Introduction
Genetic studies of ankylosing spondylitis (AS) have over the past decade provided major insights into the etiopathogenesis of the disease that have led to major therapeutic innovations. Some of these new treatments have already entered clinical practice, and others are in trials and undergoing development. It has long been known that susceptibility to and ‘severity’ of AS are largely genetically determined. Extensive progress has been made identifying susceptibility alleles in the disease, with 113 established loci identified contributing roughly 10% of the heritability of AS, over and above the major effect of HLA-B27, which determines ~20% of the genetic risk. Studies of the genetics of clinical manifestations of AS such as the extent of bony ankylosis or presence of anterior uveitis have been more challenging, though some genes have been found to influence uveitis risk beyond their effects on the risk of AS-itself. This article seeks to present the current state of understanding of the genetic influences in AS, focusing on more recent advances and their contribution to understanding mechanisms of disease.
Major Histocompatibility Complex (MHC) and Ankylosing Spondylitis
Large scale case-control studies of HLA and other MHC genes in AS have demonstrated that the genetic associations at this locus are far more complex than initially thought. Since the discovery of the association of HLA-B27 with AS, there have been many studies suggesting the presence of additional MHC associated variants 1–5. With the exception of the association of HLA-B60 with AS 6,7, until recently none of those have been convincingly replicated.
The MHC is under marked genetic selection pressure and HLA frequencies vary substantially between ethnic groups. The development of methods of HLA-typing using imputation from dense SNP genotyping together with the availability of large reference sets of subjects genotyped at both HLA loci and MHC SNPs has enabled analysis of HLA and MHC associations in large case-control cohorts. Another methodologic advance in recent studies is principal components analysis; population stratification can be identified and controlled for, making the findings robust to differences in allele frequencies due to ethnic variation rather than disease affection status. Two such studies have now been published, one in subjects of European-ancestry 8, and the other in Koreans 9. Both show convincingly that there are additional HLA-B variants associated with AS, and also other HLA Class I and II variants (Table 1). While these studies do not exclude the presence of other non-HLA MHC genetic associations, they do indicate that it is unlikely that variants of large effect exist within the MHC once the associations of HLA variants are accounted for.
Table 1.
Association of HLA-B alleles with susceptibility to AS in European descent patients. Findings are presented for consecutive conditional analyses, where for rounds 2 and onwards the test conditioned on the previous alleles.
| Round | HLA-B Allele | Odds Ratio (95% CI) | P-value |
|---|---|---|---|
| 1 | 27:05 | 62.41 | < 10−321 |
| 2 | 27:02 | 43.41 | 1.07 × 10−122 |
| 3 | 07:02 | 0.82 | 5.04 × 10−6 |
| 4 | 57:01 | 0.75 | 5.13 × 10−4 |
| 5 | 51:01 | 1.33 | 2.14 × 10−3 |
| 6 | 47:01 | 2.35 | 2.25 × 10−3 |
| 7 | 40:02 | 1.59 | 4.65 × 10−3 |
| 8 | 13:02 | 1.43 | 4.29 × 10−3 |
| 9 | 40:01 | 1.22 | 4.93 × 10−3 |
The additional HLA-associations likely contribute to the known association of AS with other diseases. For example, HLA-B51 which is also a risk variant for AS, is the major risk allele for Behçet’s syndrome, a condition that can be complicated by sacroiliitis. HLA-DRB1*0103 is AS-associated, and is also one of the major risk alleles for Crohn’s disease, which frequently co-occurs with AS. HLA-B7 has a major protective effect on AS, and interestingly is used as a control allele in transgenic rats. In this model, excess copy numbers of HLA-B27 induce a spondyloarthropathy, whereas rats with similar excess copy numbers of HLA-B7 remain healthy 10.
Using dense MHC SNP genotyping data, the amino-acid composition of the HLA alleles can be imputed and tested for association with disease, enabling the identification of the key amino-acids involved. This approach has been successfully used to extend the known amino-acids in the rheumatoid arthritis ‘shared epitope’, which provides a functional explanation for the association of HLA-DRB1 alleles with that disease 11. In AS, in the populations studied, the identity of the amino-acid at position 97 in HLA-B was shown to determine the direction of association of the major HLA-B alleles with the disease (see Table 2) 8. This does not mean that this amino acid alone is the sole HLA-B determinant of AS risk; rather it indicates that in the context of the HLA-B alleles involved, this amino acid is a key determinant of disease risk.
Table 2.
Association analysis of SNPs and amino acids at position 97 of HLA-B.
| Amino acid residue | Multivariate OR (95% CI) | P-value | Classical HLA-B Allele |
|---|---|---|---|
| Asparagine (N) | 16.51 (15.43–17.69) | < 1 × 10−300 | *27:02, *27:04, *27:05 |
| Threonine (T) | 1.12 (1.03–1.21) | 4.50 × 10−3 | *13:02, *39:06, *40:06, *51:01, *51:08, *52:01, *55:01, *56:01 |
| Arginine (R) | 1.00 (Reference) | 1 |
*15:01, *15:03, *15:10, *15:16, *15:17, *15:18, *18:01, *35:01, *35:02,
*35:03, *35:08, *35:12, *37:01, *38:01, *38:02, *39:01, *39:10, *40:01, *41:01, *44:02, *44:03, *44:04, *44:05, *45:01, *47:01, *49:01, *50:01, *53:01, *58:01 |
| Tryptophan (W) | 1.00 (0.89–1.12) | 0.95 | *14:01, *14:02 |
| Serine (S) | 0.86 (0.81–0.91) | 4.81 × 10−8 | *07:02, *07:05, *08:01, *15:07, *27:07, *40:02, *41:02, *48:01 |
| Valine (V) | 0.68 (0.59–0.78) | 1.41 × 10−8 | *57:01, *57:03 |
Aminopeptidases and Ankylosing Spondylitis
A key recent discovery has been the demonstration that variants of the M1-aminopeptidase gene, ERAP1, are associated with AS 12, and interact genetically with both the AS-associated HLA Class I alleles HLA-B27 and HLA-B*4001 8,13. Thus ERAP1 is only associated with AS in HLA-B27 positive cases, or HLA-B27-negative/HLA-B*4001 positive cases. The same ERAP1 haplotypes interact with HLA-Cw6 in psoriasis 14, and HLA-B51 in Behçet’s disease 15. This locus is also strongly associated with the rare ocular uveitis birdshot retinopathy, which is strongly associated with HLA-A29, though in this disease the number of subjects studied is too small to determine if the disease association is with ERAP1, the neighboring related gene ERAP2, or both 16. The ERAP2 association with AS is present in both HLA-B27-positive and –negative disease 17 suggesting some subtle difference in its functional mechanism in causing AS. For example ERAP2 may potentially affect on peptide handling by other AS-associated HLA Class I alleles for which ERAP1 peptide cleavage is less influential.
A recent paper has suggested that haplotypes of ERAP1 variants are more strongly associated with disease and have greater functional effects than individual disease-associated variants 18. However, the small sample size of this study (19 cases and 17 controls) is too few to distinguish haplotypic from single variant effects, and many of the haplotypic associations reported were not statistically significant 19. The study also reports the opposite direction of association of the key AS ERAP1 non-synonymous SNP, rs30187, compared to all other studies, which included more than 1000 times more cases and controls, in multiple ethnic groups 8,20–24. In the absence of further supportive evidence this study should be considered hypothesis generating.
Extensive studies of the functional mechanism of the associations of ERAP1 and ERAP2 variants and AS are underway. Both proteins are involved in peptide trimming in the endoplasmic reticulum, changing particularly the length but also the amino acid composition of peptides available for HLA Class I presentation 25–27,28. Proposed mechanisms of association include:
Effects on the peptidome presented by HLA-B27 and thus leading either to presentation of arthritogenic peptides 29 or failure to present disease-protective peptides 30
Effects on HLA-B27 free heavy chain expression and KIR interactions, in turn influencing activation of IL-17 producing immune cells 31
Effects on HLA-B27 folding and ER accumulation leading to ER stress reactions 32,33.
It is beyond the scope of this article to discuss these hypotheses and studies in detail. However, a common feature of each model is that variants that are disease-protective in AS exhibit reduced peptide cleavage function 34–36. This, and the fact that apart from with Behçet’s disease there is no convincing evidence in either humans or animal models that ERAP deficiency increases disease risk, has led to programs targeting these proteins as therapeutics for AS and related diseases 37.
T-Cells and Ankylosing Spondylitis
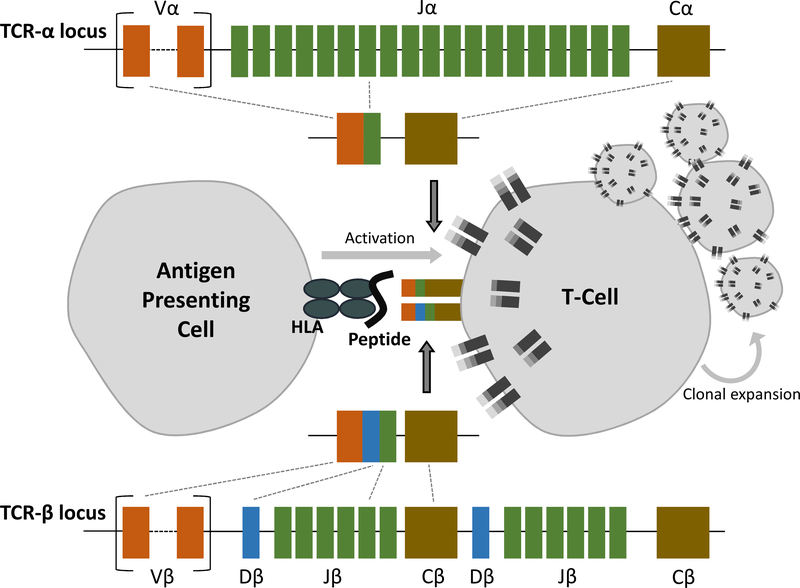
How genetic variants predisposing to immune-mediated diseases are tied to altered immune system activity is a question of primary importance. Despite being one of the first proposed mechanisms of disease development in AS, strongly supported by the robust ERAP1-HLA-B27 epistasis identified 36, it has not been conclusively demonstrated that autoreactive T-cells recognize a B27 restricted peptide in AS patients to cause disease. T-cell receptors (TCRs) develop by the process of random recombination of numerous encoded gene segments to generate highly variable and cell-specific receptor chains. Unique cell surface TCRs on each T-cell have the potential to engage different combinations and conformations of HLA bound peptides (Figure 1). Clonally expanded T-cells that have recognized an antagonistic antigen, and proliferated to hone adaptive immune responses, are a feature of many immune-mediated diseases and contribute to targeted inflammation 38,39. Little evidence of such occurrence in AS patients has been published since the work of Mamedov et al. in 2009 40, which characterized the T-cell populations of two patients and found stably expanded clones consistently representing between 5 and 50% of the profiled repertoire over a number of years. These T-cells were found to be cytotoxic and proinflammatory in nature, predominantly CD8+/CD27−/CD28− (regarded as terminally differentiated effector or cytotoxic T-cells), and expressed TCRs with homology to a number of previously reported clones in reactive arthritis 41, rheumatoid arthritis 42 and other AS patients 43. Earlier work demonstrated that a nonamer of the HLA-B*2705 molecule hypervariable region itself can be recognized by cytotoxic T lymphocytes in AS patients 44. Convergence of TCR sequences used by these nonamer-responsive T-cell populations found in the peripheral blood and synovial fluid of numerous AS patients suggested a mechanism by which self-reactive T-cells contribute to inflammation in a B27-dependent fashion. However other studies including in discordant twin pairs found no sharing of CD4+ or CD8+ peripheral T cell receptor Vβ repertoire45. TCR involvement in AS has also been looked at from the point of view of germline variable region genes, but neither linkage nor association of the TCRA or TCRB loci have been demonstrated with AS 36,46, suggesting that these loci are not important in the familiality of the disease.
Figure 1:
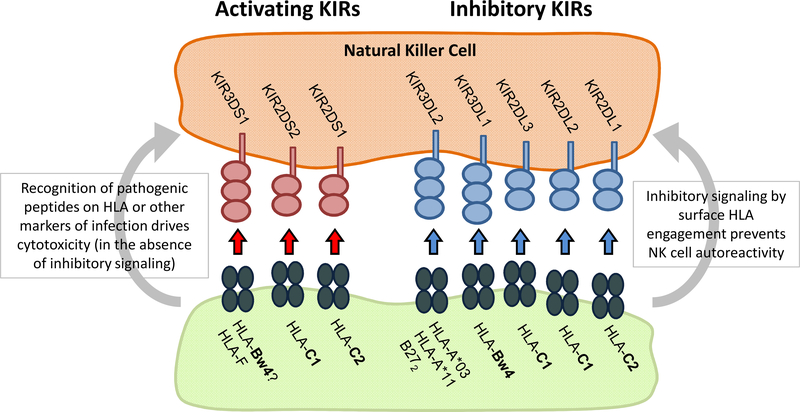
KIR gene usage and interacting partners, and their effects on NK cell activation or inhibition.
The future of immunogenetics studies in immune-mediated diseases with suspected autoreactive T-cell involvement will be in the high resolution profiling of T-cell populations. Although yet to be conducted in an AS cohort, new ‘immunosequencing’ techniques applied to a diversity of conditions, including juvenile idiopathic arthritis (JIA), has provided insight into the underlying nature of T-cell responses in disease 47. The JIA study demonstrated a restricted TCRB repertoire in the peripheral blood and synovial fluid regulatory T-cell (Treg) population of patients, with patient sharing of expanded clonotypes lacking in healthy children. Results suggested either appropriate but inefficient control of inflammatory processes by Tregs, that are typically an immensely diverse T-cell population, or perhaps pathogenicity of the expanded clones in disease. Clonal restriction of relevant T-cell populations in AS may very well also be detected with the profiling of hundreds of thousands of T-cell receptors in this fashion, providing support for the arthritogenic peptide model of disease.
Pleiotropy and Ankylosing Spondylitis
AS frequently co-occurs in individuals and families with psoriasis and inflammatory bowel disease, potentially because of shared genetic or environmental risk factors, or both. Utilizing a genotyping chip targeting immunogenetically important loci 48, the extensive role of genetic pleiotropy in these clinical associations has been demonstrated 24. This ‘cross-disease’ study showed extensive co-heritability of AS with both ulcerative colitis and Crohn’s disease, and with psoriasis albeit to a lesser extent. Utilizing this data, the investigators identified an additional 17 genome-wide significant AS-associated loci, and 65 loci associated at genome-wide significance with combinations of diseases (pleiotropic loci).
The study further strengthened the evidence of the role of the IL-23 pathway in AS pathogenesis, building on the finding of the association of IL23R variants with the disease, which initiated the development of IL-23 pathway inhibitors for the disease 12. While the exact functional mechanisms underpinning most genetic associations with AS are not yet fully understood, a high proportion of AS-associated genes influence the IL-23 pathway. At some loci multiple disease-associated variants have been identified, often with differential associations with different diseases. For example, Ellinghaus et al. identified four independent associations at IL23R, and additional variants have been identified as the primary variants associated with other immune-mediated diseases such as Behçet’s syndrome, psoriatic arthritis, and Vogt-Koyanagi-Harada syndrome 49. This suggests that differences in transcriptional regulation such as tissue specificity, or response to particular stimuli, underlie how these genes operate to cause clinically distinct diseases. Functional analysis of such variants will provide important information about the role of particular genes in disease, and inform therapeutic targeting of such genes. An excellent recent example of this has been the functional and immunogenetic dissection of the RUNX3 locus, known to be associated with AS but also celiac disease, psoriasis50 and psoriatic arthritis 51, and is a potential therapeutic target for these diseases.
Other novel gene pathways identified in the cross-disease study include:
DNA methylation: DNA methyltransferase 3a and 3b (DNMT3A, DNM3TB) are de novo methyltransferases known to be involved in genomic imprinting and X-chromosome inactivation, to influence haematopoietic stem cell development, and activation of UBE2 ubiquitin ligases, a family of genes also known to be AS-associated. The known functions of these genes raise the hypothesis that they may be involved in the male gender bias in AS, which remains unexplained.
Gut mucosal immunology: The AS-associated gene FUT2 encodes a fucosyl transferase that determines the ability to secrete blood group antigens into body fluids. This has a major effect on the gut microbiome providing further evidence that AS is a disease caused by interaction between an abnormal gut microbiome and the host immune system 52.
JAK (Janus kinase) signaling: JAK2 is the tyrosine kinase that signals from IL-23R, and is the only AS-associated JAK 24. It was therefore predictable that agents such as tofacitinib which primarily target other JAK proteins would be only moderately effective in AS 53, and suggests that more JAK2 specific inhibitors should undergo clinical trials in the disease.
Toll-like receptor signaling: The association of TLR4 with AS, which drives innate immune reactions, particularly to lipopolysaccharide (a key bacterial cell wall component), provides another non-HLA-B27 dependent pathway involved in causing the disease 24.
These findings demonstrate that further hypothesis-free genetic studies are warranted in AS; although many of the genes identified in recent studies lie in established pathways, new pathways are still being identified through new gene discoveries.
Killer Immunoglobulin-like Receptors (KIR)
Another group of genes with suspected relevance in many immune-mediated diseases are the killer immunoglobulin like receptors (KIRs) encoded within the lymphocyte receptor complex on chromosome 19. KIR disease associations have largely gone undetected by genetic studies because the biological consequences of receptor signaling on immune responses is heavily dependent on a multitude of factors that are highly variable in the population. The KIR locus is immensely polymorphic, encoding variable combinations of 17 different receptors that transduce either excitatory or inhibitory signals to natural killer (NK) and T-cells upon engagement with specific HLA class 1 or HLA-like ligands (Figure 2). Given the large degree of allelic and expression variability that exists at each gene, and that the independent HLA background of an individual governs the compatibility of receptor-ligand engagements, KIR-mediated immune response are uniquely shaped in each individual (reviewed in 54) and thus difficult to study collectively. KIR associations with AS are varied. One disease model proposes that the ability of KIR3DL2 to recognize abnormal HLA-B27 cell surface homodimers is tied to pathogenicity 55; others suggest that the balance of inhibitory and excitatory KIR receptors in AS patients is relevant in skewing inflammatory NK cell responses in disease.
Figure 2:
T-cell receptor gene rearrangement pathway determining T-cell receptor usage by individual T-cells.
Recent molecular studies have demonstrated that the KIR3DL2 receptor is up-regulated on activated CD4+ T-cells. It has been previously demonstrated that CD4+/KIR3DL2+ T-cells are found in increased numbers in AS patients relative to healthy individuals 31,56; the same research team found them to be expressed in the terminal ileum of early SpA patients 57. Engagement of KIR3DL2 with HLA-B27 homodimers or free heavy chains has been shown to provide a survival signal to these cell populations and to promote differentiation into Thelper 17 (Th17) cells, which secrete the pro-inflammatory cytokine IL-17 found at increased levels in AS patients31. The studies reporting these findings have been small, involved patients with spondyloarthropathies other than AS, and have not been replicated58. Only KIR3DL2 and KIR3DL1 usage has been studied in AS to date. A study of KIR3DL2 genetic variation found no association with AS 59. Given the potential significance of the studies of KIR3DL2 with AS, there is a clear need for independent replication of this finding, and a more comprehensive survey of KIR usage should be performed.
Genetic studies looking to compare gene dosages of differing KIR receptors in AS cases and controls have revealed a number of other genes found more or less frequently in patients, with potential ramifications for the control of NK cell immunity (Table 3). Of relevance are differences in the frequency of genes KIR3DS1 (increased in AS cohorts), and KIR3DL1 (decreased in AS cohort)60–62 given that the latter is an inhibitory receptor known to recognize HLA-Bw4 subgroups including HLA-B27. Highly homologous KIR3DS1 encodes a lymphocyte activating receptor and has been postulated to also respond to HLA-B27 ligands; KIR3DS1 co-occurrence with HLA-Bw4 is associated with slowed progression to AIDS in HIV individuals, evidence of its immune activating potential 63. Profiling the patterns of KIR and HLA inheritance in large disease cohorts is likely to be very informative in characterizing the involvement of KIR-expressing lymphocytes in AS.
Table 3.
Association studies of KIR genes in ankylosing spondylitis.
| Study | Sample numbers | Ethnicity | KIR genes typed | Main findings |
|---|---|---|---|---|
| Lopez-Larrea C, Blanco-Gelaz MA, Torre-Alonso JC, et al. Arthritis Research & Therapy. 2006;8(4) | 71 AS patients, 105 controls (HLA-B27+ve) 55 AS patients, 75 controls (HLA-B27+ve) |
Spanish Portuguese |
KIR3DL1, KIR3DS1 |
KIR3DL1 frequency decreased in patients (Spanish P < 0.0001; Azorean P < 0.003) KIR3DS1 frequency increased in patients (Spanish P < 0.0001; Azorean P < 0.003) |
| Diaz-Pena R, Blanco-Gelaz MA, Suarez-Alvarez B, et al. Human Immunology. 2008;69(7):437–442. | 42 AS patients, 30 controls (HLA-B27+ve) 30 AS patients, 16 controls (HLA-B27+ve) |
Chinese Thai |
KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL5, KIR3DL1 |
KIR3DS1, KIR2DS5 and KIR2DL5 frequencies increased in patients (Chinese P < 0.005, P < 0.001, p< 0.01 respectively; Thai P < 0.05, P < 0.05, P < 0.05 respectively) KIR3DL1/KIR3DL1 frequency decreased in patients (Chinese P < 0.05; Thai P < 0.05) KIR3DL1/KIR3DS1 frequency increased in patients (Chinese P < 0.005; That P < 0.05) |
| Jiao YL, Ma CY, Wang LC, et al. Journal of Clinical Immunology. 2008;28(4):343–349. | 119 AS patients, 128 controls | Chinese |
KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR3DL1, KIR3DL2, KIR3DL3, KIR2DP1, KIR3DP1 |
KIR3DS1 and KIR2DL5 frequencies increased in patients (P = 0.016 and P = 0.003 respectively) HLA-Cw02 frequency increased in patients (P = 0) Genotype HLA-Cw02/KIR2DS1 frequency increased in patients (P = 0.011) |
| Harvey D, Pointon JJ, Sleator C, et al. Annals of the Rheumatic Diseases. 2009;68(4):595–598. | 200 AS patients, 405 controls (KIR typing) 368 AS patients, 366 controls (KIR3DL2 typing) |
Caucasian |
KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR3DL1, KIR3DL2, KIR3DL3 |
No significant difference between KIR gene or KIR3DL2 allele frequencies between patients and controls. |
| Jiao YL, Zhang BC, You L, et al. Journal of Clinical Immunology. 2010;30(6):840–844. | 115 AS patients, 119 controls (HLA-B27+ve) | Chinese |
KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR3DL1, KIR3DL2, KIR3DL3, KIR2DP1, KIR3DP1 |
KIR2DL1 and KIR2DL5 frequencies increased in patients (P = 0.012 and P = 0.009 respectively) HLA-Cw08 frequency increased in patients (P = 0.001) |
| Zvyagin IV, Mamedov IZ, Britanova OV, et al. Cellular and Molecular Immunol. 2010;7(6):471–476. | 83 AS patients, 107 controls (HLA-B27+ve) | Russian and Caucasian |
KIR3DL1 (functional), KIR3DL1*004 (nonfunctional), KIR3DL1*005 (lowly expressed), KIR3DL1*007 (lowly expressed) KIR3DS1 |
KIR3DL1 frequency decreased in patients (P < 0.01) KIR3DS1 frequency increased in patients (P < 0.01) KIR3DL1/KIR3DL1 frequency decreased in patients (P = 0.005) KIR3DL1/KIR3DS1 frequency increased in patients (P = 0.01) KIR3DL1*F (functional alleles) frequency decreased in patients (P = 0.005) |
| Mahmoudi M, Jamshidi AR, Karami J, et al. Iranian Journal of Allergy, Asthma and Immunology. 2016;15(1):27–38 | 200 AS patients, 200 controls | Iranian |
KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DS1, KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4 KIR2DL5, KIR3DL1, KIR3DL2, KIR3DL3 KIR2DP1, KIR3DP1 |
KIR2DL3 frequency increased is patients (P = 0.005) KIR2DL5 frequency decreased in patients (P = 0.03) HLA-C2 group and HLA-B27 frequency increased in patients KIR2DL1+/HLA-CWLys+, KIR2DL2−/HLA-CWasp80−, KIR2DL3+/ HLA-CWasp80−, KIR2DS1+/HLA-CWLys+, KIR2DS2−/HLA-CWasp80− frequencies increased in patients (P = 0.0009, P = 0.01, P = 0.0008, P = 0.009, P = 0.002 respectively) KIR2DL1+/HLA-CWLys−, KIR2DL2+/HLA-CWasp80+, KIR2DL3−/HLA-CWasp80+, KIR3DL1+/HLA-Bw4Thr+, KIR2DS1−/HLA-CWLys−, KIR2DS1+/HLA-CWLys−, KIR2DS2+/HLA-CWasp80+ frequencies decreased in patients (P = 0.00008, P = 0.01, P = 0.002, P = 0.07, P = 0.02, P = 0.006, P = 0.004 respectively) |
Summary
Genetic studies have identified multiple different pathways involved in AS, and as more genes are being identified, more pathways are being uncovered. These findings are helping fill in the mystery about how HLA-B27 is involved in AS, and identifying new potential therapeutic targets. While much more will be learned from further hypothesis free studies in AS genomics, there is the research need now needs to transition increasingly to functional genomics studies to determine the mechanisms underpinning these associations and to turn the genetic discoveries into new treatments, as they have already with regard to IL-23 pathway inhibition in AS.
KEY POINTS.
Ankylosing spondylitis (AS) is a common, highly heritable inflammatory arthritis for which thus far 113 genetic associations have been identified.
HLA-B27 contributes ~20% of the heritability of AS, and non-MHC loci identified to date contribute another ~10%.
The HLA associations of AS are complex and multiple non-B27 HLA alleles have been identified as being involved.
Key pathways identified by AS genetic studies include the IL-23 and M1-aminopeptidase pathways, but multiple other pathways have been identified as increasing numbers of associations have been identified.
Preliminary evidence suggesting involvement of KIR genes in AS pathogenesis needs replication in other cohorts.
SYNOPSIS.
Ankylosing spondylitis (AS) is a common inflammatory arthritis in which genetic factors are the primary determinants of disease risk, and of disease activity and severity. Primarily through linkage disequilibrium mapping substantial progress has been made in identifying genetic pathways involved in the disease, leading to the development of IL-23 pathway inhibitors for AS, and other drug development programs, for example targeting aminopeptidase genes. Multiple other disease pathways have been identified through gene-mapping studies including pathways involving control of DNA methylation, bacterial sensing, and mucosal immunity. Additional pathways will likely be identified as a higher proportion of the genetic risk of AS is determined.
Acknowledgments
MAB has received research support, acted as an advisor or as part of the speakers’ bureau of Abbvie, Janssen, Pfizer and UCB.
Footnotes
DISCLOSURE STATEMENT
AH has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Aimee Hanson, Research Fellow, University of Queensland Diamantina Institute, Translational Research Institute, Princess Alexandra Hospital, Woolloongabba, Brisbane, Queensland, Australia..
Matthew A. Brown, Professor, Institute of Health and Biomedical Innovation, Queensland University of Technology, Translational Research Institute, Princess Alexandra Hospital, Woolloongabba, Brisbane, Queensland, Australia.
References
- 1.Brown MA, Kennedy LG, Darke C, et al. The effect of HLA-DR genes on susceptibility to and severity of ankylosing spondylitis. Arthritis Rheum. 1998;41(3):460–465. [DOI] [PubMed] [Google Scholar]
- 2.Milicic A, Lindheimer F, Laval S, et al. Interethnic studies of TNF polymorphisms confirm the likely presence of a second MHC susceptibility locus in ankylosing spondylitis. Genes Immun. 2000;1(7):418–422. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola E, Herzberg I, Laiho K, et al. Finnish HLA studies confirm the increased risk conferred by HLA-B27 homozygosity in ankylosing spondylitis. Ann Rheum Dis. 2006;65(6):775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Said-Nahal R, Miceli-Richard C, Gautreau C, et al. The role of HLA genes in familial spondyloarthropathy: a comprehensive study of 70 multiplex families. Ann Rheum Dis. 2002;61(3):201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sims AM, Barnardo M, Herzberg I, et al. Non-B27 MHC associations of ankylosing spondylitis. Genes Immun. 2007;8(2):115–123. [DOI] [PubMed] [Google Scholar]
- 6.Robinson WP, van der Linden SM, Khan MA, et al. HLA-Bw60 increases susceptibility to ankylosing spondylitis in HLA-B27+ patients. Arthritis Rheum. 1989;32(9):1135–1141. [DOI] [PubMed] [Google Scholar]
- 7.Brown MA, Pile KD, Kennedy LG, et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis. 1996;55(4):268–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortes A, Pulit SL, Leo PJ, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun. 2015;6:7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K, Bang SY, Lee S, et al. An HLA-C amino-acid variant in addition to HLA-B*27 confers risk for ankylosing spondylitis in the Korean population. Arthritis Res Ther. 2015;17:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taurog JD, Maika SD, Satumtira N, et al. Inflammatory disease in HLA-B27 transgenic rats. Immunol Rev. 1999;169:209–223. [DOI] [PubMed] [Google Scholar]
- 11.Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44(3):291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellcome Trust Case Control C, Australo-Anglo-American Spondylitis C, Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Genetics of Ankylosing Spondylitis C, Cortes A, Hadler J, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45(7):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nature Genetics. 2010;42(11):985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirino Y, Bertsias G, Ishigatsubo Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45(2):202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiper JJ, Van Setten J, Ripke S, et al. A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum Mol Genet. 2014;23(22):6081–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson PC, Costello ME, Leo P, et al. ERAP2 is associated with ankylosing spondylitis in HLA-B27-positive and HLA-B27-negative patients. Ann Rheum Dis. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves E, Colebatch-Bourn A, Elliott T, Edwards CJ, James E. Functionally distinct ERAP1 allotype combinations distinguish individuals with Ankylosing Spondylitis. Proc Natl Acad Sci U S A. 2014;111(49):17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson PC, Brown MA. ERAP1 biology and assessment in Ankylosing Spondylitis. Proc Natl Acad Sci U S A. 2015;112(15):E1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsui FW, Haroon N, Reveille JD, et al. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann Rheum Dis. 2010;69(4):733–736. [DOI] [PubMed] [Google Scholar]
- 21.Davidson SI, Liu Y, Danoy PA, et al. Association of STAT3 and TNFRSF1A with ankylosing spondylitis in Han Chinese. Annals of the rheumatic diseases. 2011;70(2):289–292. [DOI] [PubMed] [Google Scholar]
- 22.Pimentel-Santos FM, Ligeiro D, Matos M, et al. Association of IL23R and ERAP1 genes with ankylosing spondylitis in a Portuguese population. Clin Exp Rheumatol. 2009;27(5):800–806. [PubMed] [Google Scholar]
- 23.Bang SY, Kim TH, Lee B, et al. Genetic Studies of Ankylosing Spondylitis in Koreans Confirm Associations with ERAP1 and 2p15 Reported in White Patients. J Rheumatol. 2010. [DOI] [PubMed] [Google Scholar]
- 24.Ellinghaus D, Jostins L, Spain SL, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102(47):17107–17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saveanu L, Carroll O, Lindo V, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6(7):689–697. [DOI] [PubMed] [Google Scholar]
- 27.Evnouchidou I, Weimershaus M, Saveanu L, van Endert P. ERAP1-ERAP2 dimerization increases peptide-trimming efficiency. J Immunol. 2014;193(2):901–908. [DOI] [PubMed] [Google Scholar]
- 28.Sanz-Bravo A, Campos J, Mazariegos MS, Lopez de Castro JA. Dominant role of the ERAP1 polymorphism R528K in shaping the HLA-B27 Peptidome through differential processing determined by multiple peptide residues. Arthritis & rheumatology. 2015;67(3):692–701. [DOI] [PubMed] [Google Scholar]
- 29.Benjamin R, Parham P. HLA-B27 and disease: a consequence of inadvertent antigen presentation? Rheum Dis Clin North Am. 1992;18(1):11–21. [PubMed] [Google Scholar]
- 30.Kenna TJ, Brown MA. Immunopathogenesis of ankylosing spondylitis. Int J Clin Rheumatol. 2013;8(2):265–274. [Google Scholar]
- 31.Chen L, Ridley A, Hammitzsch A, et al. Silencing or inhibition of endoplasmic reticulum aminopeptidase 1 (ERAP1) suppresses free heavy chain expression and Th17 responses in ankylosing spondylitis. Ann Rheum Dis. 2016;75(5):916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenna TJ, Lau MC, Keith P, et al. Disease-associated polymorphisms in ERAP1 do not alter endoplasmic reticulum stress in patients with ankylosing spondylitis. Genes Immun. 2015;16(1):35–42. [DOI] [PubMed] [Google Scholar]
- 33.Tran TM, Hong S, Edwan JH, Colbert RA. ERAP1 reduces accumulation of aberrant and disulfide-linked forms of HLA-B27 on the cell surface. Mol Immunol. 2016;74:10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evnouchidou I, Kamal RP, Seregin SS, et al. Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. Journal of immunology. 2011;186(4):1909–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kochan G, Krojer T, Harvey D, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(19):7745–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans DM, Spencer CC, Pointon JJ, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43(8):761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zervoudi E, Saridakis E, Birtley JR, et al. Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses. Proc Natl Acad Sci U S A. 2013;110(49):19890–19895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.May E, Dulphy N, Frauendorf E, et al. Conserved TCR beta chain usage in reactive arthritis; evidence for selection by a putative HLA-B27-associated autoantigen. Tissue Antigens. 2002;60(4):299–308. [DOI] [PubMed] [Google Scholar]
- 39.de Paula Alves Sousa A, Johnson KR, Nicholas R, et al. Intrathecal T-cell clonal expansions in patients with multiple sclerosis. Ann Clin Transl Neurol. 2016;3(6):422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mamedov IZ, Britanova OV, Chkalina AV, et al. Individual characterization of stably expanded T cell clones in ankylosing spondylitis patients. Autoimmunity. 2009;42(6):525–536. [DOI] [PubMed] [Google Scholar]
- 41.Dulphy N, Peyrat M-A, Tieng V, et al. Common Intra-Articular T Cell Expansions in Patients with Reactive Arthritis- Identical b-Chain Junctional Sequences and Cytotoxicity Toward HLA-B27. The Journal of Immunology. 1998;162:3830–3839. [PubMed] [Google Scholar]
- 42.DerSimonian H, Sugita M, Glass DN, et al. Clonal Vctl2.1 + T Cell Expansions in the Peripheral Blood of Rheumatoid Arthritis Patients. Journal of Experimental Medicine. 1993;177:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duchmann R, Lambert C, May E, Hohler T, Marker-Hermann E. CD4+ and CD8+ clonal T cell expansions indicate a role of antigens in ankylosing spondylitis; a study in HLA-B27+ monozygotic twins. Clinical and experimental immunology. 2001;123(2):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frauendorf E, von Goessel H, May E, Marker-Hermann E. HLA-B27-restricted T cells from patients with ankylosing spondylitis recognize peptides from B*2705 that are similar to bacteria-derived peptides. Clin Exp Immunol. 2003;134(2):351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hohler T, Hug R, Schneider PM, et al. Ankylosing spondylitis in monozygotic twins: studies on immunological parameters. Annals of the rheumatic diseases. 1999;58(7):435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown MA, Rudwaleit M, Pile KD, et al. The role of germline polymorphisms in the T-cell receptor in susceptibility to ankylosing spondylitis. Br J Rheumatol. 1998;37(4):454–458. [DOI] [PubMed] [Google Scholar]
- 47.Henderson LA, Volpi S, Frugoni F, et al. Next Generation Sequencing Reveals Restriction and Clonotypic Expansion of Regulatory T Cells in Juvenile Idiopathic Arthritis. Arthritis & rheumatology. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou S, Du L, Lei B, et al. Genome-wide association analysis of Vogt-Koyanagi-Harada syndrome identifies two new susceptibility loci at 1p31.2 and 10q21.3. Nature genetics. 2014;46(9):1007–1011. [DOI] [PubMed] [Google Scholar]
- 50.Vecellio M, Roberts AR, Cohen CJ, et al. The genetic association of RUNX3 with ankylosing spondylitis can be explained by allele-specific effects on IRF4 recruitment that alter gene expression. Ann Rheum Dis. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apel M, Uebe S, Bowes J, et al. Variants in RUNX3 contribute to susceptibility to psoriatic arthritis, exhibiting further common ground with ankylosing spondylitis. Arthritis Rheum. 2013;65(5):1224–1231. [DOI] [PubMed] [Google Scholar]
- 52.Costello ME, Ciccia F, Willner D, et al. Intestinal dysbiosis in ankylosing spondylitis. Arthritis & rheumatology. 2014. [DOI] [PubMed] [Google Scholar]
- 53.van der Heijde D, Deodhar A, Wei J, et al. Tofacitinib in Patients with Ankylosing Spondylitis: A Phase 2, 16-Week, Randomized, Placebo-Controlled, Dose-Ranging Study. . Arthritis Rheumatol. 2015;67 (Suppl 10):5L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajalingam R Human diversity of killer cell immunoglobulin-like receptors and disease. The Korean journal of hematology. 2011;46(4):216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kollnberger S, Bird L, Sun MY, et al. Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheum. 2002;46(11):2972–2982. [DOI] [PubMed] [Google Scholar]
- 56.Chan AT, Kollnberger SD, Wedderburn LR, Bowness P. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondylarthritis. Arthritis Rheum. 2005;52(11):3586–3595. [DOI] [PubMed] [Google Scholar]
- 57.Rysnik O, McHugh K, van Duivenvoorde L, et al. Non-conventional forms of HLA-B27 are expressed in spondyloarthritis joints and gut tissue. J Autoimmun. 2016;70:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jansen DT, Hameetman M, van Bergen J, et al. IL-17-producing CD4+ T cells are increased in early, active axial spondyloarthritis including patients without imaging abnormalities. Rheumatology (Oxford). 2015;54(4):728–735. [DOI] [PubMed] [Google Scholar]
- 59.Harvey D, Pointon JJ, Sleator C, et al. Analysis of killer immunoglobulin-like receptor genes in ankylosing spondylitis. Ann Rheum Dis. 2009;68(4):595–598. [DOI] [PubMed] [Google Scholar]
- 60.Diaz-Pena R, Blanco-Gelaz MA, Suarez-Alvarez B, et al. Activating KIR genes are associated with ankylosing spondylitis in Asian populations. Hum Immunol. 2008;69(7):437–442. [DOI] [PubMed] [Google Scholar]
- 61.Lopez-Larrea C, Blanco-Gelaz MA, Torre-Alonso JC, et al. Contribution of KIR3DL1/3DS1 to ankylosing spondylitis in human leukocyte antigen-B27 Caucasian populations. Arthritis Res Ther. 2006;8(4):R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zvyagin IV, Mamedov IZ, Britanova OV, et al. Contribution of functional KIR3DL1 to ankylosing spondylitis. Cell Mol Immunol. 2010;7(6):471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434. [DOI] [PubMed] [Google Scholar]