Abstract
Isolation of high molecular weight DNA from gastropod molluscs and its subsequent PCR amplification is considered difficult due to excessive mucopolysaccharides secretion which co-precipitate with DNA and obstruct successful amplification. In an attempt to address this issue, we describe a modified CTAB DNA extraction method that proved to work significantly better with a number of freshwater and terrestrial gastropod taxa. We compared the performance of this method with Qiagen® DNeasy Blood and Tissue Kit. Reproducibility of amplification was verified using a set of taxon-specific primers, wherein modified CTAB extracted DNA could be replicated at least four out of five times but kit extracted DNA could not be replicated. In addition, sequence quality was significantly better with CTAB extracted DNA. This could be attributed to the removal of polyphenolic compounds by polyvinyl pyrrolidone which is the only difference between conventional and modified CTAB DNA extraction methods for animals. The genomic DNA isolated using modified CTAB protocol was of high quality (A260/280 ≥ 1.80) and could be used for downstream reactions even after long-term storage (more than 2 years).
Electronic supplementary material
The online version of this article (10.1007/s13205-020-2051-7) contains supplementary material, which is available to authorized users.
Keywords: CTAB, DNA extraction, Gastropods, Mucopolysaccharides, Non-marine molluscs, Polyphenols
Molecular phylogenetics uses a variety of statistical procedures to construct and understand evolutionary relationships among organisms (Sidow and Bowman 1991). These relationships are crucial in addressing questions related to delimitation of species, divergence time estimation, and study of fossils among other applications of phylogenetics (Thorne and Kishino 2002; Giribet 2003; Yang and Rannala 2010). There was an unprecedented growth in this field with the advent of various sophisticated techniques namely cloning, enzymatic amplification (Lio and Goldman 1998), whole genome sequencing (Dowell 2008), etc. Despite being diverse in nature, a common and the primary step in each of these methods is extraction of high molecular weight DNA. It is also arguably the most pivotal step, since the accuracy and quality of the final results depend to a large extent on the quality of the DNA.
Several genomic DNA extraction methods have been described for animals which include both the manual methods (Cheung et al. 1993; Winnepenninckx et al. 1993; Aljanabi and Martinez 1997; Yue and Orban 2005) and commercially available extraction kits. However, in the case of gastropod molluscs and bivalves, the presence of excessive slime (mucopolysaccharides) is detrimental due to its amplification inhibiting capacity. It co-precipitates with DNA, inhibiting the enzyme activity during PCR (Winnepenninckx et al. 1993; Sokolov 2000; Popa et al. 2007). While much has been written about bivalves and other marine molluscs in this regard (Aranishi and Okimoto 2006; Popa et al. 2007; Pereira et al. 2011), their terrestrial and freshwater counterparts have received very little attention. In this communication, we provide a detailed description of a genomic DNA extraction method that proved to work well with many terrestrial as well as freshwater gastropod families producing high-quality DNA. The extracted DNA from both fresh and old samples gave consistent results when used for PCR. We have also compared its performance with Qiagen DNeasy® Blood and Tissue Kit, since it is one of the most widely used commercially available kits for DNA extraction.
This study included 23 representative individuals belonging to 13 families, 18 genera and 23 species collected from the Western Ghats of India (Table S2). All specimens were thoroughly washed with absolute ethanol initially and kept immersed in it. For the next 1 week, ethanol was changed once every 2 days, since it became turbid due to slime discharge by the animal. In addition, ethanol was changed every time it turned turbid or yellowish over time due to slime discharge to ensure proper preservation of the animal. Prior to DNA extraction, about 20 mg of tissue (whole body excluding the shell if the animal was tiny) was kept immersed in twice the volume of absolute ethanol for another 2 days to remove any remnant slime present on the tissue.
Modified CTAB method
The alcohol-soaked tissue was crushed slightly using a pestle and transferred to 400 µL TE buffer for softening before removing excess alcohol using Kimtech® Kimwipes and incubated at room temperature (25–30 °C) with mild shaking at 400 rpm for an hour. In the case of old samples (a few years old), if not preserved properly, the slime hardens and resembles the tissue. This makes it difficult to differentiate between the two. Therefore, the treatment with TE buffer becomes crucial, since it dissolves the slime entirely leaving behind only the tissue. After incubation, the softened tissue was immersed in 400 µL of CTAB buffer pre-heated to 60 °C. The tissue was then subjected to mechanical disruption using a bead beater post adding 20 µL Qiagen® Proteinase K and a silica bead, and kept for overnight digestion at 60 °C. The suspension was extracted with 400 µL of chloroform: isoamyl alcohol (24:1) thrice at 12,000 rpm for 5 min at 4 °C. The supernatant was carefully separated to ensure the white layer remained undisturbed and precipitated with 800 µL of absolute ethanol in the presence of 40 µL 3 M Sodium acetate at 12,000 rpm for 10 min at 4 °C. The pellet was washed again with 200 µL 70% ethanol, air dried, and re-suspended in TE Buffer for long-term storage. The components and corresponding quantities of all the reagents used are summarized in Table 1. All reagents were autoclaved after preparation except lysis buffer in which CTAB was added after autoclaving. Since RNA was not found to hinder with the amplification process unlike polyphenols, no RNase treatment was done.
Table 1.
List of reagents used in modified CTAB method
| S. no. | Regent | Components | Quantity |
|---|---|---|---|
| 1 | 1M Tris–HCl | Tris Base | 121.1 g |
| HCl | Make pH 8.0 | ||
| Distilled Water | Adjust volume to 1L | ||
| 2 | 0.5 M EDTA disodium dihydrate | EDTA disodium dihydrate | 186.1 g |
| Sodium Hydroxide | Make pH 8.0 | ||
| Distilled Water | Adjust volume to 1L | ||
| 3 | 5 M Sodium chloride | Sodium Chloride | 292.2 g |
| Distilled Water | Adjust volume to 1L | ||
| 4 | Lysis Buffer | Tris–HCl (pH 8.0) | 100 mM |
| Sodium Chloride | 1.4 M | ||
| EDTA disodium dihydrate | 20 mM | ||
| Hexadecyltrimethylammonium bromide (CTAB) | 2% w/v | ||
| Polyvinyl pyrrolidone | 2% w/v | ||
| β-Mercaptoethanol | 0.2% v/v | ||
| 5 | 3 M Sodium Acetate | Sodium acetate trihydrate | 102.025 g |
| Glacial acetic acid | Adjust pH to 5.2 | ||
| Distilled Water | Adjust volume to 250 mL | ||
| 6 | TE Buffer | 1 M Tris Cl | 0.5 mL |
| 0.5 M EDTA | 100 µL | ||
| Distilled Water | 50 mL |
Qiagen DNeasy® Blood and Tissue Kit (Cat No. 69504)
Extraction of DNA was performed according to the manufacturer’s protocol with the following minor modifications. To begin with, samples were kept for overnight digestion, since foot tissues of snails do not get entirely lysed within a few hours. In addition, in the final step, DNA was eluted twice by adding 50 µL of Buffer AE instead of 200 µL to obtain a higher final concentration of DNA.
The extracted DNA samples were measured quantitatively using a spectrophotometer. All the successfully extracted samples (A260/280 ≥ 1.80) were amplified with Qiagen® Taq PCR core kit (Cat No. 201225) using 28S (Wade et al. 2006) and 16S rRNA (Palumbi et al. 2002) primers (Table S1) and sequenced. Both modified CTAB and kit extracted DNA, though comparable in quality (Table S3), differed significantly in their consistency of repeated amplification. Despite showing a high A260/280 value, the results of enzymatic amplification of kit extracted DNA could not be reliably reproduced. For instance, if a kit extracted DNA sample was amplified five times using a set of taxon-specific primers, consistent amplification was observed not more than twice. On the other hand, if the same sample was extracted using CTAB buffer, it amplified at least four out of five times. In addition, most samples extracted using DNA extraction kit either could not be amplified or gave very poor quality sequences. This efficiency of the CTAB extraction protocol was consistent for both freshly extracted (few days old) as well as stored DNA samples (over 5 years). A possible reason for this hindrance could be the presence of polyphenols.
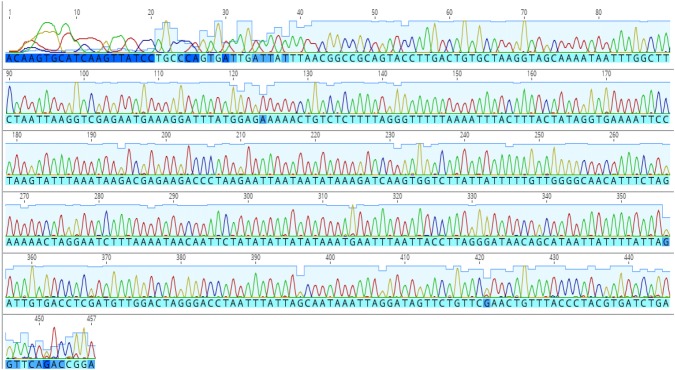
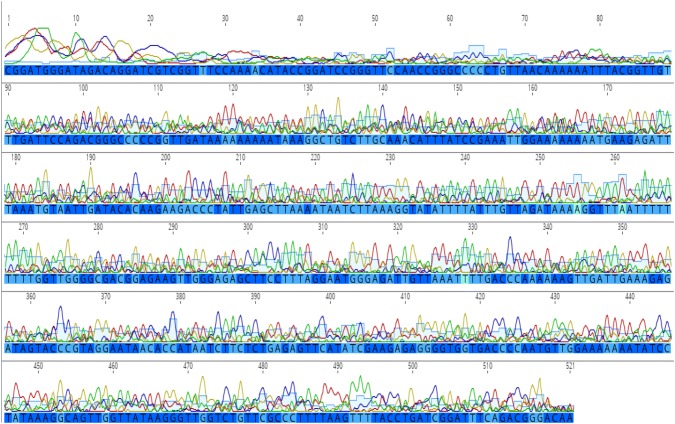
Algal association has not only been studied for marine gastropods (Geiselman and McConnell 1981; Steinberg 1988) but the herbivorous and algivorous nature of both terrestrial and freshwater snails are also well documented (Mason 1970; Butler 1976; CARTER et al. 1979; Brönmark 1989; Speiser and Rowell-Rahier 1991; Vaughn et al. 1993; Martin 2000; March et al. 2002; Chase and Knight 2006; Fink and Von Elert 2006). We have observed diatoms in the gut of Cremnoconchus sp. (Littorinidae). The preference of land snails to thrive on extremely moist areas rich in algae and considerable algal growth on the shells of most freshwater snails further strengthen this argument. This makes the presence of polyphenols in the snail tissues all the more likely. In the case of bigger animals, when only foot tissue was used for DNA extraction, the consistency of amplification was comparatively better than smaller animals, where the entire body had to be used (pers. obs.). Although the quality of extracted DNA was similar in both the cases, successful amplification and reproducibility of obtained results was the hurdle. The quality of DNA (A260/280) was not always indicative of its amplification capabilities. This is evident from chromatograms in Figs. 1 and 2 which clearly depict the quality difference between sequences of CTAB and kit extracted DNA, respectively. In addition, a comparison of average quality of sequences obtained by amplification of CTAB and kit extracted DNA is illustrated in Fig. 3. This could be attributed to the presence of increased amounts of polyphenols in the gut compared to the external foot tissue which further validates this hypothesis.
Fig. 1.
Chromatogram depicting 16S gene sequence quality in modified CTAB extracted DNA
Fig. 2.
Chromatogram depicting 16S gene sequence quality in Qiagen® DNeasy Blood and Tissue Kit extracted DNA
Fig. 3.
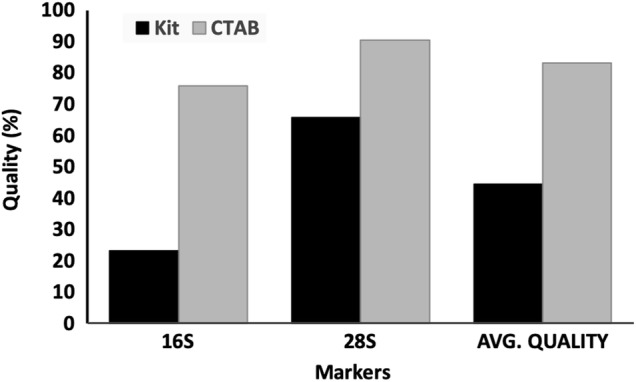
Comparison of average sequence quality between CTAB and Qiagen® DNeasy Blood and Tissue Kit extracted amplified genomic DNA
Most studies that specifically deal with DNA extraction from molluscs, almost always mention the problems of co-precipitation of mucopolysaccharides with DNA (Sokolov 2000; Skujiene and Soroka 2003; Huelsken et al. 2011; Poonam et al. 2013; Jaksch et al. 2016) but exclude the polyphenolic impurities. The prominent association of algal cells and consecutively polyphenols with snails has, invariably, been overlooked. We attribute the hindrance of inconsistent amplification to the presence of polyphenols, since the main difference between the modified CTAB extraction protocol used and the conventional method employed for animal DNA extraction was the inclusion of polyvinylpyrrolidone (PVP). Utility of PVP in plant DNA extraction has been extensively studied and its role has been identified to be the removal of polyphenols (John 1992; Porebski et al. 1997). Efficacy of PVP utilization in genomic DNA extraction of marine gastropods has also been reported earlier (Williams et al. 2003). The same proved to be true across several non-marine gastropod families included in the study. The differences between the methods discussed here have been summarized in Table 2.
Table 2.
Comparison between the two methods of DNA extraction
| S. no. | Comparison criteria | Modified CTAB | DNeasy kit |
|---|---|---|---|
| 1 | Total number of samples | 23 | 23 |
| 2 | Successfully extracted samples (A260/280 ≥ 1.90) | 19 (82%) | 13 (56%) |
| 3 | Time involved | About 15–16 h (including overnight digestion time and the time needed for evaporating excess alcohol from the pellet) | About 12 h (including overnight digestion time) |
| 4 | Cost per sample | < ₹ 100 | ₹ 250 |
The conventional CTAB DNA extraction method was also implemented for extraction of genomic DNA from gastropod molluscs, wherein the lysis buffer used consisted of CTAB, and β-mercaptoethanol (data not included). It was treated with sodium chloride (NaCl) post overnight digestion with the lysis buffer. However, the quality and quantity of DNA obtained was extremely poor. As an additional measure, the extracted DNA was kept in isopropanol overnight to improve quality. Amplifiable genomic DNA still could not be extracted. This scenario changed only with the inclusion of PVP in the lysis buffer. On addition of PVP, the second overnight treatment with isopropanol was not required. The only difference between the two protocols employed, reinforce the crucial part played by PVP in removal of polyphenolic impurities rendering the extracted DNA suitable for downstream reactions.
An added advantage of this method is the requirement of only a small amount of foot tissue and not an internal organ like liver in case of bigger animals. Unless there is a requirement for extensive morphological and anatomical studies, our modified protocol opens options for non-lethal sampling, wherein only a small part of the foot tissue is cut without the necessity of whole animal preservation. This method of sampling is especially handy when working in protected areas, since disturbance is minimal.
A few studies have mentioned a possible partial degradation of plant DNA in the presence of CTAB during extraction (Fang et al. 1992; Rowland 1993). However, in the case of snails, no such issue was encountered. On the contrary, CTAB extracted DNA was found to be very stable and could be used for amplification even after about 2 years of its extraction. Another possible shortcoming of the CTAB method could be the time and labor involved, given specialized commercially available kits like E.Z.N.A® Mollusc DNA kit which require lesser time and energy. However, the aforementioned kit is considerably expensive and overlooks the fact that polyphenolic compounds also hinder amplification and not mucopolysaccharides alone. Apart from being a relatively cheap option, our modified CTAB protocol saves the cost and effort of repeated amplification which becomes extremely important in the long run. The difference in the cost of modified CTAB extraction and kit extraction of DNA in conjunction with its added efficiency may be useful for labs with access to limited resources. The chemicals used in the CTAB extraction method are readily available as opposed to the highly specialized kits which are not always an easy option especially for projects with limited funding. In summary, this protocol can be used across non-marine molluscan taxa to successfully obtain high-quality genomic DNA without hindrance by polysaccharide or polyphenolic impurities rendering a high degree of success and consistency to enzymatic amplification reactions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors would like to thank Department of Science and Technology, Government of India (Grant No. EMR_2015/000199/v1/21385) for funding. We would also like to thank Dr. Suzanne Williams, Dr. Praveen Karanth, Dr. Maitreya Sil for help in developing the protocol.
References
- Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranishi F, Okimoto T. A simple and reliable method for DNA extraction from bivalve mantle. J Appl Genet. 2006;47:251–254. doi: 10.1007/BF03194632. [DOI] [PubMed] [Google Scholar]
- Brönmark C. Interactions between epiphytes, macrophytes and freshwater snails: a review. J Molluscan Stud. 1989;55:299–311. doi: 10.1093/mollus/55.2.299. [DOI] [Google Scholar]
- Butler AJ. A Shortage of Food for the Terrestrial Snail Helicella virgata in South Australia. Oecologia. 1976;25:349–371. doi: 10.1007/BF00345608. [DOI] [PubMed] [Google Scholar]
- Carter MA, Jeffery RCV, Williamson P. Food overlap in co-existing populations of the land snails Cepaea nemoralis (L.) and Cepaea hortensis (Müll) Biol J Linn Soc. 1979;11:169–176. doi: 10.1111/j.1095-8312.1979.tb00033.x. [DOI] [Google Scholar]
- Chase JM, Knight TM. Effects of eutrophication and snails on Eurasian watermilfoil (Myriophyllum spicatum) invasion. Biol Invasions. 2006;8:1643–1649. doi: 10.1007/s10530-005-3933-7. [DOI] [Google Scholar]
- Cheung WY, Hubert N, Landry BS. A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. Genome Res. 1993;3:69–70. doi: 10.1101/gr.3.1.69. [DOI] [PubMed] [Google Scholar]
- Dowell K (2008) Molecular phylogenetics: an introduction to computational methods and tools for analyzing evolutionary relationships. Mol Phylogenetics, pp 1–19. http://www.math.umaine.edu/~khalil/courses/MAT500/papers/MAT500_Paper_Phylogenetics.pdf. Accessed 22 Jan 2020
- Fang G, Hammar S, Grumet R. A quick and inexpensive method for removing polyssaccharides from plant genomic DNA. Biotechniques. 1992;13:52–56. doi: 10.1080/10643389.2012.728825. [DOI] [PubMed] [Google Scholar]
- Fink P, Von Elert E. Physiological responses to stoichiometric constraints: nutrient limitation and compensatory feeding in a freshwater snail. Oikos. 2006;115:484–494. doi: 10.1111/j.2006.0030-1299.14951.x. [DOI] [Google Scholar]
- Geiselman JA, McConnell OJ. Polyphenols in brown algae Fucus vesiculosus and Ascophyllum nodosum: chemical defenses against the marine herbivorous snail, Littorina littorea. J Chem Ecol. 1981;7:1115–1133. doi: 10.1007/BF00987632. [DOI] [PubMed] [Google Scholar]
- Giribet G. Molecules, development and fossils in the study of metazoan evolution; Articulata versus Ecdysozoa revisited. Zoology. 2003;106:303–326. doi: 10.1078/0944-2006-00131. [DOI] [PubMed] [Google Scholar]
- Huelsken T, Schreiber S, Hollmann M. COI amplification success from mucus-rich marine gastropods (Gastropoda: naticidae) depends on DNA extraction method and preserving agent. Mitteilungen der Dtsch Malakozool Gesellschaft. 2011;85:17–26. [Google Scholar]
- Jaksch K, Eschner A, Rintelen TV, Haring E. DNA analysis of molluscs from a museum wet collection: a comparison of different extraction methods. BMC Res Notes. 2016;9:1–12. doi: 10.1186/s13104-016-2147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acids Res. 1992;20:2381. doi: 10.1093/nar/20.9.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio P, Goldman N. Models of molecular evolution and phylogeny. Genome Res. 1998;8:1233–1244. doi: 10.1101/gr.8.12.1233. [DOI] [PubMed] [Google Scholar]
- March JG, Pringle CM, Townsend MJ, Wilson AI. Effects of freswater shrimp assemblages on benthic communities along an altitudinal gradient of a tropical stream. Freshw Biol. 2002;47:377–390. doi: 10.1046/j.1365-2427.2002.00808.x. [DOI] [Google Scholar]
- Martin SM. Terrestrial snails and slugs (Mollusca: Gastropoda) of maine. Northeast Nat. 2000;7:33–88. doi: 10.1656/1092-6194(2000)007[0033:TSASMG]2.0.CO;2. [DOI] [Google Scholar]
- Mason CF. Food, feeding rates and assimilation in woodland snails. Oecologia. 1970;4:358–373. doi: 10.1007/BF00393394. [DOI] [PubMed] [Google Scholar]
- Palumbi S, Martin A, Romano S et al (2002) The simple fool’s guide to PCR. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii
- Pereira JC, Chaves R, Bastos E, et al. An efficient method for genomic DNA extraction from different molluscs species. Int J Mol Sci. 2011;12:8086–8095. doi: 10.3390/ijms12118086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poonam Tripathi NK, Kaur P, Kumari A. Optimization of DNA isolation and PCR protocol for rapd analysis from mucopolysaccharide rich foot muscles of Lymnaea acuminata (Gastropoda: Pulmonata) from Indian Himalayan regions. Int J Zool Res. 2013;3:35–42. [Google Scholar]
- Popa OP, Murariu D, Popa LO. Comparison of four DNA extraction methods from invasive freshwater bivalve species (Mollusca, Bivalvia) in Romanian Fauna. Trav du Muséum Natl d’Histoire Nat «Grigore Antipa». 2007;50:527–536. [Google Scholar]
- Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- Rowland LJ. Use of polyethylene glycol for purification of DNA from leaf tissue of woody plants. BioTechniques. 1993;14:735–736. [PubMed] [Google Scholar]
- Sidow A, Bowman BH. Molecular phylogeny. Curr Opin Genet Dev. 1991;1:451–456. doi: 10.1016/S0959-437X(05)80191-1. [DOI] [PubMed] [Google Scholar]
- Skujiene G, Soroka M. A comparison of different DNA extraction methods for slugs (Mollusca: Pulmonata) Ekologija. 2003;1:12–16. [Google Scholar]
- Sokolov EP. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J Molluscan Stud. 2000;66:573–575. doi: 10.1093/mollus/66.4.573. [DOI] [Google Scholar]
- Speiser B, Rowell-Rahier M. Effects of food availability, nutritional value, and alkaloids on food choice in the generalist herbivore Arianta arbustorum (Gastropoda: Helicidae) Oikos. 1991;62:306–318. doi: 10.2307/3545495. [DOI] [Google Scholar]
- Steinberg PD. Effects of quantitative and qualitative variation in phenolic compounds on feeding in three species of marine invertebrate herbivores. J Exp Mar Bio Ecol. 1988;120:221–237. doi: 10.1016/0022-0981(88)90003-2. [DOI] [Google Scholar]
- Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- Vaughn CC, Gelwick FP, Matthews WJ. Effects of algivorous minnows on production of grazing stream invertebrates. Oikos. 1993;66:119–128. doi: 10.2307/3545204. [DOI] [Google Scholar]
- Wade CM, Mordan PB, Naggs F. Evolutionary relationships among pulmonate land snails and slugs (Pulmonata. Stylommatophora) Biol J Linn Soc. 2006;87:593–610. doi: 10.1111/j.1095-8312.2006.00596.x. [DOI] [Google Scholar]
- Williams ST, Reid DG, Littlewood DTJ. A molecular phylogeny of the Littorininae (Gastropoda: Littorinidae): Unequal evolutionary rates, morphological parallelism, and biogeography of the Southern Ocean. Mol Phylogenet Evol. 2003;28:60–86. doi: 10.1016/S1055-7903(03)00038-1. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx B, Backeljau T, De Wachter R. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993;9:407. doi: 10.1016/0168-9525(93)90102-N. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian species delimitation using multilocus sequence data. Proc Nat Acad Sci. 2010 doi: 10.1073/pnas.0913022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue GH, Orban L. A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis. 2005;26:3081–3083. doi: 10.1002/elps.200410411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.