Abstract
Genome editing represents a promising strategy for therapeutic correction of COL7A1 mutations that cause recessive dystrophic epidermolysis bullosa (RDEB). DNA cleavage followed by homology-directed repair (HDR) using an exogenous template has previously been used to correct COL7A1 mutations. HDR rates can be modest and the double-strand DNA breaks that initiate HDR commonly result in accompanying undesired insertions and deletions (indels). To overcome these limitations, we applied an A•T→G•C adenine base editor (ABE) to correct two different COL7A1 mutations in primary fibroblasts derived from RDEB patients. ABE enabled higher COL7A1 correction efficiencies than previously reported HDR efforts. Moreover, ABE obviated the need for a repair template and minimal indels or editing at off-target sites was detected. Base editing restored endogenous type VII collagen expression and function in vitro. We also treated induced pluripotent stem cells (iPSCs) derived from RDEB fibroblasts with ABE. The edited iPSCs were differentiated into mesenchymal stromal cells, a cell population with therapeutic potential for RDEB. In a mouse teratoma model, skin derived from ABE-treated iPSCs showed proper deposition of C7 at the dermal-epidermal junction in vivo. These demonstrate that base editing provides an efficient and precise genome editing method for autologous cell engineering for RDEB.
INTRODUCTION
Recessive dystrophic epidermolysis bullosa (RDEB) is caused by mutations in the COL7A1 gene resulting in compromised type VII collagen (C7) peptide function. C7 is a key constituent of the dermal-epidermal junction (DEJ), and its impairment leads to a severe blistering phenotype (Mittapalli et al., 2016, Rashidghamat and McGrath, 2017). Allogenic cellular therapies for RDEB include localized fibroblast injections (Wong et al., 2008) or systemic approaches with hematopoietic cell transplant (Tolar and Wagner, 2013, Wagner et al., 2010), and/or mesenchymal stromal cells (MSC) (Conget et al., 2010). For autologous cell engineering, gene therapy and gene editing represent promising strategies.
Transposons, retroviral, or lentiviral vectors have been used to deliver the COL7A1 cDNA under the control of exogenous gene regulatory elements (Droz-Georget Lathion et al., 2015, Jackow et al., 2016, Latella et al., 2017, Sebastiano et al., 2014, Siprashvili et al., 2010, Titeux et al., 2010). The integrating properties of these vectors poses an oncogenic risk, which may be magnified in RDEB patients, who are predisposed to aggressive squamous cell carcinoma. (Demeulemeester et al., 2015, Hacein-Bey-Abina et al., 2003, Turchiano et al., 2014). Unregulated overexpression of COL7A1 may also serve as a driver for carcinoma migration and invasion (Pourreyron et al., 2014). The possibility of insertional mutagenesis, and the lack of responsiveness of vector-borne COL7A1 to the endogenous cues that regulate cellular gene expression, make locus-specific targeting for treating RDEB appealing.
Genome editing agents can be used to mediate the precise correction of mutations that cause genetic diseases (Cong et al., 2013, Komor et al., 2017, 2018, Rees and Liu, 2018). The use of nuclease-based reagents leads to a double stranded DNA break (DSB) that is resolved via non-homologous end joining (NHEJ) or homology-directed repair (HDR). NHEJ typically results in complex mixtures of small insertions and deletions (indels), and has been used for restoring COL7A1 expression (Bonafont et al., 2019, Bornert et al., 2016, Mencia et al., 2018, Takashima et al., 2019). However, depending on the indel profile resulting from the stochastic NHEJ process, in-frame deletions can be infrequent, limiting gene restoration rates. HDR can be used to modify genomic sequences from a donor template; however, the efficiency in therapeutically relevant cells is typically very low, often necessitating antibiotic resistance cassettes to enrich for corrected clones (Hainzl et al., 2017, Osborn et al., 2013, Webber et al., 2016). Further, DSB repair typically results in an excess of NHEJ/indels accompanying the desired HDR product. For therapeutic applications, the ability to achieve robust allele correction with high efficiency and minimal byproducts (e.g., indels from NHEJ) is often critical.
The adenine base editor (ABE) consists of a Cas9 nickase that does not introduce DSBs, but rather directs a fused laboratory-evolved deaminase to convert target A•T base pairs to G•C within a small window of DNA displaced by the targeting single guide RNA (sgRNA) (Gaudelli et al., 2017). Base editing offers three main advantages over HDR: (1) ABE is generally able to introduce or correct single nucleotide polymorphisms with much higher efficiency, often sufficient to avoid the need for positive selection; (2) ABE activity occurs with minimal indels; and (3) no exogenous donor DNA template is required as base editing does not rely on HDR or cell division (Rees and Liu, 2018, Yeh et al., 2018).
We investigated the potential of ABE in primary cells from two RDEB patients with distinct COL7A1 nonsense mutations. An optimized, current-generation adenine base editor (ABEmax,(Koblan et al., 2018)) was delivered as mRNA and resulted in efficient gene correction that restored C7 peptide expression in fibroblasts. 3D culture of corrected cells promoted normalized epithelial layer attachment in vitro.
ABE correction was also pursued in induced pluripotent stem cells (iPSCs) that were subsequently differentiated into mesenchymal stromal cells (MSCs), a population being used to treat RDEB patients (Rashidghamat and McGrath, 2017). ABE corrected iPSC injected in mice formed teratomas with ectodermal derived skin equivalents showing contiguous C7 deposition at the DEJ in vivo. Our data demonstrate the potential of base editing to correct endogenous COL7A1 in autologous cells as a therapeutic strategy for RDEB.
RESULTS
Targeting COL7A1 mutations with base editors
We pursued base editing in two RDEB patients with premature termination codon mutations in the COL7A1 gene. One patient possessed a homozygous c.553C>T (R185X) mutation and the second was a compound heterozygote with c.1573 C>T (R525X) and c.2005C>T (R669X) mutations (Figure 1a–c). A skin biopsy was obtained from each patient and tissue staining with a polyclonal C7 antibody, capable of detecting multiple epitopes, did not show any C7 at the DEJ demonstrating that these patients are null for C7 (Figure 1d).
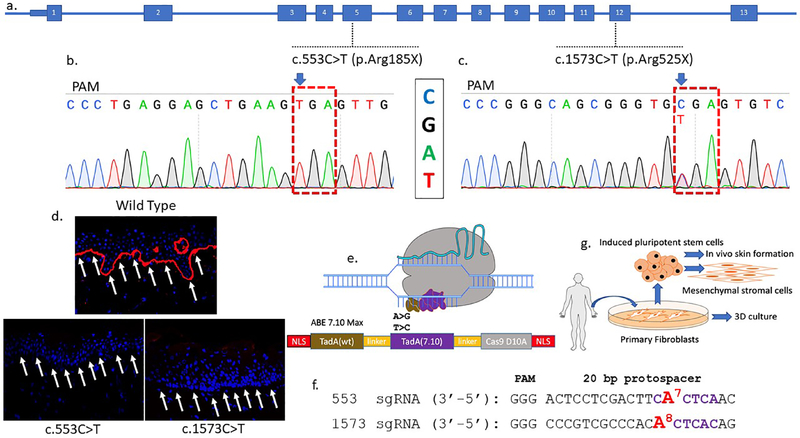
Figure 1. COL7A1 base editing experimental design.
a COL7A1 exons are numbered and the mutations targeted in this study are shown. One patient harbors a homozygous C:G>T:A mutation at nucleotide position c.553 causing an arginine to stop codon mutation in exon 5 at amino acid residue 185. The second patient was diagnosed as a compound heterozygote with a c.1573 C>T mutation in exon 12 at amino acid position 525 leading to a premature termination codon. The second allele has a c.2005C>T (R669X) mutation. b and c Sanger sequencing chromatograms of primary fibroblast samples from homozygous RDEB patients with Arg>X mutations. The sequence shown in the chromatograms corresponds to the sgRNA sequence and the NGG protospacer adjacent motif (PAM) sequence is indicated (anti-sense). d Skin biopsy immunofluorescence. Full thickness punch skin biopsies were obtained from a healthy control and the c.553 C>T and c.1573 C>T RDEB patients. Each sectioned tissue was stained with an equivalent amount of a polyclonal anti-C7 antibody and analyzed by confocal microscopy. C7 stains in red and white arrows show the DEJ in wild type but not RDEB samples. e Adenine base editor architecture. N- and C-terminal nuclear localization signals flank the E. coli TadA wildtype (WT) and evolved adenine deaminases that are separated by an XTEN linker and fused to the S. pyogenes Cas9 D10A nickase. Following Cas9 binding, the deaminases can act on the single-stranded DNA displaced by the protospacer. f c.553 and c.1573 sgRNA sequences with the target base numbered and shown in red. Purple lettering shows the ABE activity window between nucleotides corresponding to positions 4–8 of the protospacer (counting the 5’ nucleotide as position 1). g RDEB primary fibroblasts and induced pluripotent stem cells (iPSC) were obtained and corrected by electroporation of base editor mRNA and targeting sgRNAs. Fibroblasts were used in 3D organotypic cultures in vitro. Corrected iPSC were used as a platform for mesenchymal stromal cell derivation and in vivo teratoma formation that gave rise to ectoderm derived skin.
ABE consists of a single protein containing a wild-type TadA adenosine deaminase, an evolved TadA* deoxyadenosine deaminase, and a Cas9(D10A) nickase (Figure 1e) (Gaudelli et al., 2017). Depending on which DNA strand is targeted, ABE mediates A>G or T>C transitions. sgRNAs were designed that positioned the target nucleotides at position 7 (c.553C>T) or 8 (c.1573 C>T) of the protospacer (Figure 1f). The c.2005 site was not readily accessed by ABE due to the lack of an appropriately positioned protospacer adjacent motif (PAM). ABEmax mRNA and sgRNAs were delivered to primary patient fibroblasts and iPSC; the latter of which served as an engineering template for directed differentiation into MSCs and an in vivo model for skin equivalent generation from teratomas (Figure 1g).
COL7A1 restoration in null patient cells
ABE mRNA and chemically modified sgRNAs (Hendel et al., 2015) were electroporated into primary fibroblasts and base editing was assessed in genomic DNA and mRNA by Sanger and Illumina deep sequencing. c.553 C>T nonsense mutation correction rates were 24±1.0% and 45%±5.8% among the genomic DNA and mRNA, respectively (Figure S1a and Figure 2a).
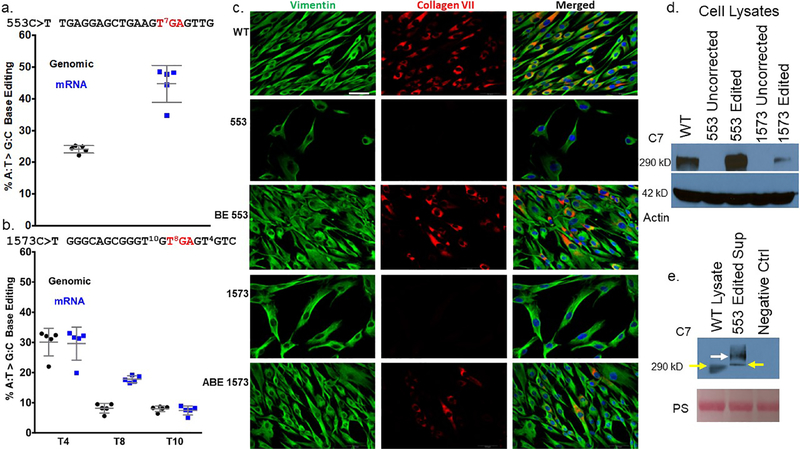
Figure 2. c.553 COL7A1 base editing in RDEB primary cells.
a and b Quantification of DNA base editing by deep sequencing. Genomic DNA and mRNA were sequenced on an Illumina MiSeq to determine the frequency of A:T>G:C editing of a c.553 cells and b c.1573 cells. Values and error bars are the mean and standard deviation from five experimental replicates. sgRNA sequences for each mutation are shown and the red lettered ‘TGA’ represents the nonsense mutation codon. Superscript numbers represent the target base for mutation correction or bystander editing and are numbered relative to the 5’ start of the sgRNA. c Immunofluorescence of primary fibroblasts. White boxes embedded in the images on the left identify samples in that row. Labels at top identify the antigen stained for in that column. Edited and uncorrected cells were stained simultaneously with equivalent amounts of anti-vimentin and anti-collagen type VII polyclonal antibodies. The images for each fluorescent channel were merged with DAPI nuclear stain that is shown at right. Images are representative of three independent experiments. Scale bar=50 μm (lower right in WT vimentin image). d and e C7 Western blotting. d Cell lysates from uncorrected 553 and 1573 cells were analyzed in parallel with base edited 553 and 1573 cells using a polyclonal anti-C7 antibody. Wild type (WT) cells are from a healthy donor. The C7 lane shows the ~290 kD C7 band and actin was used as a loading control. e Secreted C7 from cell supernatant. C7 was detected in the supernatant of c.553 edited cells that were plated in serum free media. WT is a healthy donor lysate sample and negative control (ctrl) is from cells that have a COL7A1 mutation that inactivates the gene. Pro-collagen type VII is shown at 290 kD with yellow arrows. A larger molecular weight species is shown with a white arrow representing the non-reduced C7 ultrastructural trimeric polypeptide. The Ponceau S loading control is shown and is labeled ‘PS.’
Sequencing of the c.1573C>T site showed 50% wild type (WT) sequence in unedited cells, as expected for a compound heterozygote (Figure S1b). This locus also has putative “bystander” adenine bases capable of being modified by ABE at positions 4 and 10 of the sgRNA. ABE activity resulted in detectable modification of all three possible bases in PCR-amplified genomic DNA and mRNA (Figure S1b and Figure 2b). The frequency of the desired nucleotide at position 8 in place of the stop codon, subtracting out the 50% contribution from the other allele that has a WT sequence at this site, was 8.2±1.6% (Figure 2b). Position 10 was edited to approximately the same extent, while position 4 was the most efficiently edited site with a frequency of 30±4.6% (Figure 2b). Deep-sequencing of COL7A1 mRNA showed that the frequency of edits at positions 4 and 10 matched the editing efficiency in genomic DNA. Transcripts with the nonsense mutation corrected at position 8 were enriched in mRNA to 17.8±1.1% (Figure 2b).
We next assessed whether the observed base editing resulted in restored C7 peptide expression. Wild type, RDEB c.553 and c.1573 C>T untreated, and ABE-treated fibroblasts were analyzed by immunofluorescence for vimentin and C7. As expected, all of the cells showed expression of the vimentin cytoskeletal protein (Figure 2c). In agreement with the biopsy sample analysis that was null for C7, staining with a polyclonal anti-C7 antibody showed that untreated cells were completely devoid of C7 expression (Figure 1d and 2c). ABE treatment resulted in C7 restoration with the expression frequency correlating with the observed molecular rates of base editing (Figures 2a–c). To show that the immunoreactive C7 observed in cells represented the full length C7 peptide, we performed Western blot analysis. Cell lysates from uncorrected cells did not show any C7, and base editing restored the ~290 kD full length C7 (Figure 2d). Edited c.553 cells also showed secretion of ~290 kD C7 as well as larger fragments consistent with previous reports (Christiano et al., 1996, Shinkuma et al., 2016, Titeux et al., 2010) representing the putative non-reduced C7 polypeptide/homotrimer (Figure 2e).
We performed isotype antibody staining and short tandem repeat (STR)-based cell line validation for quality control (Figures S2 and S3). STR showed that pre- and post-ABE-modified cells were derived from the same donor, demonstrating the ability of ABE to correct COL7A1 mutations in primary cells.
Base editing allelic variance
We dissected the editing outcomes using CRISPResso2 (Clement et al., 2019) bioinformatical analysis of Illumina MiSeq sequencing data to perform quantification of each distinct allele in the genomic DNA and mRNA PCR amplicons. As expected, the homozygous null patient harboring the c.553C>T mutation showed the uniform presence of the mutant thymine nucleotide in unedited cells. Allele-specific analysis showed three unique amplicons were present following ABE treatment. The unedited, disease causing thymine, and the edited cytosine were by far the most common alleles, while a T>C transition at position three leading to a V186A alteration was present at a frequency <0.5% (Figure 3a).
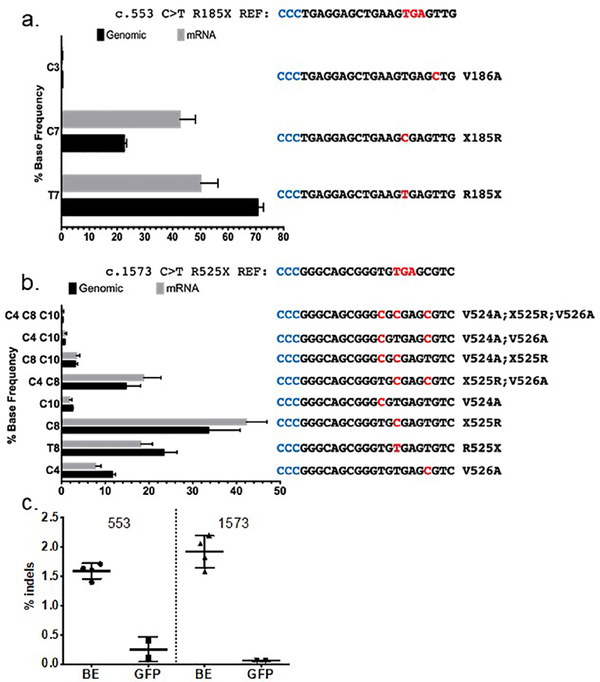
Figure 3. Allele frequencies following ABE treatment.
a COL7A1 c.553 C>T allelic analysis. The mean percentage of each individual edited DNA and mRNA sequence is shown. Amino acid changes and base alterations corresponding to the observed outcomes are shown at right (3’−5’) with PAM (anti-sense) colored in blue. b Allele distribution in c.1573 C>T cells following base editing. The mutation reference sequence is shown at top. A bar graph of edited cells and the frequency of alleles observed in genomic or mRNA derived cDNA is shown. At right are the individual allele sequences (3’−5’) identified following base editing, with altered bases highlighted in red and PAM in blue. Allelic variants occurring at less than 0.2% frequency, approximately the frequency of sequencing errors, were not included in the analysis and therefore the values for the graphs are <100%. c COL7A1 locus insertions and deletions from base editing. The percentage of deep sequencing reads with indels are shown for treated (BE) and control cells transfected with GFP for the c.553 and c.1573 mutation genomic DNA, respectively. Data for each are from 5 replicates and mean and standard deviation are graphed.
Two major alleles were observed at the c.1573C>T target site in unedited cells from the c.1573C>T and c.2005C>T compound heterozygous patient (Figure S1b). Following editing, eight different alleles were all detected at frequencies above 0.2% (Figure 3b). Bystander edits resulted in conservative Val→Ala amino acid changes (Figure 3b), but their effect on C7 function or immunogenicity is not known. Further, the frequency of the purely WT sequence in this amplicon pool containing no stop codon or bystander mutation decreased from 50% in untreated genomic DNA to only 34±6.5% following editing, indicating that the non-targeted allele was also edited at bystander positions, despite a mismatch with the sgRNA.
ABE activity can lead to the introduction of low levels of indels at the target site, thought to be caused by the nick used to direct DNA repair to the non-edited strand (Koblan et al., 2018). We observed 1.5% indels for the c.553C>T editor and 1.9% for the c.1573C>T editor (Figure 3c); rates consistent with previous work (Koblan et al., 2018). Collectively, these data demonstrate the greater purity of ABE editing that can be expected when only a single target nucleotide is present in the editing window, as is the case for the c.533C>T mutation, compared to multiple target nucleotides in the editing window, as is the case for the c.1573C>T mutation. We also observed a general enrichment of nonsense-corrected transcripts in the mRNA pool, suggesting corrected mRNA population is stabilized relative to the mutant one that is potentially subjected to nonsense-mediated decay.
Base editing is precise with a low incidence of editing at off-target sites
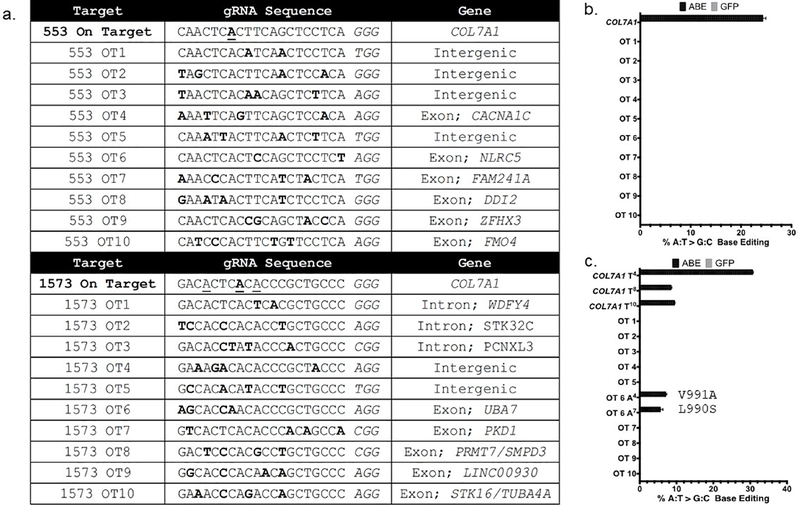
The specificity of genome editing via Cas9 depends on how well a given sgRNA is able to precisely recognize its unique target sequence in the genome. To assess the OT profile of the two ABE sgRNAs employed, we used CIRCLE-seq (Tsai et al., 2017), an unbiased method using cell free DNA, and the CRISPOR in silico predictive algorithm (Haeussler et al., 2016) (Figures S4 and S5 and Figure 4a). We amplified twenty identified loci from ABE edited cells, then used Illumina deep-sequencing to assess the frequency of OT editing. Off-target editing was not observed using the c.553C>T reagent at any of the sites assessed (Figure S4b and Figure 4b). The c.1573C>T reagent yielded A>G editing at one of the twenty evaluated sites, which fell in an exon encoding the ubiquitin modifying enzyme UBA7 (Figure 4c and Figure S4c).
Figure 4. Off target analysis.
a CRISPOR identified off target (OT) sites for the c.553 and c.1573 guide RNAs and associated genomic OT loci. Letters in bold represent mismatches between the on and off target sites. The underlined bases in the on target sequences are those capable of being edited by ABE. b and c High throughput sequencing to assess OT base editing in c.553 or c.1573 cells. Data are from three independent biological replicates of edited or control cells treated with GFP mRNA and error bars show standard deviation.
iPSC base editing and MSC derivation
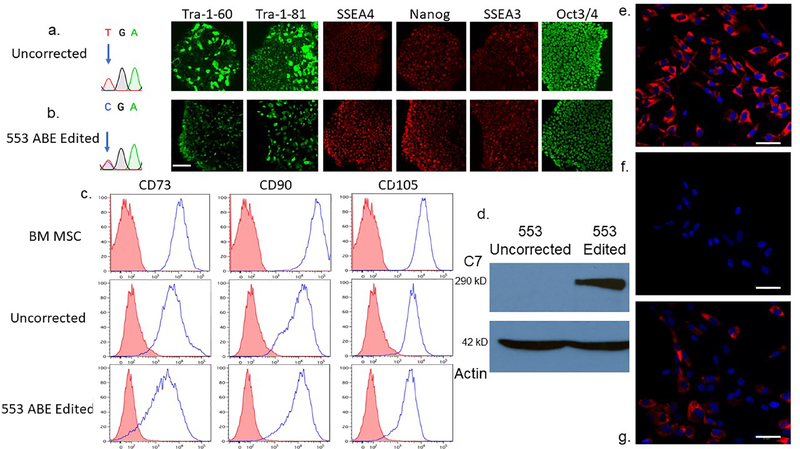
iPSCs represent a potentially inexhaustible source of cells for regenerative medicine. Following co-electroporation of iPSC with ABE mRNA and sgRNA, we isolated colonies and sequenced the COL7A1 c.553 region to assess editing. Corrected and unedited clones were characterized for iPSC markers and pluripotency was unaltered by base editing (Figure 5a and b and Figure S6).
Figure 5. RDEB induced pluripotent stem cell base editing and directed mesenchymal stromal cell differentiation and characterization.
a and b iPSC editing. Sanger chromatogram of uncorrected c.553 C>T; R553X iPSC and a representative base edited clone are shown with arrow showing the mutant/target base. B Pluripotency immunofluorescence marker analysis. Antibodies against the pluripotency markers: podocalyxin TRA-1–60/TRA-1–81, Stage-specific embryonic antigen 4 (SSEA4), SSEA3, Nanog and OCT3/4 were used to detect expression levels. c Mesenchymal stromal cell characterization. Adult bone marrow from normal donors or iPSC derived MSCs were analyzed for the cell surface markers CD73, CD90, and CD105. The isotype staining control peaks are shown in pink. d iPSC derivative MSC Western blot analysis. MSCs derived from uncorrected c.553 C>T iPSCs were compared with those corrected by ABE in a Western blot using a polyclonal anti-C7 antibody. A ~290 kD band is shown with a 42 kD actin loading control below. e-g C7 immunostaining of chamber slides containing e wild type, bone marrow derived MSCs, f unedited 553 patient RDEB iPSC-derived MSCs, and g ABE edited 553 iPSC derivative MSCs. All cells were stained at the same time with the equivalent amount of polyclonal anti-C7 primary and secondary antibodies. Scale bar=50 μm.
We differentiated c.553 uncorrected and edited iPSC clones into MSCs and they expressed the MSC antigens CD90, CD73, and CD105 similar to primary MSCs derived from normal adult bone marrow (Figure 5c). Corrected, but not uncorrected, MSCs expressed C7 as detected by immunofluorescence and full length C7 was observed following Western blotting (Figure 5 d–g). These data show that COL7A1 base edited iPSCs can differentiate into MSCs in vitro, representing a renewable cell population for RDEB treatment.
Deposition of type VII collagen in vitro and in vivo
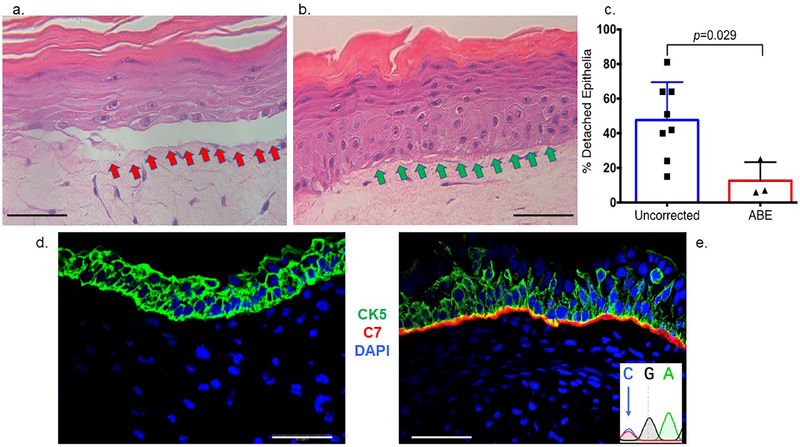
To assess the architectural properties of base edited C7, we performed an in vitro 3D epithelial tissue attachment assay (Dabelsteen et al., 2009, Dickson et al., 2000). The use of C7 null, uncorrected cells resulted in structural failure at the DEJ and detached epithelia while base edited cells restored normalized tissue architecture (Figures 6a–c). To further validate the ability of base edited cells to contribute to the DEJ, we used the ectodermal differentiation capabilities of iPSCs to form skin equivalents following injection into immune deficient mice (Osborn et al., 2013). Teratomas from both base-edited and uncorrected iPSCs showed the presence of human cytokeratin 5, while only base-edited cells deposited C7 at the DEJ in vivo (Figure 6d and e and Figure S7). These data show that base editing results in C7 that is functionally competent for fulfilling its role at the DEJ.
Figure 6. In vitro three dimensional organotypic culture and in vivo expression of type VII collagen.
a-c 3D organotypic culture. Uncorrected or base edited fibroblasts from c.553 patient cells were layered with transformed keratinocytes on a supportive matrix and stained by hematoxylin and eosin. a Uncorrected fibroblast cell 3D culture. The red arrows show the detached epithelial layer due to a fragile dermal epidermal junction that is structurally deficient when the culture contains uncorrected fibroblasts. b ABE corrected cell 3D culture. Improved structural integrity without epithelial layer detachment (green arrows) was observed when ABE corrected cells with restored C7 were employed. c Epithelium detachment quantification. The percent of detached epithelia observed by microscopy and scored by an expert was quantified for eight and three experimental replicates for uncorrected and ABE corrected fibroblasts, respectively. Mean and standard deviation are graphed and p value from Student’s t-test are shown. d-e In vivo teratoma. iPSC were injected into immune deficient mice and representative images of d COL7A1 defective and e base edited iPSC teratoma derived skin equivalents are shown. Both RDEB null and base edited samples were stained with polyclonal anti-human type VII collagen (red) and anti-human cytokeratin (CK5; green) antibodies as well as DAPI nuclear stain. Scale bars=50 μm. At inset, lower right is the base edited nucleotide that was observed following amplification of human COL7A1 DNA from the in vivo teratoma tissue.
DISCUSSION
In this study we sought to determine the potential of base editors to correct COL7A1 gene mutations. Cells from two patients with nonsense mutations were used, both of which were completely unreactive to a polyclonal C7 antibody. This suggests that mutant COL7A1 mRNA, or any produced peptide fragments, are degraded and the total absence of C7 is a crucial aspect to this study. Many RDEB patients show residual/partial C7 expression levels that can confound analyses (Wagner et al., 2010). Therefore, these two C7 null patient samples were ideal to assess and optimize COL7A1 DNA base editing in RDEB.
Base editors can convert C>T, G>A, A>G, or T>C (Gaudelli et al., 2017, Komor et al., 2016) without need for a donor DNA template, typically with much higher efficiencies than HDR, and with dramatically reduced indels (Koblan et al., 2018, Rees and Liu, 2018) The current optimized version of the adenine base editor, ABEmax (Koblan et al., 2018) was co-delivered with sgRNAs designed for opposite-strand targeting converting T>C at protospacer position 7 (c.553) or 8 (c.1573). The average T>C mutation correction rates in primary fibroblasts were 23.8% at the c.553 target and 8.2% for c.1573 in genomic DNA. Deep-sequencing following RT-PCR showed an increased presence of corrected transcripts averaging 45% and 17.8% in c.553 or c.1573 patient cells, respectively. Enrichment of T>C modified transcripts suggests that the edited mRNA molecules were stabilized while the mutant transcripts were subjected to nonsense mediated decay.
The homozygous c.553 target could be efficiently edited with little or no bystander mutations. In contrast, the compound heterozygous c. 1573 target had two bystander nucleotides within the editing window, one edited at a much greater efficiency than the disease causing mutation. ABE also modified the non-targeted, c.2005C>T allele containing a mismatch to the protospacer and introduced C4 and/or C10 bystander edits; however, these mutations on the already null allele should not have any negative impact. Nevertheless, for other applications or patients with hypomorphic mutations, the possibility of introducing additional mutations in the non-targeted allele despite a mismatch with the protospacer should be carefully considered. Each of the bystander edits results in Val→Ala amino acid changes in the C7 protein and their impact on C7 function requires further assessment prior to translational application of the c.1573C>T reagent.
The potential for off-target editing is a key consideration for the use of genome editing agents as potential therapeutics. The Cas9 nickase in ABE mitigates off-target concerns in comparison to previous methods employing nucleases because indels and translocations are much less likely to result from off-target ABE activity. Indeed, the on-target indel frequency was <2%, consistent with previous reports (Koblan et al., 2018). To identify off-target sites, we used an experimental methodology (CIRCLE-seq) and a predictive software tool (CRISPOR). The top 10 sites for each method were analyzed by deep sequencing and no off-target editing was observed using the c.553C>T sgRNA. A single off-target site in the UBA7 gene was detected for the c.1573C>T sgRNA. UBA7 has a potential role as a tumor suppressor in lung cancer (Yin et al., 2009); however, the impact on fibroblasts and MSCs is unknown. Interestingly, the computational identification method identified this bona fide off-target site while CIRCLE-seq did not. Similar to the on-target activity, the off-target site had multiple edits within the sgRNA sequence leading to V991A and L990S substitutions in UBA7. Collectively, the on- and off-target data highlight the benefits of choosing a base editor and protospacer combination with only a single editable nucleotide in the target window. This approach is favorable not only because it leads to more uniform mutation correction at the target site, but also because it decreases the likelihood of introducing non-synonymous changes at off-target loci.
C7 peptide expression was rescued in fibroblasts from both patients following ABE editing. C7 was detected by immunofluorescence and full length C7 was observed in cell lysates and supernatant via Western blotting. Corrected fibroblasts are of immediate therapeutic benefit and have shown efficacy in RDEB patients (Petrof et al., 2013). MSCs have also been used to treat RDEB patients (Petrof et al., 2015); however, MSC numbers decline with age and they are prone to senescence, making primary MSC engineering applications challenging (Serakinci et al., 2008, Stolzing et al., 2008). To circumvent these issues, we performed base editing on c.553 RDEB patient iPSCs, which in principle represent an abundant MSC source following directed differentiation. iPSCs exposed to MSC induction media acquired cell surface markers present on bone marrow derived MSCs and showed restored, full length C7 expression.
In vitro and in vivo assays were performed in order to demonstrate the functionality of the base edited cells. Using a 3D organotypic in vitro skin culture system, we observed normalized attachment of the epithelial layer when using base edited cells but not uncorrected controls. A hallmark of iPSC is their ability to form tissues of all three germ layers and we have previously observed that the ectodermal component forms skin equivalents in mice in vivo (Osborn et al., 2013). Similarly, we observed cytokeratin 5-positive skin structures with a contiguous band of C7 at the DEJ in base-edited, but not C7 null, iPSC-treated animals.
Collectively, our study shows the feasibility of autologous cellular engineering using base editing to correct COL7A1 mutations in cell populations currently employed clinically for RDEB. ABE mRNA electroporation facilitated gene correction without selection or need of a repair template and with minimal off target effects. We successfully base edited fibroblasts and iPSCs, and showed rescued C7 expression and function. These findings suggest that base editing represents a promising potential strategy for autologous RDEB cell therapy.
MATERIALS AND METHODS
Cell culture and gene transfer
RDEB fibroblasts were obtained following written informed patient consent, IRB approval, and in accordance with the Declaration of Helsinki Principles. Culture conditions were as previously reported (Tolar et al., 2014). Sendai virus reprogramming was performed to create iPSCs that were characterized as was done previously (Tolar et al., 2014, Webber et al., 2016). Polyadenylated ABEmax mRNA with a 5’ cap-1 motif was produced by Aldevron (Fargo, ND) and Neon transfection was used for electroporation (fibroblasts: 1500 V, 20 ms pulse width, 1 pulse iPSC: 1100 V, 20 ms, and 1 pulse) with 2 μg of ABEmax mRNA and 1 μg of phosphorothioate modified (Hendel et al., 2015) sgRNA R185X target: 5’-CAACUCACUUCAGCUCCUCA-3 ‘ or R525X target 5’-GACACUCACACCCGCUGCCC-3’ (Synthego, Menlo Park, CA). PCR primers are listed in Supplement.
Three dimensional organotypic cultures were performed as described previously and scored and quantified by an expert reviewer (Dabelsteen et al., 2009, Dickson et al., 2000).
Immunodetection
C7 and iPSC immunofluorescence and isotype staining was performed as previously described (Takashima et al., 2019, Tolar et al., 2014) and images were taken using confocal microscopy (Olympus BX61, Olympus Optical, Japan). Western blotting was performed with a polyclonal anti-human C7 (Chen et al., 2002) or actin antibody.
High throughput sequencing
PCR amplicons for on and off targets were generated using the primers in Supplementary material and were sequenced on an Illumina MiSeq (San Diego, CA). Bioinformatic analysis was performed with CRISPResso2 (Clement et al., 2019).
iPSC
MSCs were differentiated from iPSC using the STEMdiff™ Mesenchymal Progenitor Kit (STEMCELL Technologies, Cambridge, MA) following the manufacturer’s instructions and stained at day 28 with mouse anti-human CD73, CD90 (BD Pharmingen, San Jose, CA) and CD105 antibodies (ThermoFisher, Waltham, MA) and data was acquired on a BD FACSAria (San Jose, CA). iPSC were injected into the flank of immune deficient NSG mice and teratomas were harvested for C7 immunofluorescence as above.
DATA AVAILABILITY STATEMENT
Datasets related to this article can be found in the Supplemental material or by request from the corresponding author.
Supplementary Material
Figure S1. Sanger sequencing of primary RDEB cell PCR products. Sanger sequencing chromatograms of genomic DNA and mRNA of uncorrected and base edited a COL7A1 c.553 C>T and b COL7A1 c.1573C>T primary fibroblast cells. The bases amenable to base editing are underlined and enumerated in relation to the 5’ end of the guide RNA.
Figure S2. Fibroblast immunofluorescence controls. Edited and untreated RDEB fibroblasts were stained with isotype control antibodies for vimentin (chicken) and C7 (rabbit). DAPI nuclear stain and merged images are shown at right. scale bar=50 μm.
Figure S3. Cell line authentication. Genomic DNA from fibroblasts pre and post gene editing were submitted for validation via STR analysis. The analyzed loci are shown at left and the ‘pre’ sample was archived at the time of the initial experimental onset. ‘Post’ cells were analyzed from the same genomic DNA used for Sanger and high throughput sequencing following ABE mRNA delivery. Analytics were performed at the same time on duplicate pre- and post-samples by IDEXX BioAnalytics (Columbia, MO). Green ‘X’ indicates that the STR pattern was identical for pre- and post-ABE treated samples.
Figure S4. CIRCLE-seq Off target analysis. a CIRCLE-seq was performed and the top 10 identified off target genomic loci are shown with highlighted bases representing differences between the off target site and the COL7A1 target. Gene name and location in relation to coding or non-coding sites is indicated. b and c Illumina MiSeq sequencing to assess OT base editing in c.553 or c.1573 RDEB cells. The CIRCLE-seq identified off target sites were amplified and deep sequenced to assess off target editing in primary patient fibroblasts treated with ABE mRNA. Data are from three independent biological replicates of edited cells. GFP mRNA treated patient cells were used as a control.
Figure S5. Primary data of sequences identified by CIRCLE-seq that were deep sequenced for OT analysis. Guide RNA sequences are shown at top for the 553 site (left) and 1573 site (right). The alignments of the protospacers to the off-target sites detected by CIRCLE-seq are shown below. Nucleotides that match the protospacer are indicated with a dot and nucleotides that differ are shown for each site. Dashes indicate a skipped nucleotide in the alignment and small nucleotide letters indicate an insertion relative to the protospacer. The read count of the observed sequence is written at right along with the ID used in Supplemental Figure 4, if sequencing was possible. Brackets indicate two different alignments of the protospacer to the same off-target site. “ND” indicates that editing at that off-target site was not determined either because the site could not be amplified or because the control sample showed sequence heterogeneity, indicating that the same primers amplified homologous sites elsewhere in the genome.
Figure S6. iPSC and MSC immunofluorescence controls. a-b Uncorrected or ABE edited RDEB iPSC cells were stained with isotype (iso) control antibodies in the indicated fluorescent channel. DAPI nuclear stain for each condition is shown at bottom for each cell population. c-e MSC isotype antibody controls. c wild type, d uncorrected, and e ABE edited 553 iPSC derived MSCs were stained with C7 rabbit isotype control as well as DAPI nuclear stain. Scale bars=20 μm for iPSC and 50 μm for MSC.
Figure S7. Teratoma immune fluorescence. a Collagen type VII null or b ABE edited RDEB iPSC cells were stained with primary and isotype control antibodies for the indicated antigen. The individual image slices represent the un-merged experimental images that are merged and shown in Figure 6. Scale bar=50 μm.
Figure S8. Raw sequence date from CIRCLE-seq. Excel files show the raw data set and the tabs labeled ‘read/seq’ are consolidated to show only the read counts and associated off target sgRNA for both the c.553 and c.1573 mutations.
ACKNOWLDEGMENTS
This work was supported by funding from the Mighty Mimi St. Baldrick’s Foundation Scholar Award, the Richard M. Schulze Family Foundation, US NIH R01 AR063070, R01 HL56067, DebRA International, Jackson Gabriel Silver Fund, Epidermolysis Bullosa Medical Research Fund, KidzlstFund, U.S. NIH U01 AI142756, RM1 HG009490, and R35 GM118062, the St. Jude’s Collaborative Research Consortium, the Helen Hay Whitney Foundation, and the Howard Hughes Medical Institute.
The authors are grateful to Drs Mei Chen & David Woodley for the C7 antibody and Dr James Rheinwald for organotypic culture keratinocytes.
Work was performed in Minneapolis, MN, Boston, MA, USA, and Copenhagen, Denmark
ABBREVIATIONS
- RDEB
recessive dystrophic epidermolysis bullosa
- HDR
homology-directed repair
- indels
insertions and deletions
- ABE
adenine base editor
- iPSCs
induced pluripotent stem cells
- C7
type VII collagen peptide
- MSC
mesenchymal stromal cells
- DSB
double stranded DNA break
- NHEJ
non-homologous end joining
- sgRNA
single guide RNA
- PAM
protospacer adjacent motif
- WT
Wild type
- OT
off-target
- DEJ
Dermal-epidermal junction
- CIRCLE-seq
Circularization for In vitro Reporting of CLeavage Effects by sequencing
Footnotes
CONFLICT OF INTEREST.
G.A.N. has filed patents relating to base editor use.
D.R.L. is a consultant and cofounder of Editas Medicine, Pairwise Plants, and Beam Therapeutics, companies that use genome editing.
B.R.B. is cofounder of Tmunity.
CRediT statement.
Conceptualization: MJO, GAN, BRB, DRL, & JT; methodology: MJO, GAN, FK, ANM, SCN, MJR, LX, WC, CRE, BRW, & SD.; formal analysis: MJO, GAN, & FK; investigation: MJO, GAN, FK, ANM, SCN, MJR, LX, WC, & SD; resources: MJO, GAN, HHW, BRB, SD, DRL, & JT.; writing: all authors; funding acquisition: MJO, GAN, HHW, SD, BRB, DRL, & JT
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bonafont J, Mencia A, Garcia M, Torres R, Rodriguez S, Carretero M, et al. Clinically Relevant Correction of Recessive Dystrophic Epidermolysis Bullosa by Dual sgRNA CRISPR/Cas9-Mediated Gene Editing. Mol Ther 2019;27(5):986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornert O, Kuhl T, Bremer J, van den Akker PC, Pasmooij AM, Nystrom A. Analysis of the functional consequences of targeted exon deletion in COL7A1 reveals prospects for dystrophic epidermolysis bullosa therapy. Mol Ther 2016;24(7):1302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Costa FK, Lindvay CR, Han YP, Woodley DT. The recombinant expression of full-length type VII collagen and characterization of molecular mechanisms underlying dystrophic epidermolysis bullosa. J Biol Chem 2002;277(3):2118–24. [DOI] [PubMed] [Google Scholar]
- Christiano AM, Anton-Lamprecht I, Amano S, Ebschner U, Burgeson RE, Uitto J. Compound heterozygosity for COL7A1 mutations in twins with dystrophic epidermolysis bullosa: a recessive paternal deletion/insertion mutation and a dominant negative maternal glycine substitution result in a severe phenotype. Am J Hum Genet 1996;58(4):682–93. [PMC free article] [PubMed] [Google Scholar]
- Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 2019;37(3):224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339(6121):819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conget P, Rodriguez F, Kramer S, Allers C, Simon V, Palisson F, et al. Replenishment of type VII collagen and re-epithelialization of chronically ulcerated skin after intradermal administration of allogeneic mesenchymal stromal cells in two patients with recessive dystrophic epidermolysis bullosa. Cytotherapy 2010;12(3):429–31. [DOI] [PubMed] [Google Scholar]
- Dabelsteen S, Hercule P, Barron P, Rice M, Dorsainville G, Rheinwald JG. Epithelial cells derived from human embryonic stem cells display p16INK4A senescence, hypermotility, and differentiation properties shared by many P63+ somatic cell types. Stem Cells 2009;27(6): 1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeulemeester J, De Rijck J, Gijsbers R, Debyser Z. Retroviral integration: Site matters: Mechanisms and consequences of retroviral integration site selection. Bioessays 2015;37(11):1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 2000;20(4): 1436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droz-Georget Lathion S, Rochat A, Knott G, Recchia A, Martinet D, Benmohammed S, et al. A single epidermal stem cell strategy for safe ex vivo gene therapy. EMBO Mol Med 2015;7(4):380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551(7681):464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003;302(5644):415–9. [DOI] [PubMed] [Google Scholar]
- Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 2016;17(1):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl S, Peking P, Kocher T, Murauer EM, Larcher F, Del Rio M, et al. COL7A1 Editing via CRISPR/Cas9 in Recessive Dystrophic Epidermolysis Bullosa. Mol Ther 2017;25(11):2573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 2015;33(9):985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackow J, Titeux M, Portier S, Charbonnier S, Ganier C, Gaucher S, et al. Gene-Corrected Fibroblast Therapy for Recessive Dystrophic Epidermolysis Bullosa using a Self-Inactivating COL7A1 Retroviral Vector. J Invest Dermatol 2016;136(7):1346–54. [DOI] [PubMed] [Google Scholar]
- Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol 2018;36(9):843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017;168(1–2):20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. Editing the Genome Without Double-Stranded DNA Breaks. ACS Chem Biol 2018;13(2):383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533(7603):420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella MC, Cocchiarella F, De Rosa L, Turchiano G, Goncalves M, Larcher F, et al. Correction of Recessive Dystrophic Epidermolysis Bullosa by Transposon-Mediated Integration of COL7A1 in Transplantable Patient-Derived Primary Keratinocytes. J Invest Dermatol 2017;137(4):836–44. [DOI] [PubMed] [Google Scholar]
- Mencia A, Chamorro C, Bonafont J, Duarte B, Holguin A, Illera N, et al. Deletion of a Pathogenic Mutation-Containing Exon of COL7A1 Allows Clonal Gene Editing Correction of RDEB Patient Epidermal Stem Cells. Mol Ther Nucleic Acids 2018;11:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittapalli VR, Madl J, Loffek S, Kiritsi D, Kern JS, Romer W, et al. Injury-Driven Stiffening of the Dermis Expedites Skin Carcinoma Progression. Cancer Res 2016;76(4): 940–51. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, et al. TALEN-based gene correction for epidermolysis bullosa. Mol Ther 2013;21(6): 1151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof G, Lwin SM, Martinez-Queipo M, Abdul-Wahab A, Tso S, Mellerio JE, et al. Potential of Systemic Allogeneic Mesenchymal Stromal Cell Therapy for Children with Recessive Dystrophic Epidermolysis Bullosa. J Invest Dermatol 2015;135(9):2319–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof G, Martinez-Queipo M, Mellerio JE, Kemp P, McGrath JA. Fibroblast cell therapy enhances initial healing in recessive dystrophic epidermolysis bullosa wounds: results of a randomized, vehicle-controlled trial. Br J Dermatol 2013;169(5):1025–33. [DOI] [PubMed] [Google Scholar]
- Pourreyron C, Chen M, McGrath JA, Salas-Alanis JC, South AP, Leigh IM. High levels of type VII collagen expression in recessive dystrophic epidermolysis bullosa cutaneous squamous cell carcinoma keratinocytes increases PI3K and MAPK signalling, cell migration and invasion. Br J Dermatol 2014;170(6): 1256–65. [DOI] [PubMed] [Google Scholar]
- Rashidghamat E, McGrath JA. Novel and emerging therapies in the treatment of recessive dystrophic epidermolysis bullosa. Intractable Rare Dis Res 2017;6(1):6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018;19(12):770–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V, Zhen HH, Haddad B, Bashkirova E, Melo SP, Wang P, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med 2014;6(264):264ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serakinci N, Graakjaer J, Kolvraa S. Telomere stability and telomerase in mesenchymal stem cells. Biochimie 2008;90(1):33–40. [DOI] [PubMed] [Google Scholar]
- Shinkuma S, Guo Z, Christiano AM. Site-specific genome editing for correction of induced pluripotent stem cells derived from dominant dystrophic epidermolysis bullosa. Proc Natl Acad Sci U S A 2016;113(20):5676–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siprashvili Z, Nguyen NT, Bezchinsky MY, Marinkovich MP, Lane AT, Khavari PA. Long-term type VII collagen restoration to human epidermolysis bullosa skin tissue. Hum Gene Ther 2010;21(10):1299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzing A, Jones E, McGonagle D, Scutt A. Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech Ageing Dev 2008;129(3):163–73. [DOI] [PubMed] [Google Scholar]
- Takashima S, Shinkuma S, Fujita Y, Nomura T, Ujiie H, Natsuga K, et al. Efficient Gene Reframing Therapy for Recessive Dystrophic Epidermolysis Bullosa with CRISPR/Cas9. J Invest Dermatol 2019. [DOI] [PubMed] [Google Scholar]
- Titeux M, Pendaries V, Zanta-Boussif MA, Decha A, Pironon N, Tonasso L, et al. SIN retroviral vectors expressing COL7A1 under human promoters for ex vivo gene therapy of recessive dystrophic epidermolysis bullosa. Mol Ther 2010;18(8):1509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, McGrath JA, Xia L, Riddle MJ, Lees CJ, Eide C, et al. Patient-specific naturally gene-reverted induced pluripotent stem cells in recessive dystrophic epidermolysis bullosa. J Invest Dermatol 2014;134(5):1246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Wagner JE. Allogeneic blood and bone marrow cells for the treatment of severe epidermolysis bullosa: repair of the extracellular matrix. Lancet 2013;382(9899):1214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods 2017;14(6):607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchiano G, Latella MC, Gogol-Doring A, Cattoglio C, Mavilio F, Izsvak Z, et al. Genomic analysis of Sleeping Beauty transposon integration in human somatic cells. PLoS One 2014;9(11):e112712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JE, Ishida-Yamamoto A, McGrath JA, Hordinsky M, Keene DR, Woodley DT, et al. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med 2010;363(7):629–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber BR, Osborn MJ, McElroy AN, Twaroski K, Lonetree CL, DeFeo AP, et al. CRISPR/Cas9-based genetic correction for recessive dystrophic epidermolysis bullosa. NPJ Regen Med 2016; 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T, Gammon L, Liu L, Mellerio JE, Dopping-Hepenstal PJ, Pacy J, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol 2008;128(9):2179–89. [DOI] [PubMed] [Google Scholar]
- Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR. In vivo base editing of post-mitotic sensory cells. Nat Commun 2018;9(1):2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Cong X, Yan M, Zhang DE. Deficiency of a potential 3p21.3 tumor suppressor gene UBE1L (UBA7) does not accelerate lung cancer development in K-rasLA2 mice. Lung Cancer 2009;63(2):194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sanger sequencing of primary RDEB cell PCR products. Sanger sequencing chromatograms of genomic DNA and mRNA of uncorrected and base edited a COL7A1 c.553 C>T and b COL7A1 c.1573C>T primary fibroblast cells. The bases amenable to base editing are underlined and enumerated in relation to the 5’ end of the guide RNA.
Figure S2. Fibroblast immunofluorescence controls. Edited and untreated RDEB fibroblasts were stained with isotype control antibodies for vimentin (chicken) and C7 (rabbit). DAPI nuclear stain and merged images are shown at right. scale bar=50 μm.
Figure S3. Cell line authentication. Genomic DNA from fibroblasts pre and post gene editing were submitted for validation via STR analysis. The analyzed loci are shown at left and the ‘pre’ sample was archived at the time of the initial experimental onset. ‘Post’ cells were analyzed from the same genomic DNA used for Sanger and high throughput sequencing following ABE mRNA delivery. Analytics were performed at the same time on duplicate pre- and post-samples by IDEXX BioAnalytics (Columbia, MO). Green ‘X’ indicates that the STR pattern was identical for pre- and post-ABE treated samples.
Figure S4. CIRCLE-seq Off target analysis. a CIRCLE-seq was performed and the top 10 identified off target genomic loci are shown with highlighted bases representing differences between the off target site and the COL7A1 target. Gene name and location in relation to coding or non-coding sites is indicated. b and c Illumina MiSeq sequencing to assess OT base editing in c.553 or c.1573 RDEB cells. The CIRCLE-seq identified off target sites were amplified and deep sequenced to assess off target editing in primary patient fibroblasts treated with ABE mRNA. Data are from three independent biological replicates of edited cells. GFP mRNA treated patient cells were used as a control.
Figure S5. Primary data of sequences identified by CIRCLE-seq that were deep sequenced for OT analysis. Guide RNA sequences are shown at top for the 553 site (left) and 1573 site (right). The alignments of the protospacers to the off-target sites detected by CIRCLE-seq are shown below. Nucleotides that match the protospacer are indicated with a dot and nucleotides that differ are shown for each site. Dashes indicate a skipped nucleotide in the alignment and small nucleotide letters indicate an insertion relative to the protospacer. The read count of the observed sequence is written at right along with the ID used in Supplemental Figure 4, if sequencing was possible. Brackets indicate two different alignments of the protospacer to the same off-target site. “ND” indicates that editing at that off-target site was not determined either because the site could not be amplified or because the control sample showed sequence heterogeneity, indicating that the same primers amplified homologous sites elsewhere in the genome.
Figure S6. iPSC and MSC immunofluorescence controls. a-b Uncorrected or ABE edited RDEB iPSC cells were stained with isotype (iso) control antibodies in the indicated fluorescent channel. DAPI nuclear stain for each condition is shown at bottom for each cell population. c-e MSC isotype antibody controls. c wild type, d uncorrected, and e ABE edited 553 iPSC derived MSCs were stained with C7 rabbit isotype control as well as DAPI nuclear stain. Scale bars=20 μm for iPSC and 50 μm for MSC.
Figure S7. Teratoma immune fluorescence. a Collagen type VII null or b ABE edited RDEB iPSC cells were stained with primary and isotype control antibodies for the indicated antigen. The individual image slices represent the un-merged experimental images that are merged and shown in Figure 6. Scale bar=50 μm.
Figure S8. Raw sequence date from CIRCLE-seq. Excel files show the raw data set and the tabs labeled ‘read/seq’ are consolidated to show only the read counts and associated off target sgRNA for both the c.553 and c.1573 mutations.
Data Availability Statement
Datasets related to this article can be found in the Supplemental material or by request from the corresponding author.