Abstract
We have identified a molecular interaction between the reversibly oxidized form of PTP1B and 14-3-3ζ that regulates PTP1B activity. Destabilizing the transient interaction between 14-3-3ζ and PTP1B, prevented PTP1B inactivation by ROS and decreased EGFR phosphorylation. Our data suggest that destabilizing the interaction between 14-3-3ζ and the reversibly oxidized and inactive form of PTP1B may establish a path to PTP1B activation in cells.
Protein tyrosine phosphatases (PTPs) are transiently inactivated in response to regulated and localized rises in reactive oxygen species (ROS) in cells1, 2. It is increasingly apparent that controlling the catalytic activity of members of the PTP family by reversible oxidation facilitates phospho-dependent signaling and that dysregulated PTP inactivation by oxidation contributes to the etiology of a spectrum of human diseases2–5. Protein tyrosine phosphatase 1B (PTP1B) has given us important insights on the catalytic function and redox regulation of members of the PTP family6, 7. The reversible oxidation of PTP1B leads to profound structural changes at the active site8, 9 that can be taken advantage of by conformation sensor antibodies (scFv45) to stabilize its inactive form 10. Since stabilization of the oxidized, inactive form of PTP1B (PTP1B-OX) by scFv45 perturbs the normal role of PTP1B on signaling pathways, we enquired whether a protein having a similar function existed in vivo.
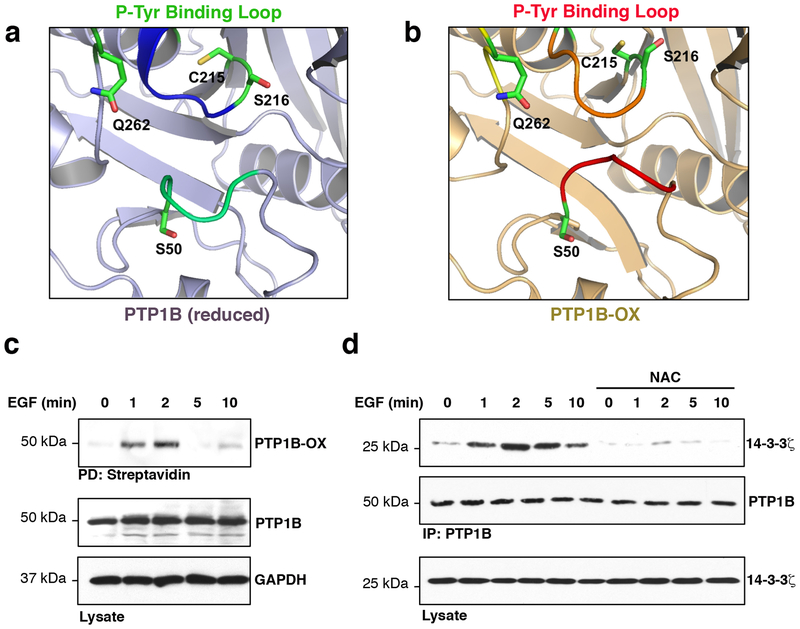
A comparative study between the crystal structure of the inactive sulphenyl-amide species of PTP1B-OX and reduced PTP1B revealed that a sequence of 10 amino acids of the phospho-tyrosine (pTyr) recognition loop, reported to bind scFv4511, were newly exposed to the cytosol in the inactive form of PTP1B in addition to the changes in the PTP loop10 (Fig. 1a, b, Supplementary Fig. 1a). We calculated the surface accessible area of these cytosol-exposed amino acids (Supplementary Fig. 1b) and we hypothesized that this loop could recruit novel redox-specific PTP1B-binding proteins in cells. Therefore, a peptide comprising PTP1B amino acids Lys41 to Ser50 was synthetized and used as bait in cells. We used isobaric mass tags and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantify and identify those proteins that interacted with the pTyr loop-derived-peptides and pulled down from cellular extracts. 14-3-3ζ was one of the most abundant proteins, and the most abundant 14-3-3 isoform, found interacting with the pTyr loop-derived-peptide when compared to the control beads (Supplementary Tables 1–2, iTRAQ: NP vs C lane). Enrichment of 14-3-3ζ was even more pronounced when a PTP1B derived-peptide comprising the sequence from Lys41 to Ser50 was phosphorylated on Ser50 and utilized as bait in cellular extracts.
Fig. 1.
The exposed phospho-tyrosine recognition loop of PTP1B-OX interacts with 14-3-3ζ. (a) Close-up view of the pTyr recognition loop of reduced PTP1B (PDB code: 2HNQ) and (b) of PTP1B-OX (sulphenyl-amide species, PDB code 1OEM). (c) Reversible oxidation of PTP1B in cells treated with EGF (100 ng/ml) for the indicated times. This experiment was repeated three independent times with representative data shown. (d) Effect of scavenging ROS on the PTP1B–14-3-3ζ association in cells incubated with the antioxidant NAC (1 mM, 1 h) and stimulated with EGF for the indicated times. This experiment was repeated two independent times with representative data shown. Uncropped images are shown in Supplementary Fig. 12.
The crystal structure of 14-3-3ζ revealed a conserved amphipathic groove that allows 14-3-3s to associate with proteins in a phosphorylation-dependent and -independent manner to perform a range of functions that include altering their activity, their association with other molecules or their subcellular localization12, 13. Optimal 14-3-3 binding motifs contain an arginine residue in the – 2 to – 5 position from the phospho-serine/threonine residue and a proline residue in the + 2 position13. Upon examination of the pTyr recognition loop sequence, we identified a non-canonical 14-3-3 binding motif containing an arginine residue in the – 3 position and a proline in the +1 position (Supplementary Fig. 2). While the pTyr recognition loop is one of the conserved motifs that define the PTP family, the residues flanking this motif vary between members of the PTP family14; in particular, the Ser residue (Ser50 in PTP1B) is only preserved in three members of the PTP family (Supplementary Fig. 3). To assess the ability of 14-3-3ζ to interact with PTP1B-OX in cells, we tested whether wild type PTP1B co-immunoprecipitated with 14-3-3ζ following epidermal growth factor receptor (EGFR) activation. Using this well-characterized signaling pathway that leads to a rapid increase in ROS production and PTP1B inactivation3, 15, we first established that PTP1B was reversibly oxidized using a cysteinyl-labeling assay that converts reversible oxidation of the catalytic cysteine of PTPs to a modification by biotin that can be visualized by immunoblotting after a biotin-streptavidin purification step (Fig. 1c, Supplementary Fig. 4)16, 17. As shown in figure 1c, minimal PTP1B oxidation was detected in resting cells, however, PTP1B biotinylation was detected in a transient manner at 1 and 2 minutes upon EGFR activation. Supporting our MS data, immunoprecipitations revealed a transient association between PTP1B and 14-3-3ζ between 2 to 5 minutes following EGFR activation (Supplementary Fig. 5a, b). This transient interaction between PTP1B and 14-3-3ζ was detected in co-immunoprecipitations of either endogenously (Supplementary Fig. 6) or exogenously expressed PTP1B and 14-3-3ζ from lysates of cells stimulated with EGF. Detection of some association between endogenous PTP1B and 14-3-3ζ in unstimulated cells (Supplementary Fig. 6) is likely the result from increased protein input in the co-immunoprecipitations to compensate for a decreased dynamic range in the assay. Redox-dependent association between PTP1B and 14-3-3ζ was confirmed by pre-incubating cells with the antioxidant N-acetylcysteine (NAC) prior to EGF stimulation (Fig. 1d). Interestingly, the mitochondrial antioxidant SS-31 did not prevent the association between PTP1B and 14-3-3ζ, and a mutant PTP1B that adopts an oxidized conformation [PTP1BCASA 10] interacted with 14-3-3ζ despite antioxidant pre-treatments (Supplementary Fig. 7). Thus, 14-3-3ζ is the first protein shown to specifically interact with an oxidized PTP in cells.
To further understand the function of the interaction between PTP1B-OX and 14-3-3ζ, and to determine whether this interaction regulates the redox status of PTP1B, we directly prevented 14-3-3ζ binding to PTP1B using the pan 14-3-3 inhibitor peptide R1818. This peptide binds the amphipathic groove of 14-3-3s and competes with endogenous ligands. As expected, inhibiting 14-3-3s by exposing cells to R18 prevented the interaction between 14-3-3ζ and PTP1B (Fig. 2a). However, the cysteinyl-labeling assay also revealed that R18 prevented biotinylation of PTP1B, which reflects the reversible oxidation of PTP1B in cells (Fig. 2b). Since Ser50 of the pTyr recognition loop of PTP1B is buried in the structure of the reduced enzyme (Fig. 1a, b, Supplementary Fig. 1), we hypothesized that when the pTyr recognition loop becomes exposed to the cytosol as a consequence of PTP1B oxidation, Ser50 becomes accessible and readily phosphorylated by AKT/Protein Kinase B, a protein kinase known to phosphorylate PTP1B on this residue19. Hence, we investigated whether the reversible oxidation of PTP1B occurred as a consequence of Ser50 phosphorylation and the subsequent association with 14-3-3ζ. Exposing cells to an AKT inhibitor (i.e. AKT Inhibitor V/API-220) prevented phosphorylation of AKT Ser473 and PTP1B Ser50 following EGF stimulation (Supplementary Fig. 8a). Similar to our previous observation using a 14-3-3 inhibitor, preventing PTP1B phosphorylation on Ser50 impaired the interaction between PTP1B and 14-3-3ζ and compromised the reversible oxidation of PTP1B (Supplementary Fig. 8b, c). This is consistent with a mechanism in which the interaction between 14-3-3ζ and PTP1B stabilizes PTP1B-OX in cells.
Fig. 2.
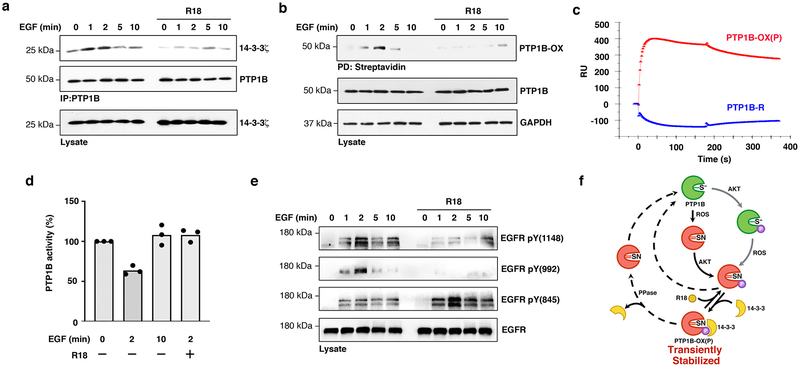
Destabilizing the association between 14-3-3ζ and PTP1B-OX prevents PTP1B inactivation and decreases EGFR phosphorylation. (a) Effect of exposing cells to the 14-3-3 inhibitor peptide R18 (25 μM, 90 min) on the transient interaction between PTP1B and 14-3-3ζ. Uncropped images are shown in Supplementary Fig. 13a. This experiment was repeated three independent times with representative data shown. (b) Reversible-oxidation of PTP1B was measured following R18 pretreatment using the cysteinyl-labeling assay. Uncropped images are shown in Supplementary Fig. 13b. This experiment was repeated three independent times with representative data shown. (c) Comparative SPR sensorgram of the interaction between 14-3-3ζ (5000 nM) and either PTP1B-OX(P) (red) or reduced PTP1B (PTP1B-R) (blue) showing direct association between PTP1B-OX(P) and 14-3-3ζ. (d) Quantitative analysis of PTP1B catalytic activity from immune-complexes, showing the effects of preventing PTP1B association with 14-3-3ζ using R18 before cell exposure to EGF for the indicated times. The average readout value of technical replicates is represented by the dot-plot bar graph. N=3 independent experiments are shown. (e) Effect of pre-treating cells with R18 on EGFR tyrosine phosphorylation. Phosphorylation of EGFR on PTP1B sites (pTyr992 and pTyr1148) was decreased whereas phosphorylation of EGFR on Tyr845 was increased. Uncropped images are shown in Supplementary Fig. 14. This experiment was repeated three independent times with representative data shown. (f) The proposed mechanism leading to transient stabilization of PTP1B oxidation in cells is indicated with black arrows. In resting cells, PTP1B is active (green) and possesses a reactive catalytic cysteine residue (S−). Following EGFR activation, a ROS-producing stimulus, PTP1B becomes rapidly and transiently inactivated by ROS (red). Key conformational changes of the inactive sulphenyl-amide (SN) species of PTP1B-OX include one element of the active site, the phospho-tyrosine recognition loop containing the sequence Lys41 to Ser50. Upon PTP1B oxidation, this loop adopts a cytosol-exposed position and becomes phosphorylated on Ser50 by AKT, which leads to 14-3-3 binding. In turn, binding of 14-3-3 to PTP1B-OX(P) transiently stabilizes the inactive, oxidized form of the enzyme and allows phosphorylation of its substrates. Alternatively, preventing the transient interaction between 14-3-3 and PTP1B-OX using R18, perturbed the redox cycle of PTP1B and effectively prevented PTP1B inactivation by ROS. Reactivation mechanisms for PTP1B, involving an unidentified phospho-Ser50 phosphatase, Trx24 or cellular thiols25, and alternative inactivation mechanisms are indicated with dashed and grey arrows respectively.
In order to gain some insight on the nature of the interaction between PTP1B and 14-3-3ζ, we next tested whether the interaction between 14-3-3ζ and PTP1B was a direct protein-protein interaction. As a first step, we performed an in vitro pull-down experiment and observed that while 14-3-3ζ interacted with PTP1B-OX when phosphorylated on Ser50 (PTP1B-OX(P)), no interaction occurred with reduced PTP1B (Supplementary Fig. 9a, b). We then analyzed the molecular interaction between 14-3-3ζ and PTP1B-OX(P) by surface plasmon resonance (SPR) and confirmed that 14-3-3ζ specifically binds to PTP1B in its oxidized and phosphorylated inactive state but not to the active reduced form of the enzyme (Fig. 2c), consistent with the interaction observed in cells. The dependency on phosphorylation of Ser50 for 14-3-3ζ binding was further confirmed by SPR using immobilized phosphoSer50-pTyr loop-derived-peptide (Supplementary Fig. 9c). Moreover, real-time binding of increasing concentrations of 14-3-3ζ and a fixed amount of PTP1B-OX(P) indicated that binding occurred with high affinity (KD: 79 nM) and a relatively slow off-rate (Koff = 4.58×10–3 s–1) (Supplementary Fig. 9d). These SPR results support a model in which 14-3-3ζ directly binds to the pTyr-recognition loop of PTP1B in a pSer50-dependent fashion, and reveal that the interaction between 14-3-3ζ and the unphosphorylated pTyr recognition loop peptide that was previously observed by MS analysis (Supplementary Tables 1–2) occurs indirectly via a protein intermediate.
The cysteinyl-labeling assay is an approach that allows us to measure PTP1B inactivation by reversible oxidation (Supplementary Fig. 4). Decreased biotinylation of PTP1B observed in the cysteinyl-labeling assay when cells are exposed to R18 could either reflect the absence of PTP1B oxidation, or irreversible oxidation of the catalytic cysteine residue to sulfinic or sulfonic forms. To determine whether disrupting the interaction between 14-3-3ζ and PTP1B decreased reversible oxidation of PTP1B, we measured PTP1B catalytic activity in lysates from EGF-stimulated cells that were pretreated with R18 or not. PTP1B was immunoprecipitated from lysates in oxygen-free conditions to minimize post-lysis oxidation of the phosphatase and the catalytic activity of PTP1B was then measured using the pTyr analog, pNPP, as substrate. As expected from previous studies, PTP1B activity was greatly decreased in lysates from cells exposed to EGF for 2 minutes (36.3 % ± 5.6), and mostly recovered after 10 minutes (Fig. 2d). However, PTP1B maintained maximal catalytic activity after 2 minutes of EGF stimulation in cells that were pretreated with R18, indicating that PTP1B remains active in R18-treated cells, when the PTP1B-OX(P)–14-3-3ζ complex is disrupted. Measuring the activity of PTP1B from the same lysates in reducing conditions confirmed that decreased enzymatic activity upon EGF stimulation occurred as consequence of reversible oxidation (Supplementary Fig. 10).
Given that R18 hindered binding of 14-3-3ζ onto PTP1B-OX(P) and maintained PTP1B in an active state, we tested whether preventing transient PTP1B oxidation had an impact on EGFR tyrosine phosphorylation and activation. Immunoprecipitation of pTyr proteins and immunoblotting with anti-EGFR antibodies showed that pretreating cells with R18 prior to EGF exposure did not affect the overall phosphorylation of EGFR (Supplementary Fig. 11). Interestingly, EGFR phosphorylation at tyrosine 992 and tyrosine 1148, two sites previously reported to be direct substrates of PTP1B21, were markedly dephosphorylated in EGF-stimulated cells pretreated with R18 (Fig. 2e). On the other hand, EGFR phosphorylation at tyrosine 845, a SRC kinase phosphorylation site22 was increased, consistent with PTP1B-mediated SRC activation23. Collectively, our data support that destabilizing protein interaction between the oxidized and phosphorylated, inactive form of PTP1B and 14-3-3 with appropriate therapeutic molecules allows rapid reactivation of PTP1B and may be used to limit excessive growth factor signaling pathways involving PTP1B (Fig. 2f). The concept that stabilization of PTP1B oxidation is necessary to maintain the enzyme in an inactive form may be broadly applicable to redox-regulated enzymes and offer a paradigm for drugs that activate protein tyrosine phosphatases.
Online methods
Materials
Anti-EGFR, GAPDH and 14-3-3ζ were from Santa Cruz Biotechnology. Anti-Pan-AKT, anti-AKT pSer473, anti phosphoEGFR antibodies were from Cell Signaling Technology. HA-peroxidase and anti-PTP1B (FG6) was from Millipore. PT-66-agarose-conjugated beads, anti-FLAG M2 beads, and anti-HA beads and anti-Flag M2 peroxidase were purchased from Sigma. Anti-PTP1B pSer50 (Ab62320) were from Abcam. Streptavidin-HRP was from GE Healthcare. HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. Protease inhibitor mixture tablets were from Roche. Catalase and superoxide dismutase were from Calbiochem. Surfact-Amps Nonidet P-40, zeba desalt spin columns, EZ-Link biotin-iodoacetyl-PEG2 (biotin-IAP), and iodoacetic acid were from ThermoScientific. The pTyr loop-derived peptide (CKNRNRYRDVS) and phospho-Ser50 pTyr loop-derived peptide (CKNRNRYRDVpS) were from GenScript USA Inc. BIACore sensor NTA and Streptavidin chips were from GE healthcare.
Cell culture
HEK293T cells were maintained in culture at 40–90% confluency in EMEM (Eagle’s Minimum Essential Medium, 1000 mg/L glucose, ATCC) containing 100 U/ml penicillin, 100 μg/ml streptomycin and 10% FBS at 5% CO2 and 37°C. 4 μg of plasmid DNA was routinely transfected using TurboFect (ThermoFisher) on 80% confluent cells. Transient expression was allowed to progress for 48 hours. Cells were serum starved overnight using EMEM without serum.
Assay of PTP oxidation
The cysteinyl-labeling assay was performed as described16, 17. In brief, cells were starved for 16 hours in serum-free EMEM. For EGF stimulations, cells were stimulated with 100 ng/ml EGF for indicated times and lysed in degassed lysis buffer (50 mM Sodium Acetate (pH 5.5), 150 mM NaCl, 1% NP40, 10% (v/v) glycerol) supplemented with 25 μg/ml aprotinin, 25 μg/ml leupeptin, 10 mM IAA, 250 U/ml catalase and 125 U/ml superoxide dismutase after which alkylation was allowed for 1hour at room temperature. Lysates were cleared by centrifugation at 10,000 rpm for 10 minutes, and buffer exchanged with (1 mM) TCEP-containing lysis buffer using Zeba columns (Pierce). Lysates were reduced for 30 minutes, and supplemented with (5 mM) a biotin-labeled iodoacetic acid probe (Pierce). Labeled PTP1B was pulled down with streptavidin-Sepharose beads, boiled for 2 minutes in sample buffer and used for immunoblotting.
Immunoprecipitation and immunoblotting
FLAG-PTP1B, HA-14-3-3ζ and EGFR were immunoprecipitated as follows. Cells were grown to 80% confluence in 10-cm plates, transfected for 48 hours and serum-starved for 16 hours. Following serum-starvation, cells were pretreated (R18, 25 μM; AKT inh V, 15 μM, 60 min; NAC, 1 mM, 1 h) or not and stimulated with EGF to activate EGFR for the indicated times. After treatment, the plates were transferred on ice, washed with cold PBS and extracted in 800 μl of a lysis buffer consisting of 20 mM HEPES pH 7,4, 150 mM NaCl, 1% NP40, 1 mM EDTA, 10 mM NaF, 25 μg/ml aprotinin, 25 μg/ml leupeptin, 1 μM microcystin, 100 nM okadaic acid. 1 mM Na3VO4 was also supplemented to the lysis buffer in experiments assessing EGFR phosphorylation. All subsequent steps were carried out on ice or at 4 °C. Cells were lysed on a rotating wheel at 4°C for 30 minutes. Cell debris were centrifuged at 14,000 x g for 10 minutes, and protein concentrations were determined. 500 μg of protein was diluted in cold lysis buffer and precleared for 20 minutes with sepharose beads. The supernatants were incubated for 3 hours on a rotating wheel with appropriate beads precoupled to -HA, -FLAG, anti-EGFR antibodies or to anti-pTyr antibodies (PT-66). The immune complexes were pelleted at 3000 x g for 5 minutes and washed three times with cold lysis buffer. The beads were resuspended in 20 μl of 4X Laemmli sample buffer and heated at 95 °C for 2 minutes. Proteins were separated by SDS-PAGE and detected by immunoblotting.
Protein expression and in vitro Ni-NTA precipitation assay
The interaction between PTP1B and 14-3-3ζ was performed in vitro using purified proteins. Bacterially expressed His-PTP1B (residues 1–321) and GST-14-3-3-ζ were purified by Ni-NTA (nitrilotriacetic acid) and agarose glutathione, respectively, and PTP1B was stored in a 1 mM TCEP solution to prevent post-purification oxidation. PTP1B was then reversibly oxidized as previously performed10, and phosphorylated. Briefly, purified PTP1B (50 nM) was reversibly oxidized with 250 μM H2O2 in HEPES buffer (50 mM HEPES, 100 mM NaCl, pH 7.0) for 10 minutes at room temperature. H2O2 was then removed by buffer exchange, using desalting columns equilibrated with a modified Kinase Assay buffer (50 mM Tris, 10 mM MgCl2, 5 mM ATP, pH 7.4) containing no reducing agent. Phosphorylation of PTP1B-OX was then initiated by the addition of AKT (in a 1:200, AKT:PTP1B-OX molar ratio) and incubated at 30°C for 16 hours. Optimal phosphorylation of PTP1B Ser50 by AKT was established by analyzing levels of PTP1B-Ser50 phosphorylation following incubations of 1, 3, 6 and 16 hours. Direct interaction between PTP1B-OX-(P) and 14-3-3-ζ, was performed by incubating 50 nM PTP1B-OX(P) or PTP1B-R with 5 μM 14-3-3ζ in Binding buffer (20 mM HEPES, 150 mM NaCl, 0.05% BSA, 0.05% Tween, pH 7.4) for 2 hours at 4°C on a clinical rotator. Glutathione agarose beads were added and incubated with the protein complexes for 1 hour at 4°C and protein complexes bound to glutathione agarose beads were centrifuged and washed (3X) 5 minutes at 4°C with binding buffer. Protein complexes were further separated by SDS-PAGE, transferred onto nitrocellulose membranes and blotted for PTP1B using anti-PTP1B (FG6) antibody, and for 14-3-3-ζ using an anti-GST antibody. Protein amounts of PTP1B were scaled up a 1000 fold to generate sufficient PTP1B-OX(P) for Surface Plasmon Resonance experiments.
Preparation of protein chips for surface plasmon resonance (SPR) experiments
SPR binding assays were performed to assess binding of 14-3-3ζ to the phospho-tyrosine recognition loop-derived peptide (Cysteine-K41NRNRYRDVS50: linked to biotin-IAP), phosphoPTP1B-OX (PTP1B-OX(P)) and reduced PTP1B (PTP1B-R) using a BIAcore 3000 (GE healthcare, Uppsala, Sweden) operated using BIAcore 3000 control and BIAevaluation software (version 4.0.1). To obtain kinetic data for Cys-K41NRNRYRDVS50/K41NRNRYRDVpS50 and 14-3-3ζ interactions, biotinylated Cys-K41NRNRYRDVS50 and Cys-K41NRNRYRDVpS50 were immobilized to streptavidin chip based on the manufacturer’s protocol. In brief, 20 μl solution of Biotin-Cys-K41NRNRYRDVS50 or Biotin-Cys-K41NRNRYRDVpS50 (3.69 mM) was injected over flow cells 2 and 3 (FC2 and FC3) of the streptavidin chip at a flow rate of 10 μl/min. The successful immobilizations of biotinylated Cys-K41NRNRYRDVS50 and Cys-K41NRNRYRDVpS50 were confirmed by the observation of a 521 and 389 resonance unit (RU) increase, respectively on the sensor chip. The control flow cell (FC1) was prepared by 1 minute injection with saturated biotin.
To obtain kinetic data for PTP1B and 14-3-3ζ interactions, his-tagged PTP1B-OX(P) and PTP1B-R were immobilized on a NTA sensor chip, which is designed to bind histidine-tagged molecules by relying on a NTA-chelated nickel atom, according to standard protocol (GE healthcare, Uppsala, Sweden). Briefly, NTA surface was activated by injection pulse of 10 μl (flow rate, 10 μl/min) of 500 μM NiCl2 in HBS-N buffer (0.01 M HEPES, 0.15 M NaCl, pH 7.4). Following activation, 20 μl of his-tagged PTP1B-OX(P) (2.7 μM) and PTP1B-R (2.7 μM) were injected over the activated biosensor surface. The successful immobilization of PTP1B-OX(P) and PTP1B-R were confirmed by the observation of a 460 and 4050 resonance unit (RU) increase, respectively on the sensor chip. A reference flow cell was used with the NTA surface without NiCl2 activation
Measurement of interactions using BIAcore
14-3-3ζ was diluted in HBS-N buffer (0.01 M HEPES, 0.15 M NaCl, pH 7.4). Diluted 14-3-3ζ samples (indicated concentrations) were injected at a flow rate of 30 μl/min. At the end of the sample injection, HBS-N buffer as running buffer was flowed over the sensor surface to facilitate dissociation. After a 3-minute dissociation time, the sensor surface was regenerated by injecting with 30 μl of 2 M NaCl to get fully regenerated surface. The response was monitored as a function of time (sensorgram) at 25 °C.
PTP activity assay
PTP1B activity assay was performed on HEK293T cells. 60–80% confluent cells were transfected with PTP1B-FLAG and 14-3-3ζ-HA for 48 hours. Cells were serum-starved 32 hours post-transfection for a 16 hours period and stimulated with EGF (100 ng/ml) for the indicated times, in presence, or absence of R18 (25 μM, 90 min). Cells were lysed under strict hypoxic conditions, on ice following with a cold degassed lysis buffer consisting of 25 mM HEPES pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Surfact-Amps NP-40. Degassing of the buffer is a critical step for this assay. The degassing steps were based on a method previously described16, 17). Cells were lysed on a rotating wheel at 4°C for 30 minutes. Cell debris were centrifuged at 14,000 x g for 10 minutes, and protein concentrations were determined. The supernatants (200 μg) were incubated 4°C for 3 hours on a rotating wheel with 10 μl anti-FLAG Dynabeads (Life technologies). The immune complexes were then washed with degassed lysis buffer and resuspended in pNPP assay buffer (20 mM HEPES pH 7.4, 100 mM NaCl and 0.05% w/v BSA, 20 mM pNPP), with or without DTT (5 μM). The beads were protected from light on rotator at room temperature and the converted substrate was measured following a 30 min incubation. 80 μl of the supernatant from the enzymatic reaction was stopped with 20 μl of 2M NaOH and absorbance was measured at 405 nm using a spectrophotometer (SpectraMax, Molecular Devices).
pTyr loop derived-peptide pulldown for MS
HEK293T cells were lysed in a lysis buffer consisting of 20 mM HEPES pH 7,4, 150 mM NaCl, 1% NP40, 1 mM EDTA, 10 mM NaF, 25 μg/ml aprotinin, 25 μg/ml leupeptin, 1 μM microcystin and 100 nM okadaic acid. Protein content of lysate was measured by Bradford assay, and equal amounts of lysates were incubated with the pTyr loop-derived peptide (C-KNRNRYRDVS), a phospho-Ser50 pTyr loop-derived peptide (C-KNRNRYRDVpS) or L-Cys coupled to UltraLink Iodoacetyl Resin (ThermoFisher) for 90 minutes at 4 °C. The resin was centrifuged and washed three times with lysis buffer and three additional times with PBS. The beads were incubated with 50 μl of 10 mM pTyr loop-derived peptide and pSer50-pTyr loop-derived peptide in PBS at RT for 20 min to elute pTyr loop binding proteins. The supernatants were collected and subjected to tryptic digestion and iTRAQ labeling.
Tryptic digestion, iTRAQ labeling and dimensional fractionation
1 X sample volume of 10% SDS was added to eluted proteins to bring final concentration of SDS to 5%. Next, TCEP was added to a final concentration of 5 mM and samples were heated to 55°C for 20 minutes and allowed to cool to room temperature. Methyl methanethiosulfonate (MMTS) was added to a final concentration of 10 mM and samples were incubated at room temperature for 20 minutes to complete blocking of free sulfhydryl groups. An S-Trap protocol was adapted and used to digest proteins26. Briefly, lysates were acidified with phosphoric acid to a final concentration of 1.2% and added to an S-Trap™ containing 6X lysate volume of S-trapping buffer (90% Methanol, 100 mM TEAB). S-Trap™ was spun down at 4000 x g for 30 seconds to remove buffer, washed with 200 μL of S-trapping buffer and spun again to remove all buffer. 2 μg of sequencing grade trypsin (Promega) in 125 μl of 50 mM TEAB was then added to the S-Trap™ and they were digested overnight at 37°C. After digestion the peptides were eluted from the column with subsequent applications of 40 μl 50 mM TEAB, 40 μl 0.2% formic acid in water and 40 μl 0.2% formic acid in 50% acetonitrile. Peptides were dried in vacuo. Peptides were then reconstituted in 50 μl of 0.5 M TEAB/70% ethanol and labeled with 4-plex iTRAQ reagent for 1 hour at room temperature27. Labeled samples were then acidified to pH 4 using formic acid, combined and concentrated in vacuo until ~10 μl remained. For dimensional fractionation, peptides were fractionated using a Pierce High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific) according to the manufacturer’s instructions with slight modifications. Briefly, peptides were reconstituted in 150 μl of 0.1% TFA, loaded onto the spin column and centrifuged at 3000 x g for 2 minutes. Column was washed with water and then peptides were eluted with the following percentages of acetonitrile (ACN) in 0.1% triethylalmine (TEA): 5%, 7.5%, 10%, 12.5%, 15%, 20%, 30% and 50%. Each of the 8 fractions was then separately injected into the mass spectrometer using capillary reverse phase LC at low pH.
Mass spectrometry
An Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific), equipped with a nano-ion spray source was coupled to an EASY-nLC 1200 system (Thermo Scientific). The LC system was configured with a self-pack PicoFrit™ 75-μm analytical column with an 8-μm emitter (New Objective, Woburn, MA) packed to 25 cm with ReproSil-Pur C18-AQ, 1.9 μM material (Dr. Maish GmbH). Mobile phase A consisted of 2% acetonitrile; 0.1% formic acid and mobile phase B consisted of 90% acetonitrile; 0.1% formic Acid. Peptides were then separated using the following steps: at a flow rate of 200 nl/minute: 2% B to 6% B over 1 minute, 6% B to 30% B over 84 minutes, 30% B to 60% B over 9 minutes, 60% B to 90% B over 1 minute, held at 90% B for 5 minutes, 90% B to 50% B over 1 minute and then flow rate was increased to 500 nl/min as 50% B was held for 9 minutes. Eluted peptides were directly electrosprayed into the Orbitrap Fusion Lumos mass spectrometer with the application of a distal 2.3 kV spray voltage and a capillary temperature of 300°C. Full-scan mass spectrum (Res=60,000; 400–1600 m/z) were followed by MS/MS using the “Top Speed” method for selection. High-energy collisional dissociation (HCD) was used with the normalized collision energy set to 35 for fragmentation, the isolation width set to 1.2 and a duration of 10 seconds was set for the dynamic exclusion with an exclusion mass width of 10 ppm. We used monoisotopic precursor selection for charge states 2+ and greater, and all data were acquired in profile mode.
Database searching
Peaklist files were generated by Mascot Distiller (Matrix Science). Protein identification and quantification was carried using Mascot 2.628 against the UniProt human sequence database (93,799 sequences; 37,184,134 residues). Methylthiolation of cysteine and N-terminal and lysine iTRAQ modifications were set as fixed modifications, methionine oxidation and deamidation (NQ) as variable. Trypsin was used as cleavage enzyme with one missed cleavage allowed. Mass tolerance was set at 30 ppm for intact peptide mass and 0.2 Da for fragment ions. Search results were rescored to give a final 1% FDR using a randomized version of the same Uniprot Human database. Protein-level iTRAQ ratios were calculated as intensity weighted, using only unique peptides with expectation values < 0.0004828. As this was a protein IP experiment, no global ratio normalization was applied.
Surface accessible area assessment
The structures of reduced PTP1B (pdb code: 2HNQ) and PTP1B-OX (pdb code: 1OEM) were used for the accessible surface area calculation. The accessible surface areas for the amino acids of PTP1B phospho-tyrosine recognition loop are calculated using the program surface implemented in ccp4 suite29.
Supplementary Material
Acknowledgments:
We thank H. Fu (Emory University) for providing the 14-3-3ζ plasmid. This research was supported by NIH grant HL138605 and American Heart Association grant 17GRNT33700265 to BB and by NIH grant GM55989 to NKT. BB is also grateful for support from the following foundations: Heart and Stroke Foundation of Canada and SUNY Research Foundation. BB is a FRQS Research Scholar and AB was the recipient of a scholarship from the FRQS.
Footnotes
Data availability
The structures of reduced PTP1B (pdb code: 2HNQ) and PTP1B-OX (pdb code: 1OEM) were used for the accessible surface area calculation. The mass spectrometry data in Supplemental Tables 1 and 2 will be made available in the PRoteomics IDEntifications (PRIDE) database.
Competing Interests: The authors declare that they have no competing interests.
References
- 1.Finkel T Signal transduction by reactive oxygen species. J Cell Biol 194, 7–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostman A, Frijhoff J, Sandin A & Böhmer FD Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem. 150, 345–356 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Tonks NK Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell. Biol 7, 833–846 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Karisch R et al. Global proteomic assessment of the classical protein-tyrosine phosphatome and “Redoxome”. Cell 146, 826–840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown DI & Griendling KK Regulation of Signal Transduction by Reactive Oxygen Species in the Cardiovascular System. Circ Res 116, 531–549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonks NK PTP1B: from the sidelines to the front lines! FEBS Lett 546, 140–148 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Feldhammer M, Uetani N, Miranda-Saavedra D & Tremblay ML PTP1B: a simple enzyme for a complex world. Crit Rev Biochem Mol Biol 48, 430–445 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Salmeen A et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769–773 (2003). [DOI] [PubMed] [Google Scholar]
- 9.van Montfort RL, Congreve M, Tisi D, Carr R & Jhoti H Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423, 773–777 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Haque A, Andersen JN, Salmeen A, Barford D & Tonks NK Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell 147, 185–198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan N et al. Harnessing insulin- and leptin-induced oxidation of PTP1B for therapeutic development. Nat Commun 9, 283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Meyerkord CL, Du Y, Khuri FR & Fu H 14-3-3 proteins as potential therapeutic targets. Semin Cell Dev Biol 22, 705–712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhardt HC & Yaffe MB Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol 14, 563–580 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Andersen JN et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol 21, 7117–7136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SR, Kwon KS, Kim SR & Rhee SG Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem 273, 15366–15372 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Boivin B, Zhang S, Arbiser JL, Zhang ZY & Tonks NK A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proc. Natl. Acad. Sci. U.S.A. 105, 9959–9964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boivin B, Yang M & Tonks NK Targeting the reversibly oxidized protein tyrosine phosphatase superfamily. Sci. Signal 3, pl2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Ravichandran LV, Chen H, Li Y & Quon MJ Phosphorylation of PTP1B at Ser(50) by Akt impairs its ability to dephosphorylate the insulin receptor. Mol Endocrinol 15, 1768–1780 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Yang L et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res 64, 4394–4399 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Milarski KL et al. Sequence specificity in recognition of the epidermal growth factor receptor by protein tyrosine phosphatase 1B. J Biol Chem 268, 23634–23639 (1993). [PubMed] [Google Scholar]
- 22.Biscardi JS et al. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem 274, 8335–8343 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Lessard L, Stuible M & Tremblay ML The two faces of PTP1B in cancer. Biochim Biophys Acta 1804, 613–619. [DOI] [PubMed] [Google Scholar]
- 24.Dagnell M et al. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-β receptor tyrosine kinase signaling. Proc Natl Acad Sci U S A 110, 13398–13403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons ZD & Gates KS Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry 52, 6412–6423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zougman A, Selby PJ & Banks RE Suspension trapping (STrap) sample preparation method for bottom‐up proteomics analysis. Proteomics 14, 1006–11010 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Ross PL et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3, 1154–1169 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Perkins DN, Pappin DJ, Creasy DM & Cottrell JS Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.