Abstract
Recent studies have implicated the nucleosome acidic patch in the activity of ATP-dependent chromatin remodeling machines. We employed a photocrosslinking-based nucleosome profiling technology - ‘photoscanning’- to identify a conserved basic motif within the catalytic subunit of ISWI remodelers, SNF2h, which engages this nucleosomal epitope. This region of SNF2h is essential for chromatin remodeling activity in a reconstituted biochemical system and in cells. Our studies suggest that the basic motif in SNF2h plays a critical role in anchoring the remodeler to the nucleosomal surface. We also examine the functional consequences of several cancer-associated histone mutations that map to the nucleosome acidic patch. Kinetic studies employing physiologically relevant heterotypic nucleosomal substrates (‘Janus’ nucleosomes) indicate that these cancer mutations can disrupt regularly spaced chromatin structure by inducing ISWI-mediated unidirectional nucleosome sliding. These results imply a potential mechanistic link between oncogenic histones and alterations to the chromatin landscape.
Introduction
The organization of eukaryotic genomes into chromatin creates an inherent barrier to essential cellular processes, providing a means to functionally silence genetic material. Access to genetic information, therefore, requires the ability to maneuver and/or disassemble nucleosomes, the fundamental repeating units of the chromatin polymer1. ATP-dependent chromatin remodeling complexes (remodelers, hereafter) are molecular machines that carry out these operations, their main functions consisting of chromatin access, assembly, and editing. In this way, remodelers gate entry to the genome, playing critical roles in fundamental processes such as gene transcription, DNA replication, and DNA damage repair2–5.
The ATP-dependent chromatin assembly factor (ACF)6–8 is a member of the mammalian ISWI remodeler family that is required for gene silencing9, and is essential for embryogenesis10. ACF consists of an ATPase unit, SNF2h, and an ancillary subunit, ACF1. The ACF complex and the isolated SNF2h subunit have been shown to promote histone octamer sliding6–9,11. Cumulative studies highlight domains flanking the translocase domain in SNF2h that regulate remodeler function by engaging different nucleosomal features, including histone tails (Fig. 1a)12–17. In addition, SNF2h and the ACF complex are thought to function in a dimeric fashion in which the motors constantly operate on both sides of the two-fold pseudo-symmetric nucleosome disc to facilitate the generation of the regularly spaced nucleosomes18,19.
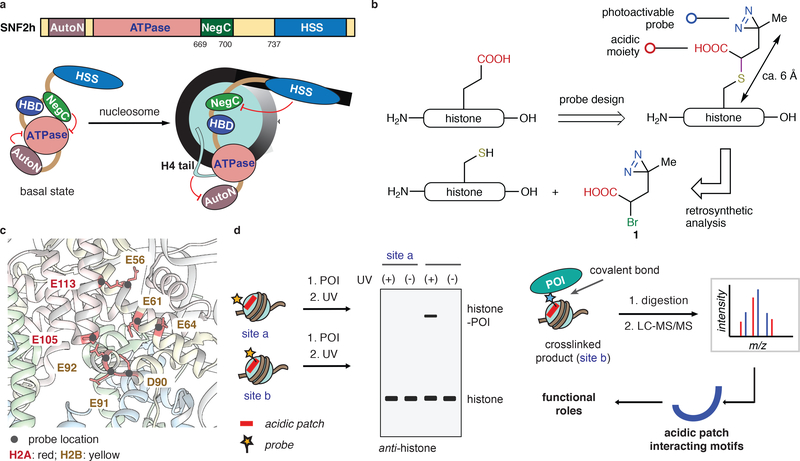
Figure 1 |. Site-directed photocrosslinking strategy used to explore SNF2h engagement with the nucleosome acidic patch.
(a) Schematic showing functional interplay between SNF2h domains and nucleosomal epitopes (unlabeled arrow points to the dyad axis). HBD: a hypothetical histone binding domain5. (b) Design, synthesis, and installation of a functional diazirine probe 1 on to histones. (c) Locations of photoactivable probes used in this study mapped on to the nucleosome structure (pdb, 1KX5). Each modified nucleosome contains one probe at a specific site as indicated. (d) Schematic showing ‘photoscanning’ of the acidic patch to identify the acidic patch interacting motifs. POI: protein of interest.
Recent studies have implicated a nucleosomal epitope termed the ‘acidic patch’ in the activity of several remodeler enzymes, including SNF2h and ACF20–22. This negatively charged structural feature is present on each face of the nucleosome disc within the globular core domains of histones H2A and H2B (Supplementary Fig. 1a,b)23,24. Importantly, the acidic patch has emerged as a key hotspot for nucleosome binding (reviewed in ref. 23,24). Moreover, a clear paradigm for nucleosome binding has emerged in which an arginine side-chain inserts into a pocket formed from a triad of residues in H2A (E61, D90 & E92) - known as the arginine anchor motif (Supplementary Fig. 1c)24–30. We have shown that alterations in and around the acidic patch, disrupt the activity of several remodelers21. In agreement with these findings, Narlikar and co-workers observed, using single-molecule FRET experiments, that acidic patch mutants alter the duration of discrete steps in SNF2h-mediated nucleosome translocation22. These results suggest that ISWI physically interacts with the acidic patch to regulate nucleosome sliding. It remains to be established which region(s) of SNF2h directly bind the acidic patch and how this engagement is coupled to sliding activity.
In the current study, we use a site-directed photocrosslinking approach to identify a conserved basic motif within SNF2h that is essential for remodeling activity. We also find that cancer-associated histone mutations that map to the acidic patch can either activate or inhibit ISWI remodeling activity, depending on the location. Motivated by these observations, we go on to show that asymmetric nucleosomes support unidirectional nucleosome sliding in biochemical assays. These studies have important implications for the regulation of chromatin structure in cells expressing oncogenic histones.
Results
Photoscanning reveals an acidic patch-binding motif
We hypothesized that a specific region within SNF2h directly engages the nucleosome acidic patch. To identify this putative binding epitope, we developed a site-directed photocrosslinking approach in which a reactive probe was scanned throughout the acidic patch – we refer to this strategy as photoscanning. Key residues in histones H2A and H2B were individually mutated to cysteine and subsequently alkylated with electrophilic reagent 1 that contains both a diazirine moiety and a carboxylic acid (Fig. 1b,c). Importantly, the alkylation reaction generates a side-chain structure that retains the negative charge of the native acidic residue. Thus, we imagined that incorporation of the probe into chromatin would lead to minimal perturbation of the acidic patch, allowing productive crosslinking to factors (e.g. SNF2h) that engage this region (Fig. 1d). With this in mind, each of the modified histones was successfully incorporated into nucleosomes (Supplementary Fig. 2).
Before moving to SNF2h, we first tested the viability of the photoscanning approach with factors known to bind the acidic patch, namely LANA peptide25, RCC126 and the Sir3-BAH domain27. In each case, we observed UV-dependent crosslinks between the probe-bearing histone and the ligand (Supplementary Fig. 3). Mutation of key binding residues, as expected, abolished the crosslinking. Moreover, the efficiency of UV-crosslinking was dependent upon the site of probe installation, highlighting the advantage of our systematic screening approach.
We next turned to identification of the putative binding motif within SNF2h. A restriction enzyme accessibility assay (REAA) was first used to show that all of the nucleosomes bearing our probes were substrates of SNF2h (Supplementary Fig. 4a,b). Crosslinking experiments were then conducted by incubating SNF2h with each modified nucleosome in the absence of ATP, followed by UV-irradiation and analysis by western blot (Fig. 2a, Supplementary Fig. 4c). The amount of histone-SNF2h crosslinks generated varied as a function of probe location. For example, we observed robust crosslinks when the probe was placed at position E91 in H2A, which is adjacent to the arginine-anchor pocket, whereas incorporation at H2AE56 led to lower levels of crosslinking. In the former case, the efficiency of photocrosslinking was reduced in the presence of the LANA peptide (Supplementary Fig. 4d). Moreover, no crosslinks were observed when the probe was introduced directly within the arginine anchor pocket, a finding that is consistent with the lower levels of remodeling activity (Supplementary Fig. 4b)21.
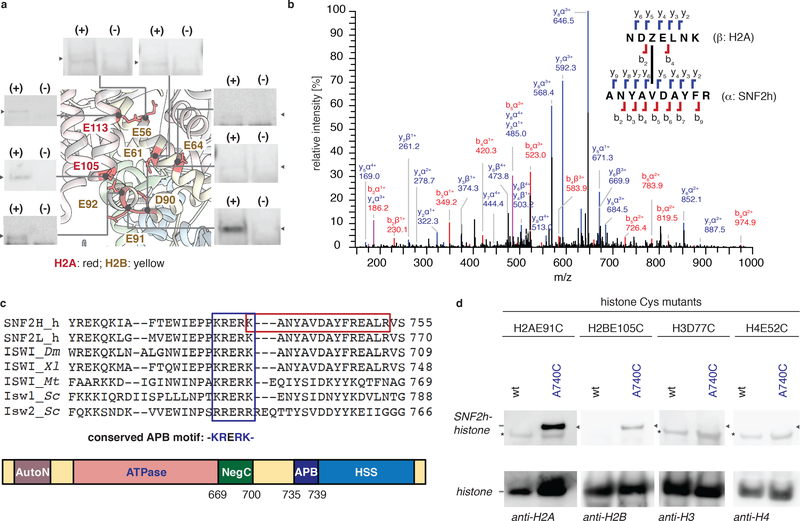
Figure 2 |. Photoscanning of SNF2h reveals a conserved basic motif that interacts with the nucleosome acidic patch.
(a) Summary of crosslinking data. Nucleosomes containing the diazirine probe at indicated positions (pdb, 1KX5) were incubated with SNF2h and UV irradiated. Mixtures were then analyzed by western blot using anti-H2A or H2B antibodies. UV-dependent (+/−) crosslinks were observed as a function of probe location. Uncropped figures are shown in Supplementary Fig. 4c. (n=2 independent experiments). (b) Mass spectrometry analysis of the crosslinking reaction between H2A containing the crosslinker at position E91 and SNF2h. Shown is a representation MS/MS spectrum annotated using the StavroX analysis program. Z = the diazirine probe. (single experiment). (c) Top, alignment of ISWI orthologs reveals a conserved basic motif (KRERK -the acidic patch binding (APB) motif, blue box) proximal to the site of crosslinking (red box). Bottom, domain structure of SNF2h showing the APB motif. The APB motif is located at the boundary of the NegC-HSS linker region and HSS. (d) Disulfide crosslinking. Nucleosomes containing the indicated histone cysteine mutant were incubated under oxidizing conditions with either wild-type SNF2h or a mutant containing a cysteine adjacent to the APB (SNF2h-A740C). Mixtures were resolved by non-reducing SDS-PAGE and analyzed by western blot using the indicated anti-histone antibodies. * = Nonspecific bands. Unlabeled arrows represent the location of 150 kD marker. Uncropped figures are shown in Supplementary Fig. 4h. (n=2 independent experiments for H2BE105C, H3D77C, H4E52C; n=3 independent experiments for H2AE91C).
Based on the photoscanning results, we selected H2AE91 as the optimal probe location. Accordingly, in-gel tryptic digestion of crosslinked bands, followed by LC-MS/MS, and data analysis using StavroX31 and pLink232 led to the identification of ten (47% of the hits using StavroX) and fifteen (88% of the hits using pLink2) crosslinked peptides that, with high confidence, map to a specific region of SNF2h (Fig. 2b; Supplementary Fig. 4e; Supplementary Table 1a,b). An analogous crosslinking experiment employing the ACF complex yielded consistent results (Supplementary Fig. 4f; Supplementary Table 2a,b). This highly conserved basic motif −735KRERK- of SNF2h is located at the boundary of the NegC-HSS linker region and HSS (Fig. 2c). Remarkably, this motif closely resembles known epitopes that engage the arginine-anchor pocket (Fig. 2c; Supplementary Fig. 4g).
A disulfide crosslinking approach was used to validate the putative interaction between the basic motif within SNF2h and the acid patch. Specifically, cysteine residues were strategically incorporated into each binding partner, SNF2h-A740C and either H2AE91C or H2BE105C, and the resulting SNF2h-nucleosome complex incubated under oxidizing conditions. Analysis by non-reducing SDS-PAGE followed by western blot, revealed the presence of a disulfide-linked SNF2h-histone crosslink (Fig. 2d; Supplementary Fig. 4h). Importantly, no crosslink was observed in a control experiment where the cysteine residue was moved to positions outside the acidic patch, H3D77C and H4E52C. Additional evidence for a direct interaction comes from a double-mutant cycle analysis where we observe positive coupling energy between the mutants on the basic sequence in SNF2h (hereafter, the acidic patch binding (APB) motif) and the acidic patch (Supplementary Fig. 4i–l). Collectively, these data are consistent with a direct interaction between the APB motif and the acid patch.
The APB motif is required for ISWI remodeling activity
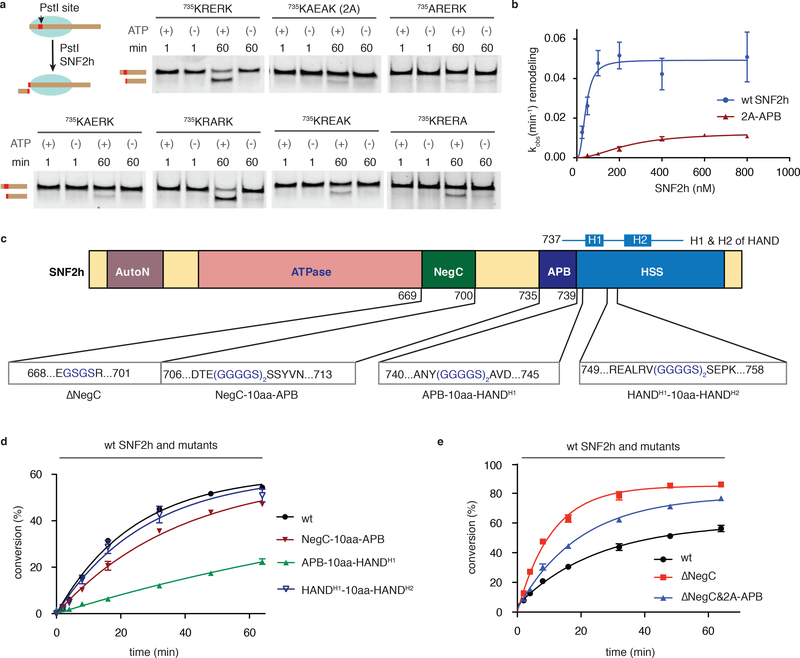
To examine the functional importance of the APB motif, we first conducted alanine scanning mutagenesis of individual residues within the region, as well a combination thereof (R736A-R738A, termed 2A-APB) (Supplementary Fig. 5a). Mutation of any basic residue within the motif markedly diminished the nucleosome sliding activity of SNF2h as monitored by REAA (up to eight-fold), while replacing Glu737 with Ala did not significantly alter remodeling activity (Fig. 3a, Supplementary Fig. 5b,c; Supplementary Table 3). Notably, the effect is more pronounced at the N-terminus of the motif (K735A and R736A). Similar inhibition was observed when the remodeling reactions were followed using an electrophoretic mobility shift assay (EMSA) (Supplementary Fig. 5d). As expected, mutation of the APB in SNF2h led to reduced levels of photocrosslinking (Supplementary Fig. 5e). Note, we also carried out mutagenesis of another basic sequence in SNF2h located just N-terminal to the APB motif (residues 720REKQK, Fig. 2c). In this case, we observed a much more modest reduction in the rate of sliding (~2-fold vs. ~8-fold for the APB mutants) (Supplementary Fig. 5c,f; Supplementary Table 3). The lysine/arginine-to-alanine mutants within the APB motif showed identical baseline ATPase activity to the wild-type protein, suggesting that the APB motif is not directly involved in ATP hydrolysis in the basal state (Supplementary Fig. 5g). Consistent with previous studies22, the presence of nucleosomes stimulated the ATPase activity of wild-type SNF2h (Supplementary Fig. 5h). This stimulation was slightly muted in the case of the R736A-APB mutant, implying that the APB motif may participate (at least in part) in controlling ATPase activity during translocation. We also measured the apparent Km (Kmapp) for both the wild-type and 2A-APB forms of SNF2h, finding that Kmapp increases ~5.5-fold upon mutation of the basic motif (Fig. 3b).
Figure 3 |. The ABP motif is required for SNF2h-mediated nucleosome remodeling.
(a) Remodeling activity of wild-type SNF2h and APB mutants (indicated in bold) was assessed by REAA. Schematic of the assay shown at top right; PstI = restriction endonuclease. Reactions (± ATP) were analyzed at indicated times by native polyacrylamide gel with SYBR® gold staining. Uncropped figures are shown in Supplementary Fig. 5b. (n=3 independent experiments). (b) Plot of observed remodeling rates, as measured by REAA, versus SNF2h concentration for both wild-type and the 2A-APB mutant. Data were fit to a Hill slope model yielding Kmapp values of 43 ± 7 nM and 243 ± 56 nM for wt SNF2h and 2A-APB, respectively. Errors = s.e.m (n=3 independent experiments). (c) Schematic showing deletion and insertion mutants in SNF2h designed to explore the interplay between the APB and the flanking NegC and HSS domains. The H1 and H2 helices in the HAND domain are indicated above. (d,e) Kinetics of remodeling as measured by REAA for the indicated SNF2h mutants relative to wild-type. Errors = s.e.m (n=3 independent experiments). For b,d and e data are represented as the mean of experimental replicates.
We wondered whether the APB motif has any functional interactions with the flanking regulatory domains in SNF2h. To explore this possibility, we inserted a flexible linker - (GGGGS)2 - at various positions in SNF2h (Fig. 3c, Supplementary Fig. 5a)33. Placement of the linker between the APB motif and HSS markedly diminished remodeling activity (Fig. 3d; Supplementary Fig. 6a,b; Supplementary Table 3). Notably, we observed no significant change in remodeling kinetics when the linker was inserted into the HAND sub-domain (Fig. 3d; Supplementary Table 3) 34. Similarly, the addition of the linker between NegC and the APB region also led to a modest reduction in sliding rate (Fig. 3d; Supplementary Fig. 6a,b; Supplementary Table 3), a result that is broadly in line with the work of Ludwigsen et al.33. To explore this further, we generated a version of SNF2h lacking the NegC domain (SNF2h-ΔNegC). Consistent with previous studies35, removal of this inhibitory domain stimulated SNF2h remodeling activity in REAA (Fig. 3e; Supplementary Fig. 6c,d; Supplementary Table 3), although it had less of an effect in the context of the ACF complex (Fig. 4a; Supplementary Fig. 6e). The nucleosome remodeling activity of SNF2h-ΔNegC was reduced when the APB was also mutated (Fig. 3e; Supplementary Fig. 6c,d; Supplementary Table 3). Taken together, these mutational studies indicate that the close apposition of the APB motif and the HSS domain is a critical determinant of SNF2h sliding activity.
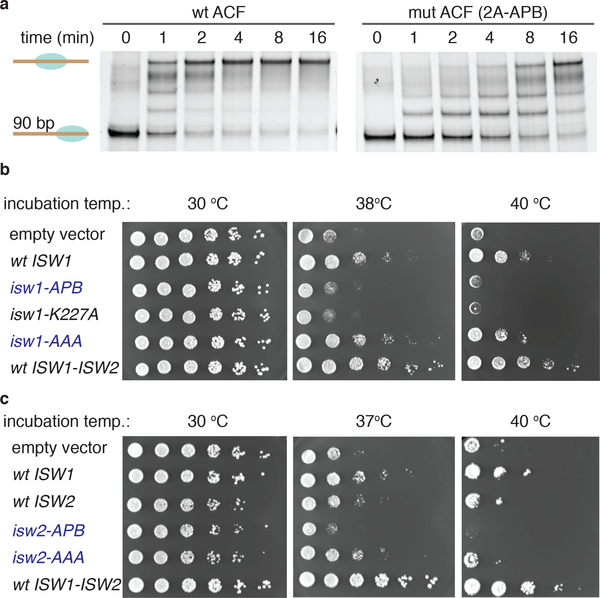
Figure 4 |. The APB motif is essential for ACF remodeling activity, and yeast viability.
(a) Remodeling activity of wild-type ACF complex (left) and the mutant ACF complex containing SNF2h 2A-APB (right) as read out by EMSA. ACF complexes (20 nM) were incubated with an end-positioned mono-nucleosome (10 nM) containing a 90 bp overhang in the presence of ATP. Aliquots of the reaction mixtures were analyzed at various time-points by native gel with SYBR gold staining. (n=3 independent experiments). Uncropped data are shown in Supplementary Fig. 17. (b,c) S. cerevisiae viability assay. Genomic copies of ISW1 and ISW2 were replaced by plasmid encoded copies of either ISW1 or mutant (panel b) or ISW2 or mutant (panel c). Yeast were then grown at indicated temperatures. Empty vector = pRS416 vector. wt ISW1 = pRS416 vector carrying wild-type ISW1 gene. isw1-APB = pRS416 vector carrying isw1-768AAEAA mutant gene (basic motif mutant). isw1-K227A = pRS416 vector carrying isw1-K227A mutant gene (known to abolish ATPase activity). isw1-AAA = pRS416 vector carrying isw1-751AAA mutant gene. wt ISW1-ISW2 = RS416 vector carrying wild-type ISW1-ISW2 genes. Wt ISW2: pRS416 vector carrying wild-type ISW2 gene. isw2-APB = pRS416 vector carrying isw2-743AAEAAA mutant gene (basic motif mutant). isw2-AAA = pRS416 vector carrying isw2-728AAA mutant gene. For b,c: two biologically independent samples, n=6 independent experiments.
We also assessed the importance of the APB motif in the context of the ACF complex. In line with our results on the isolated ATPase, mutation of the basic sequence significantly impaired the function of ACF (Fig. 4a, Supplementary Fig. 6e). This result encouraged us to validate our results in a cellular context. For this, we turned to S. cerevisiae, whose SNF2h homologues (Isw1 and Isw2) both contain the APB motif (Fig. 2c). We employed a yeast genetic system in which the genomic copies of ISW1 and ISW2 were replaced with plasmid-encoded versions thereof36. Yeast viability at elevated growth temperatures was restored when a wild-type copy of ISW1 was expressed, but not with a mutant version (isw1-K227A) that lacks ATPase activity (Fig. 4b). In line with our biochemical results, mutation of the APB motif also abolished the ability of Isw1 to support growth at elevated temperatures. An analogous result was obtained when wild-type vs. APB-mutant versions of Isw2 were employed in the growth assay (Fig. 4c). Consistent with our biochemistry data (Supplementary Fig. 5f), mutation of 720REKQK homologous sequence in Isw1 and Isw2 did not significantly alter the yeast viability (Fig. 4b,c). These results indicate that the APB motif is critical for yeast Isw1 and Isw2 mediated chromatin regulation.
Nucleosome desymmetrization leads to altered ISWI activity
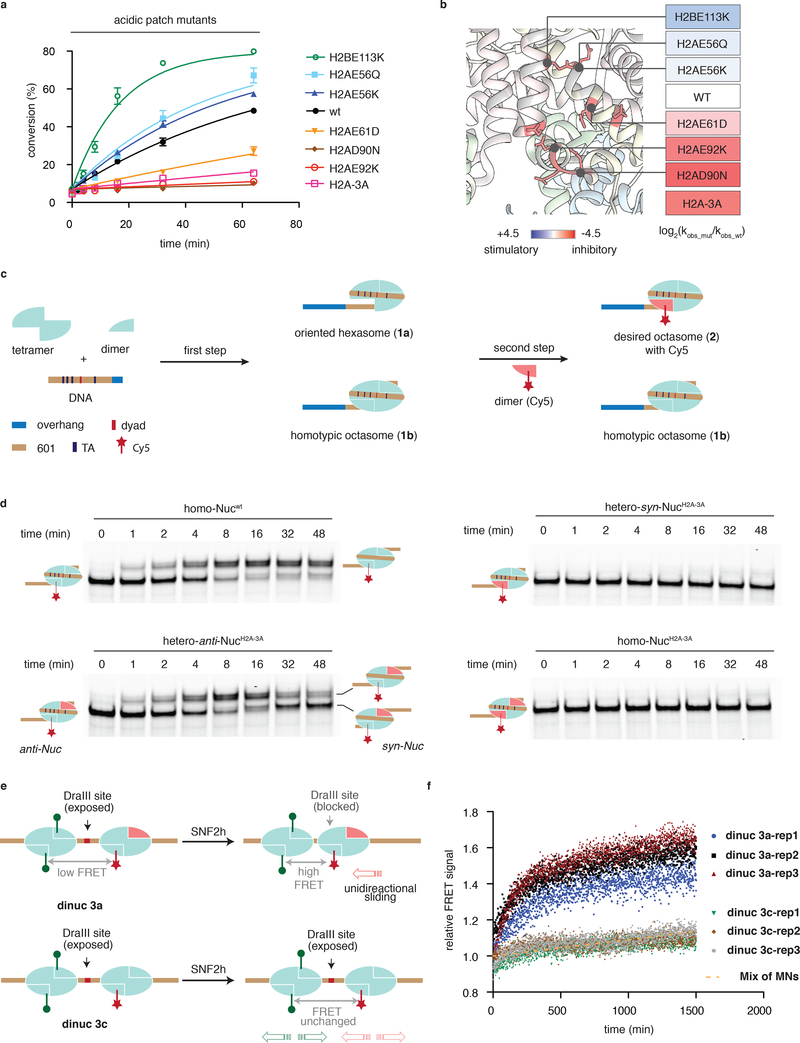
The nucleosome acidic patch has recently emerged as a mutational hotspot in a variety of cancers37–39. While the functional impact of these cancer-associated histone mutants is unexplored, it seems reasonable to expect some disruption of biochemical processes that rely on acidic-patch engagement, including chromatin remodeling. To investigate this possibility, we generated a series of nucleosomes harboring some of the most common acidic patch onco-mutants and subjected these to ISWI remodeling assays (Supplementary Fig. 7a). Strikingly, we found that depending upon the location, cancer-associated mutations in the acidic patch had the ability to either inhibit or activate SNF2h-mediated nucleosome remodeling (Fig. 5a,b, Supplementary Fig. 7b,c). Not surprisingly, mutations within the arginine-anchor pocket (H2AE92K, H2AD90N, and even H2AE61D) inhibited sliding activity. By contrast, nucleosomes bearing mutations located at more distal sites to the arginine anchor pocket (H2AE56Q, H2AE56K and H2BE113K) actually promoted the process. Analogous trends were observed in ACF complex-mediated remodeling, albeit to a slightly lesser degree (Supplementary Fig. 7d).
Figure 5 |. Nucleosome desymmetrization leads to altered ISWI activity.
(a) Kinetics of remodeling as measured by REAA for the indicated acidic patch mutants relative to wild-type. Errors = s.e.m (n=3 independent experiments). H2A-3A: H2A E61A, D90A, E92A. Data are represented as the mean of experimental replicates. (b) Heatmap representation of the kinetic data from panel a showing location of each mutant on the nucleosome surface (pdb, 1KX5). (c) Schematic of the two step-synthesis used to prepare heterotypic nucleosomes: step one, generation of oriented hexasomes (1a) by varying ratio of DNA, H3-H4 tetramer and H2A-H2B dimer. Step 2, formation of the desired heterotypic nucleosomes (2) carrying a fluorescent label (Cy5, red star). (d) Remodeling activity of SNF2h on indicated homotypic and heterotypic nucleosome substrates. SNF2h was incubated with an end-positioned mono-nucleosome containing a 45 bp overhang in the presence of ATP. Aliquots of the reaction mixtures were analyzed at various time-points by EMSA and monitoring the Cy5 label. (n=3 independent experiments). Uncropped figures are shown in Supplementary Fig. 18. (e) Schematic of the FRET-based remodeling assays employed oriented dinucleosomes containing Cy3 and Cy5 dyes and either heterotypic mutant acidic patch in one monomer (dinuc 3a) or the wild-type acidic patch in both (dinuc 3c). (f) Remodeling activity of SNF2h on dinuc 3a, dinuc 3c and a mixture of mononucleosomes as assessed by the FRET assay (n=3 independent experiments). SNF2h was incubated with indicated substrates in the presence of ATP. Change in normalized FRET signal (see Methods) as a function of time as shown for three replicates of each. Yellow dotted line: curve fitting for change in FRET signal of a mixture of pre-ligated mono-nucleosomes (n=2 independent experiments).
Histones H2A and H2B are each encoded on multiple genes, meaning that cancer cells expressing a mutant allele of one of these histones will likely contain heterotypic nucleosomes, i.e. containing a wild-type copy and a mutant copy of the protein within the histone octamer. By contrast, homotypic mutant nucleosomes in which each copy of histone harbors the mutation are statistically much less likely to occur, assuming that wild-type H2A/H2B dimers (in great excess) and mutant H2A/H2B dimers are stochastically incorporated into chromatin40. The presence of wild-type and mutant copies of a histone within a heterotypic nucleosome, necessarily leads to a desymmetrization of the structure. In the case of the acidic patch mutants, one face of the nucleosomal disc will present a native structure while the other face will be altered. Given this is likely the physiologically relevant situation in cancer cells expressing the mutant histones, we asked how ISWI remodelers interpret such ‘Janus’ substrates.
To examine this question, we initially compared remodeling activity between two sets of nucleosomes, namely: (i) mixtures of homotypic nucleosomes containing either a wild-type acidic patch or a mutated arginine anchor pocket (H2A E61AD90AE92A, termed H2A-3A) at different ratios, and (ii) a mixture of stochastic nucleosomes in which the core histone octamer was reconstituted using equimolar amounts of wild-type H2A and H2A-3A (Supplementary Fig. 8a,b). Analysis of the remodeling activity using SNF2h and the ACF complex revealed that the stochastic mix exhibited a similar kinetic profile to the 50/50 mixture of homotypic nucleosomes (Supplementary Fig. 8c,d). These results are surprising since ~75% of the stochastic mix should contain at least one copy of the mutant histone (Supplementary Fig. 8b). This suggests that the two heterotypic species in the stochastic mixture, hetero-syn-Nucmut and hetero-anti-Nucmut – which differ based on whether the mutant acidic patch is on the same side or opposite side of the DNA overhang (Supplementary Fig. 8e) - are not functionally equivalent. This intriguing idea motivated us to pursue more sophisticated methods to prepare and monitor remodeling reactions with heterotypic nucleosomes.
To generate heterotypic nucleosomes, we adapted the two-step protocol of Bowman and co-workers such that a Fluorescent dye was strategically incorporated to the second H2A/H2B dimer (Fig. 5c, Supplementary Fig. 9)41. This refinement provided a facile route to the desired oriented, heterotypic nucleosomes and, critically, allowed us to exclusively follow the action of the remodeler on the desired substrate by exploiting the dye signal within an EMSA format. The above streamlined procedure was used to prepare four substrates for remodeling assays; two heterotypic, fluorescently-tagged nucleosomes in which an inhibitory H2A/H2B dimer (i.e. containing H2A-3A) was positioned on the disc face either proximal (hetero-syn-NucH2A−3A) or distal (hetero-anti-NucH2A−3A) to the DNA overhang, as well as two control fluorescently-tagged nucleosomes containing wild-type H2A/H2B dimers (homo-Nucwt) or mutant dimers (homo-NucH2A−3A) on both faces (Supplementary Fig. 10). Analysis of the SNF2h-mediated remodeling using EMSA revealed that the two oriented, heterotypic nucleosomes are not functionally equivalent (Fig. 5d). Thus, while the hetero-anti-NucH2A−3A substrate had a similar sliding rate to homo-Nucwt, the hetero-syn-NucH2A−3A was completely inactive, similar to homo-NucH2A−3A. We note that the sensitivity of SNF2h to perturbations in the acidic patch differs substantially from that observed for the Chd1 remodeler41.
Our experiments also revealed an important distinction between the homo-Nucwt and hetero-anti-NucH2A−3A substrates (Fig. 5d, top left vs. bottom left). While both substrates behaved similarly over the initial phase of the assay, leading to the generation of a centered nucleosome species, the hetero-anti-NucH2A−3A substrate subsequently continued to be remodeled and eventually “returned” to an end-positioned state. By contrast, the wild-type nucleosome remained mostly centered, consistent with the ability of the remodeler to operate on either side of the homo-Nucwt substrate, i.e. an equilibrium state is reached. Since the remodeler can only function on one side of the hetero-anti-NucH2A−3A substrate, we propose that resulting unidirectional sliding moves the nucleosome from one end position to the center and then to the other end position (hetero-syn-NucH2A−3A). Indeed, we observed that SNF2h moved heterotypic nucleosomes bearing H2A-3A or inhibitory onco-mutant (H2AE92K) dimers from the center of DNA sequences towards the end positions (Supplementary Fig. 11; Supplementary Fig. 12a,b). Interestingly, we also observed a significant loss of heterotypic nucleosome signals after the generation of the end positions, suggesting the instability of these heterotypic substrates upon remodeling (Supplementary Fig. 12). The appearance of the fluorescent signal in the quenching plasmid DNA (Cy5 scan) and the formation of species away from the end position (SYBR scan) is consistent with the proposed unidirectional movement of the inhibitory heterotypic nucleosome to create a longer overhang on the mutant side (Supplementary Fig. 12c). Importantly, the remodeling activity of the ACF complex was similarly sensitive to the presence of Janus nucleosome substrates carrying an inhibitory mutation (H2AE92K) in the acidic patch (Supplementary Fig. 12d).
Next, we asked whether any functional imbalance between the two acidic patches within a nucleosome would lead to a bias in the sliding direction. Accordingly, a fluorescently-tagged heterotypic nucleosome carrying a stimulatory dimer (H2BE113K; hetero-syn-NucH2BE113) was prepared together with control wild type nucleosomes (Supplementary Fig. 13). Gratifyingly, SNF2h moved the hetero-syn-NucH2BE113 substrate quicker than the wild-type towards the centered position due to the stimulatory effect of the H2BE113 mutant (Supplementary Fig. 13a,b). As expected, the opposite was true for the inverse orientation, hetero-anti-NucH2BE113 (Supplementary Fig. 13c,d). Furthermore, this effect was not restricted to cancer-mutations as heterotypic nucleosomes containing H2A.Z, a histone variant that is known to simulate ISWI activity28, also exhibited biased sliding using the EMSA format (Supplementary Fig. 13e–j).
Finally, we asked whether a breakdown in nucleosome symmetry would lead to a change in ISWI-mediated inter-nucleosome spacing, a prediction of our unidirectional sliding model. To approach this question, we developed a FRET-based assay employing ‘designer’ di-nucleosomes (Figure 5e). A DNA ligation step employed DraIII sticky ends allowed us to place a heterotypic nucleosome containing a single mutant acidic patch in a defined orientation next to a wild-type nucleosome (Supplementary Fig. 14). SNF2h-mediated remodeling of these dinucleosomes was assayed by monitoring FRET between incorporated Cy3 and Cy5 dyes (Fig. 5f, Supplementary Fig. 15a,b). Remarkably, treatment of mutant dinucleosomes 3a with the remodeler led to a substantially larger time-dependent increase in FRET as compared to wild-type dinucleosomes 3c. This is consistent with closer nucleosome spacing in the former compared to the latter. Additional support for this interpretation came from a restriction enzyme accessibility assay exploiting a strategically placed DraIII site in the linker region between the nucleosomes (Supplementary Fig. 15c,d). Collectively, these data indicate that a functional imbalance within a nucleosome can lead to aberrant nucleosome spacing by SNF2h.
Discussion
In this study, we have identified a conserved basic motif (APB) within SNF2h, which is required for efficient chromatin remodeling. Collectively, our biochemical studies lead to a model in which the APB motif plays a critical role in coupling ATPase activity to DNA translocation (Supplementary Fig. 16a). We propose that the APB motif communicates with HSS to facilitate the DNA movement from the entry site into the nucleosome42. Single-molecule FRET studies reveal that there is a lag time between the entry and exit of DNA into and out of the nucleosome22. It is thought that ATPase mediated DNA movement from superhelical location 2 (SHL2) to the dyad is absorbed by DNA twist defects. Accumulated DNA translocation from the entry site then triggers DNA movement to the exit site18,43. Yet, how ATPase activity at SHL2 leads to DNA being drawn into nucleosome from the entry site is poorly understood33. We propose that binding of the APB motif to the acidic patch could play a role in facilitating the DNA shift by anchoring HAND-SANT domains to the histone octamer42. In addition, anchoring the HAND-SANT domain to the acidic patch might aid in unpeeling DNA from the histone surface to promote efficient DNA movement33. Recently, Chen and co-workers reported a series of the cryo-EM structures of yeast ISWI in complex with mononucleosome substrates44. Unfortunately, the C-terminal part of the protein, including the entire APB-HSS region, was not resolved in any of the reported structures. The reason(s) for this is unclear, but could to be taken as evidence that this region of ISWI engages in more dynamic interactions with the nucleosome as compared with the translocase domain. Indeed, we stress that our model does not require that the APB motif engage the acidic patch throughout the translocase cycle. Moreover, it is conceivable that other basic regions within SNF2h including AutoN and NegC (or other ISWI subunits) could also interact with the acidic patch at one or more steps in remodeling, as has previously been suggested22.
Recent genome-wide sequencing efforts have revealed a large number of histone missense mutations in human cancers37–39,45,46. With few exceptions38,45,46, the functional implications of these mutations have not been studied. Of particular relevance here are those cancer-associated mutations that map to the nucleosome acid patch, which we note occur at high frequency39. Remarkably, we find that these mutations can either inhibit (E61D, D90N, E92K of H2A) or stimulate (H2AE56Q, H2AE56K and H2BE113K) ISWI remodeling activity. Interestingly, the latter group includes a recurrent mutation (H2AE56Q) within human carcinosarcomas that has previously been linked to a more invasive cellular phenotype37.
Our initial survey of these missense cancer mutations employed homotypic nucleosome substrates. However, because histones H2A and H2B are encoded on multiple genes, heterotypic nucleosomes will predominate. The implications of this heterotypic incorporation are especially intriguing when it comes to acidic patch mutations since only one face of the nucleosome disc structure will be altered - creating what can be thought of as a Janus-like substrate. We have shown that this desymmetrization has important implications for ISWI-mediated remodeling. Specifically, our biochemical remodeling data shows that modification of one acidic patch within an asymmetric nucleosome creates a functional imbalance within the substrate that leads to a net unidirectional movement of the histone octamer from the center position. We note that during the revision of our manuscript, Bowman and co-workers published a report in which they reached the same conclusion, in this case employing asymmetric mononucleosomes carrying alanine mutations47. In our study, we show that this biased sliding phenomenon extends to patient-derived cancer mutations and that, remarkably, this occurs regardless of whether inhibitory or stimulatory acidic patch mutations are asymmetrically incorporated. Furthermore, our study revealed that the direction of motion is dictated by which face of the nucleosome disc presents the mutation. Thus, ISWI moves heterotypic nucleosomes carrying an inhibitory cancer mutation unidirectionally to create a longer linker DNA proximal to the mutated face, while the opposite is true for stimulatory heterotypic nucleosomes (Supplementary Fig. 16b). Conceivably, this bias in sliding rates could lead to aberrant nucleosome spacing in cancer cells, possibly leading to dysregulation of genomic processes. Supporting this idea, we deployed a FRET-based designer chromatin assay to show that inter-nucleosome spacing by ISWI is indeed affected by the presence of a heterotypic mutant nucleosome.
The impact of nucleosome desymmetrization on ISWI remodeling may have broader implications48,49. This is because both histone variants and histone PTMs can also alter the structural and electronic properties of the acidic patch. An interesting example of this involves the histone variant H2A.Z, which is enriched at the +1 nucleosome relative to gene promoters49,50. Nucleosomes containing H2A.Z have an extended acidic patch, which leads to stimulation of ISWI activity20. Pugh and coworkers have shown that +1 nucleosomes are heterotypic in nature and that the variant is disproportionally deposited distal to the nucleosome depleted region within the promoter49. Our data imply that heterotypic nucleosomes containing H2A.Z are remodeled by ISWI such that the DNA on the side distal to the variant becomes longer. It remains to be seen if this type of unidirectional sliding plays a role in defining promoter architecture.
We have recently shown that ISWI remodeling rates are sensitive to a range of histone PTMs, including those that localize to the acidic patch21. Similar to the onco-mutations, both inhibitory and stimulatory effects are observed. It is thus intriguing to speculate that nucleosome desymmetrization through face-specific incorporation of histone PTMs around the acidic patch also plays a role in regulating ISWI chromatin remodeling. Indeed, Bowman and co-workers have shown that asymmetric nucleosomes containing ubiquitylated histone H2B stimulate nucleosome sliding by the yeast Chd1 remodeler41.
In summary, we have shed new light on the role of the nucleosome acidic patch in ISWI-mediated chromatin remodeling. The tools, reagents, and assays developed as part of this study are likely to have broad utility, particularly since other remodeler families appear to utilize the acidic patch as part of their function. Finally, we propose that desymmetrization of the nucleosome acidic patch by heterotypic incorporation histone mutants, variants or PTMs, plays an underappreciated role in chromatin remodeling with potential ramifications for the regulation of genome architecture.
ONLINE METHODS
General information
Chemical reagents and solvents were purchased from Sigma-Aldrich or Thermo Fisher Scientific. Cloning enzymes were purchased from New England BioLabs. Primers were purchased from Integrated DNA Technologies and Sigma-Aldrich. All plasmid sequences were verified by GENEWIZ. Mutagenesis was performed using the Inverse PCR method, and PCR purification kits were obtained from Qiagen. Size-exclusion chromatography was performed on an AKTA FPLC GE Healthcare system coupled with a P-920 pump and a UPC-900 monitor. Analytical reversed-phase HPLC was performed on an Agilent instrument (1200 series) with a Vydac C18 column (4 × 150 mm, 5 μm), using solvent A (0.1% trifluoroacetic acid (TFA) in water), and solvent B (90% acetonitrile, 0.1% TFA in water) as the mobile phases. Semi-preparative scale purifications were performed on an Agilent 1100 series using a Vydac C18 semipreparative column (10 mm × 250 mm, 12 μm) at 4 ml/min. A MicrOTOF-Q II ESI-Qq-TOF mass spectrometer from Bruker Daltonics was used for ESI–MS analysis. SDS PAGE gel, native gel and western blot images were taken with an ImageQuant® LAS 4000 S5 imager (GE Healthcare), and a LI-COR Odyssey Infrared Imager. Agar plates were imaged using a GE ImageQuant. ATPase assays were monitored using a VersaMax tunable microplate reader from Molecular Devices. A Fluorolog3–11 fluorimeter (Horiba) was used for fluorescence measurements related to the ensemble FRET assay. Cell lysis was carried out using a S-450D Branson Digital Sonifier, an Emulsiflex-C3 homogenizer, or a Dounce homogenizer. UV irradiation was performed using a MAXIMA™ ML-3500S lamp from Spectroline. Images of the structure was obtained using the UCSF Chimera package. Crosslinked peptide MS analysis was conducted using the StavroX31, pLink232 software tools. Sequence alignment analysis was performed using Cluster Omega51. Flash silica gel chromatography was performed using Fisher Chemical silica gel (Grade 60, particle size 230–400 Mesh). NanoBay 300 MHz instrument was used to obtain NMR spectra (calibrated using residual undeuterated solvent as an internal reference CHCl3 @ 7.26 ppm for 1H-NMR and 77.16 ppm for 13C-NMR). The following abbreviations is used in 1H NMR data: s = singlet, d = doublet, m = multiplet.
Expression and purification of recombinant histones and mutants
Recombinant human histones, variants, and mutants were expressed in and purified from E. coli following previously described proctotol21.
Synthesis of reagent 1
Reagent 1 was synthesized in 3 steps from levulinic acid according to a previously described protocol52. See Supplementary Note 1 for the detailed synthesis. See Supplementary Fig. 19 for characterization data.
Installation of the probe on histone via alkylation of cysteine residues
Approximately 1.5 mg (ca. 100 nmol, 1 eq.) of a histone mutant (i.e. containing an acidic patch residue mutated to a cysteine) was dissolved in degassed alkylating buffer (400 μL; 1 M HEPES, pH 7.75, 6 M guanidine hydrochloride, 10 mM TCEP) in a small vial equipped with a stir bar. A solution of 0.5 μL of probe reagent (3:1 mixture of 1 and 2, ca. 20 eq. of reagent 1) in alkylating buffer (50 μL) was added. The tube was sealed and stirred at 25 °C without exposure to light for 9 hrs. After that, an additional probe solution (0.25 μL of reagent (ca. 10 eq.) in alkylating buffer (25 μL)) was added, and the reaction mixture was then stirred for additional 9 hrs at 25 °C. The reaction progress was monitored by analytical RP-HPLC and ESI-MS, and stopped with TFA (50%, ca. 20 μL). Alkylated histones were purified by semi-preparative RP-HPLC using a 30–70% HPLC solvent B gradient to obtain ~0.5 mg of pure product (ca. 30% isolated yield). The modified protein was analyzed using analytical RP-HPLC and ESI-MS. See Supplementary Fig. 20.
Labeling of histone
Wild-type H2A and the acidic patch mutant thereof (i.e. 3A_mut) were labeled with Cyanine-5 maleimide (abcam), Cy3 maleimide (GE Healthcare) at position T120C according to a previously described protocol with some modifications53. The protein was characterized using analytical HPLC and ESI-MS - see Supplementary Fig. 20.
Histone dimer, tetramer and octamer assembly
Histone octamers, tetramers and dimers were assembled using an established method from lyophilized histones21,54. Histones were combined in following molar ratios: H2A:H2B:H3:H4 = 1.1:1.1:1:1 for octamer assembly; H3:H4 = 1:1 for tetramer assembly; H2A:H2B = 1:1 for dimer assembly.
DNA preparation
DNA fragments containing a modified Widom 601 sequence were generated via PCR. See Supplementary Note 2 for further information.
Nucleosome reconstitution
Method 1: Synthesis of homotypic nucleosomes from octamers and 601 DNAs
Method 2: Two-step synthesis of heterotypic nucleosomes
The two-step synthesis of a mixture of homotypic and heterotypic nucleosomes followed the protocol of Levendosky, R. F. et al41 with some modifications.
Step 1: To prepare a mixture of the oriented hexasome and the undesired homotypic octasome Supplementary Fig. 9)
Generally, a 601 DNA fragment was mixed with a similar volume of KCl in water (4 M) to make a 2 M KCl solution. Then, a preformed histone tetramer, and a dimer were added (ratio of DNA:tetramer:dimer = 0.8:1:1, Supplementary Fig. 9). The refolding buffer (2 M NaCl, 10 mM Tris, 0.5 mM EDTA, 1 mM DTT, pH 7.8 at 4 °C) was used to adjust the final concentration of nucleosome to 0.5–1 μM. The subsequent salt gradient dialysis steps are identical to the reported method for reconstitution of nucleosomes21. The final mixture of hexasome and octasome was in 10 TEK buffer (10 mM Tris, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, pH 7.5). The ratio of the homotypic octasome, the oriented hexasome, and the free DNA was assessed by native polyacrylamide gel electrophoresis (5–6% acrylamide gel, 0.5×TBE, 150 V, 1 h) via SYBR® gold staining. The hexasome species migrated quicker than the octasome (variable depending on the nucleosome variant). If necessary, the mixture of nucleosomes was concentrated to achieve higher concentration using Vivaspin 500 centrifugal filter units (10 kDa MW cutoff, Vivaproducts). A typical successful hexasome preparation contains about 30–50% of free DNA.
Step 2: To generate a mixture containing the desired labelled heterotypic nucleosome.
To the mixture of the oriented hexasomes and undesired octasomes from the step 1 (total DNA 0.1–0.5 μM, 1 eq., 1 vol) was added 1/10 vol of 2M TEK buffer (10 mM Tris, 2 M KCl, 0.1 mM EDTA, 1 mM DTT, pH 7.5) to adjust to the final concentration of KCl to c.a 200 mM. The second H2A/H2B dimer (Fluorescent labelled, 5 eq.) was then added and mixed quickly. The mixture was then incubated at 37 °C for 30 min, followed by centrifugation at 4°C, 17,000g for 10 min. The supernatant was dialyzed against 10 TEK buffer (2×4 h). The quality of the nucleosome mixture was assessed by native polyacrylamide gel electrophoresis (5–6% acrylamide gel, 0.5×TBE, 150 V, 1 h) via SYBR® gold staining. The incorporation of Cy5 conjugated dimer was visualized using Image Studio Lite (LI-COR, 700 nm channel).
Preparation of dinucleosomes
Homotypic nucleosome Nuc 1 was synthesized via Method 1, and nucleosome mixture (containing heterotypic Nuc 2) was synthesized via Method 2 using corresponding histones and pre-digested sticky end DNAs. Nuc 1 (100 nM) and Nuc 2 (100 nM) were mixed in ligation buffer (100 mM Tris, pH 7.5, 8 mM MgCl2, 1 mM ATP, 100 mM DTT, ligase 20 U/uL) on ice at 4 °C. The mixture was incubated at 16 °C for 30 min. The ligated dinucleosome was then dialyzed in 10 TEK buffer (2×4 hrs). MgCl2 (200 mM) was then added to the final concentration of 12 mM. The mixture was incubated at rt for 15 mins following by centrifugation at 17,000g for 15 min at 4 °C. The precipitate was then dissolved in 10 TEK buffer, followed by dialysis in 10 TEK buffer (2×4 hrs). The quality of the dinucleosome mixture was assessed by native polyacrylamide gel electrophoresis (5.5% acrylamide gel, 0.5×TBE, 150 V, 1 h) via imaging using Cy3 and Cy5 channels. If necessary, the mixture of nucleosomes was concentrated to achieve higher concentration using Vivaspin 500 centrifugal filter units (10 kDa MW cutoff, Vivaproducts).
Expression and purification of chromatin remodelers in Sf9 cells
The SNF2h ATPase subunit, The ACF1 subunit and the ACF complex (SNF2h and ACF1) were produced in Sf9 cells using a baculovirus expression vector system as described previously21.
Expression and purification of SNF2h and mutants in E. coli
SNF2h expression in E. coli and purification were modified from a previously reported protocol55. The SNF2h construct was cloned into a pET vector containing a His6Sumo insert to make pET-His6Sumo-linker-SNF2h (linker sequence: AGSA). Cultures were grown in 2xLB (20g tryptone, 10g yeast extract, 10g NaCl for 1L culture) media to OD600 ca. 0.3 at 37 °C, then switched to 18 °C and grown until they reached an OD600 of ca. 0.75. Protein expression was induced at 18˚C by 0.4 mM IPTG for 18 hrs. Cell pellets were resuspended in Lysis Buffer (25 mM HEPES, pH 8, 300 mM KCl, 7.5 mM imidazole, 10% v/v glycerol, 2 mM β-mercaptoethanol, Protease Inhibitor Cocktail (1 tablets/8 L culture), and 1 mM PMSF), and lysed by high pressure with an Emulsiflex-C3 homogenizer. The Sumo-SNF2h was purified using Ni-NTA column (2 mL of resin/1 L culture). His6-Sumo tag was cleaved off by treating with Ulp1 protease. The mixture was then passed through Ni-NTA beads twice to remove His6-Sumo, and His6-Ulp1. SNF2h proteins was then further purified by size-exclusion chromatography on a Superdex 200 10/300 column (GE Healthcare) using degassed SEC buffer (25 mM Hepes, pH 7.6, 300 mM KCl, 2 mM β-mercaptoethanol). Relevant fractions were then dialyzed overnight in Final Buffer (25 mM Hepes, pH 7.5, 200 mM KCl, 15% v/v glycerol, 2 mM β-mercaptoethanol, 0.5 mM PMSF). SNF2h samples were flash-frozen in liquid nitrogen and stored at −80 °C. Concentrations of SNF2h proteins were determined by SDS-PAGE with BSA protein standards and staining with Coomassie blue staining. SDS-PAGE data is shown in Supplementary Fig. 5a. Note, the activity of wild-type SNF2h produced in E. coli was found to be identical that produced and purified from SF9 cells.
Post-expression preparation of the wild-type and mutant ACF complexes
The wild-type and mutant ACF complexes were made by mixing wild-type ACF1 and SNF2h/mutant at an equimolar ratio in the REA buffer (20 mM HEPES, 4 mM Tris, pH 7.75, 60 mM KCl, 10 mM MgCl2, 10% glycerol, and 0.02% (v/v) IGEPAL CA-630) for 15 minutes prior to the remodeling assay.
Protocol for the photocrosslinking reaction
Photoactivable nucleosomes (5–15 pmol, 1 eq., 0.5 μM), chromatin factors (1–3 eq.) or peptides (10 eq.) were combined in a fifty-microliter reaction (20 mM HEPES, 4 mM Tris, 7.75, 60 mM KCl, 10 mM MgCl2 and 0.02% (v/v) IGEPAL CA-630). Other additives -ATP (2 mM), AMP-PCP (2 mM) and LANA (25 μM)-were added as indicated. The reaction mixture was incubated at 30 °C for 30 mins, then switched to 4 °C for 10 min. The photocrosslinking reaction was triggered by UV irradiation at 4 °C for 10 min with 365 nm UV light (MAXIMA™ ML-3500S). Protein mixtures were resolved on SDS-PAGE, and crosslinked products were assessed using western-blot.
Photocrosslinking-MS/MS experiment
Crosslinked products obtained from photocrosslinking reactions of the selected photoactivable nucleosome (probe at H2AE91) and SNF2h or ACF complex were resolved on SDS-PAGE gel (8% bis-tris gel). Crosslinked bands were excised from the gel and pooled into a single tube (in the case of the ACF complex, both ACF1 and SNF2h crosslinked bands were pooled into one tube). In-gel tryptic digestion was performed overnight in 400 μL of NH4HCO3 (50mM), Trypsin Gold (0.5 μg/100uL) at 37 °C. Samples were then acidified with formic acid (final 1%), followed by peptide extraction. The extracted solution was then dried in a speedvac and resuspended with 20 μL of 0.1% formic acid pH 3. Injection of samples (2 μL per run) was conducted using an Easy-nLC 1200 UPLC system. Samples were loaded directly onto a 45cm long 75 μm inner diameter nano capillary column packed with 1.9 μm C18-AQ (Dr. Maisch, Germany) mated to metal emitter in-line with an Orbitrap Fusion™ Lumos™ (Thermo Scientific™, USA). The mass spectrometer was operated in data-dependent mode with the 120,000 resolution MS1 scan (400–1500 m/z) in the Orbitrap followed by MS/MS scans with CID fragmentation in the ion trap. Dynamic exclusion list was invoked to exclude previously sequenced peptides for 60s if sequenced within the last 30s and a maximum cycle time of 3s was used. Raw files were converted to mzXML format using MSconvert with default settings. mzXML files were analyzed using either the StavroX or pLink2 software tools. The crosslinker residue (i.e. cysteine alkylated with reagent 1) was defined as an amino acid with composition C8H11N3O3S1 (229.05211183) and Cys-Diazirine was defined as the crosslinker (composition –N2 (−28.006158); Site 1: Z; Site 2: Any AA). A FASTA file consisting of the modified histone sequence, SNF2h and ACF1 subunits included in the experiment was utilized.
Disulfide crosslinking experiment
Cysteine mutant nucleosomes (5 pmol, 1 eq.) and SNF2h-A740C (0.8 eq.) were combined in a fifty-microliter reaction in the REA buffer (20 mM HEPES, 4 mM Tris, pH 7.75, 60 mM KCl, 10 mM MgCl2 and 0.02% (v/v) IGEPAL CA-630). The reaction mixture was incubated at 30 °C for 30 mins, then transferred to a dialysis unit and the mixture was dialyzed against reaction buffer containing 2 mM of L-glutathione oxidized (GSSG), 1mM PMSF (final pH 7.9) at 4 °C for 4 hrs. Protein mixtures were resolved on non-reducing SDS-PAGE, and crosslinked products were assessed using western-blot.
Restriction enzyme accessibility assay (REAA)
REAA experiments were conducted under single turnover conditions (an excess amount of SNF2h and mutants vs. nucleosome) as described previously with some modifications21. Briefly, 10 nM of nucleosomes was incubated in REA buffer (20 mM HEPES, 4 mM Tris, pH 7.75, 60 mM KCl, 10 mM MgCl2, 10% glycerol, and 0.02% (v/v) IGEPAL CA-630) in the presence of 2 U/μl PstI (NEB). Reactions were incubated at 30 °C for 10 min to digest the majority of free DNA. SNF2h (200–400 nM) was then added. Reactions were initiated by addition of ATP (2 mM), and timepoints were taken and quenched using 1.5×vol of REAA quenching buffer (10% glycerol, 20 mM HEPES, 4 mM Tris, pH 7.75, 2% SDS, 0.2 mg/ml bromophenol blue, 50U/ml proteinase K (NEB)) Samples were deproteinized at 37 °C for 1 h, then resolved by native PAGE (7% polyacrylamide, 0.5× TBE). Gels were stained with SYBR® Gold, and imaged on an ImageQuant® LAS 4000. Densitometry measurements were conducted using Image Studio Lite (LI-COR). Data were fitted to a single exponential decay equation using GraphPad Prism.
(y0 :fraction cut at t = 0; kobs: observed rate constant; t: reaction time; p: fraction cut at plateau). Reactions of SNF2h and mutants were fit constrained to a common y0 and p. The ACF complex (2–20 nM) mediated reactions were performed similarly under the sub-catalytic concentration of enzymes. For measurement of the apparent Km (Kmapp), SNF2h and mutants at different concentrations (25–800 nM) were used. These data were fit to a Hill slope model (GraphPad Prism). All REAA data with wild type nucleosomes, the acidic patch mutants, heterotypic nucleosomes were taken from at least two different nucleosome samples. All REAA data with SNF2h mutants were taken from at least two different batches of enzymes, except ARERK, KAERK, KRARK, KRERA, ΔNegC, ΔNegC&KAEAK, APB-10aa-HANDH1.
Electrophoretic mobility shift assay (EMSA)
These assays used an electrophoretic mobility shift-based strategy described previously with some modifications56. Remodeling reactions involving SNF2h and mutants thereof (250–600 nM) were performed under single turnover conditions, while the ACF complex (3–20 nM) mediated reactions were performed at a sub-stoichiometric concentration of enzyme relative to the substrate. Briefly, 20–25 nM of nucleosomes and enzymes were incubated at 30 °C in the REA buffer. Reactions were initiated with the addition of ATP (2 mM), and timepoints were quenched with 1×vol of pre-chilled EMSA quenching buffer (0.7 mg/mL DNA, 25 mM ADP, 10% sucrose in 10TEK). Timepoints were kept on ice and resolved by native PAGE (7% polyacrylamide, 0.5× TBE). After Staining with SYBR® Gold, gels were imaged on ImageQuant® LAS 4000 (GE Healthcare). For the reaction with nucleosomes carrying dyes, gels were directly imaged using a LI-COR Odyssey Infrared Imager by scanning for fluorescent labels (700 channel). All EMSA data with wild type nucleosomes, the acidic patch mutants, heterotypic nucleosomes were taken from at least two different nucleosome samples. All EMSA data with SNF2h mutants were taken from at least two different batches of enzymes, except ARERK, KAERK, KRARK, KRERA, ΔNegC, ΔNegC&KAEAK, APB-10aa-HANDH1. Densitometry measurements of starting nucleosome band (e.g. end-position) were performed using Image Studio Lite. Data were fitted to a single exponential decay equation using GraphPad Prism.
(y0 :the signal of starting MN at t = 0; kobs: observed rate constant; t: reaction time; p: starting MN at plateau). Reactions of SNF2h and mutants were fit constrained to a common p.
Free energy change for catalysis in the double-mutant cycle
The observed rate constant (kobs) of individual mutant step was determined using EMSA (Supplementary Fig. 4l). The difference in free energy for catalysis of each mutant step (e.g. step I to step II: ΔGI-II) was determined by:
The change in coupling energy (ΔΔGint) was determined by:
ATPase assay
Malachite green phosphate assay kit was purchased from Sigma-Aldrich. The ATPase activity was performed as described previously with minor modifications13. Briefly, 400 nM of SNF2h, mutants thereof or BSA (control), and nucleosomes (0–180 nM) were incubated at 30 °C for 10 min in REA buffer. Reactions were initiated by addition of ATP (2 mM), and timepoints were diluted with 10×vol of MiliQ water, and immediately put in an ethanol-dry ice bath (−80 °C). Standard phosphate solutions and working reagent were prepared according to manufacturer protocol. Timepoints were thawed at room temperature, then 80 μL of each sample or phosphate standard solutions were mixed with 20 μl of the working reagent in on Fisherbrand™ 96-well plate. After incubation at room temperature for 15 min, the D650nm absorbance was measured on a VersaMax tunable microplate reader from Molecular Devices. Rates were determined using GraphPad Prism by fitting to a linear regression function. ATPase assay measurements were taken from the same batch of enzymes.
Ensemble FRET assay
Dinucleosomes (20 nM) and SNF2h (200 nM) were incubated in REA buffer at 30 °C for 10 min. The reactions were initiated by addition of ATP (2 mM). Samples were excited at 520 (slit ±5 nm) and FRET signal was collected at 660 (slit ± 10 nm) for 1500 s (integration time 0.1s). Signals were normalized to t = 0 (defined as measured FRET signal prior to the addition of ATP.)
Yeast growth experiments
Growth and manipulation of yeast strains were carried out according to standard procedures57. Yeast strains (YTT227, YTT420–424, Supplementary Table 4)36 used in this study were gifts from Toshio Tsukiyama (Division of Basic Sciences, Fred Hutchinson Cancer Research Center). The YTT227 strain lacks the URA3 gene that encodes an enzyme involved in uracil synthesis and can only grow in media supplemented with uracil. pRS416 is a yeast vector with a wildtype URA3 marker. Briefly, pRS416 plasmids were isolated from YTT420–424, followed by site-directed mutagenesis (via inverse PCR) to generate iswi mutants (isw1_APB and isw2_APB) and SNF2h homologous motifs (KKK to AAA in ISW1 and QKK to AAA in ISW2) and the final constructs were verified by sequencing. The S. cerevisiae strain YTT227 lacking ISW1, ISW2, and CHD1 genes have a growth defect at 37°C. We used this strain to perform growth experiments and transformed the YTT227 strain via a standard lithium acetate method with pRS416 plasmids carrying various derivatives of ISW1 and ISW2 under control of the endogenous promoter. Colonies were selected for 3 days at 30°C on SC-URA plates. Single colonies were picked and grown to OD600 of 0.4, serially diluted 5-fold, spotted onto synthetic minimal media plates lacking uracil and grown at 30°C, 37–39°C. wt ISW1, wt ISW2, wt ISW1-ISW2, isw1-APB, isw2-APB, isw1-K227A data were collected from two biological replicates, n=6 independent experiments.
Uncropped blots and gels
Relevant uncropped blots and gels are shown in Supplementary Fig. 21–39. These figures are called out from the corresponding Supplementary Figure legend.
Data Availability
The authors declare that data supporting the finding of this study are available within the article and its Supplementary Information. Additional data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
The authors thank R. E. Thompson, K. Diehl, J. D. Bagert, K. Jani and other members of the Muir laboratory for valuable discussions; S. Tharan from the Proteomics & Mass Spectrometry core. Yeast strains (YTT 227, YTT 420-424) were a gift from T. Tsukiyama; H.T.D. was funded by a postdoctoral fellowship from the Jane Coffin Childs Memorial fund; This work was supported by the U.S. National Institutes of Health (NIH grants R37-GM086868, and PO1- CA196539).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Jiang C & Pugh BF Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clapier CR & Cairns BR The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 78, 273–304 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Becker PB & Workman JL Nucleosome Remodeling and Epigenetics. Cold Spring Harb. Perspect. Biol. 5, a017905–a017905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomew B Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 83, 671–696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clapier CR, Iwasa J, Cairns BR & Peterson CL Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama T, Becker PB & Wu C ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367, 525–532 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Pazin MJ, Kamakaka RT & Kadonaga JT ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science 266, 2007–2011 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Deuring R et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Molecular Cell 5, 355–365 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Poot RA et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19, 3377–3387 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stopka T & Skoultchi AI The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. U.S.A. 100, 14097–14102 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Fan H-Y, Narlikar GJ & Kingston RE Human ACF1 alters the remodeling strategy of SNF2h. J. Biol. Chem. 281, 28636–28647 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Clapier CR, Längst G, Corona DF, Becker PB & Nightingale KP Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21, 875–883 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clapier CR & Cairns BR Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492, 280–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamiche A, Kang JG, Dennis C, Xiao H & Wu C Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. U.S.A. 98, 14316–14321 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang W, Kagalwala MN & Bartholomew B Regulation of ISW2 by Concerted Action of Histone H4 Tail and Extranucleosomal DNA. Mol. Cell. Biol. 26, 7388–7396 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JG, Madrid TS, Sevastopoulos E & Narlikar GJ The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 13, 1078–1083 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Deindl S et al. ISWI Remodelers Slide Nucleosomes with Coordinated Multi-Base-Pair Entry Steps and Single-Base-Pair Exit Steps. Cell 152, 442–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blosser TR, Yang JG, Stone MD, Narlikar GJ & Zhuang X Dynamics of nucleosome remodelling by individual ACF complexes. Nature 462, 1022–1027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Racki LR et al. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature 462, 1016–1021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman JA, Garlick JD & Kingston RE Chromatin Remodeling by Imitation Switch (ISWI) Class ATP-dependent Remodelers Is Stimulated by Histone Variant H2A.Z. J. Biol. Chem. 285, 4645–4651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dann GP et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamarra N, Johnson SL, Trnka MJ, Burlingame AL & Narlikar GJ The nucleosomal acidic patch relieves auto-inhibition by the ISWI remodeler SNF2h. eLife Sciences 7, 34270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K & Hansen JC The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. Royal Soc. Interface 10, 20121022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGinty RK & Tan S Nucleosome Structure and Function. Chem. Rev. 115, 2255–2273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbera AJ et al. The Nucleosomal Surface as a Docking Station for Kaposi’s Sarcoma Herpesvirus LANA. Science 311, 856–861 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Makde RD, England JR, Yennawar HP & Tan S Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armache K-J, Garlick JD, Canzio D, Narlikar GJ & Kingston RE Structural Basis of Silencing: Sir3 BAH Domain in Complex with a Nucleosome at 3.0 Å Resolution. Science 334, 977–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato H et al. A Conserved Mechanism for Centromeric Nucleosome Recognition by Centromere Protein CENP-C. Science 340, 1110–1113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinty RK, Henrici RC & Tan S Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan MT et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Götze M et al. StavroX—A Software for Analyzing Crosslinked Products in Protein Interaction Studies. J. Am. Soc. Mass Spectrom. 23, 76–87 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Yang B et al. Identification of cross-linked peptides from complex samples. Nature Methods 9, 904–906 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Ludwigsen J, Klinker H & Planitz FM No need for a power stroke in ISWI‐mediated nucleosome sliding. EMBO reports 14, 1092–1097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grüne T et al. Crystal Structure and Functional Analysis of a Nucleosome Recognition Module of the Remodeling Factor ISWI. Molecular Cell 12, 449–460 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Hwang WL, Deindl S, Harada BT & Zhuang X Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA. Nature 512, 213–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukiyama T, Palmer J, Landel CC, Shiloach J & Wu C Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13, 686–697 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao S et al. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 113, 12238–12243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arimura Y et al. Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic Acids Res. 46, 10007–10018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nacev BA; Feng L; Bagert JD; Lemiesz A E; Gao JJ; Soshnev AA; Kundra R; Schultz N; Muir TW; Allis CD “The expanding landscape of ‘oncohistone’ mutations in human cancers.” Nature, 2019. doi: 10.1038/s41586-019-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alabert C, Jasencakova Z, Groth A (2017) Chromatin Replication and Histone Dynamics In: Masai H, Foiani M (eds) DNA Replication. Advances in Experimental Medicine and Biology, vol 1042 Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- 41.Levendosky RF, Sabantsev A, Deindl S & Bowman GD The Chd1 chromatin remodeler shifts hexasomes unidirectionally. eLife Sciences 5, 3302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hota SK et al. Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat. Struct. Mol. Biol. 20, 222–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabantsev A, Levendosky RF, Zhuang X, Bowman GD & Deindl S Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat Commun 10, 1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan L, Wu H, Li X, Gao N & Chen Z Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 26, 258–266 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Lewis PW et al. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levendosky RF, Bowman GD Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. eLife Sciences 8, (2019). DOI: 10.7554/eLife.45472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voigt P et al. Asymmetrically Modified Nucleosomes. Cell 151, 181–193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhee HS, Bataille AR, Zhang L & Pugh BF Subnucleosomal Structures and Nucleosome Asymmetry across a Genome. Cell 159, 1377–1388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nekrasov M et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol. 19, 1076–1083 (2012). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 51.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ikeda Y & Behrman EJ Improved Synthesis of Photo-leucine. Syn. Commun. 38, 2276–2284 (2008). [Google Scholar]
- 53.Debelouchina GT, Gerecht K & Muir TW Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat. Chem. Biol. 13, 105–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM & Richmond TJ Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Leonard JD & Narlikar GJ A Nucleotide-Driven Switch Regulates Flanking DNA Length Sensing by a Dimeric Chromatin Remodeler. Molecular Cell 57, 850–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou CY & Narlikar GJ Analysis of Nucleosome Sliding by ATP-Dependent Chromatin Remodeling Enzymes. Meth. Enzymol. 573, 119–135 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Brachmann CB et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the finding of this study are available within the article and its Supplementary Information. Additional data are available from the corresponding author upon reasonable request.