Abstract
Purpose
We aim to reveal the disease-causing mutations in 15 Chinese families with optic atrophy (OA).
Methods
In total, 15 families with OA were recruited in the present study. Medical histories were carefully reviewed and comprehensive ophthalmic examinations were received by all recruited patients. Targeted next-generation sequencing (NGS) was selectively performed on all probands for mutation detection. Intrafamilial cosegregation and in-silico analyses were subsequently applied to predict the potential pathogenic effects of identified mutations.
Results
All included patients presented bilateral vision loss. Their fundus photographs showed temporal or total pallor of the optic discs. Fourteen mutations in the optic atrophy 1 (OPA1) gene were revealed as disease-causing mutations for the 15 families, including eight novel (c.968A>G, c.193C>G, c.1071dupT, c.987_988del, c.2012+2T>G, c.1036–1G>C, c.2126A>G, and c.1036_1038del) and six recurrent (c.1499G>A, c.1800C>A, c.1034G>A, c.2873_2876del, c.112C>T, and c.804_805del) mutations.
Conclusions
In conclusion, our study expands the mutational spectrum for the OPA1 gene and implies targeted NGS as an effective approach for the genetic diagnosis of OA, which might help to improve the clinical diagnosis for patients with OA.
Introduction
Inherited optic atrophy (OA) is characterized by the degeneration of retinal ganglion cells (RGCs) and other neuronal populations, which results in thinning of the retinal nerve fiber layer (RNFL) [1]. Typical clinical features of OA include progressive loss of bilateral vision (usually occurs within the first decade), central or paracentral scotomas, tritanopia, and pallor of the optic discs [1-3]. Familial history, temporal or diffuse pallor of the optic discs disclosed by eye fundus, and a reduced amplitude of the waveform detected by visually evoked potentials (VEP) may contribute to the clinical diagnosis of OA [2]. According to the inheritance pattern, OA can be categorized into autosomal dominant optic atrophy (ADOA) and mitochondrial inherited Leber hereditary optic neuropathy (LHON). ADOA, showing a worldwide prevalence of 1:50,000, is the most common form of inherited optic neuropathy [3].
ADOA shows genetic heterogeneity [2,4]. To date, two genes, optic atrophy 1 (OPA1, OMIM_605290) and OPA3 (OMIM_606580), and three loci, OPA4 (OMIM _605293), OPA5 (OMIM _610708), and OPA8 (OMIM_616648), are reported to be correlated with ADOA. Among all, OPA1 is the most common causative gene for ADOA [5]. OPA1 consists of 31 coding exons, and it encodes dynamin-like guanosine triphosphates (GTPase) localized to the mitochondria. Protein encoded by OPA1 targets the external face of the mitochondrial inner membrane, controls the structural integrity of mitochondrial cristae, and keeps their junctions tight during apoptosis [6]. To date, over 400 variants in OPA1 have been identified (mitodyn). About 20% of all identified variants associate with the “ADOA plus” syndrome [7,8]. Patients with “ADOA plus” syndrome present OA in childhood, followed by the subsequent onset of chronic progressive external ophthalmoplegia (PEO), ptosis, sensorineural deafness, peripheral neuropathy, and myopathy in adult life [9]. Most reported OPA1 mutations are missense mutations located in the GTPase domain and dynamin central region, which interrupt the protein function [2,10].
Next-generation sequencing (NGS), an approach that enables sequencing of a panel of candidate genes, has been revealed as an efficient tool for the molecular diagnosis of inherited retinal dystrophies (IRDs) [2,10]. Because no effective clinical therapy has been developed for OA, NGS-based molecular diagnosis will not only help to improve its clinical diagnosis, but is also essential for its prenatal diagnosis. Herein, by means of a targeted NGS approach, we reveal novel OPA1 mutations in 15 Chinese families with OA.
Methods
Participants and clinical investigations
Our study complied with the Declaration of Helsinki, and it was approved and prospectively reviewed by the ethics committee of the First Affiliated Hospital of Nanjing Medical University (2017-SRFA-034). Written informed consent was obtained from all participants or their legal guardians before enrollment. All participants underwent routine ophthalmic examinations, including best-corrected visual acuity (BCVA), funduscopy, slit-lamp examination, and fundus photography. The VEP test was selectively performed on patients OA02-II:1, OA08-II:1, and OA12-II:1. Another 150 unrelated Chinese controls were also included, each of whom received basic ophthalmic examinations to exclude major ocular problems. Peripheral blood samples from all participants were collected in EDTA tubes. DNA extraction from leukocytes was performed using a RelaxGene Blood DNA System (Qiagen, Valencia, CA) per the manufacturer’s protocols.
Targeted gene capture, NGS, bioinformatics analyses, and Sanger sequencing
Targeted sequence capture microarrays that could capture the coding and exon-intronic boundary regions of all known retinal disease genes were used in this study. Details of the targeted genes of the commercial array have been previously described [11-13]. Sequence capture, enrichment, elution, and NGS were conducted in cooperation with BGI-Shenzhen or MyGenostics-Beijing, as previously stated [14,15].
All detected variants were further filtered against the following six single nucleotide polymorphism (SNP) databases, including the dbSNP144, HapMap project, 1000 Genome Project, YH database, Exon Variant Server, and ExAC databases. Variants with a minor allele frequency value of over 0.001 were discarded. Sanger sequencing was subsequently performed for a mutation validation, intrafamilial genotype–phenotype cosegregation analysis, and prevalence test in 150 controls. Information of primers are listed in Appendix 1.
In-silico analyses
Evolutionary conservation of the mutated amino acids was analyzed through the alignment of OPA1 orthologous protein sequences from the following species: Homo sapiens (ENSP00000354681), Pan troglodytes (ENSPTRP00000027094), Bos taurus (ENSBTAP00000026013), Mus musculus (ENSRNOP00000002338), Gallus gallus (ENSGALP00000042204), Danio rerio (ENSDARP00000095031), Drosophila melanogaster (FBpp0086700), and Caenorhabditis elegans (D2013.5). Online predictive software, including SIFT [16], PolyPhen-2 [17], and PROVEN [18], was applied to evaluate the potential pathogenicity of identified mutations. The Splicing Regulation Online Graphical Engine (SROOGLE) online prediction software was used to determine whether splice site variations would alter the regular splicing sites.
Results
Clinical manifestations
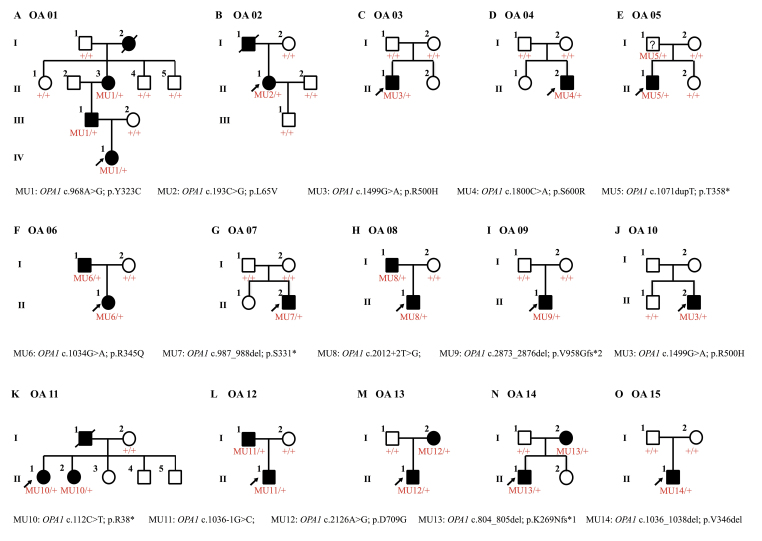
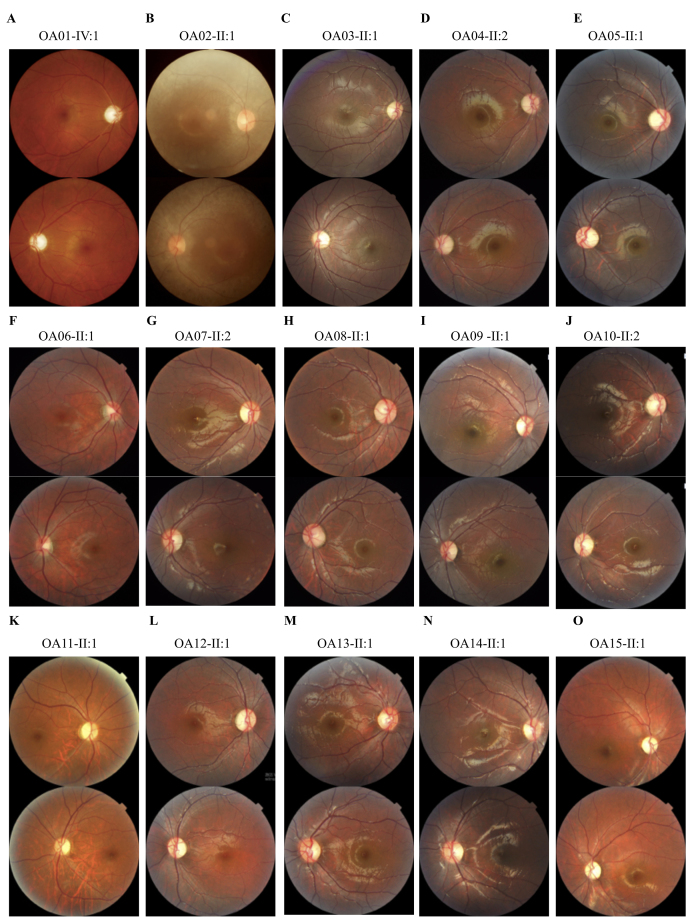
Family pedigrees are shown in Figure 1. All included patients presented typical OA symptoms, and their detailed clinical data are summarized in Table 1. Briefly, the onset ages varied from infant to 12 years old. Patient OA02-II:2 had vision defects since infancy, while patients from families OA06 and OA11 were completely unaffected until 10 to 12 years old. Most patients reported having bilateral visual impairments since their first decade of life, which was consistent with previous reports. The disease progression also varied. For example, patients OA06-I:1 and OA11-II:1 shared a similar age and onset age. However, the BCVAs for patient OA06-I:1 are 0.5 OD and 0.4 OS, while patient OA11-II:1 had much poorer visual conditions (0.1 OU). Rapid disease progression was also noticed in patient OA02-II:2. She also presented with asymmetric visual defects, with her BCVAs being 0.25 OD and 0.02 OS. Most patients had a relatively slow and stable disease progression. Nystagmus was noticed in all three patients from family OA01. Fundus presentations of all patients demonstrated bilateral optic nerve pallor (Figure 2). VEP results were attainable in five patients, including OA02-II:2, OA08-I:1, OA08-II:1, OA12-I:1, and OA12-II:1. All patients showed bilateral diminished VEP presentations. None of the patients presented systemic abnormalities.
Figure 1.
Family pedigrees. The sex (male, square; female, circle), phenotype (unaffected, empty symbol; clinically affected, black-filled symbol), and genotype (unaffected, +/+; genetically affected, MU/+) of all recruited members are shown. Probands are indicated by arrows.
Table 1. Clinical features of recruited patients.
| Patient ID | Age (years) /Sex | Onset Age (years) |
BCVA (logMAR) |
Optic disc | VEP | |
|---|---|---|---|---|---|---|
| OD | OS | |||||
| OA01-II:3 |
60/F |
3 |
0.02 |
0.01 |
Pale |
NA |
| OA01-III:1 |
38/M |
8 |
0.1 |
0.1 |
Pale |
NA |
| OA01-IV:1 |
16/F |
5 |
0.05 |
0.05 |
Pale |
NA |
| OA02-II:2 |
30/F |
Infant |
0.25 |
0.02 |
Pale |
Diminished |
| OA03-II:1 |
8/M |
3 |
0.2 |
0.3 |
Pale |
NA |
| OA04-II:2 |
10/M |
6 |
0.5 |
0.5 |
Pale |
NA |
| OA05-II:1 |
10/M |
6 |
0.4 |
0.5 |
Pale |
NA |
| OA06-I:1 |
42/M |
12 |
0.5 |
0.4 |
Pale |
NA |
| OA06-II:1 |
14/F |
11 |
0.2 |
0.1 |
Pale |
NA |
| OA07-II:2 |
9/M |
4 |
0.4 |
0.3 |
Pale |
NA |
| OA08-I:1 |
32/M |
8 |
0.3 |
0.3 |
Pale |
Diminished |
| OA08-II:1 |
9/M |
5 |
0.5 |
0.4 |
Pale |
Diminished |
| OA09-II:1 |
8/M |
5 |
0.1 |
0.1 |
Pale |
NA |
| OA10-II:2 |
9/M |
7 |
0.2 |
0.2 |
Pale |
NA |
| OA11-II:1 |
43/F |
12 |
0.1 |
0.1 |
Pale |
NA |
| OA11-II:2 |
42/F |
10 |
0.1 |
0.1 |
Pale |
NA |
| OA12-I:1 |
30/M |
8 |
0.3 |
0.3 |
Pale |
Diminished |
| OA12-II:1 |
6/M |
4 |
0.3 |
0.3 |
Pale |
Diminished |
| OA13-I:2 |
31/F |
6 |
0.4 |
0.4 |
Pale |
NA |
| OA13-II:1 |
7/M |
4 |
0.3 |
0.3 |
Pale |
NA |
| OA14-I:2 |
28/F |
6 |
0.3 |
0.4 |
Pale |
NA |
| OA14-II:1 |
6/M |
3 |
0.25 |
0.3 |
Pale |
NA |
| OA15-II:1 | 14/M | 6 | 0.2 | 0.2 | Pale | NA |
Abbreviations: OD: right eye; OS: left eye; F: female; M: male; BCVA: best corrected visual acuity; VEP: visual evoked potential; NA: not available.
Figure 2.
Fundus presentations. A-O: Fundus photographs of all probands demonstrate bilateral optic nerve head pallor.
Genetic investigation
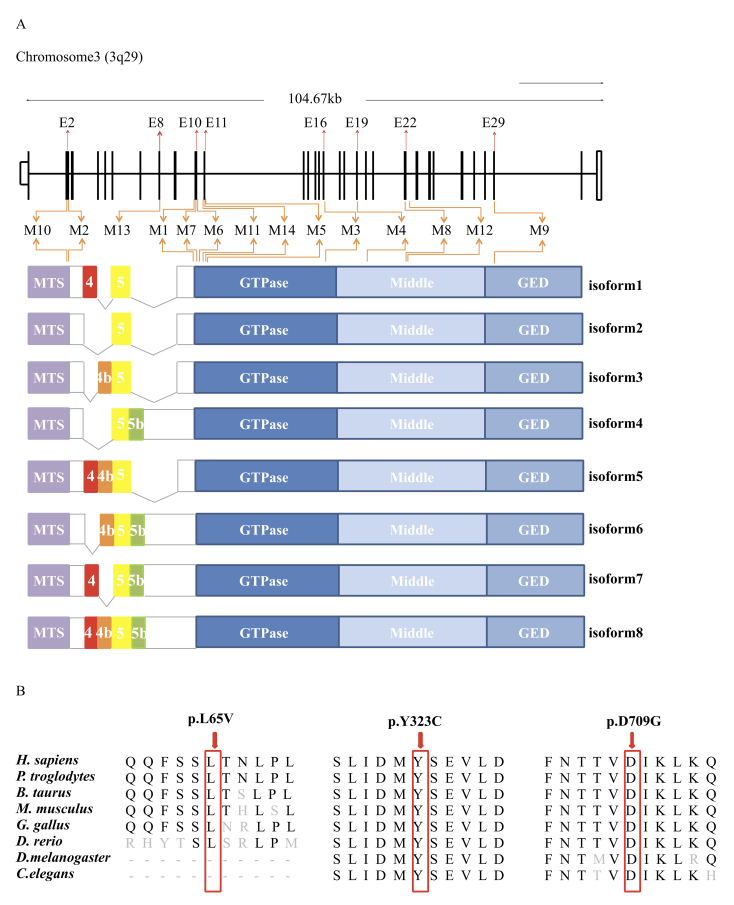
Probands from all 15 families were selected for NGS, as mentioned before. After a comprehensive genetic analysis, 14 heterozygous mutations in total in the OPA1 gene (NM_130837) were identified as potentially disease causing in the 15 families, including eight novel variants and six recurrent mutations (Figure 3 and Table 2). Eleven of the 14 mutations were located in the GTPase domain and dynamin central region (Figure 4A). All identified variants segregated the disease phenotypes in corresponding families and were absent in 150 unrelated normal controls.
Figure 3.
Chromatograms of wild-type (top) and mutant (bottom) OPA1 sequences in all recruited families.
Table 2. Characteristics of identified OPA1 mutations.
| Family ID |
Mutation |
Exon |
Bioinformatics Analysis |
Reported /Novel | MAF | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Type | Status | SIFT | PolyPhen-2 | PROVEN | ||||
| OA01 |
c.968A>G |
p.Y323C |
missense |
Het |
E10 |
DA (0.000) |
PD (1.000) |
DE (−8.03) |
Novel |
- |
| OA02 |
c.193C>G |
p.L65V |
missense |
Het |
E2 |
DA (0.016) |
B (0.278) |
N (−0.63) |
Novel |
- |
| OA03 |
c.1499G>A |
p.R500H |
missense |
Het |
E16 |
DA (0.001) |
PD (1.000) |
DE (−4.70) |
CM030379 |
- |
| OA04 |
c.1800C>A |
p.S600R |
missense |
Het |
E19 |
DA (0.001) |
PD (1.000) |
DE (−4.64) |
CM061154 |
- |
| OA05 |
c.1071dupT |
p.T358* |
nonsense |
Het |
E11 |
- |
- |
- |
Novel |
- |
| OA06 |
c.1034G>A |
p.R345Q |
missense |
Het |
E10 |
DA (0.041) |
PD (0.978) |
DE (−3.50) |
CM002636 |
- |
| OA07 |
c.987_988del |
p.S331* |
nonsense |
Het |
E10 |
- |
|
- |
Novel |
- |
| OA08 |
c.2012+2T>G |
- |
splice site |
Het |
E22–23 |
- |
|
- |
Novel |
- |
| OA09 |
c.2873_2876del |
p.V958Gfs*2 |
frameshift |
Het |
E29 |
- |
|
- |
[26] |
4/121408 |
| OA10 |
c.1499G>A |
p.R500H |
missense |
Het |
E16 |
DA (0.001) |
PD (1.000) |
DE (−4.70) |
CM030379 |
- |
| OA11 |
c.112C>T |
p.R38* |
nonsense |
Het |
E2 |
- |
- |
- |
CM024785 |
- |
| OA12 |
c.1036–1G>C |
- |
splice site |
Het |
E10–11 |
- |
- |
- |
Novel |
- |
| OA13 |
c.2126A>G |
p.D709G |
missense |
Het |
E22 |
DA (0.001) |
PD (1.000) |
DE (−6.72) |
Novel |
- |
| OA14 |
c.804_805del |
p.K269Nfs*1 |
frameshift |
Het |
E8 |
- |
- |
- |
[27] |
- |
| OA15 | c.1036_1038del | p.V346del | deletion | Het | E11 | - | - | - | Novel | - |
Abbreviations: Het: heterozygous; DA: damaging; PD: probably damaging; B: benign; DE: deleterious; N: neutral; MAF: minor allele frequency.
Figure 4.
Diagrammatic representation and conservational analyses. A: Diagrammatic representation of the 14 identified mutations in the context of genome structure (upper) and eight isoforms of the OPA1 protein (below), derived from the alternative splicing of exons 4, 4b, 5, and 5b. The OPA1 protein includes a mitochondria-targeting sequence (MTS), a GTPase domain, a middle domain, and a C-terminus GTPase effector domain (GED). B: Orthologous protein sequence alignment of OPA1 from human (H. sapiens), chimpanzees (P. troglodytes), cows (B. taurus), rats (M. musculus), chickens (G. gallus), zebrafish (D. rerio), fruit flies (D. melanogaster), and roundworms (C. elegans).
The eight novel variations comprised three missense mutations (c.193C>G in family OA02, c.968A>G in family OA01, and c.2126A>G in family OA13), one deletion (c.1036_1038del in family OA15), two nonsense mutations (c.987_988del in family OA07 and c.1071dupT in family OA05), and two splice site mutations (c.1036–1G>C in family OA12, and c.2012+2T>G in family OA08; Figure 1 and Figure 3, and Table 2). The novel missense variant c.968A>G, leading to the amino acid change from tyrosine to cysteine at residue 323 (p.Y323C), was located within the GTPase domain of the OPA1 protein (NP_570850; Figure 4A). A conservational analysis indicated that residue Tyr323 in OPA1 was evolutionarily conserved among multiple species (Figure 4B). Potential deleterious effects of this mutation was implied by all three types of online predicting software, including PROVEN (−8.03, deleterious), SIFT (0.000, damaging), and Polyphen-2 (1.000, probably damaging; Table 2). Another variant c.193C>G, causing substitution from leucine to valine at conserved residue 65 (p.L65V), was located in the basic domain of the OPA1 protein (Figure 4A,B). This variation was predicted to be damaging (0.016) by the SIFT online predicting software (Table 2). The other missense mutation, c.2126A>G, leading to the transformation from aspartic acid to glycine at residue 709 (p.D709G), was located in the dynamin central domain of the OPA1 protein (Figure 4A). Residue Asp709 was also highly conserved among all tested species (Figure 4B). The potential deleterious effects of the mutation were revealed by PROVEAN (−6.72, deleterious), SIFT (0.001, damaging), and Polyphen-2 (1.000, probably damaging).
We next used the SROOGLE online prediction software to determine whether the two splice site variations would cause abolishment of the regular splicing sites. According to our data, mutation c.1036–1G>C was found to remarkably decrease the splice site score from 4.81 to −3.26 in the Max entropy model and from 81.06 to 57.62 in the PSSM model. The regular splice site was found completely abolished by mutation c.2012+2T>G. The nonsense mutations c.987_988del and c.1071dupT would introduce immediate translation stop codons at residues 331 and 358, respectively, thus generating a truncated protein or causing nonsense-mediated mRNA decay (NMD).
Discussion
Herein, we report the identification of 14 heterozygous OPA1 variants in 15 Chinese OA families, including eight novel and six recurrent mutations. All included patients show typical OA presentation. Some of the literature suggests that gender might contribute to the severity of OA caused by an OPA1 mutation [19]. However, our data do not reveal such a correlation. The average onset age of female patients ranged from infant to 12 years old (average: 6.63 years old), and the average onset age of male patients ranged from 3 to 12 years old (average: 5.93 years old). The OA progression shows no notable difference between males and females. Of all included patients, 15 are male and eight are female. The greater number of male patients in this study is probably due to the de novo OPA1 mutations in families OA07, OA09, OA10, and OA15.
The OPA1 protein plays an important role in maintaining the mitochondrial structure and inhibiting apoptosis. The N-terminal mitochondrial localization signal sequence of OPA1 can direct protein into the mitochondria. Recent studies indicate that OPA1 proteins accumulate in the mitochondrial inner membrane and serve as anchors for mitochondrial DNA (mtDNA), contributing to its replication and distribution [20,21]. OPA1 insufficiency could disrupt oxidative phosphorylation, disturb mtDNA maintenance and replication, and further interrupt regular mitochondrial function [21]. Changes in mitochondrial genome stability would further cause ATP insufficiency, abnormal cellular function, apoptosis, and ADOA phenotype. Two frameshift, two splice-site, and three nonsense mutations are identified in this study, which are predicted to generate truncated OPA1 proteins or lead to NMD. According to previous reports, the nonsense mutation OPA1 Q285STOP would cause NMD in a murine model. This partial loss of OPA1 causes mitochondrial respiratory deficiency and a substantial resistance to endoplasmic reticulum stress-induced death [22,23]. Another frameshift mutation, OPA1 329_355del, is also found to cause a 50% reduction in the OPA1 protein in a murine model [24]. The OPA1329_355del mice are found to develop visual dysfunction due to RGC loss and an ascending optic neuropathy, while the earliest changes detected in OPA1Q285STOP mice are RGC dendritic changes leading to dysfunctional RGCs. However, it is still unclear why the two murine models display such different phenotypes. The seven heterozygous missense OPA1 mutations identified in this study are more likely to exert a dominant-negative or deleterious gain-of-function effect [25]. More experiments are still warranted to understand better the pathogenesis of OPA1 mutations.
Sometimes, clinical diagnoses are challenging in young patients with non-fully manifested phenotypes or due to the clinical heterogeneity with which hereditary retinal diseases manifest. Therefore, genetic tests should be kept be continuously researched to aid unclear clinical diagnoses and to prognosticate the disease. Genetic diagnosis also promises gene therapy or other forms of gene-specific treatments. In conclusion, we revealed eight novel and six recurrent mutations in the OPA1 gene in 15 Chinese OA families. Our findings expand the OPA1 mutational spectrum, enrich their phenotype–genotype correlations, provide crucial hints for genetic consultation in these families, and further help with better clinical management.
Acknowledgments
We thank all patients and their family members for their valuable contribution to this study. We also thank EENT Biobank for sample collection and management. This work is supported by the National Natural Science Foundation of China (81525006, 81670864 and 81730025 to C.Z., and 81700877 to X.C.); Natural Science Foundation of Jiangsu Province (BK20171087 to X.C.); Shanghai Outstanding Academic Leaders (2017BR013 to C.Z.); Open Foundation of State Key Laboratory of Reproductive Medicine (Nanjing Medical University, SKLRM-KA201607 to X.C.) and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. The co-corresponding authors are Min Wang, email: wangmin83@yahoo.com, Chen Zhao, email: dr_zhaochen@163.com, and Xue Chen, email: drcx1990@vip.163.com.
Appendix 1. Primers used in this study.
To access the data, click or select the words “Appendix 1.”
References
- 1.Chun BY, Rizzo JF., 3rd Dominant optic atrophy: updates on the pathophysiology and clinical manifestations of the optic atrophy 1 mutation. Curr Opin Ophthalmol. 2016;27:475–80. doi: 10.1097/ICU.0000000000000314. [DOI] [PubMed] [Google Scholar]
- 2.Amati-Bonneau P, Milea D, Bonneau D, Chevrollier A, Ferre M, Guillet V, Gueguen N, Loiseau D, de Crescenzo MA, Verny C, Procaccio V, Lenaers G, Reynier P. OPA1-associated disorders: phenotypes and pathophysiology. Int J Biochem Cell Biol. 2009;41:1855–65. doi: 10.1016/j.biocel.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Millet AM, Bertholet AM, Daloyau M, Reynier P, Galinier A, Devin A, Wissinguer B, Belenguer P, Davezac N. Loss of functional OPA1 unbalances redox state: implications in dominant optic atrophy pathogenesis. Ann Clin Transl Neurol. 2016;3:408–21. doi: 10.1002/acn3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belenguer P, Pellegrini L. The dynamin GTPase OPA1: more than mitochondria? Biochim Biophys Acta. 2013;1833:176–83. doi: 10.1016/j.bbamcr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Eiberg H, Kjer B, Kjer P, Rosenberg T. Dominant optic atrophy (OPA1) mapped to chromosome 3q region. I. Linkage analysis. Hum Mol Genet. 1994;3:977–80. doi: 10.1093/hmg/3.6.977. [DOI] [PubMed] [Google Scholar]
- 6.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–89. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Shi W, Song L, Zhang X, Cheng L, Wang Y, Ge X, Li W, Zhang W, Min Q, Jin ZB, Qu J, Gu F. A recurrent deletion mutation in OPA1 causes autosomal dominant optic atrophy in a Chinese family. Sci Rep. 2014;4:6936. doi: 10.1038/srep06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynier P, Amati-Bonneau P, Verny C, Olichon A, Simard G, Guichet A, Bonnemains C, Malecaze F, Malinge MC, Pelletier JB, Calvas P, Dollfus H, Belenguer P, Malthiery Y, Lenaers G, Bonneau D. OPA3 gene mutations responsible for autosomal dominant optic atrophy and cataract. J Med Genet. 2004;41:e110. doi: 10.1136/jmg.2003.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, Iommarini L, Labauge P, Figarella-Branger D, Marcorelles P, Furby A, Beauvais K, Letournel F, Liguori R, La Morgia C, Montagna P, Liguori M, Zanna C, Rugolo M, Cossarizza A, Wissinger B, Verny C, Schwarzenbacher R, Martin MA, Arenas J, Ayuso C, Garesse R, Lenaers G, Bonneau D, Carelli V. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–51. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad KE, Davis RL, Sue CM. A novel OPA1 mutation causing variable age of onset autosomal dominant optic atrophy plus in an Australian family. J Neurol. 2015;262:2323–8. doi: 10.1007/s00415-015-7849-6. [DOI] [PubMed] [Google Scholar]
- 11.Gu S, Tian Y, Chen X, Zhao C. Targeted next-generation sequencing extends the phenotypic and mutational spectrums for EYS mutations. Mol Vis. 2016;22:646–57. [PMC free article] [PubMed] [Google Scholar]
- 12.Rong W, Chen X, Zhao K, Liu Y, Liu X, Ha S, Liu W, Kang X, Sheng X, Zhao C. Novel and recurrent MYO7A mutations in Usher syndrome type 1 and type 2. PLoS One. 2014;9:e97808. doi: 10.1371/journal.pone.0097808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Liu Y, Sheng X, Tam PO, Zhao K, Rong W, Liu X, Pan X, Chen LJ, Zhao Q, Vollrath D, Pang CP, Zhao C. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2014;23:2926–39. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Zhao K, Sheng X, Li Y, Gao X, Zhang X, Kang X, Pan X, Liu Y, Jiang C, Shi H, Rong W, Chen LJ, Lai TY, Wang X, Yuan S, Liu Q, Vollrath D, Pang CP, Zhao C. Targeted sequencing of 179 genes associated with hereditary retinal dystrophies and 10 candidate genes identifies novel and known mutations in patients with various retinal diseases. Invest Ophthalmol Vis Sci. 2013;54:2186–97. doi: 10.1167/iovs.12-10967. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Sheng X, Liu X, Li H, Liu Y, Rong W, Ha S, Liu W, Kang X, Zhao K, Zhao C. Targeted next-generation sequencing reveals novel USH2A mutations associated with diverse disease phenotypes: implications for clinical and molecular diagnosis. PLoS One. 2014;9:e105439. doi: 10.1371/journal.pone.0105439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarzi E, Seveno M, Angebault C, Milea D, Ronnback C, Quiles M, Adrian M, Grenier J, Caignard A, Lacroux A, Lavergne C, Reynier P, Larsen M, Hamel CP, Delettre C, Lenaers G, Muller A. Increased steroidogenesis promotes early-onset and severe vision loss in females with OPA1 dominant optic atrophy. Hum Mol Genet. 2017;26:4764. doi: 10.1093/hmg/ddx376. [DOI] [PubMed] [Google Scholar]
- 20.Del Dotto V, Mishra P, Vidoni S, Fogazza M, Maresca A, Caporali L, McCaffery JM, Cappelletti M, Baruffini E, Lenaers G, Chan D, Rugolo M, Carelli V, Zanna C. OPA1 Isoforms in the Hierarchical Organization of Mitochondrial Functions. Cell Reports. 2017;19:2557–71. doi: 10.1016/j.celrep.2017.05.073. [DOI] [PubMed] [Google Scholar]
- 21.Elachouri G, Vidoni S, Zanna C, Pattyn A, Boukhaddaoui H, Gaget K, Yu-Wai-Man P, Gasparre G, Sarzi E, Delettre C, Olichon A, Loiseau D, Reynier P, Chinnery PF, Rotig A, Carelli V, Hamel CP, Rugolo M, Lenaers G. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21:12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushnareva Y, Seong Y, Andreyev AY, Kuwana T, Kiosses WB, Votruba M, Newmeyer DD. Mitochondrial dysfunction in an Opa1(Q285STOP) mouse model of dominant optic atrophy results from Opa1 haploinsufficiency. Cell Death Dis. 2016;7:e2309. doi: 10.1038/cddis.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams PA, Morgan JE, Votruba M. Opa1 deficiency in a mouse model of dominant optic atrophy leads to retinal ganglion cell dendropathy. Brain. 2010;133:2942–51. doi: 10.1093/brain/awq218. [DOI] [PubMed] [Google Scholar]
- 24.Williams PA, Morgan JE, Votruba M. Mouse models of dominant optic atrophy: what do they tell us about the pathophysiology of visual loss? Vision Res. 2011;51:229–34. doi: 10.1016/j.visres.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 25.Yu-Wai-Man P, Sitarz KS, Samuels DC, Griffiths PG, Reeve AK, Bindoff LA, Horvath R, Chinnery PF. OPA1 mutations cause cytochrome c oxidase deficiency due to loss of wild-type mtDNA molecules. Hum Mol Genet. 2010;19:3043–52. doi: 10.1093/hmg/ddq209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–10. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 27.Yu-Wai-Man P, Griffiths PG, Burke A, Sellar PW, Clarke MP, Gnanaraj L, Ah-Kine D, Hudson G, Czermin B, Taylor RW, Horvath R, Chinnery PF. The prevalence and natural history of dominant optic atrophy due to OPA1 mutations. Ophthalmology. 2010;117:1538–46. doi: 10.1016/j.ophtha.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]