Abstract
The discovery of the Warburg effect - the preference of cancer cells to generate ATP via glycolysis rather than oxidative phosphorylation has fostered the misconception that cancer cells become independent of the electron transport chain (ETC) for survival. This is inconsistent with the need of ETC function for the generation of pyrimidines. Along with this misconception, a large body of literature has reported numerous mutations in mitochondrial DNA (mtDNA) further fueling the notion of non-functional ETC in cancer cells. More recent findings, however, suggest that cancers maintain oxidative phosphorylation capacity and that the role of mtDNA mutations in cancer is likely far more nuanced in light of the remarkable complexity of mitochondrial genetics. This review aims at describing the various model systems that were developed to dissect the role of mtDNA in cancer, including cybrids, and more recently mitochondria-nuclear exchange (MNX) and conplastic mice. Further, we put forward the notion of mtDNA landscapes, where the surrounding non-synonymous mutations and variants can enhance or repress the biological effect of specific mtDNA mutations. Notably, we review recent studies describing the ability of some mtDNA landscapes to activate the mitochondrial unfolded protein response (UPRmt), but not others. Further, the role of the UPRmt in maintaining cancer cells in the mitohormetic zone to provide selective adaptation to stress is discussed. Among the genes activated by the UPRmt, we suggest that the dismutases SOD2 and SOD1 may play key roles in the establishment of the mitohormetic zone. Finally, we propose that using a UPRmt nuclear gene expression signature may be a more reliable readout than mtDNA landscapes given their diversity and complexity.
Keywords: mitochondria, mtDNA, UPRmt, mitohormesis, cancer, ROS, SIRT3, SOD1, SOD2, HSP60, metastasis
The role of mitochondrial DNA (mtDNA) in cancer has been the focus of extensive study over the years. The Warburg effect, characterized by a preference for glycolysis, suggests that cancer cells become unreliant on oxidative phosphorylation for the generation of ATP. Given that the activity of the mitochondrial electron transport chain (ETC) is required for the generation of pyrimidines, which are essential for the growth of rapidly dividing cells, a complete lack of ETC activity is incompatible with the growth of cancer (1). Compelling evidence for the necessity of mitochondria in tumor formation was provided by studies in which tumor cells depleted of mtDNA and engrafted in mice, only formed tumors after acquiring host mtDNA via whole mitochondria transfer(2,3). Using similar experimental systems, it was recently demonstrated that the acquisition of mitochondria by mtDNA depleted tumors cells is dependent on the acquisition of OXPHOS for pyrimidine synthesis, not ATP generation(4). Acquisition of mtDNA through exosomes or whole organelle transfer has also been shown as a resistance mechanism by breast, blood, and brain tumors in response to targeted, systemic, or radiotherapy(5–7).
Further, the confusing nature of the role of mtDNA in cancer biology is in part due to the complexity of its genetics. As a result of their bacterial origin, the mitochondria of eukaryotic cells contain their own circular genome. In humans, mtDNA is 16.5 kb in length and encodes 22 tRNAs, 2 rRNAs, and 13 proteins. These proteins encode core subunits of complexes I, III, IV, and V of the ETC. The mitochondrial genome is found in multiple copies per mitochondrion and thereby hundreds to thousands of copies per cell. The numerous copies of mtDNA per cell confer mitochondrial genetics with a level of complexity that requires specialized nomenclature. Specifically, homoplasmy denotes when the same sequence of mtDNA is present in all strains of mtDNA in a cell. In contrast, heteroplasmy, refers to a state in which mixed mtDNA sequences or mutations are present. Heteroplasmy at a specific locus is defined as a percentage of abundance relative to the wild type sequence (Cambridge reference mitochondria sequence). The genetics of mtDNA are further complicated by the fact that mitochondria are dependent on genes encoded by the nuclear genome for their function. Many subunits of the ETC are encoded in the nuclear genome. Additionally, proteins necessary for the maintenance, replication, and transcription of the mitochondrial genome are encoded by the nuclear DNA. For example, transcription of the mitochondrial genome is dependent on POLRMT, a nuclear encoded polymerase found on chromosome 19. In fact, ~99% of the mitochondrial proteome is encoded by nuclear genes. To demonstrate the influence of the mitochondrial genome alone on a given process, it must be experimentally separated from the effect of the nuclear genome.
However, the mitochondrial genome has proved difficult to edit genetically despite advanced using nucleases such as mitochondrial zinc finger nucleases (mtZFN) and mitochondrially targeted transcription activator-like effector nucleases (mitoTALENs) (8–10). The current inability to effectively deliver exogenous RNA into mitochondria has prevented mtDNA engineering from benefiting from the CRISPR revolution. Many in the field are actively trying to adapt the CRISPR/Cas9 system for mtDNA editing with focus on the introduction of gRNAs into the mitochondria(10). Further development of the molecular toolkit to edit the mitochondrial genome has the potential to transform our understanding of mtDNA and the therapeutic interventions against mitochondrial diseases.
Cybrids as a tool to test the role of mtDNA in cancer
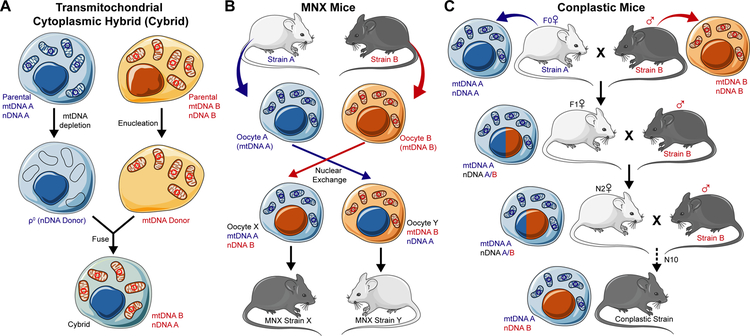
The challenges in manipulating mtDNA with the traditional approaches of molecular biology has led to the development of transmitochondrial cytoplasmic hybrid (cybrid) technology. This technique relies on the generation of ρ0 cells devoid of mtDNA by prolonged culture with ethidium bromide (Fig. 1A). The resulting cells serve as the donor of the nuclear genome. Donor cells of mtDNA are enucleated and the fusion of both creates cybrids with the nuclear and mitochondrial genomes of experimental interest (Fig. 1A).
Figure 1. Cellular and animal models to study the role of mtDNA in cancer.
A. Transmitochondrial cytoplasmic hybrid (Cybrid) technology. Cells with nuclear DNA of interest (nDNA A) have mtDNA depleted to generate ρ0 cells. Cells with mitochondrial DNA of interest (mtDNA B) are enucleated to generate an mtDNA donor. ρ0 and the mtDNA are fused to generate a cybrid with nDNA A and mtDNA B. MNX mice are generated by enucleating the oocytes of inbred mouse strains (Strain A and B, oocytes A and B). Nuclei from these oocytes are exchanged into opposite oocytes generating oocyte X with mtDNA A and nDNA B and oocyte Y with mtDNA B and nDNA A. These oocytes are placed in psuedopregnant females to generated MNX strains X and Y. C. Conplastic mice are generated by the continual backcrossing of females with mtDNA of interest (mtDNA A) to male mice with nDNA of interest (nDNA B) for at least 10 generations until conplastic mice are generated with mtDNA A and nDNA B. (modified from LES LABORATOIRES SERVIER)™ available under a Creative Commons 3.0 Unported License.
Using cybrids, the Hayashi group was the first to demonstrate that mtDNA alone could impact the metastatic behavior of cells(11). In this seminal work, cybrids were generated between non-metastatic P29 and highly metastatic A11 murine cancer cells. In tail vein injections and spontaneous metastasis assays, highly metastatic A11 mtDNA imparted a metastatic phenotype in cybrids with P29 nuclei. Conversely, non metastatic P29 mtDNA blunted the metastatic nature of A11 nuclei. The authors ascribed this effect to mutations in ND6 (G13997A and 13885insC) present in the mitochondrial genome of A11 cells. However, while frameshift and missense mutations represent a possible mechansim to explain this phenotype, additional differences in the mitochondrial genomes of the two cells cannot be excluded.
Following this original study, multiple others followed, providing countless examples of the effect of mtDNA on invasion and metastasis. Cybrids generated between invasive triple negative MDA-MB-231 breast cancer cells and fetal fibroblasts showed that the mtDNA of MDA-MB-231 confers metastasis (12). The authors postulated that the metastatic conferring capacity of MDA-MB-231 mtDNA is due to pathogenic mutations in ND4 (C12084T) and ND5 (A13966G) (12). Cybrids with the mtDNA of MDA-MB-435 cells, which carry a pathogenic A12308G mutation in the tRNALeu(CUN) gene, were also shown to promote metastasis in vivo (13). Patient derived mtDNA with an A3243T mutation in tRNALeu(UUR) conferred invasive and metastatic properties to 143 osteosarcoma cells(14). Cybrids of A549 cells paired with the mtDNA of lung adenocarcinoma patients with missense and nonsense mutations in ND6, also showed enhanced invasion, migration, and mitochondrial superoxide production(15). In addition, mtDNA of highly metastatic B82M fibrosarcoma containing a ND6 13885insC mutation conferred cybrids with metastatic capacity in vivo (16). In this study, mtDNA sequencing of NADH dehydrogenase (ND) genes in 115 patients with non-small cell lung carcinoma (NSCLC) identified 14 single nucleotide variants (SNVs) computationally predicted to decrease complex I function (16). These SNVs in ND genes were significantly more abundant in patients with distant metastasis, providing evidence that mtDNA mutations are associated with metastatsis (16). Further supporting a mechanistic link between complex I function and metastasis, is the observation that manipulation of complex I activity by subunit overexpression or knockdown affected metastsis in vivo(17). Since the majority of proteins encoded in mtDNA are subunits of complex I, a large portion of the mitochondrial genome affects complex I function. Using cybrids, it was demonstrated that mild mutations in complex I (G3460A in ND1, G11778A in ND4, and T14484C in ND6) conferred enhanced tumorgenicity in nude mice compared to parental cells(18). Cybrids with the G8363A mutation tRNALys, a rare and severe mtDNA mutation clinically implicated in Leigh syndrome, cardiomyopathy, and MERRF-like syndrome, were also generated(19). Interestingly, of all ten cybrids and cell lines generated in this study, G8363A tRNALys cybrids had the highest levels of mitochondrial superoxide but were not tumorigenic in nude mice(18). These data suggest that mild mtDNA mutations may be more potent cancer drivers than severe mtDNA mutations. One potential explanation for these observations is that on one hand, severe mtDNA mutations may compromise mitochondrial function to an extent that is incompatible with tumor formation. Milder mtDNA mutations, on the other hand, do not severely compromise mitochondrial function but still result in elevated ROS, which act as a pro-tumorigenic signaling molecule.
Clinical evidence supporting a role of mtDNA in cancer
In addition to studies using cybrids, clinical evidence further supports the influence of mtDNA mutations on cancer etiology and progression. In a cohort of 1,675 tumor samples across 30 different cancer types, 1,907 somatic mtDNA substitutions were identified (20). Similarly, somatic mtDNA mutations were found in approximately three quarters of a cohort of breast cancer patients(21). The majority of mutations described in these patients were found in genes encoding subunits of complex I (21). In renal oncocytoma patients, loss of function mtDNA mutation of complex I was identified as a driver event that proceeds chromosome 1 loss and cyclin D1 overexpression(22). In response to complex I mtDNA mutations, cancer cells of these patients showed adaptive upregulation of glutathione biosynthesis. This metabolic rewiring may function to enhance oxidative stress capacity. The compensatory changes of nuclear gene expression in response to mtDNA mutation highlight the cross talk between the mitochondria and the nucleus.
Using RNA sequencing data from 344 patients with breast cancer, some of whom had patient matched normal samples, 9,063 SNVs and 84 insertions or deletions (INDELs) were detected in tumor mtDNA(23). Of the identified somatic variants/mutations, 95% were typical of replication-coupled mtDNA substitutions. Further, the number of identified mtDNA variants/mutations was signficantly correlated with the age of patients, suggesting that replication induced mtDNA changes that accumulate with age are largely repsonsible for the mtDNA landscapes seen in breast cancer patients (23). In diffuse large B-cell lymphomas, tumor-normal pairs identified private constitutional variants and somatic mtDNA mutations in a mutational spectrum consistent with the replication associated mode of mutagenesis(24). In a panel of colorectal cancer cell lines, mtDNA mutations were found in 70% of samples(25). The majority of these somatic mutations were homoplasmic and were transtitions at purines, the preffered target of free radicals, indicating ROS induced mtDNA mutation(25). Therefore, oxidation and replication induced mutagensis may be considered to be the two main sources of mtDNA damage.
Using whole-genome sequencing of 1,916 patients with matched normal and tumor samples across 24 different cancer types, a study identified distinct mutational patterns present in the mitochondrial genomes of tumor cells (26). This study identified 2,350 tumor-specific somatic mtDNA mutations present across both the coding and non-coding regions of the mitochondrial genome, though the majority resided in RNA coding regions. When metastatic or recurrent tumor samples were compared to primary tumors, there was a significant increase in the proportion of mutations residing in RNA encoding regions of the genome (91% metastatic, 96% recurrent, 87% primary)(26). These data suggest a selection for mtDNA mutations in coding regions of the mitochondrial genome in cancer progression. Tumor specific mtDNA mutations were observed across all electron transport chain complexes, but a significant proportion of these mutations were found in genes encoding subunits of Complex I. Interestingly, several of these mutations in complex I were recurrent or mutually exclusive, suggesting certain mutations provide an adaptive advantage that promotes tumorigenesis. Analysis of the mitochondrial genomes of normal samples in this study identified 1,154 normal-cell heteroplasmies(26). The identified heteroplasmic sequences were found throughout the mitochondrial genome but the vast majority of the variants/mutations described were in the non-coding D-Loop, which is important for mtDNA replication and transcription. Compared to tumor specific mtDNA mutations, normal cell heteroplasmies were significantly less abundant in RNA encoding regions of the mitochondrial genome (69% normal, 87% primary tumor). Further, tumor mtDNA had significantly more nonsense mutations, missense mutations, insertions, and deletions than normal cells (26).
Additional insight into the changes in the mitochondrial genomes of cancer cells during metastatic progression is gained from a rapid autopsy study of 10 prostate cancer patients with primary tumors, soft tissue metastases, bone metastases, and normal tissue analyzed by mtDNA sequencing(27). This study found that bone metastases had significantly more somatic mtDNA mutations than both the primary tumor and soft tissue metastases. In 70% of patients, a G10398A missense mutation in the ND3 gene of complex I was found exclusively in bone metastases (27). This observation argues that specific mtDNA mutations provide an adaptive advantage to form secondary lesions in particular target organs. In a epidemiological study, the 10398A allele was found to be an independent risk factor for invasive breast cancer in African-American but not Caucasian women(28). Significant increased risk of breast cancer and squamous cell carcinoma of the esophagus was also seen with the 10398A in an Indian cohort with the mtDNA haplogroup N(29). Further studies using cybrids generated with the mtDNA of African Americans show that cells with the 10398A allele have resistance to apoptosis, decreased mitochondrial membrane potential, increased ROS production, enhanced colony formation capacity, and increased metastatic capacity in tail vein injection experiments(30). These data suggest that specific mtDNA mutations can significantly modulate cellular phenotypes resulting in enhanced tumorgenicity and disease progression. Other reviews discussing the influence of mtDNA in cancer provide further insight into the well-established, albeit confusing nature of this phenomenon(31–40).
Are mitochondrial unfolded protein response activating-mtDNA landscapes rather than individual mtDNA mutations the answer?
Our group recently performed mitchondrial DNA sequencing in order to determine whether common mtDNA mutations can distinguish invasive from non-invasive breast cancer cell lines (41). Specific missense mutations, A8860G in ATP8 and A15380G in CYB, were present in all cell lines at homoplasmic levels. However, no common mutations were exclusively seen in invasive cell lines. Additionally, neither the number nor location of mtDNA mutations/variations idenitfied invasive from non-invasive cell lines. Unexpectedly, we found that the only commonality in the mtDNA of invasive cells was the presence of at least one varaiant with high levels of heteroplasmy(41).
As our laboratory has a long-standing interest in the mitochondrial unfolded protein response (UPRmt), we hypothesized that perhaps the mtDNA mutational landscape of invasive cells differentially activates this pathway. The UPRmt is a multibranched signaling pathway that promotes mitochondrial fitness through induction of nuclear encoded protein quality control programs, antioxidant machinery, mitochondrial biogenesis, and mitophagy and is activated by perturbations to mitochondrial homeostasis (42–51). Similarly, others have postulated that the ability of mtDNA mutations to promote cancer initiation and progression is mediated through adaptive nuclear gene expression patterns, as reviewed elsewhere in detail(43,44,46,47,49,51–58). In agreement with our hypothesis, we found that activation of the UPRmt was a defining characteristic of invasive cell lines and inhibition of the UPRmt abolished invasion (41). Importantly, this observation was confirmed using a set of 143B/206 osteosarcoma cybrids and hybrids in which the mtDNA of CyB or COX1 cybrids was fused with mtDNA of another patient were also used in this study(59). mtDNA sequencing of cybrids and their respective hybrids reveleaded minor differences in heteroplasmy within the pairs. Despite sharing the same nuclear genome and having the same mtDNA mutation as their respective cybrid, both hybrid cells were significantly less invasvive and showed reduced activation of the UPRmt (41). These results suggest that the effects of a given ROS-generating mutation on UPRmt activation and invasion is dependent on the surrounding mtDNA lanscape in which it resides.
This observation is consisent with the finding that the 10398A allele described above is only associated with cancer in certain ethnic groups. Human populations are characterized by mtDNA haplotypes which mirror our evolution out of Africa(60). A result of sequential accumulation of mtDNA variants through maternal lineage, these haplotypes differ in polymorphisms across the mitochondrial genome. The ethnic differences in the effect of a given mtDNA mutant on clinical phenotypes is a result of the mtDNA haplotype in which that mutation residues.
The ability of mtDNA haplotype to influence cancer behavior is highlighted by a number of clinical studies. Patients with mtDNA haplotype M exhibited significant increased risk of breast cancer, with M sub-haplotype D5 having the most pronounced risk(61). Interestingly, despite increased breast cancer incidence, haplotype M had significantly lower rates of metastasis than haplotype N. This study also reported a significant increased risk of thyroid cancer in haplotype D4a(61). In hepatocellular carcinoma patients, polymorphisms in the D-Loop (315insC and 16263T) were independent predictors of increased tumor-free survival(62). In gastric cancer, mtDNA haplotype N is associated with better survival(63). In gastoenteropancreatic neuroendocine neoplasm patients, three SNVs in the D-Loop were significant predictors of survival with 16257A, 150T, and 151T alleles doing worse(64). It is not yet known how these alleles affect nuclear gene expression in these cancer types. It is likely, however that these alleles modulate nuclear gene expression. In an elegant study utilizing nuclear matched cybrids with different levels of heteroplasmy of a single mtDNA tRNALEU 3242A>G, it was demonstrated that nuclear gene expression was significnatly affected in phases corresponding to the heteroplasmic levels of the point mutant(65).
These studies highlight the ability of mtDNA landscapes to infuence clinical outcomes in cancer patients. Collectively, this body of work suggests that mild differences in mtDNA landscapces can modulate the effect of specific mtDNA mutations.
Mimicking the effect of haplotype using mice with mtDNA from different genetic backgrounds
One of the most significant advances in dissecting the effect of mtDNA in cancer has arisen from the generation of mitochondrial-nuclear exchange (MNX) mice. Using pronuclear exchange, MNX mice are generated with the nuclear genome of an inbred mouse strain in the background of mtDNA from other inbred mouse strains (Fig. 1B). When crossed to mammary specific oncogenic drivers, MNX mice with FVB/NJ nuclear DNA and BALB/cJ or C57BL/6J mtDNA had significantly different tumor latency, metastatic tumor size, and metastatic tumor burden in comparison to wildtype FVB/NJ strain mice(66,67). mtDNA backgrounds also significantly affected spontaneous tumor formation in aged mice of 9 different MNX strains(68). Genomic analyses of MNX mice generated with different mito/nuclear pairings from FVB/NJ, C3H/HeN, C57BL/6J, and BALB/cJ mice also revealed selective differential DNA methylation and nuclear gene expression in strains with identical nuclear genomes but differing mtDNA (69). The observed epigenetic and transcriptional changes in MNX mice highlight the ability of mtDNA to influence nuclear gene expression. Considering the crosstalk between UPRmt activation and chromatin and epigenetic modification(70–74), it is possible that the UPRmt may underlie the reported changes in MNX mice.
At the same time, activation of the UPRmt in the absence of underlying pathology prolongs lifespan and delays the onset of age related diseases(71,73,75,76). To reconcile the pro-longevity effect of the UPRmt and its potential ability to promote tumor progression, we hypothesize the following. In the setting of spontaneous tumor formation, MNX mice carrying mtDNA landscapes that activate the UPRmt may show a delay in aging and age related diseases, such as primary tumor formation. Once a tumor is established, however, activation of the UPRmt may inadvertently promote tumor progression, since the same beneficial effect of the UPRmt that confers longevity, may be highjacked by cancer cells to promote their growth and survival.
Additional in vivo evidence of the ability of mtDNA to influence nuclear gene expression and complex disease phenotypes like aging and cancer have emerged from conplastic mice. Taking advantage of the maternal inhertiance of mtDNA, conplastic mice are generated by crossing female mice with a mitochondrial genome of interest to male mice with the nuclear genome of interest. Female offspring are continually backcrossed to male mice with nuclear genome of interest for a minimum of ten generations to create conplastic mice with mtDNA and nuclear DNA of interest (Fig. 1C). Conplastic mice with C57BL/6 nuclear genomes paired with C57BL/6 mitochondria (BL/6C57) or NZB/OlaHsd mitochondria (BL/6NZB) were used in a comprehensive study of healthspan (77). While the divergence of mtDNA genome between these mice is comparable to that seen between Eurasian and African human mitochondrial genomes, the authors found that BL/6NZB mice had a significant extension in median but not maximal lifespan(77). Interestingly, analysis of mitochondrial ROS between conplastic strains at a young age showed that BL/6NZB actually had higher levels. However, in older BL/6NZB mice, there was no age-related increase in ROS as seen in BL/6C57 mice(77). The observation that younger BL/6NZB mice have higher ROS levels than BL/6C57 is seemingly at odds with their more youthful aging profile. This observation, however, is consistent with the phenonemon of mitohormesis which has been shown to promote health and longevity in model organisms. The findings from MNX and conplastic mice and the studies describing the influence of human mtDNA haplotype suggest that different haplotypes have different susceptibilities to cancer(66,77,78).
Mitohormesis regulates cancer metastasis
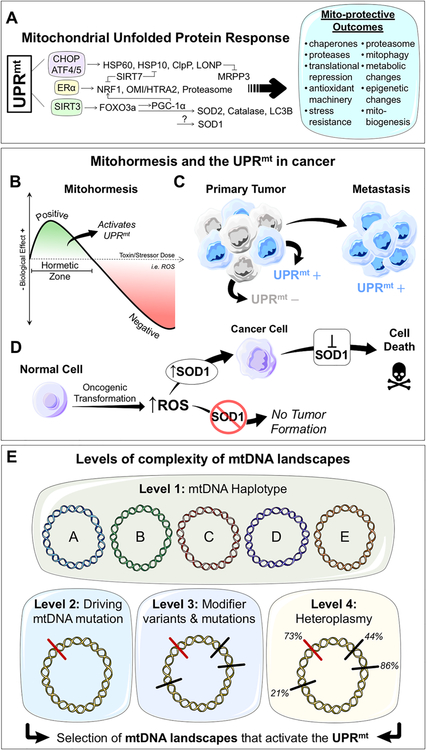
Mitohormesis describes the paradoxical effect of moderate mitochondrial stress that activates persistent cytoprotective mechanisms and results in enhanced mitochondrial stress resistance (79,80). Mitochondrial ROS has been demonstrated to be an important inducer of mitohormesis and promote longevity and healthspan in C. elegans and yeast(81–85). The UPRmt is a key effector pathway of mitohormesis and is necessary for longevity induction by mitohormesis in worms and flies (70–72,75,86–88)(Figure 2A, 2B). One possible consequence of increased ROS levels at young age in BL/6NZB mice is induction of mitohormesis and activation the UPRmt. In this scenario, mitohormetic activation of the UPRmt would protect against subsequent age related increases in ROS. However, while mitohormesis has been most extensively studied in lower model organisms such as flies, worms, and yeast, only recently has mitohormesis been demonstrated in mice. Most notably, the Shadel group generated an inducible and reversible SOD2 knockdown mouse (iSOD2-KD), and showed that mitohormesis is induced in mice by transient SOD2 knockdown during embryogenic development(89). Upon SOD2 knockdown, embryos experienced significantly more oxidative stress, which led to induction of mitohormesis that persisted when SOD2 knockdown was reversed. Mitohormetically primed mice were characterized by enhanced mitochondrial biogenesis, upregulation of antioxidant defenses, and reduced ROS. Significant overlap was seen between the signature of these mice and the mitoprotective outcomes of UPRmt activation. Taken together, these studies demonstrate that mitohormesis does in fact occur in mammals and has profound implications for health and age-related diseases. Additionally, these studies implicate ROS and the UPRmt as important components of mitohormetic signaling in mammals.
Figure 2. The mitochondrial unfolded protein response, mitohoremsis, and mtDNA landscape complexity in cancer.
A. The mammalian mitochondrial unfolded protein response (UPRmt). Different axes of the UPRmt are activated in parallel by perturbations to mitochondrial stress leading to collective mito-protective outcomes. B. Dose response curve of a mitohormetic response to a given stressor or toxin (i.e. ROS). At low levels of toxin/stressor, within the hormetic zone, positive biological responses are seen – such as activation of the UPRmt. At higher doses, negative biological responses are seen. C. Within the primary tumor, UPRmt negative (−) and positive (+) populations can be identified. UPRmt+ populations are characterized by mitohormesis and show increased invasive and metastatic capacities. D. The role of SOD1, a member of the UPRmt, in cancer. Oncogenic transformation of a normal cell results in the increase of reactive oxygen species (ROS). In the absence of SOD1, tumor formation is blocked. SOD1 inhibition selectively kills established cancer cells. E. Levels of complexity of mtDNA landscapes in cancer. Level 1 of complexity is the mtDNA haplotype with its unique set of variants and mutations. Level 2 of complexity is the acquistion of a driving mtDNA mutation in cancer. This mutation will manifest differently given the mtDNA haplotype in which it resides. Level 3 of complexity is the additional variants and mutants gained. These additional mtDNA differences can act as modifiers of both the driving mutation and the mtDNA haplotype. Level 4 of complexitiy is heteroplasmy. The heteroplasmic frequencies of each driving mutation and modifier variants/mutations will also impact the effect of mtDNA on cancer. We propose that cancer cells will select mtDNA landscapes that activate the UPRmt to promote primary tumor formation and metastasis. (modified from LES LABORATOIRES SERVIER)™ available under a Creative Commons 3.0 Unported License.
Given our interest in the UPRmt and cancer, we recently tested whether mitohormesis occurred in tumors and if so whether mitohormesis influences phenotypic cancer cell behavior or disease progression. Having demonstrated that activation of the UPRmt promotes invasion of cancer cells(41), we hypothesized that mitohormetic activation of the UPRmt may serve to promote tumor metastasis and disease progression. To test these hypotheses, the murine breast cancer model (MMTV-rtTA/NeuNT) was used. We found that primary tumors contain two sub-populations of cancer cells based on mitochondrial ROS levels. We isolated these two sub-populations and found that the ROS+ population is significantly more invasive in vitro and metastatic in vivo(90). The ROS+ population of cancer cells had significant activation of the UPRmt (Fig. 2B, 2C). Consistent with mitohormesis, basal activation of the UPRmt persisted in ROS+ cells and these cells were resistant to subsequent oxidative stress(90). These data suggest that oxidative stress in the primary tumor primes a subpopulation of cancer cells to upregulate the UPRmt through mitohormetic signaling. As a result, these cells are more invasive and metastatic. These observations were further validated in human breast cancer cell lines(90).
Mechanistically, we showed that UPRmt activation was sufficient to promote invasion and that the SIRT3 axis of the UPRmt was necessary for invasion and metastasis. This axis of the UPRmt, mediated by the mitochondrial sirtuin SIRT3, is responsible induction of antioxidant programs, mitochondrial biogenesis, and mitophagy (Fig. 2A). In this study, genetic knockdown of SIRT3 inhibited invasion and metastasis in in vitro and in vivo model systems(90). Using a 7-gene signature of the UPRmt we found that patients with UPRmt-HIGH tumors show significantly worse clinical outcomes including metastasis(90). Importantly, UPRmt-HIGH patients had gene expression programs consistent with mitohormesis. Collectively, these studies indicate that while mitohormesis promotes health and longevity in the absence of underlying pathology, it is perniciously used by cancer cells to promote tumor progression.
SOD1 as an additional key mediator of the UPRmt
Given the role of ROS in activation of mitohormesis and the UPRmt, we have shown that superoxide dismutases, SOD1 and SOD2, are upregulated by the UPRmt(90). The mitochondrial matrix localized SOD2 is part of the SIRT3 axis of the UPRmt and plays an important role in mitochondrial antioxidant capacity (Fig. 2A). The cytosolic, nuclear, and mitochondrial intermembrane space localized SOD1 is upregulated by mitochondrial stress and is part of the UPRmt, though which axis it belongs to is currently unknown (Fig. 2A). In the context of cancer, SOD2 is initially downregulated upon tumor initiation and re-activated later during cancer progression and metastasis by the SIRT3 axis of the UPRmt. We previously proposed a SOD2 to SOD1 switch during tumor initiation(91,92). We found that SOD1 levels are increased in breast cancer cells with decreased SOD2 levels(92). We showed that the majority of the superoxide activity in a panel of breast cancer cell lines is due to SOD1 activity. In addition, we found that the mitochondrial fraction of SOD1 is increased in breast cancer cells. Moreover, inhibition of SOD1 led to a selective increase in mitochondrial superoxide levels in cancer cells compared to the normal mammary epithelial cells MCF10A. SOD1 inhibition also led to an increase in mitochondrial fragmentation and altered mitochondrial morphology(92). These findings suggest that SOD1 is essential for mitochondrial integrity in the presence of increased oxidative stress in cancer cells.
We recently sought to better understand the role of SOD1 in cancer. To test the role of SOD1 in vivo, we crossed the SOD1 knockout mice with the inducible ErbB2 and Wnt transgenic models of mammary tumors. We found that in both models, SOD1 is necessary for oncogene-driven proliferation (Fig. 2D). Congruent with our previous findings that SOD1 inhibition does not affect normal mammary epithelial cells in vitro, we found that absence of SOD1 did not affect normal proliferation during different stages of mammary gland development in vivo(93). Additionally, SOD1 knockout mice do not have any phenotypic differences in other proliferative tissues such as the skin or the intestine. We hypothesized that the specific requirement for SOD1 in cancer cells but not normal highly proliferative tissues may be due to differences in mitochondrial superoxide levels. We found that the proliferation driven by pregnancy does not increase the level of mitochondrial superoxide, while tumors exhibit remarkably high levels of mitochondrial superoxide, and the high ROS sub-population from tumors show higher expression of SOD1(93). This correlated with SOD1 immunohistochemistry staining in which SOD1 was not detected in normal mammary ducts or proliferating mammary glands during pregnancy but was detected in tumors(93). Our results suggest that in the absence of SOD1, malignant cells are unable to cope with elevated superoxide levels associated with transformation and undergo oncogene-induced senescence and apoptosis. In addition, breast cancer patients with high SOD1 levels have poor clinical outcomes.
Our findings concur with other groups that have demonstrated the efficacy of inhibiting SOD1. One of the first studies linking SOD1 to cancer demonstrated that SOD1 is important for the viability of leukemia cells(94). Following this finding, the Varmus group identified SOD1 as a selective therapeutic target in a high throughput small molecules screen of approximately 200,000 compounds in human lung adenocarcinomas(95,96). Their top small molecule, LCS-1 showed efficacy at killing only lung cancer cells, and had no effect on normal lung epithelial cells in vitro. We have shown that LCS-1 treatment also selectively inhibits the growth of mammary tumor cells (93,97) . LCS-1 has also been tested in a model of neck and head cancer(98). Previously, SOD1 has also been pharmacologically inhibited with copper chelators, such as diethyldithiocarbamate (DDC) and tetrathiomolybdate (TM)(99). ATN-224, a TM derivative, and was shown to have efficacy in promoting cell death in lung cancer cells lines and an experimental model of non-small cell lung cancer(100). Collectively, these studies suggest that SOD1 is vital for cancer cells and may be a potential, selective therapeutic target.
Concluding remarks
The controversy over the role of mtDNA in cancer arises in large part from two factors. First, the misconception that cancer cells become independent of mitochondrial oxidative phosphorylation; and second, the remarkable complexity of mitochondrial genetics where in addition to heteroplasmy, distinguishing driver mutations/variants from passenger mutations/variants is extremely challenging. In light of the most recent studies, particularly those in MNX and conplastic mice, the picture that emerges is that the impact of mtDNA on cancer may be best explained if the respective contribution of the baseline (wild type) mtDNA landscape is separated from the contribution of acquired mutations and variants during transformation. This potential scenario of multiple levels of mtDNA complexity is illustrated in Figure 2E. The observations described in the experimental models described in this review suggest that different haplotypes have different susceptibilities to cancer(66,77,78). This represents Level 1 of mtDNA complexity. In other words, an individual with haplotype A has a higher risk of developing cancer than an individual with haplotype B or C (Fig. 2E). If those two individuals were to acquire the exact same mtDNA driver mutation X, the aggressiveness of the tumor would be altered by the specific haplotype in which it arises (Fig. 2E). This represents Level 2 of mtDNA complexity. Evidence for the existence of driver mtDNA driver mutations come from a study sequencing renal oncocytoma tumor (101). In this study, it was demonstrated that mtDNA mutations in complex I were early genetic events that contributed to tumor formation. While some mutations were recurrent, there were a number of ways by which these tumors mutated amtDNA encoded complex I subunit(101). This suggests that mutation of complex I, regardless of the particular mutation, is a driver mtDNA mutation. The emergence of additional stochastic mutations and variants may further modulate the impact of the mutation and haplotype on cancer, positively or negatively. These modifier variants and mutations represent Level 3 of mtDNA complexity (Fig. 2E). Level 4 of mtDNA complexity, arises from the combinatorial effect of the heteroplasmic frequencies of multiple modifiers and drivers (Fig. 2E).
Ultimately, we propose that the resulting combinatorial size and diversity of potential mtDNA landscapes can be divided into those that activate the UPRmt and those that do not (Fig. 2E). UPRmt activating landscapes enable increased mitochondrial fitness and adaptation to stress thereby providing a selective advantage to the cells carrying them. In agreement with this possibility, using a 7 gene UPRmt signature, we show that patients with UPRmt positive tumors have worst outcome and increased metastasis.
Finally, if this multi-layered model of mtDNA complexity is correct, the lack of consensus about which mtDNA mutations/variants influence metastasis may be reconciled. It is likely that all previous reports are true but the we have failed to appreciate the complex mtDNA landscapes in which these reported mtDNA mutations/variants lie. This may mean, however, that finding driver mtDNA mutations in cancer may be nearly impossible. We propose that a nuclear gene UPRmt signature can be used as a tool to bypass the complexity of mtDNA landscapes and our current inability to dissect the effect of a given mtDNA mutation/variant in cancer.
Acknowledgments:
We thank James Manfredi and Stuart Aaronson for their support through the T32 CA078207 training grant to TCK and MG. This work was funded by a NIH RO1 number CA1722046 to DG and the NIH F31 number FCA228259A to TCK.
Footnotes
Conflict of interest: The authors declare they have no conflict on interest.
References:
- 1.King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol Cell Biol. American Society for Microbiology Journals; 1992;12:480–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan AS, Baty JW, Dong L-F, Bezawork-Geleta A, Endaya B, Goodwin J, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21:81–94. [DOI] [PubMed] [Google Scholar]
- 3.Dong L-F, Kovarova J, Bajzikova M, Bezawork-Geleta A, Svec D, Endaya B, et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife. 2017;6:e22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajzikova M, Kovarova J, Coelho AR, Boukalova S, Oh S, Rohlenova K, et al. Reactivation of Dihydroorotate Dehydrogenase-Driven Pyrimidine Biosynthesis Restores Tumor Growth of Respiration-Deficient Cancer Cells. Cell Metab. Cell Press; 2019;29:399–416.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A. 2017;114:E9066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moschoi R, Imbert V, Nebout M, Chiche J, Mary D, Prebet T, et al. Protective mitochondrial transfer from bone marrow stromal cells to acute myeloid leukemic cells during chemotherapy. Blood. American Society of Hematology; 2016;128:253–64. [DOI] [PubMed] [Google Scholar]
- 7.Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. Nature Publishing Group; 2015;528:93–8. [DOI] [PubMed] [Google Scholar]
- 8.Bacman SR, Kauppila JHK, Pereira C V, Nissanka N, Miranda M, Pinto M, et al. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nat Med. Nature Publishing Group; 2018;24:1696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto M, Bacman SR, Peralta S, Falk MJ, Chomyn A, Chan DC, et al. MitoTALEN: A General Approach to Reduce Mutant mtDNA Loads and Restore Oxidative Phosphorylation Function in Mitochondrial Diseases. Mol Ther. Cell Press; 2015;23:1592–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gammage PA, Moraes CT, Minczuk M. Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized. Trends Genet. Elsevier Current Trends; 2018;34:101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–4. [DOI] [PubMed] [Google Scholar]
- 12.Imanishi H, Hattori K, Wada R, Ishikawa K, Fukuda S, Takenaga K, et al. Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PLoS One. 2011;6:e23401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009;8:1378–85. [DOI] [PubMed] [Google Scholar]
- 14.Nunes JB, Peixoto J, Soares P, Maximo V, Carvalho S, Pinho SS, et al. OXPHOS dysfunction regulates integrin- 1 modifications and enhances cell motility and migration. Hum Mol Genet. Oxford University Press; 2015;24:1977–90. [DOI] [PubMed] [Google Scholar]
- 15.Yuan Y, Wang W, Li H, Yu Y, Tao J, Huang S, et al. Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer. 2015;15:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koshikawa N, Akimoto M, Hayashi J-I, Nagase H, Takenaga K. Association of predicted pathogenic mutations in mitochondrial ND genes with distant metastasis in NSCLC and colon cancer. Sci Rep. 2017;7:15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santidrian AF, Matsuno-Yagi A, Ritland M, Seo BB, LeBoeuf SE, Gay LJ, et al. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest. 2013;123:1068–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Bermúdez A, Vallejo CG, Vicente-Blanco RJ, Gallardo ME, Fernández-Moreno MÁ, Quintanilla M, et al. Enhanced tumorigenicity by mitochondrial DNA mild mutations. Oncotarget. Impact Journals; 2015;6:13628–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virgilio R, Ronchi D, Bordoni A, Fassone E, Bonato S, Donadoni C, et al. Mitochondrial DNA G8363A mutation in the tRNALys gene: Clinical, biochemical and pathological study. J Neurol Sci. Elsevier; 2009;281:85–92. [DOI] [PubMed] [Google Scholar]
- 20.Ju YS, Alexandrov LB, Gerstung M, Martincorena I, Nik-Zainal S, Ramakrishna M, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon S, LaFramboise T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis. Oxford University Press; 2014;35:1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopal RK, Calvo SE, Shih AR, Chaves FL, McGuone D, Mick E, et al. Early loss of mitochondrial complex I and rewiring of glutathione metabolism in renal oncocytoma. Proc Natl Acad Sci U S A. National Academy of Sciences; 2018;115:E6283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weerts MJA, Smid M, Foekens JA, Sleijfer S, Martens JWM. Mitochondrial RNA expression and single nucleotide variants in association with clinical parameters in primary breast cancers. Cancers (Basel). Multidisciplinary Digital Publishing Institute; 2018;10:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng AGX, Leung ACY, Brooks-Wilson AR. Somatic Mitochondrial DNA Mutations in Diffuse Large B-Cell Lymphoma. Sci Rep. 2018;8:3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polyak K, Li Y, Zhu H, Lengauer C, Willson JKV, Markowitz SD, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. Nature Publishing Group; 1998;20:291–3. [DOI] [PubMed] [Google Scholar]
- 26.Grandhi S, Bosworth C, Maddox W, Sensiba C, Akhavanfard S, Ni Y, et al. Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection. Hum Mol Genet. 2017;26:2912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold RS, Fedewa SA, Goodman M, Osunkoya AO, Kissick HT, Morrissey C, et al. Bone metastasis in prostate cancer: Recurring mitochondrial DNA mutation reveals selective pressure exerted by the bone microenvironment. Bone. 2015;78:81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A Polymorphism and Invasive Breast Cancer in African-American Women. Cancer Res. 2005;65:8028–33. [DOI] [PubMed] [Google Scholar]
- 29.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RNK. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. Elsevier; 2007;249:249–55. [DOI] [PubMed] [Google Scholar]
- 30.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009;54:647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bussard KM, Siracusa LD. Understanding Mitochondrial Polymorphisms in Cancer. Cancer Res. 2017;77:6051–9. [DOI] [PubMed] [Google Scholar]
- 32.Hahn A, Zuryn S. The Cellular Mitochondrial Genome Landscape in Disease. Trends Cell Biol. Elsevier; 2019;29:227–40. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa K, Hayashi J-I. A novel function of mtDNA: its involvement in metastasis. Ann N Y Acad Sci. Blackwell Publishing Inc; 2010;1201:40–3. [DOI] [PubMed] [Google Scholar]
- 34.Herst PM, Rowe MR, Carson GM, Berridge MV. Functional Mitochondria in Health and Disease. Front Endocrinol (Lausanne). Frontiers; 2017;8:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace DC. Mitochondrial genetic medicine. Nat Genet. Nature Publishing Group; 2018;50:1642–9. [DOI] [PubMed] [Google Scholar]
- 36.Kalsbeek AMF, Chan EKF, Corcoran NM, Hovens CM, Hayes VM. Mitochondrial genome variation and prostate cancer: a review of the mutational landscape and application to clinical management. Oncotarget. 2017;8:71342–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheid AD, Beadnell TC, Welch DR. The second genome: Effects of the mitochondrial genome on cancer progression. Adv Cancer Res. Academic Press; 2019. page 63–105. [DOI] [PMC free article] [PubMed]
- 38.Brandon M, Baldi P, Wallace D. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–62. [DOI] [PubMed] [Google Scholar]
- 39.Vyas S, Zaganjor E, Haigis MC. Leading Edge Mitochondria and Cancer. Cell. 2016;166:555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenny TC, Hart P, Ragazzi M, Sersinghe M, Chipuk J, Sagar MAK, et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPRmt to promote metastasis. Oncogene. 2017;36:4393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenny TC, Germain D. From discovery of the CHOP axis and targeting ClpP to the identification of additional axes of the UPRmt driven by the estrogen receptor and SIRT3. J Bioenerg Biomembr. 2017;49:297–305. [DOI] [PubMed] [Google Scholar]
- 43.Kenny TC, Germain D. mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPRmt). Front cell Dev Biol. 2017;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenny TC, Manfredi G, Germain D. The Mitochondrial Unfolded Protein Response as a Non-Oncogene Addiction to Support Adaptation to Stress during Transformation in Cancer and Beyond. Front Oncol. 2017;7:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melber A, Haynes CM. UPR mt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;1–15. [DOI] [PMC free article] [PubMed]
- 46.Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol. 2018;19:109–20. [DOI] [PubMed] [Google Scholar]
- 47.Naresh NU, Haynes CM. Signaling and Regulation of the Mitochondrial Unfolded Protein Response. Cold Spring Harb Perspect Biol. 2019;11:a033944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borch Jensen M, Jasper H. Mitochondrial Proteostasis in the Control of Aging and Longevity. Cell Metab. 2014;20:214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Münch C The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. BioMed Central; 2018;16:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moehle EA, Shen K, Dillin A. Mitochondrial Proteostasis in the Context of Cellular and Organismal Health and Aging. J Biol Chem. 2018;jbc.TM117.000893. [DOI] [PMC free article] [PubMed]
- 51.Rath E, Moschetta A, Haller D. Mitochondrial function — gatekeeper of intestinal epithelial cell homeostasis. Nat Rev Gastroenterol Hepatol. Nature Publishing Group; 2018;15:497–516. [DOI] [PubMed] [Google Scholar]
- 52.Gomez M, Germain D. Cross talk between SOD1 and the mitochondrial UPR in cancer and neurodegeneration. Mol Cell Neurosci. Academic Press; 2019;98:12–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Germain D Mitochondrial UPR in Cancer. Mitochondria Cell Death. New York, NY: Springer New York; 2016. page 149–67. [Google Scholar]
- 54.He C, Hart PC, Germain D, Bonini MG. SOD2 and the Mitochondrial UPR: Partners Regulating Cellular Phenotypic Transitions. Trends Biochem. Sci. 2016. page 568–77. [DOI] [PMC free article] [PubMed]
- 55.Kenny TC, Germain D. From discovery of the CHOP axis and targeting ClpP to the identification of additional axes of the UPRmt driven by the estrogen receptor and SIRT3. J Bioenerg Biomembr. 2017;49:297–305. [DOI] [PubMed] [Google Scholar]
- 56.Bárcena C, Mayoral P, Quirós PM. Mitohormesis, an Antiaging Paradigm. Int Rev Cell Mol Biol. Academic Press; 2018;340:35–77. [DOI] [PubMed] [Google Scholar]
- 57.Higuchi-Sanabria R, Frankino PA, West J, Iii P, Tronnes SU, Dillin A. A Futile Battle? Protein Quality Control and the Stress of Aging. Dev Cell. 2018;44:139–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng P, Haynes CM. Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin Cancer Biol. 2017;47:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, et al. Heterologous mitochondrial DNA recombination in human cells. Hum Mol Genet. 2004;13:3171–9. [DOI] [PubMed] [Google Scholar]
- 60.Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell. 2015;163:33–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang H, Shen L, Chen T, He J, Ding Z, Wei J, et al. Cancer type-specific modulation of mitochondrial haplogroups in breast, colorectal and thyroid cancer. BMC Cancer. 2010;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, Wan P, Peng T, Xiao K, Su M, Shang L, et al. Associations between sequence variations in the mitochondrial DNA D-loop region and outcome of hepatocellular carcinoma. Oncol Lett. Spandidos Publications; 2016;11:3723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C, Wang Y, Wang H, Zhang R, Guo Z. Mitochondrial DNA haplogroup N is associated good outcome of gastric cancer. Tumour Biol. 2014;35:12555–9. [DOI] [PubMed] [Google Scholar]
- 64.Er L-M, Li Y, Er L, Wu M, Zheng X-L, Guo Z-J, et al. Associations between Single Nucleotide Polymorphisms in the Mitochondrial DNA D-Loop Region and Outcome of Gastroenteropancreatic Neuroendocrine Neoplasm. Ann Clin Lab Sci. Association of Clinical Scientists; 2018;48:333–7. [PubMed] [Google Scholar]
- 65.Picard M, Zhang J, Hancock S, Derbeneva O, Golhar R, Golik P, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci U S A. 2014;111:E4033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinker AE, Vivian CJ, Koestler DC, Tsue TT, Jensen RA, Welch DR. Mitochondrial Haplotype Alters Mammary Cancer Tumorigenicity and Metastasis in an Oncogenic Driver–Dependent Manner. Cancer Res. 2017;77:6941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, et al. Mitochondrial genetics regulate breast cancer Tumorigenicity and Metastatic potential. Cancer Res. 2015;75:4429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vivian CJ, Hagedorn TM, Jensen RA, Brinker AE, Welch DR. Mitochondrial polymorphisms contribute to aging phenotypes in MNX mouse models. Cancer Metastasis Rev. Springer US; 2018;37:633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vivian CJ, Brinker AE, Graw S, Koestler DC, Legendre C, Gooden GC, et al. Mitochondrial Genomic Backgrounds Affect Nuclear DNA Methylation and Gene Expression. Cancer Res. 2017;77:6202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. Elsevier Inc; 2016;165:1209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, et al. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell. Elsevier Inc; 2016;165:1197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Q, Wu X, Chen P, Liu L, Xin N, Tian Y, et al. The Mitochondrial Unfolded Protein Response Is Mediated Cell-Non-autonomously by Retromer-Dependent Wnt Signaling. Cell. Elsevier; 2018;174:870–883.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma C, Niu R, Huang T, Shao L-W, Peng Y, Ding W, et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat Cell Biol. Nature Publishing Group; 2019;21:319–27. [DOI] [PubMed] [Google Scholar]
- 75.Owusu-Ansah E, Song W, Perrimon N. Muscle Mitohormesis Promotes Longevity via Systemic Repression of Insulin Signaling. Cell. Cell Press; 2013;155:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, et al. The NAD+/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sánchez-Cabo F, Torroja C, Acín-Pérez R, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. Nature Research; 2016;535:561–5. [DOI] [PubMed] [Google Scholar]
- 78.Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, et al. Mitochondrial genetics regulate breast cancer tumorigenicity and metastatic potential HHS Public Access. Cancer Res. 2015;75:4429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yun J, Finkel T. Mitohormesis. Cell Metab. Elsevier; 2014;19:757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bárcena C, Mayoral P, Quirós PM. Mitohormesis, an Antiaging Paradigm. Int Rev Cell Mol Biol. Academic Press; 2018;340:35–77. [DOI] [PubMed] [Google Scholar]
- 81.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR Signaling Extends Chronological Life Span via Increased Respiration and Upregulation of Mitochondrial Gene Expression. Cell Metab. Cell Press; 2007;5:265–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee S-J, Hwang AB, Kenyon C. Inhibition of Respiration Extends C. elegans Life Span via Reactive Oxygen Species that Increase HIF-1 Activity. Curr Biol. Cell Press; 2010;20:2131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang W, Hekimi S. A Mitochondrial Superoxide Signal Triggers Increased Longevity in Caenorhabditis elegans. PLoS Biol. Public Library of Science; 2010;8:e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. Kim SK, editor. PLoS Genet. Public Library of Science; 2009;5:e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose Restriction Extends Caenorhabditis elegans Life Span by Inducing Mitochondrial Respiration and Increasing Oxidative Stress. Cell Metab. Cell Press; 2007;6:280–93. [DOI] [PubMed] [Google Scholar]
- 86.Dillin A, Hsu A-L, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. American Association for the Advancement of Science; 2002;298:2398–401. [DOI] [PubMed] [Google Scholar]
- 87.Feng J, Bussière F, Hekimi S. Mitochondrial Electron Transport Is a Key Determinant of Life Span in Caenorhabditis elegans. Dev Cell. Cell Press; 2001;1:633–44. [DOI] [PubMed] [Google Scholar]
- 88.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and Nuclear Accumulation of the Transcription Factor ATFS-1 Promotes OXPHOS Recovery during the UPRmt. Mol Cell. Cell Press; 2015;58:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cox CS, McKay SE, Holmbeck MA, Christian BE, Scortea AC, Tsay AJ, et al. Mitohormesis in Mice via Sustained Basal Activation of Mitochondrial and Antioxidant Signaling. Cell Metab. Elsevier; 2018;28:776–786.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kenny TC, Craig AJ, Villanueva A, Germain D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep. Elsevier; 2019;27:2292–2303.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finley LWS, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell. 2011;19:416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Papa L, Hahn M, Marsh EL, Evans BS, Germain D. SOD2 to SOD1 Switch in Breast Cancer. J Biol Chem. 2014;289:5412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gomez ML, Shah N, Kenny TC, Jenkins EC, Germain D. SOD1 is essential for oncogene-driven mammary tumor formation but dispensable for normal development and proliferation. Oncogene. Nature Publishing Group; 2019;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang P, Feng L, Oldham EA, Keating MJ, Plunkett W. Superoxide dismutase as a target for the selective killing of cancer cells. Nature. 2000;407:390–5. [DOI] [PubMed] [Google Scholar]
- 95.Somwar R, Shum D, Djaballah H, Varmus H. Identification and Preliminary Characterization of Novel Small Molecules That Inhibit Growth of Human Lung Adenocarcinoma Cells. J Biomol Screen. 2009;14:1176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Somwar R, Erdjument-Bromage H, Larsson E, Shum D, Lockwood WW, Yang G, et al. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc Natl Acad Sci. 2011;108:16375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Papa L, Manfredi G, Germain D. SOD1, an unexpected novel target for cancer therapy. Genes Cancer. 2014;5:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li S, Fu L, Tian T, Deng L, Li H, Xia W, et al. Disrupting SOD1 activity inhibits cell growth and enhances lipid accumulation in nasopharyngeal carcinoma. Cell Commun Signal. BioMed Central; 2018;16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong X, Zhang Z, Zhao J, Lei J, Chen Y, Li X, et al. The rational design of specific SOD1 inhibitors via copper coordination and their application in ROS signaling research. Chem Sci. Royal Society of Chemistry; 2016;7:6251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glasauer A, Sena LA, Diebold LP, Mazar AP, Chandel NS. Targeting SOD1 reduces experimental non–small-cell lung cancer. J Clin Invest. American Society for Clinical Investigation; 2014;124:117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gopal RK, Calvo SE, Shih AR, Chaves FL, McGuone D, Mick E, et al. Early loss of mitochondrial complex I and rewiring of glutathione metabolism in renal oncocytoma. Proc Natl Acad Sci U S A. National Academy of Sciences; 2018;201711888. [DOI] [PMC free article] [PubMed]