Significance
Biomass decomposition by microorganisms relies on a battery of enzymes, of which lytic polysaccharide monooxygenases (LPMOs) constitute a central player. LPMOs are notably able to disrupt the abundant recalcitrant carbohydrates cellulose and chitin and thus increase the overall rate of their degradation. It has recently been demonstrated that LPMOs prefer hydrogen peroxide over molecular oxygen to oxidize and cleave their substrates. Here, we explain through molecular simulations and experiments how a chitin-active LPMO activates hydrogen peroxide and catalyzes substrate hydrogen abstraction. Thereby, we reveal the molecular details of this unusual bioinorganic chemistry, which is of high relevance for the understanding of pathogenesis-defense mechanisms and biomass turnover.
Keywords: LPMO, peroxygenase, biomass, chitin, QM/MM
Abstract
Lytic polysaccharide monooxygenases (LPMOs) are a recently discovered class of monocopper enzymes broadly distributed across the tree of life. Recent reports indicate that LPMOs can use H2O2 as an oxidant and thus carry out a novel type of peroxygenase reaction involving unprecedented copper chemistry. Here, we present a combined computational and experimental analysis of the H2O2-mediated reaction mechanism. In silico studies, based on a model of the enzyme in complex with a crystalline substrate, suggest that a network of hydrogen bonds, involving both the enzyme and the substrate, brings H2O2 into a strained reactive conformation and guides a derived hydroxyl radical toward formation of a copper–oxyl intermediate. The initial cleavage of H2O2 and subsequent hydrogen atom abstraction from chitin by the copper–oxyl intermediate are the main energy barriers. Stopped-flow fluorimetry experiments demonstrated that the priming reduction of LPMO–Cu(II) to LPMO–Cu(I) is a fast process compared to the reoxidation reactions. Using conditions resulting in single oxidative events, we found that reoxidation of LPMO–Cu(I) is 2,000-fold faster with H2O2 than with O2, the latter being several orders of magnitude slower than rates reported for other monooxygenases. The presence of substrate accelerated reoxidation by H2O2, whereas reoxidation by O2 became slower, supporting the peroxygenase paradigm. These insights into the peroxygenase nature of LPMOs will aid in the development and application of enzymatic and synthetic copper catalysts and contribute to a further understanding of the roles of LPMOs in nature, varying from biomass conversion to chitinolytic pathogenesis-defense mechanisms.
The landscape of bioinorganic chemistry has recently been enriched by the discovery that oxidative monocopper enzymes named lytic polysaccharide monooxygenases (LPMOs) can use H2O2 as a cosubstrate (1), whereas O2 has long been invoked in all other established paradigms of copper-based enzymatic catalysis (2). Another unique feature of LPMOs is their ability to catalyze the oxidative cleavage of glycosidic chains found in complex, recalcitrant environments such as the crystalline lattices formed by chitin (3), cellulose (4–6), or copolymeric polysaccharide structures (7, 8). LPMO action disrupts the surface of recalcitrant polysaccharides (9, 10), in turn enhancing depolymerization by canonical glycoside hydrolases (3, 11–14). LPMOs are today classified as auxiliary activities (AA) in the carbohydrate-active enzyme database (15), in families AA9 to 11 and AA13 to 16. LPMOs are abundant in nature, notably in wood-decaying fungi (16, 17), and are key elements of current commercial enzymatic mixtures for the processing of lignocellulosic biomass (18–20). Recent developments indicate that chitin-active LPMOs may play roles beyond biomass decomposition, such as acting as virulence factors in bacterial infections (21–24). On a similar note, a 2016 study showed that a “chitin-binding protein” (Tma12) is expressed by ferns as an insecticidal agent against whitefly (25). Importantly, although not recognized as such in this study, Tma12 looks like typical, active LPMOs (SI Appendix, Fig. S1).
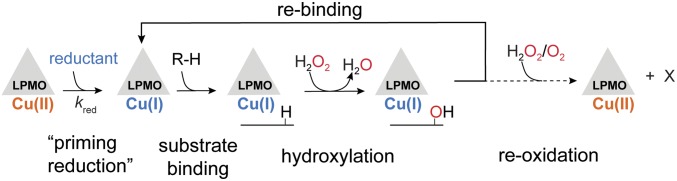
Despite substantial progress within the last decade, the mode of action of these monocopper enzymes is not yet fully mechanistically characterized (26–29). It is clear that LPMO action needs a reductant and that various small-molecule reducing compounds, including commonly used ascorbic acid (AscA), as well as redox enzymes such as cellobiose dehydrogenase, can reduce LPMOs (17). When acting on a carbohydrate substrate (R-H), and in the presence of excess reductant, a single oxygen atom derived from O2 is introduced into the final product (3, 30), explaining why LPMOs have been widely recognized as monooxygenases. However, the monooxygenase reaction (R-H + O2 + 2e− + 2H+ → R-OH + H2O) requires a second electron [the first one being stored as Cu(I)] and protons during catalysis. These catalytic events are highly puzzling considering 1) the monocopper, organic cofactor-free nature of LPMOs and 2) that access of reductants to the active site is limited when bound to a polysaccharide substrate. The finding that LPMOs efficiently incorporate oxygen atoms from H2O2 in the reaction product (as shown by, e.g., experiments with 18O-labeled H2O2) (1), thus making LPMOs a new kind of copper-dependent peroxygenase (R-H + H2O2 → R-OH + H2O; Fig. 1) (1, 31, 32), provides a straightforward answer to the “second electron conundrum,” since H2O2 brings the oxygen, electron, and proton equivalents necessary for a complete catalytic cycle.
Fig. 1.
Simplified reaction scheme of SmAA10A-catalyzed peroxygenation of chitin. Substrate binding is likely preceded by reduction of the ground-state LPMO–Cu(II) to LPMO–Cu(I), which binds with greater affinity to substrate (41–43). Binding of the polymeric substrate leads to a highly confined active site (40) that is ready to react swiftly and precisely with H2O2, resulting in hydroxylation of the substrate and release of a water molecule (1). The hydroxylated product (either at the C1 or C4 carbon) is unstable and undergoes an elimination reaction, inducing glycosidic bond cleavage (30, 36). At the end of the catalytic cycle, the Cu(I) state is conserved, and the enzyme could potentially enter a new catalytic cycle or be oxidized via nonproductive reactions with oxidants. X denotes the reaction products (e.g., hydroxyl radicals, superoxide) originating from such oxidation reactions, which, notably, may lead to enzyme damage.
The proposed peroxygenase chemistry of these copper enzymes sparks interest in unraveling the underlying mechanisms. Although previous studies attempting to decipher the molecular details of the LPMO mechanism were carried out in a monooxygenase context, some conclusions drawn at that time are likely still valid but deserve to be reinvestigated. Notably, a quantum mechanical (QM) study (33), followed by several other computational studies (34, 35), suggested that the most likely species responsible for hydrogen atom abstraction (HAA) is a copper(II)–oxyl ([CuO]+) species. A similar downstream intermediate has been proposed for the H2O2 reaction mechanism (1), where [CuO]+ is obtained after reaction of H2O2 with the LPMO–Cu(I) state, concomitant with H2O release. Wang et al. (36) recently carried out a computational assessment of the peroxygenase reaction using the crystal structure of a C4-oxidizing AA9 in complex with a water-soluble cellotriose molecule (37), showing that the peroxygenase mechanism is plausible and likely involves formation of a [CuO]+ species. However, there are no supporting biochemical data on H2O2 reactivity for that AA9. Experimental steady-state kinetic data for H2O2-driven substrate oxidation are only available for a C1-oxidizing chitin-specific LPMO from the soil bacterium Serratia marcescens (SmAA10A, also known as CBP21) (38). SmAA10A has a catalytic constant (kcat) of 6.7 s−1 and an apparent Michaelis constant (Km) for H2O2 in the low micromolar range (ca. 3 µM), resulting in a kcat/Km ≈ 106 M−1⋅s−1. These values are similar to those reported for well-characterized heme-dependent peroxygenases (39).
Building on an experimentally supported molecular model of SmAA10A in complex with crystalline chitin (40), we aimed to decipher the molecular details of the newly proposed H2O2 reaction mechanism of LPMOs. We employed multiple computational methods to find minimum energy paths and to calculate all energy barriers associated with the reaction coordinate. As experimental support, we used pre-steady-state kinetic methods to examine SmAA10A reduction and single catalytic event reactions of SmAA10A, isolated in the Cu(I) form, with both H2O2 and O2, in the presence and absence of substrate. Calculations suggested a key role in catalysis for Glu60, a highly conserved second-shell residue, and the role of this residue was interrogated experimentally by site-directed mutagenesis. Collectively, these results provide a strong foundation for establishing chitinolytic LPMOs as unusual biological peroxygenases achieving substrate-assisted catalysis.
Methods
A complete description of the experimental details is available in SI Appendix, including details on materials, site-directed mutagenesis, production and purification of recombinant LPMOs, LPMO activity and binding assays, (stopped-flow) fluorimetry experiments, and computational studies. Notably, derived force-field parameters for H2O2 are presented in SI Appendix, Table S1 and primers used for site-directed mutagenesis are given in SI Appendix, Table S2. The data generated during the current study are available from the corresponding authors on request.
Results
H2O2 Is the Cosubstrate Sustaining SmAA10A Catalysis under Steady-State Turnover Conditions.
Steady-state kinetic analyses have shown that H2O2 is an efficient cosubstrate in SmAA10A-catalyzed oxidation of chitin (38). However, the question remains whether SmAA10A catalysis also relies on H2O2 in conditions where H2O2 is not added, that is, just in the presence of ambient O2 concentrations and excess reductant. The latter conditions are standard practice in the LPMOs field. We have argued previously that these conditions promote the in situ formation of H2O2 and that, therefore, most reported LPMO rates likely reflect a H2O2-limited peroxygenase activity rather than a monooxygenase activity (1, 29).
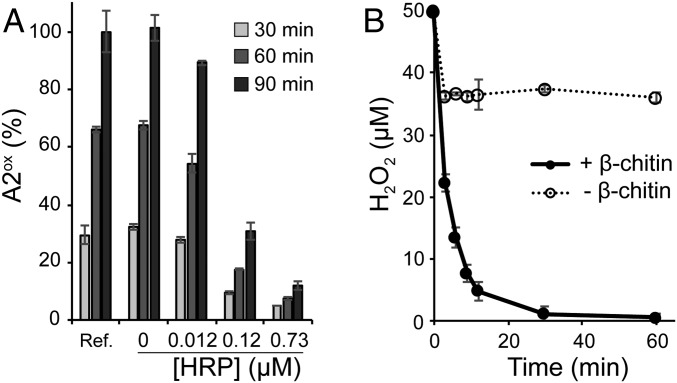
A competition experiment in the presence of a peroxidase (Fig. 2A) showed that, in the presence of ambient O2 (no added H2O2) and 1 mM reductant (AscA), 1 μM SmAA10A can be almost totally inhibited by the H2O2-scavenging activity of horseradish peroxidase (HRP), suggesting that H2O2 is produced in situ and is the main (if not only) oxidant used by SmAA10A. We then determined reaction conditions that allowed reliable monitoring of H2O2 consumption during catalysis (SI Appendix, Figs. S2 and S3). Fig. 2B shows that in reactions with added H2O2 (and substoichiometric amounts of added reductant) rapid consumption of the latter only occurs if substrate is present, indicative of a clear relationship between H2O2 consumption and substrate degradation (also discussed below).
Fig. 2.
Assessing the underlying role of H2O2 in SmAA10A-catalyzed oxidation of chitin. (A) The graph shows time courses for release of soluble oxidized products (chitobionic acid; A2ox) from β-chitin (10 mg·mL−1) by SmAA10A (1 µM) in the presence of AscA (1 mM), O2 (ca. 200 µM at atmospheric pressure), different amounts of HRP, and AmplexRed (200 µM, a cosubstrate of HRP). Quantities are expressed as a percentage of the amount of A2ox detected in the reference reaction (Ref., i.e., without HRP and AmplexRed) after 90-min reaction. (B) Time course for consumption of H2O2 (50 µM at t0) by SmAA10A (50 nM) in a reaction containing AscA (20 µM) and ethylenediaminetetraacetic acid (50 µM, needed to limit nonenzymatic background reactions; see SI Appendix, Fig. S2 for details on the experimental conditions) in the presence and absence of β-chitin (10 mg·mL−1). Note that the fast initial consumption of H2O2 in the reaction without chitin is a background reaction that is not due to LPMO activity (see SI Appendix, Fig. S3 for more details). All reactions were carried out in sodium phosphate buffer (50 mM, pH 7.0) at 40 °C in a thermomixer (1,000 rpm). Error bars show ± SD (n = 3 independent experiments).
QM/Molecular Mechanics Study of the Chitinolytic Peroxygenase Reaction.
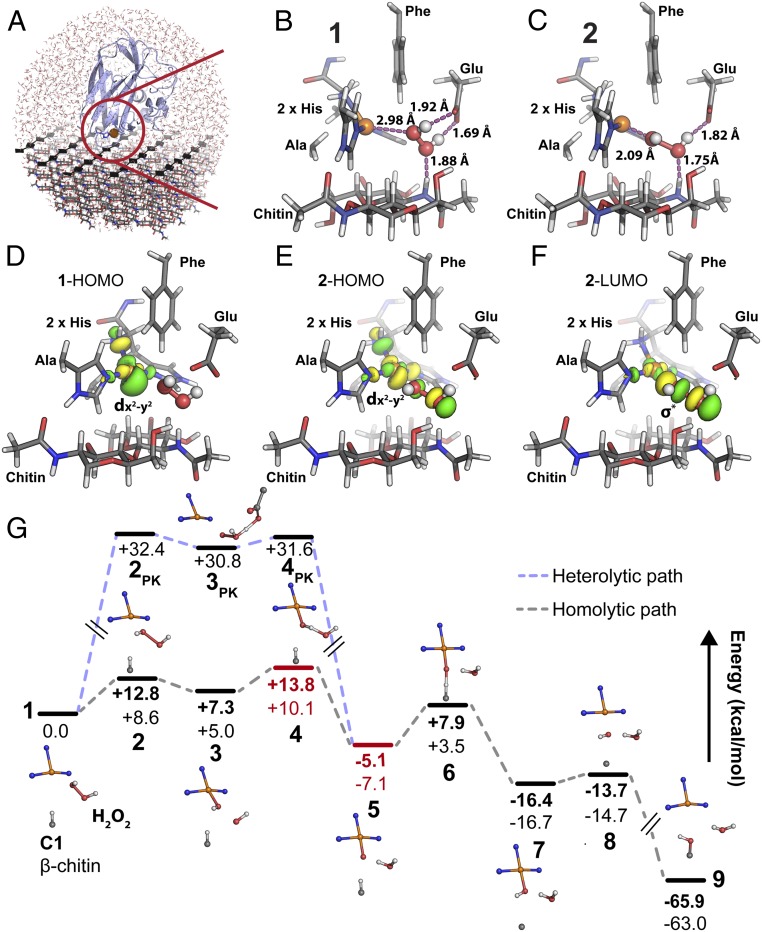
Intriguingly, neither strict C1-oxidizing LPMOs nor crystalline polysaccharide substrates have been the subject of QM/molecular mechanics (MM)-assisted investigations of the LPMO mechanism (33–36, 41, 42). Also, in the few available reports (36, 42), the initial steps of H2O2 access, positioning, and one-electron reduction by Cu(I) at the enzyme active site have not been agreed upon (discussed below). Starting from a previously published biochemically and spectroscopically supported model of SmAA10A in complex with crystalline β-chitin (Fig. 3A and SI Appendix, Fig. S4) (40), we performed QM/MM/molecular dynamics (MD) and QM/MM calculations to unravel the molecular details underlying the peroxygenase reaction (R-H + H2O2 → R-OH + H2O) catalyzed by SmAA10A–Cu(I).
Fig. 3.
QM/MM study of the peroxygenase mechanism of SmAA10A on crystalline β-chitin. (A) The QM/MM model containing ∼24,000 atoms derived from experimentally supported models of SmAA10A in complex with chitin (40). See SI Appendix, Fig. S5 for a definition of the QM region. (B) In the initial state of the reaction (1), the H2O2 molecule is aligned toward the Cu(I) ion in a strained conformation involving hydrogen bonding with both substrate and amino acid side chains. (C) The first transition state (2) displays an elongated H2O2 O–O bond and the reactive hydroxyl radical to be formed is stabilized by H bonds from Glu60 and a substrate amide H atom. (D–F) The electron transfer from Cu(I) to H2O2 can be visualized by plotting the quasi-restricted orbitals of 1 and 2. Before reaction with H2O2, the state 1 HOMO (D) exhibits delocalized dx2-y2 character, indicating a Cu(I) state ready to react with H2O2. The state 2 (2-HOMO to 2-LUMO transition); E and F indicates that the Cu(I) electron moves from the delocalized dx2-y2 like orbital to a σ* antibonding orbital localized between the H2O2 O atoms, promoting bond cleavage. (G) Complete reaction path where states 1 through 9 indicate energy minima and transition states (relative energies in kilocalories per mole). Between states 3 and 6, spin cross-overs occur, and the triplet states are shown in red and the open-shell singlet states are indicated in black. The energy minima and transition states for activation of H2O2 via a PK-like heterolytic cleavage (2PK–4PK) are indicated by the light blue path. See SI Appendix, Fig. S6 for the geometries of each state. Energies in bold type were obtained by the B3LYP functional, while energies in normal type were obtained by TPSSh. All carbons in the protein residues and chitin (only a dimer is shown in B–F) are gray. H2O2 is shown as white and red sticks and balls. For the sake of clarity, the MM region is not shown.
QM/MM/MD simulations (SI Appendix, Fig. S5 and Movie S1), followed by nudged elastic band (NEB) minimum energy path calculations, resulted in starting models for calculating the energies of the different states (1 through 9) of the SmAA10A-catalyzed reaction (Fig. 3G). A global observation is that the overall reaction is thermodynamically favorable, with a relative energy difference of −65.9 kcal⋅mol−1 between the initial and final states. State 1 represents the SmAA10A–Cu(I) bound to β-chitin substrate with a H2O2 molecule confined in the reaction cavity and no water molecules coordinated to the reduced copper ion. Hydrogen bonds from the Glu60 side chain and a substrate amide hydrogen align the H2O2 molecule toward the Cu(I) ion at a distance of 2.98 Å (Fig. 3B). The H2O2 O–O bond length of 1 is 1.44 Å, which is equal to the value obtained by geometry optimization of H2O2 in vacuum (1.44 Å). Interestingly, the dihedral bond angle of H2O2 in 1 is 53°, far from the equilibrium angle [113.6° in gaseous state (43), 90.2° in solid state (44)], indicating that the protein and substrate introduce a strained conformation (+4.1 kcal⋅mol−1) of the peroxide moiety.
The SmAA10A catalytic cycle is triggered by electron transfer from Cu(I) to H2O2, and the transition state of this step is represented by 2 (Fig. 3C). The H2O2 bond is now elongated (1.76 Å), the dihedral bond angle has relaxed to 94°, and the proximal peroxide O atom is only 2.1 Å away from the copper ion. The energy barrier of this step is estimated to be 12.8 kcal⋅mol−1 and 8.6 kcal⋅mol−1 by the density functional theory (DFT) functionals B3LYP (45) and TPSSh (46), respectively. State 2 is best described as an open-shell singlet, a conclusion that could only be reached after detailed analysis of the electronic structure (SI Appendix, Supplementary Results). The latter analysis allowed us to plot the quasi-restricted orbitals that best describe the transition from state 1 to state 2 in Fig. 3 D–F. The highest occupied molecular orbital (HOMO) of state 1 has mainly Cu-dx2-y2 and ligand σ* antibonding character (Fig. 3D, 1-HOMO), and the largest lobe points directly toward the H2O2 molecule. Reaching the transition state geometry (state 2), the HOMO orbital becomes slightly delocalized to include the H2O2 molecule (Fig. 3E, 2-HOMO). According to frontier molecular orbital theory, the lowest unoccupied molecular orbital (LUMO) indicates where the Cu(I) electron will go. Here, the state 2 LUMO displays a decreased density at the copper d-orbital, while a σ* antibonding orbital appears between the H2O2 O atoms (Fig. 3F, 2-LUMO), indicative of homolytic cleavage of the O–O bond.
The first local minimum in the reaction path is state 3 (Fig. 3G and SI Appendix, Fig. S6A), where the calculations suggest a Cu-bound hydroxide with an adjacent hydroxyl radical. This state collapses to an open-shell singlet solution due to strong antiferromagnetic coupling between Cu(II) and the hydroxyl radical (see SI Appendix, Supplementary Results for more details). Direct substrate H-atom abstraction by this H2O2-derived hydroxyl radical was investigated by QM/MM NEB calculations but was not feasible due to steric constraints in the reaction cavity of our model caused by the copper-bound hydroxyl. Hydrogen bonds from both the substrate and the protein appear to confine and orient the reactive hydroxyl radical in the reaction cavity, yielding a “precision-guided HO•” poised for HAA from the Cu-bound hydroxide with a barrier of 6.5 kcal⋅mol−1 (B3LYP; state 4, SI Appendix, Fig. S6A). Of note, Glu60 seems to play a crucial role in positioning the hydroxyl radical (Fig. 3C). At this point, the total spin of the system has changed from a singlet to a triplet state, the triplet state being favored by 0.8 kcal/mol for B3LYP and 0.5 kcal/mol for TPSSh, thus displaying spin cross-over behavior. The resulting state 5 (Fig. 3G and SI Appendix, Fig. S6A) is best described as a triplet copper–oxyl [CuO]+ species, the triplet state being favored by 3.5 kcal/mol for B3LYP and 3.1 kcal/mol for TPSSh, where the oxyl O atom, which is only 2.08 Å away from the substrate H atom that is to be abstracted, is hydrogen-bonded to the water molecule formed in the previous reaction step. In this highly reactive state, a Löwdin spin population analysis reveals Cu and oxyl spins of 0.61 and 1.1, respectively (see SI Appendix, Table S3 for all values along the reaction path), indicative of a reactive radical on the oxygen atom.
The energy barrier of the substrate H-abstraction step (state 6, Fig. 3G and SI Appendix, Fig. S6A) is estimated to be 13.0 and 10.6 kcal⋅mol−1 by the B3LYP and TPSSh functionals, respectively. At this point of the reaction the system has changed back to the singlet energy surface, indicating that the spin cross-over transitions take place between states 3 and 6 in the reaction path. The hydroxide moiety formed by the H-abstraction step partly recombines with the copper ion (Cu–O distance of 1.91 Å) in a local energy minimum (state 7, Fig. 3G and SI Appendix, Fig. S6A). Finally, the Cu-interacting hydroxide passes through a low-barrier (+2.6 kcal⋅mol−1 for B3LYP) rebound-intermediate (state 8, Fig. 3G and SI Appendix, Fig. S6A) and recombines with the substrate radical to form the final hydroxylated product (state 9, Fig. 3G and SI Appendix, Fig. S6A). Note that the Cu(I) form of the enzyme has been regenerated at the end of the reaction cycle. Importantly, it appears that along the reaction path hydrogen bonds involving the enzyme Glu60 side chain and a strategically positioned substrate amide hydrogen, which could be analogous to the C2-OH in cellulose, control the positioning of several H2O2-derived reactive oxygen species (SI Appendix, Fig. S6A), facilitating the LPMO reaction. In summary, our calculations indicate that the overall reaction pathway contains two main energy barriers, corresponding to the H2O2 cleavage and H abstraction from chitin, and that spin cross-over events and a dynamic hydrogen bond network play crucial roles in this catalysis.
Heme-iron peroxidases are known to catalyze the heterolytic cleavage of H2O2 and the reaction path has been described by the Poulos–Kraut (PK) mechanism (47, 48). Here, we considered a similar mechanism as an alternative path to generate the copper–oxyl species (state 5). Since experimental data clearly show that the Cu(II) state does not react with H2O2, heterolytic cleavage was considered for the Cu(I) state. Assuming that the Glu60 side chain could act as a base, we modeled the reaction Cu(I)–O(H)–OH (state 1) → Cu(I)–O–OH2 → Cu(II)–O• + OH2 (state 5) using the same computational methodology as for the homolytic cleavage reaction path described above. The migration of the proximal hydrogen to the distal oxygen of the H2O2 was assisted by the Glu60 side chain, and a stable Cu(I)–O–OH2 intermediate with a metal–oxygen distance of 2.94 Å (state 3PK) was found to have an energy of 30.8 kcal⋅mol−1 (TPSSh) higher than state 1 (Fig. 3G and SI Appendix, Fig. S6B). The energy barrier of the formation of this intermediate, represented by state 2PK, was calculated to be 32.4 kcal⋅mol−1 relative to state 1, while the further heterolytic cleavage of the O–O bond of Cu(I)–O–OH2 to water and the copper–oxyl intermediate had an energy barrier of 0.8 kcal⋅mol−1 (shown as state 4PK) relative to state 3PK. Thus, our results indicate that the heterolytic path of H2O2 cleavage is unfavorable compared to the homolytic path, displaying initial energy barriers of 32.4 and 8.6 kcal⋅mol−1 (TPSSh), respectively.
In a comparative manner, we investigated the energetics of substrate hydrogen abstraction using O2 as cosubstrate, exchanging H2O2 with O2 but otherwise using the same starting model. For the latter reasons, we only evaluated a scenario in which the LPMO–Cu(I) and O2 react without requiring further delivery of electrons and protons, the origin of which would have been purely arbitrary (SI Appendix, Fig. S7). The main conclusions are that substrate binding alters the orientation of the copper-bound superoxide and that H abstraction by a copper superoxide intermediate is thermodynamically plausible but kinetically very unlikely (energy barrier of 35 kcal⋅mol−1) (SI Appendix, Supplementary Results).
The Reductive Priming Reaction Is Fast and Monophasic.
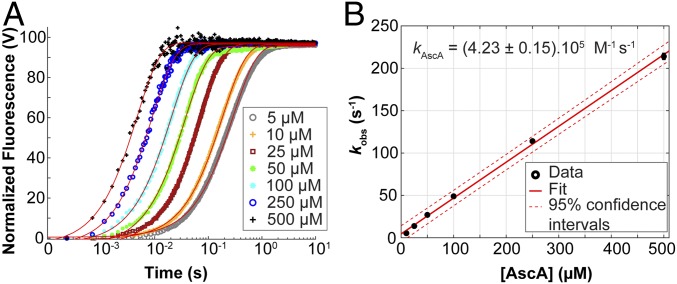
To gain insight into a potential rate-limiting role of the priming reduction, that is, the one-electron reduction of the resting state LPMO–Cu(II) to the active LPMO–Cu(I) form (1, 49), we investigated how fast such reductive reaction occurs and which amount of reductant is required to achieve complete LPMO reduction. We have previously shown (50) that the fluorescence signal of SmAA10A can be used as a reporter of its redox state (SI Appendix, Fig. S8), the Cu(II) and Cu(I) forms having respectively low and high fluorescence responses, both linear within the concentration range studied (SI Appendix, Fig. S9). Following rapid stopped-flow mixing of SmAA10A–Cu(II) with 1 eq. AscA (5 μM each) under anaerobic conditions, a monophasic increase in fluorescence was observed. The experiment was subsequently repeated with AscA present in pseudo-first-order excess (≥10 eq.). The total fluorescence change remained the same, suggesting that reduction was complete with as little as 1 eq. of AscA. The reduction reaction remained monophasic but increased in rate with increasing concentrations of AscA (Fig. 4A). Each fluorescence-versus-time curve could be fit with a single exponential function, indicative of a pseudo-first-order reaction with respect to AscA. The pseudo-first-order rate constant (kobs) varied linearly with [AscA], yielding a second-order rate constant kAscA = 4.2 × 105 M−1⋅s−1 (Fig. 4B). Such a high rate indicates that in most reaction conditions the initial reductive priming is not rate-limiting (see Discussion).
Fig. 4.
Kinetics of reduction of SmAA10A–Cu(II) by AscA. (A) SmAA10A-Cu(II) (final concentration, 5 μM) was anaerobically mixed with varying concentrations of AscA and the change in fluorescence was monitored as a function of time. The reactions were carried out in potassium phosphate buffer (50 mM, pH 7.1) at 25 °C. The final concentrations of AscA are indicated in the figure. Data were fit with single exponential functions to give observed rate constants (kobs) at each substrate concentration. Each experiment was performed in triplicate. For the sake of clarity, only the trace of one replicate is shown for each condition. (B) Plot of pseudo-first-order kobs as a function of AscA concentration. Error bars show ± SD (n = 3 independent experiments).
We have previously shown that hexa-N-acetyl-chitohexaose (NAG6) is cleaved by SmAA10A (40), and below we show that NAG6 can be used as a substrate to monitor single catalytic events, while also pointing at some limitations in Discussion. Studies of the rate of LPMO reduction by AscA in the presence of 1 mM NAG6 yielded a second-order rate constant of 5.4 × 105 M−1⋅s−1 (SI Appendix, Fig. S10). This value is only 1.26-fold higher than that obtained in absence of NAG6, suggesting that the presence of a soluble and flexible substrate does not strongly modulate the rate of reduction of the SmAA10A–Cu(II) by AscA.
The Reaction between SmAA10A–Cu(I) and O2 Is Exceedingly Slow.
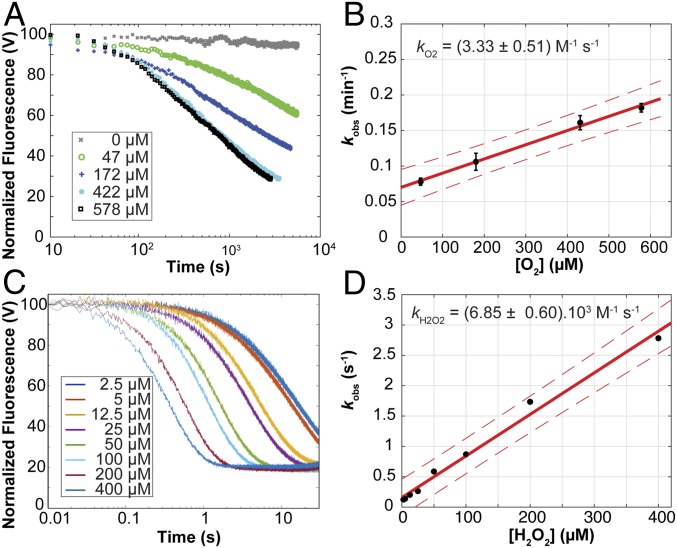
To further understand the potential competition between environmentally available H2O2 and ambient O2, we first monitored oxidation of 2 μM SmAA10A–Cu(I) (produced anaerobically by reduction of the oxidized enzyme with ascorbate, followed by removal of excess reductant) by O2 under conditions that resulted in single oxidative events (i.e., in the absence of reductant) (Fig. 5A). The change in fluorescence in the presence of 1 eq. O2 could not be discriminated from the background noise (not shown). Using pseudo-first-order concentrations of O2 ([O2] at least in a 10-fold excess relative to [SmAA10A–Cu(I)]), reoxidation of Cu(I) to a Cu(II) species was observed over a period of minutes, at a rate that was dependent on [O2] (Fig. 5B). When fitting the kinetic data, a number of models were explored (SI Appendix, Figs. S11 and S12). Although we have based the analysis of the reoxidation curves on a simple model, that is, a single exponential function, (SI Appendix, Figs. S11 and S14B), the residuals of the fits indicate that the reoxidation of SmAA10A–Cu(I) by O2 is a complex reaction. Strikingly, a complex reaction model where O2 binds and is reduced by a binuclear copper cluster formed by a SmAA10A–Cu(I) dimer results in a perfect fit (SI Appendix, Fig. S13 and Scheme S1). In such an arrangement, the SmAA10A–Cu(I) dimer binuclear active site could resemble the active sites of well-studied binuclear type-3 copper proteins such as tyrosinases and hemocyanins (2). This dimer hypothesis is currently being explored in a separate study. Nonetheless, applying the single exponential model as best simple approximation shows that the kobs varied linearly with [O2], yielding a second-order rate constant kO2 = 3.3 M−1⋅s−1 (Fig. 5B). Of note, this value shows that reoxidation of the reduced LPMO by O2 is very slow.
Fig. 5.
Reoxidation of SmAA10A–Cu(I) by (A and B) O2 or (C and D) H2O2 under single oxidative event conditions and in the absence of substrate. SmAA10A–Cu(I) (final concentration 2 μM) was anaerobically mixed with solutions containing varying concentrations of (A) O2 or (C) H2O2 (final concentrations indicated by the color code in the figure) and the changes in fluorescence associated with reoxidation of the LPMO–Cu(I) were monitored as a function of time. The reactions were carried out in sodium phosphate buffer (50 mM, pH 7.0) at 25 °C. Data were fit to single exponential functions (SI Appendix, Figs. S11, S12B, and S14) to give observed rate constants (kobs) at each cosubstrate concentration, which were plotted to obtain second-order rate constants for (B) O2 or (D) H2O2. In A and C, for the sake of clarity, only one trace for each condition is shown (experiments were performed at least in triplicate). In B and D, error bars show ± SD (n = 3 independent experiments); red solid lines show the best linear fit and the red dashed lines the 95% confidence interval.
The Reaction between SmAA10A–Cu(I) and H2O2 Is Monophasic and Faster than the Reaction with O2.
Mixing H2O2 with LPMO–Cu(I) showed decrease in fluorescence, with a stoichiometry of 1:1 (SI Appendix, Fig. S15). Moreover, spin-trap experiments (SI Appendix, Fig. S16) showed that next to Cu(II) species (as shown by the fluorescence signal) this reaction produces radicals (80% yield, when assuming that only OH• is produced). Previous studies on fluorescent transition-metal complexes indicate that the formation of an open-shell product from a closed-shell reactant (e.g., a change in total spin from 0 to 1/2) quenches fluorescence efficiently (51). Thus, we suggest that loss of the LPMO–Cu(I) fluorescence signal that we observe takes place at the first electron transfer from Cu(I) to H2O2 (state 1 to state 2 or 3 in the proposed mechanism). Therefore, we suggest that off-cycle [e.g., SmAA10A–Cu(II)] as well as on-cycle species such as Cu(II)–OH–OH• (states 3 and 4), Cu(II)–O• (state 5), or Cu(II)–OH (states 6 and 7) all quench the fluorescence signal observed with Cu(I) and participate in varying proportions to the observed fluorescence. However, it is so far not possible to discriminate these Cu(II) species by fluorescence. Monitoring reoxidation of SmAA10A–Cu(I) (2 μM) by H2O2 at varying concentrations showed changes in fluorescence consistent with complete reoxidation of the copper within seconds to milliseconds (Fig. 5C). The data can be described by single exponential functions (SI Appendix, Fig. S14), indicative of a pseudo-first-order reaction with respect to H2O2. Plots of the fitted pseudo-first-order rate constants as a function of [H2O2] could be fitted to a straight line, yielding a second-order rate constant kH2O2 = 6.9 × 103 M−1⋅s−1 (Fig. 5D). Hence, at comparable concentrations of oxidant, oxidation of the LPMO with H2O2 is three orders of magnitude faster than oxidation with O2.
We also explored the catalytic potential of SmAA10A–Cu(I) reacted with H2O2 in the absence of substrate prior to addition of NAG6 substrate. If a stable reactive enzyme–oxygen intermediate formed in the absence of substrate we would expect to observe amounts of oxidized products proportional to the intermediate lifetime when adding substrate after different incubation times with H2O2. We observed a time-dependent decay in the amount of oxidized products formed (SI Appendix, Fig. S17) that corresponded perfectly to the decay in the amounts of available reactants [i.e., SmAA10A–Cu(I) and H2O2] predicted on the basis of the second-order rate constant, kH2O2, of 6.9 × 103 M−1⋅s−1 determined above for H2O2-driven oxidation of the LPMO. Next to adding confidence to the interpretation of the stopped-flow experiments described above, this observation indicates that if a productive intermediate resulting from the reaction of substrate-free SmAA10A–Cu(I) with H2O2 would be formed then its lifetime is much shorter than the time required for a catalytic turnover in the presence of substrate. Also, we show that reacting free SmAA10A–Cu(I) with H2O2 yields only a very low loss in activity (SI Appendix, Fig. S18). This proves that under single-oxidative-event conditions we monitor reoxidation of SmAA10A–Cu(I) to SmAA10A-Cu(II) (SI Appendix, Supplementary Discussion) rather than LPMO inactivation, a process which apparently requires multiple turnovers for it to become clearly noticeable.
The Substrate Accelerates the Reaction of SmAA10A–Cu(I) with H2O2, but Not with O2.
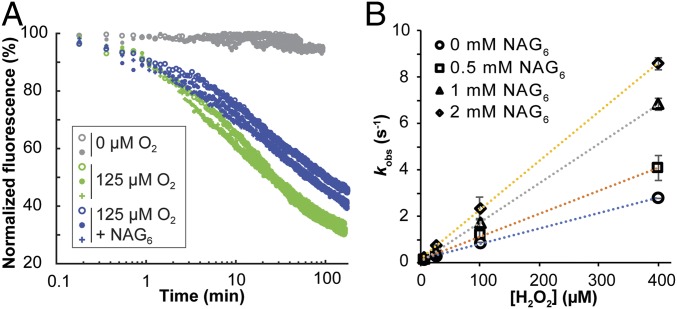
QM/MM calculations predict that several hydrogen bonds are established between H2O2, the LPMO, and the substrate during H2O2 activation (Fig. 3B), and biochemical data indicate that turnover of H2O2 is much faster in the presence of chitin (Fig. 2B). Following the procedures outlined above, we thus assessed the impact of substrate on the reoxidation event (corresponding to steps 1→3; Fig. 3G) which, on the basis of the energy profiles (Fig. 3G), is likely to determine the overall reaction rate. For these experiments, we used (NAG)6, which is a low-affinity substrate but which is compatible with fluorimetry experiments. SI Appendix, Fig. S19 shows that SmAA10A–Cu(I) supplied with H2O2 indeed is fully engaged into NAG6 oxidation. Fig. 6A shows that in the presence of 0.5 mM NAG6, reoxidation of SmAA10A–Cu(I) by O2 was slightly slower compared to the reaction without substrate. In contrast, using 0.5 mM NAG6, reoxidation of SmAA10A–Cu(I) by H2O2 was faster in the presence of substrate and displayed monophasic time-course profiles similar to experiments without substrate (SI Appendix, Fig. S20). The experiment was repeated with higher concentrations of NAG6, showing a proportional increase in reoxidation rates by H2O2, with second-order rate constants up to kH2O2 = 21 × 103 M−1⋅s−1 for the reaction with 2 mM NAG6 (Fig. 6B). A complementary spin-trapping experiment showed that the presence of NAG6 reduced the amount of detected radicals (SI Appendix, Fig. S16D). Bearing in mind that the monitored fluorescence signal likely includes on- and off-cycle Cu(II) species (discussed above), we interpret this result as follows: The observed [NAG]6-dependent increase of the global reoxidation rate (including on and off pathways) underpins an increased rate of the oxidative on-cycle reoxidation of the LPMO–Cu(I)–NAG6 complex, an interpretation in agreement with the QM/MM-based prediction that substrate binding increases reactivity with H2O2.
Fig. 6.
Reoxidation of SmAA10A–Cu(I) by (A) O2 and (B) H2O2 in the presence of NAG6. (A) Time curves for reoxidation of SmAA10A–Cu(I) (2 μM) by 125 µM O2 in the presence (0.5 mM) or absence of NAG6. All given concentrations are concentrations after mixing. The graph shows two or three experiments per condition. (B) Second-order rate plot of SmAA10A–Cu(I) reoxidation by H2O2 in the presence (0.5 to 2 mM) or absence of NAG6 (see SI Appendix, Fig. S20 for time curves obtained with 0.5 mM NAG6). Error bars in B show ± SD (n = 3 independent experiments).
It is noteworthy that the observed decrease in fluorescence signal of the LPMO–Cu(I) in the presence of NAG6 and in excess of H2O2 (SI Appendix, Fig. S20) seems to indicate that the Cu(I) form is not regenerated in these conditions. As further developed in SI Appendix, Supplementary Results, this apparent contradiction with the proposed mechanism (Fig. 1) indicates a partition between Cu(I) regenerating and nonregenerating reactions. In essence, we show that, in the presence of a flexible, soluble substrate such as NAG6, the geometric constrains and active site confinement that were ensured by the LPMO–chitin association are no longer operational, allowing the reaction to derail along the productive reaction path. Thus, the regeneration of the Cu(I) species was prevented while NAG6 oxidation was still allowed to occur (SI Appendix, Fig. S21 and Supplementary Results).
H2O2 Access to the Active Site.
Previous studies indicated the formation of a water tunnel in the enzyme–polysaccharide complex interspace, connecting bulk solvent to the monocopper active site (40). Here, we investigated the energetics associated with H2O2 diffusion into the active site by MD simulations combined with umbrella sampling, estimating free energy profiles of H2O2 moving in this interspace (SI Appendix, Supplementary Results and Figs. S22 and S23 and Movie S2). The free energy barrier associated with diffusion of H2O2 into the reaction cavity from the bulk solvent was estimated to be less than 2 kcal/mol. The simulations showed that Glu60 and Asn185 may restrict H2O2 access to the active-site cavity. While the Glu60 side chain mainly populates two conformations, pointing toward the copper ion or the bulk solvent, the Asn185 side chain is restricted by the protein backbone and always points toward the bulk solvent. Despite the small energetic cost associated with H2O2 entering the confined reaction cavity, such confinement, and, in particular, the resulting precise interaction with Glu60 (Fig. 3B), is likely important.
Glu60 Is Important for Efficient Peroxygenase Activity.
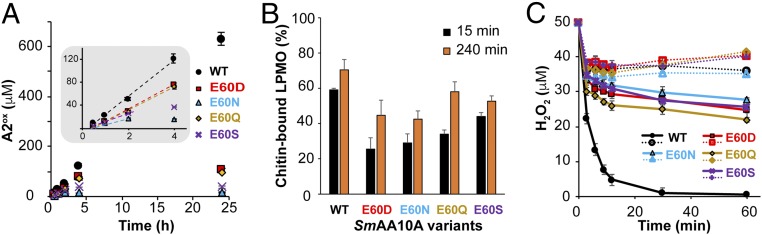
The QM/MM and MD calculations presented above suggest that Glu60 plays an important role also in positioning and constraining H2O2 and the hydroxyl radical (Fig. 3 B and C). This residue is structurally conserved throughout the entire LPMO superfamily, either as a glutamate or as a glutamine (52). In support of an important role for this residue, previous studies have shown that mutation of Glu60 in SmAA10A to alanine is detrimental for activity (53, 54). We generated additional variants of SmAA10A with mutations at position 60 (Glu → Gln, Asn, Asp, or Ser) and carried out a detailed analysis of the mutational effects. Fig. 7A shows that under standard conditions (1 mM reductant, no added H2O2) all mutations led to drastically decreased product yields after 24 h of reaction, while effects on initial enzyme rates were more moderate, in particular for the E60Q and E60D variants (Fig. 7 A, Inset). So, clearly, the mutations lead to an increased rate of enzyme inactivation. These drastic changes in enzyme performance did not correlate with changes in substrate binding, which were modest (Fig. 7B and SI Appendix, Fig. S24), but did correlate with a decreased ability to utilize externally added H2O2 in reactions with substrate (Fig. 7C). In the absence of substrate, and under single-oxidative-event conditions, the reactivity of the various mutants with H2O2 was equivalent or slightly lower than that of the wild type (WT) (SI Appendix, Fig. S25 and Table S4). Thus, in accordance with the mechanistic conclusions described above and depicted in Fig. 3, it would seem that mutation of Glu60 impairs the ability of substrate-bound LPMOs to turn over H2O2, while increasing the chance that the formation of a now less-confined hydroxyl radical leads to enzyme damage.
Fig. 7.
Biochemical characterization of SmAA10A and Glu60 mutants. (A and B) Time courses for (A) the release of soluble chitobionic acid (A2ox) from β-chitin in the presence of 1 mM AscA and (B) binding to β-chitin, 1 µM enzyme (see SI Appendix, Fig. S24 for entire time courses). (A, Inset) A zoom-in view of the first 4 h showing the initial oxidative rate (dotted lines) for each variant, amounting to 0.53, 0.34, 0.15, 0.33, and 0.26 min−1 for SmAA10A-WT, E60D, N, Q and S, respectively. (C) H2O2 consumption (50 µM at t0) by SmAA10A WT and mutants thereof (50 nM) in the presence (solid lines) or absence (dotted lines) of β-chitin. Reactions were initiated by addition of AscA (20 µM). Note that for comparative purposes the data shown in B for SmAA10A-WT are a reproduction of data displayed in Fig. 2B. All reactions were carried out in sodium phosphate buffer (50 mM, pH 7.0) with β-chitin (10 g⋅L−1) and incubated at 40 °C in a thermomixer (1,000 rpm). The color/symbol code is provided in each panel. Error bars show ± SD (n = 3 independent experiments).
Discussion
When LPMOs were identified as monocopper enzymes, it soon became apparent that they may work differently than hitherto characterized copper enzymes due to their peculiar structure and lack of additional redox-active cofactors. The special character of these enzymes became even more apparent after the discovery that they can effectively catalyze a peroxygenase reaction (1, 38). In this study, we present computational data supported by a series of experiments that shed light on the peroxygenase reaction. Importantly, while cellulose-active LPMOs have received most of the attention so far, our study proposes an atomistic-level description of the mode of action of solid-state chitin-active LPMOs whose functionalities in nature, notably in pathogenesis mechanisms (21, 24, 25, 55), only are now being revealed.
Fig. 2 shows that, like cellulose oxidation, chitin oxidation by LPMOs depends mainly on H2O2 availability in standard aerobic conditions. Two recent computational (QM/MM) studies have addressed the reaction between H2O2 and the LPMO-bound Cu(I) (36, 42), both starting with a crystal structure of the AA9 C4-oxidizer from Lentinus similis (LsAA9A) obtained in complex with cellotriose (Protein Data Bank ID code 5ACF) (37). While we suggest that the torsional strain imposed on the H2O2 molecule (dihedral angle = 53°) in SmAA10A in state 1 is important for the progress of the reaction, QM/MM models obtained by Wang et al. (36) suggest a torsional unstrained H2O2 molecule (dihedral angle = 116°) that is 2.8 Å away from the active site Cu(I), while hydrogen-bonded to a glutamine side chain (analogous to Glu60 in SmAA10A) and a water molecule. This and other differences between the various studies could be due to real variation in the second-sphere environments between AA10 and AA9s but are more likely due to conspicuous differences in the definition of the QM regions (SI Appendix, Supplementary Discussion and Fig. S26). Wang et al. (36) identified a transition state for electron transfer to H2O2 akin to state 2 presented in this work with an energy barrier of 5.8 kcal⋅mol−1 (B3LYP). This value is lower than the one calculated for the SmAA10A–chitin model [12.8 kcal⋅mol−1 (B3LYP) and 8.6 kcal⋅mol−1 (TPSSh)]. The two previous computational studies suggest substrate H-abstraction energy barriers (states 5 to 6; Fig. 3) of 5.5 kcal⋅mol−1 (B3LYP) (36) and 16.5 kcal mol−1 (TPSS) (42), whereas we obtained values of 13.0 kcal⋅mol−1 (B3LYP) and 10.6 kcal⋅mol−1 (TPSSh) for the SmAA10A–chitin model. Not only may the use of different DFT functionals explain the discrepancies observed between the 2 different LsAA9A-cellotriose models (36, 42), but it is also apparent that the cellotriose ligand has been repositioned during model preparation, resulting in starting models different from the crystal structure (SI Appendix, Fig. S26A).
While Wang et al. (36) reported, based on their QM/MM calculations, that all of the steps of the LPMO-catalyzed peroxygenase reaction take place on a singlet energy surface, Hedegård and Ryde (42) identified two intermediates with triplet ground states that resemble states 5 and 7 in Fig. 3G. Although the QM/MM/MD simulation (Movie S1) of the SmAA10A–chitin model, which provided the underpinning molecular geometries for our mechanistic approach, also progressed on a singlet surface, subsequent refinement provided more details. We found that states 4 and 5 have triplet ground states, and this was confirmed by unrestricted corresponding orbital (UCO) analysis (zero UCO overlap). Supporting our findings for state 5, a previous study, employing a higher level of theory (CASSFC/CASPT2) than DFT, showed that copper–oxyl complexes of various (not LPMO-related) model compounds have triplet ground states (56). It thus seems that LPMO-catalyzed chemistry relies on spin cross-over transitions (here in a singlet–triplet–singlet sequence), which will reduce energetic barriers of reactions involving unpaired electrons (2). Despite differences in calculated transition-state energy barriers and spin states, available modeling data collectively show that the LPMO peroxygenase reaction is thermodynamically and kinetically feasible for LPMOs belonging to AA9 and AA10 families. Our work indicates that the peroxygenase reaction is kinetically more favorable than the monooxygenase reaction. Importantly, this claim is strongly supported by the kinetic experimental data presented here.
In the experimental part of this study we took advantage of the fluorophore properties of the SmAA10A–Cu(I)/Cu(II) redox couple (50) and used an experimental approach that allowed separation of priming reduction and reoxidation catalytic events. Our data indicate that the off-cycle priming reduction is likely not a rate-limiting step of the global LPMO catalysis. Considering the second-order rate constant for reduction by AscA, even the lowest AscA concentration employed here (20 μM) would yield a rate (8 s−1) that is similar to the kcat of 6.7 s−1 determined for SmAA10A (38). The reduced SmAA10A–Cu(I) is the precursor for all suggested LPMO catalytic pathways (1, 26, 27). Since the LPMO–Cu(I) form binds more strongly to substrates than LPMO–Cu(II) (49, 57, 58) and since access to the active site is severely hindered when the LPMO is bound to substrate (40), LPMO reduction is likely to involve substrate-free LPMO molecules. In agreement, our stopped-flow experiments indicated that the rate of SmAA10A reduction is not significantly influenced by the presence of carbohydrate substrate (SI Appendix, Fig. S10).
Depending on the substrate concentration and substrate affinity, reduced LPMOs will react with available O2 and H2O2. Under the conditions used here, the second-order rate constant for the reaction with O2, kO2 = 3.33 M−1⋅s−1, is surprisingly low and indicates a rate at ambient O2 concentration on the order 0.05 min−1. This result is in contrast with a rate of >0.15 s−1 that has been reported for reoxidation of a cellulose-active AA9 C1/C4 oxidizer from Thermoascus aurantiacus (TaAA9A) (41). This difference could reflect differences in experimental conditions (such as different pH) but could also reflect fundamental differences between AA9s and AA10s with respect to O2 activation pathways (SI Appendix, Supplementary Discussion). In the frame of the Marcus theory, we can predict the rate of outer-sphere electron transfer between LPMO–Cu(I) and O2, as outlined by Kjaergaard et al. (41) (see SI Appendix, Supplementary Discussion for details). Thereby, we estimate an electron transfer (ET) rate kET, O2 = 3.39 M−1⋅s−1, a value remarkably similar to the experimentally determined kO2 = 3.33 M−1⋅s−1. This result implies that reduction of O2 would be subject to an outer-sphere reduction, in contrast to an inner-sphere reduction suggested by data on TaAA9A (41). An outer-sphere reduction entails that O2 would be first reduced in a “prebound” configuration and only then the resulting O2•− and Cu(II) would form a covalent bond. Such prebound O2 has been suggested by neutron protein crystallography and DFT calculations for an AA9 (59), but not for any AA10 yet.
Comparing the kO2 value of SmAA10A with values obtained for other “classical” monooxygenases provides some interesting insights. Indeed, the rate constants for reoxidation by O2 are several orders of magnitude higher (105 to 107 M−1⋅s−1) for binuclear copper enzymes such as tyrosinase (2) and dopamine monooxygenase (60), for monocopper amine oxidase (61) and galactose oxidase (62), as well as for certain multicopper laccases (63, 64). In light of this comparison, together with steady-state kinetics data showing that the peroxygenase reaction (38) is orders of magnitude faster than the monooxgenase reaction (32, 37) and together with the QM/MM results presented here, we conclude that SmAA10A has not been evolved to act as a O2-using oxygenase.
The second-order rate constant for the reaction of SmAA10A–Cu(I) with H2O2, 6.85 × 103 M−1⋅s−1, is three orders of magnitude higher compared to the corresponding reaction with O2. Also, in the second-order rate constant plots (Fig. 5 B and D), the y intercept is close to zero when using H2O2 as oxidant but not when using O2. This suggests that the reaction with H2O2 is irreversible, while there may be some reversibility in the reaction with O2. Importantly, using spin trapping, we show that the reaction with H2O2 generates radicals (SI Appendix, Fig. S16) while consuming equimolar amounts of SmAA10A–Cu(I) and H2O2 (SI Appendix, Fig. S15). Remarkably, it appears that the reaction between SmAA10A–Cu(I) and H2O2 is slower in the absence of substrate than in the presence of substrate. If we compare the maximum velocity (Vmax) determined by Kuusk et al. (38) for reactions with chitin (at saturating [H2O2] of 20 μM) with the Vmax calculated from the second-order rate constant kH2O2 determined here for reoxidation of SmAA10A–Cu(I) in the absence of substrate, it appears that the reaction with H2O2 is accelerated by about 50 times in the presence of substrate (kcat × [E]/kH2O2 × [E] × [H2O2] = 6.7 s−1/6.85 × 103 M−1⋅s−1 × 20 μM = 48.9). Repeating reoxidation experiments under single-catalytic-event conditions in the presence of NAG6 showed that reoxidation by O2 was slightly slowed down whereas reoxidation by H2O2 was boosted in a NAG6 concentration-dependent manner. Thus, the presence of substrate further promotes the peroxygenase reaction, relative to the monooxygenase reaction.
In our QM/MM models, the roles of the enzyme-substrate interactions appear to be 3-fold along the reaction coordinates: 1) Together with Glu60, the substrate H bond assists H2O2 molecule positioning in the reaction cavity (state 1; Fig. 3B) and 2) helps stabilizing the reactive “precision-guided HO•” (states 2 through 4); 3) these interactions seem to position the H2O2-derived water molecule that contributes with an H bond to the reactive species in the final stages of the reaction mechanism (states 5 through 9) (SI Appendix, Fig. S7). The cellulose “analog” of the chitin amide group is the 2′-OH moiety, which could well act as a hydrogen bond donor. The resulting conclusion that peroxygenase catalysis by LPMOs is substrate-assisted is supported by kinetic studies (38). In accordance with the results from the modeling studies, mutation of Glu60, a functionally conserved residue in all LPMOs known to be important for LPMO activity (12, 53, 54, 65), revealed that this residue likely is important for both H2O2 activation and controlling hydroxyl radical reactivity. In that respect, it is fair to consider whether LPMOs may employ the PK mechanism seen in heme peroxidases (47, 48). In the latter, the resting Fe(III) state reacts with H2O2 to form compound I using a catalytic base (e.g., a His) for deprotonating the proximal oxygen (i.e., heterolytic cleavage of H2O2). Here, the analogous resting SmAA10A–Cu(II) form does not react with H2O2 (refs. 1 and 38 and SI Appendix, Fig. S19) and we therefore considered the Cu(I) state. Our calculations indicate that a PK-like mechanism entailing a heterolytic cleavage of H2O2, where Glu60 acts as a catalytic base, is unfavorable (32.4 kcal/mol, TPSSh) compared to the homolytic cleavage of H2O2 (8.6 kcal/mol, TPSSh) (Fig. 3B) and is thus unlikely.
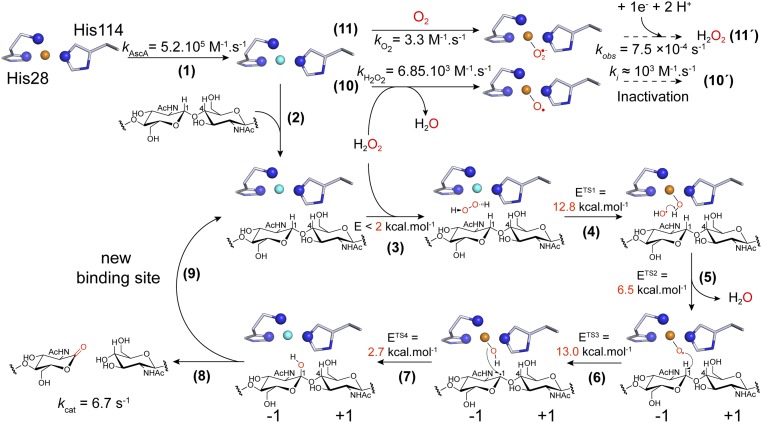
The rate-limiting step in the peroxygenase reaction remains unknown. In their modeling study, Wang et al. (36) proposed that (nonenzymatic) hydrolysis of the hydroxylated carbohydrate (i.e., state 9 in Fig. 3G) is the rate-limiting step. This suggestion was based on a predicted high transition-state energy barrier for the glycosidic bond cleavage reaction (18.2 kcal/mol) that coincided with the observed catalytic constant (kcat = 0.11 s−1) determined under ambient O2 (i.e., uncontrolled, limiting H2O2) conditions. Despite limitations when attempting to relate energy barriers to catalytic constants (SI Appendix, Supplementary Discussion), this possibility needs to be considered while awaiting experimental validation and may hold for LPMOs acting on soluble substrates, in particular when using O2 as cosubstrate (32). For polysaccharide-active LPMOs, both from AA9 and AA10 families, experimental evidence suggests that the apparent LPMO rate under ambient conditions is limited by the rate of in situ production or supply of H2O2 (1, 32, 38, 66). Therefore, in natural settings, where low steady-state concentrations of H2O2 are expected (29), the rate of LPMO catalysis will be limited by the probability of encounters between H2O2 and a substrate-bound prereduced LPMO. As summarized in Fig. 8, we propose the following order of events for the LPMO peroxygenase reaction. Step 1: The substrate-free enzyme is reduced at high rate. Step 2: The LPMO binds to its substrate, where it is positioned to facilitate formation of a ternary complex. Step 3: H2O2 diffuses freely into the reaction cavity via a tunnel access and is guided into a strained position, optimal for H2O2 reduction, by specific hydrogen bonding interactions. Steps 4 through 7: The reaction progresses as indicated by states 1 to 9 shown in Fig. 3G. Steps 8 and 9: Glycosidic bond cleavage (via an elimination reaction) and enzyme dissociation from the substrate occur in an order that remains to be clarified. Steps 10 and 10′: In the absence of substrate, the LPMO may become reoxidized, which may lead to inactivation processes if there is free H2O2 in the system. It is interesting to note that kH2O2 is nearly one order of magnitude higher than the apparent inactivation rate previously determined in the presence of excess of reductant (103 M−1⋅s−1) (38), indicating that not every reaction with H2O2 (step 10) leads to inactivation, as is apparent from experimental data shown in SI Appendix, Fig. S18. Steps 11 and 11′: Substrate-free LPMOs can also engage into the conversion of O2 to H2O2. In this latter process, the reactivity of SmAA10A–Cu(I) with O2 seems to be the rate-limiting step since the second-order rate of 3.3 M−1⋅s−1 yields a turnover of 8⋅10−4 s−1 at ambient [O2], a value strikingly similar to 7.5⋅10−4 s−1, the apparent H2O2 production rate under these conditions (29).
Fig. 8.
Proposed molecular mechanism of chitin oxidation by SmAA10A and associated rate constants or energy barriers. Step 1: The resting LPMO–Cu(II) is reduced to the LPMO–Cu(I) active state (4.23 × 105 M−1⋅s−1; i.e., 423 s−1 or 8.5 s−1 when using 1 mM or 20 µM AscA, respectively). Step 2: Polysaccharide binding. Step 3: H2O2 diffusion to the active-site cavity through a tunnel access. Step 4: Reaction of H2O2 with Cu(I), inducing homolytic bond cleavage yielding a Cu(II)–OH species and a hydroxyl radical. Step 5: The hydroxyl radical reacts with Cu(II)–OH, yielding a water molecule and a [CuO]+ intermediate. Step 6: [CuO]+ catalyzes HAA from the substrate. Step 7: Hydroxylation of the C1 carbon via an oxygen-rebound mechanism (33). Step 8: The hydroxylated product undergoes an elimination reaction (30) suggested to be enzyme-independent (36), inducing glycosidic bond cleavage. Step 9: The LPMO–Cu(I) can enter a new catalytic cycle. Step 10: Substrate-free LPMO–Cu(I) can react with H2O2 (6.85 × 103 M−1⋅s−1), which may lead to enzyme inactivation (step 10′, ca. 103 M−1⋅s−1) (38). Step 11: The LPMO–Cu(I) can also react with O2 (3.3 M−1⋅s−1; i.e., 8.10−4 s−1 at 250 µM [atmospheric] O2), leading in fine to H2O2 production (step 11′, 7.5.10−4 s−1) (29). Note that steps 10′ and 11′ proceed via mechanisms that remain to be fully elucidated.
The experimental conditions of the LPMO reaction will affect the outcome of the chain of events. With low-affinity substrates and high concentrations of H2O2 the number of turnovers may be low, as nonproductive enzyme reoxidation, with a concomitant risk of enzyme inactivation will be prominent. At optimal conditions, when reducing equivalents and H2O2 are delivered in a timely fashion in the presence of saturating amounts of a suitable substrate, multiple turnovers will occur (49, 66). The proposed substrate-assisted mechanism provides a competitive advantage to polysaccharide-bound LPMOs for turning over H2O2 compared to the reaction of substrate-free LPMOs with H2O2 that leads to enzyme inactivation via formation of hazardous radicals.
Conclusions
In the above, we show how LPMOs harness the intrinsic oxidative power of H2O2 in a controlled and substrate-assisted manner to oxidize crystalline chitin in what is a unique peroxygenase reaction. Part of the uniqueness of the LPMO chemistry comes from the formation of a confined, catalytically competent active site reaction cavity only upon association of the enzyme with the polysaccharide. LPMO catalysis could perhaps be described as an enzyme-guided Fenton reaction, as we suggested when first describing the peroxygenase reaction (31). In the context of DFT, we presented in great details the electronic features of the reactivity of H2O2 with LPMO–Cu(I). The occurrence of spin cross-over events along the reaction coordinate suggests that intermediates detectable by spectroscopy may be trapped and characterized in future studies. Importantly, the experimental part of this study provides kinetics data of single oxidative event reactions of LPMO–Cu(I) with O2 and H2O2. Together with steady-state kinetics and computational simulations, our data show that the use of O2 as cosubstrate by SmAA10A to oxidize chitin is unlikely and support the peroxygenase paradigm. No doubt assessing the universality of this mechanism across LPMOs families will be a topic of intense investigations for the coming years.
We foresee that insight into the molecular basis of the unusual, copper-based chemistry catalyzed by LPMOs will inspire and assist the design of new model complexes for oxy-functionalization of inert C–H bonds. Furthermore, witnessing strong indications of a role of chitin-active LPMOs in pathogenesis and defense mechanisms, we believe our study may eventually contribute to a better understanding of biologically important questions.
Supplementary Material
Acknowledgments
This work was supported by Research Council of Norway grants 240967 (Å.K.R.) and 269408 (V.G.H.E.) and National Science Foundation grant MCB1715176 (J.L.D.). Computational work was performed on the Saga and Abel Clusters, owned by the Norwegian Metacenter for High Performance Computing (project NN1003K) and the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant ACI-1548562, through allocation TG-MCB090159 to G.T.B. This work was authored in part by Alliance for Sustainable Energy, LLC, the manager and operator of the National Renewable Energy Laboratory for the US Department of Energy (DOE) under contract DE-AC36-08GO28308. Funding was provided to G.T.B. by the US DOE Office of Energy Efficiency and Renewable Energy Bioenergy Technologies Office.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.M.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904889117/-/DCSupplemental.
References
- 1.Bissaro B., et al. , Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13, 1123–1128 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Solomon E. I., et al. , Copper active sites in biology. Chem. Rev. 114, 3659–3853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaaje-Kolstad G., et al. , An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Quinlan R. J., et al. , Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U.S.A. 108, 15079–15084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg Z., et al. , Cleavage of cellulose by a CBM33 protein. Protein Sci. 20, 1479–1483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langston J. A., et al. , Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 77, 7007–7015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frommhagen M., et al. , Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol. Biofuels 8, 101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couturier M., et al. , Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat. Chem. Biol. 14, 306–310 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Villares A., et al. , Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci. Rep. 7, 40262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermaas J. V., Crowley M. F., Beckham G. T., Payne C. M., Effects of lytic polysaccharide monooxygenase oxidation on cellulose structure and binding of oxidized cellulose oligomers to cellulases. J. Phys. Chem. B 119, 6129–6143 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Eibinger M., Sattelkow J., Ganner T., Plank H., Nidetzky B., Single-molecule study of oxidative enzymatic deconstruction of cellulose. Nat. Commun. 8, 894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris P. V., et al. , Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Payne C. M., et al. , Fungal cellulases. Chem. Rev. 115, 1308–1448 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Vaaje-Kolstad G., Horn S. J., van Aalten D. M. F., Synstad B., Eijsink V. G. H., The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floudas D., et al. , The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Kracher D., et al. , Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352, 1098–1101 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Müller G., Várnai A., Johansen K. S., Eijsink V. G. H., Horn S. J., Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol. Biofuels 8, 187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannella D., Hsieh C. W. C., Felby C., Jørgensen H., Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol. Biofuels 5, 26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen K. S., Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 44, 143–149 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wong E., et al. , The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 8, e1002373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loose J. S. M., Forsberg Z., Fraaije M. W., Eijsink V. G. H., Vaaje-Kolstad G., A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett. 588, 3435–3440 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Chiu E., et al. , Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization. Proc. Natl. Acad. Sci. U.S.A. 112, 3973–3978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agostoni M., Hangasky J. A., Marletta M. A., Physiological and molecular understanding of bacterial polysaccharide monooxygenases. Microbiol. Mol. Biol. Rev. 81, e00015–e00017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla A. K., et al. , Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 34, 1046–1051 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Walton P. H., Davies G. J., On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr. Opin. Chem. Biol. 31, 195–207 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Beeson W. T., Vu V. V., Span E. A., Phillips C. M., Marletta M. A., Cellulose degradation by polysaccharide monooxygenases. Annu. Rev. Biochem. 84, 923–946 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Meier K. K., et al. , Oxygen activation by Cu LPMOs in recalcitrant carbohydrate polysaccharide conversion to monomer sugars. Chem. Rev. 118, 2593–2635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bissaro B., Varnai A., Røhr Å. K., Eijsink V. G. H., Oxidoreductases and reactive oxygen species in conversion of lignocellulosic biomass. Microbiol. Mol. Biol. Rev. 82, e00029-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beeson W. T., Phillips C. M., Cate J. H. D., Marletta M. A., Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134, 890–892 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Bissaro B., et al. , Fenton-type chemistry by a copper enzyme: Molecular mechanism of polysaccharide oxidative cleavage. bioRxiv 10.1101/097022 (27 December 2016).
- 32.Hangasky J. A., Iavarone A. T., Marletta M. A., Reactivity of O2 versus H2O2 with polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 115, 4915–4920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S., Ståhlberg J., Sandgren M., Paton R. S., Beckham G. T., Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc. Natl. Acad. Sci. U.S.A. 111, 149–154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedegård E. D., Ryde U., Targeting the reactive intermediate in polysaccharide monooxygenases. J. Biol. Inorg. Chem. 22, 1029–1037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertini L., et al. , Catalytic mechanism of fungal lytic polysaccharide monooxygenases investigated by first-principles calculations. Inorg. Chem. 57, 86–97 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Wang B., et al. , QM/MM studies into the H2O2-dependent activity of lytic polysaccharide monooxygenases: Evidence for the formation of a caged hydroxyl radical intermediate. ACS Catal. 8, 1346–1351 (2018). [Google Scholar]
- 37.Frandsen K. E. H., et al. , The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat. Chem. Biol. 12, 298–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuusk S., et al. , Kinetics of H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J. Biol. Chem. 293, 523–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofrichter M., Ullrich R., Oxidations catalyzed by fungal peroxygenases. Curr. Opin. Chem. Biol. 19, 116–125 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Bissaro B., Isaksen I., Vaaje-Kolstad G., Eijsink V. G. H., Røhr Å. K., How a lytic polysaccharide monooxygenase binds crystalline chitin. Biochemistry 57, 1893–1906 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Kjaergaard C. H., et al. , Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111, 8797–8802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedegård E. D., Ryde U., Molecular mechanism of lytic polysaccharide monooxygenases. Chem. Sci. (Camb.) 9, 3866–3880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baraban J. H., Changala P. B., Stanton J. F., The equilibrium structure of hydrogen peroxide. J. Mol. Spectrosc. 343, 92–95 (2018). [Google Scholar]
- 44.Busing W. R., Levy H. A., Crystal and molecular structure of hydrogen peroxide: A neutron-diffraction study. J. Chem. Phys. 42, 3054–3059 (1965). [Google Scholar]
- 45.Becke A. D., Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993). [Google Scholar]
- 46.Tao J., Perdew J. P., Staroverov V. N., Scuseria G. E., Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids. Phys. Rev. Lett. 91, 146401 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Poulos T. L., Kraut J., The stereochemistry of peroxidase catalysis. J. Biol. Chem. 255, 8199–8205 (1980). [PubMed] [Google Scholar]
- 48.Derat E., Shaik S., The Poulos-Kraut mechanism of compound I formation in horseradish peroxidase: A QM/MM study. J. Phys. Chem. B 110, 10526–10533 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Kuusk S., et al. , Kinetic insights into the role of the reductant in H2O2-driven degradation of chitin by a bacterial lytic polysaccharide monooxygenase. J. Biol. Chem. 294, 1516–1528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bissaro B., et al. , Fueling biomass-degrading oxidative enzymes by light-driven water oxidation. Green Chem. 18, 5357–5366 (2016). [Google Scholar]
- 51.Lochenie C., Wagner K. G., Karg M., Weber B., Modulation of the ligand-based fluorescence of 3d metal complexes upon spin state change. J. Mater. Chem. C Mater. Opt. Electron. Devices 3, 7925–7935 (2015). [Google Scholar]
- 52.Vaaje-Kolstad G., Forsberg Z., Loose J. S., Bissaro B., Eijsink V. G., Structural diversity of lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 44, 67–76 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Vaaje-Kolstad G., Houston D. R., Riemen A. H. K., Eijsink V. G. H., van Aalten D. M. F., Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280, 11313–11319 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Loose J. S. M., et al. , Multipoint precision binding of substrate protects lytic polysaccharide monooxygenases from self-destructive off-pathway processes. Biochemistry 57, 4114–4124 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Sabbadin F., et al. , An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 9, 756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huber S. M., et al. , Generating Cu(II)-oxyl/Cu(III)-oxo species from Cu(I)-alpha-ketocarboxylate complexes and O2: In silico studies on ligand effects and C-H-activation reactivity. Chemistry 15, 4886–4895 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kracher D., Andlar M., Furtmüller P. G., Ludwig R., Active-site copper reduction promotes substrate binding of fungal lytic polysaccharide monooxygenase and reduces stability. J. Biol. Chem. 293, 1676–1687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hangasky J. A., Marletta M. A., A random-sequential kinetic mechanism for polysaccharide monooxygenases. Biochemistry 57, 3191–3199 (2018). [DOI] [PubMed] [Google Scholar]
- 59.O’Dell W. B., Agarwal P. K., Meilleur F., Oxygen activation at the active site of a fungal lytic polysaccharide monooxygenase. Angew. Chem. Int. Ed. Engl. 56, 767–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brenner M. C., Klinman J. P., Correlation of copper valency with product formation in single turnovers of dopamine β-monooxygenase. Biochemistry 28, 4664–4670 (1989). [DOI] [PubMed] [Google Scholar]
- 61.Mukherjee A., et al. , Inner-sphere mechanism for molecular oxygen reduction catalyzed by copper amine oxidases. J. Am. Chem. Soc. 130, 9459–9473 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whittaker M. M., Ballou D. P., Whittaker J. W., Kinetic isotope effects as probes of the mechanism of galactose oxidase. Biochemistry 37, 8426–8436 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Cole J. L., Solomon E. I., Ballou D. P., Spectroscopic characterization of the peroxide intermediate in the reduction of dioxygen catalyzed by the multicopper oxidases. J. Am. Chem. Soc. 113, 8544–8546 (1991). [Google Scholar]
- 64.Andréasson L. E., Reinhammar B., Kinetic studies of Rhus vernicifera laccase. Role of the metal centers in electron transfer. Biochim. Biophys. Acta 445, 579–597 (1976). [DOI] [PubMed] [Google Scholar]
- 65.Span E. A., Suess D. L. M., Deller M. C., Britt R. D., Marletta M. A., The role of the secondary coordination sphere in a fungal polysaccharide monooxygenase. ACS Chem. Biol. 12, 1095–1103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Müller G., Chylenski P., Bissaro B., Eijsink V. G. H., Horn S. J., The impact of hydrogen peroxide supply on LPMO activity and overall saccharification efficiency of a commercial cellulase cocktail. Biotechnol. Biofuels 11, 209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.