Abstract
Bromodomains (BrDs), a conserved structural module in chromatin-associated proteins, are well known for recognizing ε-N-acetyl lysine residues on histones. One of the most relevant BrDs is BRD4, a tandem bromodomain containing protein (BrD1 and BrD2) that plays a critical role in numerous diseases including cancer. Growing evidence shows that the two BrDs of BRD4 have different biological functions; hence selective ligands that can be used to study their functions are of great interest. Here, as a follow-up of our previous work (Gacias et al., Chem Biol. 2014; 21:841-854), we first provide a detailed characterization study of the in silico rational design of Olinone as part of a series of five tetrahydro-pyrido indole-based compounds as BRD4 BrD1 inhibitors. Additionally, we investigated the molecular basis for Olinone’s selective recognition by BrD1 over BrD2. Molecular dynamics simulations, free energy calculations and conformational analyses of the apo-BRD4-BrD1|2 and BRD4-BrD1|2/Olinone complexes showed that Olinone’s selectivity is facilitated by five key residues: Leu92 in BrD1|385 in BrD2 of ZA loop, Asn140|433, Asp144|His437 and Asp145|Glu438 of BC loop, and Ile146|Va149 of helix C. Furthermore, the difference in hydrogen bonds number and in mobility of the ZA and BC loops of the acetyl-lysine binding site between BRD4 BrD1/Olinone and BrD2/Olinone complexes also contribute to the difference in Olinone’s binding affinity and selectivity towards BrD1 over BrD2. Altogether, our computer-aided molecular design techniques can effectively guide the development of small-molecule BRD4 BrD1 inhibitors, explain their selectivity origin, and further open doors to the design of new therapeutically-improved derivatives.
Keywords: Bromodomains, Molecular Dynamics Simulations, Free Energy Calculations, Conformational Analysis, Selectivity, Structure-Based Drug Design
INTRODUCTION
Lysine acetylation is well recognized as an important post-translational modification (PTM) that regulates several cellular functions and disease biology.1,2 In particular, histone lysine acetylation plays an active role in epigenetic control of gene transcription. Studies of histone biology show that specific proteins add PTM marks (writers), recognize marks (readers) and remove marks (erasers).3,4 The proteins associated with acetylated lysine (AcK) are well described: histone acetyltranferases (HATs) add the acetyl group, histone deacetylases (HDACs) remove the acetyl group, and bromodomains (BrDs), which are a conserved structural module present in numerous chromatin-associated proteins and HATs, recognize AcK acting as “readers” of lysine acetylation state.5
There are 61 human BrDs embedded in 46 proteins,6 which are divided into eight subfamilies with distinctive features based on sequence similarities.7 One subfamily is the BET (bromodomain and extra-terminal domain) proteins composed of four members in humans: BRD2, BRD3, BRD4 (bromodomain-containing proteins 2-4) and BRDT (bromodomain testis-specific protein), each containing tandem bromodomains (BrD1 and BrD2). Bromodomains adopt a conserved fold of a left-handed bundle of four α helices (αZ, αA, αB, and αC) with inter-helical ZA and BC loops of variable length and sequence.8 These loops form a hydrophobic pocket serving to stabilize acetyl-lysine binding. The amino acids residues in the acetyl-lysine binding pocket are highly conserved, with over 90% sequence identity between the two BrDs in each BET protein. Among the BET proteins, BRD4 has been arguably the most broadly investigated. Recent studies have implicated BRD4 in numerous diseases including cancer,6 obesity,9 kidney malfunction,10 and inflammation.11
Growing evidence shows that the biological functions of BRD4 are mediated through its two BrDs,10,11 however, the specific role of each bromodomain is still poorly understood. An important strategy for understanding the function of each BrD of BRD4 is to use small-molecule probes that selectively disrupt the interaction of BRD4 BrD1|2 with their native histones AcK. Nevertheless, developing a selective inhibitor towards either BrD1|2 of BRD4 is challenging due to the high sequence similarity of the acetyl-lysine binding sites. Thus, despite several potent BRD4 inhibitors reported in previous findings,12 at the time our research was conducted, there was only one published inhibitor that had been validated to differentiate between the two BrDs of BRIM.13 This compound, MS436 (Figure 1A), is a diazobenzene-based inhibitor with an estimated Ki, of 30-50 nM for BRD4 BrD1 and a 10-fold selectivity over BrD2, which is achieved through water-mediated intermolecular interactions.13 Recently, our group reported another selective inhibitor of BRD4 BrD1. This compound, MS402 (Figure 1A), is a cyclopentene-based BrD inhibitor with an estimated Ki of 77 nM for BRD4 BrD1 and a nine-fold selectivity over BrD2 of BRD4.14 In addition, in 2014 we reported a more selective inhibitor of BRD4 BrD1, but with a different chemical scaffold, tetrahydro-pyrido indole.15 This compound, Olinone (Figure 1A), has a Kd of 3.4 μM against BRD4 BrD1 and is about 100-fold selective over BrD2. Olinone displays preferred BrD1 binding over BrD2 for all three BET proteins BRD4, BRD3, and BRD2, while exhibiting nearly no detectable binding to other bromodomain-containing proteins.15 Olinone has been shown to accelerate the progression of mouse primary oligodendrocyte progenitors towards differentiation, while inhibition of both bromodomains of BET proteins hindered differentiation.15
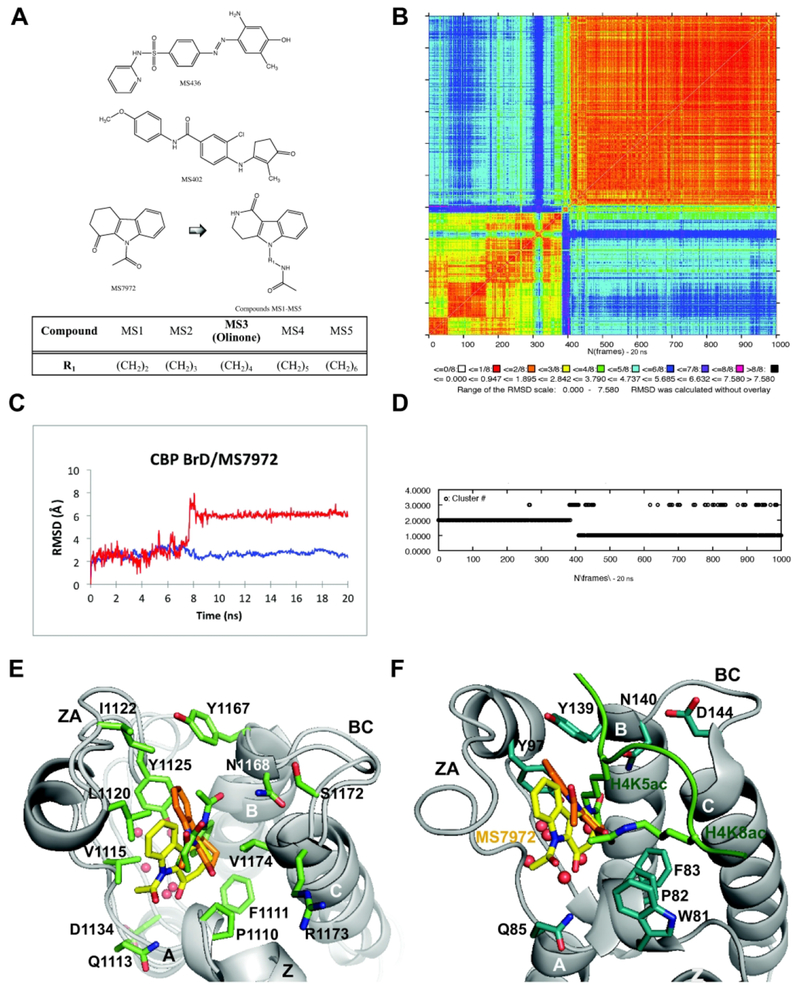
Figure 1. Structure-guided design of BET-BrD inhibitors.
(A) 2D ligand structures: MS436 (top), MS402 (middle), MS7972 (bottom) and newly designed small-molecule inhibitors MS1 to MS5 of BRD4 BrD1 (bottom). The substituent R1 represents a methylene unit increment for each of the MSi compound. (B) 2D-RMSD map for the 20 ns MD simulation including all atoms of the CBP BrD/MS7972 complex. (C) All atoms RMSD for the AcK binding site (blue) and the ligand (red) of the CBP BrD/MS7972 complex as function of time, both calculated with respect to the NMR structure. (D) K-means cluster analysis. (E) Representative structures of CBP BrD/MS7972 complex for the 20 ns of the MD simulation: Cluster 1 (orange, ~40% and ~1.5 Å backbone RMSD with respect to the minimized NMR structure of MS7972, PDB ID 2D8216, bound to CBP bromodomain), Cluster 2 (green, ~60%, ~2.2 Å backbone RMSD), minimized NMR structure of MS7972 (yellow). The water molecules are depicted as red sphere. (F) Superimposition of the NMR structure of MS7972 (yellow, PDB ID 2D8216; orange, Cluster 1 from MD simulation depicted in E) bound to CBP bromodomain and the X-ray crystal structure of the histone H4K5ac/K8ac peptide (green) bound to BRD4 BrD1 (gray, PDB ID 3UVW19). The pictures in (E) and (F) were rendered using PyMOL program.34
In the present paper, as a follow-up of our previous work,15 we first provide a detailed characterization study of our in silico design rationale of Olinone(MS3)15 as part of a series of five tetrahydro-pyrido indole-based compounds (MS1 to MS5, MSi) modulators of BRD4 BrD1, and second explain the molecular basis for Olinone’s selectivity towards BRD4 BrD1 over BrD2. The first part of this computational study was contemporaneously completed with the experimental characterization of Olinone.15 Our design rationale used as starting point compound MS7972, an inhibitor of the structurally related bromodomain of the CREB-binding protein (CBP), that had been previously identified by NMR-based screening in our laboratory.16 The in silico study presented herein indicates that Olinone15 has the strongest binding affinity for BRD4 BrD1 out of the five molecules originally designed as BRD4 BrD1 inhibitors. This result was validated experimentally as the MSi compounds were synthesized and their binding affinity towards BRD4 BrD1 measured.15 Moreover, we explain the molecular basis for Olinone15 binding to BRD4 BrD1 and the potential origin of its selectivity for BrD1 over BrD2. Towards this end, we present Molecular Dynamics (MD) simulations of BRD4 BrD1/Olinone X-ray crystal structure15 and BRD4 BrD2/Olinone complexes, free energy calculations as well as conformational analyses. The origin of Olinone’s selectivity for BrD1 over BrD2 seems to be related to the most favorable energetic contribution to the binding free energy of acetyl-lysine binding site gatekeeper residues Ile146 in BrD1 compared to Val439 in BrD2 together with four other residues Leu92|385, Asn140|433, Asp144|His437, and Asp145|Glu438 in BrD1|BrD2. Our study also revealed that the binding free energy is mainly driven by van der Waals interactions, and the potential of modifying the amide group of the piperidone ring of Olinone and the amide group of the N-acetamidoalkyl chain for developing new derivatives with improved affinity towards BRD4 BrD1, while keeping its selectivity over BrD2.
METHODS AND MODELS
Molecular Systems
The CBP BrD in complex with MS7972 system was built from the NMR structure (PDB ID 2D8216) placed in a truncated octahedron periodic box with 5,307 water molecules and four sodium ions to neutralize the simulation cell. The initial structures of the MSi compounds, designed by the Marvin program17, were used for docking studies and parameterization (see below Parameterization of MS Compounds). The initial poses of MSi compounds bound to BRD4 BrD1 were obtained from docking calculations using MOE18 (Molecular Operating Environment). Each compound was docked against the BRD4 BrD1 X-ray structure (PDB ID 3UVW19), from which the H4K5ac/K8ac peptide had been removed. Docking ligands MS1, MS2, MS3 and MS4 produced only one predominant pose and it was chosen to build the respective models (see below Molecular Docking sections). Their acetamidoalkyl chains pointed inside the acetyl-lysine binding pocket (see Figure S1 Supporting Information) as in the histone H4K5ac/K8ac peptide bound to the BRD4 BrD1 (see Figure 1F). For ligand MS5, two different poses were selected: 1) with the acetamidoalkyl chain pointing inside the acetyl-lysine binding pocket (MS5.1) and 2) with the tetrahydro-pyrido indole scaffold pointing inside the acetyl-lysine binding pocket (MS5.2) (see Figure S1 Supporting Information). Thus, six different systems were built as described above: BRD4 BrD1 in complexes with the different designed molecules (MS1, MS2, MS3, MS4, MS5.1 and MS5.2). In addition, in order to study the selectivity of Olinone (MS3) towards BRD4 BrD1 over BrD2, four additional systems were built. Two of these were built using the complex of Olinone bound to BRD4 BrD1 X-ray crystal structure (PDB ID 4QB315): apo-BRD4 BrD1 and in complex with Olinone (MS3). The two other systems were built using the second bromodomain (BrD2) of BRD4 X-ray crystal structure (PDB ID 2OUO19): apo-BRD4 BrD2 and in complex with Olinone (MS3) (see Molecular Systems in Methods and Models in Supporting Information for further details).
Molecular Docking
The H4K5ac/K8ac peptide from the PDB structure 3UVW of BRD4 BrD1 was used to correct the 3D structure (e.g., terminal amino acids and protonation states) with the Structure Preparation application in MOE. The LigX of MOE was invoked to protonate the BRD4 BrD1 structure using the 3D protonation application. Water molecules farther than 4.5 Å were removed. Finally, the energy of the retrieved protein molecule (BRD4 BrD1) was minimized using the default parameters of MOE energy minimization algorithm (gradient: 0.1 kcal/mol/Å2, force field: MMFF94X). A MOE database with all MSi (MS1 to MS5) compounds was created with the MMFF94x.20 The Rigid Receptor with flexible ligands docking protocol was used for the molecular docking of the MSi ligands to BRD4 BrD1.
At the end of docking, the top scoring poses were selected for MD simulations. The initial models were minimized (see Setup of the Simulations) using Amber 12.0 program21 and rescored using the Pose Rescoring protocol in MOE Dock program. The minimized structure of BRD4 BrD1/MS3(Olinone)15 is very similar to the X-ray crystal structure (see Table S1 in Supporting Information). In addition, the MOE score of the BRD4 BrD1/MS3(Olinone) docking model (−8.04 kcal/mol) is very similar to that of the X-ray crystal structure (−8.23 kcal/mol). Thus, we conclude that the docked poses are appropriate as initial models.
Setup of the Simulations
We performed 20 ns MD simulations of the CBP BrD/MS7972 complex and 25 ns MD simulations of each model BRD4 BrD1/MSi complex to select the best binder of the MSi series. Afterwards, we conducted 200 ns MD simulation of each BRD4 BrD1/Olinone complex and without ligand, and 400 ns MD simulations of each BRD4 BrD2/Olinone complex and without ligand to study Olinone’s selectivity towards BrD1 over BrD2 (a total of ~1.4 microseconds MD simulation). Based on pKa calculations using Maestro 16-1,22 His437 was considered charged. The all-atom FF99SB Amber force field21 and the TIP3P water model23 were used for all first seven systems (i.e., CBP BrD/MS7972 and BRD4 BrD1/MS1-5.1&5.2). The all–atom FF14SB Amber force field,21,24 and the TIP3P water model23 were used for all other four systems calculations (i.e., apo-BRD4 BrD1, BRD4 BrD1/Olinone X-ray crystal structure, apo-BRD4 BrD2 and BRD4 BrD2/Olinone complex) because during the project an updated and improved Amber force field for protein modeling, FF14SB, became available. The system was initially minimized using the Steepest Descent and Conjugate Gradient methods to remove unfavorable steric interactions. Then, it was heated at a constant rate to 300 K for 100 ps. Heating was followed by a sequence of equilibrations with positional restraints on the protein and the ligand, which were gradually reduced from 25 kcal/mol until the system was allowed to move freely. Twin–range non–bonded cutoffs of 10 Å and 12 Å were used for the Lennard–Jones potentials and electrostatic interactions were calculated using particle–mesh Ewald. The production MD simulations were carried out using the NPT ensemble. In the production stage the temperature was maintained using Langevin dynamics with a temperature coupling parameter of 5 ps and a collision frequency of 5 ps−1. The length of all bonds involving hydrogen atoms was kept fixed with the SHAKE algorithm.25 The pressure was kept fixed at 1 atm. The equations of motion were integrated with a time–step of 2 fs. The first six systems involved in the in silico rational design (BRD4 BrD1/MSi) were solvated using Grand Canonical ensemble Monte Carlo simulations (GCMC).26,27 The remaining four systems involved in the selectivity study of Olinone were solvated using the solvateOct command in the LEaP program.28 Truncated octahedron periodic boundary unit cell was used throughout (see Setup of the Simulations in Methods and Models in Supporting Information for further details). The coordinates were saved every 5 ps. All MD simulations and analysis were performed using the Amber21, Simulaid29, the Metropolis Monte Carlo (MMC)30 and VIDA31 programs.
Parameterization of MS Compounds
The force field parameters for the ligands MS7972 and MS1 to MS5, which are neutral, were obtained by using the following protocol: 1) the geometry of the molecules, drawn using Marvin17 and VIDA31 programs, was optimized using Quantum Mechanics Semi-empirical (QMS) minimization; 2) all ligands atomic partial charges were computed with the AM1-BCC semi-empirical method as implemented in the Amber 12.0 or 14.0 Antechamber programs;21,32 and 3) the parameterization of the ligands were done using the GAFF force field.21 Molecular Operating Environment (MOE)18, Visual Molecular Dynamics (VMD)33, PyMOL34, Marvin17 and VIDA31 programs were used for further structure visualization and drawing. The charges and other parameters are available upon request. The molecular mechanics Generalized Born (GB)35,36 surface area (MM/GBSA) method was used to calculate the free energy of binding between a small molecule (a ligand, L, MS1 to MS5) and a macromolecule (the receptor, R, BRD4 BrD1 and BRD4 BrD2) forming a receptor–ligand complex (RL, BRD4 BrD1/MSi and BRD4 BrD2/Olinone; see MM/GBSA Method in Methods and Models in Supporting Information for further details).
Water Molecules Calculations
The water molecules inside the acetyl-lysine binding pocket and close to the MSi ligands were determined by using the following protocol: 1) the average number of water molecules in the first shell of 3.4Å of the N-alkylated chain (MS1, MS2, MS3, MS4 and MS5.1) or the tricyclic moiety (MS5.2) were obtained using the watershell module in Amber 12.0 or 14.0 Antechamber;21,32 2) the closest water (~8) molecules obtained in Step 1 were written onto an Amber MD trajectory file; and 3) a hydrogen bonding analysis of the MD trajectory was computed in VMD33 to identify the BRD4 BrD1 residues that form hydrogen bonds with water molecules and/or with the MSi ligands. For MS1, MS2, MS3, MS4 and MS5.1 the residues selected were Pro82, Gln85, Tyr97, Met105, Met132, and for MS5.2 the same residues were selected as for the other MSi ligands plus Asn135. In the case of BRD4 BrD2/MS3 the residues selected were Pro375, Lys378, Tyr390, Met398, Met425, 4) the average number of water molecules in the first shell (closer than 3.4Å) about the residues selected in Step 3 was obtained using the watershell module in Amber 12.0 or 14.0 Antechamber,21,32 and 5) the closest water molecules obtained in Step 4 were written onto an amber MD trajectory file that was later used to perform the MM/GBSA calculations.
RESULTS AND DISCUSSION
A). IN SILICO DESIGN OF BRD4 BrD1 INHIBITORS
Molecular Dynamics Simulations of CBP BrD/MS7972-Lead Compound
The rationale of our in silico design of BET-specific BrD inhibitors started from MS7972, which we had previously identified in NMR-based screening as a modest inhibitor (PDB ID 2D8216) (Figure 1A). MD simulations of the NMR structure of the CBP BrD/MS7972 complex identified two stable bound poses of MS7972: 1) a NMR-like orientation16 with a population of −40% and 2) a flipped orientation by −90° along the long axis of the molecule with a population of −60% (~6.1±0.3 Å all ligand atoms root-mean-squared deviations, RMSD) (Figures 1B–1E). MS7972 forms a network of hydrophobic and aromatic interactions with Pro1110, Phe1111, Vai1115, Pro1117, Leu1120, Ile1122, Tyr1167 and Vai1174 (Figure 1E) as well as with polar residues (Gln1113, Tyr1125, Asp1134, Tyr1167, Asn1168, Ser1172 and Arg1173) in the ZA and BC loops. As in all bromodomains a few water molecules are located deep in the acetyl-lysine binding pocket (Figure 1E). Based on these findings, and given the structural similarities of the bromodomains of CBP and BET proteins we used MS7972 as a starting point in our search for selective BRD4 BrD1 inhibitors.
Structure-Guided Rational Design and Development of Selective BRD4 BrDs Inhibitors
Our in silico design rationale for a BRD4 BrD1 inhibitor relied on three elements: 1) ZA and BC surface loops because they include polar residues and exhibit considerable mobility; 2) the hydrophobic acetyl-lysine binding pocket and the structured water molecules near the ligand; and 3) the residue-based energetic contributions of the BRD4 BrD1/ligand interaction. Thus, we first superimposed the representatives NMR-like and flipped orientations of MS7972 bound to CBP bromodomain resulting from the MD simulation on the X-ray crystal structure of histone H4K5ac/K8ac peptide bound to the BRD4 BrD1 (PDB ID 3UVW19, Figures 1E and 1F). This strategy helped us identify key structural features on MS7972, such as the potential change in position of the C=O group, to enhance its derivative interactions with the BRD4 BrD1 AcK binding pocket. Secondly, our analysis focused mainly on the surface of the polar ligand acetyl-lysine binding vestibule. We aimed at taking advantage of Gln85, Tyr97 and Asp106 of the ZA loop, and Tyr139, Asn140, Asp144 (unique to BRD4 BrD1) and Asp145 of the BC loop in BRD4 BrD1, which do not interact with MS7972 directly, but could enhance potency towards BRD4 BrD1 by introducing groups in MS7972 that could provide additional electrostatic interactions. Thirdly, we paid attention to several trapped water molecules in the acetyl-lysine hydrohobic pocket proximal to Tyr97 and Asp106 in BRD4 BrD1 (i.e., correspoding to Tyr1125 and Asp1134 in CBP BrD). Similar waters are observed in other BrDs bound to their native peptides or small-molecule inhibitors.5,13,15 These structured water molecules in the acetyl-lysine binding pocket often extend into a channel formed by the ZA loop, called the ZA channel, which can be used to enhance potency and selectivity as describe previuosly.37–41 It could be expected that if these waters are not strongly bound, introducing groups that displace the waters and interact better with the protein will enhance the ligand’s affinity.37–41 Thus, on this basis, we designed the family of structurally related 1-substituted-2,3,4,5-tetrahydro-pyrido-[4,3-b]indol-1-ones as chemical modulators of BRD4 BrD1 (Figure 1A). These molecules retain the key features of MS7972 and contain a longer acetamidoalkyl substituent at the N-indole. The C=O group of MS7972 cis to the acetyl group that does no interact with the protein is converted to an endocyclic amide that points in the opposite direction to enhance possible interactions with the ZA and BC loops of BrDs. The MSi compounds connect the N-acetamido group to the ring N with varying length linkers (n=2,3,4,5,6) to probe the depth of the binding pocket and potentially displacing the inner waters (Figure 1A). To probe the relation of the linker’s length and the affinity of the ligand we conducted MM/GBSA calculations.
MM/GBSA Calculations
The results of a typical MM/GBSA calculation for the binding of each of the designed MSi compound (MS1 to MS5) are given in Table S1 in Supporting Information. The ensemble of structures from the MD simulations of each complex occupied a single basin as indicated by the RMSD with respect to the minimized initial structure (see Figure S2 in Supporting Information) so the average energies can be considered as representative. In addition, the number of water molecules close to ligand is nearly constant varying between 7 and 8 (see Table S2 in Supporting Information). Thus, it is reasonable to consider them as part of the protein in the MM/GBSA calculations.
The results show that MS3 and MS5 are predicted to bind to BRD4 BrD1 with the highest affinities in good agreement with the docking binding energy calculations (see Table S1 in Supporting Information). Indeed, MS3(Olinone) experimentally showed the highest affinity of the series (Kd = 3.4 μM) towards BRD4 BrD1 using isothermal titration microcalorimetry (ITC).15 Our calculated values for binding free energies are in very good agreement in trend with their corresponding experimental values (see Table S1 in Supporting Information). In addition, we found that the length of the N-alkylated chain of the MSi ligands plays an important role in binding to BRD4 BrD1.15 The optimal length of the TV-alkylated chain seems to be four methylene units, which corresponds to Olinone. This length allows Olinone to stay in an extended T-like conformation in the bound state and be anchored between Asn140 (a highly conserved residue in BrDs) and Asp144 in BRD4 BrD1. Later it was confirmed experimentally that MS3 (Olinone) not only binds BRD4 BrD1 with the highest affinity within the series but also has a high selectivity towards BRD1 over BrD2.15 Thus, the success of predicting Olinone as the best binder of the series makes the above in silico strategy feasible to be used next for further selectivity studies.
B). SELECTIVITY ORIGIN OF MS3 (OLINONE)
To gain better understanding of the origin of the selectivity of Olinone for BRD4 BrD1 over BrD2 we performed longer MD simulations of unbound proteins: 1) apo-BRD4 BrD1 (200 ns) and 2) apo-BRD4 BrD2 (400 ns); and of bound complexes: 3) BRD4 BrD1 in complex with Olinone using its X-ray crystal structure15 as initial structure (200 ns) and 4) BRD4 BrD2 in complex with Olinone (400 ns). As initial structure we used the X-ray crystal structure of the second bromodomain (BrD2) of BRD4 (PDB ID 2OUO19) and the Olinone was taken from the superimposed BRD4 BrD1/Olinone. We analyzed each of the four simulations using RMSD (Figure 2) and two-dimension-RMSD (2D-RMSD; see Figure S3 in Supporting Information), as well as root-mean-squared fluctuations (RMSF) and average correlations between the motions of residues as function of protein residue and simulation time, respectively. Finally, we conducted MM/GBSA free energy calculations and pairwise decomposition of interaction energies for each complex.
Figure 2. Backbone atoms RMSD for protein, AcK binding site and ligand of the BRD4 BrD1|2 as function of time.
(A) BRD4 BrD1 without ligand, (B) BRD4 BrD1/Olinone complex, (C) BRD4 BrD2 without ligand, and (D) BRD4 BrD2/Olinone.
Molecular Dynamics Simulations
Protein dynamics in ligand binding is well recognized to be an important property that affects the thermodynamics in a complex way. On the one hand, the loss of vibrational flexibility upon ligand binding to protein (e.g., BrDs) could have an unfavorable entropic effect. On the other hand, the conformational mobility and sequence variability of the ZA and BC loops, which define the ligand acetyl-lysine binding site, can adapt to the ligand to optimize the enthalpic contribution to selectivity.42–45 Indeed, the BrD loops can accommodate the specific shape of their native p53-AcK382 and histone H4K5ac/K8ac peptides as well as small molecules that inhibit peptide binding.40,41,43,45–49 Therefore, we have examined the binding free energy of the BRD4 BrD1/Olinone and BRD4 BrD2/Olinone complexes and their ZA and BC loops mobility before and upon ligand binding along the MD simulation time.
The apo-BRD4 BrD1 MD simulation as well as its binding pocket stabilized after 13 ns with an average RMSD of 1.8±0.2 Å with respect to the minimized initial structure backbone atoms (see Figure 2A). The core protein structure (i.e., the left-handed bundle of four α helices) is the part that confers stability to the protein. The BRD4 BrD1/Olinone complex stabilizes from the beginning of the MD simulation with average backbone binding site RMSD of 0.8±0.2 Å relative to the minimized initial structure, which is about 2-fold lower than for tfpo-BRD4 BrD1 binding site (see Figures 2A and 2B). In contrast, the apo-BRD4 BrD2 (see Figure 2C) and in complex with Olinone show much more structural variability (see Figure 2D). With regard to the binding site of the apo-BRD4 BrD2, its backbone atom RMSD relative to the minimized initial structure show high fluctuations along the MD simulation with an average value of 1.9±0.5 Å (see Figure 2C). In case of BRD4 BrD2/Olinone complex, the binding site backbone atoms RMSD show less fluctuations than the respective unbound systems with an average value of 0.8±0.2 Å, respectively. Olinone binding mode to BRD4 BrD2 has higher heavy atoms RMSD value (relative to the minimized initial structure) compared to its counterpart BRD4 BrD1, on average 3.1±1.4 Å (last 200 ns: 2.6±0.9 Å; see Figure 2D) and 1.1±0.4 Å (see Figure 2B), respectively. All in all, BRD4 BrD2, either with or without the ligand, shows more fluctuations than BrD1. This may have implications on Olinone’s selectivity towards BRD4 BrD1 over BrD2 as discussed below in Conformational Analysis | ZA and BZ Loops Mobility section.
Olinone Acetyl-Lysine Binding Site
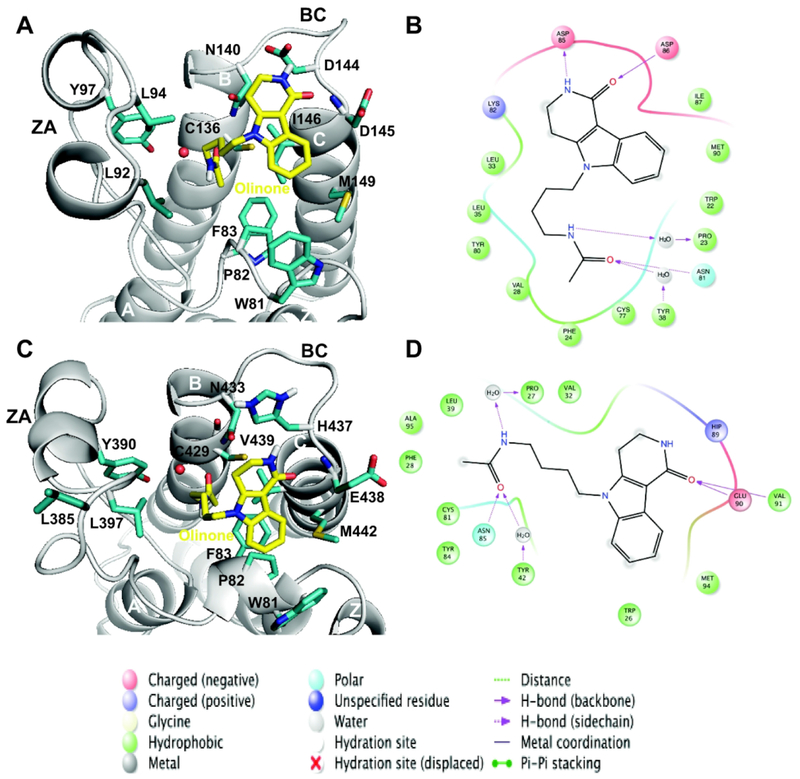
We have analyzed the Olinone binding site for each of the complexes formed by BRD4 BrD1|2; this site overlaps with the binding pocket of the acetyl-lysine. Olinone forms on average an extended T-like conformation in the bound state. Their N-acetylated chain intercalates into the hydrophobic pocket between the ZA and BC loops (Figure 3). The Olinone heterocyclic moiety packs 1) against the side chain of Trp81, Pro82 and Phe83 (WPF lipophilic shelf residues)39,40,50 in the one-turn helix Z, and the lie 146 (gatekeeper residue)39,40,50 in helix C for BRD4 BrD1 (Figures 3A and 3B), and 2) Trp374, Pro375, Phe376 (WPF lipophilic shelf residues) in the one-turn helix Z, and the Val439 (gatekeeper residue) in helix C for BRD4 BrD2 (Figures 3C and 3D). Olinone is trapped in a network of hydrophobic and aromatic interactions that includes 1) Trp81, Pro82, Phe83, Va187, Leu92, Leu94, Tyr97, Cys136, Tyr139, Ile146 and Met149 in BRD4 BrD1 protein, and 2) Trp374, Pro375, Phe376, Va1380, Leu387, Leu390, Tyr497, Asn433, His437, Glu438, Val439 and Met442 in BRD4 BrD2 protein. As described previously, many BET bromodomain ligands derive their affinity from effective hydrophobic interactions with these residues including the hydrophobic WPF shelf.43
Figure 3. Main interactions for the AcK binding site of BRD4 BrD1|2.
Three–dimensional representation of representative structures of the BRD4 BrD1|2/Olinone complexes: (A) BRD4 BrD1/Olinone and (C) BRD4 BrD2/Olinone (rendered using PyMOL program34). Each representative structure was obtained using the average-linkage algorithm within the cluster command in cpptraj in AMBER. The whole MD simulation for each ligand was grouped together to produce one cluster using the pairwise RMSD between frames as a metric comparing the all acetyl-lysine binding site and MSi ligand atoms. The water molecule for each complex is depicted as red sphere. Schematic diagram of main ligand interactions: BRD4 BrD1|2/Olinone complexes: (B) BRD4 BrD1/Olinone and (D) BRD4 BrD2/Olinone (rendered using Ligand Interaction Diagram in Maestro 2016-122). In case of BRD4 BrD1 (B), to align the residues numbering depicted in the figure to the ones in BRD4 BrD1/H4K5ac/K8ac X-ray crystal structure (PDB ID 3UVW19), 59 units should be added to the figure residues numbering. In case of BRD4 BrD2 (D), to align the residues numbering depicted in the figure to the ones in BRD4 BrD2 X-ray crystal structure (PDB ID 2OUO19), 348 units should be added to the figure residues numbering.
The MD simulation of the BRD4 BrD1/Olinone complex reveals five direct hydrogen bonds between the protein and Olinone. These hydrogen bonds are between 1) the side chain of the sulfhydryl -SH group of Cys136 in the BRD4 BrD1 protein and the carbonyl oxygen of the acetyl group of Olinone, 2) the side chain -NH2 group of Asn140 in the BRD4 BrD1 protein (Asn140 is a highly conserved asparagine in BrDs) and the carbonyl oxygen of the acetyl group of Olinone, 3) the side chain carbonyl oxygen of Asp 144 and the amide group of the piperidone ring of Olinone, 4) the nitrogen backbone of Asp 145 and the amide group of the piperidone ring of Olinone, and 5) the nitrogen backbone of Ile146 and the amide group of the piperidone ring of Olinone (Figure 3B). Furthermore, the carbonyl oxygen of the acetyl group of Olinone interacts with the side chain hydroxyl group of Tyr97 through a water molecule (Figure 3B). To a lesser extent, the -NH2 group of the amide group of the N-acetylated chain of Olinone interacts with the backbone carbonyl oxygen group of Pro82 through a water molecule as well (see Figure 3B, and Tables S3 and S4 in Supporting Information). Table 1 summarizes the main hydrogen bonds formed during the MD simulation using a distance cutoff between the heavy atoms of 3.5 A and the angle cutoff between the acceptor and donor atoms of 120°. The occupancy of the hydrogen bond between 1) Cys136, 2) Asn140, 3) Asp144, 4) Asp145, and 5) Ile146 and Olinone are about 28%, 92%, 32%, 30% and 37% along the last 100 ns of MD simulation within a distance cutoff of 3.5 Å, respectively. For the same portion of the MD simulation, the occupancy of the water-mediated hydrogen bonds between the side chain hydroxyl group of Tyr97 and the carbonyl oxygen of the acetyl group of Olinone, and the backbone carbonyl oxygen group of Pro82 and the amide group of the N-acetylated chain Olinone are −84% and −20% along the last 100 ns of MD simulation within a distance cutoff of 3.5 Å, respectively (Tables S3 and S4 in Supporting Information).
Table 1.
Hydrogen bonds analysis for the BRD4 BrD1|2/Olinone complexes
| Complex | Acceptor | Donor | % Occupied | Distance (Å) | Angle (°) |
|---|---|---|---|---|---|
|
BRD4
BrD1/Olinone X-ray crystal 100-200 ns |
MS3@OA* | Cys136@SH | 28.10 | 3.29±0.13 | 25.58±12.63 |
| MS3@OA | Asn140@ND2 | 91.80 | 2.98±0.18 | 34.71±10.95 | |
| Asp144@OD1 | MS3@NR† | 16.40 | 3.14±0.21 | 36.36±12.96 | |
| Asp144@OD2 | MS3@NR | 15.80 | 3.15±0.21 | 34.98±13.87 | |
| MS3@OR† | Asp145@N | 30.10 | 3.06±0.19 | 41.03±12.02 | |
| MS3@OR | Ile146@N | 37.20 | 3.19±0.18 | 29.83±12.67 | |
|
BRD4
BrD2/Olinone Model 200-400ns |
MS3@OA | Cys429@SH | 36.47 | 3.28±0.13 | 25.45±12.00 |
| MS3@OA | Asn433@ND2 | 93.80 | 2.98±0.17 | 34.35±11.12 | |
| MS3@OR | Glu438@N | 18.63 | 3.02±0.19 | 43.03±11.01 | |
| MS3@OR | Val439@N | 17.87 | 3.14±0.20 | 26.71±13.00 | |
OA represents the carbonyl oxygen of the acetyl group of Olinone.
OR and NR represent the carbonyl oxygen and nitrogen of the amide group of the piperidone ring of Olinone, respectively. Hydrogen bonds were determined via the distance between the heavy atoms using a cutoff of 3.5 Å and the angle between the acceptor and donor atoms using a cutoff of 120°.
Analysis of the MD simulation of BRD4 BrD2/Olinone complex shows four direct hydrogen bonds between the protein and ligand. These four hydrogen bonds have their corresponding ones in the BRD4 BrD1/Olinone complex: between 1) the side chain -SH group of Cys429 in the BrD2 protein and the carbonyl oxygen of the acetyl group of Olinone, 2) the side chain -NH2 group of a highly conserved residue Asn433 in the BrD2 protein and the carbonyl oxygen of the acetyl group of Olinone, 3) the nitrogen backbone of Glu438 and the amide group of the piperidone ring of Olinone, and 4) the nitrogen backbone of Val439 and the amide group of the piperidone ring of Olinone. Furthermore, the carbonyl oxygen of the acetyl group of Olinone interacts with the side chain hydroxyl group of Tyr390 through a water molecule (Figures 3C and 3D). To a lesser extent, the amide group of the N-alkylated chain of Olinone interacts with the backbone carbonyl oxygen group of Pro375 through a water molecule as well (see Figure 3D, and Tables S3 and S4 in Supporting Information). Table 1 summarizes the main hydrogen bonds formed along the MD simulation using a distance cutoff of 3.5 Å and the angle cutoff between the acceptor and donor atoms of 120°. The occupancy of the hydrogen bond between 1) Cys429, 2) Asn433, 3) Glu438, and 4) Val439 and Olinone are about 36%, 94%, 19%, and 18% along the last 200 ns of the MD simulation, respectively. The occupancy of the water-mediated hydrogen bonds between the side chain hydroxyl group of Tyr390 and the carbonyl oxygen of the acetyl group of Olinone, and the backbone carbonyl oxygen group of Pro375 and the amide group of the N-alkylated chain Olinone are −88% and −17% along the last 200 ns of MD simulation, respectively (Tables S3 and S4 in Supporting Information).
Altogether, the BRD4 BrD1/Olinone complex exhibits five direct hydrogen bonds between the protein and the ligand in contrast to four hydrogen bonds in the BRD4 BrD2/Olinone complex. The hydrogen bond involving the highly conserved Asn140|433 in BrD1|2 and Olinone, for the two complexes, maintains the highest occupancy of ~92% among all shown directly hydrogen bonds along the whole MD simulations. In addition, the occupancy of the hydrogen bonds between Asp145|Ile146 and Olinone in BrD4 BrD1/Olinone complex are about two fold than the corresponding residues Glu438|Val439 in BrD4 BrD2/Olinone complex. A notable difference between the two complexes is the hydrogen bond formed by Asp144 and Olinone in BRD4 BrD1/Olinone, which it is not displayed for the corresponding residue His437 in BRD4 BrD2/Olinone. This hydrogen bond, and the difference in number and population of such direct bonds between BRD4 BrD1|2/Olinone complexes could be in part responsible for the origin of the selectivity of Olinone for BRD4 BrD1 over BrD2.
Conformational Analysis | ZA and BC Loops Mobility
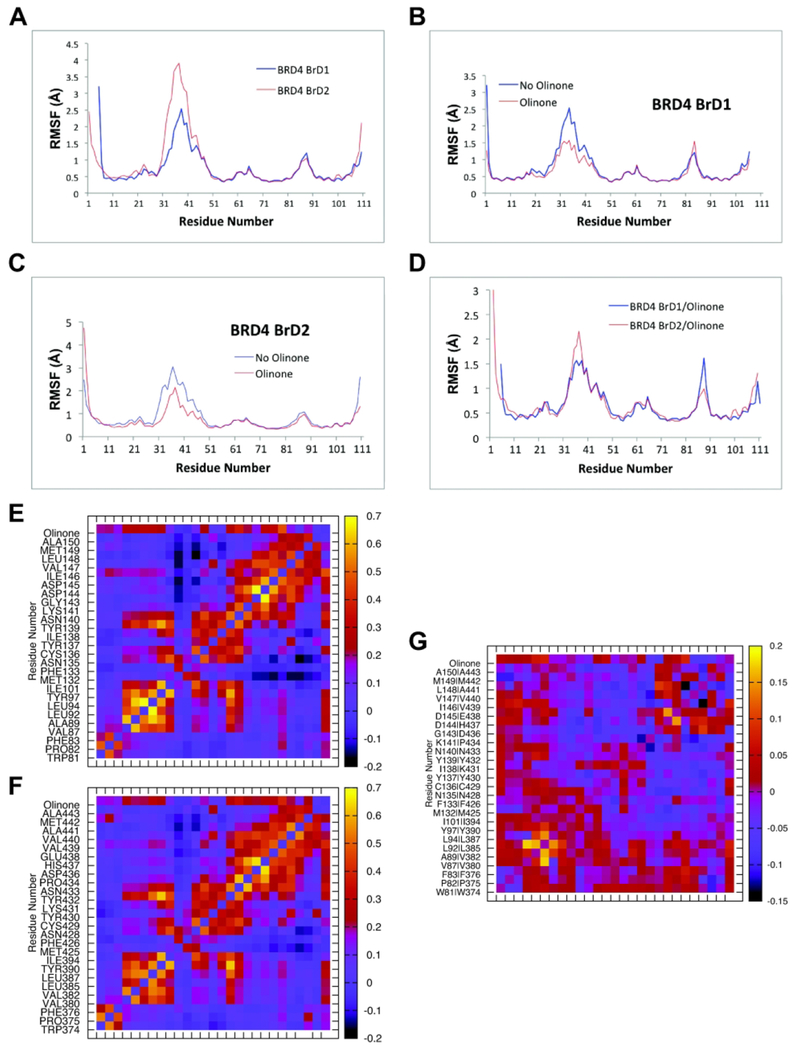
Figure 4 shows the root-mean-square fluctuations as function of residue number of apo-BRD4 BrD1 and BrD2, and their complexes with Olinone. It is clear that the overall patterns of the RMSF are similar; the ZA loop has more mobility in BrD2 in both the apo-form and in the complex (see Figures 4A and 4D). Another clear pattern in the RMSF is the reduction of the ZA loop mobility upon Olinone binding regardless of the specific complex (see Figures 4B and 4C).6,42–45,51 Interestingly, the mobility of the BC loop shows an opposite behavior. It is higher in both the apo-BRD4 BrD1 and in the complex with Olinone compared to BrD2 (see Figures 4A and 4D). Furthermore, the ligand increases the mobility of the BRD4 BrD1 BC loop (see Figure 4B) but not of BrD2. Considering that the change in mobility of the ZA loop is larger in BrD2 this could be in part responsible for the selectivity of Olinone for BRD4 BrD1 over BrD2 due to a larger loss of configurational entropy in BrD2 contributing unfavorably to its binding affinity. The average correlations between the motions of residues within the binding site can shed some light onto this phenomenon as next further discussed (see Figures 4E–4G).
Figure 4. RMSF values and average correlations between motions of residues collected from the starting structures during the MD simulations.
RMSF: (A) apo-BRD4 BrD1|2, (B) apo-BRD4 BrD1 and with Olinone, (C) apo-BRD4 BrD2 and with Olinone, and (D) BRD4 BrD1/Olinone and BRD4 BrD2/Olinone. Correlation between the motions of residues: (E) BRD4 BrD1/Olinone, (F) BRD4 BrD2/Olinone and (G) correlation difference: BRD4 BrD1/Olinone – BRD4 BrD2/Olinone.
Although the motion correlation patterns overall seem similar for the binding sites of BRD4 BrD1|2/Olinone complexes, as depicted in Figure 4, they are more correlated in BrD1 than in BrD2. For example, in case of BRD4 BrD1 (see Figure 4E), the motion of Leu92 of ZA loop and Ile146 in helix C is correlated. The motion of Ile146 is also highly correlated with residues spanning Trp81 to Tyr97 in the ZA loop. Interesting, the motion of residues in the vicinity of Ile146 have low correlation with ZA loop residues, except for Asp144 and Asp145 of BC loop that show low correlation with residues Ala89, Leu92, Leu94, and Tyr97 of the ZA loop. In addition, the motion of residue Met132 in helix B is anti-correlated with residues spanning Asn140 to Asp 145 of BC loop and Ile146 to Ala150 in helix C. The motion of Asn135 is also anti-correlated or non-correlated with residues ranging Gly143 to Ala150. In case of BRD4 BrD2 (see Figure 4F), all these correlations or anti-correlations occur in lesser extent compared to BrD1 (see Figure 4G). These differences in intensity of the correlation patterns motions for the binding site of BRD4 BrD1 compared to those for BrD2 could be the origin of locally less losses of configurational entropy and thus less penalization on binding affinity.52 In addition, the motion of Olinone is more correlated with Leu92 of the ZA loop, Asp144 and Asp 145 of the BC loop, and lie 146 in helix C in BRD4 BrD1 (all key residues contributors to the energetics of binding; see MM/GBSA Calculations and Energetics of BRD4 BrD1|2/Olinone Interactions sections below) than with the corresponding ones Leu385 of the ZA loop, His437 and Glu438 of the BC loop, and Val439 in helix C in BRD4 BrD2, respectively (see Figure 4G). All in all, these differences in correlation motions of the ZA and BC loops of BRD4 BrD1 compared to the ones of BRD4 BrD2 as well as Olinone correlation motions with BRD4 BrD1 or BrD2 shed some light on Olinone’s selectivity.
Our modeling indicates that BRD4 BrD1 and BrD2 can both accommodate Olinone, as it can enlarge or compress the AcK binding pocket via the ZA and BC loops motion. This is also in agreement with experimental results showing several kinds of inhibitor with different molecular scaffold (shape and size) that can bind to AcK binding pocket40,41,43,45–49 In addition, although it is outside of the scope of this study, binding kinetics (i.e., binding/unbinding processes) could play an important role in the Olinone’s selectivity towards BRD4 BrD1 over BrD2 as it has been described for the binding of (+)-JQ1 or (−)-JQ1 to BRD4 BrD1.51,53 Kuang et al.51 have showed that (+)-JQ1 binds stronger than (−)-JQ1 because the binding of (+)-JQ1 into the pocket is favorable, which leads to its achievable binding kinetics and consistency with its good inhibitory effect (IC50:BRD4 BrD1 = 77 nM). Conversely, (−)-JQ1 finds difficulty in its path entering into the binding pocket and leads to a worse inhibitory effect as it has been reported (IC50:BRD4 BrD1 = 10,000 nM).51 Thus, based on our conformational analysis, Olinone entrance to the AcK binding pocket in BRD4 BrD1 could occur easier than to the AcK binding pocket in BRD4 BrD2 due to in part to 1) the gatekeepers isoleucine (Ile146, BrD1) or valine (Val439, BrD2) residues that can impose spatial constraints on Olinone size gaining access to the WPF shelf and other important residues on the ZA and BC loops for ligand recognition, and allowing Olinone to attain the perfect shape complementary to the AcK binding pocket, 2) the mobility of the ZA and BC loops, 3) the correlation between the motions of residues within the AcK binding pocket such as the correlated motion between Leu92 and lie146, which are key residues for the selectivity of Olinone towards BrD1 over BrD2 (see below MM/GBSA Calculations and Energetics of BRD4 BrD1|2/Olinone Interactions sections below), 4) the more pronounced anti-correlated motion between Met132 and residues spanning Gly143 to Ala150 in BRD4 BrD1 than between Met425 and residues spanning Asp436 to Ala443 in BRD4 BrD2, which could result in an enhanced plasticity of the binding pocket, and 5) the higher dynamics correlation of Olinone with Leu92 of the ZA loop, Asp144 and Asp145 of the BC loop, and Iie146 in helix C in BRD4 BrD1 than with the corresponding ones in BRD4 BrD2, which facilitates the creation of the AcK binding pocket to accommodate Olinone and triggering an enhanced selectivity towards BRD4 BrD1 over BrD2.
MM/GBSA Calculations
There are 8 water molecules in the first shell (closer than 3.4 Å) of the acetyl-lysine binding site residues (see Water Molecules Calculations in Methods and Models section). They are included in the MM/GBSA calculations as part of the protein (see Tables S5A and S5B in Supporting Information). The MM/GBSA calculations for both systems, BRD4 BrD1/Olinone and BRD4 BrD2/Olinone, were carried out on portions of the MD simulations where they were equilibrated and stabilized: 100-200 ns for BRD4 BrD1/Olinone and 200-400 ns for BRD4 BrD2/Olinone (see Figures 2B and 2D). Table 2 shows the main energy terms (ΔEelect, ΔEvdW, ΔGsolv, ΔGnp, and TΔSMM) and the calculated total free energy of binding (ΔGcalc). The main contribution to the total binding free energy comes from the combined van der Waals and nonpolar solvation, the combined electrostatic and polar solvation, and the configurational entropy terms. In comparison, the combined electrostatic and the polar solvation energy contribution is more positive in BRD4 BrD1 than in BrD2. In addition, the combined van der Waals and nonpolar solvation energy contribution is less negative in BRD4 BrD1 than in BrD2. This result is in agreement with the fact that the electrostatic potential of the surface area around the acetyl-lysine binding site of BRD4 BrD2 is more polar than the one of BrD1 (see Figure 5), In fact, the acetyl-lysine binding site of BRD4 BrD2 is slightly more negatively charged (see Figure 5B) than the one of BrD1 (see Figure 5A).19 This difference in electrostatic potential surface between the two BrDs could also play an important role on Olinone binding and selectivity towards BrD1 over BrD2 since the slightly larger extent of negative potential on the Olinone surface (see Figure 5C).
Table 2.
Free energy calculations of BRD4 BrD1|2/Olinone complexes using MM/GBSA approach†
| BRD4 BrD1 | BRD4 BrD2 | |
|---|---|---|
| 100-200 ns | 200-400 ns | |
| ΔEvdW | −32.07±0.08 | −27.94±0.11 |
| ΔEelect | −16.97±0.17 | −27.38±0.22 |
| ΔGsolv | 23.50±0.12 | 31.76±0.21 |
| ΔGnp | −4.65±0.01 | −4.22±0.01 |
| ΔEvdW+ΔGnp | −36.72±0.08 | −32.16±0.11 |
| ΔEelect+ΔGsolv | 6.53±0.21 | 4.38±0.30 |
| ΔEMM/GBSA* | −30.19±0.11 | −27.77±0.12 |
| TΔSMM | −22.64±0.09 | −21.50±0.09 |
| ΔGcalc | −7.55±0.14 | −6.27±0.15 |
ΔEMM/GBSA, the MM/GBSA energy of binding, is obtained from molecular mechanics calculations of van der Waals (EvdW), electrostatic (Eelect), as well as solvation (Gsolv), and nonpolar (Gnp) energy contributions (see Eqs. 1 and 2 in MM/GBSA Method in Methods and Models in Supporting Information). TΔSMM is obtained as result of the addition of the translational (TΔStrans), rotational (TΔSrot) and vibrational (TΔSvib) terms of the configurational entropy of the complex (i.e., TΔSMM = TΔStrans + TΔSrot + TΔSvib).
The uncertainties were determined by calculating the SD, standard deviation over 1,000 snapshots (N) after equilibration and dividing by the square root of 1,000 snapshots, σ = SD/N1/2. The uncertainty of ΔGcalc was determined by calculating the square root of the addition of the uncertainties square of ΔEMM/GBSA and TΔSMM for each system. The MM/GBSA calculations were carried out on portions of the MD simulation where both systems, BRD4 BrD1/Olinone and BRD4 BrD2/Olinone, were equilibrated and stabilized (see Figures 2B and 2D).
Figure 5. Electrostatic potential surface visualization.

(A) BRD4 BrD1/Olinone, (B) BRD4 BrD2/Olinone, and (C) Olinone (calculated using the Adaptive Poisson-Boltzmann Solver (APBS) and the associated software package PDB2PQR62–64, and rendered using PyMOL program34). The electrostatic potential of the molecules are shown from −3 to +3 kT/e highlighting the charged nature of the surface comprising the AcK binding site.
Based on the MM/GBSA free energy calculations, Olinone has the lower total MM/GBSA energy of binding value towards BRD4 BrD1 (30.19 kcal/mol) than towards BrD2 (−27.77 kcal/mol) with a difference of over 2 kcal/mol (see Table 2). The lower mobility of the ZA loop in the BRD4 BrD1/Olinone complex together with a slightly higher mobility of its BC loop, compared to their corresponding ones in the BRD4 BrD2/Olinone complex as mentioned earlier (see above Conformational Analysis | ZA and BC Loops Mobility section), are reflected on a higher average entropic penalty of the BRD4 BrD1/Olinone complex (~ −23 kcal/mol, see Table 2) compared to the BRD4 BrD2/Olinone complex (~ −22 kcal/mol, see Table 2). This entropy penalty is compensated by a higher MM/GBSA energy binding of Olinone for BRD4 BrD1 compared to BRD4 BrD2. As a result, the binding of Olinone towards BRD4 BrD1 (−7.55 kcal/mol, Kd = 3.2 μM) is much stronger than towards BRD4 BrD2 (BRD4 BrD2 −6.27 kcal/mol, Kd = 27.1 μM) hence contributing to the selectivity of Olinone for BrD1 over BrD2, which is in agreement with experimental results (see Table 2).15 However, these calculations predict that Olinone binds about 10-fold stronger to BRD4 BrD1 than to BrD2, whereas experimental results show a factor of 100 (see Tables S5A and S5B in Supporting Information). This could be possible due to the fact that BRD4 BrD2 has more mobility than BrD1 as showed earlier in Conformational Analysis | ZA and BZ Loops Mobility section. Thus, it is also possible that the MD simulations do not sample the configurational space correctly for BrD2. As a result, this would have lowered the calculated free energy of binding for BrD2.
Altogether, if the MM/GBSA energy and entropy terms of both complexes are compared, it is revealed that the difference in MM/GBSA energy contribution between the two complexes is about 10%. However, the difference in entropy contribution between the two complexes is about 5%. Therefore, this result would make the MM/GBSA energy contribution two fold more responsible than entropy for the selectivity of Olinone towards BRD4 BrD1 over BrD2.
Energetics of BRD4 BrD1|2/Olinone Interactions
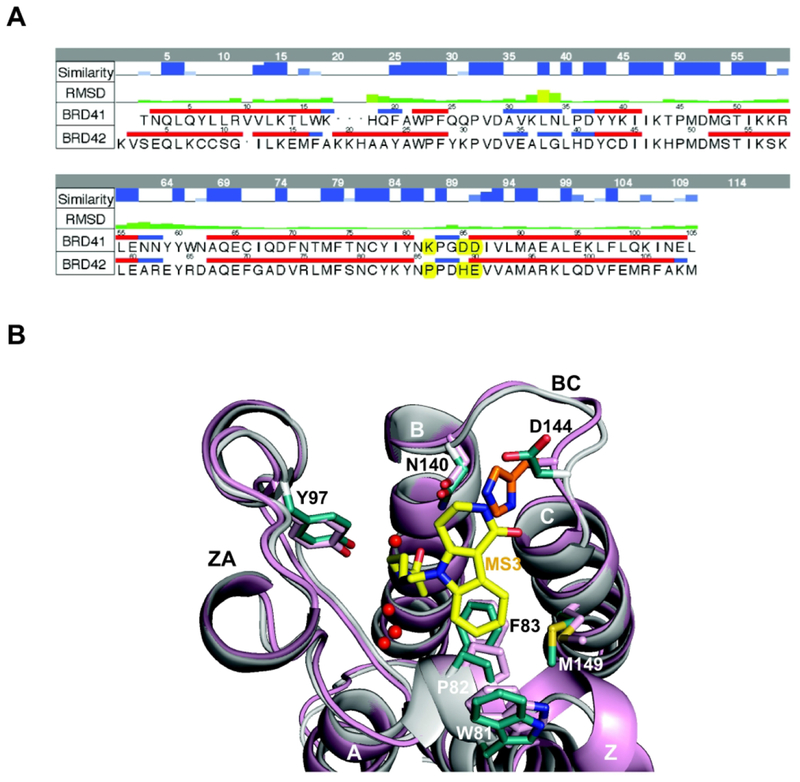
To further explore the interaction between BRD4 BrD1 and BRD4 BrD2 acetyl-lysine binding pocket and Olinone aimed at better explaining the selectivity of Olinone for BrD1 over BrD2, MM/GBSA energy decomposition was conducted. Selected residue contributions to the interaction energy between BRD4 BrD1|2 and Olinone, including water molecules, are shown in Figure 6, and Tables S6–S8 in Supporting Information. In case of BRD4 BrD1/Olinone, Trp81, Pro82, Phe83 (WPF lipophilic shelf residues), Val87, Leu92, Leu94, Tyr97, Cys136, Tyr139, Asn140, Asp144, Asp145, and Ile146 (gatekeeper residue) contribute favorably to the free energy of binding within a hydrophobic and aromatic network that traps Olinone. Asn140 (highly conserved asparagine in BrDs), Asp144 and Asp145 of the BC loop, and Ile146 in helix C are clearly the most favorable residues interacting with Olinone. In case of BRD4 BrD2/Olinone, Trp374, Pro375, Phe376 (WPF lipophilic shelf residues), Val380, Leu387, Leu390, Tyr497, Cys429, Tyr432, Asn433, His437, Glu438, and Val439 (gatekeeper residue) contribute favorably to the free energy of binding within a hydrophobic and aromatic network that traps Olinone. Asn433 (highly conserved asparagine in BrDs), His437 and Glu438 of the BC loop, and Val439 in helix C are the most favorable residues interacting with Olinone. If the BC loop sequences of BRD4 BrD1 and BrD2 are aligned (see Figure 7A), it is found that Asp144 is one of the few residues that are different between BRD4 BrD1 and BrD2 in the acetyl-lysine binding site. In BrD2, its corresponding residue His437 could clash with the indole moiety of Olinone based on their X-ray structures (Figure 7B) thus preventing its binding. Asp144 in BRD4 BrD1 forms a unique hydrogen bond interaction between the amide group of the piperidone ring Olinone and the side chain carbonyl oxygen of Asp144 contributing favorably to the energetics of binding (see Table S7 in Supporting Information). However, its corresponding residue His437 in BRD4 BrD2 also contributes favorably to the interaction with Olinone, but not through hydrogen bonding as Asp144 in BRD4 BrD1 (see Table S8 in Supporting Information). Hence, from an energetic point of view, although these two residues are different, Asp144 in BRD4 BrD1 and His437 in BRD4 BrD2, they seem not to directly explain by themselves the origin of the selectivity of Olinone towards BrD1 over BrD2 as hypothesized previously by Gacias et al.15 However, if we pay attention to the energetic contribution of the two acetyl-lysine binding pocket gatekeeper residues, Ile146 in BrD1 and Val439 in BrD2, towards their Olinone binding, Ile146 contributes about 1.5-fold more favorable than Val439 (see Figure 6 and Tables S6–S8 in Supporting Information). Thus, the most favorable contribution of Ile146 in the binding of Olinone towards BrD1, compared to Val439 in BrD2, could also contribute in part to the selectivity of Olinone for BrD1 over BrD2 together with less contributing residues Asp144 in BrD1 and His437 in BrD2, respectively. The importance of gatekeeper residues (e.g., Ile146 in BrD1 and Val439 in BrD2) and WPF shelf residues on achieving selectivity in BET bromodomains has been also previously highlighted.39,40,50,54 In addition, another residue that could contribute in part to the selectivity of Olinone towards BRD4 BrD1 over BrD2 is Leu92 in BrD1. Compared to its counterpart Leu385 in BrD2, Leu92 in BrD1 contributes above 14-fold more favorable towards Olinone binding than Leu385 in BrD2. Two other residues show slightly stronger interactions in BRD4 BrD1 than in BrD2: Asn140 and Asp145 in BrD1 and corresponding Asn433 and Glu438 in BrD2, which could also contribute to the selectivity of Olinone towards BrD1 over BrD2 (see Tables S6–S8 in Supporting Information). All in all, from the energetic decomposition analysis, there are five residues involve in the molecular mechanism helping on attaining the selectivity of Olinone towards BRD4 BrD1 over BrD2: Leu92|385, Asn140|433, Asp144|His437, Asp145|Glu438, and Ile146|Val49.
Figure 6.
Total energetic contributions of selected residues of the BRD4 BrD1|2/Olinone complexes.
Figure 7. Selectivity binding of Olinone to BRD4 BrD1 versus BRD4 BrD2.
(A) Sequence alignment between the two BRD4 BrDs, BrD1 and BrD2 (Some amino acids that are different in both BrDs, including the Asp144 (BrD1) and His437 (BrD2) are highlighted in yellow). To align the residues numbering of both X-ray crystal structures, 59 units should be added to the figure residue numbering for BrD1 and 348 units for BrD2. The top two rows in the table represent the similarity and RMSD between the two structures, respectively. The alignment was made using MOE18. (B) Structural comparison of BRD4 BrD1 (gray, PDB ID 3UVW19) and BRD4 BrD2 (pink, PDB ID 2OUO19,50) X-ray crystal structures, showing steric clash between MS3 (yellow) and His437 in BRD4 BrD2 (orange) rendered using PyMOL program.34 Side chains of key amino acid residues at the ligand-binding site in the protein are color-coded by atom type. Water molecules in the ligand-binding site are shown as red spheres.
On the other hand, regarding the significance of water molecules towards Olinone ligand binding free energy, and although the watershell calculations provides about 8 waters close to Olinone for each complex BRD4 BrD1/Olinone and BRD4 BrD2/Olinone, the energetic decomposition shows that mainly one water molecule strongly contributes favorably to the free energy of binding for each ligand as mentioned above (see Tables S5–S8 in Supporting Information). This energetic contribution is similar on average for BRD4 BrD1/Olinone and BRD4 BrD2/Olinone (see Tables S6–S8 in Supporting Information). Such water molecule is mediating the interaction between the carbonyl oxygen of the acetyl group of Olinone ligand and the hydroxyl group of Tyr97 in BRD4 BrD1 and Tyr390 in BRD4 BrD2 (see Tables S6–S8 in Supporting Information). This kind of water mediating interaction has been largely found among BET bromodomains inhibitors.40,43,47 In addition, this stable structured water molecule has been reported by Huang et al.38 to be displaced by cosolvent m(ethanol) in apo structures. However, this is not the case for our BRD4 BrD1|2/Olinone complexes. Based on Huang et al. study38, we could take advantage of stable structured waters that can be displaced by cosolvent m(ethano) during ligand design by incorporating hydroxyl substituents in the ligand so as to map them. Thus, we could take advantage of the remaining 7 structured waters to design more potent binders. It is also worth mentioning that the number of waters for apo-BRD4 BrD1 is similar on average than the BRD4 BrD1/Olinone complex (i.e., 8). However, in the case of BRD4 BrD2, the number of waters of apo-BRD4 BrD2 is on average 0.6 larger than in the BRD4 BrD1/Olinone complex (see Tables S5B in Supporting Information). Thus, since overall the binding pocket of the apo-BRD4 BrD2 system has more mobility than the apo-BRD4 BrD1 system, the binding of Olinone could contract the binding pocket causing an entropic cost of about 1 kcal/mol in the free energy of binding. This would help explain why Olinone is better binder towards BRD4 BrD1 (−7.55 kcal/mol) than to BRD4 BrD2 (−5.27 kcal/mol) taking into consideration such 1 kcal/mol correction (see Table 2).
Overall, it is also worth mentioning that our calculations reproduce quite well many crystal structures of potent BET inhibitors bound to BRD4 BrD1, such as 3MXF, (+)-JQ1 | IC50:BRD4 BrD1 = 77 nM6; 3P5O, I-BET762 | IC50:BRD4 BrD1 = 631 nM54,55 and; and 3ZYU, I-BET151 | IC50:BRD4 BrD1 = 790 nM56 in terms of the formation of 1) a direct hydrogen bond between the triazole ring (e.g., (+)-JQ1 or I-BET762, currently under clinical development) or the isoxazole moiety (e.g., I-BET151), which both chemotypes occupy the same position as the acetyl group in histone tail peptides or our Olinone inhibitor, and the amide side chain of the highly conserved Asn140 in BRD4 BrD1, and 2) a water-mediated hydrogen bond to the hydroxyl side chain of Y97 in BRD4 BrD1 through the triazole ring and isoxazole oxygen and nitrogen atoms, respectively. Additionally, in I-BET151 the quinolone nitrogen atom forms a water-mediated bridging hydrogen bond to the backbone carbonyl group of Pro82 in BRD4 BrD1 as it is also present in our Olinone inhibitor. These latter compounds (i.e., (+)-JQ1, I-BET762, and I-BET151) use triazole ring and isoxazole moiety respectively to mimic the acetyl-lysine moiety present in histone tail peptides and in our Olinone inhibitor. Other BET inhibitors with different chemotypes that reproduce our results include the X-ray crystal structures of BRD4 BrD1 bound to PFI-1, a dihydroquinazoline-based compound, (PDB ID: 4E96, IC50:BRD4 BrD1 = 220 nM57), MS436, a diazobenzene-based compound, (PDB ID: 4NUD, Ki = 30-50 nM13), MS402, cyclopentene-based compound (PDB ID: 5ULA, Ki = 77 nM14), RVX-208, quinazoline-based compound, (PDB ID: 4MR4, Kd = 1.1 μM58), 8302, sulfonamide-based compound (PDB ID: 4LR6, IC50:BRD4 BrD1 = 11 μM48).
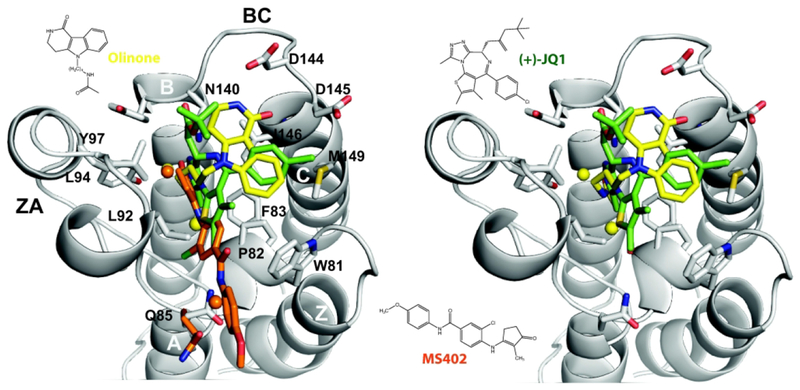
Furtheremore, the overlapping of the X-ray crystal structures of our Olinone inhibitor, and the potent inhibitors (+)-JQ1 and MS402 all bound to BRD4 BrD1 (see Figure 8) reveals a conserved binding mechanism of BET inhibitors which include three key area of interaction: the AcK binding pocket, the WPF shelf, and the structurally conserved ZA channel.41 Besides the presence of the critical hydrogen bond with the side chain amide group of the highly conserved Asn140, and the ZA channel water-mediated hydrogen bond with the side chain hydroxyl group of Tyr97 as decribed in this study, (+)-JQ1 and MS402 display extensive hydrophobic interactions. Among them are π–π stacking and van der Waals interactions between the dimethyl-substituted thieno ring and Pro82 and Leu92 in the ZA loop in case of (+)-JQ1, and between the chlorobenzene and Pro82 and Leu92 in the ZA loop in the case of MS402. Although Olinone inhibitor also has a benzene ring that occupies a similar position to the 4-chlorobennzene substituent of (+)-JQ1, it lacks several of these stabilizing hydrophobic interactions presented in (+)-JQ1 and MS402. Additionally, opposite to our Olinone, MS402 exhibits two additional electrostatic interactions: a direct hydrogen bond with backbone carbonyl group of Pro82 of the WPF shelf and a water-mediated hydrogen bond to the side chain carbonyl oxygen of Gln85. Olinone also forms an intermittently water-mediated hydrogen bond with the backbone carbonyl group of Pro82 of the WPF shelf, but does not engage with Gln85. (+)-JQ1 and MS402 also participate in van der Waals interactions with the gatekeeper reside I1466,14 like Olinone. The lack of the mentioned above hydrophobic interactions of Olinone when bound to the AcK binding pocket in BRD4 BrD1 could explain its weaker binding affinity compared to (+)-JQ1 (IC50:BRD4 BrD1 = 77 nM) and MS402 (Ki = 77 nM), which in turn provides room to improve potency while maintatining its selectivty for BRD4 BrD1 over BrD2.
Figure 8. Superimposition of two potent BET inhibitors and Olinone in complex with BRD4 BrD1.
Left Panel: Olinone (yellow), and the potent inhibitors (+)-JQ1 (green) and MS402 (orange). Right Panel: Olinone (yellow), and the potent inhibitors (+)-JQ1 (green). Water molecules are shown as yellow (Olinone and (+)-JQ1) or orange (MS402) spheres.
CONCLUSIONS
As a follow-up of our previous work,15 we first provided a detailed characterization study of the in silico rational design of a series of five tetrahydro-pyrido indole-based compounds (MS1-MS5, MSi) as BRD4 BrD1 inhibitors15 using as a starting point compound MS7972, a CBP bromodomain inhibitor previously identified in our laboratory, and second the molecular basis for the selectivity of the most potent ligand of such series, MS3 (Olinone), towards BRD4 BrD1 over BrD2.15 Based on the computational analysis of the MD simulations as well as on the energetic calculations using the MM/GBSA approach, the Olinone compound was predicted to be the ligand with the highest affinity towards BRD4 BrD1 among the family of compounds designed in this study.15 Olinone, once its rational designed was completed, experimentally showed not only the highest affinity of the series but to have high selectivity for BRD4 BrD1 over BrD2.15 Our in silico study through the analysis of the MD simulations of apo-BRD4 BrD1|2 and BRD4 BrD1|2/Olinone complexes together with the MM/GBSA calculations also shed some light on the origin of this selectivity. First, the binding free energy calculations showed that Olinone binds stronger to BRD4 BrD1 than to BrD2, where five residues play key roles on that: Leu92|385 of ZA loop, Asn140|433 (highly conserved BrDs asparagine), Asp144|His437 (unique to respectively BrD1|2) and Asp145|Glu438 of BC loop, and Ile146|Val49 (gatekeeper residue) of helix C in BrD1|BrD2. Second, the difference in electrostatic potential surface between the AcK binding site of BRD4 BrD and BrD2 due to the slightly larger extent of negative electrostatic potential on the Olinone surface. Third, the difference in number of direct hydrogen bonds (HBonds) between BRD4 BrD1/Olinone (i.e., 5 HBonds) and BRD4 BrD2/Olinone (i.e., 4 HBonds) complexes. Fourth, the difference in mobility of the ZA and BC loops upon ligand binding to the AcK binding pocket between BRD4 BrD1|2/Olinone complexes due to changes in configurational entropy contribution, the coupling of proteins and Olinone motions, the correlation between the motions of residues within the AcK binding pocket such as the correlated motion of Leu92 and Ile146, key residues for the energetics of binding of Olinone towards BrD1 over BrD2, and the more pronounced anti-correlated motion between Met132 and residues spanning Asn140 to Ala150 in BRD4 BrD1 than the corresponding ones in BRD4 BrD2, which could result in an enhanced plasticity of the binding pocket. And fifth, the contribution to ligand affinity of a structured water molecule found to be tightly bound to Olinone, which mediates their interaction with side chain hydroxyl group of Tyr97 in BRD4 BrD1 (or Tyr390 in BRD4 BrD2), together with the remaining seven structured waters included in our analysis, but not directly to Olinone’s selectivity towards BRD4 BrD1 over BrD2.
This study also found that none of the 8 structured water molecules located deep inside in the acetyl-lysine binding cavity were displaced by Olinone or any of the MSi compounds. Based on our second design principle “getting insight into the properties inside of the ligand acetyl-lysine binding pocket,” this latter result also leaves room to continue exploring new ways to improve Olinone potency and selectivity by displacing weakly bound water molecules with Olinone derivative groups that could interact better with BRD4 BrD1. As stated by Huang et al.38, the position of weakly bound structured water molecules in the ZA channel could be used to design hydroxyl substituents of Olinone. For example, the water mediating hydrogen bond between Pro82|Pro375 in BRD4 BrD1|BrD2 and Olinone with both low ~20% effective occupancy could be a good candidate for such design, but not the more stable bound water mediating the hydrogen bond interaction between Tyr97|Tyr390 in BRD4 BrD1|2 and Olinone with both high ~90% effective occupancy. This kind of modification in lead optimization could improve potency by up to two orders of magnitude as reported previously.37–41,59–61 The understanding of all these physical forces and the molecular mechanisms of ligand-protein interactions provide a good starting point for a successful structure-based rational design of Olinone analogs with enhanced affinity-selectivity profiles. For instance, by modifying the Olinone’s amide group of the piperidone ring and the amide group of the N-acetamidoalkyl chain its affinity towards BRD4 BrD1 could be enhanced, while keeping its selectivity over BrD2. As mentioned above, there is an intermittent hydrogen bond interaction between the amide group of the piperidone ring Olinone and the side chain carbonyl oxygen of Asp144 (32% effective occupancy). This could provide more room for enhancing Olinone potency and selectivity. Olinone analogs of BRD4 BrD1 could be of invaluable aid to keep unraveling the mechanistic insights underlying BRD4 functions in human biology and disease and evaluate their potential as therapeutic targets. A valuable example of the potential of such selective BRD4 BrD1 inhibitors is the finding in which Olinone was shown to accelerate the progression of mouse primary oligodendrocyte progenitors towards differentiation, whereas chemical inhibition of both bromodomains of BET proteins hindered differentiation.15
Supplementary Material
ACKNOWLEDGMENT
Y.R. thanks Dr. Mercedes Martín-Martínez (Instituto de Química Médica (IQM), Spanish National Research Council (CSIC) - Madrid, Spain) and Dr. Mihaly Mezei (Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai) for critically reading the manuscript and suggesting improvements. This work was supported in part by the Research Supplements to Promote Diversity in Health-Related Research Program from the National Cancer Institute of the National Institutes of Health (NIH, to Y.R.), PSC CUNY Research Award (PSC CUNY 63571-00 41 to Y.R.) and by research grants from NIH (R01 AI124465 and R01 HG004508 to M.-M.Z.). Computations were supported in part through the computational resources and staff expertise provided by the Scientific Computing Facility at the Icahn School of Medicine at Mount Sinai.
ABBREVIATIONS
- BrD
bromodomain
- BRD2, BRD3, BRD4
bromodomain-containing proteins 2-4
- BRDT
bromodomain testis-specific protein
- BrD1 and BrD2
tandem bromodomains
- HDAC
histone deacetylases
- HAT
histone acetyltranferases
- HDAC
histone deacetylases
- ITC
isothermal titration microcalorimetry
- AcK
acetylated lysine
- MD
Molecular dynamics
- MM/GBSA
molecular mechanics Generalized–Born surface area
- PTM
post-translational modification
- RMSD
root–mean–square deviation
- RMSF
root–mean–square fluctuations
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Allfrey VG; Faulkner R; Mirsky AE, Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Nat Acad Sci US. 1964; 51:786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T, Acetylation: a regulatory modification to rival phosphorylation? The EMBO J. 2000; 19:1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrowsmith CH; Bountra C; Fish PV; Lee K; Schapira M, Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012; 11:384–400. [DOI] [PubMed] [Google Scholar]
- 4.Merrick CJ; Huttenhower C; Buckee C; Amambua-Ngwa A; Gomez-Escobar N; Walther M; Conway DJ; Duraisingh MT, Epigenetic dysregulation of virulence gene expression in severe Plasmodium falciparum malaria. J Infect Dis 2012; 205:1593–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez R; Zhou MM, The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009; 12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 6.Filippakopoulos P; Qi J; Picaud S; Shen Y; Smith WB; Fedorov O; Morse EM; Keates T; Hickman TT; Felletar I; Philpott M; Munro S; McKeown MR; Wang Y; Christie AL; West N; Cameron MJ; Schwartz B; Heightman TD; La Thangue N; French CA; Wiest O; Kung AL; Knapp S; Bradner JE, Selective inhibition of BET bromodomains. Nature. 2010; 468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidler LR; Brown N; Knapp S; Hoelder S, Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem. 2012; 55:7346–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhalluin C; Carlson JE; Zeng L; He C; Aggarwal AK; Zhou MM, Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999; 399:491–496. [DOI] [PubMed] [Google Scholar]
- 9.Belkina AC; Denis GV, BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012; 12:465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G; Liu R; Zhong Y; Plotnikov AN; Zhang W; Zeng L; Rusinova E; Gerona-Nevarro G; Moshkina N; Joshua J; Chuang PY; Ohlmeyer M; He JC; Zhou MM, Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012; 287:28840–28851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B; Yang XD; Zhou MM; Ozato K; Chen LF, Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009; 29:1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidler LR; Filippakopoulos P; Fedorov O; Picaud S; Martin S; Tomsett M; Woodward H; Brown N; Knapp S; Hoelder S, Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening. J Med Chem. 2013; 56:8073–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G; Plotnikov AN; Rusinova E; Shen T; Morohashi K; Joshua J; Zeng L; Mujtaba S; Ohlmeyer M; Zhou MM, Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains. J Med Chem. 2013; 56:9251–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung K; Lu G; Sharma R; Vincek A; Zhang R; Plotnikov AN; Zhang F; Zhang Q; Ju Y; Hu Y; Zhao L; Han X; Meslamani J; Xu F; Jaganathan A; Shen T; Zhu H; Rusinova E; Zeng L; Zhou J; Yang J; Peng L; Ohlmeyer M; Walsh MJ; Zhang DY; Xiong H; Zhou MM, BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc Natl Acad Sci U S A. 2017; 114:2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gacias M; Gerona-Navarro G; Plotnikov AN; Zhang G; Zeng L; Kaur J; Moy G; Rusinova E; Rodriguez Y; Matikainen B; Vincek A; Joshua J; Casaccia P; Zhou MM, Selective Chemical Modulation of Gene Transcription Favors Oligodendrocyte Lineage Progression. Chem Biol. 2014; 21:841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachchidanand; Resnick-Silverman L; Yan S; Mutjaba S; Liu WJ; Zeng L; Manfredi JJ; Zhou MM, Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem Biol. 2006; 13:81–90. [DOI] [PubMed] [Google Scholar]
- 17.Marvin. Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions, Marvin 5.12.0. ChemAxon; 2013: Budapest, Hungary: http://www.chemaxon.com. [Google Scholar]
- 18.MOE Chemical Computing Group: Montreal, Quebec, Canada: http://www.chemcomp.com. [Google Scholar]
- 19.Filippakopoulos P; Picaud S; Mangos M; Keates T; Lambert JP; Barsyte-Lovejoy D; Felletar I; Volkmer R; Muller S; Pawson T; Gingras AC; Arrowsmith CH; Knapp S, Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012; 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteiger J; Marsili M, Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron. 1980; 36:3219–3228. [Google Scholar]
- 21.Case DA; Cheatham TE 3rd; Darden T; Gohlke H; Luo R; Merz KM Jr.; Onufriev A; Simmerling C; Wang B; Woods RJ, The Amber biomolecular simulation programs. J Comput Chem. 2005; 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrödinger Release 2016-1, Maestro SR, Schrödinger, LLC, New York, NY, 2016. [Google Scholar]
- 23.Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML, Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983; 79:926–935. [Google Scholar]
- 24.Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput. 2015; 11;3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryckaert JP; Ciccotti G; Berendsen HJC, Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J Comput Phys. 1977; 23:327–341. [Google Scholar]
- 26.Adams DJ, Grand canonical ensemble Monte Carlo for a Lennard-Jones fluid. Molecular Physics. 1975; 29:307–311. [Google Scholar]
- 27.Mezei M Grand-canonical ensemble Monte Carlo study of dense liquids: Lennard-Jones, soft spheres and water. Mol Phys. 1987; 61:565–582. erratum: 67:1207 (1989). [Google Scholar]
- 28.Schafmeister CEAF; Ross WS; Romanovski V, LEaP. University of California, San Francisco. LEAP was developed in Peter A. Kollman’s laboratory at the University of California-San Francisco 1995.
- 29.Mezei M Simulaid: a simulation facilitator and analysis program. J Comput Chem. 2010; 31:2658–2668. [DOI] [PubMed] [Google Scholar]
- 30.Mezei M, MMC: Monte Carlo program for molecular assemblies. 2008: New York, NY: http://inka.mssm.edu/~mezei/mmc. [Google Scholar]
- 31.VIDA 4.3.0.4: OpenEye Scientific Software, Santa Fe, NM: http://www.eyesopen.com. [Google Scholar]
- 32.Wang JM; Wang W; Kollman PA, Antechamber: An accessory software package for molecular mechanical calculations. Abstr Pap Am Chem S. 2001; 222:U403. [Google Scholar]
- 33.Humphrey W; Dalke A; Schulten K, VMD: visual molecular dynamics. J Mol Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 34.The PyMOL Molecular Graphics System, Version 1.8.2.3 Schrödinger, LLC. [Google Scholar]
- 35.Gohlke H; Case DA, Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J Comput Chem. 2004; 25:238–250. [DOI] [PubMed] [Google Scholar]
- 36.Bashford D; Case DA, Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000; 51:129–152. [DOI] [PubMed] [Google Scholar]
- 37.Pan C; Mezei M; Mujtaba S; Muller M; Zeng L; Li J; Wang Z; Zhou MM, Structure-guided optimization of small molecules inhibiting human immunodeficiency virus 1 Tat association with the human coactivator p300/CREB binding protein-associated factor. J Med Chem. 2007; 50:2285–2288. [DOI] [PubMed] [Google Scholar]
- 38.Huang D; Rossini E; Steiner S; Caflisch A, Structured water molecules in the binding site of bromodomains can be displaced by cosolvent. ChemMedChem. 2014; 9:573–579. [DOI] [PubMed] [Google Scholar]
- 39.Crawford TD; Tsui V; Flynn EM; Wang S; Taylor AM; Cote A; Audia JE; Beresini MH; Burdick DJ; Cummings R; Dakin LA; Duplessis M; Good AC; Hewitt MC; Huang HR; Jayaram H; Kiefer JR; Jiang Y; Murray J; Nasveschuk CG; Pardo E; Poy F; Romero FA; Tang Y; Wang J; Xu Z; Zawadzke LE; Zhu X; Albrecht BK; Magnuson SR; Bellon S; Cochran AG, Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains. J Med Chem. 2016; 59:5391–5402. [DOI] [PubMed] [Google Scholar]
- 40.Romero FA; Taylor AM; Crawford TD; Tsui V; Cote A; Magnuson S, Disrupting Acetyl-Lysine Recognition: Progress in the Development of Bromodomain Inhibitors. J Med Chem. 2016; 59:1271–1298. [DOI] [PubMed] [Google Scholar]
- 41.Hewings DS; Rooney TP; Jennings LE; Hay DA; Schofield CJ; Brennan PE; Knapp S; Conway SJ, Progress in the development and application of small molecule inhibitors of bromodomain-acetyl-lysine interactions. J Med Chem. 2012; 55:9393–9413. [DOI] [PubMed] [Google Scholar]
- 42.Steiner S; Magno A; Huang D; Caflisch A, Does bromodomain flexibility influence histone recognition? FEBS Lett. 2013; 587:2158–2163. [DOI] [PubMed] [Google Scholar]
- 43.Brand M; Measures AR; Wilson BG; Cortopassi WA; Alexander R; Hoss M; Hewings DS; Rooney TP; Paton RS; Conway SJ, Small molecule inhibitors of bromodomain-acetyl-lysine interactions. ACS Chem Biol. 2015; 10:22–39. [DOI] [PubMed] [Google Scholar]
- 44.Spiliotopoulos D; Caflisch A, Molecular dynamics simulations of bromodomains reveal binding-site flexibility and multiple binding modes of the natural ligand acetyl-lysine. Isr J Chem. 2014; 54:1084–1092. [Google Scholar]
- 45.Spiliotopoulos D; Caflisch A, Fragment-based in silico screening of bromodomain ligands. Drug Discov Today Technol. 2016; 19:81–90. [DOI] [PubMed] [Google Scholar]
- 46.Zaware N; Zhou MM, Chemical modulators for epigenome reader domains as emerging epigenetic therapies for cancer and inflammation. Curr Opin Chem Biol. 2017; 39:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z; Wang P; Chen H; Wold EA; Tian B; Brasier AR; Zhou J, Drug Discovery Targeting Bromodomain-Containing Protein 4. J Med Chem. 2017; 60:4533–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen BK; Mehta S; Ember SWJ; Zhu JY; Schonbrunn E; Ayad NG; Schurer SC, Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations. ACS Omega. 2017; 2:4760–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang G; Smith SG; Zhou MM, Discovery of Chemical Inhibitors of Human Bromodomains. Chem Rev. 2015; 115:11625–11668. [DOI] [PubMed] [Google Scholar]
- 50.Chung CW; Coste H; White JH; Mirguet O; Wilde J; Gosmini RL; Delves C; Magny SM; Woodward R; Hughes SA; Boursier EV; Flynn H; Bouillot AM; Bamborough P; Brusq JM; Gellibert FJ; Jones EJ; Riou AM; Homes P; Martin SL; Uings IJ; Toum J; Clement CA; Boullay AB; Grimley RL; Blandel FM; Prinjha RK; Lee K; Kirilovsky J; Nicodeme E, Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem. 2011; 54:3827–3838. [DOI] [PubMed] [Google Scholar]
- 51.Kuang M; Zhou J; Wang L; Liu Z; Guo J; Wu R, Binding Kinetics versus Affinities in BRD4 Inhibition. J Chem Inf Model. 2015; 55:1926–1935. [DOI] [PubMed] [Google Scholar]
- 52.Chang CE; Chen W; Gilson MK, Ligand configurational entropy and protein binding. Proc Natl Acad Sci U S A. 2007; 104:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magno A; Steiner S; Caflisch A, Mechanism and Kinetics of Acetyl-Lysine Binding to Bromodomains. J Chem Theory Comput. 2013; 9:4225–4232. [DOI] [PubMed] [Google Scholar]
- 54.Nicodeme E; Jeffrey KL; Schaefer U; Beinke S; Dewell S; Chung CW; Chandwani R; Marazzi I; Wilson P; Coste H; White J; Kirilovsky J; Rice CM; Lora JM; Prinjha RK; Lee K; Tarakhovsky A, Suppression of inflammation by a synthetic histone mimic. Nature. 2010; 468:1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirguet O; Gosmini R; Toum J; Clement CA; Barnathan M; Brusq JM; Mordaunt JE; Grimes RM; Crowe M; Pineau O; Ajakane M; Daugan A; Jeffrey P; Cutler L; Haynes AC; Smithers NN; Chung CW; Bamborough P; Uings IJ; Lewis A; Witherington J; Parr N; Prinjha RK; Nicodeme E, Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013; 56:7501–7515. [DOI] [PubMed] [Google Scholar]
- 56.Dawson MA; Prinjha RK; Dittmann A; Giotopoulos G; Bantscheff M; Chan WI; Robson SC; Chung CW; Hopf C; Savitski MM; Huthmacher C; Gudgin E; Lugo D; Beinke S; Chapman TD; Roberts EJ; Soden PE; Auger KR; Mirguet O; Doehner K; Delwel R; Burnett AK; Jeffrey P; Drewes G; Lee K; Huntly BJ; Kouzarides T, Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011; 478:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picaud S; Da Costa D; Thanasopoulou A; Filippakopoulos P; Fish PV; Philpott M; Fedorov O; Brennan P; Bunnage ME; Owen DR; Bradner JE; Taniere P; O’Sullivan B; Muller S; Schwaller J; Stankovic T; Knapp S, PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013; 73:3336–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picaud S; Wells C; Felletar I; Brotherton D; Martin S; Savitsky P; Diez-Dacal B; Philpott M; Bountra C; Lingard H; Fedorov O; Muller S; Brennan PE; Knapp S; Filippakopoulos P, RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A. 2013; 110:19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lafleur K; Huang D; Zhou T; Caflisch A; Nevado C, Structure-based optimization of potent and selective inhibitors of the tyrosine kinase erythropoietin producing human hepatocellular carcinoma receptor B4 (EphB4). J Med Chem. 2009; 52:6433–6446. [DOI] [PubMed] [Google Scholar]
- 60.Zhao H; Dong J; Lafleur K; Nevado C; Caflisch A, Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics. ACS Med Chem Lett. 2012; 3:834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lafleur K; Dong J; Huang D; Caflisch A; Nevado C, Optimization of inhibitors of the tyrosine kinase EphB4. 2. Cellular potency improvement and binding mode validation by X-ray crystallography. J Med Chem. 2013; 56:84–96. [DOI] [PubMed] [Google Scholar]
- 62.Jurrus E; Engel D; Star K; Monson K; Brandi J; Felberg LE; Brookes DH; Wilson L; Chen J; Liles K; Chun M; Li P; Gohara DW; Dolinsky T; Konecny R; Koes DR; Nielsen JE; Head-Gordon T; Geng W; Krasny R; Wei GW; Holst MJ; McCammon JA; Baker NA, Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018; 27:112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dolinsky TJ; Czodrowski P; Li H; Nielsen JE; Jensen JH; Klebe G; Baker NA, PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007; 35:W522–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker NA; Sept D; Joseph S; Holst MJ; McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001; 98:10037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.