Abstract
Despite the prevalence and clinical importance of influenza, its long-term effect on lung immunity is unclear. Here we describe that following viral clearance and clinical recovery, at one month post-influenza, mice are better protected from Streptococcus pneumoniae infection due to a population of monocyte-derived alveolar macrophages (AMs) which produce increased IL-6. Influenza-induced monocyte-derived AMs have a surface phenotype similar to resident AMs but display a unique functional, transcriptional and epigenetic profile which is distinct from resident AMs. In contrast, influenza-experienced resident AMs remain largely similar to naive AMs. Thus, influenza changes the composition of the AM population to provide prolonged antibacterial protection. Monocyte-derived AMs persist over time but lose their protective profile. Our results help to understand how transient respiratory infections, a common occurrence in human life, can constantly alter lung immunity by contributing monocyte-derived, recruited cells to the AM population.
Introduction
Influenza A virus is a common respiratory pathogen that infects 10% of the global population annually. Therefore, most humans will experience influenza once or several times over their lifetime, as one of many diverse respiratory infections. While prior infections play a crucial role in maturation and memory of adaptive immunity, examples of sustained changes in innate immune cells have also been demonstrated1–3. However, it is largely unclear by which mechanisms prior infections confer such a long-term imprint in the lung.
Most current murine models of infectious disease fail to reflect the frequent pathogen exposure experienced by humans, or account for the effect this has on immune system reactivity4,5. We therefore established a model to study the long-term consequences of influenza on lung immunity, including the response to an unrelated pathogen, to investigate infectious disease in a setting that more closely resembles the human situation of sequential polymicrobial exposure.
Alveolar macrophages (AM) represent plausible targets for retaining a tissue-specific imprint of infection, due to their location and local turnover. AMs are a major immunological constituent of the airways, important in governing pulmonary homeostasis6,7 and in regulating the immune response to respiratory challenges. The importance of origin for macrophage reactivity has been demonstrated in gut, skin, lung, peritoneum, liver, heart and pleural cavity3,8–14. In naive mice, AMs are derived from embryonic precursors and have unique longevity and self-renewal capacity6,15,16. In adult lungs, bone marrow (BM) derived monocytes are capable of differentiating into AMs, should the niche be accessible17–19. During acute influenza infection, AM numbers are drastically reduced in the lung, and must be quickly re-established to resolve infection and repair the tissue20–22. It is unknown if and how AM function changes during this re-establishment process, and what consequences this has on host immunity.
We studied lung immunity long-term post-influenza, when virus is eliminated, inflammation is resolved, and lung damage is not detectable. Influenza-experienced mice harboured an increased AM population which had incorporated BM-derived cells. At one month post-influenza, recruited macrophages transcriptionally resemble monocytes, and have high chromatin accessibility at a select number of immune-related gene loci. As a result these cells, when stimulated, produce increased IL-6 amounts and are protective in subsequent Streptococcus pneumoniae (S.p.) challenge. At two months post influenza, the recruited AM population remains abundant in the lung, but becomes transcriptionally and functionally more similar to resident AMs, and no longer provides antibacterial protection. Our results highlight that innate immunity and anti-pathogen protection in the lung dynamically reflect prior exposure history.
Results
Post-influenza mice have an increased number of alveolar macrophages which confer antibacterial protection
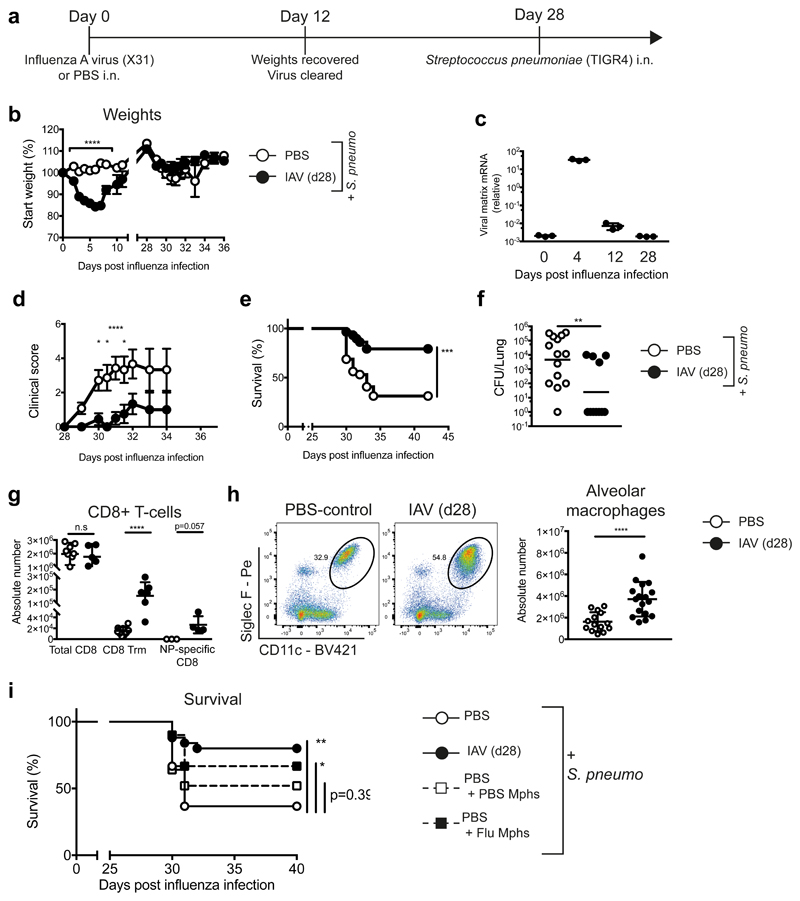
To investigate prolonged effects of a transient viral infection, we established a model in which mice were challenged with S.p. at one month post-influenza infection (Fig. 1a). Initial infection with influenza A virus (IAV) strain X31 (Fig. 1a,b) induced acute, self-limiting disease (Fig. 1b). At day 12 post-infection, virus was undetectable in the lung (Fig. 1c). On day 28 post-influenza, naive control (PBS) and influenza-experienced (IAV d28) mice were challenged with the gram-positive bacterium S.p. (serotype 4 -TIGR4). Naive mice displayed high clinical scores in response to bacterial infection (Fig. 1d), which resulted in ~70% mice reaching clinical endpoint (Fig. 1e). In contrast, post-influenza mice were significantly less susceptible to S.p. infection, displaying lower clinical scores, lower mortality and reduced bacterial loads (Fig. 1d,e,f).
Figure 1. At one month post-influenza, mice are more protected against S. pneumoniae infection, and have an increased population of alveolar macrophages which confers resistance.
a, Schematic of infection schedule. b,d-f, Female C57BL/6 mice were inoculated with 8000 TCID50 X31 (H3N2)/30μl i.n. (influenza - black circles) or PBS (white circles). 28 days later, all mice were infected with 5 x 105 CFU TIGR4/30μl i.n. (S. pneumoniae) and monitored for (b) weight loss, (d) clinical scores and (e) survival (p=0.0002). n=32 mice (PBS)/ n=30 mice (X31) from 4 independent experiments. c, qRT-PCR of influenza matrix protein of whole lung at indicated time points post-influenza infection. f, Lung colony forming units (CFUs) from whole lung at 48 hours post-S. pneumoniae. (p=0.0047). n=14 mice (PBS)/ n=11 mice (X31) from one representative experiment. g-h, Quantification of cells identified in lung by flow cytometry. n= 9 mice (PBS)/n=17 mice (X31) from 3 independent experiments. (g) n.s.=0.0789 i, Alveolar macrophages were isolated from naive or post-influenza mice and transferred (106 AMs/20μl i.t.) into naive mice. 24 hours later, mice were infected with S. pneumoniae and survival was monitored. n=25 mice (X31/PBS + PBS Mphs)/n=30 mice (PBS), n= 35 mice (PBS+ X31 Mphs) from 3 independent experiments (** = 0.0012, *=0.0128, n.s. = 0.386).
Data shown as arithmetic means ±SEM (weights) or ±SD for cell quantification and statistical significance assessed by a two-tailed test using 2-way ANOVA (weights and clinical scores), Log Rank (Mantel-Cox) test for survival or Mann-Whitney U for cell quantification and lung CFUs. Data in (c) and (f) are shown as geometric means. * p=≤0.05, ** p=≤0.01, *** p=≤0.001, **** p=≤0.0001.
To investigate changes in the lungs of influenza-experienced mice which contribute to anti-pneumococcal protection, immune cells in the lung were profiled by flow cytometry. Increased germinal centre B cells and lymphoid structures were found in the lungs of influenza-infected mice, as previously described23,24 (Supplementary Fig. 1a-c). More tissue resident memory CD4 and CD8 T cells, together with an increase in CD8 T cells specific for an influenza nucleoprotein epitope, confirmed a successful influenza infection (Fig. 1g, Supplementary Fig. 1d). However, most innate and adaptive cell populations were equally represented in naive and influenza-experienced lungs (Supplementary Fig. 1a, d, gated as shown in Supplementary Fig. 8).
A substantial increase in the frequency and absolute number of alveolar macrophages (AMs; Siglec F+CD11c+MerTK+CD64+) was observed in the lung 28 days post influenza (Fig. 1h, Supplementary Fig. 2a). AMs rapidly disappeared from the airways during influenza infection (Supplementary Fig. 2b), followed by their repopulation at day 7-11 post-infection. Post-influenza AMs were similar to naive AMs and distinct from Ly6Chi monocytes based on surface markers and morphology (Supplementary Fig. 2c,d).
To assess whether AMs contributed to the increased antibacterial protection in influenza-experienced mice, we adoptively transferred 106 AMs from d28 naive or influenza-experienced mice into naive recipients via intratracheal transfer and infected recipients one day later. The transfer of influenza-experienced macrophages conferred a significant increase in protection against pneumococcal infection, while transfer of naïve AMs did not (Fig. 1i), demonstrating that influenza infection induces a functional change in the AM population that provides prolonged protection against bacterial infection.
Increased IL-6 production by post-influenza AMs contributes to antibacterial protection
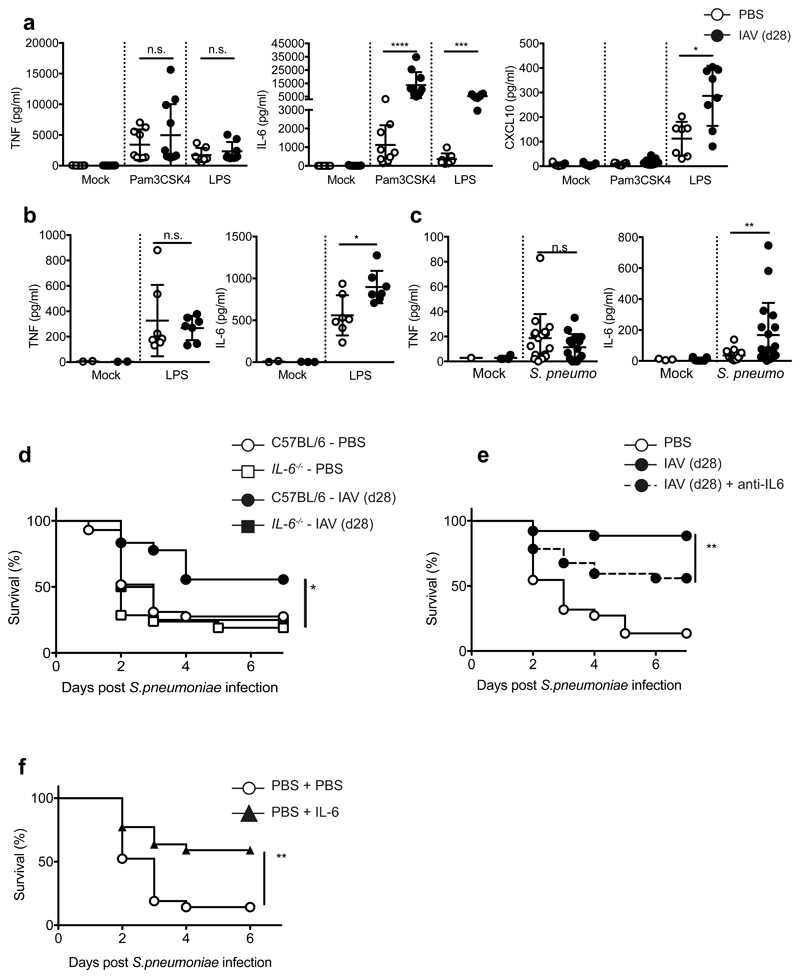
In functional tests, we found equivalent bacterial phagocytosis and reactive oxygen species production by naive and post-influenza AMs (Supplementary Fig. 2e,f). Both PBS- and influenza-experienced AMs isolated from the lung produced pro-inflammatory cytokines in response to ex vivo Toll-like receptor (TLR) stimulation (Pam3CSK4, TLR2-ligand; lipopolysaccharide (LPS), TLR4-ligand) (Fig. 2a). Notably, post-influenza AMs secreted significantly more IL-6 than naive AMs, while producing comparable amounts of TNF (Fig. 2a). This dichotomy of response was evident in other cytokines: CXCL1, CXCL2 and CXCL5 were produced in equal amounts, whereas CCL3, CCL4 and G-CSF were produced to significantly higher levels by influenza-experienced AMs (Supplementary Fig. 3a-d). These differences were observed across a titration of stimulus (Supplementary Fig. 3e) and apparent in AMs isolated from both whole lung and broncho-alveolar lavage (BAL) (Supplementary Fig. 3f). Increased CXCL10 and CCL5 production was seen in post-influenza macrophages following LPS-, but not Pam3CSK4-stimulation (Fig. 2a, Supplementary Fig. 3c). Increased IL-6 and equivalent TNF were also observed in vivo in the BAL of influenza-experienced mice following intranasal LPS administration (Fig. 2b) or S.p. infection (Fig. 2c). Together, these data reveal that post-influenza AMs display a specific response profile to TLR-stimulation or bacterial infection in vitro and in vivo, with increased production of select cytokines such as IL-6.
Figure 2. Alveolar macrophages from post-influenza mice produce increased IL-6 that confers protection from S. pneumoniae infection.
a, Alveolar macrophages isolated from the lungs of naive and post-influenza mice were stimulated ex vivo for 16 hours with 100ng/ml Pam3CSK4 or LPS. Cytokines were quantified in the supernatants by 36plex. n= 7 mice (PBS)/ n=11 mice (X31) from 2-3 independent experiments. (a, TNF– Pam3CSK4 n.s. = 0.37/LPS n.s. = 0.15) b-c, Cytokine quantification by ELISA in the broncho-alveolar lavage (BAL) of mice (b) 16 hours after in vivo administration of 1.5μg/50μl LPS i.n. (* = 0.038, n.s. = 0.8) or (c) 6 hours post infection with 5 x 105 CFU TIGR4/30μl i.n. (** = 0.0071, n.s. = 0.1915) d, Male and female C57BL/6 (PBS n=36 mice /X31 =25 mice) and IL-6-/- (PBS n = 21 mice, X31 n=13 mice) mice were infected with 5 x 105 CFU TIGR4/30μl i.n. (*=0.0171) in 5 independent experiments e, Mice were treated with 90μg anti-IL6 (MPF-20F3) in 50μl i.n. at -2 hours prior to S. pneumoniae infection and survival was monitored. n=26 mice (X31/X31 +anti-IL-6)/n=24 mice (PBS) from 3 independent experiments (**=0.0088) f, Naive mice were treated with 500ng IL6 in 50μl i.n. at -3hrs and +16hrs post-S. pneumoniae infection and survival was monitored. n=21 mice (PBS)/ n=22 mice (PBS + IL-6) from 4 independent experiments (**=0.0027).
Data shown as arithmetic means ±SD and statistical significance assessed by a two-tailed test using Mann-Whitney U (a-c) or Log Rank (Mantel-Cox) test for survival (d-f). * p=≤0.05, *** p=≤0.01, *** p=≤0.001, **** p=≤0.0001.
As IL-6-mediated antibacterial protection has been suggested previously25, we examined IL-6 effects against S.p. Naive mice produce low levels of IL-6 in response to S.p. (Fig. 2c), and IL-6 deficiency did not increase their susceptibility to pneumococcal infection. However, in influenza-experienced mice, IL-6 deficiency abrogated protection from S.p. infection (Fig. 2d). Similarly, an IL-6 blocking antibody significantly reduced the antibacterial protection of influenza-experienced mice (Fig. 2e). Administration of exogenous IL-6 to the lung conferred significant antibacterial protection to naive mice (Fig. 2f), resembling the survival phenotype observed in post-influenza mice. ELISA performed on sorted Siglec F positive and –negative cells confirmed that post influenza, the majority of lung IL-6 is produced by AMs, and the significant increase in IL-6 production post-influenza is exclusive to AMs (Supplementary Fig. 3g). Together, these results indicate that at one month post-influenza, increased IL-6 production by alveolar macrophages is crucial for protection against S.p., whereas the anti-IL-6 antibody treatment suggests that apart from IL-6, other factors contribute to enhanced protection.
CCR2-dependent monocyte recruitment is required for development of post-influenza AMs that produce increased IL-6
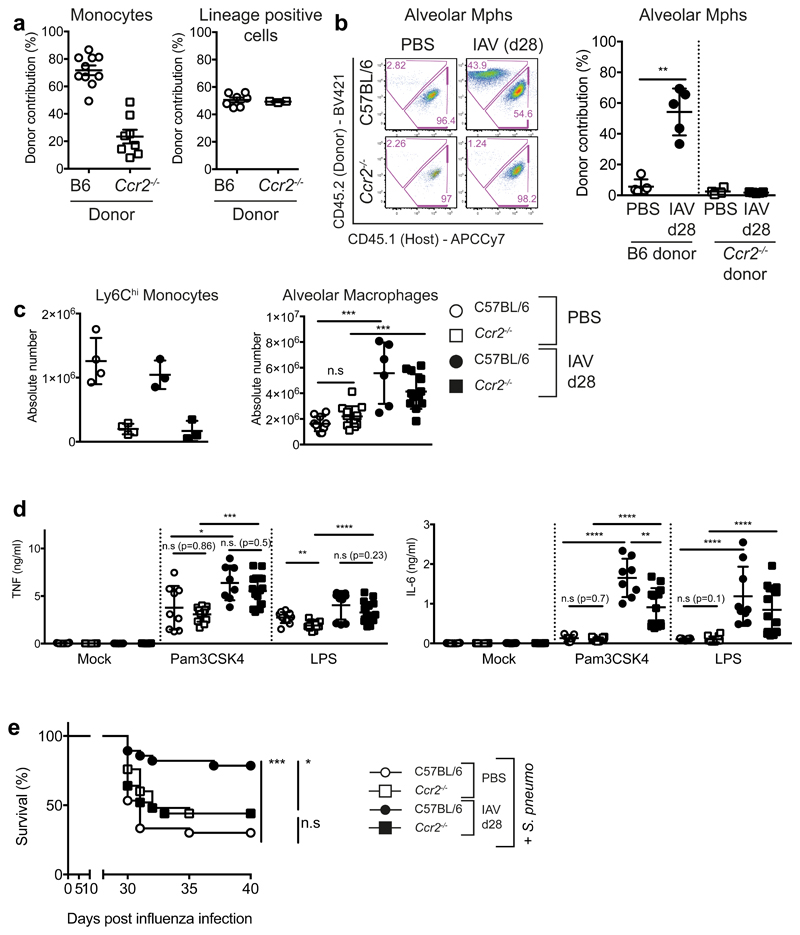
We observed a shift in the expression levels of surface markers CD11b, CD64, MHC class II, CD200R, and MARCO on post-influenza AMs in comparison to naive AMs (Supplementary Fig. 3h), suggestive of monocytic origin13,17,26. To assess the origin of alveolar macrophages post-influenza, we used the myeloablative drug Busulfan to generate bone-marrow chimeras. Busulfan usage was favoured over total body irradiation, to preserve the host peripheral cell populations, while still allowing engraftment of donor bone marrow (BM). C57BL/6J (CD45.1+) mice were used as hosts for the transfer of donor C57BL/6J (CD45.2+) or Ccr2–/– (CD45.2+) bone marrow (Supplementary Fig. 4a). A substantial but incomplete replacement of circulating innate and adaptive immune cells was found in these chimeras, regardless of donor genotype (Fig. 3a, Supplementary Fig. 4b,c). Congruent with the role of CCR2 in egress of monocytes from the BM27, mice reconstituted with Ccr2–/– BM had low frequencies of circulating donor monocytes, in contrast with mice reconstituted with Ccr2+/+ BM (Fig. 3a, Supplementary Fig. 4c). In naive mice, AMs remained predominantly of host origin (Fig. 3b), confirming that in steady state, BM-derived cells do not contribute to this population6,15. Post influenza, AMs were found to contain a significant contribution (~44%) of donor Ccr2+/+ (CD45.2+) BM-derived cells. In contrast, no donor Ccr2–/– (CD45.2+) BM-derived cells were present in the AM population post-influenza (Fig. 3b). These results confirm that a CCR2-dependent population of BM-derived cells, most likely Ly6Chi blood monocytes19, differentiate into AMs during or following influenza virus infection.
Figure 3. CCR2-dependent monocyte recruitment contributes to the population of AMs and their altered functional profile at one month post-influenza.
a,b, CD45.1 mice were treated with Busulfan and reconstituted with CD45.2 B6 or Ccr2-/- BM. a, Frequency and chimerism of blood monocytes (n=10 mice C57BL/6, n=8 mice Ccr2-/-) and lineage (CD3/NK1.1/CD19) positive cells (n=7 mice C57BL/6, n=3 mice Ccr2-/-) 40 days post-reconstitution, prior to influenza infection. 2 independent experiments. b, Chimerism of alveolar macrophages in the lung one month post-influenza (n=5 mice from two independent experiments **=0.0079). c, Quantification of cells identified in naive and post-influenza lungs of B6 (n=10 mice (PBS)/n=6 mice (X31)) and Ccr2-/- (n=13 mice (PBS)/n=12 mice (X31) mice by flow cytometry. 3 independent experiments (n.s.=0.1010, ***=0.0005). d, Alveolar macrophages were isolated from the lungs of naive and post-influenza C57BL/6 and Ccr2-/-mice and stimulated ex vivo for 16 hours with 100ng/ml Pam3CSK4 or LPS. IL-6 and TNF were quantified in the supernatants by ELISA. n=5-15 mice from 2-4 independent experiments. e, 8-10 week old female C57BL/6 (n=30 mice (PBS)/ n=28 mice (X31)) and Ccr2-/- (n=25 mice (PBS/X31) mice were infected with 8000 TCID50 X31/30μl i.n. 28 days later, all mice were infected with 5 x 105 CFU TIGR4/30μl i.n. (S. pneumoniae) and monitored for survival. 3 independent experiments (*= 0.0167, n.s = 0.08).
Data shown as arithmetic means ±SD and statistical significance assessed using two-tailed tests; Mann-Whitney U or Log Rank (Mantel-Cox) test for survival (e). * p=≤0.05, *** p=≤0.01, *** p=≤0.001, **** p=≤0.0001.
Because of incomplete chimerism, this model could not be used to test the contribution of the recruited cells to the increased antibacterial protection. To address this question, we infected Ccr2+/+ and Ccr2-/- animals and analysed their responses at one month post-influenza. In both genotypes, the infection led to an increase in the number of AMs (Fig. 3c), despite the absence of Ly6Chi monocytes in Ccr2-/- animals.
Compared to IAV d28 Ccr2+/+ AMs, the production of IL-6 was significantly reduced in IAV d28 Ccr2–/– AMs (Fig. 3d), corresponding with reduced protection against pneumococcal infection in influenza-experienced Ccr2–/– mice (Fig. 3e). In contrast, no difference in TNF production was observed (Fig. 3d).
Of note, no significant changes were seen in the cellular composition of the BM at one month post-influenza (Supplementary Fig. 4d), thus excluding the possibility that major changes in myeolopoiesis are responsible for the improved antibacterial protection, as described in other experimental systems28.
We conclude that CCR2-dependent BM derived cells are recruited to the lung during influenza, and are required for subsequent S.p. protection.
Bone marrow-derived AMs are the main contributors of protective IL-6
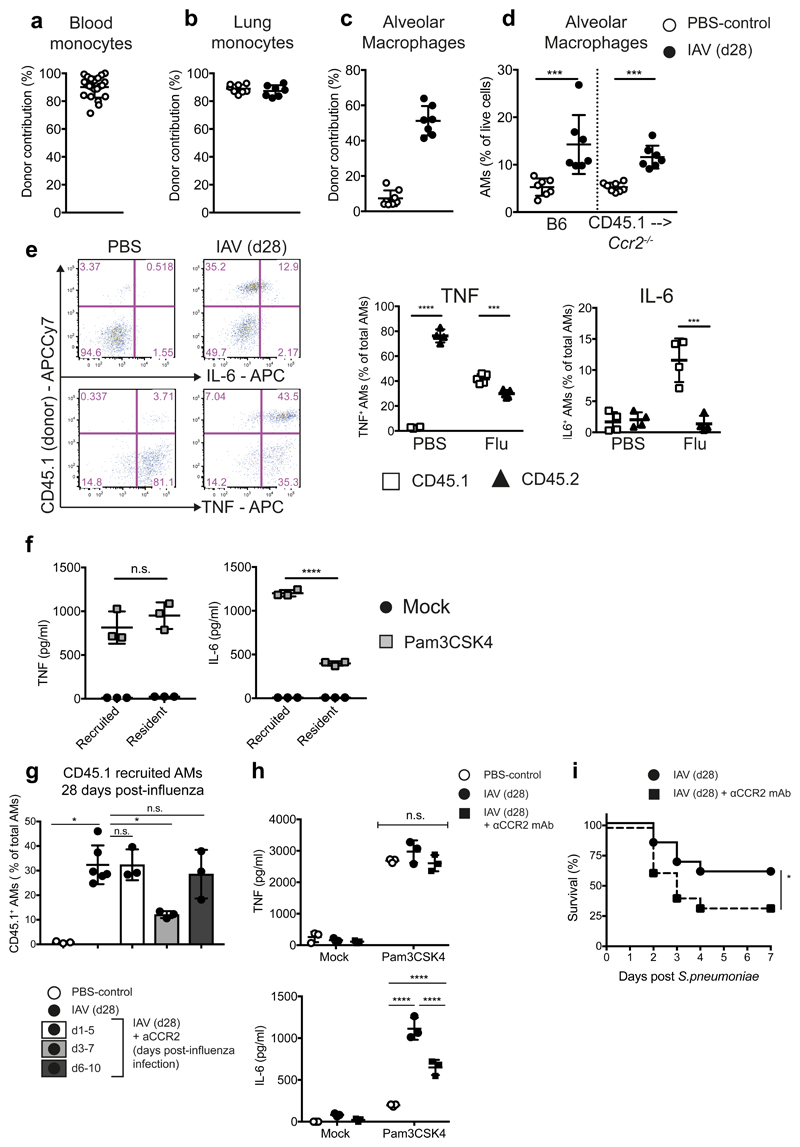
To further dissect the role of monocyte-derived AMs in antibacterial protection, we established a chimera in which macrophage origin post-influenza could be unequivocally identified. Ccr2–/– (CD45.2+) mice were treated with Busulfan and reconstituted with CCR2-competent congenically marked C57BL/6J (CD45.1+) bone marrow. A complete replacement of blood and lung monocytes was achieved in these mice (Fig. 4a,b), while retaining the host origin of AMs in naive mice (Fig. 4c). Following influenza infection, the frequency of AMs increased, reflecting the Ccr2+/+ situation (Fig. 4d). Of these cells, ~55% were identified by CD45.1 expression to be of BM origin (Fig. 4c).
Figure 4. Quantifying the contribution of CCR2-dependent monocytes to alveolar macrophages and their cytokine production at one month post-influenza.
Ccr2-/- mice were treated with Busulfan and reconstituted with CD45.1 WT BM. a, Chimerism of blood monocytes 30 days post-reconstitution and prior to influenza infection (n=22 mice). b,c, Chimerism of (b) lung monocytes and (c) alveolar macrophages in naïve (n=8 mice) and influenza-experienced (n=7 mice) mice. d, Frequency of alveolar macrophages in naive and influenza experienced WT and chimeric mice. (b-d) Represents two independent experiments. e, Naive and IAV (d28) alveolar macrophages were isolated from the lungs of chimeric mice and stimulated ex vivo with 100ng/ml Pam3CSK4. ICCS for IL6 (top panels) and TNF (bottom panels) n=4 mice from 1 experiment. f, Donor (CD45.1) and host (CD45.2) alveolar macrophage populations from post-influenza mice were FACS-sorted and stimulated for 16hr with 100ng/ml Pam3CSK4. IL-6 and TNF were quantified by ELISA in the supernatants. n=3 mice from one experiment. (n.s.= 0.74) g, Chimeras as above were treated with anti-CCR2 mAb (MC21) at the indicated time points post-influenza and the BM-derived (CD45.1) cell contribution to AMs measured at d28 post-influenza. n=3 (all groups except for n=6 mice (X31)) from 1 experiment. n.s. = 0.9, *=0.02
h,i, Female C57BL/6 mice were influenza-infected and administered anti-CCR2 mAb or PBS at d3-7. h, Alveolar macrophages were isolated on d28 from whole lung and stimulated ex vivo for 16 hours with 100ng/ml Pam3CSK4. Cytokines were quantified in the supernatants by ELISA. n=3 mice from 1 experiment. i, At d28 post-influenza, mice were infected with 5 x 105 CFU TIGR4/30μl i.n. (S. pneumoniae) and monitored for survival. n=25 mice (X31)/n=24 mice (X31 + anti-CCR2) from three independent experiments *=0.0479.
Data shown is arithmetic means ±SD and statistical significance assessed by a two-tailed test using Mann-Whitney U or Log Rank (Mantel-Cox) test for survival (i). * p=≤0.05, *** p=≤0.001, **** p=≤0.0001.
AMs were isolated from the lungs of chimeric mice post-influenza, and stimulated ex vivo to compare cytokine production by resident CD45.2+ (Ccr2–/–) and recruited CD45.1+ (Ccr2+/+) macrophages. Intracellular cytokine staining demonstrated that recruited, BM-derived (CD45.1+) AMs were responsible for the majority of the detectable IL-6 produced following stimulation (Fig. 4e, Supplementary Fig. 4e). Both resident (CD45.2+) and recruited (CD45.1+) AMs produced equivalent amounts of TNF, regardless of origin or infection history (Fig. 4e). This was confirmed by flow-sorting recruited (CD45.1+) and resident (CD45.2+) AMs post-influenza, and measuring the amounts of IL-6 and TNF by ELISA following ex vivo stimulation of these populations (Fig. 4f).
To define the time window when CCR2-dependent monocyte recruitment into the infected lung is required for AM repopulation, we treated Busulfan chimeras at three different time intervals during influenza with αCCR2 blocking antibody. We found that inhibition of CCR2 on days 3-7 significantly reduced the monocyte contribution to the AM pool (Fig. 4g), and prevented the increased production of IL-6 by AMs, but did not alter TNF (Fig. 4h). Using this model also allowed us to discount confounding factors of monocyte absence during S.p. infection in constitutively CCR2-deficient mice: treatment with anti-CCR2 blocking antibody at days 3-7 of influenza infection led to reduced protection during S.p. infection 3 weeks later (Fig. 4i), replicating the phenotype of the Ccr2–/– mouse (Fig. 3e).
To determine the impact of the inflamed environment on monocyte differentiation in the infected lung, we investigated the phenotype of recruited AMs which developed in a non-inflammatory setting. Resident AMs were depleted with clodronate-loaded liposomes. One month post-liposome treatment, no increase in IL-6 production by AMs was seen (Supplementary Fig. 4f), demonstrating that only monocytes recruited during the inflammatory context of acute influenza sustain a phenotype of increased IL-6 production.
Transcriptomic and chromatin profiles of AMs reveal that origin, not infection history, determines cellular identity
To characterise the differences between recruited and resident AMs more globally, we performed RNA-sequencing of AMs which were flow-sorted (based on congenic markers) from naive and post-influenza lungs of Busulfan chimeras, generated as described above (Fig 4). To exclude that the CCR2 deficiency conditions the transcriptional profile of resident AMs in the BM chimeras, we compared these samples to resident AMs from naive Ccr2+/+ mice and found no significant effect of Ccr2 genotype on resident AMs (Supplementary Fig. 5a), validating our chimera approach. Resident CD45.2+ macrophages from naive and post-influenza lungs were found to be strikingly similar (Fig. 5a). In contrast, there were substantial transcriptional differences between recruited CD45.1+ (Ccr2+/+) and resident CD45.2+ (Ccr2–/–) AMs that were isolated from the same chimeric lung post-influenza (Fig. 5a and Supplementary Fig. 5b,d). These data demonstrate a significant impact of origin on the transcriptional profile of AMs at one month post-influenza, while infection history is less important, as also reflected in the Principal Component Analysis (PCA) of the transcriptional data (Fig. 5b).
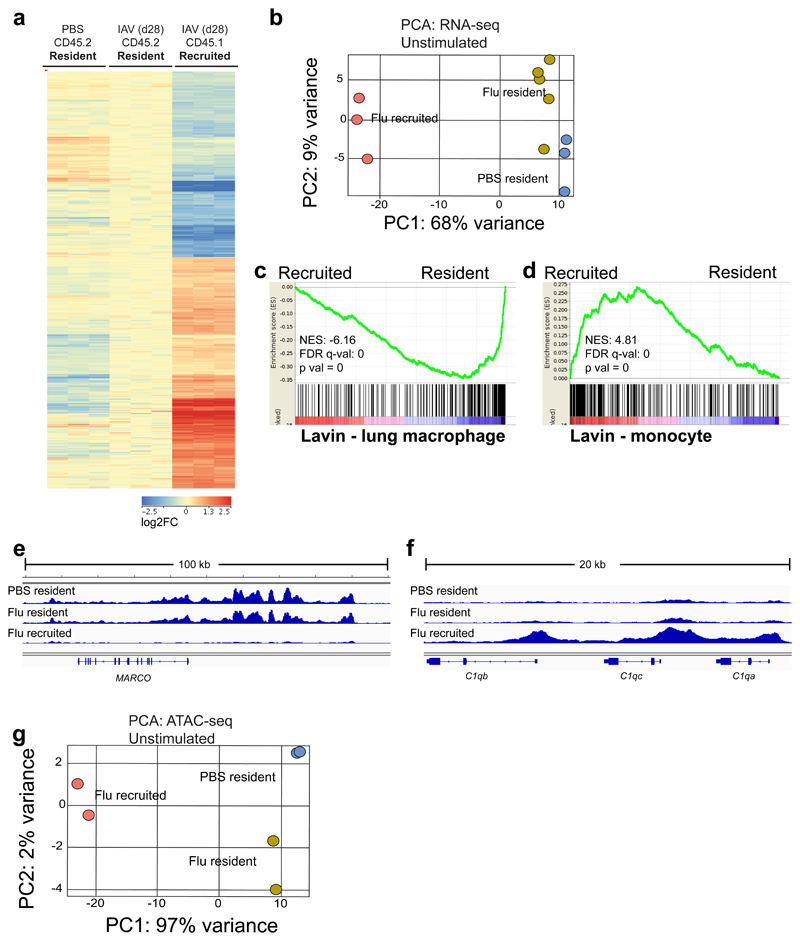
Figure 5. Recruited macrophages are distinct in their expression and chromatin profile, relative to resident macrophages at one month post-influenza.
RNA-seq and ATAC-seq of AMs FACS-sorted from the lungs of naïve and post-influenza Busulfan chimeras. a, Heat map of genes differentially expressed (FC>1.5, 1-way ANOVA with Benjamini-Hochberg correction, p<0.05) between CD45.2 (Ccr2-/-) resident and CD45.1 recruited AMs. b, Principal component analysis (PCA) plot of RNA-seq data from resident and recruited macrophages from post-influenza and naive lungs of Ccr2-/- Busulfan chimeras. c,d, Gene set enrichment analysis (GSEA) of Flu resident (CD45.2) vs Flu recruited (CD45.1) macrophages using clusters from Lavin et al.30 representing (c) lung macrophages and (d) blood monocytes. e,f, IGV tracks from ATAC-seq data; depicting chromatin in unstimulated resident and recruited macrophages from naïve and post-influenza lungs of Ccr2-/- Busulfan chimeras. All tracks were group-autoscaled to enable comparison. g, PCA plot of ATAC-seq data from unstimulated resident and recruited macrophages from naïve and post-influenza lungs.
Pairwise comparisons confirm the similarity between resident AMs from naive versus post-influenza mice with only 60 genes differentially expressed between these two groups, while recruited AMs differed from each of the resident populations in the expression of 400-500 genes (Supplementary Fig. 5b). These pairwise comparisons were further interrogated by Ingenuity Pathway Analysis (IPA) to identify canonical pathways in which the AM subsets differ (Supplementary Fig. 5d, first three columns). We found inflammation- and STAT3-related pathways most prominent when recruited AMs were compared to any of the resident AMs. The few differences between resident AMs from naive and post-influenza mice mapped to prostanoid and eicosanoid synthesis and, interestingly, to Chronic Obstructive Pulmonary Disease (COPD).
We then used gene set enrichment analysis (GSEA) to investigate genes differentially expressed between resident and recruited AMs. In a comparison with published datasets, we found that resident (CD45.2+) AMs were enriched for signatures corresponding to lung macrophages of embryonic origin from several independent studies (Fig. 5c and Supplementary Fig. 5e)17,29–31. Conversely, a Ly6Chi monocyte signature was enriched in recruited (CD45.1+) macrophages (Fig. 5d and Supplementary Fig. 5f). This is in line with the notion that CCR2-dependent recruited AMs are of Ly6Chi monocyte origin and indicates that, despite their surface phenotype and morphology, monocyte-derived AMs still retain a large proportion of the transcriptional profile of their progenitors.
Overlap of our transcriptional dataset with others17,29–31 allowed us to retrieve common genes related to a resident, embryonic origin of AMs, such as Marco. ATAC-seq analysis revealed regions of open chromatin at the Marco locus in resident AMs from naive and influenza-experienced mice (Fig. 5e), while this locus was completely inaccessible in recruited macrophages. Conversely, the C1q locus was accessible in recruited AMs post-influenza, but closed in resident populations (Fig. 5f). Further GSEA, using publicly available databases, showed that recruited AMs were enriched for pathways related to cell cycle, as observed in other organs8 (Supplementary Fig. 5g,h).
PCA clustering of ATAC-seq data revealed that the PC1 axis which separated resident from recruited macrophages represented almost the entirety (97%) of total variance (Fig. 5g). In contrast, resident macrophages clustered closely together (PC2: 2%) independently of prior influenza exposure (Fig. 5g). Therefore, at one month post-influenza, the major determinant of variance observed in the transcriptional and chromatin profile of AMs is not infection history, but origin.
We then compared gene expression (Fig. 6a, Supplementary Fig. 6a) and chromatin accessibility (Fig. 6b) in unstimulated and Pam3CSK4-stimulated resident (CD45.2+) and recruited (CD45.1+) AMs one month post-influenza. Although stimulation was seen to account for the majority (PC1: 60%) of the transcriptional variance (Fig. 6a), origin was still a powerful criterion to separate these groups (PC2: 24%), while infection history remained largely irrelevant. In contrast, chromatin accessibility as measured by ATAC-seq was less influenced by the short-term in vitro TLR-stimulus, and the bulk of variance (92%) was explained by origin (Fig. 6b), again with little impact of influenza exposure history.
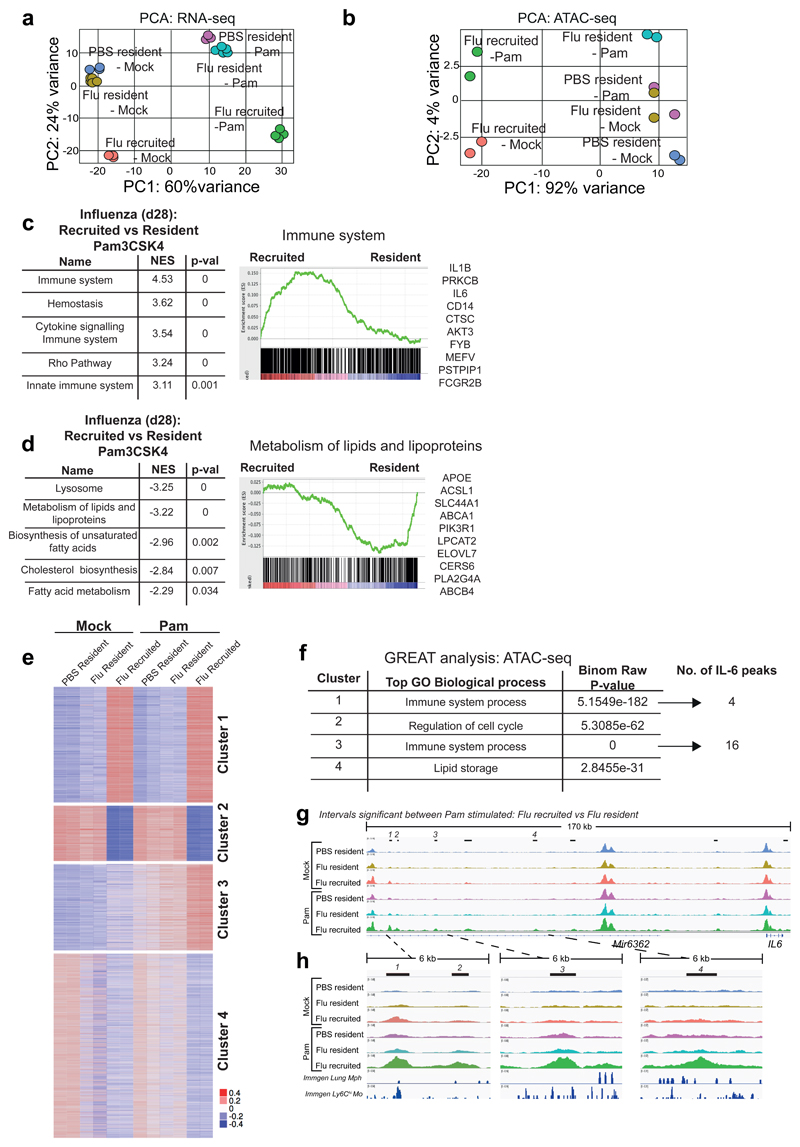
Figure 6. Following TLR stimulation, origin determines gene expression, chromatin profile and IL-6 gene accessibility.
AMs were FACS-sorted based on congenic markers from the lungs of naïve and post-influenza Busulfan chimeras, and stimulated ex vivo with Pam3CSK4. PCA plot of (a) RNA-seq data and (b) ATAC-seq data from unstimulated and Pam3CSK4-stimulated CD45.2 (Ccr2-/-) resident and CD45.1 recruited AMs. c,d, Top 5 pathways (FDR q-val <0.01) from GSEA of Pam3CSK4 stimulated AMs from post-influenza lungs. NES = Normalized Enrichment Score. Representative GSEA plots are shown, with the top 10 highest ranked genes from each GSEA listed. e, ATAC-seq data as in (b) shown as heatmap of mean centred k-means clustering of rlog values generated by DESeq2 for differentially accessible intervals found to be significant (FDR ≤ 0.01) in at least one pairwise comparison. f, GREAT analysis of ATAC-seq-derived intervals from each cluster in (e). g,h, IGV tracks of ATAC-seq data for AMs, showing the IL-6 locus and upstream peaks. Significantly differentially accessible intervals in the comparison Flu_resident_Pam vs Flu_recruited_Pam are marked using bars. ImmGen chromatin tracks from lung macrophages and blood Ly6Chi monocytes are shown underneath (h). All tracks were group-autoscaled to enable comparison.
Interrogation of the RNA-seq data by GSEA revealed an enrichment of pathways associated with the immune response in stimulated, recruited (CD45.1+) AMs (Fig. 6c). IL-6 was identified as a top gene in these pathways, confirming our previous in vitro and in vivo observations. Il1b was also seen to increase at the mRNA level, but no protein was detectable (Supplementary Fig. 3d). Conversely, resident (CD45.2+) macrophages were enriched for pathways associated with metabolism of lipids, including genes such as Apoe (Fig. 6d and Supplementary Fig. 5h).
K-means clustering of ATAC-seq data (using 4 clusters) (Fig. 6e), led to the identification of biological states similar to the RNA-seq. Cluster 1 and cluster 2/4 denoted sites which were accessible to a greater degree in macrophages of recruited or resident origin respectively, regardless of stimulation. Cluster 3 represented sites that opened in response to Pam3CSK4 stimulation, and to a greater degree in recruited (CD45.1+) AMs post-influenza. To assign biological meaning to these regions, we used GREAT (Genomic Regions Enrichment of Annotations Tool). GREAT analysis of these clusters revealed GO biological processes that were parallel to the pathways identified by GSEA of RNA-seq data (Fig. 6f, extended list in Supplementary Fig. 6b). In clusters where recruited AMs show higher chromatin accessibility (cluster 1 and 3), immune function was the top pathway assigned. Interestingly, cluster 1 shows that many immune genes are more accessible already in unstimulated recruited AMs, indicating that these cells are prone to mount a vigorous immune response. Within the top GO biological process (Immune system process) identified by GREAT analysis in cluster 3, sixteen IL-6 gene–associated genomic intervals were differentially accessible in recruited AMs following Pam3CSK4-stimulation (Fig. 6f). 5 of these peaks were identified within 100-200kb upstream of the IL-6 transcription start site (Fig. 6g,h), a feature often observed in enhancer regions32. Some of these intervals were confirmed by DESeq2 to be significantly more accessible in recruited AMs than in resident AMs following Pam3CSK4 stimulation (see top bars in Fig. 6g,h, comparing Pam3CSK4-stimulated recruited to resident AMs from Flu-experienced mice). These chromatin regions were noticeably more open already in the unstimulated recruited macrophages, while this locus remained largely closed in resident macrophages, even following stimulation (Fig. 6h).
We overlaid our peaks with publicly available ATAC-seq data obtained from the ImmGen database (Fig. 6h), corresponding to “Lung macrophages”, or “Ly6Chi blood monocytes”. Regions of chromatin which were accessible in “Ly6Chi monocytes”, yet closed in “Lung macrophages”, were found to intersect with open chromatin surrounding the IL-6 locus in recruited, but not resident AMs (Fig. 6h). This demonstrates that open chromatin in recruited macrophages upstream of the IL-6 locus may be related to their monocyte origin, and likely promotes increased expression of this gene following TLR stimulation.
Monocyte-derived AMs persist in the lung at two months post-influenza, with an altered phenotype
We then used Busulfan chimeras to understand the persistence of antibacterial protection and AM profiles. At two months post-influenza infection, the frequency of total AMs in the lung remained high (Fig. 7a), and the contribution of bone-marrow derived cells increased (Fig. 7b). To exclude that this contribution is due to a constant influx of bone-marrow derived monocytes into post-influenza lungs, we administered Busulfan to wild-type C57BL/6J (CD45.2+) mice on day 28 post influenza, followed by transfer of congenically marked (CD45.1+) bone marrow, and monitored the appearance of BM-derived cells in the AM pool for 20 days. No such cells were found (Supplementary Fig. 6c), further confirming our previous finding (Fig. 4g) that the BM-derived cells contributing to the AM pool are recruited in a limited time window during influenza infection and subsequently become part of the locally self-renewing AM population, without further contribution from the bone marrow.
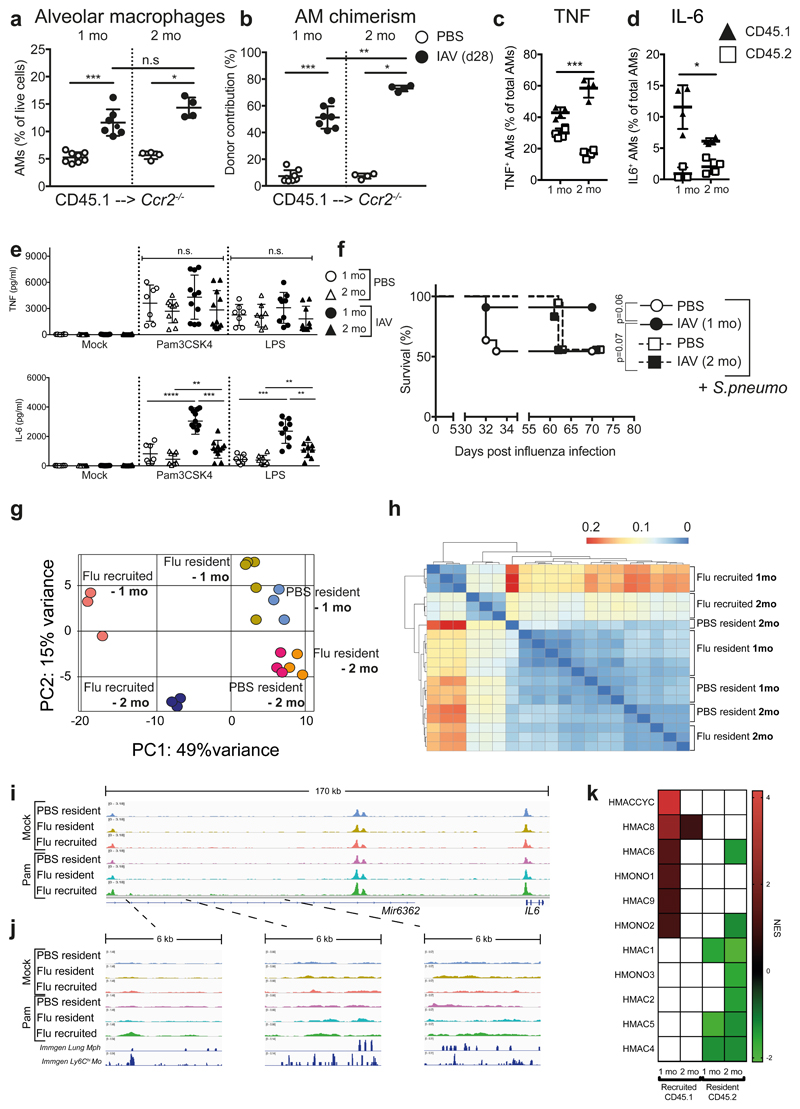
Figure 7. Monocyte-derived alveolar macrophages persist for 2 months post-influenza, but do not produce increased IL-6 and afford bacterial protection.
a,b, Frequency (a) and chimerism (b) of alveolar macrophages in the lungs of CD45.1 --> Ccr2-/- Busulfan chimeras at one (n=8 mice PBS/n=7 mice X31) and two months (n=4 mice) post-influenza infection. From 1-2 separate experiments. c,d, Alveolar macrophages were isolated from the lungs of CD45.1 --> Ccr2-/- Busulfan chimeras at one and two months post-influenza infection and stimulated ex vivo with Pam3CSK4. TNF (c) and IL-6 (d) were assessed by intracellular cytokine staining. n=3-5 mice from 1 experiment. e, Alveolar macrophages were isolated from the lungs of naive and 2-mo post-influenza C57BL/6 mice and stimulated ex vivo for 16 hours with 100ng/ml Pam3CSK4 or LPS. IL-6 and TNF were quantified by ELISA in the supernatants. n=7-16 mice from 4 independent experiments. f, At one or two months post-influenza infection, mice were infected with 5 x 105 CFU TIGR4/30μl i.n. (S. pneumoniae) and monitored for survival. n=11 mice (PBS 1 mo/X31 1mo) n=12 mice (X31 2 mo) n=18 mice (PBS 2 mo) from 2-3 independent experiments. n.s.= 0.0734 g, PCA plot of RNA-seq data from unstimulated resident (CD45.2) and recruited (CD45.1) AMs from the lungs of post-influenza and naive Ccr2-/- Busulfan chimeras at one and two months post-infection. h, Hierarchical clustering of RNA-seq data using Pearson dissimilarity (1-correlation) of the samples’ rlog values generated by DESeq2. i,j, IGV tracks from ATAC-seq of unstimulated and Pam3CSK4-stimulated resident (CD45.2) and recruited (CD45.1) AMs from the lungs of post-influenza and naive Ccr2-/-Busulfan chimeras at two months post-infection. The intervals chosen are identical to those in Fig. 6g,h. The scale on corresponding tracks are identical to Fig. 6g,h. to to enable comparison. k, Heatmap showing the NES of GSEA performed using human macrophage, monocyte and dendritic cell datasets obtained from Zilionis et al.34.
Data shown as arithmetic means ±SD and statistical significance assessed by two-tailed tests using 2-way ANOVA or Log Rank (Mantel-Cox) test for survival. * p=≤0.05, ** p=≤0.01, *** p=≤0.001.
We tested the cytokine profile of recruited AMs at two months post infection by intracellular cytokine staining. The amount of TNF produced was reflective of the degree of chimerism (Fig. 7b), demonstrating that on a per-cell basis, both resident (CD45.2+) and recruited (CD45.1+) AMs produced equivalent amounts of this cytokine (Fig. 7c). However, the amount of IL-6 production by recruited (CD45.1+) AMs was significantly reduced at two months post-influenza (Fig. 7d). This was also evident by bulk analysis of all post-influenza AMs at two months from wild-type mice (Fig. 7e), suggesting that the cytokine profile characteristic of recruited cells is slowly waning to resemble that of embryonic-derived resident AMs. The reduction in IL-6 production by AMs at two months post-influenza is accompanied by a loss of protection against S.p. infection (Fig. 7f), indicating that the increased protection is sustained by recruited AMs as long as they retain an altered cytokine profile.
In comparison to the significant differences seen in the transcriptional profile of recruited AMs at one month post-influenza, recruited AMs at two months post-influenza clustered more closely to the resident AMs in PCA (Fig. 7g). Pearson correlation (Fig. 7h), unsupervised gene clustering (Supplementary Fig. 6d), and IPA of the pairwise comparisons (Supplementary Fig. 5d, last three columns to the right) all confirmed greater similarity between recruited AMs and resident AMs at 2 months than at one month.
To analyse globally the changes over time that lead to progressively increasing resemblance between recruited and resident AMs, we grouped the data points shown in Fig. 7g into three sample clusters: 1-month recruited (cluster 1), 2-month recruited (cluster 2) and all resident (cluster 3) AMs. We statistically filtered the genes against the null condition that they are the same across all three clusters (DESeq2 LRT test, FDR<0.05), then classified genes based on their progressive upregulation (cluster1<2<3) or downregulation over time (cluster3<2<1, Supplementary Fig. 6e). Pathways associated with the genes from these two classifications were obtained using GSEA (Supplementary Fig. 6f). Genes which were upregulated with increased residence time, are associated with TNF- and IL1-related immune pathways, while genes which were progressively downregulated map to inflammatory and IL-6-related pathways (Supplementary Fig. 6f). This indicates that loss of some but not all immune reactivity characterises the progressive change of recruited AMs.
We then investigated the accessibility of the chromatin at two months post-influenza by ATAC-seq. Loci upstream of IL-6, which at one month were markedly open (Fig. 6h), were no longer identified by DESeq2 to be differentially accessible at two months, even after Pam3CSK4 stimulation (Fig. 7i,j).
Single cell studies of human lung immune cells have demonstrated the broad range of myeloid cell phenotypes which exist both at steady state and in disease, such as lung cancer33,34, but have not addressed macrophage origin. To understand if origin may explain some of the characteristics described in these studies, we used our datasets to perform GSEA against macrophage, dendritic cell and monocyte gene sets identified by scRNA-seq in human and mouse lung cancer34. We find recruited and resident AMs post-influenza to represent distinct human subsets (Fig. 7k, nomenclature from ref. 34). The gene sets which characterise steady-state “alveolar macrophages”, present also in healthy lung34 (mMac4, hMac4, hMac5), are highly enriched in our lineage marker-defined resident AMs (Fig 7k, Supplementary Fig. 7a-c). In contrast, monocyte gene sets (mMono1, mMono2, mMono3, hMono1, hMono2) are significantly represented in our recruited AMs at one month, but not two months. Strikingly, all subsets identified as strong cytokine producers in these studies (mMac1, hMac8 and 9) map to our expression profile of recruited AMs. This is also in line with another study showing that tumor macrophages produced significantly more IL-6 than macrophages derived from non-involved lung tissue33, although macrophage origin was not identified in that study. Overall, our data strongly suggest that, across species and across disease states in the lung, monocyte-derived AM subsets can be considered the most immunologically reactive.
Together, this study shows a profound impact of monocyte legacy in the transcriptional profile and chromatin accessibility of recruited alveolar macrophages for an extended period after resolved influenza infection, which offers a molecular explanation for the functional responses of these cells. Over time, the transcriptional, chromatin and functional profile of these recruited AMs becomes more similar to that of resident AMs, and the non-specific protection due to previous influenza exposure is lost. In contrast to the dynamic changes in recruited AMs due to previous influenza infection, the exposure history has remarkably little effect on chromatin accessibility, transcription and function of resident AMs.
Discussion
Influenza is a common respiratory disease which annually infects over 800 million people worldwide. Despite its prevalence, the prolonged impact of this ubiquitous infection on lung immunity is not understood. We have shown that a previous, resolved influenza infection confers extended protection against the antigenically distinct bacterium S.pneumoniae. Central to this non-specific protection is a population of recruited, monocyte-derived AMs. Recruited AMs have a similar surface phenotype to resident AMs, but differ in their functional, transcriptional and epigenetic profile which is beneficial during S.p. infection.
To precisely address AM origin and its implications on function, we developed a chimeric model utilising Busulfan6,15. Functional assays, RNA- and ATAC-seq all revealed that recruited, but not resident, AMs resemble the profile of monocytes. Thus, recruited AMs retain an epigenetic legacy of their monocyte origin, which facilitates induction of select immune genes to provide a specific immune response. Our definition of an epigenetic legacy in recruited macrophages is therefore an alternative to previously described mechanisms of “trained immunity” in resident innate immune cells, which refer to the induction of de novo histone modifications following stimulation35,36.
The recruitment of CCR2-dependent monocytes is central to the prolonged antibacterial protection. With the use of blocking mAbs we have identified day 3 to day 7 as the time window during acute influenza when monocyte recruitment is crucial for the later contribution to the AM pool. We find no evidence of monocyte engraftment following resolution of influenza, as the niche is no longer accessible18. We demonstrate, using CCR2 blocking antibodies and clodronate liposomes, that recruitment during this period of acute inflammation is central to the prolonged ability of monocyte-derived AMs to produce IL-6. Therefore, we hypothesise that this inflamed lung environment promotes the retention of the Ly6Chi monocyte profile observed in recruited AMs, while a non-inflammatory lung promotes terminal differentiation into steady-state resident AMs. When Ly6Chi monocytes differentiate into AMs under non-inflammatory conditions in the newborn lung, these cells transcriptionally resemble their embryonically derived counterparts after 6 weeks19. On the contrary, following pro-inflammatory bleomycin treatment, monocyte-derived macrophages differentially express ~5000 genes at 14 days post recruitment, which is reduced to 330 genes at 10 months post-injury12. Furthermore, in a non-inflamed liver, monocyte-derived Kupffer cells transcriptionally resemble their embryonic counterparts after 30 days37. The minimal differences between steady-state and post-influenza resident AMs may be explained by lower plasticity due to terminal differentiation of these populations. This is in line with the observation that differentiated tissue macrophages from other organs, such as liver, peritoneum or colon, were not able to develop into AMs following transfer to the lungs19.
The determining factor that results in monocyte recruitment versus macrophage proliferation during infection is not known. Local signals can instruct the proliferation of resident alveolar macrophages under certain conditions10,38. During influenza, we observe an almost complete disappearance of AMs from the airways, and monocyte recruitment that is typical for inflammatory processes39. Thus, the scarcity of AMs in combination with readily available monocytes may influence the source of AM replacement. In cases of less dramatic AM depletion there may be less monocyte recruitment, and local macrophage proliferation may be sufficient to replenish the lung niche15. Since we find no major effect of influenza on the functional or transcriptional profile of resident AMs, we speculate that a prolonged change will occur only if the insult is severe enough to induce monocyte recruitment15. This may explain why in other influenza studies, increased AM reactivity was not found2.
We have demonstrated that monocyte-derived macrophages are beneficial in the context of S.p. infection. However there may be instances in which a higher pro-inflammatory output by these cells could contribute to a detrimental outcome, such as in the development of chronic lung diseases including COPD and fibrosis. IL-6 has been implicated in tumor invasiveness and in several diseases including rheumatoid and juvenile idiopathic arthritis, arteritis 40, and other conditions where infections are implicated in inflammatory flares. The mechanisms described here might provide an explanation for how infections can induce cells with a high propensity for production of pro-inflammatory cytokines, and thereby promote inflammation. Studies have shown monocyte-derived macrophages in the lung to be responsible for driving a fibrotic phenotype following bleomycin, and for reducing the degree of allergic asthma in a situation of chronic latent gammaherpes virus infection3,12.
As we demonstrate monocyte-derived macrophages to be responsible for functional changes in lung immunity, it is important to determine the contribution of these cells in human disease. We found similarities between monocyte-derived macrophages post-influenza, and tumor-associated macrophages in human studies, in particular with regards to immune reactivity: the most cytokine-prone tumor-associated macrophages overlay with monocyte-derived AMs in our study33,34, implicating a broad role for these cells in human disease.
Overall, we have demonstrated that a single, transient viral infection changes lung immunity for a prolonged period, and have provided a cellular and molecular explanation for changes in lung immunity. Recruited alveolar macrophages retain chromatin accessibility and transcriptional profiles derived from their monocyte precursors that are consistent with altered immune reactivity and prove beneficial to host survival during subsequent bacterial infection. Monocyte-derived AMs become more similar to resident AMs over time, and are no longer capable of providing enhanced antibacterial protection. Our results suggest that in humans, the life-long sequence of respiratory infections will constantly re-shape immune reactivity, responsiveness to pathogens and inflammatory conditions in the lung.
Methods
Mice and infections
All experiments used mice at 6-12 weeks of age bred at the MRC-National Institute for Medical Research (NIMR) in Mill Hill (2013-2016), and subsequently at the Francis Crick Institute (2016-2019) under specific pathogen free conditions. All animal experiments were approved by the Home Office, UK, under the project licence 70/7643 add new licence and carried out in accordance with the Animals (Scientific Procedures) Act 1986 and the GSK Policy on the Care, Welfare and Treatment of Animals. All genotypes used were bred on a C57BL/6J background, and maintained as homozygous lines. Genotypes used were C57BL/6J, CD45.1 (B6.SJL.CD45.1), Ccr2–/– (kindly made available by Dr. Jean Langhorne and Dr. Anne O’Garra, The Francis Crick Institute), Il6-/- (Kindly given by Dr. Caetano Reis e Sousa, The Francis Crick Institute, originally generated by Manfred Kopf and Georges Kohler and imported from Jackson, Stock No 002650).
X31 (kind gift from Dr J. Skehel, MRC-NIMR) were grown in the allantoic cavity of 10 day-embryonated hen’s eggs and were free of bacterial, mycoplasma and endotoxin contamination. All viruses were stored at - 80°C and titrated on MDCK cells. Virus was quantified in infected lungs by qPCR for the influenza matrix gene on cDNA from whole lungs, normalized to the housekeeping gene Hprt..
Pre-infection body weights were recorded, and mice were weighed daily (at similar times of day), and monitored for clinical symptoms. Mice reached clinical endpoint following the loss of 25% of their initial starting weight, or at a clinical score of >5. Clinical scores were determined by (1 point each) piloerection, hunched posture, partially closed eyes, laboured breathing, hypothermia, decreased movement, movement only on provocation or (2 points) absence of movement on provocation or (5 points) middle ear infection (disrupted balance).
Mice were infected with Influenza (X31) and Streptococcus pneumoniae (TIGR4) under light anaesthesia (3% isoflurane). Substances were diluted in PBS (phosphate buffered saline) and administered in 30μl intranasally. With the exception of chimeras, mice were infected at 8-10 weeks of age with influenza (X31). At 28 days (1 month) or 60 days (2 months) following influenza infection, mice were challenged with S.p. (TIGR4)(a kind gift from Klaus Okkenhaug, Babraham Institute).
Mouse treatments
Mouse treatments such as LPS (1.5μg/50μl) (EnzoLifeSciences), IL-6 and anti-IL-6) were delivered under light anaesthesia (3% isoflurane) in 50μl intranasally (i.n.) to ensure delivery to the lower airways. IL-6 (500ng/50μl) (R&D) was delivered at -3hr and +16hrs relative to S.p. infection. α-IL-6 (Clone MP5-20F3) was a kind gift from Dr. Stefan Rose-John (University of Kiel) and delivered at 90μg/50μl 1 hour prior to S.p. infection. For anti-CCR2 antibody treatments, mice were treated with 20μg/ 200μl of anti-CCR2 mAb (MC-21, provided by Matthias Mack, University of Regensburg) intraperitoneally (i.p.) for 5 consecutive days on the indicated days. Mice were depleted of alveolar macrophages by administering 40μl liposomes (Liposoma) containing clodronate (5mg/ml) or control PBS via oropharyngeal route twice every 24 hours.
Busulfan chimeras
6-12 week old mice were treated with 10mg/kg Busilvex (Pierre Fabre) intraperitoneally for 2 consecutive days with a 24 hour interval. 24 hours after the final dose, 1-2 x 106 donor bone marrow cells were delivered intravenously into the tail vein in 100μl PBS. Donor bone marrow was processed by collecting and cleaning the femur and tibia of both legs from donor mice, and crushing in RPMI-1640 (BioWhittaker) using a mortar and pestle. Cells were washed through a 70μm strainer, a short red blood cell lysis with 0.83% ammonium chloride. B-cells, T-cells, NK-cells and neutrophils were removed by incubating cells at a concentration of 1 x 107 cells in 70μl of MACS buffer and FC-block prior to incubation with biotinylated α-CD3, α-CD19, α-NK1.1, α-Ly6G (eBioscience). Cells were subsequently incubated with Streptavadin microbeads (Miltenyi Biotech) and separated using an LS-column (Miltenyi Biotech) on the magnetic field of a MACS Separator (Miltenyi Biotech). The negative fraction was collected, counted, and injected. Mice were reconstituted for a minimum of 40 days.
Flow cytometry
Leukocytes from the lung were quantified using flow cytometry. In brief, lungs were excised from mice and digested with 20μg/ml Liberase TL (Roche) and 50μg/ml DNAse 1 (Sigma) and further homogenised with the gentleMACS (Miltenyi) prior to passage through a 70μm strainer. Cells were pre-incubated with anti-FcgRIII/II (Fc block) in FACS buffer before a 30-min incubation with one or more of fluorochrome-labelled antibodies, then analysed using a Fortessa X20 (Becton Dickinson). The following mAbs were used (name / clone / cat# / supplier / dilution: CCR2 MC-21 M. Mack; CD3-APC 17A2 #20-0032 Tonbo 1/100; CD3-biotin 145-2C11 #13-0031-85 eBioscience 1/100; CD3-BV785 17A2 #100232 Biolegend 1/100; CD4-APC GK1.5 #100412 Biolegend 1/100; CD4-PECy7 GK1.5 #25-0041-82 eBioscience 1/100; CD4-PerCP/Cy5.5 RM4-5 #100540 1/100; CD8a-biotin 53-67 #13-0081-85 eBioscience 1/100; CD8a-APC 53-67 #100712 Biolegend 1/100; CD8a-BV510 53-67 #100752 Biolegend 1/100; CD11b-BV711 M1/70 #101242 Biolegend 1/400; CD11b-PECy7 M1/70 #101216 Biolegend 1/2000; CD11c-BV605 N418 #117334 Biolegend 1/400; CD11c-PE N418 #117307 Biolegend 1/200; CD16/32-APCCy7 93 #101328 Biolegend 1/100; CD19-BV785 6D5 #115543 Biolegend 1/100; CD19-PECy7 1D3 #25-0193-82 eBioscience 1/100; CD34-FITC RAM34 #553733 BD Pharmigen 1/100; CD44-APC IM7 #17-0441-83 eBioscience 1/200; CD45.1-APCCy7 A20 #110716 Biolegend 1/200; CD45.2-PB 104 #109820 Biolegend 1/200; CD48-APCCy7 HM48-1 #103432 Biolegend 1/100; CD62L-FITC MEL-14 #11-0621-85 eBioscience; CD64-APC X54-5/7.1 #139306 Biolegend 1/200; CD103 2E7 #121422 Biolegend 1/100; CD115 (c-fms)-FITC AFS98 #35-1152 Biolegend 1/100; CD117(cKit)-PECy7 2BG #25117182 eBioscience 1/100; CD135 (Flt3)-PE A2F10.1 #553842 BD Pharmigen 1/100; CD150-BV605 TC15-12F12.2 #115927 Biolegend 1/100; Blue fluorescent reactive dye #L34962 Invitrogen 1/400; IL-6-APC MP5-20F3 #561367 BD Pharmigen 1/100; Lineage Cocktail-PB 17A2/RB6-8C5/RA3-6B2/Ter-119/M1/70 #133311 Biolegend 1/100; Ly6C-BV785 #128041 HKl.4 Biolegend 1/400; Ly6G-FITC 1A8 #127606 Biolegend 1/200; Ly-6A/E(Sca-1)-APC #17598183 D7 eBioscience 1/100; MerTK-FITC 2B10C42 #151503 Biolegend 1/200; MHCII (I-A/I-E) M5/114.15.2 #107614 Biolegend 1/400; NK1.1-PE PK136 #108710 Biolegend 1/100; Influenza NP Pentamer-PE H-2Db ASNENMETM #NP/5491-19 Pro-immune 1/10; Siglec F-APCCy7 E50-2440 #565527 BD Pharmigen 1/600; Siglec F-PE E50-2440 #552126 BD Pharmigen 1/200; TNFa-APC MP6-Xt22 #17-7321-82 lnvitrogen 1/100; ZombieAqua #77143 Biolegend 1/400.
For intracellular staining of IL-6 and TNF, alveolar macrophages from naive or influenza-experienced mice were stimulated in vitro for 6 hours with 100ng/ml Pam3CSK4 (Enzo Life sciences) and then cultured for 12-16 hours with 100ng/ml Pam3CSK4 and 10μg/ml Brefeldin (Sigma). Following a surface stain, cells were fixed in methanol-free PFA (Polysciences) for 30 minutes at room temperature, and then stained with intracellular antibodies (Table 1) in Permabilisation buffer (BD biosciences) for 30 minutes before being washed and resuspended in PBS.
Net fluorescence: The geometric mean of a fluorophore signal from an “unstained” control (stained only with the necessary colours to permit cell identification) was subtracted from the fluorophore signal generated by the complete antibody panel. This was termed “Net fluorescence”, and was used to account for variations in auto-fluorescence between samples.
For bone-marrow precursor stainings, the following lineage cocktail to exclude lin+ cells was used: 17A2;RB6-8C5;RA3-6B2;Ter-119;M1/70 (anti-mouse CD3; Ly-6G/Ly-6C; CD45R/B220, TER-119; CD11b respectively). Precursor subsets were identified as follows28,41: Long-term hematopoietic stem cells (LT-HSCs):Lin-Sca1+cKit+CD48-CD150+; Short-term HSCs (ST-HSCs): Lin-Sca1+cKit+CD48-CD150-; Multipotent progenitors (MPPs): Lin-Sca1+cKit+CD48+CD150-; Common myeloid progenitors (CMPs): Lin-ScaI-cKit+CD16/32-CD34+; Granulocyte macrophage progenitors (GMPs): Lin-ScaI-cKit+CD16/32+CD34+; Monocyte-macrophage DC progenitors (MDPs): cKit+CD115+CD135+Ly6C−; common monocyte progenitors (cMoPs): cKit+CD115+CD135−Ly6C+.
Cell isolation by magnetic separation
Whole lung was processed into a single cell suspension as described in the Flow cytometry section above. Cells were counted and resuspended at a concentration of 1 x 107 cells in 70μl of MACS buffer (PBS + 2% Fetal Calf serum + 2mM EDTA), and FC-block for 4 minutes at 4°C. Cells were incubated with biotinylated α-Siglec F (Miltenyi biotech) for 20 minutes, washed, and streptavidin microbeads (Miltenyi Biotech) were added for 20 minutes. Cells were run through an LS-Column (Miltenyi Biotech) on the magnetic field of a MACS Separator (Miltenyi Biotech) at 4°C. The positively selected fraction was collected, and cells were plated at a density of 100,000 cells/well in a flat-bottomed 96-well plate in RPMI 1640 (BioWhittaker)(Supplemented with: 10% fetal calf serum, L-glutamine, penicillin, streptomycin, and β-Mercaptoethanol). Alveolar macrophages were adhered for a minimum of 2 hours and washed in warm RPMI. Cells were stimulated with a final concentration of 100ng/ml Pam3CSK4 (EnzoLifeSciences) or LPS (EnzoLifeSciences) for 16 hours at 37°C, 5% CO2.
Quantification of cytokines
To quantify protein in the airways, mice were culled using 600mg/kg Pentobarbital + 16mg/kg Mepivacaine. Broncho-alveolar lavage (BAL) fluid was collected in 400-500μl PBS and centrifuged at 1400 rpm for 5 minutes at 4°C. Supernatants of BAL and cells were collected and stored at -20°C.
IL-6 and TNF were measured using enzyme-linked immunosorbent assay (ELISA) mouse eBioscience Ready-set-Go kits as per the manufacturer’s instructions and read on a Safire II plate reader (Tecan). The ProcartaPlex Cytokine & Chemokine Mouse 36-Plex (eBioscience) was used to assess the concentrations of 36 cytokines (GM-CSF, IFN-gamma, IL-1 beta, IL-12p70, IL-13, IL-18, IL-2, IL-4, IL-5, IL-6, TNF, ENA-78, G-CSF, IFN-alpha, IL-1 alpha, IL-15/IL-15R, IL-28, IL-3, IL-31, LIF, M-CSF, IL-10, IL-17A, IL-22, IL-23, IL-27, IL-9, Eotaxin, GRO-alpha, IP-10, MCP-1, MCP-3, MIP-1 alpha, MIP-1 beta, MIP-2, RANTES) and read on a Luminex 100 (BioRad).
Adoptive transfer
Mice were inoculated with influenza or PBS. At one month post-infection, lungs from naive and post-influenza mice were collected and processed into a single cell suspension as for flow cytometry. Alveolar macrophages were enriched by positive magnetic selection using α-Siglec F, and 1 x 106 macrophages were transferred per recipient. Transfer was done in 20μl PBS by intra-tracheal delivery into mice anaesthetised with a solution (0.025ml/10g body weight) containing Fentanyl (0.05mg/kg), Midazolam (5mg/kg) and Medetomidine (0.5mg/kg) delivered by intraperitoneal injection. Host alveolar macrophages were not depleted prior to transfer, to avoid bystander inflammation. Mice were revived with a solution (0.085ml/10g body weight) which contained Naloxone (1.2mg/kg), Flumazenil (0.5mg/kg) and Atipamezole (2.5mg/kg) and was delivered by intraperitoneal injection.
ATAC-seq
Library-level analysis
Sequencing was performed on the Illumina HiSeq 4000 platform and typically generated ~95 million 101bp paired-end reads per library. Raw reads from each sample were adapter-trimmed using cutadapt (version 1.9.1) 42 with parameters “-a CTGTCTCTTATA -A CTGTCTCTTATA --minimum-length=25 –quality-cutoff=20 -u -50 -U -50”. BWA (version 0.6.2) 43 with default parameters was used to perform genome-wide mapping of the adapter-trimmed reads to the mouse mm10 genome assembly downloaded from the UCSC 44. Read group addition, duplicate marking and insert size assessment was performed using the picard tools AddOrReplaceReadGroups, MarkDuplicates and CollectMultipleMetrics, respectively (version 2.1.1). Reads mapped to mitochondrial DNA were removed using the pairToBed command from BEDTools (version 2.26.0)45. Additional filtering was performed to only include uniquely mapped, properly-paired reads with insert size <= 2kb, and mismatches <= 1 in both reads. SAMtools (version 1.3.1) 46 was used for bam file sorting and indexing.
Merged library-level analysis
The filtered alignments from each library were merged at the sample-level using the picard MergeSamFiles command. Duplicate marking and removal was re-performed on the merged alignments. BedGraph coverage tracks representing the accessibility signal per million mapped paired-reads were generated using BEDTools genomeCoverageBed with the parameters “-bg -pc -scale <SCALE_FACTOR>”. BedGraph files were converted to bigWig using the wigToBigWig binary available from the UCSC with the "-clip" parameter 47.
Regions of chromatin accessibility were identified genome-wide using MACS2 callpeak (version 2.1.1.20160309) 48 with the parameters “--gsize=mm --keep-dup all –f BAMPE --nomodel --broad”. A union set of intervals were obtained by merging the regions identified across all samples.
Merged replicate-level analysis
Fragment counts relative to the union set of intervals were obtained using the featureCounts tool from the Subread package (version 1.5.1) 49. The parameters used were “-O --minOverlap 1 --primary --ignoreDup -p -B -C”. Differential chromatin accessibility sites were obtained using the DESeq2 package (version 1.12.3) 50 within the R programming environment (version 3.3.1). Differential sites with an FDR <= 0.01 and fold-change >= 2 were kept for further analysis. The annotatePeaks.pl program from HOMER (version 4.8) 51 was used to annotate the differential sites relative to mm10 RefSeq features downloaded from the UCSC on 11th December 2017.
RNA-seq
Following reconstitution and infection of Busulfan Ccr2–/– chimeras, alveolar macrophages were enriched by α-Siglec F magnetic selection and sorted into CD45.1+ and CD45.2+ AMs using a FACS Aria III (BD Biosciences). Cells were then stimulated with 100ng/ml Pam3CSK4 for 16 hours. RNA was prepared from these cells using the RNeasy Mini Kit or RNeasy Micro Kit (Qiagen).
RNA sequencing was carried out on the Illumina HiSeq 4000 platform and typically generated ~35 million 76bp or 101bp unstranded single-end reads per sample. Adapter trimming was performed with cutadapt (version 1.9.1) 42 with parameters “--minimum-length=25 --quality-cutoff=20 -a AGATCGGAAGAGC”. The RSEM package (version 1.3.0) 52 in conjunction with the STAR alignment algorithm (version 2.5.2a) 53 was used for the mapping and subsequent gene-level counting of the sequenced reads with respect to mm10 RefSeq genes downloaded from the UCSC Table Browser 44 on 11th December 2017. The parameters used were “--star-output-genome-bam --forward-prob 0.5”. Differential expression analysis was performed with the DESeq2 package (version 1.12.3) 50 within the R programming environment (version 3.3.1). Unless otherwise indicated, an adjusted p-value of ≤ 0.01 was used as the significance threshold for the identification of differentially expressed genes.
Gene set enrichment analysis (Supplementary Fig. 6f) for the genes from two classes (cluster1<2<3; cluster3<2<1) in Supplementary Fig. 6e was performed by Pathway Maps using GeneGo MetaCore (https://portal.genego.com/).
Luminol
1 x 105 alveolar macrophages isolated from the lungs of naive, and post-influenza mice, were plated on a white 96-well flat-bottom plate in 100μl and allowed to settle for 1 hour at room temperature 50μM of luminol (Sigma) and 1.2U/ml Horseradish Peroxidase (Sigma) were added with 15μl of TIGR4 (1 x 104 CFU). The plate was spun to collect stimulus and luminescence was immediately read on a Safire II plate reader (Tecan).
Phagocytosis assay
To measure phagocytosis of S.p. by alveolar macrophages, a protocol from the Fleischman lab was adapted. 1 x 109 CFU/ml TIGR4 was labelled with 5μM/ml CFSE (Sigma) for 1 hour in the dark at room temperature while rocking. Alveolar macrophages were isolated from naive and post-influenza lungs as outlined in the Flow cytometry section. 1 x 105 alveolar macrophages were incubated with CFSE labelled, or unlabelled bacteria for 1 hour in the dark in a U-bottomed plated at 37°C, 5% CO2. Cells were washed with PBS three times, and then stained with appropriate antibodies for 30 minutes at 4°C. Immediately prior to acquiring cells on an LSR Fortessa, Trypan blue (1/4 dilution) was added to cells to quench signal from external CFSE. The background from the samples incubated with unlabelled bacteria was subtracted from the samples incubated with labelled bacteria to determine CFSE fluorescence.
Imagestream
Samples were acquired on an ImageStreamx MKII at 40x magnification, low flow rate, with the 405nm, 488nm, 561nm, 642nm and 785nm lasers un-shuttered and set at 120mW, 350mW, 200mW, 150mW and 0.9mW, respectively. ImageStream data was acquired using INSPIRE and analysed using IDEAS (both Amnis, Seattle). PLS-DA analysis was performed using R3.4.2 and RStudio.
Statistical Analysis
Data shown as the means ±s.d. Sample sizes were designed to give statistical power, while minimizing animal use. All statistical comparisons were performed using Prism 6 (GraphPad). Figure legends denote the specific statistical tests used for each experiment. Statistical significance was determined as p<0.05.
Supplementary Material
Acknowledgements
We are grateful to Prof. S. Rose-John (Kiel University) for the kind gift of anti IL-6 mAb, to Drs. C. Reis e Sousa and G. Stockinger for reading the manuscript, and Dr. A. Warnatsch for initial help with reactive oxygen species measurements. This work benefitted from data assembled by the ImmGen consortium. We thank the Advanced Sequencing, Flow Cytometry, Biological Research, and Histopathology facilities of the Crick for excellent support. This study was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001206), the UK Medical Research Council (FC001206) and the Wellcome Trust (FC001206). Support by MRC grant U117597139 (SC, AW) and a BBSRC-GSK-funded case studentship BB/L502315/1 (HA) is gratefully acknowledged.
Footnotes
Data availability
Sequencing data are available in GEO under accession code GSE120543.
Author contributions
H.A, S.C., J.K. and A.W. designed the experiments; H.A., J.K. and S.C. did the experiments; H.A., J.K., S.C., H.P. and G.K. analysed data; H.A., S.C., H.P., E.M.H., S.B. and A.W. interpreted data; M.M. provided vital reagents; S.C., H.P., E.M.H., S.B. contributed to manuscript writing; H.A. and A.W. wrote the manuscript.
Competing Interests statement
E.M.H. and S.B. were employees of GSK at the time of this study. The other authors declare that they have no conflict of interest.
References
- 1.Dahl ME, Dabbagh K, Liggitt D, Kim S, Lewis DB. Viral-induced T helper type 1 responses enhance allergic disease by effects on lung dendritic cells. Nat Immunol. 2004;5:337–343. doi: 10.1038/ni1041. [DOI] [PubMed] [Google Scholar]
- 2.Didierlaurent A, et al. Sustained desensitization to bacterial Toll-like receptor ligands after resolution of respiratory influenza infection. J Exp Med. 2008;205:323–329. doi: 10.1084/jem.20070891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machiels B, et al. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat Immunol. 2017;18:1310–1320. doi: 10.1038/ni.3857. [DOI] [PubMed] [Google Scholar]
- 4.Beura LK, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reese TA, et al. Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe. 2016;19:713–719. doi: 10.1016/j.chom.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guilliams M, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Abdelmalak B, Inoue Y, Culver DA. Pulmonary alveolar proteinosis in adults: pathophysiology and clinical approach. Lancet Respir Med. 2018;6:554–565. doi: 10.1016/S2213-2600(18)30043-2. [DOI] [PubMed] [Google Scholar]
- 8.Bain CC, et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat Commun. 2016;7 doi: 10.1038/ncomms11852. ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merad M, et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol. 2002;3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misharin AV, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molawi K, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211:2151–2158. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw TN, et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J Exp Med. 2018;215:1507–1518. doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soucie EL, et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 2016;351 doi: 10.1126/science.aad5510. aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbings SL, et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood. 2015;126:1357–1366. doi: 10.1182/blood-2015-01-624809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilliams M, Scott CL. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol. 2017;17:451–460. doi: 10.1038/nri.2017.42. [DOI] [PubMed] [Google Scholar]
- 19.van de Laar L, et al. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity. 2016;44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Halstead ES, et al. GM-CSF overexpression after influenza a virus infection prevents mortality and moderates M1-like airway monocyte/macrophage polarization. Respir Res. 2018;19:3. doi: 10.1186/s12931-017-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang FF, et al. GM-CSF in the lung protects against lethal influenza infection. Am J Respir Crit Care Med. 2011;184:259–268. doi: 10.1164/rccm.201012-2036OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sever-Chroneos Z, et al. GM-CSF modulates pulmonary resistance to influenza A infection. Antiviral Res. 2011;92:319–328. doi: 10.1016/j.antiviral.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GeurtsvanKessel CH, et al. Dendritic cells are crucial for maintenance of tertiary lymphoid structures in the lung of influenza virus-infected mice. J Exp Med. 2009;206:2339–2349. doi: 10.1084/jem.20090410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyron-Quiroz JE, et al. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat Med. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 25.van der Poll T, et al. Interleukin-6 gene-deficient mice show impaired defense against pneumococcal pneumonia. J Infect Dis. 1997;176:439–444. doi: 10.1086/514062. [DOI] [PubMed] [Google Scholar]
- 26.Plantinga M, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 28.Mitroulis I, et al. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 2018;172:147–161 e112. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider C, et al. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 32.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Lavin Y, et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765 e717. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zilionis R, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019;50:1317–1334 e1310. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 36.Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott CL, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7 doi: 10.1038/ncomms10321. 10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minutti CM, et al. Local amplifiers of IL-4Ralpha-mediated macrophage activation promote repair in lung and liver. Science. 2017;356:1076–1080. doi: 10.1126/science.aaj2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang N, et al. Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J Exp Med. 2019;216:1291–1300. doi: 10.1084/jem.20182024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schett G. Physiological effects of modulating the interleukin-6 axis. Rheumatology (Oxford) 2018;57:ii43–ii50. doi: 10.1093/rheumatology/kex513. [DOI] [PubMed] [Google Scholar]
- 41.Hettinger J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14:821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 42.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet,journal. 2011;17:10–12. [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.