 10.1172/JCI90429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paquet-Fifield S., Koh S.L., Cheng L., Beyit L.M., Shembrey C., Molck C., Behrenbruch C., Papin M., Gironella M., Guelfi S., et al. Tight Junction Protein Claudin-2 Promotes Self-Renewal of Human Colorectal Cancer Stem-like Cells. Cancer Res. 2018;78:2925–2938. doi: 10.1158/0008-5472.CAN-17-1869. [DOI] [PubMed] [Google Scholar]
- 87.Zheng H.C. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8:59950–59964. doi: 10.18632/oncotarget.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang K., Li T., Xu C., Ding Y., Li W., Ding L. Claudin-7 downregulation induces metastasis and invasion in colorectal cancer via the promotion of epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2019;508:797–804. doi: 10.1016/j.bbrc.2018.10.049. [DOI] [PubMed] [Google Scholar]
- 89.Tabaries S., Siegel P.M. The role of claudins in cancer metastasis. Oncogene. 2017;36:1176–1190. doi: 10.1038/onc.2016.289. [DOI] [PubMed] [Google Scholar]
- 90.Zhang L., Wang Y., Zhang B., Zhang H., Zhou M., Wei M., Dong Q., Xu Y., Wang Z., Gao L., et al. Claudin-3 expression increases the malignant potential of lung adenocarcinoma cells: role of epidermal growth factor receptor activation. Oncotarget. 2017;8:23033–23047. doi: 10.18632/oncotarget.14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Q., Zhang Y., Zhang T., Han Z.G., Shan L. Low claudin-6 expression correlates with poor prognosis in patients with non-small cell lung cancer. Onco Targets Ther. 2015;8:1971–1977. doi: 10.2147/OTT.S85478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Landers K.A., Samaratunga H., Teng L., Buck M., Burger M.J., Scells B., Lavin M.F., Gardiner R.A. Identification of claudin-4 as a marker highly overexpressed in both primary and metastatic prostate cancer. Br. J. Cancer. 2008;99:491–501. doi: 10.1038/sj.bjc.6604486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hwang T.L., Changchien T.T., Wang C.C., Wu C.M. Claudin-4 expression in gastric cancer cells enhances the invasion and is associated with the increased level of matrix metalloproteinase-2 and -9 expression. Oncol. Lett. 2014;8:1367–1371. doi: 10.3892/ol.2014.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohtani S., Terashima M., Satoh J., Soeta N., Saze Z., Kashimura S., Ohsuka F., Hoshino Y., Kogure M., Gotoh M. Expression of tight-junction-associated proteins in human gastric cancer: downregulation of claudin-4 correlates with tumor aggressiveness and survival. Gastric Cancer. 2009;12:43–51. doi: 10.1007/s10120-008-0497-0. [DOI] [PubMed] [Google Scholar]
- 95.Stebbing J., Filipovic A., Giamas G. Claudin-1 as a promoter of EMT in hepatocellular carcinoma. Oncogene. 2013;32:4871–4872. doi: 10.1038/onc.2012.591. [DOI] [PubMed] [Google Scholar]
- 96.Zhou B., Moodie A., Blanchard A.A., Leygue E., Myal Y. Claudin 1 in Breast Cancer: New Insights. J. Clin. Med. 2015;4:1960–1976. doi: 10.3390/jcm4121952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang M., Li Y., Ruan Y., Lu Y., Lin D., Xie Y., Dong B., Dang Q., Quan C. CLDN6 enhances chemoresistance to ADM via AF-6/ERKs pathway in TNBC cell line MDAMB231. Mol. Cell. Biochem. 2018;443:169–180. doi: 10.1007/s11010-017-3221-8. [DOI] [PubMed] [Google Scholar]
- 98.Shang X., Lin X., Manorek G., Howell S.B. Claudin-3 and claudin-4 regulate sensitivity to cisplatin by controlling expression of the copper and cisplatin influx transporter CTR1. Mol. Pharmacol. 2013;83:85–94. doi: 10.1124/mol.112.079798. [DOI] [PubMed] [Google Scholar]
- 99.Yoshida H., Sumi T., Zhi X., Yasui T., Honda K., Ishiko O. Claudin-4: a potential therapeutic target in chemotherapy-resistant ovarian cancer. Anticancer Res. 2011;31:1271–1277. [PubMed] [Google Scholar]
- 100.Zhao Z., Li J., Jiang Y., Xu W., Li X., Jing W. CLDN1 Increases Drug Resistance of Non-Small Cell Lung Cancer by Activating Autophagy via Up-Regulation of ULK1 Phosphorylation. Med. Sci. Monit. 2017;23:2906–2916. doi: 10.12659/MSM.904177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akizuki R., Maruhashi R., Eguchi H., Kitabatake K., Tsukimoto M., Furuta T., Matsunaga T., Endo S., Ikari A. Decrease in paracellular permeability and chemosensitivity to doxorubicin by claudin-1 in spheroid culture models of human lung adenocarcinoma A549 cells. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:769–780. doi: 10.1016/j.bbamcr.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 102.Philip R., Heiler S., Mu W., Buchler M.W., Zoller M., Thuma F. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget. 2015;6:2046–2063. doi: 10.18632/oncotarget.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoggard J., Fan J., Lu Z., Lu Q., Sutton L., Chen Y.H. Claudin-7 increases chemosensitivity to cisplatin through the upregulation of caspase pathway in human NCI-H522 lung cancer cells. Cancer Sci. 2013;104:611–618. doi: 10.1111/cas.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lechpammer M., Resnick M.B., Sabo E., Yakirevich E., Greaves W.O., Sciandra K.T., Tavares R., Noble L.C., DeLellis R.A., Wang L.J. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod. Pathol. 2008;21:1320–1329. doi: 10.1038/modpathol.2008.116. [DOI] [PubMed] [Google Scholar]
- 105.Osunkoya A.O., Cohen C., Lawson D., Picken M.M., Amin M.B., Young A.N. Claudin-7 and claudin-8: immunohistochemical markers for the differential diagnosis of chromophobe renal cell carcinoma and renal oncocytoma. Hum. Pathol. 2009;40:206–210. doi: 10.1016/j.humpath.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 106.Yang L., Sun X., Meng X. Differences in the expression profiles of claudin proteins in human gastric carcinoma compared with nonneoplastic mucosa. Mol. Med. Rep. 2018;18:1271–1278. doi: 10.3892/mmr.2018.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Danzinger S., Tan Y.Y., Rudas M., Kastner M.T., Weingartshofer S., Muhr D., Singer C.F., kConFab I. Differential Claudin 3 and EGFR Expression Predicts BRCA1 Mutation in Triple-Negative Breast Cancer. Cancer Investig. 2018;36:378–388. doi: 10.1080/07357907.2018.1499934. [DOI] [PubMed] [Google Scholar]
- 108.Karabulut M., Alis H., Bas K., Karabulut S., Afsar C.U., Oguz H., Gunaldi M., Akarsu C., Kones O., Aykan N.F. Clinical significance of serum claudin-1 and claudin-7 levels in patients with colorectal cancer. Mol. Clin. Oncol. 2015;3:1255–1267. doi: 10.3892/mco.2015.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sabatier R., Finetti P., Guille A., Adelaide J., Chaffanet M., Viens P., Birnbaum D., Bertucci F. Claudin-low breast cancers: clinical, pathological, molecular and prognostic characterization. Mol. Cancer. 2014;13:228. doi: 10.1186/1476-4598-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nissinen L., Siljamaki E., Riihila P., Piipponen M., Farshchian M., Kivisaari A., Kallajoki M., Raiko L., Peltonen J., Peltonen S., et al. Expression of claudin-11 by tumor cells in cutaneous squamous cell carcinoma is dependent on the activity of p38delta. Exp. Dermatol. 2017;26:771–777. doi: 10.1111/exd.13278. [DOI] [PubMed] [Google Scholar]
- 111.Dias K., Dvorkin-Gheva A., Hallett R.M., Wu Y., Hassell J., Pond G.R., Levine M., Whelan T., Bane A.L. Claudin-Low Breast Cancer; Clinical & Pathological Characteristics. PLoS ONE. 2017;12:e0168669. doi: 10.1371/journal.pone.0168669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Upadhaya P., Barhoi D., Giri A., Bhattacharjee A., Giri S. Joint detection of claudin-1 and junctional adhesion molecule-A as a therapeutic target in oral epithelial dysplasia and oral squamous cell carcinoma. J. Cell. Biochem. 2019;120:18117–18127. doi: 10.1002/jcb.29115. [DOI] [PubMed] [Google Scholar]
- 113.Chau C.H., Steeg P.S., Figg W.D. Antibody-drug conjugates for cancer. Lancet. 2019;394:793–804. doi: 10.1016/S0140-6736(19)31774-X. [DOI] [PubMed] [Google Scholar]
- 114.Shuptrine C.W., Surana R., Weiner L.M. Monoclonal antibodies for the treatment of cancer. Semin. Cancer Biol. 2012;22:3–13. doi: 10.1016/j.semcancer.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mailly L., Xiao F., Lupberger J., Wilson G.K., Aubert P., Duong F.H.T., Calabrese D., Leboeuf C., Fofana I., Thumann C., et al. Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat. Biotechnol. 2015;33:549–554. doi: 10.1038/nbt.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fukasawa M., Nagase S., Shirasago Y., Iida M., Yamashita M., Endo K., Yagi K., Suzuki T., Wakita T., Hanada K., et al. Monoclonal antibodies against extracellular domains of claudin-1 block hepatitis C virus infection in a mouse model. J. Virol. 2015;89:4866–4879. doi: 10.1128/JVI.03676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fofana I., Krieger S.E., Grunert F., Glauben S., Xiao F., Fafi-Kremer S., Soulier E., Royer C., Thumann C., Mee C.J., et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953–964, 964. e4. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 118.Colpitts C.C., Tawar R.G., Mailly L., Thumann C., Heydmann L., Durand S.C., Xiao F., Robinet E., Pessaux P., Zeisel M.B., et al. Humanisation of a claudin-1-specific monoclonal antibody for clinical prevention and cure of HCV infection without escape. Gut. 2018;67:736–745. doi: 10.1136/gutjnl-2016-312577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Offner S., Hekele A., Teichmann U., Weinberger S., Gross S., Kufer P., Itin C., Baeuerle P.A., Kohleisen B. Epithelial tight junction proteins as potential antibody targets for pancarcinoma therapy. Cancer Immunol. Immunother. 2005;54:431–445. doi: 10.1007/s00262-004-0613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Romani C., Cocco E., Bignotti E., Moratto D., Bugatti A., Todeschini P., Bandiera E., Tassi R., Zanotti L., Pecorelli S., et al. Evaluation of a novel human IgG1 anti-claudin3 antibody that specifically recognizes its aberrantly localized antigen in ovarian cancer cells and that is suitable for selective drug delivery. Oncotarget. 2015;6:34617–34628. doi: 10.18632/oncotarget.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cherradi S., Ayrolles-Torro A., Vezzo-Vie N., Gueguinou N., Denis V., Combes E., Boissiere F., Busson M., Canterel-Thouennon L., Mollevi C., et al. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J. Exp. Clin. Cancer Res. 2017;36:89. doi: 10.1186/s13046-017-0558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fujiwara-Tani R., Sasaki T., Luo Y., Goto K., Kawahara I., Nishiguchi Y., Kishi S., Mori S., Ohmori H., Kondoh M., et al. Anti-claudin-4 extracellular domain antibody enhances the antitumoral effects of chemotherapeutic and antibody drugs in colorectal cancer. Oncotarget. 2018;9:37367–37378. doi: 10.18632/oncotarget.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sahin U., Schuler M., Richly H., Bauer S., Krilova A., Dechow T., Jerling M., Utsch M., Rohde C., Dhaene K., et al. A phase I dose-escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro-oesophageal junction cancer. Eur. J. Cancer. 2018;100:17–26. doi: 10.1016/j.ejca.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 124.Singh P., Toom S., Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J. Hematol. Oncol. 2017;10:105. doi: 10.1186/s13045-017-0473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shrestha A., Uzal F.A., McClane B.A. The interaction of Clostridium perfringens enterotoxin with receptor claudins. Anaerobe. 2016;41:18–26. doi: 10.1016/j.anaerobe.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Romanov V., Whyard T.C., Waltzer W.C., Gabig T.G. A claudin 3 and claudin 4-targeted Clostridium perfringens protoxin is selectively cytotoxic to PSA-producing prostate cancer cells. Cancer Lett. 2014;351:260–264. doi: 10.1016/j.canlet.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 127.Lal-Nag M., Battis M., Santin A.D., Morin P.J. Claudin-6: a novel receptor for CPE-mediated cytotoxicity in ovarian cancer. Oncogenesis. 2012;1:e33. doi: 10.1038/oncsis.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kominsky S.L., Vali M., Korz D., Gabig T.G., Weitzman S.A., Argani P., Sukumar S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am. J. Pathol. 2004;164:1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cocco E., Shapiro E.M., Gasparrini S., Lopez S., Schwab C.L., Bellone S., Bortolomai I., Sumi N.J., Bonazzoli E., Nicoletti R., et al. Clostridium perfringens enterotoxin C-terminal domain labeled to fluorescent dyes for in vivo visualization of micrometastatic chemotherapy-resistant ovarian cancer. Int. J. Cancer. 2015;137:2618–2629. doi: 10.1002/ijc.29632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pahle J., Menzel L., Niesler N., Kobelt D., Aumann J., Rivera M., Walther W. Rapid eradication of colon carcinoma by Clostridium perfringens Enterotoxin suicidal gene therapy. BMC Cancer. 2017;17:129. doi: 10.1186/s12885-017-3123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pahle J., Aumann J., Kobelt D., Walther W. Oncoleaking: Use of the Pore-Forming Clostridium perfringens Enterotoxin (CPE) for Suicide Gene Therapy. Methods Mol. Biol. 2015;1317:69–85. doi: 10.1007/978-1-4939-2727-2_5. [DOI] [PubMed] [Google Scholar]
- 132.Vecchio A.J., Stroud R.M. Claudin-9 structures reveal mechanism for toxin-induced gut barrier breakdown. Proc. Natl. Acad. Sci. USA. 2019;116:17817–17824. doi: 10.1073/pnas.1908929116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Becker A., Leskau M., Schlingmann-Molina B.L., Hohmeier S.C., Alnajjar S., Murua Escobar H., Ngezahayo A. Functionalization of gold-nanoparticles by the Clostridium perfringens enterotoxin C-terminus for tumor cell ablation using the gold nanoparticle-mediated laser perforation technique. Sci. Rep. 2018;8:14963. doi: 10.1038/s41598-018-33392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Becker A., Lehrich T., Kalies S., Heisterkamp A., Ngezahayo A. Parameters for Optoperforation-Induced Killing of Cancer Cells Using Gold Nanoparticles Functionalized With the C-terminal Fragment of Clostridium Perfringens Enterotoxin. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20174248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Torres J.B., Knight J.C., Mosley M.J., Kersemans V., Koustoulidou S., Allen D., Kinchesh P., Smart S., Cornelissen B. Imaging of Claudin-4 in Pancreatic Ductal Adenocarcinoma Using a Radiolabelled Anti-Claudin-4 Monoclonal Antibody. Mol. Imaging Biol. 2018;20:292–299. doi: 10.1007/s11307-017-1112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kuwada M., Chihara Y., Luo Y., Li X., Nishiguchi Y., Fujiwara R., Sasaki T., Fujii K., Ohmori H., Fujimoto K., et al. Pro-chemotherapeutic effects of antibody against extracellular domain of claudin-4 in bladder cancer. Cancer Lett. 2015;369:212–221. doi: 10.1016/j.canlet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 137.Rabinsky E.F., Joshi B.P., Pant A., Zhou J., Duan X., Smith A., Kuick R., Fan S., Nusrat A., Owens S.R., et al. Overexpressed Claudin-1 Can Be Visualized Endoscopically in Colonic Adenomas In Vivo. Cell. Mol. Gastroenterol. Hepatol. 2016;2:222–237. doi: 10.1016/j.jcmgh.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hollandsworth H.M., Lwin T.M., Amirfakhri S., Filemoni F., Batra S.K., Hoffman R.M., Dhawan P., Bouvet M. Anti-Claudin-1 Conjugated to a Near-Infrared Fluorophore Targets Colon Cancer in PDOX Mouse Models. J. Surg. Res. 2019;242:145–150. doi: 10.1016/j.jss.2019.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Evans M.J., von Hahn T., Tscherne D.M., Syder A.J., Panis M., Wolk B., Hatziioannou T., McKeating J.A., Bieniasz P.D., Rice C.M. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 140.Romani C., Comper F., Bandiera E., Ravaggi A., Bignotti E., Tassi R.A., Pecorelli S., Santin A.D. Development and characterization of a human single-chain antibody fragment against claudin-3: A novel therapeutic target in ovarian and uterine carcinomas. Am. J. Obstet. Gynecol. 2009;201:70 e71–e79. doi: 10.1016/j.ajog.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kono T., Kondoh M., Kyuno D., Ito T., Kimura Y., Imamura M., Kohno T., Konno T., Furuhata T., Sawada N., et al. Claudin-4 binder C-CPE 194 enhances effects of anticancer agents on pancreatic cancer cell lines via a MAPK pathway. Pharmacol. Res. Perspect. 2015;3:e00196. doi: 10.1002/prp2.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zheng A., Yuan F., Li Y., Zhu F., Hou P., Li J., Song X., Ding M., Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yamamoto T., Oshima T., Yoshihara K., Yamanaka S., Nishii T., Arai H., Inui K., Kaneko T., Nozawa A., Woo T., et al. Reduced expression of claudin-7 is associated with poor outcome in non-small cell lung cancer. Oncol. Lett. 2010;1:501–505. doi: 10.3892/ol_00000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim C.J., Lee J.W., Choi J.J., Choi H.Y., Park Y.A., Jeon H.K., Sung C.O., Song S.Y., Lee Y.Y., Choi C.H., et al. High claudin-7 expression is associated with a poor response to platinum-based chemotherapy in epithelial ovarian carcinoma. Eur. J. Cancer. 2011;47:918–925. doi: 10.1016/j.ejca.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 145.Takigawa M., Iida M., Nagase S., Suzuki H., Watari A., Tada M., Okada Y., Doi T., Fukasawa M., Yagi K., et al. Creation of a Claudin-2 Binder and Its Tight Junction-Modulating Activity in a Human Intestinal Model. J. Pharmacol. Exp. Ther. 2017;363:444–451. doi: 10.1124/jpet.117.242214. [DOI] [PubMed] [Google Scholar]