Abstract
Background:
Usually, chemoradiotherapy can be used for the treatment of locally advanced colorectal cancer (CRC) before surgery. On the other hand, some studies have shown that fractional radiation of tumor cells leads to chemoresistance. The aim of this study was to evaluate the chemoresistance of radioresistant sub-line (RR sub-line)
Methods:
This study was done in Hamadan University of Medical Sciences in 2017-2018. MTT assay and sub-G1 fraction analysis by flow cytometry were used to evaluate cross-resistance of RR sub-line to gefitinib and regorafenib. Real-time PCR was used to investigate the role of four miRNAs and their target genes in the cross-resistance of RR sub-line. The t test and repeated measures test were used for the assessment of statistical significance between groups
Results:
The IC50 of gefitinib and regorafenib for RR sub-line were significantly higher than those of the parental cell line. On the other hand, the resistance index of RR sub-line for gefitinib and regorafenib were 1.92 and 1.44, respectively. The sub-G1 fraction of RR sub-line following treatment with gefitinib and regorafenib was significantly lower than that of the parental cell line (P=0.012 and P=0.038, respectively). The expression of miR-9, Let-7e, and Let-7b in RRsub-line was significantly lower than that of the parental cell line. However, NRAS, IGF1R, NFKB1, and CCND1 found to be upregulated in RR sub-line in comparison with the parental cell line
Conclusion:
We can conclude that the acquired RR sub-line was cross-resistance to gefitinib and regorafenib. Furthermore, miR-9/NFKB1, let-7b/CCND1, let-7e/NRAS, and IGF1R played essential roles in the chemoradioresistance of CRC
Keywords: Colorectal neoplasms, Drug resistance, MicroRNAs, gefitinib, regorafenib
What’s Known
Chemoresistance of colorectal cancer occurs as the result of genetic and epigenetic modifications of genes that are crucial for colorectal cancer cells’ response to therapeutic agents.
Dysregulation of miRNAs, as the main components of the epigenome, regulates the resistance of colorectal cancer to several therapeutic drugs.
What’s New
The acquired radioresistant sub-line was cross-resistant to gefitinib and regorafenib.
MiR-9/ NFKB1, let-7b /CCND1, let-7e/ NRAS, and IGF1R played essential roles in chemoradioresistance of colorectal cancer.
Introduction
Colorectal cancer (CRC) is one of the most widespread malignancies worldwide. There are different methods of treatment for CRC including surgery, radiation therapy, and chemotherapy. 1 The recent use of targeted therapeutic drugs such as multi-tyrosine kinase inhibitors (regorafenib and Sorafenib) and epidermal growth factor receptor (EGFR) inhibitors (gefitinib and Erlotinib) has improved CRC outcome. 2 , 3 As a Sorafenib analog, regorafenib suppresses numerous kinases such as vascular endothelial growth factor receptors (VEGFRs), fibroblast growth factor receptor 1 (FGFR1), tyrosine kinase with immunoglobulin and epidermal growth factor homology domain 2 (TIE2), platelet-derived growth factor receptor beta (PDGFR-β), KIT proto-oncogene receptor tyrosine kinase (KIT), ret proto-oncogene (RET), and B-Raf Proto-Oncogene, Serine/Threonine Kinase. Therefore, regorafenib suppresses the metastasis, angiogenesis, and growth in colorectal tumors. 4 gefitinib inhibits the kinase activity of EGFR by binding to its intracellular domain. However, blockade of EGFR by gefitinib leads to the inhibition of CRC growth mediated by downstream signaling pathway of EGFR. 5
Chemotherapy and radiotherapy are the two main therapeutic methods that are often combined for the treatment of locally-advanced rectal cancers prior to surgery. Neoadjuvant chemoradiotherapy leads to tumor downstaging; therefore, it increases the efficiency of treatment. 6 Unfortunately, most colorectal tumors are intrinsically resistant to chemotherapeutic drugs and radiation. Furthermore, fractionated radiation during radiotherapy may lead to acquired chemoresistance. 7
Recent studies have shown that chemoresistance of tumor cells originates from different processes including enhanced DNA repair capability, reduced drug absorption, modifications of intracellular targets of anticancer drugs, cell metabolism alteration, and apoptosis inactivation. Nevertheless, based on the findings of recent studies, another proposed mechanism for chemoresistance of CRC could be cancer stem cell (CSC). 8
Moreover, numerous studies have shown that chemoresistance of CRC could happen as the result of genetic and epigenetic modifications of genes that are crucial for CRC cells’ response to therapeutic agents. 9 Micro RNAs (miRNAs), as the main components of the epigenome, control several biological processes including proliferation, invasion, apoptosis, survival, and metastasis. MiRNAs are small non-coding RNAs (19–24 nt) that downregulate protein-coding genes post-transcriptionally by binding to their 3’-untranslated region (3’ UTR). 10 Down regulation or upregulation of miRNAs, which function as a tumor suppressor or oncogene, regulates the resistance of CRC to therapeutic drugs. 11
Based on the evidence of cross-resistance of radioresistant tumor cells to chemotherapeutic agents, we hypothesized that the acquired radioresistant CRC cell lines also would not respond to chemotherapeutic agents. In order to evaluate this hypothesis, we established a radioresistant sub-line (RR sub-line) and evaluated the cross-resistance of this cell line to gefitinib and regorafenib. In spite of the progressive studies on the molecular mechanism of chemoresistance of CRC, the accurate role of miRNAs, as a critical epigenetic factor in cross-resistance of CRC, is still unclear. Therefore, based on the results of similar studies, some miRNAs that were significantly dysregulated after acute radiation exposure were selected for further evaluation. These miRNAs included: MiR-9 (Accession number: MIMAT0000441), 12 miR625 (Accession number:MIMAT0003294), 13 let-7e (Accession number: MIMAT0000066), 14 and let-7b (Accession number: MIMAT0000063). 15
The aim of this study was to estimate the resistance index of RR sub-line. Moreover, the study was aimed to evaluate the expression level of four miRNAs, including miR-9, miR-625, let-7b, and let-7e and their candidate target genes in RR sub-line and parental cells.
Materials and Methods
This study was done in the Research Center for Molecular Medicine, Hamadan University of Medical Sciences, 2017-2018.
Cell Lines
The CRC cell line HCT116 was obtained from Pasteur Institute, Iran. The radioresistant CRC cell line (RR-HCT116) was established by fractional radiation. The radioresistance of RR sub-line had already been validated in our previous study. 16 The cell lines were cultured in DMEM medium (Gibco, USA) with 10% FBS (Gibco, USA) and 1% Penicillin-Streptomycin (Gibco, USA). These cells were incubated at 37 °C, with high relative humidity and in 5% CO2.
Drugs
regorafenib and gefitinib were obtained from Shaanxi YuanTai Biological Technology Co. (China), Ltd, and Santa Cruz Biotechnology Inc. (USA), respectively. Each drug was dissolved in DMSO (Sigma, USA) to make a 4mM stock solution.
MTT Assay
3-(4, 5-dimethylthiazol-2-yl)-2, 5- diphenyltetrazolium bromide (MTT) assay was used to evaluate the cytotoxicity effects of gefitinib and regorafenib on RR and parental HCT116 cell line. Briefly, 4×103 cells were seeded in each well of a 96-well plate and incubated overnight. Afterward, cells were treated with different concentrations of each drug (the final volume of each well was 200 µL) for 72 hours. Then 10 μl of MTT solution (5 mg/mL) was added to each well and the plate was incubated for 4 hours at 37 °C. After removing the medium and adding DMSO for dissolving formazan crystals, the absorbance was measured at 570 nm by a microplate reader. The IC50 values of gefitinib and regorafenib for each cell were calculated using Compusyn software based on the following equation:
log (fa/fu)=m log (D)-m log (Dm) 17
Sub-G1 Fraction Analysis
Fluorochrome propidium iodide (PI) can bind and label DNA. Therefore, PI can be used for the quick and precise evaluation of cellular DNA content and subsequent identification of sub-G1 fraction as an apoptotic population of cells that are characterized by DNA fragmentation. 18 Parental and RR cells were seeded at a density of 4×105 cell per well of a 6-well plate. Cells were treated with gefitinib (final concentration 0 and 10 µM), regorafenib (final concentration 0 and 45 µM) for 72 hours. Afterward, the wells were washed, fixed, and stained using methods suggested by Pozarowski and Darzynkiewicz, which is described in our previous study. 19 Briefly, cells were stained with 50 µg/ml Propidium iodide (Sigma, USA) solution containing 100 µg/ml RNaseA (Sigma, USA). Following incubation at 37 °C for 20 min, fluorescence emission was analyzed by a flow cytometer (Partec GmbH, Munster, Germany).
MicroRNA Target Prediction
In this study, we used microT CDS (http://diana.imis.athena-innovation.gr), Rna22 (https://cm.jefferson.edu/rna22/Interactive/), RNAhybrid (https://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid), and Miranda (http://cbio.mskcc.org/) softwares for the target prediction of mir-9, let-7e, let-7b, and mir-625. We selected the target genes with high prediction scores, which could play an essential role in apoptosis, cell cycle control, and proliferation.
Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
Total RNA extraction was performed by RNX-Pluskit (CinnaGen, Tehran, Iran) based on the suggested protocol by the manufacturer. The quality of extracted RNA was verified by agarose gel electrophoresis. Purity (OD 260/280 nm) and concentration of RNA were evaluated by NonoDrop (Thermo Fisher Scientific, Waltham, Massachusetts, United States). CDNA for miRNA expression analysis was synthesized using miRCURY LNA™ Universal RT cDNA Synthesis Kit (Exiqon, Vedbaek, Denmark); cDNA for mRNA expression analysis was synthesized using First Strand cDNA synthesis Kit (Fermentas, Waltham, Massachusetts, United States) according to the manufacturer’s recommendations. The miRNAs and mRNAs expression levels were evaluated using Roche LightCycler® 96 system (Roche Diagnostic, Mannheim, Germany). ExiLENT SYBR® Green master mix (Exiqon, Vedbaek, Denmark) and miRNA specific LNA™ PCR primers (Exiqon, Vedbaek, Denmark) for each miRNA and U6 as a reference gene were used for the amplification of miRNAs. SYBR® Premix ExTaq™ II Kit (Takara, Japan), specific primer pairs of each target gene, and beta-actin (ACTB), as a reference gene, were used for the amplification of target genes mRNA. Primer pairs were designed by Alleleid 6 and the specificity of each primer pair was evaluated by NCBI primer blast (table 1). The measurement of real-time PCR efficiency for each gene was close to 100%; therefore, we used the method for relative gene expression. 16
Table1.
Primer pairs sequences of genes used in the qRT-PCRa
| Gene | Gene ID | Forward primer | Reverse primer |
|---|---|---|---|
| NRAS1 | 4893 | TATTCATCTACAAAGTGGTTCTGG | CGGCTGTGGTCCTAAATCTG |
| NFKB1 | 4790 | GAAGGTGGATGATTGCTAAG | TGCTGGAGTTCAGGATAAC |
| IGF1R | 3480 | CGGTAATAGTCTGTCTCATAG | GCCAATAAGTTCGTCCAC |
| CCND1 | 595 | TTCTGTTCCTCGCAGACCTCC | CGATGCCAACCTCCTCAACG |
| ACTB | 60 | AAGATCAAGATCATTGCT | TAACGCAACTAAGTCATA |
Quantitative reverse transcription PCR
Statistical Analysis
All statistical analyses were done using SPSS software, version 16.0. Significant differences between the mean of the two groups were determined using independent sample t test and repeated measures test (by considering the normal distribution data based on the Kolmogorov-Smirnov test). P<0.05 was considered statistically significant.
Results
Evaluation of Cross-Resistance of RR Cell Sub-Line by MTT Assay
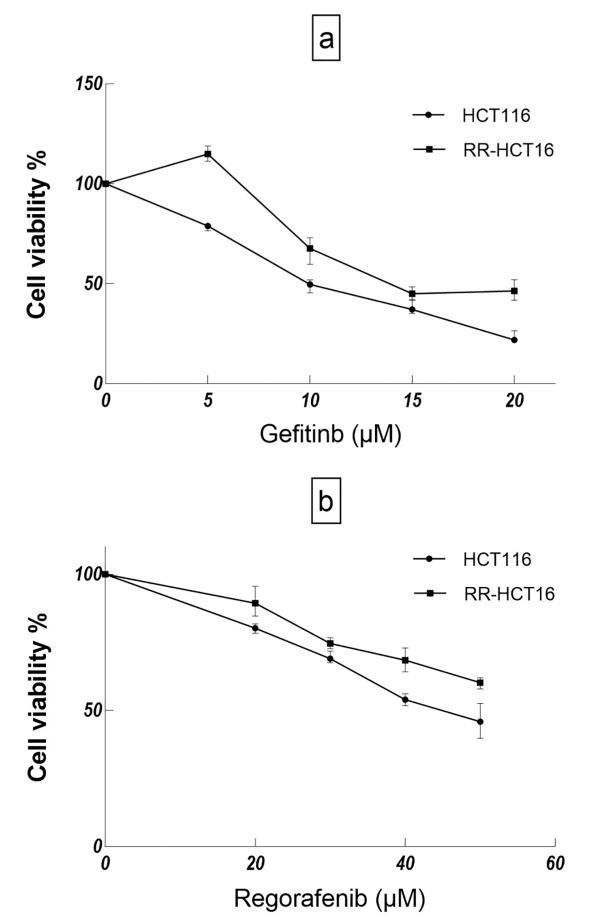
The cross-resistance of RR sub-line to gefitinib and regorafenib was evaluated by MTT assay. As could be seen in figure 1, the viability of RR sub-line after treatment with different concentrations of gefitinib (5-20 µM) is significantly higher than that of the parental cell line (P<0.001 for 5, 10, 15, and 20 µM, respectively). Similarly, the viability of RR sub-line after treatment with different concentrations of regorafenib (20-50 µM) is significantly higher than that of the parental cell line (P<0.001 for 20, 30, 40, and P=0.015 for 50 µM, respectively); the P values of repeated measures test for gefitinib and regorafenib were 6.64*10-14 and 0.000039, respectively. Resistance index (RI, IC50 drug dose in RR-HCT 116/drug dose in HCT 116 of RR sub-line for gefitinib and regorafenib were 1.92 and 1.44, respectively. 20 Furthermore, the IC50 of gefitinib and regorafenib for RR sub-line were higher than those of the parental cell line (table 2).
Figure1.
Growth inhibitory effects of gefitinib and regorafenib on RR-HCT116 and parental cell line were evaluated by MTT assay. a) The cell viability% of HCT116 cells was significantly lower than RR-HCT116 under treatment with different doses of gefitinib. b) The cell viability% of HCT116 cells was significantly lower than RR-HCT116 under treatment with different doses of regorafenib.
Table2.
Results of cytotoxicity for gefitinib and regorafenib in RR-HCT116 and parental HCT116
| Drug | IC50 (µM) | Resistance index b | |
|---|---|---|---|
| RRa sub-line | HCT116 | ||
| gefitinib | 19.53±0.98 | 10.16±0.21 | 1.92 |
| regorafenib | 64.99±1.32 | 44.95±1.92 | 1.45 |
Radioresistant;
Ratio of IC50 drug dose in resistant sub-line to that in parental
Apoptosis Assay
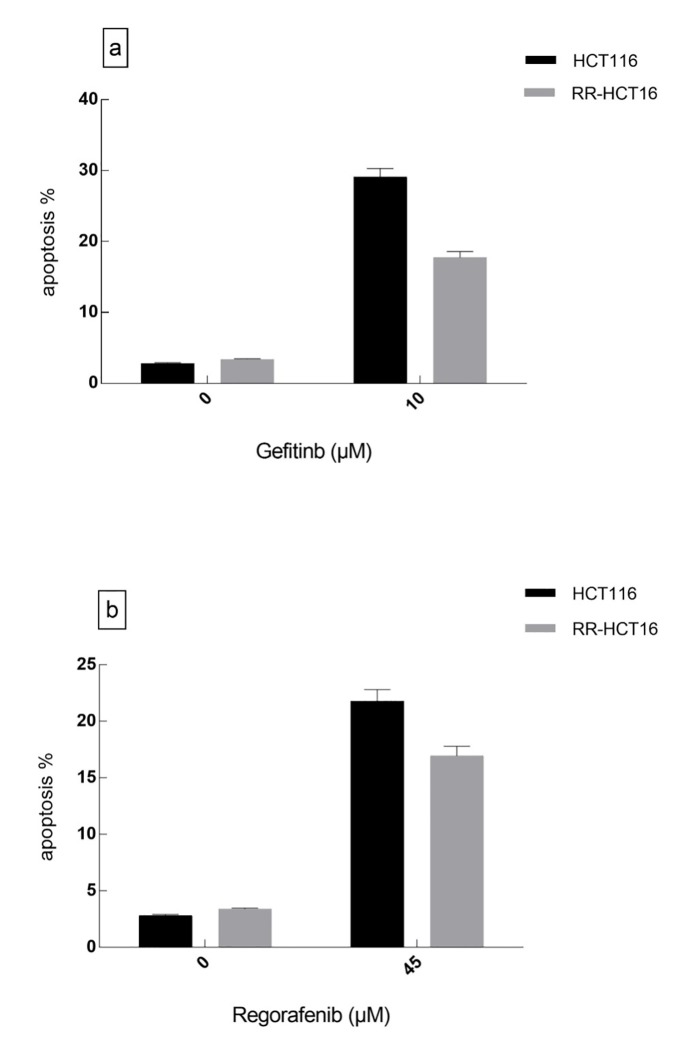
As shown in figure 2, the sub-G1 fraction of RR sub-line at 72 hours following treatment with gefitinib (10 µM) and regorafenib (45 µM) was significantly lower than that of the parental cell line (P=0.012 and P=0.038, respectively).
Figure2.
Sub-G1 fractions of RR-HCT116 and parental cell line under treatment with gefitinib and regorafenib were evaluated by flow cytometry. a) The gefitinib induced more apoptosis in HCT116 rather than RR-HCT116 cells (P=0.012). b) regorafenib induced more apoptosis in HCT116 rather than RR-HCT116 cells (P=0.038).
In Silico miRNA Target Prediction Results
MiRNA target prediction was performed by four different online softwares with a specific prediction algorithm. In miRanda, Diana-microT, and RNAhybrid softwares, complementarity, and free energy binding were accounted for final score calculation. In Rna22 software, pattern recognition and folding energy were accounted for final score calculation. 21
The candidate target of each miRNA was selected based on the software prediction score and the role each miRNA played in chemoradioresistance (table 3). In summary, IGF1R (insulin-like growth factor 1 receptor), nuclear factor-kappa B1 (NFKB1), CCND1 (Cyclin D1), and NRAS (neuroblastoma RAS viral oncogene homolog) were selected as predicted targets of miR-625, miR-9, let-7b, and let-7e, respectively. These miRNA targets were validated by real-time PCR.
Table3.
Results of prediction scores of miRNA target genes
| miRanda(mirSVRaScore) | Rna22 (folding energy for heteroduplex (Kcal/mol)) | DIANA microTb-CDSc(miTGd Score) | RNAhybrid 2.2 (mfe)e | |
|---|---|---|---|---|
| miR-625/IGF1R | -0.1746 | -17.20 | 9.00 | -30.8 |
| miR-9/NFKB1 | -0.4032 | -15.70 | 5.52 | -28.2 |
| Let-7b/CCND1 | -0.0278 | -15.60 | 24.86 | -29.2 |
| Let-7e/NRAS | -0.2772 | -14.50 | 15.97 | -26.4 |
miRNA support vector regression
MicroRNA target
Coding sequence
miRNA target gene
Mean free energy
The Association of Cross-Resistance of CRC Cell Line with Dysregulation of MiR-9, MiR-625, Let-7e, and Let-7b
Considering the insignificant difference in U6 expression between parental HTC116 and RR-HTC116, U6 was used as a reference gene for the normalization of each miRNA. figure 3a shows the expression level of miRNAs. The expressions of MiR-9, let-7e, and let7b were significantly lower (P=0.005, P=0.031, and P=0.028) in RR, which was cross-resistant to regorafenib and gefitinib than in parental cell line (-2.02, -1.74, and -1.80-fold, respectively).
Figure3.
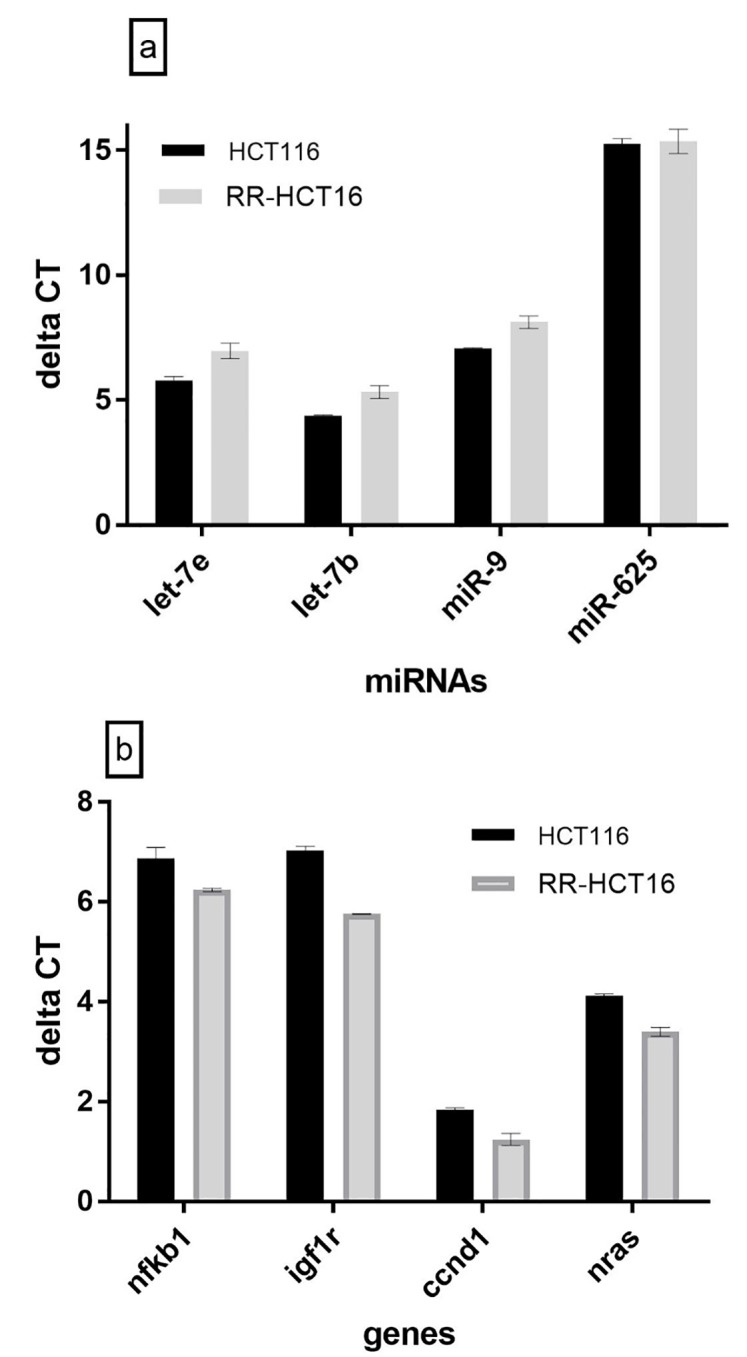
miRNAs and target genes expression levels on RR-HCT116 and parental cell line were evaluated by real-time PCR. a) The expression levels of miR-9, let-7b, and let 7ein RR-HCT116 sub-line were significantly lower than HCT-116 cell line (P=0.005, P=0.028, and P=0.031). b) The expression levels of nfkb1, igf1r, ccnd1, nras in RR-HCT116 sub-line were significantly higher than HCT-116 cell line (P=0.045, P=0.003, P=0.011, and P=0.033). Delta CT values have a reverse relationship with miRNAs and target genes expression levels.
MiRNAs Target Genes Validation by Real-Time PCR
The evaluation of beta-actin expression showed that there was no significant difference in the expression of this gene between the two cell lines. Therefore, beta-actin was used as a reference gene. As seen in figure 3b, the expressions of CCND1, NRAS, IGF1R, and NFKB1 were significantly higher (P=0.011, P=0.033, P=0.003, and P=0.045) in RR than in the parental cell line (1.51, 1.65, 2.42, and 1.55-fold, respectively).
Discussion
The results of the current study indicated that the RR sub-line had higher viability after treatment with different concentrations of gefitinib and regorafenib than the parental cell line. On the other hand, the IC50 of gefitinib and regorafenib for RR sub-line were higher than those of parental cell line. The results of the first study conducted in this field by Moulder and colleagues indicated that the acquired radioresistance of tumor cells due to fractionated radiation-induced chemoresistance. 7 In their study, Servidei and colleagues showed that cisplatin-resistant tumor cells were cross-resistant to other therapeutic agents such as etoposide and carboplatin. 22 On the other hand, the results of a study done by Mutlu and colleagues showed that multidrug-resistant myeloma cell lines were cross-resistant to cobalt-60 (γ radiation). 23 These results are consistent with the results of the present study, which showed that the acquired radioresistant cells were cross-resistant to regorafenib and gefitinib.
Furthermore, in the present study, the apoptotic percentage of RR sub-line following treatment with gefitinib and regorafenib was significantly lower than that of the parental cell line. The findings of recent studies have indicated that escaping apoptosis is one of the crucial mechanisms of tumor cell resistance to therapeutic agents. 24 Thus, the cross-resistance of radioresistant cells to gefitinib and regorafenib may be due to the evasion from apoptosis.
In the present study, let-7e and let-7b were significantly downregulated in the cross-resistant sub-line compared with the parental cell line. Let-7 family of miRNA is involved in the chemosensitivity of several cancers. 25 Shimizu and colleagues showed that let-7 family could sensitize hepatocellular carcinoma cell line to sorafenib (analog of regorafenib) by inducing apoptosis. 26 Stahlhut and colleagues have shown that synergic treatment with let-7b and miR-34 sensitizes non-small-cell lung cancer to erlotinib (analog of gefitinib). 27 In our study, NRAS and CCND1, targets of let-7e and let-7b, respectively, were significantly upregulated in the cross-resistant sub-line compared with the parental cell line. Deregulation of CCND1 as a proto-oncogene was associated with the resistance of tumor cells to different therapeutic agents such as gefitinib. 28 NRAS, as an oncogene, is a small GTPase protein that controls cellular processes such as proliferation, survival, migration, and invasion. 29 Milosevic and colleagues showed that RAS-MAPK-ERK signaling pathway inhibition could lead to the sensitization of tumor cells to therapeutic agents. 30 Eberlein and colleagues revealed that NRAS mutation was common in gefitinib-resistant cell lines. 31 Considering these results, we can conclude that let-7b and let-7e regulate the cross-resistance of RR sub-line by targeting CCND1 and NRAS, respectively. MiR-625, which suppresses proliferation, migration, and invasion of tumor cells, was downregulated in several types of cancers. 32 In our study, the expression of miR-625 was lower in cross-resistant sub-line than in the parental cell line; however, the difference was not significant. Salendo and colleagues showed that miR-625 expression level was lower in chemoradioresistant cell lines than in the sensitive cell lines. 33 MiR-625 targets IGF1R, a trans-membrane receptor tyrosine kinase that affects processes such as proliferation and apoptosis in signaling pathways, leading to chemoresistance of CRC. 34 The results of our study indicated that IGF1R was upregulated in the RRsub-line compared with the parental cell line. Findings from other similar studies show that IGF1R signaling pathway affects the sensitivity of tumor cells to EGFR inhibitors. 35 Therefore, IGF1R might regulate the cross-resistant of RR sub-line independent of miR-625 regulation. MiR-9, as a tumor suppressor miRNA, inhibits proliferation, invasion, and metastasis of gastrointestinal tumors. 36 The findings of our study indicated that the expression of miR-9 was significantly lower in cross-resistant cell line than in the parental cell line. Recent studies have revealed that miR-9 overexpression sensitizes tumor cells to several therapeutic agents. 37 As a target of miR-9, NFKB1 transcription factor regulates several genes that are involved in different critical mechanisms, such as survival, proliferation, and inflammation, in cellular pathways. 38 Our study indicated that NFKB1 was significantly upregulated in cross-resistant sub-line compared with the parental cell line. Andersen and colleagues have shown that NFKB1 overexpression NFKB1 can sensitize CRC to several therapeutic agents by regulating the expression of multidrug-resistant 1 (MDR1). 39 Therefore, miR-9, by targeting the NFKB1, plays an essential role in regulating the cross-resistance of CRC.
The present study, for the first time, investigated the role of miR-9, let-7b, and let-7e in cross-resistance of radioresistant CRC cells to gefitinib and regorafenib. The main limitation of this study is that the exact mechanism through which miR-9, let-7b, and let-7e regulate resistance to gefitinib and regorafenib was not identified.
Conclusion
In summary, we conclude that the acquired RR sub-line, established by fractionated radiation, is cross-resistant to chemotherapeutic agents including gefitinib and regorafenib. Further study is required to examine the cross-resistance of RR sub-line to other therapeutic agents. The results of our study also indicated that miR-9/NFKB1, let-7b/CCND1, let-7e/NRAS, and IGF1R had critical roles in the chemoradioresistance of CRC. However, further investigation of each miRNAs and their target genes by mimicking miRNA transfection is required to define these roles.
Acknowledgement
This study was part of a Ph.D. dissertation completed and defended by Saeid Afshar at Hamadan University of Medical Sciences. The study was funded by Vice-chancellor for Research and Technology, Hamadan University of Medical Sciences (No. 9412257365).
Conflict of Interest: None declared.
References
- 1.Geng L, Wang J. Molecular effectors of radiation resistance in colorectal cancer. Precis Radiat Oncol. 2017;1:27–33. doi: 10.1002/pro6.5. [DOI] [Google Scholar]
- 2.Bronte G, Rolfo C, Giovannetti E, Cicero G, Pauwels P, Passiglia F, et al. Are erlotinib and gefitinib interchangeable, opposite or complementary for non-small cell lung cancer treatment? Biological, pharmacological and clinical aspects. Crit Rev Oncol Hematol. 2014;89:300–13. doi: 10.1016/j.critrevonc.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Carr BI, Cavallini A, Lippolis C, D’Alessandro R, Messa C, Refolo MG, et al. Fluoro-Sorafenib (regorafenib) effects on hepatoma cells: growth inhibition, quiescence, and recovery. J Cell Physiol. 2013;228:292–7. doi: 10.1002/jcp.24148. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majithia N, Grothey A. regorafenib in the treatment of colorectal cancer. Expert Opin Pharmacother. 2016;17:137–45. doi: 10.1517/14656566.2016.1118054. [DOI] [PubMed] [Google Scholar]
- 5.Yar Saglam AS, Alp E, Elmazoglu Z, Menevse S. Treatment with cucurbitacin B alone and in combination with gefitinib induces cell cycle inhibition and apoptosis via EGFR and JAK/STAT pathway in human colorectal cancer cell lines. Hum Exp Toxicol. 2016;35:526–43. doi: 10.1177/0960327115595686. [DOI] [PubMed] [Google Scholar]
- 6.Williamson JS, Jones HG, Williams N, Griffiths AP, Jenkins G, Beynon J, et al. Extramural vascular invasion and response to neoadjuvant chemoradiotherapy in rectal cancer: Influence of the CpG island methylator phenotype. World J Gastrointest Oncol. 2017;9:209–17. doi: 10.4251/wjgo.v9.i5.209. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulder JE, Hopwood LE, Volk DM, Davies BM. Radiation induction of drug resistance in RIF-1: correlation of tumor and cell culture results. Int J Radiat Oncol Biol Phys. 1991;20:213–6. doi: 10.1016/0360-3016(91)90092-i. [DOI] [PubMed] [Google Scholar]
- 8.Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8:59950–64. doi: 10.18632/oncotarget.19048. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crea F, Nobili S, Paolicchi E, Perrone G, Napoli C, Landini I, et al. Epigenetics and chemoresistance in colorectal cancer: an opportunity for treatment tailoring and novel therapeutic strategies. Drug Resist Updat. 2011;14:280–96. doi: 10.1016/j.drup.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Singh PK, Campbell MJ. The Interactions of microRNA and Epigenetic Modifications in Prostate Cancer. Cancers (Basel) 2013;5:998–1019. doi: 10.3390/cancers5030998. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 12.Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG, Ru G, et al. Role of miR-100 in the radioresistance of colorectal cancer cells. Am J Cancer Res. 2015;5:545–59. [ PMC Free Article ] [PMC free article] [PubMed] [Google Scholar]
- 13.He J, Hua J, Ding N, Xu S, Sun R, Zhou G, et al. Modulation of microRNAs by ionizing radiation in human gastric cancer. Oncol Rep. 2014;32:787–93. doi: 10.3892/or.2014.3246. [DOI] [PubMed] [Google Scholar]
- 14.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–43. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed FE, Vos PW, Jeffries C, Wiley JE, Weidner DA, Mota H, et al. Differences in mRNA and microRNA microarray expression profiles in human colon adenocarcinoma HT-29 cells treated with either Intensity-modulated Radiation Therapy (IMRT), or Conventional Radiation Therapy (RT) Cancer Genomics Proteomics. 2009;6:109–27. [PubMed] [Google Scholar]
- 16.Khoshinani HM, Afshar S, Pashaki AS, Mahdavinezhad A, Nikzad S, Najafi R, et al. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Jpn J Radiol. 2017;35:664–72. doi: 10.1007/s11604-017-0679-y. [DOI] [PubMed] [Google Scholar]
- 17.Lombardo T, Anaya L, Kornblihtt L, Blanco G. Median effect dose and combination index analysis of cytotoxic drugs using flow cytometry. Flow Cytometry-Recent Perspectives. London : IntechOpen; 2012. [Google Scholar]
- 18.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1:1458–61. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 19.Pozarowski P, Darzynkiewicz Z. Analysis of cell cycle by flow cytometry. Methods Mol Biol. 2004;281:301–11. doi: 10.1385/1-59259-811-0:301. [DOI] [PubMed] [Google Scholar]
- 20.Servidei T, Riccardi A, Mozzetti S, Ferlini C, Riccardi R. Chemoresistant tumor cell lines display altered epidermal growth factor receptor and HER3 signaling and enhanced sensitivity to gefitinib. Int J Cancer. 2008;123:2939–49. doi: 10.1002/ijc.23902. [DOI] [PubMed] [Google Scholar]
- 21.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical Aspects of microRNA Target Prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Servidei T, Riccardi A, Sanguinetti M, Dominici C, Riccardi R. Increased sensitivity to the platelet-derived growth factor (PDGF) receptor inhibitor STI571 in chemoresistant glioma cells is associated with enhanced PDGF-BB-mediated signaling and STI571-induced Akt inactivation. J Cell Physiol. 2006;208:220–8. doi: 10.1002/jcp.20659. [DOI] [PubMed] [Google Scholar]
- 23.Mutlu P, Baran Y, Ural AU, Avcu F, Dirican B, Beyzadeoglu M, et al. Effect of cobalt-60 (gamma radiation) on multidrug-resistant multiple myeloma cell lines. Cell Biol Int. 2011;35:721–5. doi: 10.1042/CBI20100061. [DOI] [PubMed] [Google Scholar]
- 24.Hopper-Borge EA, Nasto RE, Ratushny V, Weiner LM, Golemis EA, Astsaturov I. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert Opin Ther Targets. 2009;13:339–62. doi: 10.1517/14712590902735795. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao M, Cai J, Cai L, Jia J, Xie L, Zhu Y, et al. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J Ovarian Res. 2017;10:24. doi: 10.1186/s13048-017-0321-8. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Stahlhut C, Slack FJ. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle. 2015;14:2171–80. doi: 10.1080/15384101.2014.1003008. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kothari V, Mulherkar R. Inhibition of cyclin D1 by shRNA is associated with enhanced sensitivity to conventional therapies for head and neck squamous cell carcinoma. Anticancer Res. 2012;32:121–8. [PubMed] [Google Scholar]
- 29.Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007;1773:1177–95. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Milosevic Z, Pesic M, Stankovic T, Dinic J, Milovanovic Z, Stojsic J, et al. Targeting RAS-MAPK-ERK and PI3K-AKT-mTOR signal transduction pathways to chemosensitize anaplastic thyroid carcinoma. Transl Res. 2014;164:411–23. doi: 10.1016/j.trsl.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Eberlein CA, Stetson D, Markovets AA, Al-Kadhimi KJ, Lai Z, Fisher PR, et al. Acquired Resistance to the Mutant-Selective EGFR Inhibitor AZD9291 Is Associated with Increased Dependence on RAS Signaling in Preclinical Models. Cancer Res. 2015;75:2489–500. doi: 10.1158/0008-5472.CAN-14-3167. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang W, Fan Y, Fa Z, Xu J, Yu H, Li P, et al. microRNA-625 inhibits tumorigenicity by suppressing proliferation, migration and invasion in malignant melanoma. Oncotarget. 2017;8:13253–63. doi: 10.18632/oncotarget.14710. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salendo J, Spitzner M, Kramer F, Zhang X, Jo P, Wolff HA, et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 2013;108:451–7. doi: 10.1016/j.radonc.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 34.Shali H, Ahmadi M, Kafil HS, Dorosti A, Yousefi M. IGF1R and c-met as therapeutic targets for colorectal cancer. Biomed Pharmacother. 2016;82:528–36. doi: 10.1016/j.biopha.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Tang N, Thompson RC, Mobley BC, Clark SW, Sarkaria JN, et al. InsR/IGF1R Pathway Mediates Resistance to EGFR Inhibitors in Glioblastoma. Clin Cancer Res. 2016;22:1767–76. doi: 10.1158/1078-0432.CCR-15-1677. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park YR, Lee ST, Kim SL, Liu YC, Lee MR, Shin JH, et al. MicroRNA-9 suppresses cell migration and invasion through downregulation of TM4SF1 in colorectal cancer. Int J Oncol. 2016;48:2135–43. doi: 10.3892/ijo.2016.3430. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Zhao L, Li N, Miao Y, Zhou H, Jia L. miR-9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. Oncol Rep. 2017;37:2193–200. doi: 10.3892/or.2017.5464. [DOI] [PubMed] [Google Scholar]
- 38.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–43. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 39.Andersen V, Vogel U, Godiksen S, Frenzel FB, Saebo M, Hamfjord J, et al. Low ABCB1 gene expression is an early event in colorectal carcinogenesis. PLoS One. 2013;8:e72119. doi: 10.1371/journal.pone.0072119. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]