Significance
Phospholipid membranes often contribute a large fraction of the environment to which integral membrane proteins are exposed. As a result, the behavior of these proteins is commonly affected by solubilization in detergent micelles or even by the particular chemical composition of the lipid bilayer in which they are embedded. In this work, we report cryoelectron microscopy structures of the bacterial pentameric ligand-gated ion channel ELIC embedded in nanodiscs formed by palmitoyl-oleoyl-phosphatidylcholine (POPC) molecules alone, a lipid composition that reduces this channel’s activity to a large extent. Comparison of the unliganded and agonist-bound structural models suggests that POPC membranes disrupt the strict coupling between binding-site affinity and pore permeability that underlies the operation of all ligand-gated ion channels.
Keywords: Cys-loop receptors, structure-function relationships, lipid membranes, molecular dynamics, allosteric proteins
Abstract
The lipid dependence of the nicotinic acetylcholine receptor from the Torpedo electric organ has long been recognized, and one of the most consistent experimental observations is that, when reconstituted in membranes formed by zwitterionic phospholipids alone, exposure to agonist fails to elicit ion-flux activity. More recently, it has been suggested that the bacterial homolog ELIC (Erwinia chrysanthemi ligand-gated ion channel) has a similar lipid sensitivity. As a first step toward the elucidation of the structural basis of this phenomenon, we solved the structures of ELIC embedded in palmitoyl-oleoyl-phosphatidylcholine- (POPC-) only nanodiscs in both the unliganded (4.1-Å resolution) and agonist-bound (3.3 Å) states using single-particle cryoelectron microscopy. Comparison of the two structural models revealed that the largest differences occur at the level of loop C—at the agonist-binding sites—and the loops at the interface between the extracellular and transmembrane domains (ECD and TMD, respectively). On the other hand, the transmembrane pore is occluded in a remarkably similar manner in both structures. A straightforward interpretation of these findings is that POPC-only membranes frustrate the ECD–TMD coupling in such a way that the “conformational wave” of liganded-receptor gating takes place in the ECD and the interfacial M2–M3 linker but fails to penetrate the membrane and propagate into the TMD. Furthermore, analysis of the structural models and molecular simulations suggested that the higher affinity for agonists characteristic of the open- and desensitized-channel conformations results, at least in part, from the tighter confinement of the ligand to its binding site; this limits the ligand’s fluctuations, and thus delays its escape into bulk solvent.
Phospholipid membranes often contribute a large fraction of the environment to which integral membrane proteins are exposed. As a result, it is not surprising to find that the conformational free-energy landscapes of these proteins are affected by solubilization in detergents or even by the particular chemical composition of the lipid bilayer in which they are embedded. Among ion channels, for example, the fact that some detergent-solubilized, agonist-bound neurotransmitter-gated ion channels have been imaged in what is very likely to be the open-channel conformation is, in and of itself, evidence for this lipid sensitivity. Indeed, if we have learned anything about these membrane proteins in the last few decades, it is that agonist binding biases the mixture of closed, open, and desensitized conformations toward the desensitized state, and that the equilibrium is attained in a few tens of seconds, at the longest. It seems inescapable to propose that the lack of a membrane environment may explain this disconnect between what is expected from a membrane-embedded channel and what is actually observed in structural studies upon detergent solubilization.
One well-known example of an ion channel that displays marked lipid sensitivity is the muscle-type nicotinic acetylcholine receptor (AChR) from the electric organ of Torpedo fish. It has been suggested that specific protein–lipid interactions are required for this receptor-channel to undergo the low-affinity ⇌ high-affinity interconversion of the (extracellular) ACh-binding sites (e.g., refs. 1–8), as well as the nonconductive ⇌ ion-conductive interconversion of the transmembrane pore (e.g., refs. 5–12). The molar fraction of cholesterol, the type of phospholipid polar head group, and the length and degree of saturation of the acyl chains have been found to be variables of a complex relationship between lipid-bilayer chemical composition and receptor-channel function. Intriguingly, certain membrane compositions led to a behavior consistent with the disruption of the strict coupling between binding-site affinity and pore permeability (5, 6, 9) that is central to the operation of all ligand-gated ion channels embedded in native lipid bilayers (13, 14).
The lipid dependence of the AChR from Torpedo has long been recognized, and one of the most consistent experimental observations is that, when reconstituted in membranes formed by zwitterionic phospholipids alone, agonist binding fails to elicit ion-flux activity (5, 6, 10). More recently, it has been suggested that the bacterial homolog ELIC (Erwinia chrysanthemi ligand-gated ion channel) (15, 16), may have a similar lipid sensitivity, failing to form an agonist-gated ion channel when reconstituted in palmitoyl-oleoyl-phosphatidylcholine- (POPC-) only vesicles (17). The inability of linear acyl-chain PC- or linear acyl-chain phosphatidylethanolamine-only bilayers to support ion-channel function has also been reported for the bacterial K+ channel KcsA (18–20), bacterial homologs of voltage-dependent Na+ channels (21), and human inward-rectifying K+ channels (22). Similar observations have been made for Na+-coupled amino acid transporters (23) and class-A G protein-coupled receptors (24), for example. Intriguingly, a branched acyl-chain phosphatidylcholine (diphytanoyl-phosphatidylcholine) has recently been reported to support the function of a noninactivating mutant of KcsA (E71A) in the absence of any other lipid (25).
The mechanisms underlying the lipid dependence of the Torpedo AChR and ELIC remain unknown, as are the structures they adopt in membranes of different composition. Furthermore, it is likely that this sensitivity to lipids is not unique to these two receptor-channels, but rather, that it extends to others members of the pentameric ligand-gated ion-channel (pLGIC) superfamily as well. Thus, as a first step toward the elucidation of the structural aspects of this phenomenon, we solved the structures of ELIC embedded in POPC-only nanodiscs in both the agonist-bound and unliganded states using single-particle cryoelectron microscopy (cryo-EM). Despite some noted differences (26), ELIC is, undoubtedly, a bona fide pLGIC, and POPC is one of the most abundant phospholipids in the plasma membrane of HEK-293 cells (27), a cell type that fully supports the function of heterologously expressed ELIC (e.g., ref. 26).
Comparison of the structural models of agonist-bound and unliganded POPC-embedded ELIC revealed that the largest differences occur at the level of loop C—at the agonist-binding sites—and the loops at the interface between the extracellular and transmembrane domains (ECD and TMD, respectively). On the other hand, the transmembrane pore is occluded in a remarkably similar manner in both structures although some rearrangements of the transmembrane α-helices can be observed. A straightforward interpretation of these findings is that POPC-only membranes frustrate the ECD–TMD coupling in such a way that the “conformational wave” of liganded-receptor gating (28) takes place in the ECD and the interfacial M2–M3 linker but fails to penetrate the membrane and propagate into the TMD. Moreover, analysis of the structural models and molecular dynamics (MD) simulations suggested that the higher affinity for agonists characteristic of the open- and desensitized-channel conformations results, at least in part, from the tighter confinement of the ligand to its binding site. This increased degree of “caging” dampened the fluctuations of the bound ligand about its mean position, thereby delaying its escape into bulk solvent. Further analysis of these simulations also revealed the formation of state-dependent electrostatic contacts between ionized side chains of ELIC and the charged moieties of POPC’s polar head group that may contribute to the observed lipid-dependence of function.
Results
ELIC Structures in POPC Nanodiscs.
We reconstituted detergent- (n-undecyl-β-d-maltoside-) solubilized wild-type ELIC in nanodiscs of pure POPC using the membrane-scaffolding protein MSP1E3D1 (29) (SI Appendix, Fig. S1). These discs are expected to contain at most one copy of ELIC pentamer, two copies of MSP1E3D1, and ∼200 molecules (∼100 per monolayer) of POPC per particle, and to be 100 to 120 Å in diameter. Detergent solubilization did not seem to compromise ELIC’s ability to form functional channels, inasmuch as electrophysiological recordings from membrane patches excised from cell-sized proteoliposomes prepared with asolectin (a mixture of soybean lipids) displayed the basic hallmarks of function observed for this pLGIC when expressed heterologously in eukaryotic cells (Fig. 1 A and B). Remarkably, this similarity extended to a large patch-to-patch variability in the desensitization time course (Fig. 1C), an intriguing property of ELIC and animal pLGICs (e.g., ref. 30 for the human α1 GlyR) that is frequently—and, now we know, perhaps wrongly—ascribed to the vagaries of eukaryotic cell-based expression systems. Given the relatively slow and markedly multiexponential desensitization time course of asolectin-embedded ELIC, we anticipate a complex mixture of conformations in this lipid even upon prolonged incubations with saturating concentrations of agonist. Moreover, we anticipate that such marked patch-to-patch variability will complicate efforts to clearly define the lipid dependence of this channel.
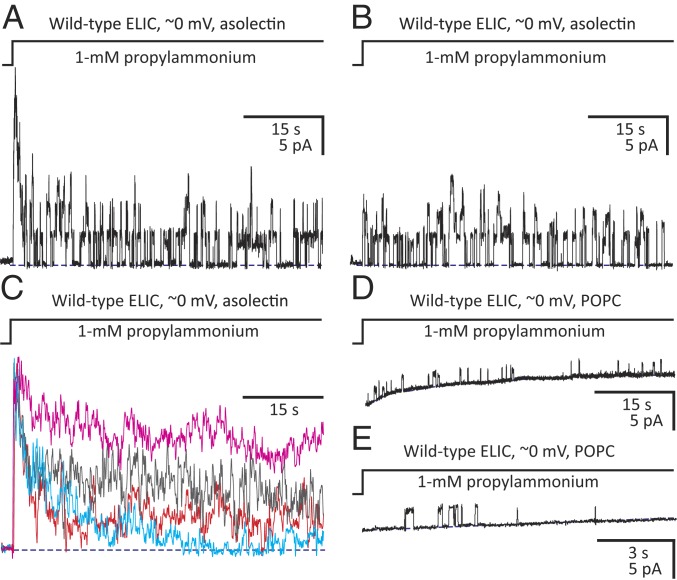
Fig. 1.
Lipid dependence of ELIC ion-channel function. Outward currents recorded under asymmetrical ∼150-mM/5-mM (inside/outside) K+-concentration conditions upon stepping the concentration of extracellular propylammonium from zero to 1 mM. Openings are upward deflections. (A and B) Currents through ELIC–asolectin inside-out patches containing (at least) six (A) and three (B) active channels. (C) Currents through ELIC–asolectin inside-out patches containing (at least) 14 (red trace), 17 (gray), 18 (cyan), and 28 (magenta) active channels. These traces were normalized to their peak values to emphasize the differences between their decay time courses. (D and E) Currents through ELIC–POPC inside-out patches. The low open probability of these traces precluded the estimation of the number of active channels in each patch. Most successful patches excised from ELIC–POPC proteoliposomes, however, were electrically silent. Note that the single-channel current amplitude was lower in POPC than it was in asolectin, not an unexpected finding for a cation-selective channel embedded in a zero net-charge membrane versus a negatively charged one. Note also that the traces in D and E are displayed on two different time scales. The curved shape of the zero-current baseline is an artifact of the ligand application (there is a liquid-junction current on switching the concentration of propylammonium chloride from zero to 1 mM in a solution that is, otherwise, only ∼5-mM in KCl) that becomes more prominent as seals become leakier. In each panel, a blue dashed line denotes the zero-current baseline. The pipette potential was zero, and the +2.7-mV liquid-junction potential between the pipette and bath solutions was offset. As a result, the membrane potential was close to zero, thus approximating the conditions used for cryo-EM image acquisition. Each displayed trace was recorded from a different patch of membrane. s: seconds; pA: picoAmperes.
Patch-clamp experiments performed on cell-sized proteoliposomes prepared with POPC, instead, failed to produce currents comparable to those recorded from asolectin vesicles irrespective of the side of the membrane to which the agonist was applied. Certainly, only short-lived, isolated single-channel openings could be recorded (Fig. 1 D and E), and the vast majority of successful seals were electrically silent even though the extent of ELIC association with POPC membranes was similar to that achieved with asolectin (SI Appendix, Fig. S1B). Electrophysiological recordings from Xenopus oocytes injected with ELIC-reconstituted POPC-only proteoliposomes led to similar findings (17), although the interpretation of negative results from such an indirect approach is not straightforward.
To establish a “baseline” for the effect of the lipid bilayer on ELIC’s ion-channel activity, we set out to characterize its structure in membranes of well-defined and simple composition. Thus, we imaged ELIC-containing POPC nanodiscs in the absence and in the presence of a saturating concentration of the full agonist propylammonium (100 mM) (31) using single-particle cryo-EM. The global resolution of the respective reconstructions [expressed as the reciprocal of the spatial frequency at which the Fourier shell correlation (FSC) (32) between two independently refined “half maps” first crosses the 1/7 (∼0.143) threshold (33; see refs. 34 and 35 for other proposed cutoff values and criteria)] was 4.1 Å for the unliganded channel and 3.3 Å for the agonist-bound channel (SI Appendix, Figs. S2 and S3 and Table S1). As a means to validate the fitted atomic models, FSC curves were also computed between the full experimental density maps and noise-free maps calculated from the atomic coodinates; these plots crossed the 0.5 threshold at 4.3 Å and 3.3 Å for unliganded and agonist-bound ELIC, respectively (SI Appendix, Figs. S2E and S3E). These values are nearly identical to the estimated resolutions (see above), as expected from the theoretical relationship between full maps, half maps, noiseless maps, and correctly built atomic models (33). Map resolutions were also estimated in a local, voxel-by-voxel manner using a method (MonoRes) (36) that does not make use of FSC curves (SI Appendix, Fig. S4). For both reconstructions, the density was weaker for the β8–β9 loop (one of the ECD–TMD interfacial loops; residues 149 to 159) and the M1–M2 linker (residues 225 to 230), and it was weakest for the post-M3 stretch (that is, the M3–M4 linker and the M4 segment; residues 285 to 317). This feature suggested that these regions may be more flexible, and thus, conformationally more heterogeneous.
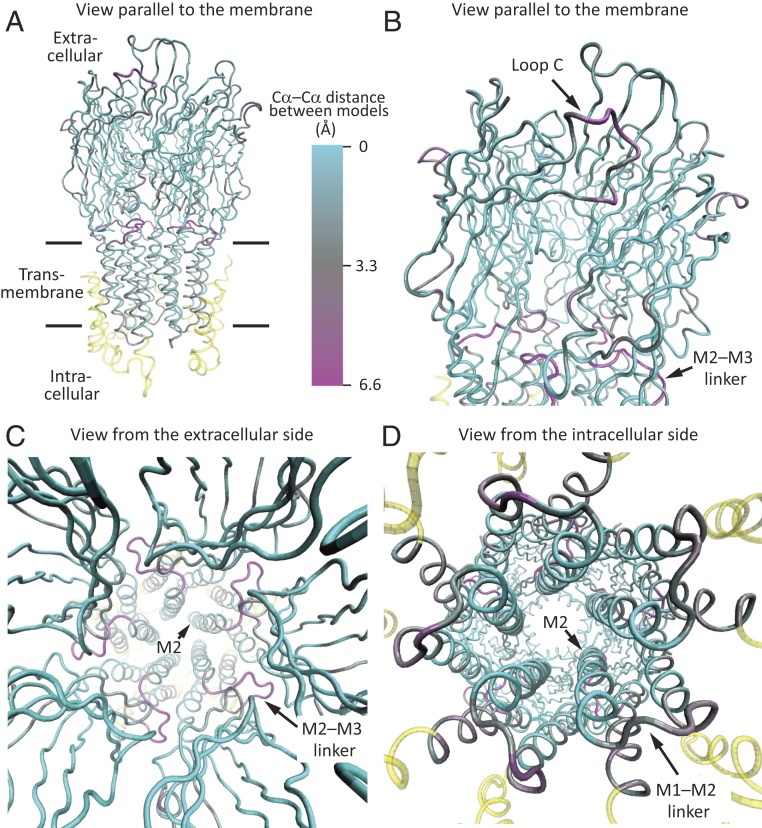
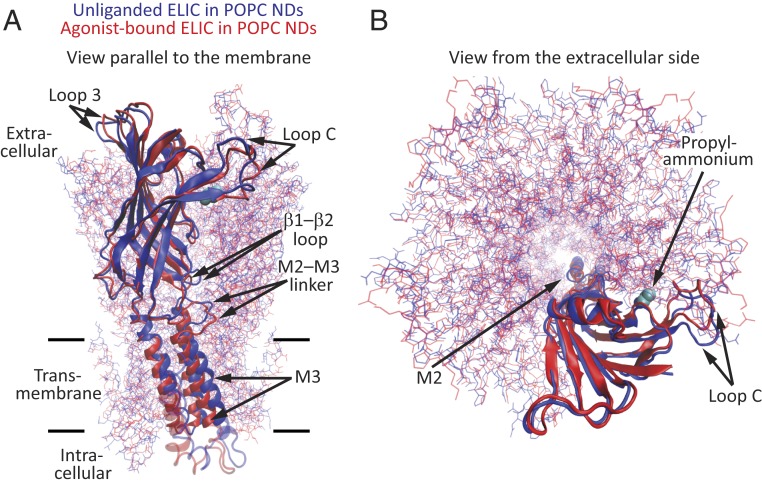
Global superposition of whole pentamers of the unliganded and agonist-bound models of POPC-embedded ELIC (Figs. 2 and 3 and SI Appendix, Fig. S5) revealed that the largest Cα–Cα distances—disregarding the regions of ambiguous density—are in the M2–M3 linker (6.4 Å at Pro-257), the most extracellular α-helical turn of M2 (5.3 Å at Ile-252), the β9–β10 loop (“loop C,” 5.2 Å at Pro-183), the β1–β2 loop (4.1 Å at Leu-29), M3 (∼3.6 Å at residues 272 to 280), and M1 (3.4 Å at Leu-214). Comparing our agonist-bound model with the unliganded model of detergent-solubilized ELIC obtained from X-ray crystallography (PDB ID code 2VL0; 3.3-Å resolution) (16), instead, revealed essentially the same differences: The regions that differ the most between the two models (measured as Cα–Cα distances) are the M2–M3 linker and the most extracellular α-helical turn of M2 (5.3 to 5.6 Å at residues 252 to 257), loop C (5.1 Å at Pro-183), and the β1–β2 loop (3.4 Å at Leu-29). In contrast, and with the exception of their most extracellular α-helical turns, the pore-lining M2 α-helices of unliganded and agonist-bound models superposed with smaller Cα–Cα deviations (<3.0 Å at residues 231 to 250) whether we take our POPC-embedded model or the X-ray crystal-structure model as the unliganded structure. It is worth noting here that X-ray crystal structures of agonist-bound ELIC were found to be nearly indistinguishable from that of its unliganded counterpart (31) or to only show an outward (rather than inward; Figs. 3 and 4) displacement of loop C (38). Very likely, these findings reflect the effects of crystal-lattice forces or the lack of a membrane.
Fig. 2.
Cryo-EM structures of unliganded and propylammonium-bound ELIC embedded in POPC-only nanodiscs. The two models were superposed “globally” in such a way as to minimize the root mean-square Cα–Cα distance between identically numbered residues using the stretch ranging from Pro-11 (the first amino acid in the structure) to Ile-280 (at the C terminus of M3) of all five subunits. Shown is only the propylammonium-bound model, displayed in ribbon representation, and colored according to Cα deviations from the model of POPC-embedded unliganded ELIC. (A and B) Views parallel to the plane of the membrane. The enlarged view in B emphasizes the extracellular domain and the ECD–TMD interface. (C) View perpendicular to the plane of the membrane, from the extracellular side. (D) View perpendicular to the plane of the membrane, from the intracellular side. The cryo-EM density for the M1–M2 linkers was weak. The color code in A is the same for all panels. Regions not included in the superposition are colored yellow. Molecular images were prepared with VMD (37).
Fig. 3.
Global superposition of whole pentameric models of unliganded and propylammonium-bound ELIC. (A) View parallel to the plane of the membrane. (B) View perpendicular to the plane of the membrane, from the extracellular side. For clarity, only one subunit is displayed in cartoon representation with its corresponding orthosteric-site–bound propylammonium molecule shown in van der Walls representation (cyan). All other chains are displayed in lines representation. The color code is the same for both panels. Molecular images were prepared with VMD (37). NDs: nanodiscs.
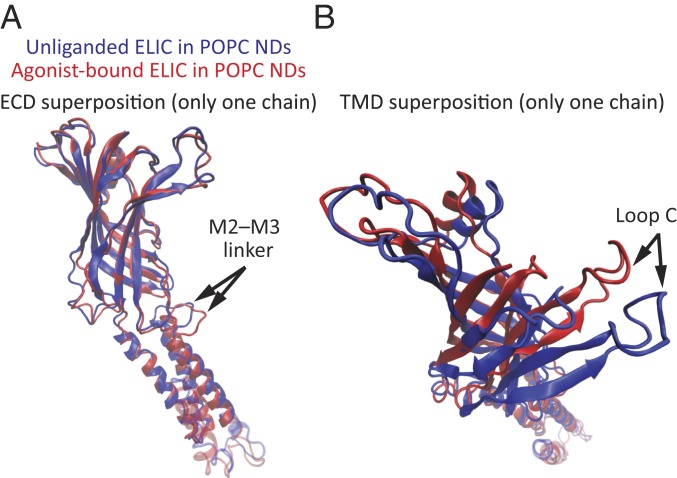
Fig. 4.
Partial superpositions of individual subunits of unliganded and propylammonium-bound ELIC. (A and B) One subunit from each of the structural models of POPC-embedded ELIC was extracted from the pentameric assembly, and the two were superposed partially using either the ECD (residues 11 to 199; A) or the TMD (excluding the M3–M4 linker and M4; residues 200 to 280; B) as the fragments over which Cα–Cα distances were minimized. The models are displayed in cartoon representation. The color code is the same for both panels. Molecular images were prepared with VMD (37). NDs: nanodiscs.
We also compared pairs of models interpreted to represent the unliganded–closed and agonist-bound–open models of other pLGICs: The bacterial GLIC (PDB ID codes 4NPQ and 4HFI) (39, 40), the invertebrate α1 glutamate-gated Cl− channel (GluCl; PDB ID codes 4TNV and 3RIF) (41, 42), and the human serotonin-type 3A receptor (5-HT3AR; PDB ID codes 6BE1, 6DG8, and 6HIN) (43, 44). In all three cases, global superposition of whole pentamers also indicated that the M2–M3 linker is one of the structural elements that differs the most between the two states (4.6 Å in GLIC, 7.0 Å in GluCl, and—depending on the particular agonist-bound model—3.6 Å or 6.9 Å in the 5-HT3AR) followed by loop C (4.2 Å in GLIC, 2.7 Å in GluCl, and 5.0 in the 5-HT3AR).
To better illustrate the conformational differences between the models of unliganded and agonist-bound ELIC in POPC membranes at the level of individual subunits, we superposed single subunits using either the ECD (residues 11 to 199) (Fig. 4A) or the TMD (excluding the M3–M4 linker and M4; residues 200 to 280) (Fig. 4B) as the fragments over which Cα–Cα distances were minimized. When using the ECD for this “partial” superposition, the largest Cα–Cα differences occurred at the M2–M3 linker (7.2 Å at Pro-257) and the most extracellular turn of M2 (6.0 Å at Ile-252). When using the TMD, the largest difference was observed at loop C, being as large as 12.7 Å (at Gln-182). This analysis indicated that, in addition to rigid-body displacements, changes in tertiary structure contribute to the overall differences between the two structures of ELIC examined here.
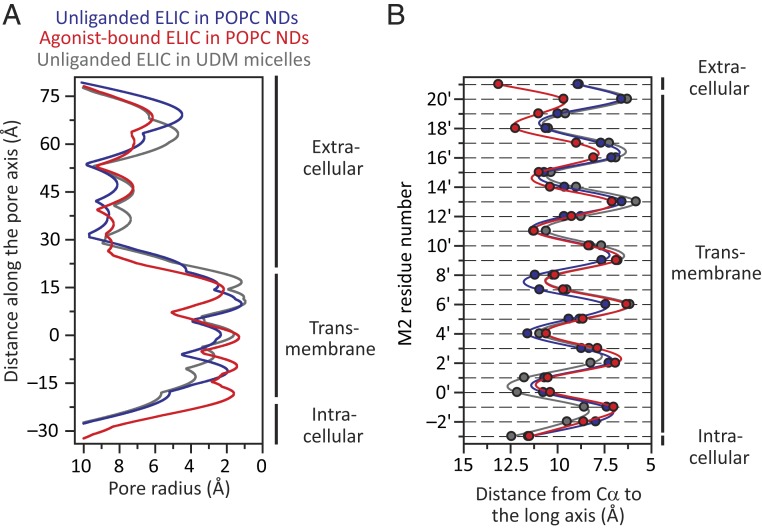
The narrowest constriction of the transmembrane pore (Fig. 5A) in the unliganded model is at Phe-247 (position 16ʹ) and is 1.2 Å in radius; in the crystal-structure model, it is 1.0 Å and occurs at the same position. In the agonist-bound model, on the other hand, the narrowest constriction occurs at Leu-240 (position 9ʹ) and is only a bit wider: 1.3 Å in radius. Considering that the radii of Na+ and K+ with their first hydration shells are 3.1 Å (46) and 3.5 Å (47), respectively, it follows that both pore conformations of this cation-selective channel are nonconductive. To compare the orientations of the pore-lining M2 α-helices in these two cryo-EM models—while removing the uncertainty associated with the modeled conformation of some side chains—we computed the distances between the M2 Cα atoms and the long axis of the pore. Inspection of these “Cα profiles” (Fig. 5B) indicated that, when embedded in POPC-only membranes, the pore lining of ELIC adopts a remarkably similar conformation whether unliganded or bound to agonist. The largest difference occurs in the last α-helical turn of M2 (positions 20ʹ and 21ʹ), which is pulled outwardly by the M2–M3 linker in the agonist-bound model in what seems to be an attempt to open the pore. For positions 20ʹ and 21ʹ, the Cα distances to the long axis increase by 3.1 Å and 4.2 Å, respectively; somehow, however, the rest of M2 does not follow. On the other hand, the differences displayed by the M2 α-helices of unliganded–closed and liganded–open pLGIC model pairs are much larger (SI Appendix, Fig. S6). As for the differences between the models of detergent-solubilized (X-ray) and POPC-embedded (cryo-EM) unliganded ELIC, a comparison of M2-segment Cα profiles (Fig. 5B) reveals a narrower intracellular “mouth” and a straighter, less-kinked, α-helix for the membrane-reconstituted channel.
Fig. 5.
Pore profiles of POPC-embedded unliganded and propylammonium-bound ELIC models. For comparison, the profile corresponding to the X-ray crystal structure of detergent-solubilized unliganded ELIC [PDB ID code 2VL0 (16)] is also shown. (A) Pore-radius profiles estimated using HOLE (45). In the case of agonist-bound ELIC, the molecules of propylammonium were removed. The zero value along the y axis corresponds to the mean position of all five Leu-240 (position 9ʹ) Cα atoms along the axis of ion permeation. (B) Distances between the axis of ion permeation and the Cα atoms for residues in the pore-lining M2 α-helices. Along the y axis, M2 residues are denoted using the prime-numbering system. The color code is the same for both panels. NDs: nanodiscs; UDM: n-undecyl-β-d-maltoside.
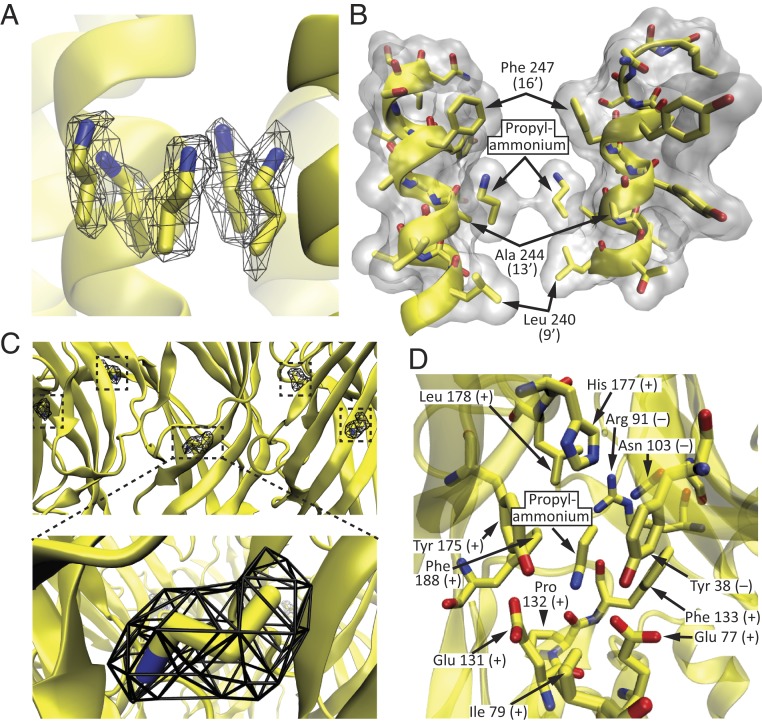
The 3D reconstruction obtained in the presence of a saturating concentration of propylammonium showed additional density at several positions. In 10 of these—two per subunit—the shape of the density was consistent with the presence of propylammonium. Five such positions correspond to the extracellular half of the transmembrane pore (Fig. 6 A and B), and the other five correspond to the well-known orthosteric agonist-binding sites shared by other pLGICs (Fig. 6 C and D). At the transmembrane sites (Fig. 6 A and B), each molecule of propylammonium lies parallel to a different M2 α-helix, next to the short side chain of Ala-244 (13ʹ), filling up the cavity created by the lumen-facing side chains of the flanking Phe-247 (position 16ʹ) and Leu-240 (position 9ʹ). We modeled the ammonium moiety pointing toward the extracellular side simply because this orientation minimizes the distance between the positively charged end of the ligand and the aromatic ring of Phe-247, assuming that they are appropriately positioned to establish cation–π interactions (48). The distance between the nitrogen atom of propylammonium and the center of the aromatic side chain of Phe-247 was ∼5.5 Å. Binding of propylammonium to these pore-lumen sites is likely to be devoid of physiological relevance, and it may simply reflect the high concentration of propylammonium chloride that we applied to the grids to ensure binding to the orthosteric agonist-binding sites.
Fig. 6.
Propylammonium-binding sites. (A and B) Pore-lumen sites. (C and D) Agonist-binding sites. The protein backbone is displayed in cartoon representation, whereas selected residues and propylammonium are displayed in licorice representation. In B, only two (nonadjacent) subunits are shown, for clarity. In this panel, the protein and the two molecules of propylammonium are also displayed in surface representation (colored white) using a probe radius of 1.4 Å. In D, all amino acid residues with atoms within ∼5 Å of any atom of propylammonium are displayed; the view is from the membrane toward the extracellular side. Residues belonging to the principal side of the agonist-binding interface are denoted with a “+” sign, whereas those belonging to the complementary side are denoted with a “−” sign. In A and C, the cryo-EM density is displayed as a black mesh contoured to an isovalue of 0.05. For all panels, carbon atoms are colored yellow; nitrogens, blue; and oxygens, red. Molecular images were prepared with VMD (37).
To estimate the extent to which the presence of agonist molecules bound to the pore-lumen sites accounts for the observed differences between the structural models of unliganded and agonist-bound ELIC, we performed all-atom MD simulations and focused on the nearby M2–M3 linker. Because neither the occupancy of the pore sites nor the protonation state of these propylammonium molecules in close proximity are known, we compared simulations under three extreme pore-site situations intended to cover a range of possible scenarios. These were: 1) five molecules of propylammonium bound (that is, the unmodified agonist-bound structural model); 2) five molecules of (neutral) propylamine bound; and 3) no molecules of propylammonium or propylamine bound. Analysis of these three trajectories (SI Appendix, Fig. S7) supported the notion that the observed structural changes between the models of unliganded and agonist-bound ELIC are largely due to the occupancy of the orthosteric sites. As indicated above, the Cα–Cα distance between the two atomic models is largest at Pro-257, in the M2–M3 linker, with a value of 6.4 Å. Averaging the last 50 ns of the three 500-ns simulations, this distance was 6.5 Å with five pore-lumen–bound propylammonium molecules, 5.4 Å with five pore-lumen–bound propylamine molecules, and 4.6 Å without propylammonium or propylamine bound to the pore sites. Thus, although occupancy of the pore-lumen sites did have an effect on the conformation of the M2–M3 linker, the Cα atom of Pro-257 in the simulated models remained far from its counterpart in the unliganded model throughout the simulations regardless of whether these sites were vacant or bound.
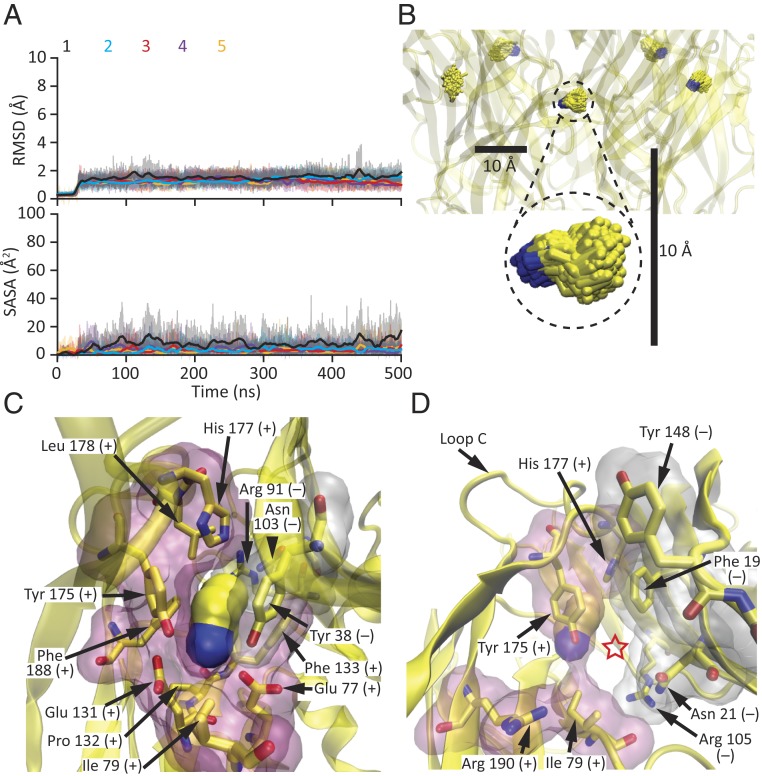
The cryo-EM density at the orthosteric agonist-binding sites (Fig. 6C) was stronger than that at the pore-lumen sites (Fig. 6A), perhaps because of a higher occupancy. As is the case for all homomeric members of the superfamily, these extracellular sites are contributed by residues lying at the interface between any two adjacent subunits. On the “principal” side, these residues (defined here as amino acids with atoms within 5 Å from any atom of the agonist) are: Glu-77 and Ile-79, in the β3–β4 loop (“loop A”); Glu-131, Pro-132, and Phe-133, in the β7–β8 loop (“loop B”); and Tyr-175, His-177, Leu-178, and Phe-188, in loop C. On the “complementary” side, these residues are: Tyr-38 in β2 (“loop D”), Arg-91 in β5, and Asn-103 in β6 (“loop E”) of the (–) adjacent subunit (Fig. 6D). Chemical intuition suggested that the positively charged ammonium moiety must be pointing “downward,” toward the membrane, so as to minimize its distance to the carboxylates of Glu-77 and Glu-131 and maximize its distance to the guanidinium group of the adjacent subunit’s Arg-91. All-atom MD simulations starting with either orientation of the ligand—that is, with its nitrogen atom pointing toward (Fig. 7 A and B) or away from the binding-site glutamates (SI Appendix, Fig. S8)—amply confirmed the choice of the downward orientation as the more stable one. After relieving the restraints, all five agonist molecules remained very close to their experimentally determined densities—irrespective of their starting orientations—for the rest of the simulation (Fig. 7A and SI Appendix, Fig. S8A). Moreover, inspection of bound propylammonium both in the structural model and throughout the simulations revealed that binding-site residues form a tight “cage” around it, leaving little room for it to fluctuate and escape from the protein (Fig. 7 C and D). Quantitatively, this was manifest as a relatively low solvent-accessible surface area for bound propylammonium (Fig. 7 A, Lower).
Fig. 7.
Agonist-binding sites in the agonist-bound state. (A and B) All-atom MD simulation starting from the cryo-EM model of propylammonium-bound ELIC. The root mean-square deviation (RMSD) of the heavy atoms of each of the five propylammonium molecules relative to the initial frame, as well as their solvent-accessible surface areas (SASA), were calculated every 10 ps and are displayed (thin lines) in the Upper and Lower panels of A, respectively, using the same y-axis scale as for their counterparts in Fig. 8A and SI Appendix, Fig. S8A; thick lines represent moving averages. In B, the five molecules of propylammonium in the 51- to 500-ns simulation interval are displayed at one frame per nanosecond, in licorice representation. Carbon atoms are colored yellow and nitrogens are colored blue. The scale bars are the same as those in Fig. 8B. (C) Propylammonium-bound agonist-binding site viewed from the membrane, as in Fig. 6D. Propylammonium is displayed in surface representation using a probe radius of 1.4 Å. The inward movement of loop C tightens the agonist-binding cavity (compare to Fig. 8C). (D) View of the window leading from each agonist-binding site to bulk solution (indicated with a red star). The movement of loop C narrows this opening (compare to Fig. 8D). In C and D, amino acid residues lining the agonist-binding sites are also displayed in surface representation (principal side in pink and complementary side in white) using a probe radius of 1.4 Å. Residues belonging to the principal side of the agonist-binding interface are denoted with a “+” sign, whereas those belonging to the complementary side are denoted with a “−” sign. Molecular images were prepared with VMD (37).
Structural Changes Underlying the Affinity Change.
Because propylammonium binding to ELIC favors channel opening (31), it follows that this ligand binds to the open-channel conformation with higher affinity than it does to the closed state; this is a theoretical necessity, and as a general concept it is one of the cornerstones of molecular pharmacology. Although we do not know yet how well the ECD of our unliganded and agonist-bound models approximate the low-affinity–closed and high-affinity–open states of the liganded channel’s ECD, respectively, we assumed here that they indeed are good approximations. With this idea in mind, we set out to compare the binding sites of our two models in search for the structural changes that underlie the agonism—that is, the differential affinity—of propylammonium on ELIC. Global superposition of whole pentamers (Figs. 2 and 3) revealed that His-177, in loop C, changes the most, its Cα atom moving 4.4 Å; Tyr-175, also in loop C, shows the second largest change (2.6 Å for the Cα atom). In contrast, the Cα atoms of the propylammonium-binding side chains of Glu-77 and Glu-131 move less than 1.0 Å. These observations seemed to be robust inasmuch as the superposition of our propylammonium-bound model to that of the detergent-solubilized unliganded ELIC (16) yielded the same qualitative results: His-177 changes the most (3.5 Å), followed by Tyr-175 (1.5 Å), whereas Glu-77 and Glu-131 move less (1.1 Å and 0.6 Å, respectively). His-177’s side chain is likely to be largely neutral at pH 7.4 (in both conformations), and its closest distance to the positively charged end of propylammonium is 6.9 Å. Thus, the major reorganization of the agonist-binding sites between its low- and high-affinity forms seems to involve residues that are not engaged in direct interactions with the ligand.
In the propylammonium-bound model, His-177 is surrounded by several residues, including three aromatic amino acids with atoms within 5 Å: Tyr-175 from the same subunit, and Phe-19 and Tyr-38 from the adjacent subunit (SI Appendix, Fig. S9A). On the other hand, in the unliganded model, the histidine has moved away from this microenvironment, mainly as a result of the outward movement of loop C. In this alternative position, the side chain of His-177 remains close to (that is, within 5 Å of) the aromatic side chains of Tyr-175 from the same subunit and Phe-19 from the adjacent subunit, but exchanges the side chain of Tyr-38 for that of Tyr-148 (of the adjacent subunit) as interaction partner (SI Appendix, Fig. S9B). To assess the relevance of the changing interactions involving His-177’s imidazole group for channel function in the context of all of the other bonds that form and break elsewhere in the protein as the channel binds ligand and changes conformations, we studied the functional effect of truncating its side chain. Electrophysiological recordings showed that mutating this histidine to alanine slows down both activation and the time course of entry into desensitization in the presence of a saturating concentration of propylammonium (SI Appendix, Fig. S10 A and B), and speeds up channel deactivation (SI Appendix, Fig. S10 C and D). These three observations are consistent with a lower low-affinity–closed ⇌ high-affinity–open equilibrium constant for the alanine mutant, which indicates that a histidine at position 177 stabilizes the high-affinity–open state relative to the low-affinity–closed state more than does an alanine. Nevertheless, regardless of quantitative details, we would like to emphasize that channel function was remarkably well-preserved in the H177A mutant in keeping with the low sequence conservation of loop C among pLGICs, and the idea that this loop's functional role is distributed among several residues.
Caging Delays Agonist Escape.
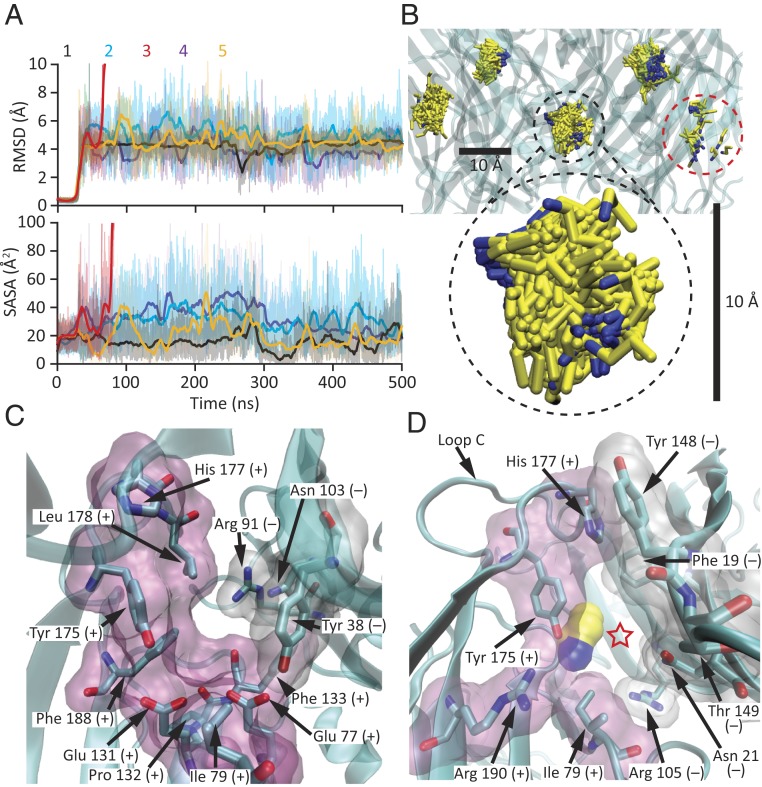
As discussed above, all-atom MD of the propylammonium-bound model showed that—upon relieving the restraints—all five agonist molecules remained very close to their experimentally determined densities for the rest of the simulation (Fig. 7). During the last 450 ns of the trajectories, the root mean-square fluctuation of propylammonium about its mean position (averaged over the five molecules) was 0.6 Å for the nitrogen atom and 1.2 Å for the most distal carbon. We assume, here, that this structural model corresponds to the high-affinity state. Furthermore, assuming that the unliganded model provides a good approximation for the conformation of the agonist-binding sites in the agonist-bound, low-affinity state, we modeled propylammonium molecules in these five sites, taking their coordinates from the agonist-bound model, and simulated the system using all-atom MD. Compared to the simulation of the agonist-bound model, the five molecules of propylammonium modeled in the unliganded binding sites experienced much larger fluctuations and had a much larger solvent-accessible surface area; one of them even escaped the protein during the simulation (molecule #3, shown in red in Fig. 8 A and B). The root mean-square fluctuation of propylammonium about its mean position (averaged over the four molecules that remained within the protein in the last 450 ns) was as much as 1.8 Å for the nitrogen atom and 2.6 Å for the most distal carbon.
Fig. 8.
Agonist-binding sites in the unliganded state. (A and B) All-atom MD simulation starting from the cryo-EM model of unliganded ELIC with five propylammonium molecules modeled at their positions in the agonist-bound model. The heavy-atom RMSD (relative to the initial frame) and SASA values of each of these molecules were calculated every 10 ps and are displayed as in Fig. 7A. Molecule #3, in red, escaped from the protein at ∼80 ns, and thus, its RMSD and SASA values are not reported beyond this point. In B, the five molecules of propylammonium in the 51- to 500-ns simulation interval are displayed at one frame per nanosecond, as in Fig. 7B. Propylammonium molecule #3, present only in the first few displayed frames, is highlighted with a red, dashed-line circle. The scale bars are the same as those in Fig. 7B. (C) Unliganded agonist-binding site viewed from the membrane, as in Figs. 6D and 7C. The outward movement of loop C opens up the agonist-binding cavity (compare to Fig. 7C). (D) View of the window traversed by propylammonium molecule #3 (displayed in surface representation using a probe radius of 1.4 Å) as it escaped from the protein into bulk solution during the simulation (indicated with a red star). The movement of loop C widens this opening (compare to Fig. 7D). In C and D, amino acid residues lining the agonist-binding sites are also displayed in surface representation (principal side in pink and complementary side in white) using a probe radius of 1.4 Å. Residues belonging to the principal side of the agonist-binding interface are denoted with a “+” sign, whereas those belonging to the complementary side are denoted with a “−” sign. Molecular images were prepared with VMD (37).
The larger fluctuations and faster dissociation of propylammonium observed in simulations of the unliganded model are consistent with the presumed lower affinity of this conformation for the ligand, and thus with the well-established agonism of small alkylammonium cations on ELIC. These fluctuations correspond to what could be interpreted as repeated unbinding–rebinding events of propylammonium that continue until the ligand finally gets to escape into bulk solvent (“dissociate”). As long as rebinding is faster than the channel’s conformational transitions, this “geminate recombination”—as this concentration-independent rebinding phenomenon (49) is commonly referred to in the literature of gas-binding heme proteins (e.g., refs. 50 and 51)—would effectively prolong the mean lifetime of the agonist–channel complex in the low-affinity state. In the high-affinity state, on the other hand, the tighter cage formed around propylammonium dampened these fluctuations and delayed its dissociation from the protein even further. Comparison of the two structural models offers an explanation for the simulation results: The unliganded state (taken here to represent the low-affinity conformation) features a wider binding-site cavity (Fig. 8C) and a larger “window” leading to bulk solution (Fig. 8D) than does the ligand-bound state (taken here to represent the high-affinity conformation) (Fig. 7 C and D). A mechanism in which steric considerations—such as size and shape complementarity—are important determinants of ligand affinity may be of general relevance for small-molecule agonists that (like propylammonium) have few functional groups, and thus do not offer alternative contact points with the surrounding protein as a means to increase or decrease their affinities in a conformation-dependent manner. Intriguingly, different degrees of ligand caging have been proposed to also underlie the different affinities of gas-binding heme proteins for NO, CO, and O2 (e.g., ref. 52).
Gratifyingly, free-energy perturbation calculations of the binding of propylammonium to the orthosteric ligand-binding sites also were consistent with the proposed higher affinity of the agonist-bound model. Certainly, the estimated difference between the standard Gibbs free energies of association of propylammonium to the agonist-bound and unliganded protein structures was −9.3 ± 0.3 kcal mol−1 per site. This value compares favorably with experimental estimates obtained for the association of ACh to the open and closed conformations of the muscle AChR (ΔΔG° = −5.2 kcal mol−1 at the α1-δ subunit interface, −5.6 kcal mol−1 at the α1-ε interface, and ΔΔG° = −7.9 kcal mol−1 at the α1-γ interface, at 22 °C) (53), the only pLGIC for which experimental values of this variable have been reported.
State-Dependent Lipid–Protein Interactions.
At least in the case of the Torpedo AChR, it has been clearly established that the electrostatic properties of a phospholipid’s polar head group are a key determinant of ion-channel function (e.g., ref. 8). With this idea in mind, we reanalyzed some of the obtained MD trajectories of the unliganded and agonist-bound atomic models of ELIC focusing on the attractive interactions between ionized amino acid side chains and the charged moieties of POPC’s polar head. This analysis revealed that ionized side chains from the extracellular β6–β7 and β8–β9 loops and the intracellular M3–M4 linker contribute many of these contacts, and that their number differs between the unliganded and agonist-bound models throughout the simulations (SI Appendix, Fig. S11); averaging the last 50 ns, the unliganded conformation made ∼15 more lipid–protein contacts per pentamer. Because of their state-dependence, these electrostatic interactions are poised to affect the channel’s closed–open–desensitized equilibrium constants, thus hinting at a possible link between membrane chemical composition and ion-channel function.
Discussion
Our understanding of the relationship between structure and function in pLGICs is not thorough enough yet to allow us to infer functional states solely on the basis of structural features. Indeed, we can state with confidence that the transmembrane pore is in a similar nonconductive conformation in both the unliganded and agonist-bound structural models of ELIC presented here, but we cannot discern whether this conformation corresponds to the closed state or to one of the many desensitized states that pLGICs have been inferred to adopt (e.g., ref. 54). Similarly, although it is clear that the agonist-binding sites are vacant in one model and occupied in the other, we do not know whether the latter represents a conformation with high affinity for the agonist (characteristic of the open and desensitized states) or a conformation with low affinity (characteristic of the closed state). However, the marked similarity between the unliganded and agonist-bound models at the level of the pore-lining M2 α-helices suggested that the pore is in the same nonconductive conformation, probably the “resting,” closed-state conformation. In addition, the observation that the structural differences between the unliganded and agonist-bound models at the level of the binding-site loop C and the interfacial ECD–TMD loops are of similar magnitude to those observed upon pairwise comparison of the closed- and open-state models of other pLGICs led us to favor the idea that the binding sites of the propylammonium-bound model of ELIC represent the high-affinity conformation and that its interfacial loops have moved in an attempt to open the pore. Thus, according to this interpretation, agonist-bound ELIC embedded in POPC membranes adopts a hybrid conformation having a high-affinity open/desensitized conformation in the ECD—fully supported by our MD simulations and free-energy calculations—but a closed-state conformation in the transmembrane pore. Clearly, this conformational “chimera” need not exist in membranes that support the ion-channel activity of ELIC, and instead, may simply represent the effect of a pure POPC bilayer frustrating the propagation of the gating conformational change through the protein. Therefore, in POPC membranes, the strict coupling between binding-site affinity and pore permeability that underlies the operation of all ligand-gated ion channels (13, 14) would be deficient, and the short and infrequent single-channel openings observed (Fig. 1 D and E) would represent rare instances in which the gating conformational change propagates successfully. We wonder whether this loss-of-function mechanism also holds for other function-incompetent lipid compositions and for other lipid-sensitive pLGICs. In principle, the situation seems to be different for the AChR from Torpedo: Although POPC-only membranes have been proposed to also stabilize a closed-state–like conformation of the pore domain, POPC has been suggested to stabilize the ACh-binding sites in a low-affinity, closed-state–like conformation (7).
There does not seem to be any reason why the loss of activity in electrophysiological assays should be accompanied by the loss of coupling between the agonist binding-site affinities and the conductive versus nonconductive state of the transmembrane pore. Indeed, one can imagine that maneuvers that diminish the electrical activity of a channel, such as mutations, may simply stabilize the entire protein in the low-affinity–closed-channel conformation, in which case the ECD–TMD coupling would be fully preserved. At this point, it is unclear what properties of a pure-POPC bilayer hinder the propagation of the gating conformational change of ELIC into its TMD and how these properties change in going from POPC to a complex mixture of lipids, such as asolectin, for the latter to restore this coupling. Much more generally, it is also unclear how the properties of a piece of plasma membrane adhered to the interior of a glass patch-clamp micropipette (e.g., ref. 55) compare to those of a protein-wrapped flat disk of symmetrical lipid bilayer with enough room for only a single copy of the channel, and whether one can be used to learn about the other and, ultimately, about membranes of the size and shape of cells.
Many other questions remain and other interpretations of our structural models in terms of their corresponding functional states are, certainly, conceivable. We regard the current work with POPC-only membranes as a “foundation” or “baseline” on top of which future work with increasingly complex lipid bilayers and animal pLGICs will likely stand. We hope this line of investigation will get us closer to a thorough understanding of the lipid dependence of pLGIC function (5–10, 56), one of the earliest noted—yet, most elusive—properties of this superfamily of ion channels.
Materials and Methods
ELIC was overexpressed and purified as previously described (16, 57). Purified ELIC was reconstituted into POPC (Avanti Polar Lipids) nanodiscs following established protocols (58), and into POPC or asolectin (Cat. No. 11145; Sigma) cell-sized proteoliposomes following the “dehydration–rehydration” method (59, 60). Currents from proteoliposomes or transiently transfected HEK-293 cells were recorded in the inside-out and whole-cell patch-clamp configurations, respectively. Agonist-concentration jumps were applied using a piece of double-barreled glass “θ-tubing.” The flow of solution through the θ-tube was controlled using a gravity-fed system, and the movement of the θ-tube was achieved using a piezo-electric arm. In the inside-out configuration, the pipette solution was: 142 mM KCl, 5.4 mM NaCl, 1.8 mM CaCl2, 1.7 mM MgCl2, and 10 mM Hepes/KOH, pH 7.4. In the whole-cell configuration, the pipette solution was: 110 mM KCl, 40 mM KF, and 5 mM Hepes/KOH, pH 7.4. For both configurations, the bath solution was: 5 mM KCl, 250 mM mannitol, and 5 mM Hepes/KOH, pH 7.4. The two extracellular solutions applied through the θ-tube barrels were bath solution with and without 1- or 10-mM agonist (propylammonium-chloride/propylamine; pKa = 10.71). In a subset of patch-clamp recordings from POPC–ELIC proteoliposomes, the agonist was also added to the pipette solution. Unliganded and propylammonium-bound ELIC cryo-EM grids were prepared using Spotiton 1.0 (61). Images were acquired by a Titan Krios microscope (ThermoFisher Scientific) operated at 300 kV using a Gatan K2 direct electron detector (Gatan) with an energy filter and in counting mode. Particle stacks were imported into RELION2.1 (62) and were processed through several rounds of 2D and 3D classification. Selected classes were then processed for high-resolution 3D refinement. Structural models of both unliganded and propylammonium-bound ELIC were built using PHENIX (63, 64), Coot (65), and MD flexible fitting (66–68). Data-collection, processing, and model-refinement statistics are listed in SI Appendix, Table S1. NAMD (69) was used to perform MD flexible fitting, MD simulations, and free-energy perturbation calculations using the CHARMM forcefield (70, 71). Visual molecular dynamics (VMD) (37) was used for most data visualization and analyses. Additional details are described in SI Appendix, SI Materials and Methods.
Data Availability.
The cryo-EM maps and atomic coordinates reported in this paper have been deposited in the Electron Microscopy Databank (EMDB) and Protein Data Bank (PDB), respectively, under the following ID codes: EMD-20986 and PDB 6V0B for unliganded ELIC in POPC-only nanodiscs, and EMD-20968 and PDB 6V03 for the ELIC–propylammonium complex in POPC-only nanodiscs.
Supplementary Material
Acknowledgments
Part of this work was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247) and the US NIH/National Institute of General Medical Sciences (GM103310), with additional support from Agouron Institute (F00316) and NIH (OD019994). Molecular simulations were performed using supercomputers from Blue Waters (ACI-1713784) (to E.T.) and Extreme Science and Engineering Discovery Environment (XSEDE) (TG-MCA06N060) (to E.T.). Blue Waters is supported by the National Science Foundation (OCI-0725070 and ACI-1238993) and the State of Illinois. XSEDE is supported by the National Science Foundation (ACI-1548562). This work was also supported by NIH Grants P41-GM104601, U54-GM087519, and R01-GM123455 (to E.T.) and R01-NS042169 (to C.G.), and a Richard and Margaret Romano Professorial Scholarship (to C.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The cryoelectron microscopy maps and atomic coordinates reported in this paper have been deposited in the Electron Microscopy Databank, https://www.ebi.ac.uk/pdbe/emdb/ (ID codes: EMD-20986 for unliganded ELIC in POPC-only nanodiscs, and EMD-20968 for the ELIC-propylammonium complex in POPC-only nanodiscs) and Protein Data Bank, https://www.wwpdb.org/ (ID codes 6V0B for unliganded ELIC in POPC-only nanodiscs and 6V03 for the ELIC-propylammonium complex in POPC-only nanodiscs).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906823117/-/DCSupplemental.
References
- 1.Franklin G. I., Potter L. T., Studies of the binding of α-bungarotoxin to membrane-bound and detergent-dispersed acetylcholine receptors from Torpedo electric tissue. FEBS Lett. 28, 101–106 (1972). [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. B., Weber M., Changeux J.-P., Effects of local anesthetics and calcium on the interaction of cholinergic ligands with the nicotinic receptor protein from Torpedo marmorata. Mol. Pharmacol. 10, 904–932 (1974). [PubMed] [Google Scholar]
- 3.Chang H. W., Bock E., Structural stabilization of isolated acetylcholine receptor: Specific interaction with phospholipids. Biochemistry 18, 172–179 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Heidmann T., Sobel A., Changeux J.-P., Conservation of the kinetic and allosteric properties of the acetylcholine receptor in its Na cholate soluble 9 S form: Effect of lipids. Biochem. Biophys. Res. Commun. 93, 127–133 (1980). [DOI] [PubMed] [Google Scholar]
- 5.Criado M., Eibl H., Barrantes F. J., Functional properties of the acetylcholine receptor incorporated in model lipid membranes. Differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J. Biol. Chem. 259, 9188–9198 (1984). [PubMed] [Google Scholar]
- 6.Fong T. M., McNamee M. G., Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry 25, 830–840 (1986). [DOI] [PubMed] [Google Scholar]
- 7.daCosta C. J. B., Baenziger J. E., A lipid-dependent uncoupled conformation of the acetylcholine receptor. J. Biol. Chem. 284, 17819–17825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.daCosta C. J. B., et al. , Anionic lipids allosterically modulate multiple nicotinic acetylcholine receptor conformational equilibria. J. Biol. Chem. 284, 33841–33849 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalziel A. W., Rollins E. S., McNamee M. G., The effect of cholesterol on agonist-induced flux in reconstituted acetylcholine receptor vesicles. FEBS Lett. 122, 193–196 (1980). [DOI] [PubMed] [Google Scholar]
- 10.Kilian P. L., et al. , Reconstitution of acetylcholine receptor from Torpedo californica with highly purified phospholipids: Effect of α-tocopherol, phylloquinone, and other terpenoid quinones. Biochem. Biophys. Res. Commun. 93, 409–414 (1980). [DOI] [PubMed] [Google Scholar]
- 11.Lindstrom J., et al. , Purification of acetylcholine receptors, reconstitution into lipid vesicles, and study of agonist-induced cation channel regulation. J. Biol. Chem. 255, 8340–8350 (1980). [PubMed] [Google Scholar]
- 12.Quesada O., et al. , Uncovering the lipidic basis for the preparation of functional nicotinic acetylcholine receptor detergent complexes for structural studies. Sci. Rep. 6, 32766 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson M. B., Perfection of a synaptic receptor: Kinetics and energetics of the acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 86, 2199–2203 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosman C., Auerbach A., The dissociation of acetylcholine from open nicotinic receptor channels. Proc. Natl. Acad. Sci. U.S.A. 98, 14102–14107 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasneem A., Iyer L. M., Jakobsson E., Aravind L., Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 6, R4 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilf R. J., Dutzler R., X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Carswell C. L., Sun J., Baenziger J. E., Intramembrane aromatic interactions influence the lipid sensitivities of pentameric ligand-gated ion channels. J. Biol. Chem. 290, 2496–2507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heginbotham L., Kolmakova-Partensky L., Miller C., Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111, 741–749 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valiyaveetil F. I., Zhou Y., MacKinnon R., Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 41, 10771–10777 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Marius P., et al. , Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys. J. 94, 1689–1698 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Avanzo N., et al. , Differential lipid dependence of the function of bacterial sodium channels. PLoS One 8, e61216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng W. W. L., D’Avanzo N., Doyle D. A., Nichols C. G., Dual-mode phospholipid regulation of human inward rectifying potassium channels. Biophys. J. 100, 620–628 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin G. R., McCormick J. I., Dhe-Paganon S., Silvius J. R., Johnstone R. M., Role of specific acidic lipids on the reconstitution of Na+-dependent amino acid transport in proteoliposomes derived from Ehrlich cell plasma membranes. Biochemistry 29, 4575–4581 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Yoshida K., et al. , Phospholipid membrane fluidity alters ligand binding activity of a G protein-coupled receptor by shifting the conformational equilibrium. Biochemistry 58, 504–508 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Winterstein L.-M., et al. , Reconstitution and functional characterization of ion channels from nanodiscs in lipid bilayers. J. Gen. Physiol. 150, 637–646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Gutierrez G., Grosman C., The atypical cation-conduction and gating properties of ELIC underscore the marked functional versatility of the pentameric ligand-gated ion-channel fold. J. Gen. Physiol. 146, 15–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., et al. , High throughput lipidomic and transcriptomic analysis to compare SP2/0, CHO and HEK-293 mammalian cell lines. Anal. Chem. 89, 1477–1485 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Grosman C., Zhou M., Auerbach A., Mapping the conformational wave of acetylcholine receptor channel gating. Nature 403, 773–776 (2000). [DOI] [PubMed] [Google Scholar]
- 29.McLean M. A., Gregory M. C., Sligar S. G., Nanodiscs: A controlled bilayer surface for the study of membrane proteins. Annu. Rev. Biophys. 47, 107–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papke D., Grosman C., The role of intracellular linkers in gating and desensitization of human pentameric ligand-gated ion channels. J. Neurosci. 34, 7238–7252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann I., Dutzler R., Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS Biol. 9, e1001101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harauz G., van Heel M., Exact filters for general geometry three dimensional reconstruction. Optik (Stuttg.) 73, 146–156 (1986). [Google Scholar]
- 33.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Malhotra A., et al. , Escherichia coli 70 S ribosome at 15 A resolution by cryo-electron microscopy: Localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280, 103–116 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Saxton W. O., Baumeister W., The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127, 127–138 (1982). [DOI] [PubMed] [Google Scholar]
- 36.Vilas J. L., et al. , MonoRes: Automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Spurny R., et al. , Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. U.S.A. 109, E3028–E3034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauguet L., et al. , Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. U.S.A. 111, 966–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauguet L., et al. , Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. EMBO J. 32, 728–741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Althoff T., Hibbs R. E., Banerjee S., Gouaux E., X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hibbs R. E., Gouaux E., Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basak S., Gicheru Y., Rao S., Sansom M. S. P., Chakrapani S., Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polovinkin L., et al. , Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Rempe S. B., Pratt L. R., The hydration number of Na+ in liquid water. Fluid Phase Equilibr. 183–184, 121–132 (2008). [Google Scholar]
- 47.Whitfield T. W., et al. , A theoretical study of aqueous solvation of K+ comparing ab initio, polarizable, and fixed-charge models. J. Chem. Theory Comput. 3, 2068–2082 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallivan J. P., Dougherty D. A., Cation-π interactions in structural biology. Proc. Natl. Acad. Sci. U.S.A. 96, 9459–9464 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C., Dynamics of ligand binding to myoglobin. Biochemistry 14, 5355–5373 (1975). [DOI] [PubMed] [Google Scholar]
- 50.Brunori M., Gibson Q. H., Cavities and packing defects in the structural dynamics of myoglobin. EMBO Rep. 2, 674–679 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silkstone G., Jasaitis A., Vos M. H., Wilson M. T., Geminate carbon monoxide rebinding to a c-type haem. Dalton Trans. 21, 3489–3494 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Andrew C. R., et al. , The dynamics behind the affinity: Controlling heme-gas affinity via geminate recombination and heme propionate conformation in the NO carrier cytochrome c’. ACS Chem. Biol. 11, 3191–3201 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Nayak T. K., Vij R., Bruhova I., Shandilya J., Auerbach A., Efficiency measures the conversion of agonist binding energy into receptor conformational change. J. Gen. Physiol. 151, 465–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elenes S., Auerbach A., Desensitization of diliganded mouse muscle nicotinic acetylcholine receptor channels. J. Physiol. 541, 367–383 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suchyna T. M., Markin V. S., Sachs F., Biophysics and structure of the patch and the gigaseal. Biophys. J. 97, 738–747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hénault C. M., et al. , A lipid site shapes the agonist response of a pentameric ligand-gated ion channel. Nat. Chem. Biol. 15, 1156–1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Gutierrez G., et al. , Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc. Natl. Acad. Sci. U.S.A. 109, 6331–6336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie T. K., et al. , Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keller B. U., Hedrich R., Vaz W. L. C., Criado M., Single channel recordings of reconstituted ion channel proteins: An improved technique. Pflugers Arch. 411, 94–100 (1988). [DOI] [PubMed] [Google Scholar]
- 60.Cuello L. G., Cortes D. M., Perozo E., The gating cycle of a K+ channel at atomic resolution. eLife 6, e28032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Razinkov I., et al. , A new method for vitrifying samples for cryoEM. J. Struct. Biol. 195, 190–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine P. V., et al. , New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trabuco L. G., Villa E., Mitra K., Frank J., Schulten K., Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singharoy A., et al. , Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. eLife 5, 61–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., et al. , Constructing atomic structural models into cryo-EM densities using molecular dynamics—Pros and cons. J. Struct. Biol. 204, 319–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klauda J. B., et al. , Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang J., et al. , CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps and atomic coordinates reported in this paper have been deposited in the Electron Microscopy Databank (EMDB) and Protein Data Bank (PDB), respectively, under the following ID codes: EMD-20986 and PDB 6V0B for unliganded ELIC in POPC-only nanodiscs, and EMD-20968 and PDB 6V03 for the ELIC–propylammonium complex in POPC-only nanodiscs.