Abstract
Purpose:
The robust and reliable utilization of proton MRSI at ultra-high fields is hampered by several key technical difficulties, including contamination from extracranial lipids. To that end, this work presents novel lipid suppression sequences for proton MRSI in the human brain utilizing elliptical localization with pulsed second order fields (ECLIPSE).
Methods:
Two lipid suppression methods were implemented with the ECLIPSE gradient insert. One method is a variable power, four-pulse sequence optimized to achieve outer volume suppression (OVS) and compared against a standard, eight-slice OVS method. The second ECLIPSE method is implemented as an inversion recovery (IR) sequence with elliptical inner volume selection (IVS) and compared against a global IR method.
Results:
The ECLIPSE-OVS sequence provided a 116-fold mean lipid suppression (range 104–134), while an optimized 8-slice OVS sequence achieved 15-fold suppression (range 13–18). Furthermore, the superior ECLIPSE-OVS suppression was achieved at 30% of the RF power required by 8-slice OVS. The ECLIPSE based IR sequence suppressed skull lipids by 155-fold (range 122–257), compared to 16-fold suppression (range 14–19) achieved with IR.
Conclusion:
OVS and IVS executed with ECLIPSE provide robust and effective lipid suppression at reduced RF power with high immunity to variations in B1 and T1.
Keywords: Outer Volume Suppression, ECLIPSE, human brain MRSI, lipids
Introduction
Proton Magnetic Resonance Spectroscopic Imaging (MRSI) is a powerful technique that can map the metabolic profile in the human brain, non-invasively. The ability of MRSI to detect alterations in the neurochemical profile has seen applications, among others, in brain tumor classification (1), traumatic brain injury (2), epilepsy (3), multiple sclerosis (4), and neurodegenerative diseases such as Alzheimer’s disease (5).
Moving towards high and ultra-high magnetic fields (≥ 3T), MRSI provides increased SNR and spectral dispersion, thus improving the number of metabolites that can be quantified, and quantification accuracy. However, MRSI at ≥ 3T presents with technical challengers such as increased B0/B1 field heterogeneity, and increased power deposition as quantified by the specific absorption rate (SAR), in addition to lipid and water contamination. Extracranial lipids and water signals are the most dominant contaminants that affect reliable and accurate metabolite quantification in the brain regardless of field strength. These technical challenges, despite its potential, have impeded MRSI from becoming a widespread imaging modality in the clinical setting.
Water contamination in MRSI can be effectively addressed with water suppression methods such as WET (water suppression enhanced through T1 effects 6) or VAPOR (variable power RF pulses with optimized relaxation delays 7) modules, metabolite cycling (8,9), or with post processing techniques such as the Hankel singular value decomposition (HSVD) algorithm (10,11). Extracranial lipid suppression techniques demonstrated for in vivo brain MRSI, have their own set of advantages and disadvantages based on SAR requirements, reliability and robustness, level of lipid suppression achievable, and brain coverage (12,13). Popular and emerging extracranial lipid suppression techniques for MRSI include: i) cubical inner volume selection (IVS) based on single-volume MRS techniques, such as PRESS (point resolved spectroscopy 14), STEAM (stimulated echo acquisition mode 15), or LASER/semi-LASER (localization by adiabatic spin-echo refocusing 16), ii) outer volume suppression (OVS) of extracranial lipids by saturating the skull region with multi-slice (typically eight) excitation pulses (17), iii) T1-based nulling tailored for extracranial lipids (18,19), iv) RF shimming based localization (20,21), and v) dedicated crusher coils (22). Recently, extracranial lipid suppression with post-processing based methods (23,24) such as L2-regularization (24) is gaining popularity (13,25), primarily due to the absence of any additional RF pulses in the sequence, which is beneficial at ultra-high field applications. L2-regularization however only provides up to ~10-fold in lipid suppression (13,24), thus is typically combined with increased k-space coverage (26) to exploits spatially improved point-spread function in tandem with SNR gains achievable at ultra-high fields (13,25).
The conventional OVS method, in which eight saturation bands are placed around the skull, is an attractive alternative to cubical IVS ROI methods (see Figure 1A) to maximize anatomical coverage. However, due to unavoidable overlapping of slice regions, and finite time requirements for RF pulses and spoiler gradients, all eight saturation slices cannot be executed in a way that avoids spatially differential relaxation of lipids. The incorporation of two-cycles of the 8-slice OVS can: i) minimize spatially differential relaxation of lipids effects due to overlapping slices, and ii) improve immunity to B1+ inhomogeneity and immunity to different T1 species inherent to lipids. Previous studies (27,28) have shown improvements in the performance of 8-slice OVS (mean lipid suppression of ~17-fold and ~25-fold in the two studies, respectively) by optimizing inter-pulse delays and RF pulse powers for improved extracranial lipid suppression and reduce chemical shift displacements (CSD). However, a 16-RF pulse (8 × 2) based OVS module becomes SAR intensive and can be prohibitive moving towards ultra-high fields; as demonstrated by using TR > 4500 ms for MRSI applications reported for ≥ 7T studies (28–30).
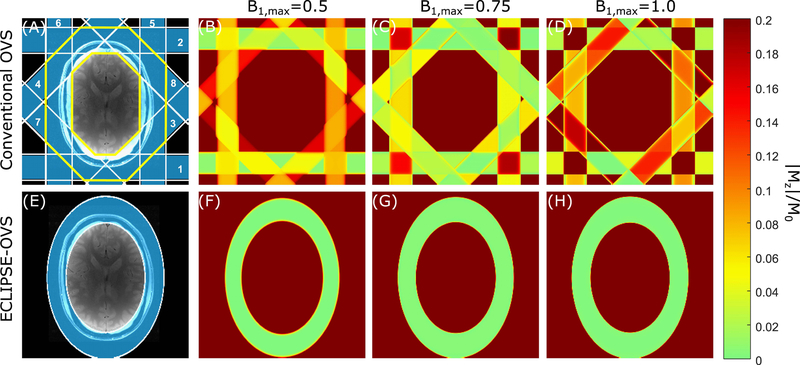
FIG. 1.
Comparison of conventional-OVS and ECLIPSE-OVS methods. (A, E) ROI placement for the conventional-OVS and ECLIPSE-OVS methods, respectively, which are overlaid on an axial brain slice. (A) The conventional-OVS method was implemented with 30mm thick slices at fixed 45° increments and two cycles placed around the scalp. The order in which slices were played out is marked. The extracranial ROI considered in optimizations is marked by the yellow boundary. (B, C, D) illustrate spatially differential relaxation of lipid resonances (T1=380 ms) due to non-instantaneous OVS pulses, and regions that experience/do not experience overlapping slices, for B1,max = 0.5, B1,max = 0.75, and B1,max = 1.0, corresponding to a − 33.33%, 0%, and + 33.33% variation in B1, respectively. (F, G, H) illustrate lipid suppression (< 0.02) with ECLIPSE-OVS, where no differential relaxation of lipid resonances is seen due to single-shot outer volume selection, while being insensitive to ± 33.33% B1 variation.
Elliptical localization with pulsed second order fields (ECLIPSE) for MRSI was previously demonstrated (31) with IVS of an elliptical ROI, providing improved axial coverage relative to a cubical ROI and reduced number of RF pulses relative to conventional OVS. The objectives of this work were to develop ECLIPSE-based OVS and inversion recovery (IR) sequences with highly effective and robust lipid suppression combined with low SAR, for applications in high and ultra-high field MRSI.
Methods
MR system
All MR experiments were performed on a 4T magnet (Magnex Scientific Ltd.) interfaced to a Bruker Avance III HD spectrometer running ParaVision 6 (Bruker, Billerica, MA, USA). The system contains actively shielded gradients capable of switching 30mT/m in 1150μs, and provides up to third order shimming. A within-brain B1+ optimized, 8-element Tx/Rx volume coil embedded in the ECLIPSE gradient coil was used for the study in a fixed phase configuration. The 8-element Tx/Rx volume coil has ± 30% and ± 60% B1+ variation within the brain and extracranial region, respectively.
The ECLIPSE gradient system
The ECLIPSE system (31) is a home-built, unshielded gradient insert consisting of Z2, X2Y2, and XY second order spherical harmonic magnetic fields with efficiencies of 5.48, 2.58 and 2.76 Hz/cm2/A, respectively, driven by three independent 100A Techtron 7780 current amplifiers (AE Techtron, Elkhart, IN, USA) interfaced to a home-built multi-channel gradient controller (32). The minimum rise time of the ECLIPSE magnetic fields is 660 µs, which was increased to 1150μs to match the system gradient rise time for convenience with pulse sequence timings. The gradient rise time is fixed, independent of amplitude.
The Z2, X2Y2, and XY gradient coils on the ECLIPSE system, in combination with the linear X, Y, Z system gradients are used to generate magnetic fields for an elliptical ROI that can be translated and rotated as detailed in the original study (31). On an axial slice , the spatially varying gradient field can be expressed in the following form: , where and are offsets in and directions. and are radii along and directions, and and are constants, which are determined by the Z2 and X2Y2 field amplitudes, RF pulse bandwidth, and RF pulse carrier frequency to select an elliptical disk or ring ROI (31). An elliptical disk can be used for inner volume selection (IVS), and a ring ROI can be used for single-shot OVS for extracranial lipid suppression (see Figure 1E).
Switching the ECLIPSE gradient coil resulted in small eddy currents in terms of time-varying B0 field variations (1.3Hz/A with a 260ms decay constant). These modulations were virtually eliminated by an empirically determined gradient pre-pulse applied with opposite polarity. No post-processing eddy current corrections were required.
For all applications of ECLIPSE in this work, the Z2 gradient amplitude [A] was set such that: , thereby generating a radially decreasing gradient field in the X-Y plane. This convention ensures that a skull ROI selected for water on an anatomical MRI generates a smaller ROI for up-field lipid signals that then fully encompasses the inner lining of extracranial lipids.
A MATLAB (MathWorks, Natick, MA, USA) script was created to allow interactive placement of the elliptical ROI superimposed on an anatomical axial image. The script then generates the required Z2, X2Y2, XY, X, Y gradients, and frequency offset of the RF pulse to select the desired ROI, based on the RF pulse bandwidth, and previously determined spatial B0 field distributions of the gradient fields of the ECLIPSE system per unit current.
Optimized lipid and water suppression
The building block for optimizing lipid/water suppression in simulation, includes an instantaneous RF pulse with a flip-angle governed by the B1 field (B1+ will be labeled as B1 hereafter, since no reference to the receive field is used) followed by an instantaneous spoiler gradient to mimic a situation whereby no coherent transverse magnetization remains, and finally an inter-pulse delay to allow longitudinal relaxation, as previously described (6) for water suppression. If longitudinal relaxation of a spin species is T1, and longitudinal magnetization before the nth water/lipid suppression pulse (with flip angle θn as governed by the B1 field amplitude) is longitudinal magnetization after a time delay of τn is given by:
where is the equilibrium magnetization (6,27). This process can be repeated for as many RF pulses and delays until the desired level of lipid/water suppression is achieved over a required T1 and B1 range. A least-squares minimization was carried out (to minimize by optimizing , ), using the Levenberg-Marquardt method built into MATLAB, for a given range of T1 and B1 values, as previously described for water suppression with WET (6) and VAPOR (7), and subsequently extended for 8-slice OVS (hereafter referred to as conventional-OVS) lipid suppression (27,33). To avoid convergence to sub-optimum local minima, the minimization was repeated for random initial conditions till a satisfactory solution was found.
All water suppression modules were performed with Gaussian pulses (length =10 ms, bandwidth = 200 Hz), and OVS/IR modules used an adiabatic full passage (AFP) hyperbolic secant (HS4 (34)) modulated RF pulse (length = 6.66 ms, bandwidth = 3,000 Hz) that provides frequency-selective excitation at lower RF amplitudes, and frequency-selective inversion above a minimum threshold RF amplitude (Figure 2C–D); as previously exploited (35). The non-sinusoidal relationship between and for an AFP HS4 pulse was numerically characterized with Bloch simulations and used in all optimizations. The minimum B1 value corresponding to Mz/M0 = −0.99 is taken to represent a complete inversion (180°, or 99.5% inversion efficiency), and is assigned the value ‘1’ in the normalized B1 amplitude scale used throughout this work (see Figure 2C–D).
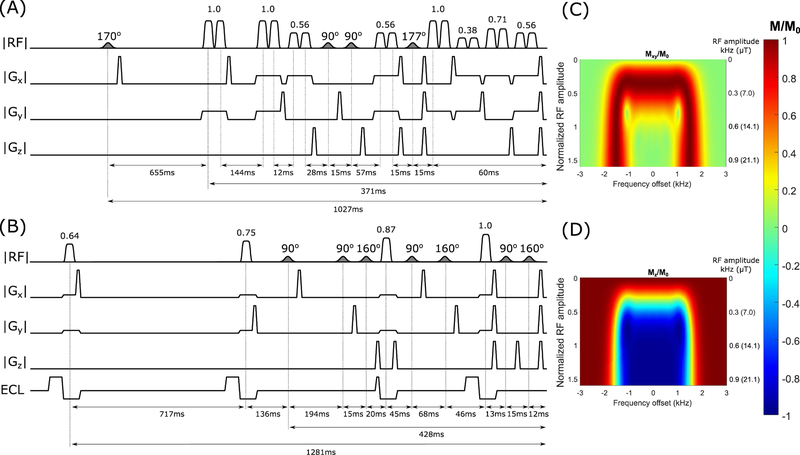
FIG. 2.
(A–B) Pulse sequence diagrams for conventional-OVS, and ECLIPSE-OVS modules. In all pulse sequence diagrams, water suppression RF pulses are shaded, and OVS RF pulses are open. Most inter-pulse delays are labeled along with total module lengths. Unmarked inter-pulse timings represent minimum timing delays and can be calculated by knowing that water suppression pulses, OVS pulses, and spoiler gradients are 10 ms, 6.66 ms, and 3.3 ms in duration, respectively. The marked OVS RF pulse amplitude ratios are scaled to B1,max. (A) Water suppression comprises of four RF pulses and three flip angle settings (170°, 90°, and 177°) intertwined with the conventional-OVS component. Parallel slice pairs are played out back-to-back with no spoilers in between. (B) The optimized ECLIPSE-OVS module with four OVS pulses and optimized VAPOR style water suppression pulses. Each OVS pulse is played out with an ECLIPSE gradient (labeled as ECL), which includes an opposite polarity pre-pulse with an empirically determined 70% higher amplitude to eliminate eddy current induced time varying B0 field modulations. The gradient waveforms for Z2, X2Y2 and XY channels are identical, apart from their respective amplitudes. Typically, the X2Y2 and XY amplitudes are ~ 50% and ~ 10% of the Z2 amplitude, respectively. (C–D) All modules developed were constructed with an adiabatic full passage (AFP) hyperbolic secant (HS4 (34)) modulated RF pulse, with a near-flat inversion profile vs B1 as shown; (C) represents the Mxy/M0 profile and (D) represents the Mz/M0 profile. The RF amplitude is normalized to 1 when the AFP pulse achieves 99.5% inversion efficiency, also the corresponding B1 value in kHz and µT are reported on the right side (C–D).
8-slice Outer Volume Suppression and Water Suppression Module Optimization
A conventional-OVS module as shown in Figure 1A was optimized with two cycles and used as the reference against an ECLIPSE-based OVS module. The global T1 value of extracranial lipids was determined to be ~380 ms at 4T in vivo. The conventional-OVS module optimization was carried out over a T1 range spanning from 370–390 ms, and a B1 range spanning ± 40% with four allowed RF power settings to be allocated among the 2 × 8 = 16 AFP pulses. Extending the optimization beyond the stated B1 and T1 ranges leads to detrimental OVS performance. Since placement of the 16 OVS pulses was prioritized, the last water suppression pulse had to be placed 75 ms before the start of the spin-echo sequence (see Figure 2A); this constraint had an impact on the water suppression performance with a seven-pulse VAPOR style sequence with two flip angle settings in simulation. Instead, an optimized four RF pulse-based water suppression sequence with three variable flip angle settings (170°, 90°, and 177°), was intertwined within the 16 OVS pulses.
ECLIPSE based outer volume suppression
ECLIPSE based OVS is achieved by repeatedly exciting and dephasing an elliptical ROI as illustrated in Figure 1E. Since each ECLIPSE based OVS pulse uniformly affects the entire in-plane extracranial lipid region, spatially differential relaxation as associated with conventional-OVS is avoided. As a result, ECLIPSE-OVS lipid suppression can be optimized in close analogy to water suppression in which the spatial component does not play a direct role. Feasibility of ECLIPSE based OVS was evaluated in simulation with two-pulse, three-pulse, and four-pulse variants with variable powers and delays. Furthermore, efficacy of ECLIPSE-OVS was evaluated in simulation for applications at 7T MRSI based on lipid species with T1 values spanning ~300–900ms reported in the literature (19,28). Lipid suppression in the order of >100-fold (T1 range 300–500 ms) can be achieved with a three-pulse ECLIPSE-OVS variant, with immunity to ± 20% variation in B1. Reducing the number of pulses to two greatly reduces immunity to B1 inhomogeneity and T1 variation, such that this sequence is not recommended for practical in vivo lipid suppression for MRSI. The four RF pulse-based ECLIPSE-OVS module was the only method (out of the three) capable of being immune to a ± 60% variation in B1, while covering T1 species from 300–400 ms. A seven-pulse VAPOR water suppression sequence was re-optimized to be used in conjunction with the four-pulse ECLIPSE-OVS module (Figure 2B).
Lipid-nulled MRSI with global inversion recovery (IR) and ECLIPSE based inner volume selection + IR
Extracranial lipid suppression with global inversion recovery (IR) was implemented as a single, AFP inversion pulse. A seven-pulse VAPOR water suppression sequence was re-optimized to account for the effect of the IR global inversion pulse (see Supporting Information Figure S1).
All the above described OVS and IR modules were followed with the spin-echo sequence illustrated in Supporting Information Figure S2A (detailed below).To improve extracranial lipid suppression of the global IR module (Supporting Information Figure S1), the excitation pulse in the spin-echo component described in Supporting Information Figure S2A was modified with ECLIPSE magnetic fields, thereby selecting an elliptical ROI for IVS as shown in Supporting Information Figure S2B. This sequence utilizes a global IR, with an ECLIPSE-IVS excitation pulse, herein termed the ECLIPSE-IVS + IR module.
The AFP RF pulse amplitude ratios for OVS/IR modules, marked in Figure 2A–B and Supporting Information Figure S1, are scaled by B1,max (maximum B1 amplitude) to arrive at their respective normalized B1 amplitudes (the B1 amplitude in kHz or μT corresponding to the normalized B1 amplitude is illustrated in Figure 2C–D). Variations in B1,max is then used to evaluate and optimize water/lipid suppression modules with respect to B1 inhomogeneity.
The spin-echo with MRSI
The two spin-echo variants used (Supporting Information Figure S2A–B) have the following parameters in common: TE = 15 ms spectral width= 5.0 kHz, 1024 points, and a 204 ms acquisition to capture the entire FID of metabolites in vivo. The excitation pulse in Supporting Information Figure S2A is achieved with a Shinnar-Le Roux (36) pulse (length = 2.0 ms, bandwidth = 2.8 kHz), and refocusing is achieved with Hermite refocusing pulses (length = 2.44 ms, bandwidth = 1.4 kHz). Both excitation and refocusing pulses are z-selective for a 10mm slab, and phase encoding gradients superimposed on the last spoiler. For the spin-echo sequence in Supporting Information Figure S2B, the excitation pulse was applied during an IVS selecting ECLIPSE gradient, and the refocusing pulse was z-selective for a 10mm slab. To minimize in-plane chemical shift displacement, the Shinnar-Le Roux excitation pulse was shortened to 1.24 ms, to obtain a 4.5 kHz bandwidth. The inherent requirement for a polarity reversed rewinding gradient following the excitation pulse gradient was sufficient (as a pair) to eliminate eddy current induced temporal B0 shifts with ECLIPSE, thus no additional Z2 pre-pulse was used (see Supporting Information Figure S2B).
The MRSI phase-encoding gradients were set to sample an elliptical portion of k-space with a field of view 170 × 210 mm2 (17 × 21 matrix) resulting in a nominal 1 mL volume resolution. To meet SAR requirements for the conventional-OVS MRSI sequence, the TR was set to 3000 ms leading to a 12.25-minute MRSI acquisition. Though the ECLIPSE-OVS MRSI method has a lower SAR requirement than conventional-OVS MRSI, it was also executed with a TR = 3000 ms to allow quantitative comparisons. The SAR requirement for both IR and ECLIPSE-IVS + IR MRSI sequences allowed for a TR < 3000 ms, and since IR based lipid suppression with and without ECLIPSE was a separate analysis, both these IR sequences were executed with a TR = 2000 ms leading to an 8.17-minute MRSI acquisition.
Human subjects and preparation
Three (n = 3) healthy volunteers participated in the study to compare ECLIPSE-OVS and conventional-OVS, and the same three volunteers participated in a separate session to compare the IR and ECLIPSE-IVS + IR methods. All volunteers provided written informed consent prior to each MRI session, and all procedures were previously approved by the Yale University Institutional Review Board. Efforts were made to match placement of the MRSI slab during the two sessions for all three volunteers.
Multi-slice gradient-echo MRIs were used to plan the MRSI slice location in the z-direction (superior to the corpus callosum), and a single-slice GE MRI was used to interactively plan OVS and ECLIPSE ROI’s. High-resolution proton density weighted axial MR images were acquired (TR/TE = 500/6.5 ms, FA= 15°, resolution 1 × 1 × 4 mm) for anatomical referencing of the MRSI grid. The magnetic field homogeneity of the 10 mm thick MRSI slice was optimized with first-and-second-order shim fields using a vendor supplied B0 field mapping method, resulting in water linewidths of ~12Hz over the entire slice. A water reference MRSI was acquired with lipid and water suppression pulses off, and ECLIPSE gradients turned off with other parameters identical to the metabolite-MRSI acquisition. No manual RF power adjustments were made for water suppression, OVS or ECLIPSE following the vendor-supplied, slice-based RF power calibration.
Data processing
A home-written MATLAB based software toolkit, NMRWizard, was used for all data processing. The following steps were employed on all datasets. 1) The acquired water reference MRSI was used to compute optimal weightings from each RF coil element per voxel basis. 2) A spatial 2D Fourier transform without apodization was performed, followed by 3) 3 Hz Lorentzian apodization, Fourier transformation and zero-order phase correction. 4) Metabolic maps were generated by integrating over a 0.2 ppm range centered on the respective metabolite peak from the raw spectra following frequency alignment.
Measurement of lipid and water suppression performance
The reference water MRSI was also used to evaluate residual water and lipid fractions with the methods presented, as quantified by residual magnetization (Mz/M0). For lipid suppression comparisons, the water signal was removed from the reference water MRSI with a HSVD algorithm to minimize baseline related errors. The residual lipid fractions were calculated by taking the ratio of the integral over the 0.8 – 1.6 ppm region for the method under test, and the water removed reference MRSI. The residual water fraction was calculated by taking the ratio between the maximum amplitude of water in the 4.5 – 4.8 ppm range for the method under test, and the raw water reference MRSI.
Results
Lipid and water suppression in simulation
The optimized conventional-OVS module with inter-pulse delays is illustrated in Figure 2A. The mean |Mz|/M0 factor over the extracranial ROI (defined by the solid yellow line in Figure 1A), as a function of B1,max and T1 is illustrated in Figure 3A, where |Mz|/M0 < 0.05 is achieved in the T1 range of 340 – 400 ms within a B1 variation of ± 25%. Though the mean |Mz|/M0 appears acceptable in this B1 range, there are regions of ‘hotspots’ that arise. For example, when the B1 intensity is increased by ± 33%, hot spots due to spatially differential relaxation occur, as illustrated in Figure 1B–D with (Mz/M0)max ~0.22. The optimized conventional-OVS sequence can handle up to ± 20% variation in B1 with hotspots (Mz/M0)max < 10%, comparable to previous reports of optimized conventional OVS sequences in simulation (27). The water suppression performance as a function of T1 and B1 (represented as nutation angle for a 90° excitation pulse) is shown in Figure 3B.
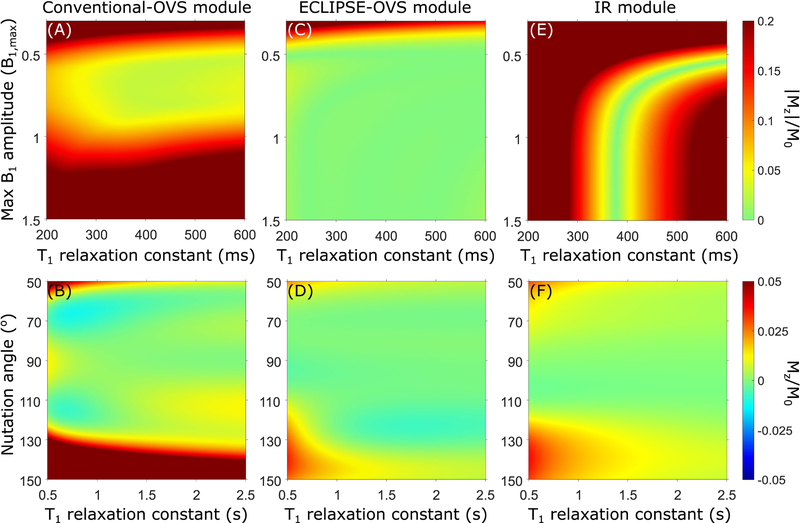
FIG. 3.
(A, C, and E) Simulated mean residual lipid fraction (|Mz|/M0) as a function of T1 and B1,max, for conventional-OVS, ECLIPSE-OVS, and IR modules, respectively. (B, D, and F) Simulated residual water fraction (Mz/M0) as a function of T1 and B1 (represented as nutation angle for a 90° excitation pulse) for conventional-OVS, ECLIPSE-OVS, and IR modules, respectively. Water suppression was optimized to tolerate a 33.33% variation in B1 (60°−120° range in flip angle), and a T1 range spanning 500–3500 ms. The mean residual lipid fraction (|Mz|/M0) for the conventional-OVS module illustrated in (A) is calculated by averaging over the extracranial ROI associated with the ROI marked in yellow in Figure 1A.
The optimized ECLIPSE-OVS module illustrated in Figure 2B provides excellent lipid suppression [(Mz/M0)max < 0.03] in simulation, given a minimum B1 amplitude is met and is effective for T1 species spanning 200–600 ms (Figure 3C), with no hotspots for ± 33% variation in B1,max amplitudes (see Figure 1F–H). Superior water suppression performance is seen for ECLIPSE-OVS (Figure 3D) relative to conventional-OVS (Figure 3C) due to the reduced number of OVS pulses (4 vs 16), which improves flexibility in placement of water suppression pulses during optimization.
The IR module in simulation is effective for a narrow band of T1 species centered at 380 ms (see Figure 3E), and is ineffective outside this regime, as expected. Since an AFP inversion pulse is used, suppression of lipid resonances with T1 = 380 ms is achieved for B1,max ≥ 1 (corresponding to a full inversion). Water suppression performance in simulation, as a function of T1 and B1 (represented as nutation angle for a 90° excitation pulse) is shown in Figure 3F.
Table 1 summarizes lipid suppression and water suppression performance for the three modules in simulation. Mean/median/std statistics were calculated for a B1,max span of 0.375–1.125 and T1 span of 300–400 ms. Residual water fractions were calculated in the flip angle span from 60°−120° and T1 span of 500–3000 ms; for the two IR methods, a B1,max span of 0.6–1.8, and T1 span of 500–3000 ms was used.
Table. 1.
Summary of water suppression, and lipid suppression performance of the developed OVS/IR modules in simulation.
| Conventional-OVS (mean/median/std) | ECLIPSE-OVS (mean/median/std) | IR (mean/median/std) | |
|---|---|---|---|
| Residual lipid fraction (Mean lipid suppression factor) | 0.073/0.052/0.045 (13.7-fold) |
0.008/0.004/0.015 (125-fold) |
0.076/0.068/0.056 (13.2-fold) |
| Residual water fraction (Mean water suppression factor) | 0.0038/0.0027/0.0034 (263-fold) |
0.0017/0.0013/0.0015 (588-fold) |
0.0023/0.0014/0.0023 (434-fold) |
The optimized ECLIPSE-OVS module for applications in 7T MRSI (results not shown), has an overall duration of 1100ms, thus is compatible with a TR = 2s sequence, and provides excellent lipid suppression [(Mz/M0)max < 0.03, and mean lipid suppression factor of 89-fold], calculated for a B1,max span of 0.375–1.125 and T1 span of 300–1000 ms. The minimum cumulative power required by the ECLIPSE-OVS module in simulation in this case is equivalent to 0.6 times the power required by one AFP inversion pulse with 99.5% inversion efficiency).
Outer volume suppression for in vivo MRSI
From simulations and preliminary findings in vivo, it was determined that B1,max = 0.75 provides optimal lipid suppression for both conventional-OVS and ECLIPSE-OVS, where both modules operate in the non-adiabatic regime. This power setting was used in all in vivo studies without further modification.
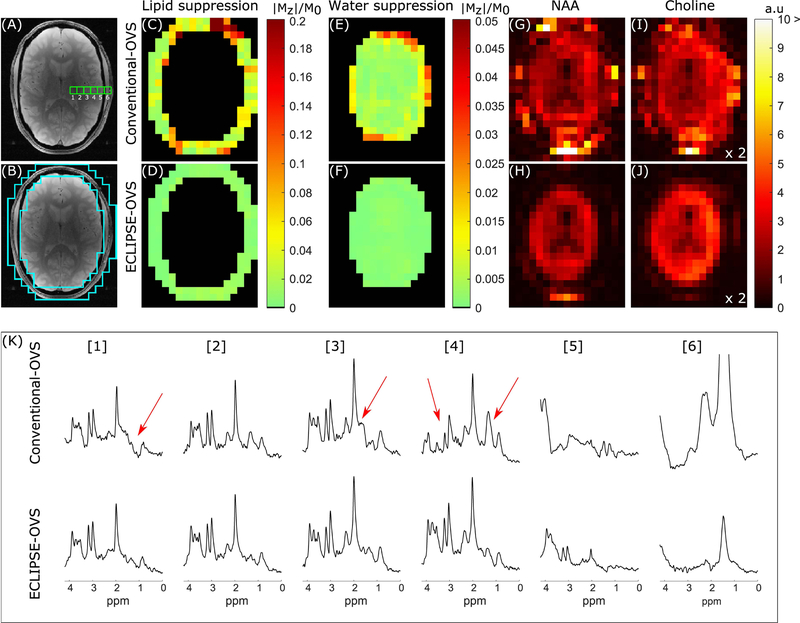
Lipid and water suppression performance, and the overall MRSI data quality from one volunteer for conventional-OVS and ECLIPSE-OVS are summarized in Figure 4. The mean lipid suppression factor for this volunteer was 18-fold for conventional-OVS; comparable in mean suppression performance with previous OVS methods described in the literature (27,28), with the exception of isolated hotspots (Mz/M0)max ~0.3 due to the high experimental B1 inhomogeneity of the RF coil. In comparison, ECLIPSE-OVS shows excellent lipid suppression (Figure 3D), where (Mz/M0)max < 0.02 with a mean suppression factor of ~135-fold. Residual water fraction for the two OVS sequences within the brain is illustrated in Figure 4 E–F, with mean water suppression factors of ~137-fold and ~870-fold for conventional-OVS and ECLIPSE-OVS, respectively. Figure 4G–J illustrates NAA and choline maps, obtained with conventional- OVS (Figure 4G/I) and ECLIPSE-OVS (Figure 4H/J) methods. Note that metabolic maps are not intensity corrected for the Tx/Rx profiles of the coil, and choline maps have been scaled up by a factor of two relative to the NAA maps for clarity. A clear outline of the ventricles is seen with hypo intensities in both NAA and choline maps with ECLIPSE-OVS, demonstrating minimal lipid and water contamination. Choline maps for the conventional-OVS method do not show a clear delineation between brain and extracranial space primarily due to residual water contamination around the scalp. Additional metabolic maps for creatine and Glx, and ROI placement for conventional-OVS and ECLIPSE-OVS is illustrated in Supporting Information Figure S3. Figure 4K shows MR spectra for both OVS methods, from the locations indicated in Figure 4A. While conventional-OVS achieves reasonable lipid suppression, essentially every spectrum displays contamination from extracranial lipids (arrows). In contrast, ECLIPSE-OVS spectra show a very consistent macromolecular signature with clear variations in the metabolite signature moving from WM (left) to GM (right)-dominant voxels. Though it may appear that a water baseline is present in spectra in Figure 4K, no noticeable baseline shift with either method due to insufficient water suppression was present; see spectra in Supporting Information Figure S5, which are identical to spectra in Figure 4K spanning up to 6ppm. Close inspection of spectra show that conventional-OVS contains voxels with the water peak ~ 7 times higher than that of NAA, and ECLIPSE-OVS to contain voxels with the water peak comparable to the NAA peak.
FIG. 4.
Summary of conventional-OVS and ECLIPSE-OVS sequence performance on one volunteer. (A) Anatomical axial slice corresponding to the region used for MRSI illustrating six adjacent MRS voxel locations compared in (K). (B) The ROI used to calculate extracranial lipid suppression in (C–D). (C–D) Residual extracranial lipid fraction maps for the conventional-OVS and ECLIPSE-OVS sequences, respectively. The conventional-OVS method shows hotspots as predicted in simulation, with mean suppression of ~18-fold (mean/median |Mz|/M0 of 0.054/0.034, respectively). ECLIPSE-OVS in comparison, demonstrates excellent lipid suppression with a mean suppression factor of ~135-fold (mean/median |Mz|/M0 of 0.0074/0.0073, respectively) with no hotspots. (E–F) Mean water suppression factors for conventional-OVS within the brain were ~137-fold (mean/median |Mz|/M0 of 0.0073/0.0041, respectively), with hotspots ~0.05. In comparison, the ECLIPSE-OVS sequence demonstrates superior water suppression with a mean suppression factor ~870-fold (mean/median |Mz|/M0 of 0.0011/0.0010, respectively). (G–H) NAA and (I–J) choline maps calculated for the two methods. (K) Spectra from the 6 adjacent voxels shown in (A) were selected to start from predominantly a ventricular area (voxel 1), moving towards white/gray matter (voxel 3, 4), and finally moving out to the extracranial lipid region (voxels 5, 6). Voxels 1–4 with conventional-OVS show prominent metabolites (NAA, creatine, and choline) with noticeable artifacts (marked with red arrows); in comparison no such contaminants are observed from voxels 1–4 with ECLIPSE-OVS. Lipid contaminants in voxel 6 is seen to be below the amplitude of NAA in brain for ECLIPSE-OVS, but beyond the displayed bounds for conventional-OVS.
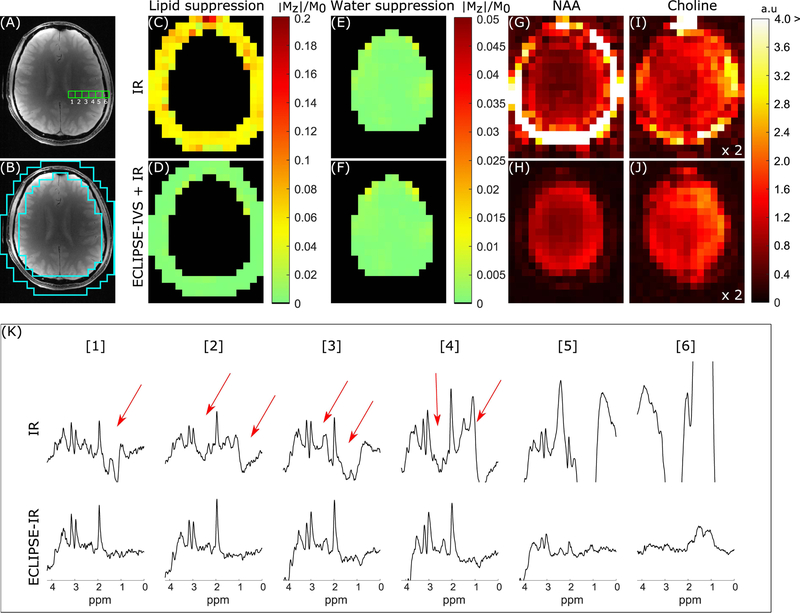
The inversion pulse used for IR has 99.5% inversion efficiency when B1,max = 1, however, was empirically set to B1,max = 1.2 to account for B1 inhomogeneities. The IR based MRSI sequence was compared against the ECLIPSE-IVS + IR sequence as summarized in Figure 5 with an identical layout to the OVS comparison in Figure 4 on a different volunteer. The mean lipid suppression factor was ~18-fold with IR, and ~257-fold with ECLIPSE-IVS + IR (Figure 5D). Both sequences had near-identical mean water suppression performance (Figure 5E–F) of ~760-fold. NAA and choline maps generated with IR and ECLIPSE-IVS + IR methods are shown in Figure 5G–H and Figure 5I–J, respectively, where an overall ~2-fold reduction in SNR relative to results in Figure 4 is seen due to the IR module, and shorter TR of 2 seconds used compared to TR of 3 seconds with OVS. Additional metabolic maps for creatine and Glx, and ROI placement for ECLIPSE-IVS + IR is illustrated in Supporting Information Figure S5. A significant proportion of extracranial lipids remain for the IR sequence that contaminates spectra as shown with arrows for voxels in Figure 5K. In comparison, ECLIPSE-IVS + IR MRSI spectra appear with minimal baseline for brain voxels. The voxel-wise mean improvement in extracranial lipid suppression (within brain) due to ECLIPSE-IVS + IR is ~14-fold relative to the IR sequence.
FIG. 5.
Summary of IR and ECLIPSE-IVS + IR sequences with identical layout to Figure 4. (A) The anatomical image corresponding to the MRSI slice illustrating six adjacent MRS voxel locations used to compare the two IR methods in (K). (B) The ROI used to calculate extracranial lipid suppression in (C–D). (C–D) Residual extracranial lipid fraction maps for the IR and ECLIPSE-IVS + IR sequences, respectively. (C) Near-uniform lipid suppression throughout the ROI is seen (relative to conventional-OVS) with ~18-fold mean suppression with IR (mean/median |Mz|/M0 of 0.057/0.054, respectively), and (D) ECLIPSE-IVS + IR in comparison, demonstrates superior lipid suppression with a mean suppression factor of ~257-fold (mean/median |Mz|/M0 of 0.0039/0.0025, respectively) with no hotspots. (E–F) Residual water fraction maps within the brain for IR and ECLIPSE-IVS + IR sequences with near identical mean water suppression performance of ~760-fold (mean/median |Mz|/M0 of 0.0013/0.0009, respectively). (G–H) NAA and (I–J) Choline maps calculated for the two methods. NAA maps with Global IR is dominated by extracranial lipids, which continues to bleed into choline maps, while no such hyper-intensities are seen with ECLIPSE-IVS + IR. The less pronounced ventricular outline in this case (relative to that of Figure 4) is primarily due to the low level of CSF in the slice. Note that the axial slices in Figure 4 and Figure 5 are from different subjects and slice positions. (K) Spectra from the 6 adjacent voxels shown in (A). Voxels 1–4 with IR show prominent metabolites (NAA, creatine, and choline) with significant lipid contaminations (marked with red arrows). In comparison minimal contaminants are observed from voxels 1–4 with ECLIPSE-IVS + IR, and lipid contaminants in voxel 6 is seen to be below the amplitude of NAA in brain, but are beyond displayed bounds for IR.
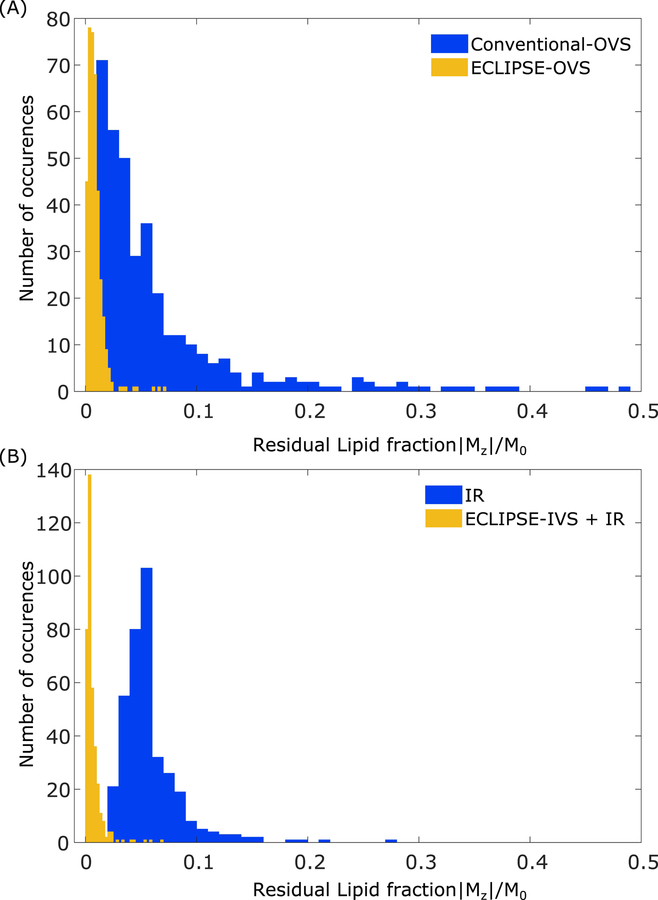
Figure 6 summarizes the residual magnetization in the extracranial regions in the form of histogram plots for the four lipid suppression methods presented, calculated over data from all three volunteers. The overall mean lipid suppression factors obtained with conventional-OVS, ECLIPSE-OVS, IR, and ECLIPSE-IVS + IR are15-fold (range 13–18), 116-fold (range 104–134), 16-fold (range 14–19), and 155-fold (range 122–257), respectively.
FIG. 6.
Residual lipid magnetization for conventional-OVS, ECLIPSE-OVS, IR, and ECLIPSE-IVS + IR sequences. Each histogram contains the combined data from all three volunteers, amounting to 375 and 370 voxels for OVS and IR-based methods, respectively. (A) The residual lipid fractions for the conventional-OVS sequence appear consistent with observations in Figure 4, and most voxels have sufficient suppression (|Mz|/M0 < 0.075), with ~10% of the voxels with |Mz|/M0 > 0.1 that span as high as ~0.48. Lipid suppression with ECLIPSE-OVS in comparison, is seen to be robust and highly effective, with 96% of the voxels with |Mz|/M0 < 0.02. The highest residual lipid fraction observed with ECLIPSE-OVS across all volunteers is 0.071. (B) The spread of residual lipid fraction with the IR sequence is less than the conventional-OVS method, with 6.5% of the voxels with |Mz|/M0 > 0.1. Lipid suppression performance of the ECLIPSE-IVS + IR sequence is similarly robust to ECLIPSE-OVS, with ~94% of the voxels with (|Mz|/M0 < 0.015). The highest lipid fraction observed with ECLIPSE-IVS + IR is 0.068.
In addition to the improved and highly robust lipid suppression provided by ECLIPSE, the results are also achieved with considerably less RF power. Table 2 summarizes the TR-averaged RF power as measured at the RF coil input for the methods presented. For conventional OVS circa 80% of the total sequence power is consumed by the 16 OVS pulses amounting to 2.0W. In comparison the four OVS pulses in ECLIPSE-OVS require 0.6W, representing only 30% of the power required by conventional OVS. The IR and ECLIPSE-IVS + IR sequences consume more power than ECLIPSE-OVS due to the use of one broadband AFP pulse calibrated at B1,max = 1.2.
Table. 2.
Summary of power requirements for each sequence, and averaged over the sequence TR. The first number corresponds to the total power, and the number in parenthesis corresponds to the power required by RF pulses of the OVS/IR module only.
| OVS TR = 3000 ms | ECLIPSE-OVS TR = 3000 ms | IR & ECLIPSE-IVS + IR TR = 2000 ms | |
|---|---|---|---|
| 2.6 ± 0.1W (2.0W) | 1.2 ± 0.1W (0.6W) | 1.7 ± 0.1W (0.9W) |
Discussion
The ECLIPSE-OVS and ECLIPSE-IR sequences were effective across all subjects studied with > 104-fold and > 122-fold mean extracranial lipid suppression, respectively. Furthermore, none of the OVS or water suppression pulse powers were manually optimized for different participants, and automated shimming was used; thus the developed ECLIPSE based MRSI methods were robust with reduced operator dependencies. Planning the ECLIPSE ROI, and loading the ECLIPSE gradient amplifier controller takes ~2 minutes, which was conducted during the vendor supplied B0 map. Automated placement of the ECLIPSE ROI, as previously reported for 8-slice OVS (45), based on an axial anatomical scan, will be incorporated in future work.
The acquisition can be significantly shortened for OVS based MRSI by utilizing a water unsuppressed metabolite cycled scheme incorporating a 2D readout such as a concentric ring approach (9). The use of metabolite cycling will further eliminate the need for a water reference that was acquired during each scan session for this work.
The current implementation of ECLIPSE-OVS has a duration of 1281ms, which in its current implementation restricts the use to TR > 1.6s MRSI. For shorter TR implementations, the ECLIPSE-OVS module can be re-optimized with no loss in performance (since at steady state, lipid suppression is less demanding with shortened TR). However for TR < 1.0s variants, SAR can be a limiting factor.
The availability of high powered and higher order shim coil inserts (37) can bring ECLIPSE localization to the wider MRI community. Extending a higher order shim insert to allow pulsing at sufficient amplitude, may require additional shielding to minimize potential eddy current induced effects. Fortunately, actively shielded Z2 shim coils are becoming more commonplace (37), thereby further opening the path towards wide-spread use of ECLIPSE. In addition, shim pre-emphasis has been demonstrated to be an effective method in mitigating effects of higher-order shim eddy currents (38).
In the current study, conventional-OVS and ECLIPSE-OVS methods were attempted to be optimized over a wide B1 range to counter experimental B1 inhomogeneity of the Tx/Rx coil used. The use of a dedicated volumetric transmitter and separate receive array, as is commonly used on clinical MRI scanners, would improve the B1 homogeneity. For a volume coil with ± 20% B1 variability, excellent lipid suppression (>100-fold suppression, T1 range 300–500 ms) can be achieved with a three-pulse ECLIPSE-OVS sequence, reducing the power requirements by an additional 40% relative to the current four-pulse ECLIPSE-OVS method. For typical B1 inhomogeneity encountered around the scalp at 7 T and 9.4T (39–42), the re-optimized four-pulse ECLIPSE method in simulation will provide robust lipid suppression, while consuming 0.6 times the power required by one equivalent AFP inversion pulse. The lower power requirement for ECLIPSE-OVS is due to operating in the non-adiabatic regime.
The use of spectral fitting would have been appropriate for the quantification of overlapping resonances, relative to spectral integration. The scope of this work however, was on describing and evaluating novel lipid suppression methods for human brain MRSI. In comparison to a spectral fitting approach, spectral integration is highly sensitive to lipid contamination and baseline shifts due to insufficient water suppression; thus served as a good tool to evaluate lipid and water contamination
The ECLIPSE-IVS + IR MRSI data in Figure 5K show inverted resonances below 2.0 ppm; given the high level of lipid suppression, these resonances can be assigned to macromolecules below 2.2 ppm with short T1 values as previously reported (19). In order to obtain macromolecule-nulled MR spectra, the sequence TI can be adjusted to predominantly suppress in-brain macromolecular resonances, while relying on ECLIPSE-IVS to selectively isolate extracranial lipid signals. Note that improved ECLIPSE-IVS is possible without extending the number of RF pulses, a feat not achievable with linear magnetic field gradients. Thus ECLIPSE-IVS can easily be incorporated into FID based MRSI sequences.
Combining ECLIPSE-OVS and IVS would clearly give the highest level of performance. However, the level of lipid suppression achievable with ECLIPSE-OVS or IVS alone is sufficiently high for most applications, such that there is rarely a need for a combined approach. An ECLIPSE-IVS based double AFP spin-echo approach, as described in the original manuscript (31), is expected to be superior to ECLIPSE-OVS, since it will provide T1 and B1 independent lipid suppression. There are however limitations with this approach: 1) A double AFP spin-echo would require > 30ms sequence TE on the 4T system, thus limiting short TE MRSI, 2) Both AFP RF pulses would need to be calibrated above the minimum B1 threshold to account for B1 inhomogeneity, thus consume > 2x SAR of the optimized 4-pulse ECLIPSE-OVS module presented.
MRSI applications where accurate NAA maps are required, an IR based ECLIPSE approach will be beneficial to minimize errors in spectral fitting from the underlying macromolecular baseline. The IR module however results in reduced SNR due to the inversion recovery component, which is unattractive when quantifying lower SNR metabolites. In such instances, the ECLIPSE-OVS approach is beneficial to maximize SNR, while relying on an accurate macromolecular signature for each voxel spectra.
The mismatched bandwidth between excitation and refocusing pulses, combined with the 1.4 kHz refocusing pulse bandwidth leads to CSD in the Z-direction; thus will uniformly affect all in-plane MRSI voxels. However, no observable loss in SNR was present, thus no further action were taken. Like any gradient-based localization method, ECLIPSE is affected by CSD. As detailed previously, the in-plane CSD with ECLIPSE manifests itself as a change in elliptical ROI size, rather than position (31). In its implementation for this work, the transition band ( Mz/M0 = 0.9 to −0.9) for ECLIPSE-OVS, is 1.25 cm, where the BW zone is 12.8 cm, resulting in a transition zone/BW ratio of 9.8%; similarly the transition band (Mz/M0 = 0.9 to 0.1) for the excitation pulse with ECLIPSE-IVS + IR is 1.22 cm, where the BW zone is 12.7 cm, resulting in a transition zone/BW ratio of 9.5%. As a result, utilization of ECLIPSE only affected the outer most brain MRSI voxels of the elliptical ROI as seen in spectra in voxel 5 (Figure 4K and Figure 5K). A 3 kHz AFP pulse was used to construct all lipid suppression modules to make fair comparisons between lipid suppression and power requirements between methods, while not reaching prohibitive SAR for conventional-OVS. The current amplitude used on any of the three ECLIPSE 2nd order gradients were < 10 A (or <10% of the maximum current), and resulted in negligible acoustic noise (as the system gradients dominate). The next iteration of ECLIPSE involves the development of GOIA (gradient-offset-independent adiabaticity (43)) or FOCI (frequency-offset-corrected inversion (44)) derived RF pulses that can provide 10-fold improvements in RF pulse bandwidth to considerably decrease axial CSD effects, which is possible since the gradient amplifiers are currently under-utilized.
The elliptical ROI generated with ECLIPSE is ideally suited for axial brain slices near-symmetrical around both x and y axes (for example, the shape of the head in Figure 4A). For axial brain slices that deviate in symmetry around the x-axis (as illustrated in Figure 5A), an elliptical ROI may not provide optimal axial slice coverage (however provides significantly improved coverage relative to cubical localization). Future work is underway to incorporate a secondary multi-coil (MC) array to supplement the ECLIPSE fields for gradient field ‘shaping’, to better approximate axial brain ROIs that are different from an elliptical shape.
The magnetic field generated by the Z2 coil has a Z2 component. As a result, the radius of the elliptical ROI selected would increasingly deviate for Z ≠ 0, thus restricting single shot whole-brain OVS utilizing the ECLIPSE setup. In its current state however, a ~ 40 mm axial slab can be selected with ECLIPSE without significantly affecting axial coverage. Whole brain MRSI is possible with a multi-slab selection approach, designed to account for overlapping OVS bands in the z-direction. Current work is in progress to explore the combination of multiple, shaped magnetic fields to achieve an improved fit to the curvature of the human head.
Conclusions
In summary, two outer volume suppression methods based on pulsed second order fields (ECLIPSE) are presented. Both methods provide > 100-fold mean lipid suppression to enable robust and short-TE MRSI of the human brain. In addition, ECLIPSE-OVS consumes 30% of the power required for an optimized, conventional-OVS sequence, while being immune to B1+ field heterogeneity. The low power requirements in combination with insensitivity to B1 and T1, makes ECLIPSE-based OVS particularly attractive for high and ultra-high field MRSI.
Supplementary Material
FIG. S1. Pulse sequence diagrams for the IR module. In the pulse sequence diagrams, water suppression RF pulses are shaded, and the IR pulse (unshaded) set such that TI = 260 ms with optimized VAPOR style water suppression pulses.
FIG. S2. (A) The spin-echo variant z-selective on both excitation and refocusing pulses used on all sequences except the ECLIPSE-IVS + IR sequence. Phase encodings for MRSI are superimposed on the last spoiler gradient. (B) The spin-echo variant used for the ECLIPSE-IVS + IR module, where the excitation pulse is selective to the ECLIPSE ROI, and the refocusing pulse is z-selective. Phase encodings for MRSI are superimposed on the last spoiler gradient.
FIG. S3 Extension of Figure 4 comparing conventional-OVS and ECLIPSE-OVS sequence performance on one volunteer. (A) Represents the conventional-OVS saturation slice placements, (B) represents the ECLIPSE-OVS ROI placement. (C–J) represent NAA, creatine, choline and Glx maps obtained with the two OVS methods, where all metabolic maps except NAA have been scaled up by a factor 2 for improved visibility.
Fig. S4 Extension of Figure 4K comparing conventional-OVS and ECLIPSE-OVS sequence performance on one volunteer. (A) Anatomical axial slice corresponding to the region used for MRSI illustrating six adjacent MRS voxel locations compared in (E). (B) The ROI used to calculate extracranial lipid suppression. (C–D) Mean water suppression factors for conventional-OVS and ECLIPSE-OVS, respectively. (E) All spectra illustrated are identical to those in Figure 4K with the exception of spanning up to 6ppm to illustrate the level of water suppression with the two methods, and to illustrate that no noticeable water baseline is present.
FIG. S5 Extension of Figure 5 comparing IR and ECLIPSE-IVS + IR sequence performance on one volunteer. (A) Represents the axial ROI, (B) represents the ECLIPSE-IVS ROI placement. (C–J) represent NAA, creatine, choline and Glx maps obtained with the two IR methods, where all metabolic maps except NAA have been scaled up by a factor 2 for improved visibility.
Acknowledgements
This work was supported by NIH grant R01-EB014861.
References
- 1.Graves EE, Nelson SJ, Vigneron DB, Chin C, Verhey L, McDermott M, Larson D, Sneed PK, Chang S, Prados MD, Lamborn K, Dillon WP. A preliminary study of the prognostic value of proton magnetic resonance spectroscopic imaging in gamma knife radiosurgery of recurrent malignant gliomas. Neurosurgery 2000;46(2):319–326; discussion 326–318. [DOI] [PubMed] [Google Scholar]
- 2.Hetherington HP, Hamid H, Kulas J, Ling G, Bandak F, de Lanerolle NC, Pan JW. MRSI of the medial temporal lobe at 7 T in explosive blast mild traumatic brain injury. Magn Reson Med 2014;71(4):1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan JW, Duckrow RB, Gerrard J, Ong C, Hirsch LJ, Resor SR Jr., Zhang Y, Petroff O, Spencer S, Hetherington HP, Spencer DD. 7T MR spectroscopic imaging in the localization of surgical epilepsy. Epilepsia 2013;54(9):1668–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suhy J, Rooney WD, Goodkin DE, Capizzano AA, Soher BJ, Maudsley AA, Waubant E, Andersson PB, Weiner MW. 1H MRSI comparison of white matter and lesions in primary progressive and relapsing-remitting MS. Mult Scler 2000;6(3):148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuff N, Amend D, Ezekiel F, Steinman SK, Tanabe J, Norman D, Jagust W, Kramer JH, Mastrianni JA, Fein G, Weiner MW. Changes of hippocampal N-acetyl aspartate and volume in Alzheimer’s disease. A proton MR spectroscopic imaging and MRI study. Neurology 1997;49(6):1513–1521. [DOI] [PubMed] [Google Scholar]
- 6.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B 1994;104(1):1–10. [DOI] [PubMed] [Google Scholar]
- 7.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med 1999;41(4):649–656. [DOI] [PubMed] [Google Scholar]
- 8.Dreher W, Leibfritz D. New method for the simultaneous detection of metabolites and water in localized in vivo 1H nuclear magnetic resonance spectroscopy. Magn Reson Med 2005;54(1):190–195. [DOI] [PubMed] [Google Scholar]
- 9.Emir UE, Burns B, Chiew M, Jezzard P, Thomas MA. Non-water-suppressed short-echo-time magnetic resonance spectroscopic imaging using a concentric ring k-space trajectory. NMR in biomedicine 2017;30(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pijnappel WWF, van den Boogaart A, de Beer R, van Ormondt D. SVD-based quantification of magnetic resonance signals. Journal of Magnetic Resonance (1969) 1992;97(1):122–134. [Google Scholar]
- 11.Cabanes E, Confort-Gouny S, Le Fur Y, Simond G, Cozzone PJ. Optimization of residual water signal removal by HLSVD on simulated short echo time proton MR spectra of the human brain. J Magn Reson 2001;150(2):116–125. [DOI] [PubMed] [Google Scholar]
- 12.de Graaf RA. In Vivo NMR Spectroscopy: Principles and Techniques: Wiley; 2018. [Google Scholar]
- 13.Hangel G, Strasser B, Povazan M, Heckova E, Hingerl L, Boubela R, Gruber S, Trattnig S, Bogner W. Ultra-high resolution brain metabolite mapping at 7 T by short-TR Hadamard-encoded FID-MRSI. Neuroimage 2018;168:199–210. [DOI] [PubMed] [Google Scholar]
- 14.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci 1987;508:333–348. [DOI] [PubMed] [Google Scholar]
- 15.Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hanicke W, Sauter R. Localized high-resolution proton NMR spectroscopy using stimulated echoes: initial applications to human brain in vivo. Magn Reson Med 1989;9(1):79–93. [DOI] [PubMed] [Google Scholar]
- 16.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med 2008;59(1):1–6. [DOI] [PubMed] [Google Scholar]
- 17.Duyn JH, Gillen J, Sobering G, van Zijl PC, Moonen CT. Multisection proton MR spectroscopic imaging of the brain. Radiology 1993;188(1):277–282. [DOI] [PubMed] [Google Scholar]
- 18.Ebel A, Govindaraju V, Maudsley AA. Comparison of inversion recovery preparation schemes for lipid suppression in 1H MRSI of human brain. Magn Reson Med 2003;49(5):903–908. [DOI] [PubMed] [Google Scholar]
- 19.Hangel G, Strasser B, Povazan M, Gruber S, Chmelik M, Gajdosik M, Trattnig S, Bogner W. Lipid suppression via double inversion recovery with symmetric frequency sweep for robust 2D-GRAPPA-accelerated MRSI of the brain at 7 T. NMR in biomedicine 2015;28(11):1413–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hetherington HP, Avdievich NI, Kuznetsov AM, Pan JW. RF shimming for spectroscopic localization in the human brain at 7 T. Magn Reson Med 2010;63(1):9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boer VO, Klomp DW, Juchem C, Luijten PR, de Graaf RA. Multislice (1)H MRSI of the human brain at 7 T using dynamic B(0) and B(1) shimming. Magn Reson Med 2012;68(3):662–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boer VO, van de Lindt T, Luijten PR, Klomp DW. Lipid suppression for brain MRI and MRSI by means of a dedicated crusher coil. Magn Reson Med 2015;73(6):2062–2068. [DOI] [PubMed] [Google Scholar]
- 23.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med 1996;35(5):678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilgic B, Chatnuntawech I, Fan AP, Setsompop K, Cauley SF, Wald LL, Adalsteinsson E. Fast image reconstruction with L2-regularization. J Magn Reson Imaging 2014;40(1):181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassirpour S, Chang P, Henning A. High and ultra-high resolution metabolite mapping of the human brain using (1)H FID MRSI at 9.4T. Neuroimage 2018;168:211–221. [DOI] [PubMed] [Google Scholar]
- 26.Hu X, Patel M, Ugurbil K. A new strategy for spectroscopic imaging. J Magn Reson B 1994;103(1):30–38. [DOI] [PubMed] [Google Scholar]
- 27.Henning A, Schar M, Schulte RF, Wilm B, Pruessmann KP, Boesiger P. SELOVS: brain MRSI localization based on highly selective T1- and B1- insensitive outer-volume suppression at 3T. Magn Reson Med 2008;59(1):40–51. [DOI] [PubMed] [Google Scholar]
- 28.Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for (1)H-MRSI of the human brain at 7 T with minimal signal loss. NMR in biomedicine 2009;22(7):683–696. [DOI] [PubMed] [Google Scholar]
- 29.Kirchner T, Fillmer A, Tsao J, Pruessmann KP, Henning A. Reduction of voxel bleeding in highly accelerated parallel (1) H MRSI by direct control of the spatial response function. Magn Reson Med 2015;73(2):469–480. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Soher BJ, Ouwerkerk R, Schar M, Barker PB. Spin-echo magnetic resonance spectroscopic imaging at 7 T with frequency-modulated refocusing pulses. Magn Reson Med 2013;69(5):1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Graaf RA, Brown PB, De Feyter HM, McIntyre S, Nixon TW. Elliptical localization with pulsed second-order fields (ECLIPSE) for robust lipid suppression in proton MRSI. NMR in biomedicine 2018;31(9):e3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nixon TW, McIntyre S, de Graaf RA. The design and implementation of a 64 channel arbitrary gradient waveform controller. Proc Int Soc Magn Reson Med. 2017;25:969. [Google Scholar]
- 33.Gu M, Spielman DM. B1 and T1 insensitive water and lipid suppression using optimized multiple frequency-selective preparation pulses for whole-brain 1H spectroscopic imaging at 3T. Magn Reson Med 2009;61(2):462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silver MS, Joseph RI, Chen CN, Sank VJ, Hoult DI. Selective population inversion in NMR. Nature 1984;310(5979):681–683. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y, de Graaf RA, DelaBarre L, Tannus A, Garwood M. BISTRO: an outer-volume suppression method that tolerates RF field inhomogeneity. Magn Reson Med 2001;45(6):1095–1102. [DOI] [PubMed] [Google Scholar]
- 36.Pauly J, Le Roux P, Nishimura D, Macovski A. Parameter relations for the Shinnar-Le Roux selective excitation pulse design algorithm [NMR imaging]. IEEE Trans Med Imaging 1991;10(1):53–65. [DOI] [PubMed] [Google Scholar]
- 37.Pan JW, Lo KM, Hetherington HP. Role of very high order and degree B0 shimming for spectroscopic imaging of the human brain at 7 tesla. Magn Reson Med 2012;68(4):1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juchem C, Nixon TW, Diduch P, Rothman DL, Starewicz P, de Graaf RA. Dynamic Shimming of the Human Brain at 7 Tesla. Concepts Magn Reson Part B Magn Reson Eng 2010;37B(3):116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adriany G, Van de Moortele PF, Ritter J, Moeller S, Auerbach EJ, Akgun C, Snyder CJ, Vaughan T, Ugurbil K. A geometrically adjustable 16-channel transmit/receive transmission line array for improved RF efficiency and parallel imaging performance at 7 Tesla. Magn Reson Med 2008;59(3):590–597. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert KM, Belliveau JG, Curtis AT, Gati JS, Klassen LM, Menon RS. A conformal transceive array for 7 T neuroimaging. Magn Reson Med 2012;67(5):1487–1496. [DOI] [PubMed] [Google Scholar]
- 41.Avdievich NI, Giapitzakis IA, Bause J, Shajan G, Scheffler K, Henning A. Double-row 18-loop transceive-32-loop receive tight-fit array provides for whole-brain coverage, high transmit performance, and SNR improvement near the brain center at 9.4T. Magn Reson Med 2019;81(5):3392–3405. [DOI] [PubMed] [Google Scholar]
- 42.Clement J, Gruetter R, Ipek O. A combined 32-channel receive-loops/8-channel transmit-dipoles coil array for whole-brain MR imaging at 7T. Magn Reson Med 2019. [DOI] [PMC free article] [PubMed]
- 43.Tannus A, Garwood M. Adiabatic pulses. NMR in biomedicine 1997;10(8):423–434. [DOI] [PubMed] [Google Scholar]
- 44.Ordidge RJ, Wylezinska M, Hugg JW, Butterworth E, Franconi F. Frequency offset corrected inversion (FOCI) pulses for use in localized spectroscopy. Magn Reson Med 1996;36(4):562–566. [DOI] [PubMed] [Google Scholar]
- 45.Yung KT, Zheng W, Zhao C, Martinez-Ramon M, van der Kouwe A, Posse S. Atlas-based automated positioning of outer volume suppression slices in short-echo time 3D MR spectroscopic imaging of the human brain. Magn Reson Med 2011;66(4):911–922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIG. S1. Pulse sequence diagrams for the IR module. In the pulse sequence diagrams, water suppression RF pulses are shaded, and the IR pulse (unshaded) set such that TI = 260 ms with optimized VAPOR style water suppression pulses.
FIG. S2. (A) The spin-echo variant z-selective on both excitation and refocusing pulses used on all sequences except the ECLIPSE-IVS + IR sequence. Phase encodings for MRSI are superimposed on the last spoiler gradient. (B) The spin-echo variant used for the ECLIPSE-IVS + IR module, where the excitation pulse is selective to the ECLIPSE ROI, and the refocusing pulse is z-selective. Phase encodings for MRSI are superimposed on the last spoiler gradient.
FIG. S3 Extension of Figure 4 comparing conventional-OVS and ECLIPSE-OVS sequence performance on one volunteer. (A) Represents the conventional-OVS saturation slice placements, (B) represents the ECLIPSE-OVS ROI placement. (C–J) represent NAA, creatine, choline and Glx maps obtained with the two OVS methods, where all metabolic maps except NAA have been scaled up by a factor 2 for improved visibility.
Fig. S4 Extension of Figure 4K comparing conventional-OVS and ECLIPSE-OVS sequence performance on one volunteer. (A) Anatomical axial slice corresponding to the region used for MRSI illustrating six adjacent MRS voxel locations compared in (E). (B) The ROI used to calculate extracranial lipid suppression. (C–D) Mean water suppression factors for conventional-OVS and ECLIPSE-OVS, respectively. (E) All spectra illustrated are identical to those in Figure 4K with the exception of spanning up to 6ppm to illustrate the level of water suppression with the two methods, and to illustrate that no noticeable water baseline is present.
FIG. S5 Extension of Figure 5 comparing IR and ECLIPSE-IVS + IR sequence performance on one volunteer. (A) Represents the axial ROI, (B) represents the ECLIPSE-IVS ROI placement. (C–J) represent NAA, creatine, choline and Glx maps obtained with the two IR methods, where all metabolic maps except NAA have been scaled up by a factor 2 for improved visibility.