Abstract
To date, over 1000 melanocytic neoplasms, spanning all stages of tumorigenesis, have been sequenced, offering detailed views into their -omic landscapes. This has coincided with advances in genetic engineering technologies that allow molecular biologists to edit the human genome with extreme precision and new mouse models to simulate disease progression. In this review, we describe how these technologies are being harnessed to provide insights into the evolution of melanoma at an unprecedented resolution, revealing that prior models of melanoma evolution, in which pathways are turned ‘on’ or ‘off’ in a binary fashion during the run-up to melanoma, are oversimplified.
Introduction
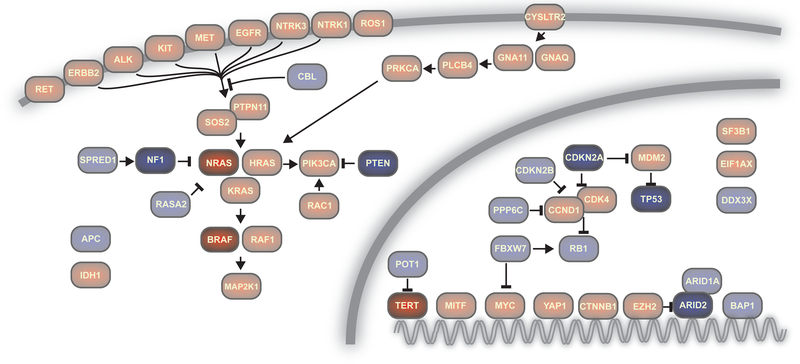
Melanoma is driven by mutations that result in the uncontrolled proliferation of a melanocyte. Melanocytes have checkpoints so that a single mutation is unable to drive their transformation into a fully malignant tumor, and thus they require approximately 5–10 pathogenic alterations, which are spread across several signaling pathways, to trigger their transformation to melanoma (Shain and Bastian 2016). To date, over 1000 melanomas have been sequenced, illuminating the key pathogenic alterations involved (Fig. 1) (Cancer Genome Atlas Network 2015; Hayward et al. 2017; Hodis et al. 2012; Krauthammer et al. 2015; Shain et al. 2015a).
Figure 1. Genes and pathways with pathogenic mutations in melanoma.
Gene products are labeled by their HUGO gene names. Red indicates gain- or change- of function mutations predominate in that gene, and blue denotes loss-of-function mutations prevail. The most commonly altered genes in cutaneous melanoma are highlighted.
Mutations that activate the Mitogen-Activated Protein-Kinase (MAPK) pathway are ubiquitous, and most melanomas also harbor somatic alterations that upregulate telomerase and disrupt cell-cycle checkpoint control. Finally, many melanomas have somatic alterations that perturb the p53 pathway, activate the Phosphatidyl-Inositol 3–kinase (PI3-kinase) signaling cascade, and disturb chromatin remodeling complexes. These constitute the main signaling pathways with genetic perturbations in melanoma and will be the main focus of this review (Fig. 1).
Other factors also help drive the progression of melanoma. In particular, signaling pathways can be rewired via non-genetic mechanisms, and cell non-autonomous factors, including the immune system, cell-cell signaling, and the composition of the local extracellular matrix play critical roles in shaping melanocytic neoplasms. These factors are not covered in this review, but are covered elsewhere (Arozarena and Wellbrock 2019).
Melanocytic neoplasms are staged by their clinical and histopathologic features. Melanocytic nevi, also known as common moles, occupy the benign end of this spectrum. It is estimated that only 1 in 10,000 nevi eventually transform into melanoma (Tsao et al. 2003). There are also intermediate lesions, defined as having worrisome clinical or histopathologic features but nonetheless falling short of an unequivocal diagnosis of melanoma. Some of the lesions in this category are likely nevi or melanomas, masquerading as something in between; however, genetic studies indicate that there are true biologically intermediate neoplasms, harboring more oncogenic alterations than common nevi but less than melanoma (Shain et al. 2018; Shain et al. 2015b). Finally, melanomas occupy the malignant end of this spectrum, and they are further staged by their thickness, ulceration, and the extent to which they have (or have not) spread to other parts of the body (Balch et al. 2009; Gershenwald et al. 2017). During the course of tumor progression, melanocytes accumulate pathogenic mutations, eliminating barriers to transformation, and allowing the ensuing neoplasms to transition through the clinical stages of melanocytic neoplasia described above.
It is tempting to describe the progression of melanoma as a series of binary events, in which specific pathways are turned “on” or “off”, ultimately producing a melanoma, but this is an oversimplification. For this review, we emphasize the specific genetic events that titrate discrete levels of activation (or inactivation) of the critical pathways during the course of progression of melanocytic neoplasms. Reviews covering other aspects of melanoma progression are elsewhere (Arozarena and Wellbrock 2019; Shain and Bastian 2016). We focus primarily on the Mitogen-Activated Protein-Kinase (MAPK) pathway and the Rb pathway because these pathways are the most thoroughly studied with respect to gene dosage.
MAPK signaling ramps up during the course of progression
Activation of the Mitogen-Activated Protein-Kinase (MAPK) pathway is likely required to form a melanocytic neoplasm. Mutations that activate BRAF and NRAS, two critical nodes of the MAPK signaling cascade, cumulatively occur in 75% of melanomas (Cancer Genome Atlas Network 2015). The term “wild-type” melanoma has been widely adopted to describe the melanomas without these mutations; however, more comprehensive genomic studies, geared specifically towards these melanomas, have found that “wild-type” melanomas are, themselves, riddled with a “long-tail” of relatively uncommon mutations in the pathway (Fig. 1) (Ablain et al. 2018; Hayward et al. 2017; Krauthammer et al. 2015; Shain et al. 2015a; Wiesner et al. 2014). Nowadays, melanomas without recognized mutations in the MAPK pathway are rare, and we propose that the ones that do seem to exist probably reflect cases in which a mutation was missed or unappreciated rather than a true biological state.
Mutations in MAPK-pathway genes are also ubiquitous in earlier stages of melanocytic neoplasia (Pollock et al. 2003). Nevertheless, the levels of pathway activation do not stay the same during the course of progression. This is reflected, in part, by changes in the zygosity of oncogenic mutations in these genes. For instance, most nevi have a heterozygous BRAFV600E mutation (Colebatch et al. 2019; Shain et al. 2015b), whereas, the gene dosage of oncogenic BRAF is typically elevated, as a result of copy number gains, in melanoma (Maldonado et al. 2003; Shain et al. 2018). Oncogenic mutations in other genes in the pathway, including NRAS, show a similar increase in dosage during the course of progression (Hélias-Rodzewicz et al. 2017; Shain et al. 2018). Altogether, increases in gene dosage of oncogenic mutations imply that MAPK-pathway output ramps up during the evolution of melanoma.
Many melanocytic neoplasms also acquire more than one mutation in the MAPK pathway during the course of progression. To be sure, some combinations of mutations are rarely observed. For example, BRAFV600E mutations do not typically coincide with NRAS codon 61 mutations, and this finding is often interpreted as evidence that these mutations are functionally redundant, precluding their selection (Haluska et al. 2006). However, there are many combinations of mutations in genes in the MAPK pathway that can co-exist. For example, hypoactive BRAF mutations, NF1 mutations, MAP2K1 mutations, and mutations associated with RASopathy syndromes commonly overlap amongst one another or alongside BRAFV600E and NRAS codon 61 mutations (Cancer Genome Atlas Network 2015; Krauthammer et al. 2015; Shain et al. 2015a). These additional mutations are acquired continuously throughout the evolution of melanoma, again supporting the notion that MAPK-pathway activation is ramped up during progression (Shain et al. 2018).
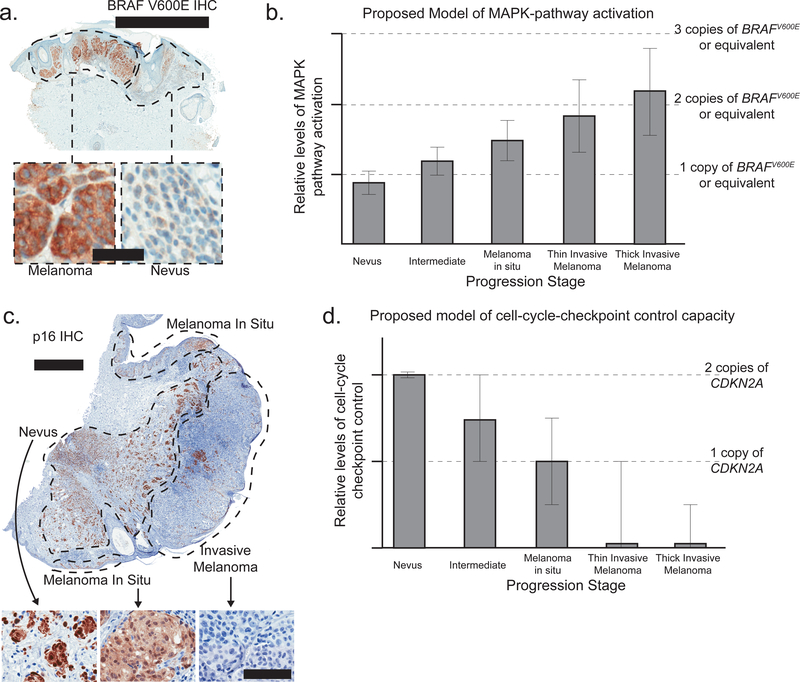
Transcriptomic and proteomic data further indicate that MAPK signaling strengthens during the evolution of melanoma. RNA-sequencing can measure total levels of gene expression as well as the expression from each allele. RNA-sequencing of nevi and melanomas indicates that the oncogenic allele of BRAF is somewhat repressed in nevi and preferentially expressed in melanoma when compared to the wild-type allele (Shain et al. 2018). This holds true in tumors without changes in BRAF copy number, suggesting that there may be epigenetic mechanisms to specifically recognize and modulate the expression of the oncogenic allele. Immunostaining with an antibody that specifically recognizes the V600E form of the BRAF protein confirms that BRAFV600E is more highly expressed in melanomas than nevi at the protein level (Fig. 2A) (Busam et al. 2013; Yeh et al. 2013).
Figure 2. The MAPK- and Rb- pathways are perturbed in an incremental fashion during the evolution of melanoma.
A. Immunostaining with an antibody specific for the V600E form of BRAF in a melanoma and its precursor nevus. Note the greater intensity staining in the melanoma compartment, implying heightened levels of pathway activation. Top scale bar = 2mm; bottom scale bar = 50um. B. A model of MAPK pathway output during the course of melanoma progression. Bars depict average levels of MAPK pathway output. Dotted lines note the expected output from one or more copies of BRAFV600E (or equivalent alterations elsewhere in the pathway). The error bars acknowledge heterogeneity with respect to pathway activation both within and across tumors. The effect sizes are estimated from genomic, transcriptomic, and proteomic observations, as described, and are intended as a model rather than to be strictly quantitative. C. Immunostaining with an antibody against p16INK4A in an invasive melanoma and its remnant precursor lesions – a melanoma in situ (MIS) and a nevus. Note the stepwise reduction of p16INK4A from nevus to MIS to invasive melanoma, implying loss of cell-cycle checkpoint control during the course of progression. Top scale bar = 1mm; bottom scale bar = 100um. D. A model of cell-cycle checkpoint control during the course of melanoma progression, plotted as in panel B. The images published in panels A and C were originally published here (Shain et al. 2015b) and are reprinted with permission.
In aggregate, genomic, transcriptomic, and proteomic data each indicate that MAPK signaling is turned ‘on’ at the initial stages of melanocytic neoplasia but progressively ramps up during the course of evolution. Based on the composite evidence, we propose a model whereby pathway activation more than doubles during the course of progression (Fig. 2B).
Molecular studies also illustrate the importance of the levels of MAPK signaling in melanocytes. In particular, there are notable phenotypic differences that are dependent on BRAFV600E expression levels in both mice and humans. In mouse models, introduction of a single allele of BrafV600E in melanocytes results in highly pigmented lesions and melanocytic hyperplasia within a month (Dankort et al. 2009; Dhomen et al. 2009; Goel et al. 2009). However, these small, papular, and pigmented lesions lack aberrant mitotic figures and no further tumor progression occurs over 20 months (Dankort et al. 2009; Dhomen et al. 2009). Interestingly, two alleles of BrafV600E produces benign melanocytic hyperplasia that is more extensive and more highly pigmented than the growth induced by a heterozygous BrafV600E mutation (Dankort et al. 2009). These observations indicate that BrafV600E gene dosage exerts a physiologically relevant impact on mouse melanocyte proliferation in vivo – an experimental conclusion that may rationalize the observation of increased copy number of oncogenic BRAF in human melanomas.
The importance of gene dosage is also reflected by in vitro studies of human cells. Ectopic expression of BRAFV600E in fibroblasts or melanocytes induces rapid growth restriction within 3–7 days (Haferkamp et al. 2009; McNeal et al. 2015; Michaloglou et al. 2005). However, this experimental approach results in supra-physiological levels of the mutant protein that is immune to endogenous transcriptional regulation. Another approach is precision engineering of a BRAFV600E mutation into its endogenous locus with CRISPR/Cas9-mediated gene editing. In contrast to overexpression, primary human melanocytes engineered in this way acquire a proliferative advantage over sibling melanocytes, which are wild-type for BRAF, and this proliferative advantage is sustained for 2 months (Zeng et al. 2018). Altogether, these observations illustrate that modulating the dosage of BRAFV600E produces different phenotypes in human melanocytes. This is important to appreciate because the mechanisms underlying the growth arrest that follows an initial period of proliferation in BRAFV600E-mutant melanocytes are poorly understood, and the molecular consequences of gaining copies of the oncogenic allele are also not known. Moving forward, accurate methods for modeling the precise changes in BRAFV600E allele dosage, both in vitro and in vivo, will be critical for understanding how gene dosage contributes to melanoma initiation and progression.
The importance of gene dosage in the context of oncogenic RAS is well established through molecular studies in other cancers. For example, an increase in KRAS-mutant-allele dosage drives both tumorigenesis and metastasis in a mouse model of pancreatic ductal adenocarcinoma (Mueller et al. 2018), and loss of the wild-type allele of KRAS in acute myeloid leukemias enhances cell growth and dependency on the MAP-kinase signaling pathway (Burgess et al. 2017). Moreover, the oncogenic potential of homozygous NRASG12D/G12D cells are increased as compared to heterozygous NRASG12D/+ or hemizygous NRAS G12D/− cells in hematopoietic transformation (Xu et al. 2013).
In melanoma, Pederson et. al. developed a mouse model of NrasG12D that can be selectively induced in melanocytes. All mice with homozygous NrasG12D/G12D mutations show a darkening of the skin within 2 months, whereas only half of mice with a heterozygous NRasG12D mutation show a darkening of skin that is also much weaker (Pedersen et al. 2013). Interestingly, when NrasG12D is expressed in a non-inducible setting, leptomeningeal melanoma forms during development and occurs earlier in homozygous NrasG12D/G12D mice than in heterozygous NrasG12D/+ mice (Pedersen et al. 2013). Given that gain of mutant-NRAS alleles has been observed during melanoma evolution (Shain 2018, Shain 2015), further investigations into the importance of physiologically relevant NRAS-mutant allele dosages are warranted in the human setting.
Disruption of cell-cycle checkpoint control occurs in a stepwise fashion
Normal cells have mechanisms to halt cell-cycle progression at the transition from the G1 to the S phase of the cell cycle, but abrogation of this checkpoint occurs in most, if not all, melanomas (Sharpless and Chin 2003). The p16INK4A protein primarily governs this regulation, and it is encoded by the CDKN2A gene, which harbors bi-allelic, loss-of-function alterations in nearly 50% of melanomas. Deletions of CDKN2A often encompass the neighboring gene, CDKN2B, which encodes p15INK4B – another critical checkpoint protein. Finally, somatic alterations also affect additional genes involved in checkpoint control, including CDK4, CCND1, PPP6C, and FBXW7, among others (Cancer Genome Atlas Network 2015).
Loss of cell-cycle-checkpoint control occurs in a stepwise manner during the progression of melanoma. Common nevi have no aberrations in checkpoint genes (Colebatch et al. 2019; Shain et al. 2018), whereas, intermediate neoplasms and melanomas in situ tend to have heterozygous mutations affecting CDKN2A or mutations affecting genes encoding less critical components of the checkpoint apparatus (Shain et al. 2018). Invasive melanomas typically harbor bi-allelic alterations disrupting the CDKN2A gene (Cancer Genome Atlas Network 2015; Shain et al. 2018). Stepwise-loss of cell-cycle-checkpoint control is further reflected by immunostaining studies, in which reduction of p16INK4A protein occurs at the transition from nevus to melanoma in situ, and complete elimination of protein occurs at the transition to invasive melanoma (Fig. 2C) (Pavey et al. 2002; Reed et al. 1995; Talve et al. 1997; Zeng et al. 2018). Based on the composite evidence, we propose a model whereby cell-cycle control is gradually lost during the course of progression (Fig. 2D).
Molecular studies also illustrate the importance of the precise levels of p16INK4A in melanocytes. Bi-allelic loss of CDKN2A promotes invasion and metastasis in vivo in genetically engineered mice as well as similar phenotypes in vitro in primary human melanocytes (Ackermann et al. 2005; Dhomen et al. 2009; Krimpenfort et al. 2001; Tyagi et al. 2017; Zeng et al. 2018). In an especially informative experiment, the contribution of loss-of-one versus loss-of-two copies of CDKN2A to metastatic potential was directly assessed using a pair of related melanoma cell lines – a parental line (WM793) and a subclone of this line (1205Lu). Genetic sequencing of the two lines revealed that complete loss of CDKN2A is the only additional pathogenic mutation in the subclone (1205Lu). Both cell lines proliferate in culture and as primary tumors when injected subcutaneously into immune-compromised mice; however only the subclone (1205Lu), which has complete ablation of the CDKN2A gene, readily metastasizes in the same setting (Krimpenfort et al. 2001; Zeng et al. 2018). These phenotypes can be toggled by re-expression of p16INK4A in the derivative line (1205Lu) or knock-down of p16INK4A in the parental line (WM793), respectively eliminating or inducing the metastatic phenotype (Zeng et al. 2018).
p16INK4A is a direct regulator of RB1 phosphorylation (Rubin 2013; Serrano et al. 1993). RB1 was the first discovered tumor suppressor gene, spawning the “two-hit hypothesis”, whereby it was proposed that tumor suppressor genes must lose both copies to form a neoplasm (Knudson 2001; Knudson 1971). This is a useful model, but in some circumstances, it is oversimplified, and it appears that the role of CDKN2A in melanoma progression is one of those circumstances. Different dosages of CDKN2A influence several melanocyte phenotypes, including growth arrest, proliferation, and invasion (Bennett 2016; Michaloglou et al. 2005; Zeng et al. 2018; Zhao et al. 2016). The mechanisms underlying how CDKN2A dosage affects these phenotypes are undefined, but they are almost certainly tied to RB1 phosphorylation. One likely mechanism is that by varying RB1 phosphorylation, key lineage-restricted transcription factors that bind to phosphorylated-RB1, such as MITF and TBX2, alter their transcriptional targets (Carreira et al. 2005; Halaban 2005; Vance et al. 2010; Zeng et al. 2018). Altogether, since CDKN2A bi-allelic loss occurs in melanomas and mono-allelic loss occurs in earlier stages of neoplasia, further investigation into how RB1 phosphorylation and RB1 binding partners are influenced by specific levels of p16INK4A will provide valuable insights into the progression of melanoma.
Other “hits” to the Rb pathway can likely cooperate with, or even substitute for, loss of CDKN2A to promote melanoma progression. In particular, p15INK4B loss was functionally shown to reverse the growth-arrest phenotype in nevus cells, resulting in progression towards melanoma (McNeal et al. 2015). Mutations in additional genes, such as CDK4, PPP6C, CCND1, and RB1 are less common, and therefore we speculate that their phenotypes are attenuated as compared to CDKN2A loss.
Perturbation of other pathways tends to also be incremental
More studies are needed to fully resolve whether other signaling pathways are disrupted in an incremental or a binary fashion during the evolution of melanoma, but genetic observations provide some clues.
The SWI/SNF chromatin-remodeling-complex is likely perturbed in an incremental fashion during the evolution of melanoma. Loss-of-function mutations affect several members of the complex, including ARID2, ARID1A, ARID1B, PBRM1, and SMARCA4 (Hodis et al. 2012). In the melanoma genome atlas project, and broadly across other cancers, mutations in these genes frequently co-occur (Cancer Genome Atlas Network 2015; Shain and Pollack 2013), though in one study, certain combinations of genes did not have overlapping mutations (Garman et al. 2017). More studies are needed to resolve the spectrum of these mutations in melanoma, but their co-occurrence implies continual selection to perturb chromatin remodeling, even after acquisition of the first mutation.
It is unclear whether the p53 pathway is disrupted incrementally during progression. TP53 mutations occur in approximately 20% of melanomas (Cancer Genome Atlas Network 2015), but the p53 pathway may be disrupted in a much higher percentage of melanomas as a result of CDKN2A loss. The CDKN2A locus encodes two protein products – p16INK4A (described above) and p14ARF, which operates in the p53 pathway (Fig. 1). In melanoma, p16INK4A is thought to be the dominant tumor suppressor, indicated by the fact that germline and somatic alterations can affect p16INK4A while sparing p14ARF (Goldstein et al. 2007), whereas mutations affecting p14ARF alone are rare (Hewitt et al. 2002). Nevertheless, most genetic alterations do impact both proteins, arguing that subsequent TP53 mutations could function as secondary hits to the pathway – this would imply incremental disruption of the pathway.
Lineage-specific transcription factors, including MITF and SOX10, are also critical regulators of melanoma, and the activity of these proteins likely increases throughout progression. MITF and SOX10 are required in the earliest stages of tumorigenesis (Seberg et al. 2017; Shakhova et al. 2012), and high-level amplification of MITF has been observed in metastases and cell lines (Garraway et al. 2005), implying selection to upregulate MITF activity in later stages of tumorigenesis. Moreover, the activity of these genes’ products are carefully modulated at the transcript and protein levels (reviewed elsewhere (Goding and Arnheiter 2019; Seberg et al. 2017)), leading to the proposition of a rheostat model (Goding 2011), whereby levels of activity from MITF and its collaborators elicit different phenotypes.
Up-regulation of telomerase is likely an exception to the pattern of incremental disruption described in other pathways. In melanoma, the TERT gene has a high frequency of somatic mutations affecting its promoter (Horn et al. 2013; Huang et al. 2013). Telomerase is ordinarily not expressed in melanocytes, but the promoter mutations create a GABP transcription-factor-binding site, enabling expression of telomerase (Bell et al. 2015). Most melanomas have a single TERT promoter mutation, without any other mutations in this pathway, arguing that telomerase is simply turned “on”, rather than incrementally upregulated. Mechanistic studies also support this view. Despite having TERT promoter mutations, melanomas have short telomeres (Hayward et al. 2017). This paradoxical observation can be explained by the fact that the TERT promoter mutation is sufficient to stave off telomeric crisis without significantly lengthening telomeres (Chiba et al. 2017). In aggregate, there seems to be no selective advantage to increase telomere lengths beyond the levels necessary to forestall telomeric crisis, and a single mutation in the TERT promoter is sufficient to do this. Overall, these findings argue that a single ‘hit’ is sufficient to turn ‘on’ telomerase and immortalize melanoma cells.
Conclusions and Next Steps
There is a prevailing sentiment that multiple mutations in the same pathway rarely co-occur in cancers because they are functionally redundant (Ciriello et al. 2012; Miller et al. 2011; Yeang et al. 2008). In melanoma, this is generally true for BRAFV600E and NRAS codon 61 mutations, but there are numerous exceptions, and the notion that multiple mutations in the same pathway cannot co-occur should not be treated as dogma. It is especially important to pay attention to the zygosity of mutations, as many mutations are themselves subject to changes in their gene dosage. The recent revolution in genetic engineering tools, most notably CRISPR/Cas9, now make possible the modeling of different combinations of pathogenic mutations or different mutant-allele dosages under endogenous transcriptional regulation. Such approaches should be considered when performing functional and mechanistic studies aimed at understanding the early stages of melanoma progression.
Finally, knowing the precise levels of pathway activation (or inactivation) that are optimal for tumor cells will help guide therapeutic interventions. The importance of this issue is exemplified by patient responses to MAPK-pathway inhibitors, as most patients have an initial response to treatment yet ultimately develop resistance – thereby illustrating how tumors are able to adapt and find their optimal levels of signaling. In extreme scenarios, tumors can even acquire dual BRAFV600E and NRASQ61L mutations (Raaijmakers et al. 2016) – a combination that is likely incompatible outside the scope of drug treatment (Petti et al. 2006). Moving forward, studying how patients with different dosages of oncogenic MAPK-pathway mutations respond to MAPK-pathway inhibitors may provide novel biomarkers of drug sensitivity, and we anticipate that new drug targets will emerge by understanding how different dosages rewire cellular signaling pathways and influence cell behavior.
In conclusion, we encourage the melanoma research community to think beyond binary models of melanoma evolution, in which pathways are either ‘on’ or ‘off’, and to consider the precise levels of pathway perturbation in their studies.
Acknowledgements
We would like to thank the following sources of funding (Shain): NCI K22 CA17997, Melanoma Research Alliance, Melanoma Research Foundation, Dermatology Foundation, and PhRMA Foundation. This work was originally presented at the joint meeting between the Pan American Society of Pigment Cell Research and the Montagna Symposium on the Biology of Skin (Gleneden Beach, Oregon). We would like to thank the following grants from NIAMS, NIA, and NIEHS for supporting this scientific retreat and defraying these publication costs: R13 AR009431-53 (Kulesz-Martin) and R13 AR074279-01 (Leachman).
Footnotes
Conflicts of Interests
The authors state no conflict of interest.
References
- Ablain J, Xu M, Rothschild H, Jordan RC, Mito JK, Daniels BH, et al. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science. 2018;362(6418):1055–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res. 2005;65(10):4005–11 [DOI] [PubMed] [Google Scholar]
- Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 2019;19(7):377–91 [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong S-J, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol 2009;27(36):6199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348(6238):1036–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC. Genetics of melanoma progression: the rise and fall of cell senescence. Pigment Cell Melanoma Res. 2016;29(2):122–40 [DOI] [PubMed] [Google Scholar]
- Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, et al. KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer. Cell. 2017;168(5):817–829.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busam KJ, Hedvat C, Pulitzer M, von Deimling A, Jungbluth AA. Immunohistochemical analysis of BRAF(V600E) expression of primary and metastatic melanoma and comparison with mutation status and melanocyte differentiation antigens of metastatic lesions. Am. J. Surg. Pathol 2013;37(3):413–20 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert M-D, Denat L, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433(7027):764–9 [DOI] [PubMed] [Google Scholar]
- Chiba K, Lorbeer FK, Shain AH, McSwiggen DT, Schruf E, Oh A, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. 2017;357(6358):1416–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22(2):398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch AJ, Ferguson P, Newell F, Kazakoff SH, Witkowski T, Dobrovic A, et al. Molecular genomic profiling of melanocytic nevi. J. Invest. Dermatol 2019; [DOI] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet 2009;41(5):544–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15(4):294–303 [DOI] [PubMed] [Google Scholar]
- Garman B, Anastopoulos IN, Krepler C, Brafford P, Sproesser K, Jiang Y, et al. Genetic and Genomic Characterization of 462 Melanoma Patient-Derived Xenografts, Tumor Biopsies, and Cell Lines. Cell Rep. 2017;21(7):1936–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–22 [DOI] [PubMed] [Google Scholar]
- Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(6):472–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding CR. Commentary. A picture of Mitf in melanoma immortality. Oncogene. 2011;30(20):2304–6 [DOI] [PubMed] [Google Scholar]
- Goding CR, Arnheiter H. MITF-the first 25 years. Genes Dev. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel VK, Ibrahim N, Jiang G, Singhal M, Fee S, Flotte T, et al. Melanocytic nevus-like hyperplasia and melanoma in transgenic BRAFV600E mice. Oncogene. 2009;28(23):2289–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J. Med. Genet 2007;44(2):99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp S, Scurr LL, Becker TM, Frausto M, Kefford RF, Rizos H. Oncogene-induced senescence does not require the p16(INK4a) or p14ARF melanoma tumor suppressors. J. Invest. Dermatol 2009;129(8):1983–91 [DOI] [PubMed] [Google Scholar]
- Halaban R Rb/E2F: a two-edged sword in the melanocytic system. Cancer Metastasis Rev. 2005;24(2):339–56 [DOI] [PubMed] [Google Scholar]
- Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin. Cancer Res 2006;12(7 Pt 2):2301s–7s [DOI] [PubMed] [Google Scholar]
- Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–80 [DOI] [PubMed] [Google Scholar]
- Hélias-Rodzewicz Z, Funck-Brentano E, Terrones N, Beauchet A, Zimmermann U, Marin C, et al. Variation of mutant allele frequency in NRAS Q61 mutated melanomas. BMC Dermatol. 2017;17(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt C, Lee Wu C, Evans G, Howell A, Elles RG, Jordan R, et al. Germline mutation of ARF in a melanoma kindred. Hum. Mol. Genet 2002;11(11):1273–9 [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A Landscape of Driver Mutations in Melanoma. Cell. 2012;150(2):251–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61 [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Natl Acad Sci U S A 1971;68(4):820–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer 2001;1(2):157–62 [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413(6851):83–6 [DOI] [PubMed] [Google Scholar]
- Maldonado JL, Fridlyand J, Patel H, Jain AN, Busam K, Kageshita T, et al. Determinants of BRAF mutations in primary melanomas. J. Natl. Cancer Inst 2003;95(24):1878–90 [DOI] [PubMed] [Google Scholar]
- McNeal AS, Liu K, Nakhate V, Natale CA, Duperret EK, Capell BC, et al. CDKN2B Loss Promotes Progression from Benign Melanocytic Nevus to Melanoma. Cancer Discov. 2015;5(10):1072–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LCW, Soengas MS, Denoyelle C, Kuilman T, van der Horst CMAM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–4 [DOI] [PubMed] [Google Scholar]
- Miller CA, Settle SH, Sulman EP, Aldape KD, Milosavljevic A. Discovering functional modules by identifying recurrent and mutually exclusive mutational patterns in tumors. BMC Med Genomics. 2011;4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavey SJ, Cummings MC, Whiteman DC, Castellano M, Walsh MD, Gabrielli BG, et al. Loss of p16 expression is associated with histological features of melanoma invasion. Melanoma Res. 2002;12(6):539–47 [DOI] [PubMed] [Google Scholar]
- Pedersen M, Küsters-Vandevelde HVN, Viros A, Groenen PJTA, Sanchez-Laorden B, Gilhuis JH, et al. Primary melanoma of the CNS in children is driven by congenital expression of oncogenic NRAS in melanocytes. Cancer Discov. 2013;3(4):458–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C, Molla A, Vegetti C, Ferrone S, Anichini A, Sensi M. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006;66(13):6503–11 [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat. Genet 2003;33(1):19–20 [DOI] [PubMed] [Google Scholar]
- Raaijmakers MIG, Widmer DS, Narechania A, Eichhoff O, Freiberger SN, Wenzina J, et al. Co-existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget. 2016;7(47):77163–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JA, Loganzo F, Shea CR, Walker GJ, Flores JF, Glendening JM, et al. Loss of expression of the p16/cyclin-dependent kinase inhibitor 2 tumor suppressor gene in melanocytic lesions correlates with invasive stage of tumor progression. Cancer Res. 1995;55(13):2713–8 [PubMed] [Google Scholar]
- Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 2013;38(1):12–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seberg HE, Van Otterloo E, Cornell RA. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res. 2017;30(5):454–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–7 [DOI] [PubMed] [Google Scholar]
- Shain AH, Bastian BC. From melanocytes to melanomas. Nat. Rev. Cancer 2016;16(6):345–58 [DOI] [PubMed] [Google Scholar]
- Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat. Genet 2015a;47(10):1194–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Joseph NM, Yu R, Benhamida J, Liu S, Prow T, et al. Genomic and Transcriptomic Analysis Reveals Incremental Disruption of Key Signaling Pathways during Melanoma Evolution. Cancer Cell. 2018;34(1):45–55.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE. 2013;8(1):e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med 2015b;373(20):1926–36 [DOI] [PubMed] [Google Scholar]
- Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat. Cell Biol 2012;14(8):882–90 [DOI] [PubMed] [Google Scholar]
- Sharpless E, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22(20):3092–8 [DOI] [PubMed] [Google Scholar]
- Talve L, Sauroja I, Collan Y, Punnonen K, Ekfors T. Loss of expression of the p16INK4/CDKN2 gene in cutaneous malignant melanoma correlates with tumor cell proliferation and invasive stage. Int. J. Cancer 1997;74(3):255–9 [DOI] [PubMed] [Google Scholar]
- Tsao H, Bevona C, Goggins W, Quinn T. The transformation rate of moles (melanocytic nevi) into cutaneous melanoma: a population-based estimate. Arch Dermatol. 2003;139(3):282–8 [DOI] [PubMed] [Google Scholar]
- Tyagi E, Liu B, Li C, Liu T, Rutter J, Grossman D. Loss of p16INK4A stimulates aberrant mitochondrial biogenesis through a CDK4/Rb-independent pathway. Oncotarget. 2017;8(34):55848–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Shaw HM, Rodriguez M, Ott S, Goding CR. The retinoblastoma protein modulates Tbx2 functional specificity. Mol. Biol. Cell 2010;21(15):2770–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Haigis KM, Firestone AJ, McNerney ME, Li Q, Davis E, et al. Dominant role of oncogene dosage and absence of tumor suppressor activity in Nras-driven hematopoietic transformation. Cancer Discov. 2013;3(9):993–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang C-H, McCormick F, Levine A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J. 2008;22(8):2605–22 [DOI] [PubMed] [Google Scholar]
- Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J. Natl. Cancer Inst 2013;105(12):917–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Jorapur A, Shain AH, Lang UE, Torres R, Zhang Y, et al. Bi-allelic Loss of CDKN2A Initiates Melanoma Invasion via BRN2 Activation. Cancer Cell. 2018;34(1):56–68.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Choi BY, Lee M-H, Bode AM, Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine. 2016;8:30–9 [DOI] [PMC free article] [PubMed] [Google Scholar]