Abstract
Covalent probes serve as valuable tools for global investigation of protein function and ligand binding capacity. Despite efforts to expand coverage of residues available for chemical proteomics (e.g. cysteine and lysine), a large fraction of the proteome remains inaccessible with current activity-based probes. Here, we introduce sulfur-triazole exchange (SuTEx) chemistry as a tunable platform for developing covalent probes with broad applications for chemical proteomics. We show modifications to the triazole leaving group can furnish sulfonyl probes with ~5-fold enhanced chemoselectivity for tyrosines over other nucleophilic amino acids to investigate, for the first time, more than 10,000 tyrosine sites in lysates and live cells. We discover that tyrosines with enhanced nucleophilicity are enriched in enzymatic, protein-protein interaction, and nucleotide recognition domains. We apply SuTEx as a chemical phosphoproteomics strategy to monitor activation of phosphotyrosine sites. Collectively, we describe SuTEx as a biocompatible chemistry for chemical biology investigations of the human proteome.
INTRODUCTION
Chemical proteomics is a powerful technology for ascribing function to the vast number of uncharacterized proteins in the human proteome1, 2. This proteomic method employs probes designed with reactive groups that exploit accessibility and reactivity of binding sites to covalently label active proteins with reporter tags for functional assignment and inhibitor development3. Selective probes resulting from competitive screening efforts serve as enabling, and often first-in-class, tools for uncovering biochemical and cellular functions of proteins (e.g. serine hydrolases4, proteases5, kinases6, phosphatases7, and glycosidases8) and their roles in contributing to human physiology and disease. The basic and translational opportunities afforded by chemical proteomics has prompted exploration of new biocompatible chemistries for broader exploration of the proteome.
Covalent probes used for chemical proteomics range from highly chemoselective fluorophosphonates for catalytic serines9 to general thiol alkylating agents and amine-reactive esters for cysteines10 and lysines11, respectively. The ability to globally measure protein functional states and selectively perturb proteins of interest has substantially augmented our basic understanding of protein function in cell and animal models1, 3. Exploration of new redox-based oxaziridine chemistry, for example, identified a conserved hyper-reactive methionine residue (Met169) in redox regulation of mammalian enolase12. Hydrazine probes revealed a novel N-terminal glyoxylyl post-translational modification on the poorly characterized protein SCRN3 (ref. 13). More recent exploration of photoaffinity probes facilitated global evaluation of reversible small molecule–protein interactions to expand the scope of proteins available for chemical proteomic profiling14.
Sulfonyl-fluorides15 (-SO2F) and fluorosulfates16, 17 (-OSO2F) have emerged as promising scaffolds for covalent probe development because of the wide range of amino acids (e.g. serine18, 19, tyrosine20, lysine21, histidine22) and diverse protein targets (proteases18, 19, kinases21, GPCRs23) available for sulfur-fluoride exchange chemistry (SuFEx24). Reactivity of SuFEx is driven largely through stabilization of the fluorine leaving group (LG) at protein sites during covalent reaction25, 26. The sensitivity of SuFEx to protein microenvironments allows, for example, the ability to target orthogonal nucleophilic residues in the same nucleotide-binding site of decapping enzymes27. The broad reactivity and context-dependent activation of SuFEx present opportunities for modulating the sulfur electrophile to target novel, and potentially functional, sites of proteins21, 25, 26, 28. The reliance on fluorine, while key for activating SuFEx chemistry, is limiting in terms of LG modifications to modify reactivity, specificity, and binding affinity at protein sites across the proteome.
Here, we introduce sulfur-triazole exchange chemistry (dubbed SuTEx) for development of phenol-reactive probes that can be tuned for tyrosine chemoselectivity in proteomes (>10,000 distinct sites in ~3,700 proteins) through modifications to the triazole LG. We use these probes to discover a subset of tyrosines with enhanced reactivity that are localized to functional protein domains and to apply SuTEx for global phosphotyrosine profiling of pervanadate-activated cells. Our findings illustrate the broad potential for deploying SuTEx to globally investigate tyrosine reactivity, function, and post-translational modification state in proteomes and live cells.
RESULTS
Design and synthesis of sulfonyl-triazole probes
We reasoned that triazoles could serve as a suitable replacement for the fluorine LG used to promote SuFEx24. Previous studies demonstrated that triazoles activate ureas for covalent protein modification with a substantial advantage of tunability29, which is not possible with fluorine as a LG by comparison. We envisioned that a sulfonyl-triazole scaffold would permit evaluation, and potentially control, of reactivity and specificity of the sulfur electrophile through structural modifications to the triazole LG (Fig. 1a). Our hybrid probe strategy is further bolstered by the broad functional group tolerance of 1,2,3- and 1,2,4-triazoles as LGs for development of covalent serine hydrolase inhibitors29, 30.
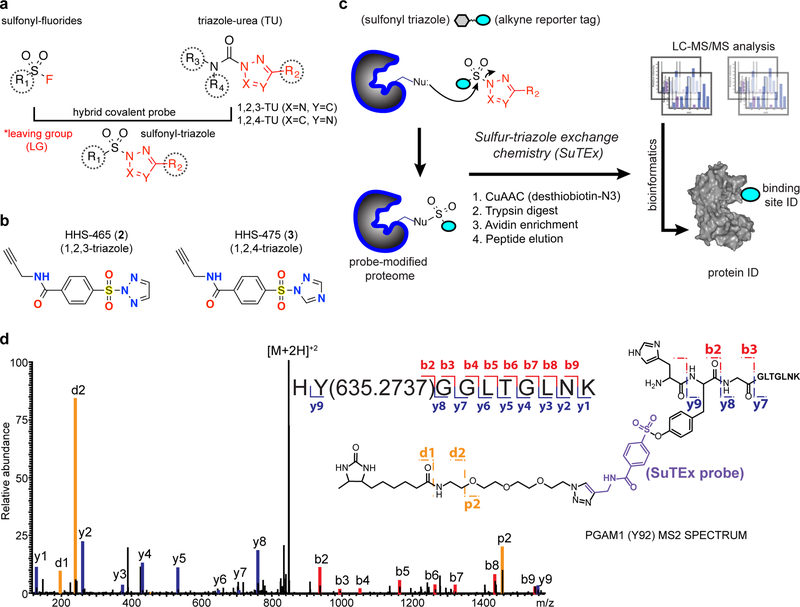
Figure 1. Development of sulfur-triazole exchange (SuTEx) chemistry for chemical proteomics.
a) Sulfonyl-triazoles are a hybrid of sulfonyl-fluoride and triazole-ureas for developing covalent probes with reactivity that can be modulated through the triazole leaving group (LG). b) Chemical structures of 1,2,3- and 1,2,4-sulfonyl triazoles HHS-465 and HHS-475, respectively. c) Proposed reaction mechanism of sulfur-triazole exchange (SuTEx) chemistry and LC-MS/MS workflow to identify proteins and corresponding binding sites from SuTEx reaction. See Online Methods for additional details. d) MS2 spectrum annotation of an HHS-475-modified tyrosine site (Tyr92) found in PGAM1. Covalent reaction with HHS-465 and HHS-475 adds +635.2737 Da to the modified amino acid (Tyr92 from PGAM1 shown as a representative example) and supports the proposed SuTEx reaction mechanism. Data shown are representative of two experiments (n=2 biologically independent experiments). Additional annotated MS2 spectra of tyrosine and lysine modified sites can be found in Supplementary Figures 4–6 and 7–8, respectively.
We developed a general strategy for synthesizing sulfonyl-triazole probes for testing in chemical proteomic assays. To add an alkyne reporter tag for downstream detection, we coupled propargyl-amine to 4-(chlorosulfonyl)benzoic acid to produce an alkyne-modified sulfonyl chloride intermediate, S1 (1), that could be further coupled to either unsubstituted or substituted triazoles (Supplementary Fig. 1). Initially, we synthesized an unsubstituted triazole analog HHS-465 (2) as a starting point for testing LG effects on proteome reactivity (Fig. 1b). The N2 isomeric state of HHS-465 was confirmed by NMR and x-ray crystallography (Supplementary Fig. 2). Purity of the N2-isomer was confirmed to be >95% as measured by HPLC. See Supplementary Note 1 for full synthetic procedures and characterization of all sulfonyl probes reported.
Chemical proteomic evaluation of SuTEx chemistry
We established a chemical proteomic method to assess the reactivity of HHS-465 with amino acid residues in proteomes. HEK293T cell proteomes were treated with HHS-465 (100 μM, 1 hr, 25 °C) followed by copper-catalyzed azide-alkyne cycloaddition (CuAAC) coupling with a desthiobiotin-azide tag. Proteomes were digested with trypsin protease and desthiobiotin-modified peptides enriched by avidin affinity chromatography, released, and analyzed by high-resolution liquid chromatography-mass spectrometry (LC-MS; Fig. 1c). Probe-modified peptide-spectrum matches (PSMs) that met our quality control confidence criteria of ≥300 Byonic score31 and <5 ppm mass accuracy were selected for further manual evaluation (see Online Methods for additional details).
We predicted, based on our proposed reaction mechanism, that amino acid residues modified by HHS-465 would be identified by differential modification with a sulfonyl-desthiobiotin adduct that is the product of SuTEx reaction (Fig. 1c). We synthesized and included a 1,2,4-triazole counterpart, HHS-475 (3), for testing to demonstrate SuTEx as a common mechanism among triazole regioisomers (Fig. 1b and Supplementary Fig. 2). Initial evaluation of our data assigned >60% of HHS-465- and HHS-475-labeled peptides as uniquely modified tyrosines (Supplementary Fig. 3). Evaluation of MS2 spectra showed confident identification of all major y-ions and a large fraction of b-ions, including fragment ions (y and b) that allowed identification of the tyrosine site of HHS-465 and HHS-475 binding (mass adduct of 635.2737 Da, Fig. 1d and Supplementary Figs. 4–6). The remaining probe-modified peptides were assigned largely to lysines, which after removal of incorrect search algorithm matches to C-terminal modified peptides represented a minor fraction of total modified residues (<25%; Supplementary Figs. 3, 7, and 8). We evaluated additional human cell proteomes to determine the number and type of tyrosines amenable to SuTEx reaction. On average, we reliably identified >2,800 tyrosines per data set and in aggregate, ~8,000 tyrosine sites from ~3,000 proteins with diverse enzymatic and non-catalytic functions across 5 cell proteomes evaluated with HHS-465- and −475 (Fig. 2a,b and Supplementary Dataset 1). A large fraction of HHS-465/475-modified sites were also annotated as phosphorylation sites as reported in the PhosphoSitePlus database32 (Fig. 2c).
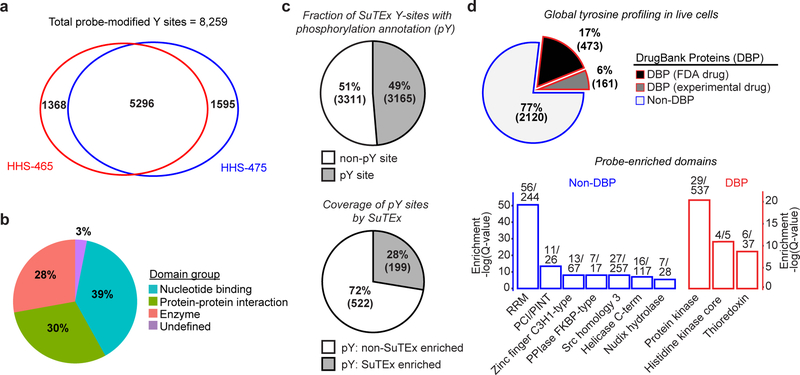
Figure 2. Functional tyrosine profiling in proteomes and live cells.
a) Comparison of HHS-465- and HHS-475-tyrosine modified sites identified from human cell proteomes (HEK293T, A549, DM93, H82, and Jurkat cells) treated with SuTEx probes (100 μM, 1 hr, 25 °C). b) Distribution of protein domain groups that are significantly overrepresented using probe-modified tyrosine sites from in situ chemical proteomic studies. Enriched domain annotations are those with a Q-value < 0.01 after Benjamini–Hochberg correction of a two-sided binomial test (see Online Methods for details). c) Top: Overlap between in situ HHS-465- and HHS-475-modified tyrosine sites that are also phosphorylation sites (number of phosphotyrosine high throughput annotation on PhosphoSitePlus (HTP score); HTP ≥1). Bottom: coverage of phospho-tyrosine sites (HTP ≥10) that were detected by in situ chemical proteomics of HEK293T and Jurkat cells (HHS-465 and −475). d) Top: Comparison of HHS-465 and HHS-475 in situ probe-modified proteins with DrugBank proteins (DBP group). The Non-DBP group consists of proteins that did not match a DrugBank entry. Bottom: probe-enriched domains from DBP and non-DBP groups. Enriched domain annotations are those with a Q-value < 0.01 after Benjamini–Hochberg correction of a two-sided binomial test. All data shown are representative of two experiments (n=2 biologically independent experiments).
We next tested whether SuTEx probes exhibit sufficient stability and cell permeability to permit global tyrosine profiling in living systems. We observed robust proteome labeling that was concentration- and time-dependent in fluorescence gel-based analyses of proteomes from HEK293T cells treated with HHS-465 or HHS-475 (Supplementary Figs. 9 and 10). Using a saturating probe labeling condition (100 μM, 2 hr, 37 °C) for our live cell studies, we consistently measured ~3,500 distinct tyrosine sites (corresponding to ~1,700 proteins), in total, across membrane and soluble fractions in each cell line tested (HEK293T, Jurkat; Supplementary Dataset 1). For comparison, recent reports using sulfonyl-fluorides showed probe modifications of ~70–130 protein targets in live cell studies21, 25. HHS-465- and HHS-475-labeled proteins from live cell profiling were largely absent from the DrugBank database33 (77%, Fig. 2d). Evaluation of probe-enriched domains (Q-values < 0.01) from the non-DrugBank protein (non-DBP) group revealed highly enriched functions that include proteins involved in RNA recognition (RRM domain34) and protein-protein interactions (PCI/PINT and SH3 domains35; Fig. 2d and Supplementary Dataset 1). By comparison, the DBP group was largely overrepresented with domains found in enzymes (kinases and redox enzymes, Fig. 2d).
Discovery of hyper-reactive tyrosines in human proteomes
Previous studies identified a subset of hyper-reactive cysteine and lysine residues that specify function and are susceptible to binding with electrophilic ligands10, 11. Whether tyrosines differ in intrinsic reactivity and the functional implications of heightened nucleophilicity remain largely underexplored on a proteome-wide scale. Here, we used HHS-465 and quantitative chemical proteomics to evaluate tyrosine reactivity directly in human cell proteomes derived from isotopically light and heavy amino acid-labeled HEK293T cells (i.e. stable isotope labeling with amino acids in cell culture; SILAC36). We measured concentration-dependent HHS-465 labeling where nucleophilic tyrosines are expected to exhibit comparable labeling intensity at low and high concentrations of HHS-465 while less nucleophilic tyrosines show concentration-dependent increases in probe labeling. We treated HEK293T proteomes with high versus low concentrations of HHS-465 (250 versus 25 μM; 10:1 comparison) for 1 hr (25 °C) and then analyzed samples by quantitative LC-MS (Supplementary Fig. 11). Tyrosine nucleophilicity was segregated into low, medium, and high groups based on their respective SILAC ratios (SR >5, 2 < SR ≤5, SR ≤ 2, respectively; Fig. 3a and Supplementary Dataset 1). We also verified in a control experiment (25 vs 25 μM) that SR values were ~1 in a 1:1 comparison (Supplementary Fig. 12).
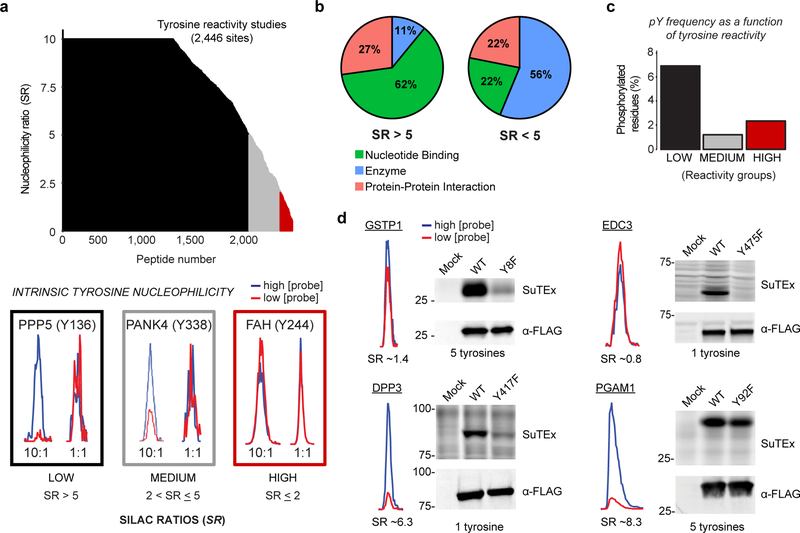
Figure 3. SuTEx-enabled discovery of intrinsically nucleophilic tyrosines in human cell proteomes.
HEK293T SILAC heavy and light soluble proteomes were treated with 250 or 25 μM HHS-465 (10:1 comparison), respectively. The resulting SILAC ratios (SR) were quantified using the area under the curve of MS1 extracted ion chromatograms (EIC) to determine tyrosine nucleophilicity. a) A waterfall plot of nucleophilicity ratio (median SR values) as a function of probe-modified tyrosine sites to quantitate tyrosine reactivity across the proteome. A MS1 EIC is shown for SR values that represent each nucleophilicity group (low, black; medium, grey; high, red). b) Distribution of protein domain groups that contain tyrosines quantified as low (SR >5) or medium/high (SR <5) reactivity. Domain annotations shown were significantly enriched (Q-value < 0.01 after Benjamini–Hochberg correction of a two-sided binomial test) with HHS-465. c) Bar plot depicting tyrosines with medium to high nucleophilicity are less likely to be phosphorylated (HTP ≥10, PhosphoSitePlus) compared with less-reactive tyrosines. d) Proteins containing a hyper-reactive tyrosine (GSTP1 Tyr8, EDC3 Tyr475) or single probe-modified tyrosine (DPP3 Tyr417) can be site-specifically labeled with SuTEx probes (50 μM, 30 min, 37 °C). Recombinant wild-type (WT) protein or corresponding tyrosine (Y)-to-phenylalanine (F) mutant HEK293T proteomes were treated with HHS-475 (GSTP1, DPP3, PGAM1) or HHS-465 (EDC3) and analyzed by gel-based chemical proteomics. Proteins that contain less-nucleophilic tyrosines (PGAM1 Tyr92) are labeled at multiple sites and show negligible differences in probe labeling between WT and tyrosine mutant. Western blots show equivalent expression of recombinant WT and mutant proteins. Full images of gels and blots are shown in Supplementary Figure 27. All data shown are representative of two experiments (n=2 biologically independent experiments).
In total, we quantified ~2,400 tyrosine residues from >1,100 proteins in soluble proteomes from HEK293T cells that showed consistent SILAC ratios across replicate experiments (n = 4, Fig. 3a). The majority of quantified tyrosines showed concentration-dependent increases in HHS-465 labeling, which is indicative of low intrinsic nucleophilicity (Fig. 3a). Similar to cysteines and lysine residues, a subset of tyrosines (~5%, 127 sites in total; Fig. 3a and Supplementary Dataset 1) demonstrated enhanced nucleophilicity (i.e. hyper-reactivity10, 11) as evidenced by SR ≤ 2 for 10:1 conditions (Fig. 3a). The majority of proteins contained a single hyper-reactive tyrosine among several tyrosines quantified (Supplementary Fig. 13). Reactive tyrosines (SR < 5) were enriched in domains of enzymes while tyrosines with lower reactivity (SR > 5) were localized at small molecule binding sites (Fig. 3b). Comparison of tyrosine reactivity and evidence of phosphorylation revealed a marked inverse correlation. Specifically, tyrosines with low reactivity (SR > 5) were significantly overrepresented for phosphotyrosine sites compared with medium- and hyper-reactive groups (SR ≤ 5, Fig. 3c).
We verified our tyrosine reactivity annotations by comparing SuTEx probe labeling of recombinant wild-type (WT) and tyrosine-to-phenylalanine mutants of human proteins with tyrosine sites identified as high (Tyr8, GSTP1; Tyr475, EDC3), low/medium (Tyr417, DPP3), or low hyper-reactivity (Tyr92, PGAM1). Proteins like glutathione S-transferase Pi (GSTP1) with a single hyper-reactive tyrosine, among several modified tyrosines, showed robust HHS-475 labeling that was largely abolished in recombinant Y8F mutant (Fig. 3d). Mutation of the hyper-reactive tyrosine in the Yjef-N domain of enhancer of mRNA decapping protein 3 (EDC3) also resulted in near-complete loss of probe labeling (Y475F, Fig. 3d). In contrast, mutation of a tyrosine with low nucleophilicity in PGAM1 resulted in negligible alterations in probe labeling (Y92F, Fig. 3d). A notable exception was dipeptidyl peptidase 3 (DPP3), which contains a single modified tyrosine (Tyr417) that, despite a low/medium nucleophilicity ratio (SR ~6), showed near-complete blockade of probe labeling in corresponding tyrosine mutants (Y417F, Fig. 3d).
Finally, we confirmed the catalytic role of GSTP1 tyrosine 8, located in the GSH binding site (G-site), by mutating this residue (Y8F) and demonstrating abolished biochemical activity (Supplementary Figs. 14a,b and 15). In comparison, recombinant DPP3 WT- and Y417F mutant-overexpressed cell lysates showed comparable catalytic activity in a peptidase substrate assay, supporting a non-catalytic role for Tyr417 (Supplementary Figs. 14c,d and 16). Future studies will focus on testing whether the moderate reactivity of the non-catalytic Tyr417 (Fig. 3d) can be exploited for DPP3 inhibitor development.
Tuning the triazole LG for tyrosine chemoselectivity
A key advantage of SuTEx technology is the capacity for modifying the triazole LG to tune chemoselectivity of resulting probes. Here, we tested whether we could enhance the specificity of HHS-465/475 for tyrosine modification through addition of functional groups to the triazole (Fig. 4a). To globally evaluate probe reactivity and specificity in parallel, we compared the total number of probe-modified sites (Y and K combined) as a function of the ratio of modified tyrosines to lysines (Y/K ratio), respectively, for each SuTEx analog. First, we synthesized a sulfonyl-fluoride counterpart to HHS-465/475, termed HHS-SF-1 (4; Fig. 4a), to directly compare fluoro- and triazole-LGs with respect to proteome specificity and reactivity. HHS-SF-1 exhibited a ~4-fold reduction in the total number of modified sites and lower specificity for tyrosine compared with HHS-465 and HHS-475 (Y/K of 2.3 versus 2.5 and 2.8, respectively; Fig. 4a and Supplementary Fig. 17).
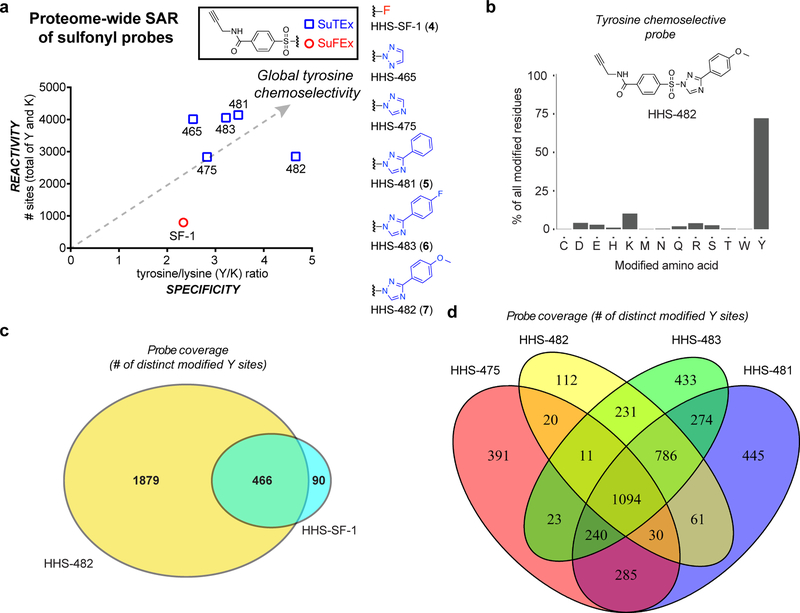
Figure 4. Tuning SuTEx probes for tyrosine chemoselectivity in cell proteomes.
HEK293T soluble proteomes were treated with SuFEx and SuTEx probes. a) Global reactivity [total number of tyrosine (Y) and lysine (K) sites] and specificity (Y/K ratio) of probe-labeled sites from LC-MS chemical proteomic experiments. A bar graph depiction of reactivity and selectivity data can be found in Supplementary Figure 17. b) Bar plot showing distribution of HHS-482-modified sites (high confidence sites; Byonic score > 600) against nucleophilic amino acid residues detected in proteomes. c) High overlap of tyrosine-modified sites from proteomes treated with sulfonyl-triazoles (HHS-482) compared with sulfonyl-fluorides (HHS-SF-1). d) Comparison of probe-modified tyrosine sites from LC-MS chemical proteomic studies using 1,2,4-sulfonyl-triazoles. Each 1,2,4-sulfonyl-triazole probe was able to modify unique tyrosine sites to increase overall tyrosine coverage. All data shown are representative of two experiments (n=2 biologically independent experiments).
In light of the improved tyrosine specificity of HHS-475, we synthesized and evaluated a series of 1,2,4-triazole analogs bearing different substituents at the R2 position (Figs. 1a and 4a). Addition of a phenyl group improved both tyrosine specificity (Y/K = 3.5) and overall proteome reactivity of the resulting HHS-481 (5) probe (~4,000 total sites; Fig. 4a). Modification of the phenyl-triazole resulted in further alterations in proteome activity of SuTEx probes. Addition of a para-fluoro substituent (HHS-483, 6) resulted in comparable reactivity and slightly lowered tyrosine specificity compared with HHS-481 (Fig. 4a). In contrast, the para-methoxy probe HHS-482 (7) showed the highest tyrosine specificity (Y/K ratio of ~5) while maintaining good overall proteome reactivity (~3,000 probe-modified sites, HHS-482; Fig. 4a). Evaluation of HHS-482 reactivity against other amino acids revealed high tyrosine specificity with ~75% of probe-modified residues assigned to tyrosines (Fig. 4b).
Comparison of tyrosine sites modified by HHS-SF-1 and HHS-482 revealed high overlap (>90%) indicating that substitution of fluorine for a triazole LG did not result in loss of tyrosine coverage (Fig. 4c). In contrast, LG modifications to 1,2,4-SuTEx probes furnished analogs that each expanded tyrosine coverage via detection of unique-modified sites (HHS-475, 391 sites; HHS-482, 112 sites; HHS-483, 433 sites; HHS-481, 445 sites; Fig. 4d). In summary, our studies highlight a key difference between sulfonyl-fluoride compared with -triazole chemistry; the latter reaction dramatically enhances overall reactivity and through LG modifications can be tuned for enhanced tyrosine chemoselectivity and coverage in proteomes (Fig. 4b,d).
Triazole LG enhances phenol reactivity of probes
Next, we compared solution reactivity of sulfonyl probes to evaluate whether the enhanced tyrosine reactivity of SuTEx is a function of the LG or protein microenvironment. We established an HPLC assay to test reactivity of SuTEx and SuFEx probes with nucleophiles that model side chain groups of tyrosine (p-cresol) and lysine (n-butylamine). We synthesized the predicted products from p-cresol (KY-2–48, 8) and n-butylamine (KY-2–42, 9) reaction with sulfonyl probes to establish HPLC conditions for monitoring this covalent reaction in solution (Supplementary Fig. 18; see Online Methods). We incubated p-cresol with a mixture of all three sulfonyl probes and monitored time-dependent reaction by depletion of respective SuTEx (HHS-475, HHS-482) and SuFEx (HHS-SF-1) probe signal. Our probe competition studies were performed with increasing tetramethylguanidine (TMG37) base to compare probe reactivity as a function of increasing phenol nucleophilicity. We also measured stability and found that all three sulfonyl probes showed negligible hydrolysis in aqueous and organic solvents even after incubation for 48 hours at room temperature (Supplementary Fig. 19).
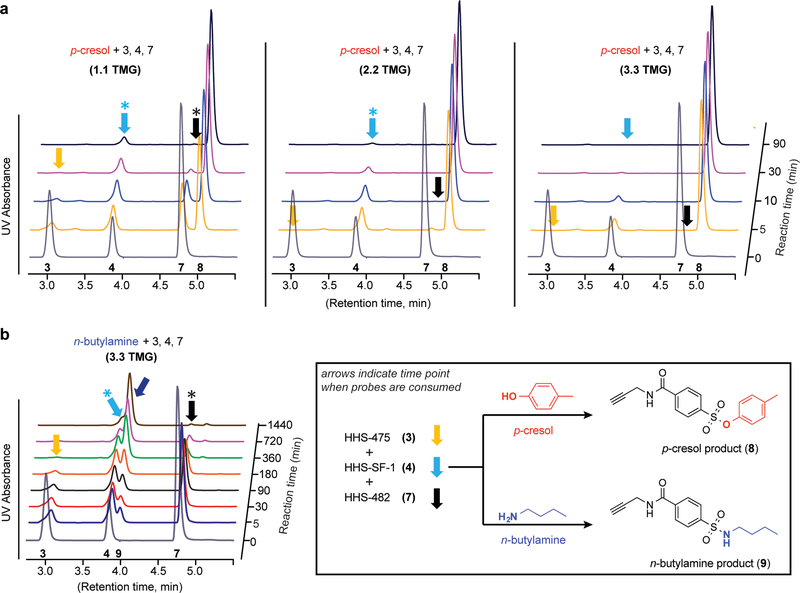
At lower TMG (1.1 equivalents), HHS-475 (3) was the most reactive probe as evidenced by consumption by 30 minutes while unreacted HHS-SF-1 (4) and HHS-482 (7) was still detectable (Fig. 5a). The difference in reactivity between SuTEx and SuFEx was apparent at higher TMG (2.2 equivalents) conditions. Both SuTEx probes (HHS-475 and HHS-482) were consumed by 10 minutes while HHS-SF-1 was still detectable even after 90 minutes of reaction (Fig. 5a); depletion of HHS-SF-1 signal was only observed at the highest TMG tested (3.3 equivalents, Fig. 5a and Supplementary Fig. 20). We also verified a similar trend in reactivity when p-cresol was incubated with individual sulfonyl probes (Supplementary Dataset 1). The reactivity of all three sulfonyl probes for n-butylamine was substantially reduced compared with p-cresol even at high TMG (3.3 equivalents) conditions (Fig. 5b). Reaction of HHS-475 with n-butylamine required 6 hours to complete and HHS-482 and HHS-SF-1 were not consumed even after 24 hours (Fig. 5b and Supplementary Fig. 20). To investigate selectivity further, we incubated sulfonyl probes with n-butylamine and p-cresol mixed in a 5:1 ratio and demonstrated minimal n-butylamine- compared with p-cresol-probe adduct formation for HHS-475 as well as HHS-482 and HHS-SF-1 (Supplementary Fig. 21).
Figure 5. Triazole LG enhances phenol reactivity of sulfonyl probes in solution.
a) A mixture of HHS-475 (3), HHS-SF-1 (4), and HHS-482 (7) was incubated with p-cresol in the presence of increasing amounts of tetramethylguanidine (TMG) base and time-dependent covalent reaction monitored by reduction of respective probe signal. Formation of the common p-cresol-probe adduct (8) was confirmed by retention time that matched our synthetic standard KY-2–48 (Supplementary Fig. 18). Colored arrows denote the time points when each respective probe was consumed, and the asterisks denote time points corresponding to substantial but not complete probe depletion. b) Reduced reactivity of n-butylamine against sulfonyl probes under high TMG conditions (3.3 equivalents). Formation of the n-butylamine-probe adduct (9) was validated by retention time that matched our KY-2–42 synthetic standard (Supplementary Fig. 18). See Online Methods for additional details. HPLC raw data can be found in Supplementary Dataset 1. Data shown are representative of three independent experiments (n=3).
Collectively, we show that the triazole LG enhances intrinsic reactivity of sulfonyl probes for phenol without compromising stability in solvents commonly used for biological experiments (i.e. DMSO). While our solution findings agree with the enhanced reactivity of SuTEx compared with SuFEx observed by proteomics, the differences in tyrosine chemoselectivity between HHS-482 and HHS-475 are likely a function of the protein microenvironment and a feature of probe reactivity that has been reported for other electrophiles38.
Chemoproteomic profiling of phosphotyrosine activation
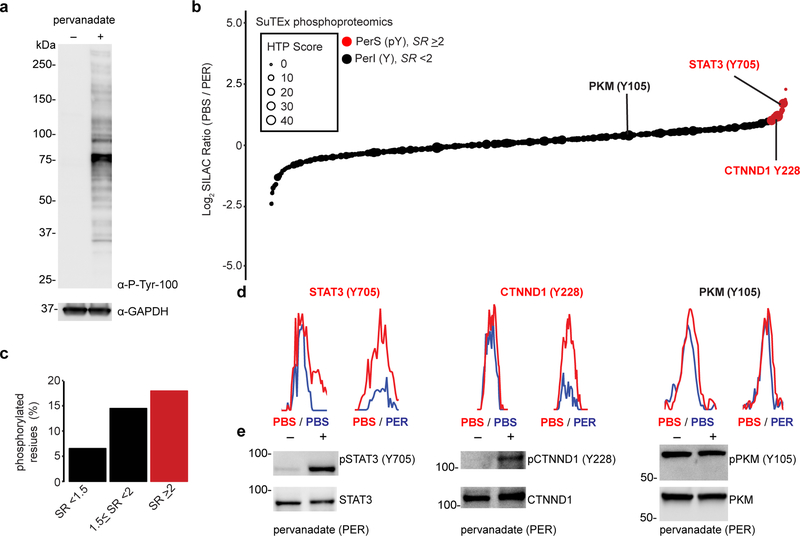
Considering the overlap of SuTEx-modified tyrosines with reported phosphotyrosine sites (pY, Fig 2c), we investigated whether we could apply this methodology for a “chemical” phosphoproteomics approach. We hypothesized that tyrosine accessibility by SuTEx probes would be inversely correlated with modification status and could be used to identify changes in pY sites (Supplementary Fig. 22). Given the low abundance of phospho-tyrosine (1%) compared with phospho-serine (88%) and phospho-threonine (11%) detected in cell39 and tissue proteomes40, we activated global phosphorylation using cell permeable tyrosine phosphatase inhibitors to increase pY signals for our LC-MS studies. Previous live cell studies demonstrated the high efficiency of pervanadate for global inhibition of tyrosine phosphatase activity41. We treated live A549 cells with pervanadate at varying concentrations (0 – 500 μM) and time (0 – 30 min) and measured global changes in tyrosine phosphorylation by western blot using a pY-specific antibody (P-Tyr-100, ref. 42). We observed robust increases in global tyrosine phosphorylation as judged by a massive increase in pY-antibody signals that appeared to saturate at 100 μM and 30 min of pervanadate treatment (Supplementary Figs. 23 and 24).
Proteomes from cells treated with our pervanadate activation conditions (100 μM, 30 min) were labeled with HHS-475 or HHS-482 (100 μM, 30 min) followed by CuAAC with desthiobiotin and quantitative LC-MS to evaluate how phosphorylation status affected SuTEx probe labeling. Pervanadate blockade of tyrosine phosphatases should activate endogenous phosphorylation and compete for SuTEx probe labeling at phosphorylated but not unmodified tyrosine sites that can be differentiated by SILAC ratios of vehicle- (light) versus pervanadate (heavy)-treated cells (Supplementary Fig. 22 and Fig. 6a). We detected in total ~2,200 probe-modified tyrosine sites across ~1,000 proteins using both HHS-475 and HHS-482 that were further separated into pervanadate-sensitive (PerS, SR ≥ 2) and -insensitive groups (PerI, SR < 2, Fig. 6b and Supplementary Fig. 25). In support of our hypothesis, the probe-modified tyrosines found in the PerS group appeared to be enriched for annotated phosphotyrosine sites (HTP ≥10 in PhosphoSitePlus, Fig. 6c and Supplementary Fig. 25) and represented only a small fraction of all unique HHS-475- and HHS-482-modified tyrosines detected by chemical proteomics (~3%, 67 sites; Supplementary Dataset 1). The overall median SR of all probe-modified tyrosines was ~1 for both HHS-475 and HHS-482 datasets (Supplementary Dataset 1), which supports tyrosine phosphorylation as a rare post-translational event and the ability of our platform to capture subtle changes in the tyrosine phosphoproteome.
Figure 6. Chemical phosphotyrosine-proteomics by SuTEx.
a) Western blot analysis confirming activation of global tyrosine phosphorylation (detected via a phospho-tyrosine monoclonal antibody, P-Tyr-100) with pervanadate treatment conditions of A549 cells (100 μM, 30 min) used for chemical proteomic studies. b) Plot of HHS-475-modified tyrosine sites (represented by individual circles) as a function of SILAC ratios (SR, light (PBS)/heavy (pervanadate or PER)). Size of circles reflect the HTP score (PhosphoSitePlus). Tyrosine sites were further segregated into pervanadate-insensitive (PerI) and pervanadate-sensitive (PerS) groups based on SR <2 or ≥2, respectively. Soluble proteomes from pervanadate-activated A549 cells were labeled with HHS-475 (100 μM) for 30 min at 37 °C. c) Bar plot showing trend towards increased number of phosphotyrosine annotations (HTP ≥10) on tyrosine sites with enhanced pervanadate sensitivity. d,e) Validation that blockade of HHS-475 labeling (d) of individual tyrosine sites on STAT3 (Tyr705), CTNND1 (Tyr228), and PKM (Tyr105) coincides with increased phosphorylation at respective sites with pervanadate activation (e). Equivalent protein loading was confirmed by western blot analysis of non-phosphorylated protein counterparts. See Online Methods for additional details of pervanadate activation and phosphotyrosine western blot procedures. See Supplementary Dataset 1 for SR values of tyrosines sites detected by chemical proteomics. Full images of blots are shown in Supplementary Figure 28. All data shown are representative of two experiments (n=2 biologically independent experiments).
To further validate our chemical phosphoproteomics strategy, we tested whether tyrosine sites identified as pervanadate-sensitive were also directly phosphorylated under the same treatment conditions. For our studies, we chose several proteins from the PerS group based on a high phosphotyrosine annotation score (HTP >100, PhosphoSitePlus) and evidence for a role in signaling in human cancer cells like A549. We identified signal transducer and activator of transcription 3 (STAT3) as a target protein with reduced SuTEx probe labeling at Y705 (SR = 2.3, Fig. 6d) that corresponded with enhanced phosphorylation at this site upon pervanadate activation (Fig. 6e). Our data are in agreement with previous findings reporting STAT3 Tyr705 as a phosphorylation site for activation by tyrosine kinases in human non-small cell lung cancer lines including A54943. We validated another tyrosine kinase-targeted site (Tyr228) on catenin δ−1 (CTNND144) and showed blockade of SuTEx probe labeling (SR = 3.3, Fig. 6d) coincided with direct phosphorylation at this tyrosine site by western blot analysis (Fig. 6e).
In contrast, we identified Tyr105 as a pervanadate-insensitive site (SR = 1.1, Fig. 6d) on pyruvate kinase (PKM) that showed negligible changes in phosphorylation at this tyrosine upon pervanadate activation (Fig. 6e). Our proteomic findings support previous reports of substantial basal levels of phosphorylated-Tyr105 on PKM in A549 cells45, which could explain why pervanadate activation did not further enhance pY levels. As a control, we showed that SuTEx probe treatment of pervanadate-activated cell proteomes did not result in non-specific displacement of phosphates from tyrosines (Supplementary Fig. 26). In summary, we applied SuTEx technology as a chemical strategy that exploits probe labeling as a site-specific readout of changes in pY levels upon global activation of the phosphoproteome.
DISCUSSION
We describe sulfur-triazole exchange chemistry for development of covalent probes that are compatible with biological systems, easily accessible via modern synthetic chemistry, and can be adapted for diverse chemical proteomic applications. We demonstrate, on a proteomic scale, that addition of a triazole LG introduces key capabilities to the sulfur electrophile including tunability for protein reaction, robust cellular activity, and capacity for directing amino acid specificity. Compared with more widely used sulfonyl-fluorides, the triazole LG dramatically enhanced overall reactivity of sulfonyl probes in solution (Fig. 5) that can, through modest structural modifications, be optimized for high tyrosine chemoselectivity in proteomes (Fig. 4a,b). Key to success is a general synthetic strategy for introducing a common mass spectrometry-stable enrichment tag (Fig. 1d) and incorporating diverse triazole LGs to enable global structure-activity relationship (SAR) studies of SuTEx probes directly in lysates and live cells (Fig. 2).
We exploited these features of SuTEx for functional studies of >10,000 unique tyrosine sites from ~3,700 protein targets detected in human cell proteomes. While previous chemical proteomic studies have shown promise for functional tyrosine profiling20, 25, 26, 46, the broad coverage of SuTEx permitted global tyrosine quantitation with unprecedented depth and breadth. A striking discovery from our studies was high enrichment of tyrosine sites in nucleotide-binding domains from in vitro and in situ probe-labeling experiments using HHS-465 and HHS-475 (Fig. 2b,d). We identified prominent labeling of tyrosines localized in RNA-recognition motifs (RRMs) of serine/arginine-rich protein splice factors (SRSF1–12, ~70% coverage of members by SuTEx; Supplementary Dataset 1) involved in regulation of mRNA splicing, export, and translation47. Several probe-labeled tyrosines including Tyr13 of SRSF3 RRM have been shown through structural studies to directly mediate RNA binding48. Combined with prominent in situ labeling at domains mediating protein-protein interactions (e.g. PCI/PINT and SH335), SuTEx offers a valuable resource for developing chemical probes against proteins that have been historically challenging to target with small molecules (Fig. 2d).
Our functional profiling studies led to the discovery of intrinsically nucleophilic tyrosines that are enriched in enzyme sites but also prominent in domains mediating protein–small molecule and protein-protein interactions (SR < 5, Fig. 3b). The rare nature of hyper-reactive tyrosines (~5% of all quantified sites) are in agreement with previous chemical proteomic studies that identified minor subsets of cysteine and lysine residues that demonstrate enhanced reactivity10, 11. We demonstrated that hyper-reactive residues like Tyr8 of GSTP1 are key for catalytic function, and mutation of this site (Y8F) abolished biochemical activity (Supplementary Fig. 14a,b). We also identified a non-catalytic tyrosine near the zinc-binding region of DPP3 (Tyr417) that exhibited moderate nucleophilicity (SR ~6) and may offer future opportunities for developing site-specific ligands (Fig. 3d and Supplementary Fig. 14d). We find it noteworthy that several arginines (Arg548 and Arg572, Supplementary Fig. 14c) are in close proximity to Tyr417 and these positively charged residues may play a role in perturbing the pKa of neighboring tyrosine residues as previously reported for alanine racemase49. In contrast with GSTP1 and DPP3 enzymes, the discovery of a hyper-reactive tyrosine (Tyr475, Fig. 3d) in the Yjef-N domain of the scaffolding protein EDC3 is intriguing given the role of this domain in assembly of cytoplasmic RNA–protein (RNP) granules known as P-bodies involved in post-transcriptional regulation50. Future studies will test whether the hyper-reactive nature of the Tyr475 site can be exploited for developing ligands to modulate EDC3 function.
We applied SuTEx for development of a chemical phosphoproteomics platform to identify and quantitatively measure tyrosine sites whose probe modification status is competed by activation of phosphorylation. As proof of concept, we studied global changes in the tyrosine phosphoproteome under pervanadate activation of A549 cells to identify pervanadate-sensitive (PerS) sites that represented putative phosphotyrosines (Fig. 6 and Supplementary Fig. 25). Across >2,000 quantified sites, we identified a small subset of PerS sites (67 sites, Supplementary Dataset 1), which is in agreement with the low frequency of tyrosine phosphorylation (1%) compared with more abundant phospho-serines and phospho-threonines39, 40. We verified that SuTEx probe labeling is anticorrelated with phosphorylation at Tyr705 and Tyr228 of STAT3 and CTNND1, respectively (Fig. 6d,e). Both sites are highly annotated phosphotyrosines and reported substrates for tyrosine kinases in cancer cell signaling43, 44. In contrast, the pervanadate-insensitive Tyr105 site of PKM did not show changes in phosphotyrosine signals with pervanadate activation and further supports the ability of SuTEx to differentiate probe labeling of tyrosines based on phosphorylation state (Fig. 6d,e). Future studies will focus on further refinement, e.g. improvements to LC-MS method and use of SuTEx probe cocktails, to expand the number and type of phosphotyrosine sites quantified.
In summary, we deployed SuTEx for development of a quantitative chemical proteomics platform to globally profile tyrosine nucleophilicity and post-translational modification state in human cell proteomes. We believe our current findings serve as a blueprint for design of activity-based probes that can be synthetically modulated to meet the proteomic demands of chemical biology applications. Expansion of our chemical phosphoproteomics to other activation paradigms should afford additional opportunities for studying and potentially targeting tyrosine post-translational modifications in future studies (Fig. 2c). The latter effort will be expedited by conversion of SuTEx probes into inhibitors or ligands to reveal the inventory of tyrosine (and potentially phospho-tyrosine) sites that are “druggable” in proteomes.
ONLINE METHODS
HPLC assay for profiling solution reactivity and stability of sulfonyl probes
The following reagents were prepared and kept at 0 °C prior to use: 0.1 M solution of caffeine in acetonitrile (ACN), 1.0 M solution of n-butylamine, p-cresol, tetramethylguanidine (TMG), acetic acid in ACN, and 10 mM solution of the probes in a mixture of DMF-ACN (v/v=10:90) are made.
-
(i)
p-Cresol reactivity against a probe mixture: A solution of p-cresol (16.5 μmol, 3.3 eq) was premixed with 1.1, 2.2, or 3.3 eq of TMG. To initiate the reaction, the p-cresol/TMG solution was added to a sulfonyl probe mixture of HHS-475/HHS-482/HHS-SF-1 (500 μL, 5 μmol, 1.0 eq each) and the reaction was kept at 0 °C. The reaction progress was monitored by taking out a 50.0 μL aliquot of the reaction mixture at various time points followed by addition of a 10 μL quenching solution of acetic acid (0.5 M final, 5.0 μmol) and the internal caffeine standard (0.05 M final, 0.5 μmol). Sample (1.0 μL) was injected and analyzed by reverse-phase HPLC on a Shimadzu 1100 Series spectrometer with UV detection at 254 nm. Reaction progress was evaluated by monitoring consumption of sulfonyl probes because all probes generate a shared p-cresol and n-butylamine product. Chromatographic separation was performed using a Phenomenex Kinetex C18 column (2.6 μm, 50 mm x 4.6 mm). Mobile phases A and B were composed of H2O (with 0.1% AcOH) and CH3CN (with 0.1% AcOH), respectively. Using a constant flow rate of 0.8 mL/min, the gradient was as follows: 0–0.5 min, 15% B; 0.5–6.5 min 15–85% B (linear gradient); 6.5–7 min 85–100% B (linear gradient); 7– 8.5 min 100% B; 8.5–9 min 100–15% B (linear gradient); 9–9.8 min 15% B.
-
(ii)
n-Butylamine reactivity against a probe mixture: Reactivity of sulfonyl probes against n-butylamine (3.3 eq) was performed as described above except the amount of TMG was fixed at 3.3 eq.
-
(iii)
Probe reactivity against a p-cresol/n-butylamine mixture: A solution of n-Butylamine (50.0 μL, 50.0 μmol, 5.0 eq), p-cresol (10.0 μL, 10.0 μmol, 1.0 eq), and TMG (5.0 μL, 5 μmol, 0.5 eq) were prepared. Probe reaction was initiated by addition of this solution to HHS-475, HHS-482, or HHS-SF-1 (10 μmol, 1.0 eq) at 0 °C. Reaction progress was monitored as described above. A control experiment was also performed where equal amounts of n-butylamine (1.0 eq) and p-cresol (1.0 eq) were mixed.
-
(iv)
Probe stability studies: Each probe was dissolved in DMSO or a solution of DMF:ACN:PBS (4:6:1 (v/v)) at the following concentrations: 20 mM of HHS-475, 20 mM HHS-SF-1, and 10 mM of HHS-482 in a final volume of 50 μL. The internal caffeine standard (0.5 μmol) was spiked into each probe sample. Probe stability was monitored at room temperature by taking 1.0 μL of sample at three time points (0, 24, and 48 hours) and analyzing probe degradation by HPLC as described above.
Cell culture
Cell lines were cultured at 37 ˚C with 5% CO2 with manufacturer recommended media supplemented with 10% fetal bovine serum (FBS, U.S. Source, Omega Scientific) and 1% L-glutamine (Fisher Scientific): HEK293T: DMEM; DM93, A549, Jurkat, H82: RPMI. Cells were harvested for experimental use when they reached ∼90% confluency. The media was aspirated, cells washed with cold PBS (2X) and scraped from plates. The cells were pelleted by centrifugation at 400 × g for 5 min, snap-frozen using liquid nitrogen and stored at −80 ˚C until further use.
SILAC cell culture
SILAC HEK293T cells were cultured at 37 ˚C with 5% CO2 in either ‘light’ or ‘heavy’ media consisting of DMEM (Fisher Scientific) supplemented with 10% dialyzed FBS (Omega Scientific), 1% L-glutamine (Fisher Scientific), penicillin/streptomycin, and isotopically-labeled amino acids. Light media was supplemented with 100 μg/mL L-arginine and 100 μg/mL L-lysine. Heavy media was supplemented with 100 μg/mL [13C615N4]L-arginine and 100 μg/mL [13C615N2]L-lysine. The cells were grown for 6 passages before use in proteomics experiments. Cells were washed with PBS (2X), harvested, snap-frozen using liquid nitrogen and stored at −80 ˚C until further use.
Transient Transfection
Recombinant protein production by transient transfection of HEK293T cells was performed as previously described51. The following plasmid constructs (human proteins) were purchased from GenScript: pcDNA3.1-GSTP1-FLAG, pcDNA3.1-DPP3-FLAG, pcDNA3.1-PGAM1-FLAG, pcDNA3.1-EDC3-FLAG. Site-directed mutagenesis of wild-type constructs was used to generate mutant plasmids: pcDNA3.1-GSTP1 (Y8F)-FLAG, pcDNA3.1-DPP3 (Y417F)-FLAG, pcDNA3.1-PGAM1 (Y92F)-FLAG, pcDNA3.1-EDC3 (Y475F)-FLAG.
Pervanadate Activation
Pervanadate (100 mM) was prepared as previously described41 by mixing 100 μL of sodium orthovanadate (100 mM Na3VO4, New England BioLabs #P0758S) with 1 μL of hydrogen peroxide (H2O2, 30% v/v in water) on ice. The mixture was incubated on ice for 15 min followed by immediate addition to cells (1:1000, 100 μM final) and incubation for 30 min at 37 ˚C with 5% CO2 for general inhibition of protein tyrosine phosphatases. After pervanadate treatment, cells were washed twice with cold PBS followed by harvest. Cell pellets were resuspended in PBS supplemented with protease and phosphatase inhibitor mini tablets (Thermo Scientific #A32959) and then lysed by sonication (3 × 1 sec pulse, 20% amplitude). For CTNND1 western blot studies, cell pellets were lysed in NP40 Cell Lysis Buffer (Invitrogen #FNN0021) supplemented with protease/phosphatase inhibitor tablets. Cell lysates were separated via centrifugation at 100,000 x g for 45 min at 4 ˚C for western blot or chemical proteomic studies. Note: pervanadate treatments are performed on live cells but SuTEx probe labeling occurs in proteomes in vitro.
Western blot analysis
Western blot analysis of recombinant protein expression was performed as previously described51. For analysis of tyrosine phosphorylation, the protocol used was the same except the nitrocellulose blot was blocked with 3% BSA instead of 5% milk in TBS-T. The following antibodies were purchased from Cell Signaling Technology (CST) for phosphotyrosine studies: Phospho-tyrosine (pY): P-Tyr-100 biotinylated, CST #9417S; pPKM: Phospho-PKM (Y105) Rabbit Ab, CST #3827S; PKM: PKM Rabbit Ab, CST #3198S; pSTAT3: Phospho-STAT3 (Y705) Rabbit mAb, CST #9145S; STAT3: STAT3 Mouse mAb, CST #9139S; pCTNND1: Phospho-Catenin δ−1 (Tyr228) Rabbit Ab, CST #2911; CTNND1: Catenin δ−1 Rabbit Ab, CST #4989; GAPDH: GAPDH Rabbit mAb, CST #2118S. The following secondary antibodies were used for fluorescence detection: Goat Anti-Rabbit IgG DyLight 550 Conjugated, Thermo Scientific, #84541; Goat Anti-Mouse IgG DyLight 650 Conjugated, Invitrogen, #84545; Streptavidin DyLight 550 Conjugated, Thermo Scientific, #84542.
Gel-based chemical proteomic assay
Cell pellets were lysed in PBS by sonication and fractionated (100,000 × g, 45 min, 4 ˚C) to generate soluble and membrane fractions. Protein concentrations were determined using the Bio-Rad DC protein assay and adjusted to 1 mg/mL in PBS. Proteome samples (49 μL aliquots) were treated with sulfonyl-triazole or -fluoride probes at the indicated concentrations (1 μL, 50x stock in DMSO) for 1 hr at room temperature. Probe-labeled samples were conjugated by copper-catalyzed azide-alkyne cycloaddition (CuAAC) to rhodamine-azide (1 μL of 1.25 mM stock; final concentration of 25 μM) using tris(2-carboxyethyl)phosphine (TCEP; 1 μL of fresh 50 mM stock in water; final concentration of 1 mM), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, 3 μL of a 1.7 mM 4:1 t-butanol/DMSO stock, final concentration of 100 μM), and copper sulfate (CuSO4, 1 μL of 50 mM stock, final concentration of 1 mM). Samples were reacted for 1 hr at room temperature, quenched with 17 μL of 4X SDS-PAGE loading buffer and beta-mercaptoethanol (βME), and quenched samples (30 μL) analyzed by SDS-PAGE gel and in-gel fluorescence scanning.
Live cell evaluation of sulfonyl-triazole probes
Cells grown to ~90% confluency in 10 cm plates were treated with DMSO vehicle or sulfonyl-triazole probe (10 μL of 1000X DMSO stock) in serum-free media for the indicated concentrations and times at 37 ˚C with 5% CO2. After treatment, cells were washed with cold PBS twice before harvesting and preparation for gel-based chemical proteomic evaluation as described above. For LC-MS studies, protein concentrations were normalized to 2.3 mg/mL and 432 μL (for 1 mg final protein amount) were used for sample preparation as detailed below.
Preparation of proteomes for LC-MS/MS analysis
Proteomes were diluted to 2.3 mg/ mL in PBS and sample aliquots (432 μL) were treated with sulfonyl-triazole or -fluoride probes at the indicated concentrations (5 μL, 100X stock in DMSO), mixed gently and incubated for 1 h at room temperature. Probe-modified proteomes were subjected to CuAAC conjugation to desthiobiotin-PEG3-azide (10 μL of 10 mM stock in DMSO; final concentration of 200 μM) using TCEP (10 μL of fresh 50 mM stock in water; 1 mM final concentration), TBTA ligand (33 μL of a 1.7 mM 4:1 t-butanol/DMSO stock, 100 μM final concentration), and CuSO4 (10 μL of 50 mM stock, 1 mM final concentration). Samples were mixed by vortexing and then incubated for 1 h at room temperature. Excess reagents were removed by chloroform-methanol extraction as previously described51. Protein pellets were re-suspended in 500 μL of 6M urea/25 mM ammonium bicarbonate followed by DTT reduction and IAA alkylation as previously described51. Excess reagents were removed by chloroform/methanol extraction as described above, and the protein pellet was re-suspended in 500 μL of 25 mM ammonium bicarbonate and then digested to peptides using trypsin/Lys-C (7.5 μg in 15 μL of ammonium bicarbonate, sequencing grade from Promega) was added to the mixture and incubated for 3 hrs at 37 ˚C. Probe-modified peptides were enriched by avidin affinity chromatography, eluted, and prepared for LC-MS analysis as previously described51.
Preparation of SILAC proteomes for LC-MS/MS analysis
Heavy and light proteomes (432 μL of each) were diluted to 2.3 mg/mL in PBS. For 10:1 comparisons, heavy and light proteomes were treated with 250 μM and 25 μM of HHS465, respectively (5 μL, 100x stock in DMSO). In a control 1:1 comparison experiment both heavy and light proteome were treated with 25 μM of HHS465. Samples were mixed gently and incubated for 1 h at room temperature. Light and heavy samples were separately conjugated to desthiobiotin-PEG3-azide as described above. Light and heavy samples were mixed during the chloroform-methanol extraction step. Probe-modified peptides were prepared for LC-MS/MS analysis as described above.
LC-MS/MS analysis of samples
Nano-electrospray ionization-liquid chromatography-mass spectrometry analyses (LC-MS/MS) were performed using an Ultimate 3000 RSLC nanoSystem-Orbitrap Q Exactive Plus mass spectrometer (Thermo Scientific) as previously described51 except LC conditions were modified to use the following gradient (A: 0.1% formic acid/H2O; B: 80% ACN, 0.1% formic acid in H2O): 0–1:48 min 1% B, 400 nL/min; 1:48 – 2:00 min 1% B, 300 nL/min; 2–90 min 16% B; 90–146 25% B; 146–147 min 95% B; 147–153 min 95% B; 153–154 min1% B; 154.0–154.1 min 1% B, 400 nL/min; 154.1–180 min 1% B, 400 nL/min. A top 10 data-dependent acquisition MS method was used.
LC-MS/MS data analysis
Identification of peptides and proteins from tandem mass spectrometry analyses was accomplished using the Byonic™ software package (Protein Metrics Inc.31). Data were searched against a modified human protein database (UniProt human protein database, angiotensin I and vasoactive intestinal peptide standards; 40,660 proteins) with the following parameters: up to 3 missed cleavages to account for a lysine probe modification, 10 ppm precursor mass tolerance, 20 ppm fragment mass tolerance, too high (narrow) “precursor isotope off by x”, precursor and charge assignment computed from MS1, maximum of 1 precursor per MS2, 0.01 smoothing width, 1% protein false discovery rate, variable (common) methionine oxidation (+15.9949 Da) and fixed cysteine carbamidomethylation (+57.021464 Da). Sulfonyl-probe modifications of tyrosine, lysine, and other amino acids were included as a variable (common) modification of +635.27374 Da. Search results were imported into R and filtered for fully tryptic peptides (except N- and C-terminally modified), a Byonic score of ≥300 (unless otherwise specified), and a precursor mass error between −5 ppm and +5 ppm. A Byonic score of 300 was applied for a more inclusive initial evaluation of the search results and thereby consider more possible probe-modified sites. We manually verified the MS1 and MS2 spectra corresponding to the highest-scoring tyrosine- and lysine (internal or non-C terminal)-modified sequences (~50–100 peptides). The next most frequently matched and high-scored probe-modified amino acid residues were C-terminal lysines or arginines, which were determined to be false positive matches based on manual analysis of MS2 spectra (top ~50 highest Byonic scored-matches). These findings are consistent with the observation from previous studies with other probes11, 51 that trypsin does not cleave after a modified lysine or arginine. Distinct peptides containing probe-modified amino acid residues (termed sites) were determined by identifying all unique razor protein and site combinations across all of the proteomes tested.
Analysis and comparison of sulfonyl probe modified amino acid sites
To compare amino acid residues modified by sulfonyl probes, protein and peptide identifications were accomplished as described above with variable (common) modification of +635.27374 Da on the following amino acid residues: cysteine, aspartic acid, glutamic acid, histidine, lysine, methionine, asparagine, glutamine, arginine, serine, threonine, tryptophan, and tyrosine. For these amino acid comparisons, carbamidomethylation (+57.021464 Da) of cysteines was searched as a variable/common modification to allow for the potential of probe modification on cysteines. Comparisons of probe-modified sites across all probes and cell lines tested were performed using the R package ggplot2 (https://ggplot2.tidyverse.org/). Venn diagrams for comparisons were generated using the VennDiagram R package52. For amino acid comparisons, a Byonic score cutoff of 600 was used to minimize false positive identifications of modified residues, which were confirmed by manual evaluation to be incorrect assignments.
Domain enrichment analysis
Probe-modified sites were compared to ProRule domain annotations (available on PROSITE, release 20.8553, http://prosite.expasy.org/) using the annotated human UniProt proteome (https://www.uniprot.org/) as a database for identifying amino acid sequences that match ProRule domains. A probe-modified site that is within a ProRule domain is considered a “hit” and is counted as enrichment of a domain by the sulfonyl probe. Several sites within the same ProRule domain annotation are a considered a single hit. If a site had several annotations each one was considered a hit; for example, a modified site within the proton acceptor region of a kinase domain would be annotated as a hit for ProRules PRU10027 and PRU00159, respectively. The database count is determined by the number of non-overlapping occurrences of the domain such that calmodulin would account for 4 EF-hand domains (PRU00448). We find the probability of the domains P(D) in the reference UniProt human database to determine how frequently they exist in nature:
Where n(D) is the number of domain occurrences in the database and N is the total number of domains in the reference database. The p-values were calculated using a binomial test previously reported for GO statistical overrepresentation test54.
Binomial test
Where K is number of domain annotation hits in the experimental data (sulfonyl probe). The p-value was then corrected for a 1% false discovery rate (FDR) using Benjamini-Hochberg correction for multiple hypothesis testing. From these statistical analyses, ProRule domains that show statistically significant overrepresentation (Q value < 0.01) are used to generate bar graphs and pie charts shown in figures. Note that a -log (Q value) is used so that positive values are shown for simplicity. In order to verify that the binomial approximation to hypergeometric probability we ensured sum of all n(D) was less than 5 percent of N and verified that using a hypergeometric test did not alter the enriched domains. The enriched domains were grouped according to their function into four categories; nucleotide binding, enzyme, protein-protein interaction and undefined based on gene ontology molecular function annotation of the respective ProRule domain. Pie charts and bar graphs were generated using the ggplot2 package in R.
Classification of protein domains
The distinction between protein-protein interaction (PPI) and nucleotide binding domains was determined by whether the interacting partner of the domain is annotated as a peptide or a nucleotide sequence. The SH2 domain (PRU00191) which interacts with proteins featuring phosphorylated tyrosines is classified as a PPI domain, and a Homeobox DNA-binding domain (PRU00108) is classified as a nucleotide binding domain. An enzyme domain is the protein subunit that has been shown to catalyze the conversion of a substrate to a product. The Ribonuclease H domain (PRU00408) functions as an endonuclease which will interact with RNA but is classified as an enzyme domain because of its nuclease activity. We applied gene ontology (GO) molecular function annotations associated with the ProRule domains that inherit the annotation for catalytic activity (GO:0003824) to determine if proteins belong to the enzyme domain group. For example, the term Ribonuclease H domain (PRU00408) has the GO annotation for endonuclease activity (GO:0004519) which has catalytic activity (GO:0003824) in its ancestor chart and is therefore classified as an enzyme.
DrugBank analysis
Proteins labeled by sulfonyl probes in live cells were compared against protein targets of FDA approved and all drugs in the DrugBank databases33 (version 5.1.1).
Phosphosite Plus analysis
Probe-labeled sites were searched for in the PhosphoSitePlus database32 either unfiltered or filtered by a high-throughput reference score of 10 or greater where specified.
Nucleophilicity data analysis (SILAC)
Peptide and protein identification was accomplished using Byonic as previously described above. SILAC samples were searched with added masses for heavy-labeled amino acids (+10.0083 Da for R, +8.0142 Da for K) and converted into mzXML (from raw data file) and mzid (exported from Byonic) format for export into Skyline-daily55 to determine SILAC ratios (SR) of light/heavy peptides as previously described51. SILAC ratios from peptides with the same probe-modified site were averaged. The SILAC ratios were then plotted using the ggplot2 package in R56. Nucleophilicity was defined as follows: hyper-reactive, SR ≤2; mild reactivity, 2 < SR ≤5; low reactivity, SR >5.
GSTP1 biochemical substrate assay
Recombinant GSTP1-HEK293T soluble cell proteomes were diluted to 1 mg/ml in assay buffer (100 mM NaH2PO4, pH 7.0). GSH stock solution (250 mM in water) was diluted to 4 mM in assay buffer and 25 μL of diluted GSH solution was added to each sample. A substrate stock solution of 75 mM 1-bromo-2,4-dinitrobenzene (DBNB) in ethanol was diluted to 2 mM in assay buffer. Samples (25 μL) were aliquoted into a 96 well plate and spun briefly via centrifuge. 50 μl of 2 mM BDNB was added to each well and the reaction was monitored in kinetic mode by measuring absorbance at 340 nm for 10 min on a BMG Labtech CLARIOstar plate reader.
DPP3 biochemical substrate assay
Substrate assays were performed on recombinant DPP3-HEK293T soluble proteomes diluted to 1 mg/mL in assay. DPP3 sample (10 μL) was diluted to 85 μL with assay buffer and transferred to a black 96-well plate. A stock solution of DPP3 substrate (Arg-Arg β-naphthylamide trihydrochloride, 0.5 mM; Sigma-Aldrich) was diluted to 100 μM in assay buffer. Substrate solution (5 μL) was added to each sample. Samples were mixed briefly by shaking and reaction monitored in kinetic mode by measuring fluorescence at 450 nm for 10 min on a BMG Labtech CLARIOstar plate reader.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Hsu Lab and colleagues at the University of Virginia for helpful discussions and review of the manuscript. We thank M. Ross for his assistance with mass spectrometry experiments and data analysis. We thank S. Campbell for assistance with molecular biology experiments. We thank D. Dickie for assistance with small molecule crystallography studies. We thank R. Rumana for assistance with cell culture studies. This work was supported by the University of Virginia (start-up funds to K.-L.H.), University of Virginia Cancer Center (A.H.L. and K.-L.H), National Institutes of Health Grants GM801868 (T.B.W.), GM007055 (J.W.B.), CA009109 (A.L.B.), and DA043571 (K.-L.H.), the Schiff Foundation (K.-L.H.), the Wagner Fellowship (A.L.B), the National Science Foundation Graduate Research Fellowship (Grant No. 2018255830 to R.L.M.), and the U.S. Department of Defense (W81XWH-17–1-0487 to K.-L.H.).
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interest.
Data Availability
All data produced or analyzed for this study are included in the published article (and its supplementary information files) or are available from the corresponding author on reasonable request. Crystallographic data for small molecules has been deposited in the Cambridge Crystallographic Data Centre and have been assigned the following deposition numbers HHS-465 (CCDC 1954297), HHS-475 (CCDC 1954298), HHS-483 (CCDC 1954299).
Code Availability
All code is available upon reasonable request from the corresponding author.
REFERENCES
- 1.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem 2008, 77: 383–414. [DOI] [PubMed] [Google Scholar]
- 2.Sadaghiani AM, Verhelst SH, Bogyo M. Tagging and detection strategies for activity-based proteomics. Curr Opin Chem Biol 2007, 11(1): 20–28. [DOI] [PubMed] [Google Scholar]
- 3.Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem 2014, 83: 341–377. [DOI] [PubMed] [Google Scholar]
- 4.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov 2012, 11(1): 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deu E, Verdoes M, Bogyo M. New approaches for dissecting protease functions to improve probe development and drug discovery. Nat Struct Mol Biol 2012, 19(1): 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 2007, 46(2): 350–358. [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, Zhou B, Liang F, Wang WQ, Huang Z, Zhang ZY. Activity-based probes for protein tyrosine phosphatases. Proc Natl Acad Sci U S A 2004, 101(21): 7943–7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vocadlo DJ, Bertozzi CR. A strategy for functional proteomic analysis of glycosidase activity from cell lysates. Angew Chem Int Ed Engl 2004, 43(40): 5338–5342. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A 1999, 96(26): 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468(7325): 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacker SM, Backus KM, Lazear MR, Forli S, Correia BE, Cravatt BF. Global profiling of lysine reactivity and ligandability in the human proteome. Nat Chem 2017, 9(12): 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S, Yang X, Jia S, Weeks AM, Hornsby M, Lee PS, et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355(6325): 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews ML, He L, Horning BD, Olson EJ, Correia BE, Yates JR 3rd, et al. Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat Chem 2017, 9(3): 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker CG, Galmozzi A, Wang Y, Correia BE, Sasaki K, Joslyn CM, et al. Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell 2017, 168(3): 527–541 e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayanan A, Jones LH. Sulfonyl fluorides as privileged warheads in chemical biology. Chem Sci 2015, 6(5): 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao B, Zhang L, Zheng Q, Zhou F, Klivansky LM, Lu J, et al. Bifluoride-catalysed sulfur(VI) fluoride exchange reaction for the synthesis of polysulfates and polysulfonates. Nat Chem 2017, 9(11): 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong J, Sharpless KB, Kwisnek L, Oakdale JS, Fokin VV. SuFEx-based synthesis of polysulfates. Angew Chem Int Ed Engl 2014, 53(36): 9466–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahrney DE, Gold AM. Sulfonyl Fluorides as Inhibitors of Esterases. I. Rates of Reaction with Acetylcholinesterase, α-Chymotrypsin, and Trypsin. Journal of the American Chemical Society 1963, 85(7): 997–1000. [Google Scholar]
- 19.Shannon DA, Gu C, McLaughlin CJ, Kaiser M, van der Hoorn RA, Weerapana E. Sulfonyl fluoride analogues as activity-based probes for serine proteases. Chembiochem 2012, 13(16): 2327–2330. [DOI] [PubMed] [Google Scholar]
- 20.Gu C, Shannon DA, Colby T, Wang Z, Shabab M, Kumari S, et al. Chemical proteomics with sulfonyl fluoride probes reveals selective labeling of functional tyrosines in glutathione transferases. Chem Biol 2013, 20(4): 541–548. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Q, Ouyang X, Wan X, Gajiwala KS, Kath JC, Jones LH, et al. Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes. J Am Chem Soc 2017, 139(2): 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang B, Wu H, Schnier PD, Liu Y, Liu J, Wang N, et al. Proximity-enhanced SuFEx chemical cross-linker for specific and multitargeting cross-linking mass spectrometry. Proc Natl Acad Sci U S A 2018, 115(44): 11162–11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Michiels TJM, de Jong C, Soethoudt M, Dekker N, Gordon E, et al. An Affinity-Based Probe for the Human Adenosine A2A Receptor. J Med Chem 2018, 61(17): 7892–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong J, Krasnova L, Finn MG, Sharpless KB. Sulfur(VI) fluoride exchange (SuFEx): another good reaction for click chemistry. Angew Chem Int Ed Engl 2014, 53(36): 9430–9448. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, et al. Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J Am Chem Soc 2016, 138(23): 7353–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortenson DE, Brighty GJ, Plate L, Bare G, Chen W, Li S, et al. “Inverse Drug Discovery” Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates. J Am Chem Soc 2018, 140(1): 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fadeyi OO, Hoth LR, Choi C, Feng X, Gopalsamy A, Hett EC, et al. Covalent Enzyme Inhibition through Fluorosulfate Modification of a Noncatalytic Serine Residue. ACS Chem Biol 2017, 12(8): 2015–2020. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Li J, Li S, Li G, Sharpless KB, Wu P. SuFEx Click Chemistry Enabled Late-Stage Drug Functionalization. J Am Chem Soc 2018, 140(8): 2919–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol 2011, 7(7): 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn K, Boehm M, Brown MF, Calloway J, Che Y, Chen J, et al. Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism. ACS Chem Biol 2016, 11(9): 2529–2540. [DOI] [PubMed] [Google Scholar]
- 31.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics 2012, Chapter 13: Unit13 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 2015, 43(Database issue): D512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018, 46(D1): D1074-D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 2018, 19(5): 327–341. [DOI] [PubMed] [Google Scholar]
- 35.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 2002, 3(3): 177–186. [DOI] [PubMed] [Google Scholar]
- 36.Shin M, Franks CE, Hsu KL . Isoform-selective activity-based profiling of ERK signaling. Chem Sci 2018, 9(9): 2419–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi EJ, Jung D, Kim JS, Lee Y, Kim BM. Chemoselective Tyrosine Bioconjugation through Sulfate Click Reaction. Chemistry 2018, 24(43): 10948–10952. [DOI] [PubMed] [Google Scholar]
- 38.Shannon DA, Banerjee R, Webster ER, Bak DW, Wang C, Weerapana E. Investigating the proteome reactivity and selectivity of aryl halides. J Am Chem Soc 2014, 136(9): 3330–3333. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey SJ, Yang G, Yang P, Fazakerley DJ, Stockli J, Yang JY, et al. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab 2013, 17(6): 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundby A, Secher A, Lage K, Nordsborg NB, Dmytriyev A, Lundby C, et al. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat Commun 2012, 3: 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hilger M, Bonaldi T, Gnad F, Mann M. Systems-wide analysis of a phosphatase knock-down by quantitative proteomics and phosphoproteomics. Mol Cell Proteomics 2009, 8(8): 1908–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song G, Chen L, Zhang B, Song Q, Yu Y, Moore C, et al. Proteome-wide Tyrosine Phosphorylation Analysis Reveals Dysregulated Signaling Pathways in Ovarian Tumors. Mol Cell Proteomics 2019, 18(3): 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene 2003, 22(27): 4150–4165. [DOI] [PubMed] [Google Scholar]
- 44.Hong JY, Oh IH, McCrea PD. Phosphorylation and isoform use in p120-catenin during development and tumorigenesis. Biochim Biophys Acta 2016, 1863(1): 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2009, 2(97): ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weerapana E, Simon GM, Cravatt BF. Disparate proteome reactivity profiles of carbon electrophiles. Nat Chem Biol 2008, 4(7): 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 2010, 24(11): 1073–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 2006, 25(21): 5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life 2002, 53(2): 85–98. [DOI] [PubMed] [Google Scholar]
- 50.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 2012, 4(9): a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franks CE, Campbell ST, Purow BW, Harris TE, Hsu KL. The Ligand Binding Landscape of Diacylglycerol Kinases. Cell Chem Biol 2017, 24(7): 870–880 e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011, 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res 2013, 41(Database issue): D344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013, 8(8): 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bereman MS, Beri J, Sharma V, Nathe C, Eckels J, MacLean B, et al. An Automated Pipeline to Monitor System Performance in Liquid Chromatography-Tandem Mass Spectrometry Proteomic Experiments. J Proteome Res 2016, 15(12): 4763–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wickham H, SpringerLink (Online service), SpringerLINK ebooks - Mathematics and Statistics (2016) ggplot2 Elegant Graphics for Data Analysis. 2nd ed. [S.l.]: Springer International Publishing; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.