Significance
The ability to study human liver disease is limited by available hepatocyte models. Primary human hepatocytes (PHH) and xenograft models suffer from limited availability, donor-to-donor variability, and high cost. Here we report two transformative advances. First, the alkaloid retrorsine improves humanization of the murine liver, which allows routine production of highly humanized mice and high-quality mouse-passaged PHH. Second, the ability to genetically modify PHH cultures and retransplant to create highly humanized mice with genetically altered grafts. When combined, these two advances open new frontiers for creating disease-specific PHH models and for performing genetic and other screens in PHH.
Keywords: liver, primary human hepatocytes, humanized mice, hepatotropic pathogens, hepatitis B virus
Abstract
Primary human hepatocytes (PHHs) are an essential tool for modeling drug metabolism and liver disease. However, variable plating efficiencies, short lifespan in culture, and resistance to genetic manipulation have limited their use. Here, we show that the pyrrolizidine alkaloid retrorsine improves PHH repopulation of chimeric mice on average 10-fold and rescues the ability of even poorly plateable donor hepatocytes to provide cells for subsequent ex vivo cultures. These mouse-passaged (mp) PHH cultures overcome the marked donor-to-donor variability of cryopreserved PHH and remain functional for months as demonstrated by metabolic assays and infection with hepatitis B virus and Plasmodium falciparum. mpPHH can be efficiently genetically modified in culture, mobilized, and then recultured as spheroids or retransplanted to create highly humanized mice that carry a genetically altered hepatocyte graft. Together, these advances provide flexible tools for the study of human liver disease and evaluation of hepatocyte-targeted gene therapy approaches.
Hepatocytes, the predominant cell type in the liver, perform multiple functions and play essential roles in metabolic reactions and drug clearance (1, 2). In addition, human hepatocytes are the exclusive host for several hepatotropic pathogens, including hepatitis B virus (HBV), hepatitis C virus, and malaria-causing Plasmodium parasites (3). Therefore, hepatocytes are central players in many of the world’s leading disease etiologies.
Primary human hepatocytes (PHHs) isolated from human liver are the gold standard for studies of hepatocyte biology. However, current PHH options suffer from donor-to-donor variability, limited availability of high-quality donors, high financial cost to isolate and procure, and failure to maintain long-term function in culture. Furthermore, the inability to isolate high-quality PHH from patients with liver disease has precluded the study of possible causal genetic variants in a physiologically relevant model.
Underpinning these limitations is the inability of PHHs to proliferate once isolated, despite their capacity for regeneration and expansion in vivo in response to injury. Renewable sources of human hepatocytes have been sought, including differentiation of pluripotent stem cells into hepatocyte-like cells (HLCs). Despite the potential of this approach, including the ability to generate patient-specific hepatocytes from induced pluripotent stem cells, current HLCs resemble fetal hepatoblasts rather than mature hepatocytes (4, 5). Protocols have been developed to expand PHH, including ex vivo growth as organoids (6) or immortalization (7, 8). A more widely adopted approach involves PHH transplantation into immunodeficient mice with various forms of liver injury to provide a niche and milieu that favors PHH engraftment and expansion. There are several widely used murine models, including immunodeficient Fah−/− NOD Rag1−/− Il2rgnull (FNRG) mice (9–11) in which liver injury is induced by withdrawing the protective drug 2-(2-nitro-4-(trifluoromethyl)benzoyl)cyclohexane-1,3-dione (NTBC) (12). While the specific stimuli remain unidentified, this expansion method is presumed to function via the provision of signals derived from the damaged mouse liver and by creating vacant space that is liberated as murine hepatocytes die. Collectively, this environment allows transplanted human hepatocytes to engraft within the damaged scaffold and proliferate to repopulate the mouse liver parenchyma. Once these chimeric livers are highly humanized, human hepatocytes can be reisolated. However, to date, these chimeric models have suffered from the same detractors that limit the use of unexpanded PHH: most PHH donors repopulate poorly, and the level of chimerism is variable between animals.
Here, we describe a protocol that, by improving chimeric mouse liver humanization, enables the expansion of the usable PHH donor pool. Specifically, we show that, via this protocol, mouse-passaged primary human hepatocytes (mpPHHs) can successfully establish long-term cultures that are suitable for drug metabolism studies and are also susceptible to human hepatotropic pathogen infections. Notably, we observe that even poorly plateable PHHs can be passaged via humanized mouse expansion and that this process seems to normalize the subsequent plateability of almost every mpPHH tested. Perhaps most significantly, mpPHH can be efficiently transduced with lentiviral vectors, mobilized, and replated for use in ex vivo applications or retransplanted to generate humanized mice that carry a genetically modified human graft, opening doors for the experimental study of human liver biology at a level that has thus far been unachievable.
Results
Retrorsine Improves Human Hepatocyte Repopulation in Liver Chimeric Mouse Models.
To reliably study PHH in vivo in liver chimeric mice or isolate the human graft for ex vivo studies, a minimum level of humanization is required. For most applications, the minimum level is ∼10%, which corresponds to 103 μg/mL serum human albumin (hAlb), an established surrogate marker for liver humanization (9, 10, 13). Unfortunately, with current protocols, PHHs from most donors engraft poorly, and few mice achieve the degree of humanization required for further use. To address this limitation, we sought to establish protocols where the majority of PHH donors consistently generate mice with over 10% chimerism.
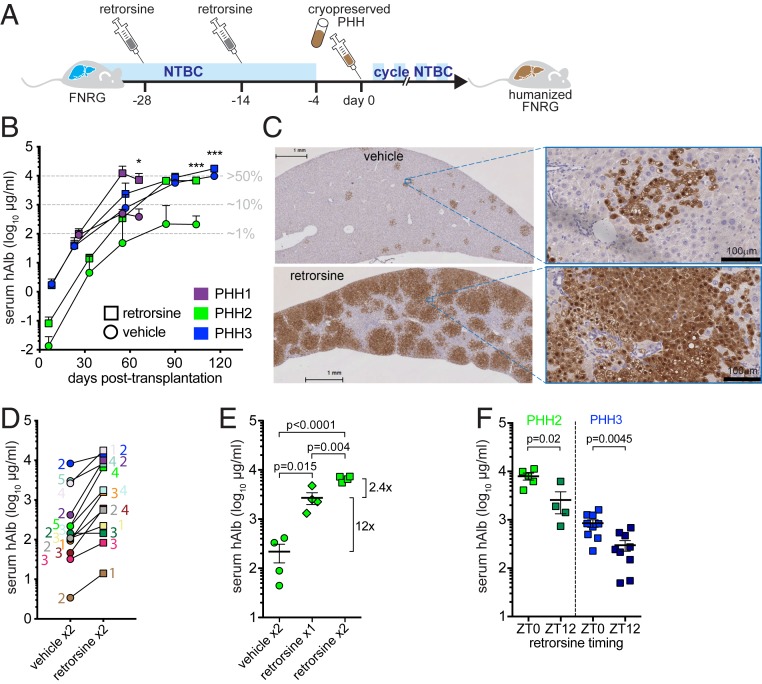
In rats, the pyrrolizidine alkaloid retrorsine enhances liver repopulation with donor hepatocytes after partial hepatectomy or carbon tetrachloride administration (14, 15). We, therefore, tested if preconditioning FNRG mice with two injections of retrorsine before PHH transplantation (Fig. 1A) enhanced hAlb levels in mouse serum. Across PHH from three donors, retrorsine improved hAlb levels (Fig. 1B) without affecting animal survival (SI Appendix, Fig. S1A). Retrorsine increased peak hAlb levels by 23- and 30-fold in mice engrafted with poor donors PHH1 and PHH2, respectively (Fig. 1B). In contrast, retrorsine had only a minor effect (twofold increase) on mice engrafted with PHH3, an efficient donor that humanizes well even with a standard protocol (vehicle control). Consistent with hAlb levels, histological staining for fumarylacetoacetate hydrolase (FAH) demonstrated that the liver parenchyma of retrorsine-treated humanized Fah−/− NOD Rag1−/− Il2rgnull (huFNRG) mice was largely replaced by human hepatocytes (Fig. 1C). We then tested 10 additional PHH donors and found that 9 of them reached higher peak hAlb levels after retrorsine preconditioning. Across 13 donors, retrorsine on average resulted in 9.5-fold higher peak serum hAlb levels than vehicle control mice (P = 0.003) (Fig. 1D). This finding confirms that retrorsine preconditioning improves engraftment across a wide range of PHH donors.
Fig. 1.
Retrorsine improves liver repopulation of FNRG mice with PHHs. (A) Experimental schematic of FNRG mice treated with retrorsine 4 and 2 wk prior to transplantation with cryopreserved PHHs and NTBC cycling. (B) Serial hAlb measurement in serum of transplanted mice shows higher peak hAlb values in retrorsine preconditioned (squares) than vehicle control mice (circles). Median ± SEM, t test of end points. PHH1 n = 2 per group, PHH2 n ≥ 4 per group, and PHH3 n ≥ 7 per group. Estimated percentage of liver humanization is shown in gray. *P < 0.05; ***P < 0.001. (C) Livers from retrorsine preconditioned PHH1 transplanted animals contain larger human areas by FAH staining than vehicle control mice. (Scale bars: Left, 1 mm; Right, 100 μm.) (D) hAlb levels in serum reached a higher plateau in retrorsine than vehicle preconditioned FNRG mice with 12 of 13 PHH donors ranging from 2- to 30-fold. Symbols are median hAlb per group of two to five transplanted mice. Color-matched numbers indicate animal numbers that reached hAlb plateau (for vehicle, numbers are shown on the left, and for retrorsine, they are on the right). (E) Retrorsine works in a dose-dependent manner, with the first dose having the largest benefit on peak serum hAlb values in PHH2 transplanted FNRG mice. Symbols are individual mice, median ± range, t test. (F) Serum hAlb values peak higher in mice receiving retrorsine at ZT0 than ZT12 prior to PHH2 transplantation (Left), while PHH3 engrafted faster in mice that received at ZT0 than ZT12 as illustrated by day 42 hAlb values (Right). Symbols are individual mice, median ± range, t test.
We then tested if retrorsine also improved humanization with fetal hepatoblasts, which require human oncostatin-M (hOSM) supplementation for efficient repopulation of FNRG livers (13). To supplement hOSM, we created AAV8-hOSM vectors that, at low doses, were not toxic and improved humanization (SI Appendix, Fig. S1B). As with PHH, we found that retrorsine preconditioning of FNRG mice improved fetal hepatoblast repopulation (SI Appendix, Fig. S1C).
The scale of retrorsine effects on PHH engraftment varied with the dose such that one dose led to a large improvement and a second dose conferred a small additional benefit (Fig. 1E). For PHH3, there was no benefit of administering a third dose (1.02-fold increase in serum hAlb, n = 11 two doses vs. n = 9 three doses, P = 0.98). In rat models, retrorsine is bioactivated by CYP450 enzymes (16, 17), many of which are under circadian regulation. We, therefore, examined whether injecting mice with retrorsine at different times of the day influences its effectiveness. Interestingly, retrorsine administered at Zeitgeber time 0 (ZT0) resulted in fivefold higher (PHH2) or faster rise in hAlb (PHH3) than at ZT12 (Fig. 1F).
We hypothesized that retrorsine impaired mouse hepatocyte proliferation, limiting restoration of the damaged mouse liver and giving PHH a repopulation advantage. In the Fah−/− model, this effect can best be observed after animals are being cycled back on NTBC, when proliferation-based liver restoration by mouse hepatocytes takes place. To test our hypothesis, livers from untransplanted FNRG mice were harvested at various times after NTBC withdrawal and reintroduction (SI Appendix, Fig. S2A). Histological examination did not show morphological differences between retrorsine and vehicle preconditioned livers at the end of NTBC withdrawal or 3 d after restarting NTBC (SI Appendix, Fig. S2B). To quantify proliferating hepatocytes, livers were stained for the cell cycle marker Ki67. As expected, few hepatocytes expressed Ki67 in mice that were off NTBC. On NTBC reintroduction, Ki67 was more frequently detected in hepatocytes of retrorsine than vehicle-treated animals, which was mostly pronounced on day 17 (SI Appendix, Fig. S2C). This did not support our hypothesis. We then tested whether retrorsine caused increased mouse hepatocyte senescence or a ductular reaction, and FNRG livers were stained for p21 or cytokeratins (CKs), respectively (18). No enhanced hepatocyte senescence or ductular reaction was observed in mice that received retrorsine compared with vehicle, making these unlikely mechanisms to explain the increased humanization phenotype. We then screened untransplanted FNRG livers for murine growth factors at the end of the NTBC withdrawal cycle (day 14) or 3 d after restarting NTBC (day 17). Numerous growth factors and receptors could be detected at the end of the NTBC withdrawal cycle, a few of which varied slightly in livers of mice that had received retrorsine from livers of mice preconditioned with vehicle (SI Appendix, Fig. S2D). These combined observations suggest that retrorsine promotes PHH repopulation through mechanisms that are distinct from the cell cycle inhibition observed in rat hepatocytes and may involve an altered growth factor milieu that is beneficial to human graft expansion.
To extend the application of retrorsine beyond the Fah−/− model, we next tested whether retrorsine also improved engraftment in two other chimeric animal models: immunodeficient thymidine kinase (TK) (19) and hemizygous urokinase plasminogen activator (uPA)+/− transgenic mice. Using the PHH2 donor for transplantation, we found that retrorsine preconditioning increased peak hAlb levels by 28-fold in TK recipient mice (SI Appendix, Fig. S1D), consistent with the Fah−/− model. In uPA+/− mice, which are known to suffer from graft loss (20) over time, the graft persisted in retrorsine-treated mice as illustrated by hAlb remaining above 103 μg/mL for months (SI Appendix, Fig. S1E). These data show that retrorsine preconditioning improved humanization in three widely used chimeric mouse models with the large majority of human donors.
Isolation of PHHs from Chimeric Mouse Livers.
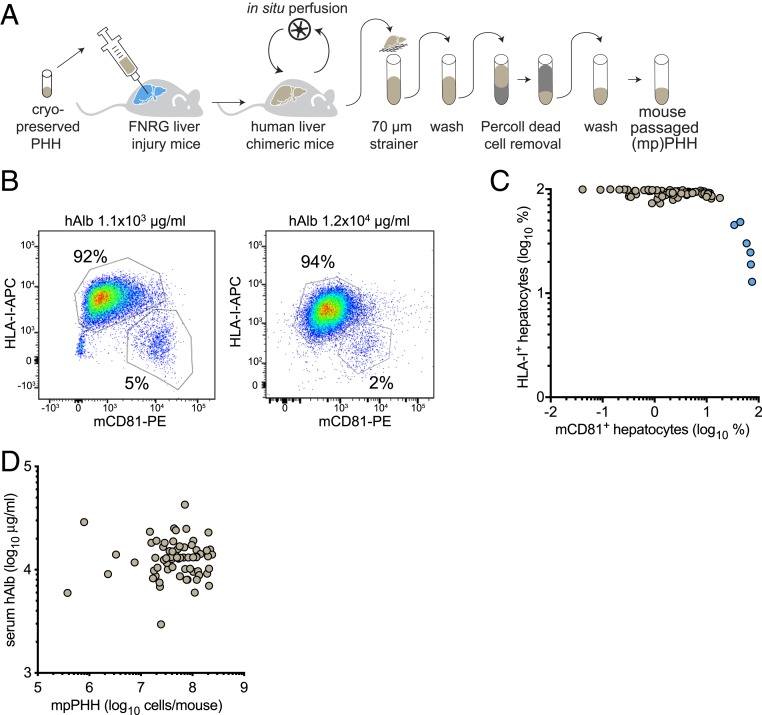
Retrorsine preconditioning greatly increased the availability of highly humanized mice. Since human hepatocytes can be isolated from chimeric livers (9, 11, 21), we established protocols to enrich for live human hepatocytes from these livers. Retrorsine preconditioned huFNRG mice were taken off the protective drug NTBC (12) at least 7 d before harvest. We perfused huFNRG livers and purified hepatocyte suspensions by Percoll and low-speed centrifugations to enrich for live hepatocytes (mpPHH) (Fig. 2A). Flow cytometry on mpPHH isolated from moderately (1.1 × 103 μg/mL hAlb) and highly (1.2 × 104 μg/mL hAlb) repopulated huFNRG mice showed that our isolation/purification protocol yielded predominantly human hepatocytes independent of the extent of liver humanization (Fig. 2B). Collectively, we perfused 84 huFNRG livers and after purification, analyzed the cells by flow cytometry to measure the expression of human and murine markers. In 78 mice (93%), this protocol yielded mostly human cells (Fig. 2C). To obtain high yields, we perfused livers from retrorsine preconditioned highly chimeric huFNRG mice based on hAlb serum levels around 104 μg/mL. Liver perfusions of 70 huFNRG mice yielded on average 75 million mpPHH per liver, ranging from 0.4 to 240 million (Fig. 2D). These data show that our isolation/purification protocol results in consistently large numbers of predominantly human hepatocytes isolated from highly chimeric huFNRG mice.
Fig. 2.
Isolation of PHHs from liver chimeric mice. (A) Graphic representation of engrafting FNRG mice with cryopreserved PHH followed by perfusion and purification to yield mpPHH suspensions. (B) Flow cytometry of mpPHH after isolation and purification from a moderately or highly huFNRG mouse. The isolation protocol yields few mouse CD81 (mCD81-PE)-positive hepatocytes and even fewer in highly engrafted huFNRG mice. (C) Percent HLA-I+ vs. mCD81+ hepatocytes from 84 individual huFNRG mice were analyzed by flow cytometry. Most mice (n = 78, brown symbols) yielded predominantly human hepatocytes. (D) Serum hAlb levels of 70 individual huFNRG mice (circles) were plotted against mpPHH yield per mouse. On average, 75 million (range from 0.4 to 240 million) viable mpPHHs were recovered from huFNRG mice.
Freshly Isolated mpPHHs Overcome Donor Variability and Form Long-Term Cultures.
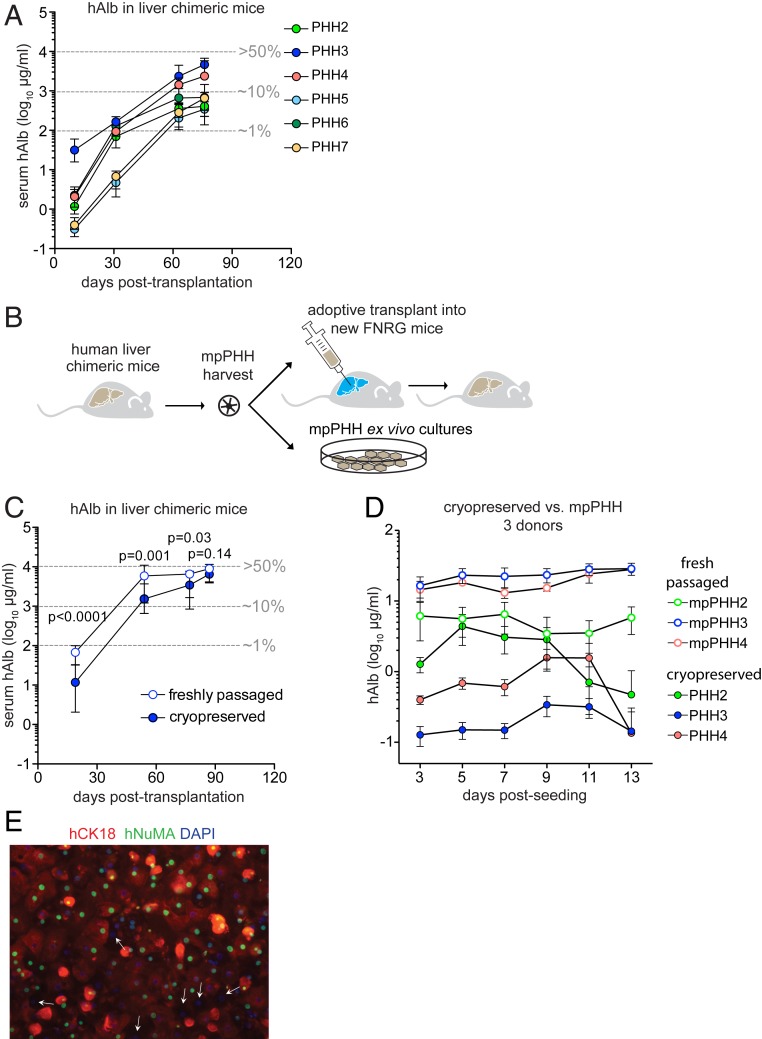
Cryopreserved PHHs from different donors vary in their ability to engraft in mice and form cultures. We transplanted six cryopreserved PHH donors (PHH2 to PHH7) into FNRG mice without preconditioning and followed repopulation by serum hAlb. Mice transplanted with PHH3 had the highest median hAlb (Fig. 3A). Based on these findings, unless otherwise indicated for subsequent experiments, we used retrorsine preconditioned FNRG mice transplanted with PHH3. Perfusion yielded on average 150 times more mpPHH than input cryopreserved PHH. To rule out that multiple cell divisions in chimeric livers affect mpPHH quality, we compared liver repopulation following adoptive transplantation and hepatocyte stability after plating to cryopreserved PHH (Fig. 3B). To test the repopulation potential as previously shown by Azuma et al. (9), equal numbers of cryopreserved PHH3 or freshly isolated mpPHH3 were transplanted into FNRG mice. mpPHH3 exhibited superior repopulation kinetics compared with cryopreserved PHH based on faster hAlb kinetics, although both groups reached similar peak values (Fig. 3C). We concluded that mpPHH retained high repopulation potential on one round of adoptive transplantation, allowing for expansion of cryopreserved PHH by over 20,000-fold.
Fig. 3.
Freshly isolated mpPHHs engraft and plate more efficiently than cryopreserved PHHs and overcome donor-to-donor variability. (A) Serum hAlb levels in FNRG mice after transplantation with six different cryopreserved donors (PHH2 to PHH7). Median ± range hAlb levels of groups of five transplanted mice. Three to five mice per group survived until day 76. Estimated percentage of liver humanization is shown in gray. (B) Graphic representation of passaging PHH through mice for adoptive transfer and plating for ex vivo cultures. (C) Comparison of cryopreserved PHH3 vs. freshly isolated mpPHH3 for repopulation of FNRG mice as shown by serum hAlb levels over time. Freshly isolated mpPHH3 repopulates faster than cryopreserved PHH3. Median ± range hAlb levels of groups of nine transplanted mice. Six to eight mice per group survived until day 87. t test between groups of individual timepoints. (D) Cryopreserved (closed circles) and freshly isolated mpPHHs (open circles) of the same three donors (PHH2: green; PHH3: blue; and PHH4: orange) were cultured for 13 d. Media were changed every 2 d, and hAlb levels were measured over time. The data represent at least five biological replicates per donor and are presented as mean ± SD. (E) Freshly isolated mpPHHs were plated. After fixation with 4% paraformaldehyde, cells were stained with human markers CK18 (red) and NuMA (green). Nuclei were stained with DAPI. Most cells were positive for one or both of the human markers, except for cells indicated by white arrows. (Magnification: 20×.)
To determine how closely mpPHH resembled cryopreserved PHH, we compared the transcriptomes of mpPHH3 suspensions from five humanized livers with two aliquots of cryopreserved PHH3. Both groups had similar expression of genes related to Phase I and Phase II detoxification pathways as well as many other liver-specific genes (SI Appendix, Fig. S3).
We then evaluated if mpPHH could form stable cultures (22) by using hAlb in culture supernatant as a marker for culture stability. Whereas cryopreserved PHH from three donors (PHH2, PHH3, PHH4) formed cultures that showed variable and modest levels of hAlb secretion that decreased over time, passaging these donors through mice yielded large numbers of hepatocytes that formed robust cultures with high and stable hAlb over time (Fig. 3D). Next, we characterized the long-term stability of mpPHH cultures. We found that denser mpPHH cultures formed confluent monolayers and were stable for at least 8 wk as shown by hAlb and 4 wk by multiple other readouts (SI Appendix, Fig. S4). We optimized these conditions for various plating formats (Methods). Because some nonplateable donors engrafted beyond 103 μg/mL hAlb levels in retrorsine preconditioned mice (SI Appendix, Fig. S5A), we cultured nonplateable PHH8 and plateable PHH2 before and after passaging through mice. Whereas plateable PHH2 formed cultures both in vitro and ex vivo, nonplateable PHH8 only attached ex vivo (SI Appendix, Fig. S5B).
Similar to the flow cytometry data on freshly isolated mpPHH (Fig. 2 B and C), the vast majority of cells after plating were human hepatocytes as assessed by human CK18 and nuclear mitotic apparatus protein (NuMA) staining (Fig. 3E).
These data show that passaging cryopreserved PHH through retrorsine preconditioned huFNRG mice yielded large numbers of mpPHH that could be plated, resulting in robust and stable cultures and hence, overcoming marked donor-to-donor variation in plateability of cryopreserved PHH.
Drug Metabolism and Infection Studies in Long-Term mpPHH Cultures.
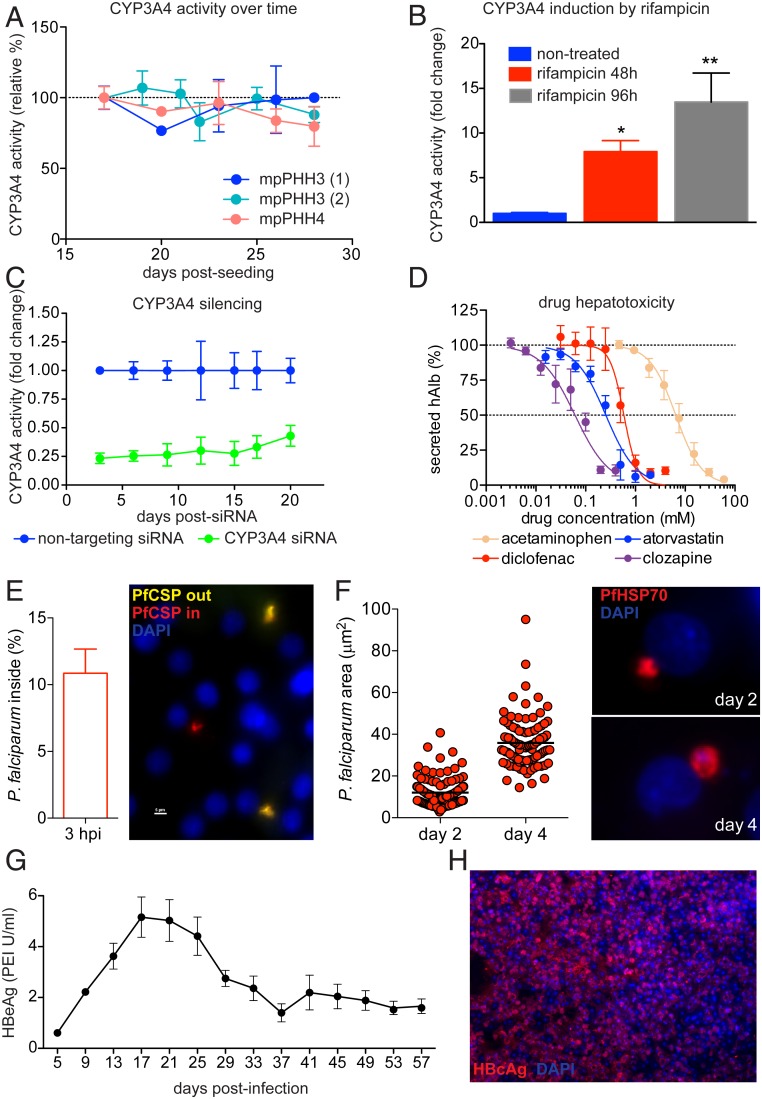
To exploit the stability of mpPHH cultures, we tested their potential for preclinical drug safety studies and evaluation of drug induction potential. First, we observed that Phase I CYP450 activity is retained in mpPHH cultures as seen by the stability of CYP3A4 levels measured in two different mpPHH donors (mpPHH3 and mpPHH4) for at least 1 mo postplating (Fig. 4A). Next, we determined whether mpPHHs maintain the ability to induce CYP450 activity after long-term culture by exposing the cells to 25 μM rifampicin, a known inducer of CYP3A4. Rifampicin treatment for 48 and 96 h induced CYP3A4 activity by 5- to 10-fold in a time-dependent manner (Fig. 4B). We then tested if enzyme activity of CYP3A4 could be modulated by RNA interference (RNAi)-mediated knockdown. To this end, mpPHHs were treated with CYP3A4-targeting small interfering RNA (siRNA) or nontargeting control for 24 h, and enzyme activity was monitored for up to 20 d posttreatment. Notably, CYP3A4 activity remained silenced during the long-term culture of mpPHH (Fig. 4C). Finally, to evaluate their use for toxicological studies, we exposed mpPHH to serial dilutions of four known hepatotoxic compounds and plotted their effect on hAlb secretion as a cell injury readout (Fig. 4D). We selected drugs that are metabolized by different enzymes (CYP3A4, CYP2E1, CYP2C9, and CYP1A2) and observed dose-dependent hepatotoxicity on a 24-h treatment. Taken together, these experiments suggest that mpPHHs are suitable for long-term drug metabolism and drug safety studies.
Fig. 4.
mpPHH cultures are suitable for long-term drug metabolism and infection studies. (A) CYP3A4 activity was stable over time in mpPHH. Freshly isolated mpPHHs derived from two donors (PHH3 and PHH4) were seeded in 96-well plates. CYP3A4 activity was measured from day 15 until day 29 postseeding, and values (mean ± SEM, n = 3) were plotted as percent activity relative to day 15. (B) Rifampicin induces CYP3A4 activity. Cells were cultured in 96-well plates and treated with 25 μM rifampicin for 48 and 96 h on day 11 or 14 postseeding. CYP3A4 activity was measured as in A. Data (mean ± SEM, n = 3) were plotted as fold change relative to untreated samples. A representative of two independent experiments is shown. *P = 0.0457 and **P = 0.0026 for 48 and 96 h, respectively; one-way ANOVA. (C) siRNA silencing of CYP3A4. Cells were cultured in 96-well plates and treated with 50 nM siRNA (either nontargeting control or targeting CYP3A4) for 24 h. Silencing efficiency was determined every 2 to 3 d by measuring CYP3A4 activity as in A. Data (mean ± SEM, n = 3) were plotted as fold change relative to the nontargeting control. P < 0.0001; two-way ANOVA. A representative of three independent experiments is shown. (D) Drug hepatotoxicity in mpPHH cultures. Cells were cultured in 96-well plates and treated with increasing concentrations of drugs for 24 h. The supernatants were harvested, and levels of secreted hAlb were measured by ELISA. Data were normalized to drug-free controls. The graph represents the mean ± SEM of three independent experiments (n = 3). The four drugs are metabolized by different CYP450 enzymes: acetaminophen (yellow), CYP2E1 and CYP3A4; atorvastatin (blue), CYP3A4; diclofenac (red), CYP2C9; and clozapine (purple), CYP1A2. (E) Cells were seeded in 96-well plates and infected 1 wk later with 10,000 Pf sporozoites per well isolated from infected mosquitoes. The percentage of invaded sporozoites at 3 h postinfection was determined with sequential immunofluorescence staining using PfCSP antibodies. Noninvaded sporozoites are detected as yellow and invaded parasites are detected as red after cell permeabilization. Values (mean ± SEM, n = 6) represent one of two independent experiments. (F) Cells infected with 40,000 Pf sporozoites per well were monitored for several days. Media were changed every 2 d, and cells were stained for PfHSP70 (red) after 2 and 4 d postinfection. Parasite intracellular development was monitored by measuring the area occupied by parasites (each dot represents a single invading parasite); 30 to 50 parasites per well were scored (n = 3). Data shown represent one of two independent experiments. (G) Cells were seeded in 96-well plates and infected with 50 GEQ per cell HBV on day 0. Media were changed every 4 d, and HBeAg levels measured and plotted over time from day 5 until day 57 postinfection. Data from three biological replicates are represented as mean ± SEM. PEI, Paul Ehrlich Institute. (H) Cells seeded in 24-well plates were infected with 300 GEQ per cell HBV. Media were changed every 2 d, and cells were fixed and stained for HBcAg after 4 wk (HBcAg: red; nuclei: blue).
Next, we tested whether mpPHH cultures could support infection and propagation of Plasmodium falciparum (Pf) and HBV, two prevalent hepatotropic pathogens that exclusively infect human hepatocytes. To determine mpPHH permissiveness to Pf, we used parasite entry assays and staining with Pf-specific antibodies with or without cell permeabilization. This allowed us to distinguish parasites that only bound to the cell surface from those that invaded the cells. On average, 11% of Pf parasites successfully invaded hepatocytes (Fig. 4E). We then monitored parasite growth over time by measuring the intracellular area occupied by individual parasites and found that parasites increased in size from days 2 to 4 postinfection (Fig. 4F). Together, these data showed that mpPHHs support efficient infection and development of Pf parasites.
The chronic phase of HBV infection cannot readily be recapitulated in cell culture due to the short lifespan of current PHH systems. We took advantage of the longevity of mpPHH cultures to monitor HBV infection parameters for up to 8 wk postinfection. Hepatitis B virus e antigen (HBeAg) was detected in the supernatant of 96-well cultures for 8 wk (Fig. 4G). In addition, at 1 mo postinfection, robust expression of hepatitis B virus core antigen (HBcAg) illustrated widespread infection of mpPHH (Fig. 4H). In summary, mpPHH supported HBV infection for at least 2 mo.
Since mpPHH were amenable to pathogen infections and drug toxicity studies, we adapted the system to a microscale 384-well format suitable for high-throughput screens. As a proof of concept, we infected cells with HBV and treated a fraction of the wells with the HBV entry inhibitor Myrcludex B. Ten days postinfection, naïve but not Myrcludex B-treated cells were efficiently infected with HBV as evidenced by secreted HBeAg and HBcAg expression (SI Appendix, Fig. S6).
These combined data illustrate that the long-term stability of mpPHH expands research into various human hepatocyte functions that cannot accurately be studied in short-term cultures.
Lentiviral Transduction of mpPHH Generates Transgenic Spheroids and Chimeric Mice.
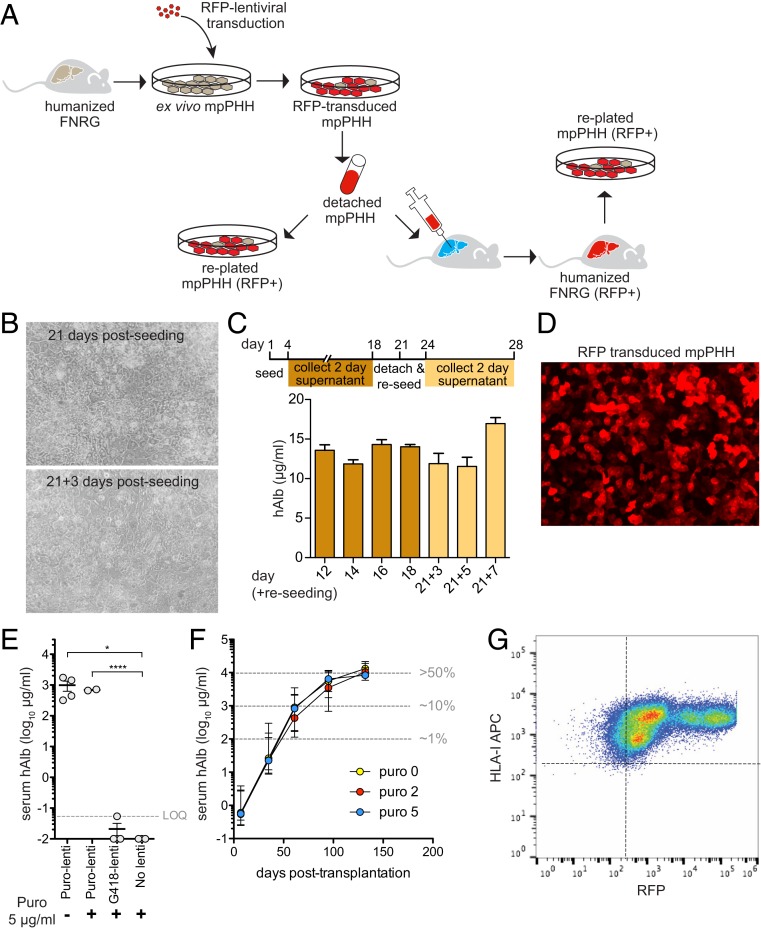
One of the major limitations of PHH is their resistance to efficient genetic manipulation. This stems in part from widespread observations that two-dimensional (2D) PHH cultures cannot survive mobilization, therefore impairing subsequent applications. Given the stability of mpPHH cultures, we aimed to establish protocols to efficiently transduce hepatocytes with lentiviral vectors using red fluorescent protein (RFP) as proof of concept. In addition, we sought to overcome the much larger hurdle of mobilizing transduced mpPHH and either reculture or retransplant these modified hepatocytes to create huFNRG mice with a genetically manipulated graft, as depicted in Fig. 5A.
Fig. 5.
Cultured mpPHH can be mobilized and either recultured or retransplanted, creating a modified human graft for in vitro and in vivo applications. (A) Schematic representation of lentivirus transduction of mpPHH and subsequent detachment for reculturing or retransplantation. Transduced cells are shown in red. (B) mpPHHs were cultured in 12-well plates, and media were changed every 2 d for 3 wk. Cells were then mobilized and reseeded in new plates. Media were changed every 2 d for an additional week. Representative images from cultures before and after detachment and replating are shown. (C) hAlb levels from supernatants from the initial cell culture (dark yellow) and the replated cells (light yellow) were measured and plotted over time. Data from six biological replicates are represented as mean ± SEM. (D) mpPHHs were transduced with RFP-expressing lentivirus and imaged after 3 d. (E) mpPHHs were cultured in six-well plates and transduced with RFP/puro-expressing lentiviruses. Untransduced or transduced with a G418-expressing lentivirus was used as controls. After 3 d, cells were treated with puromycin for 5 d. Cells were detached and transplanted into FNRG mice. Serum hAlb levels 10 wk after transplantation are displayed for individual mice. t test between groups. *P < 0.05; ****P < 0.0001. (F) mpPHHs were transduced with RFP/puro-expressing lentiviruses and after 3 d, treated with different concentrations of puromycin for 5 d. Cells were detached and transplanted into FNRG mice. Serum hAlb levels rose similarly between groups. Median ± range hAlb levels of groups of 9 to 12 transplanted mice. Three to six mice per group survived until day 132. (G) Mouse livers from E were perfused, and mpPHHs were isolated. These cells were analyzed by flow cytometry to quantify RFP expression. (Magnification: B and D, 20×.)
To this end, lentiviral-infected mpPHH cultures were monitored for 3 wk, after which cells were detached and replated. Between 60 and 80% of mpPHHs survived this process and also, successfully established healthy 2D cultures (Fig. 5B) with similar hAlb levels as before detachment (Fig. 5C). We then defined conditions at which more than 90% of mpPHHs were transduced with RFP-lentiviral vectors (Fig. 5D).
We combined these two technical advances for two applications. In the first case, we attempted to create three-dimensional (3D) spheroid hepatocyte cultures, similar to those we routinely achieve via plating mpPHH in U-bottom 96-well plates. However, we observed that lentiviral RFP transduction of preformed spheroids was consistently inefficient (SI Appendix, Fig. S7A). To overcome this problem, we first transduced mpPHH in 2D, detached the cells, and then created spheroid cultures. Following this protocol, more than 90% of the spheroid cells were RFP positive (SI Appendix, Fig. S7 B and C). In addition, by titrating the seeding density, we obtained spheroids of different size (SI Appendix, Fig. S7D). As a second application, we sought to generate liver chimeric mice with a modified human graft. To this goal, we transduced mpPHH with RFP puromycin resistance (RFP-puro-lenti) or G418 resistance-conferring lentiviral vectors. After 5 d of puromycin treatment in culture, mpPHHs were mobilized into single-cell suspensions and transplanted into FNRG mice. Five weeks posttransplantation, we detected hAlb in mice transplanted with RFP-puro-lenti transduced mpPHH regardless of puromycin selection, suggesting that mpPHH tolerated the selection process. As expected, both RFP-G418-lenti transduced and untransduced mpPHH cultures did not survive puromycin treatment as indicated by minimal serum hAlb levels 5 wk after transplantation (Fig. 5E).
To further optimize repopulation of modified mpPHH in FNRG mice, cells were transduced with concentrated RFP-puro-lenti and then treated with 0, 2, and 5 μg/mL puromycin. All mice reached high serum hAlb levels irrespective of puromycin selection (Fig. 5F). Four months after transplantation, immunohistochemistry on chimeric livers showed ∼50% human chimerism (SI Appendix, Fig. S8A), consistent with high serum hAlb values. Flow cytometry from perfused livers of these mice showed that the majority of human hepatocytes expressed RFP (Fig. 5G). Moreover, plated mpPHHs from these animals formed stable cultures and maintained RFP expression (SI Appendix, Fig. S8B).
These data show that mpPHH cultures can be efficiently transduced in 2D format, detached, and used to create highly transduced 3D spheroids or humanized mice with robust, persistent, genetically altered grafts.
Discussion
The increased demand for physiologically relevant hepatocyte systems led the field to develop methods to expand this limited resource beyond the hepatocytes isolated from human livers. Different approaches are based on in vitro expansion by transforming PHH or by differentiating stem cells to immature HLCs. In parallel, in vivo approaches rely on PHH expansion using liver injury mouse models. We show here that the latter approach efficiently expanded PHH numbers by about 150-fold per round of transplantation and reduced the donor-to-donor variability of cryopreserved PHH in terms of plating efficiency. Retrorsine enhanced this process, particularly for poor donors that otherwise cannot be studied in cell culture. We are unsure of the mechanism by which retrorsine improves humanization. In untransplanted FNRG mice cycled off NTBC, retrorsine enhanced mouse hepatocyte Ki67 expression after restarting NTBC and modestly increased intrahepatic FGF7 and EGFR protein levels but did not change p21 expression or cause a ductular reaction. These data are consistent with observations in wild-type mice (23) and distinct from those in rats (14), suggesting that mechanisms may differ from the impaired hepatocyte proliferation widely observed in rats.
Freshly isolated mpPHHs reliably formed cultures that could be maintained for months, which contrasts with previously reported short-term cultures (9). The combination of retrorsine preconditioned huFNRG mice and Percoll enrichment resulted in mpPHH cultures composed of mostly human hepatocytes. Since we did not magnetically deplete mouse cells (21), the long-term stability may, however, depend on a small fraction of undefined mouse stromal support cells. These cultures advance research across a wide spectrum of human hepatocyte biology. For example, following HBV infection and spread for 8 wk allows for studies into the molecular mechanisms by which HBV establishes chronicity. A second application is the adaptation to a 384-well microscale mpPHH format. This will facilitate high-throughput genetic and compound screens in human hepatocytes. Additionally, because yields from chimeric mice are high and mpPHH cultures are robust, plated cells were easily shipped to collaborators who then conducted drug metabolism and Plasmodium infection studies. This illustrates that mpPHH can be widely used by the research community without the need to set up logistically complex chimeric mouse colonies.
The most significant advance of our systems is the ability to efficiently genetically manipulate mpPHH. Although lentiviral manipulation of hepatocytes prior to transplantation has been reported, transduction remained very inefficient (24, 25) (in our laboratory, 5 to 10%). Once plated, PHHs do not proliferate and were generally believed to resist further applications, such as expansion in chimeric mice. Our systems to transduce mpPHH, select transduced cells with puromycin, mobilize these cells, and then either create spheroid cultures or repopulate huFNRG mice to high chimerism have many implications for the study of hepatocyte biology. It allows for the efficient modifications of mpPHH in vitro by either overexpressing or knocking down genes of interest. These modified mpPHHs can then be expanded in mice for in vivo studies, or chimeric mice can be subjected to manipulations, such as infections or diets, and their mpPHHs can subsequently be harvested for in vitro applications. This versatile system will allow for disease-specific genetic manipulations linked to liver physiology and hepatocyte-specific interactions. It will allow for the creation of disease-specific human hepatocyte systems and will advance the long-term goal of therapeutic hepatocyte transplantation for human liver conditions.
Methods
Cells.
Cryopreserved PHHs were purchased from Triangle Research Labs and Lonza or were isolated from surgical specimens and cryopreserved in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% dimethyl sulfoxide (DMSO) and 20% fetal bovine serum (FBS) (11). Human fetal hepatoblasts were isolated from fetal liver tissue (Advanced Bioscience Resources, Inc.) as described previously (13, 26). Briefly, fetal livers were incubated with collagenase containing digestion buffer, and large cells were enriched by density centrifugation before cryopreservation. The use of human materials was approved by the Rockefeller University Institutional Review Board; however, was exempted of informed consent under category 4.
Mice.
Fah−/− mice (27) were provided by Markus Grompe, Oregon Health and Science University, Portland, OR and crossed to NOD Rag1−/− Il2rgnull mice (Jackson Labs) to derive FNRG mice as previously described (11). After the first NTBC withdrawal cycle, we observed 67.3 ± 17.1% survival in FNRG females and 1.8 ± 3.6% survival in FNRG males in our facility. Therefore, only FNRG females were used for these experiments. Thymidine kinase transgenic mice on the NOD SCID Il2rgnull background (TK-NOG) (19) were obtained from Taconic Biosciences. Only males were used since female TK-NOG mice do not support high humanization (19). Immunodeficient SCID mice hemizygous for the uPA transgene were bred at Ghent University as previously described (28). Male uPA+/− mice were used, although both genders support humanization. FNRG mice were maintained on an ad libitum chow diet with amoxicillin and drinking water containing 16 mg/mL NTBC (Yecuris). TK-NOG and uPA mice were maintained on regular chow. Mice were housed under a 12-h light cycle from 7:00 AM (ZT0) to 7:00 PM (ZT12). All experiments were conducted under animal use protocols approved by Rockefeller University and Ghent University.
Mouse Preconditioning.
Retrorsine (catalog no. R0382; Sigma Aldrich) was dissolved at 20 mg/mL in 100% ethanol at 56 °C and stored in aliquots at −20 °C. For intraperitoneal injections, 100 μL (2 mg) retrorsine or 100 μL ethanol vehicle was diluted with 400 μL phosphate buffered saline (PBS) without calcium or magnesium (PBS−/−). For Fah−/− and TK-NOG experiments, retrorsine was administered at 2-wk intervals, typically the first dose at 3 to 4 wk prior to transplantation and a second dose 2 to 3 wk prior to transplantation. In uPA+/− mice, retrorsine was injected 12 to 13 d after birth at 70 mg/kg. For circadian experiments, ZT0 injections were administered between 7:00 AM and 8:00 AM, and ZT12 injections were between 7:00 PM and 8:00 PM. For hOSM, supplementation of an AAV8-hOSM was created by cloning hOSM into the pAAV-CB vector expressing hOSM driven by the cytomegalovirus enhanced chicken β-actin promoter. Seventy-two hours after cotransfecting this construct or a control vector expressing firefly luciferase with the pDG8 helper plasmid into HEK293 cells, virus was purified from cell pellets using an iodixanol density gradient protocol. Titers were determined by dot blot hybridization as described previously (29). Mice received the AAV8-hOSM vector through tail vein injections 7 to 10 d prior to transplantation of human fetal hepatoblasts.
Transplantations.
Female FNRG recipients were withdrawn from NTBC 4 to 5 d prior to surgery. Male TK-NOG mice 7 wk of age received 48 h of 0.1 mg/mL valganciclovir (Sigma Aldrich) in drinking water 14 d prior to transplantation (19). uPA+/− mice were transplanted 8 d after retrorsine injection when they were 20 to 21 d old. Human cells were transplanted as described previously (11). Briefly, under sterile conditions and using isoflurane anesthesia, the skin was cleaned with povidone iodine, and a 0.5- to 1-cm incision was created over the left flank, after which the peritoneum was mobilized and opened. The ventral tip of the spleen was mobilized onto the peritoneum, and human cells were injected into the spleen using a 28-gauge insulin syringe. After hemostasis was achieved by applying pressure to the injection site, the peritoneum was closed using 4.0 Vicryl suture (Ethicon) and the skin was closed using metal Mikron Autoclips (BD Biosciences). Mice received two doses of postoperative buprenorphine (Abbot Animal Health) 0.05 mg/kg for analgesia. Metal clips were removed 14 d after surgery.
Liver Perfusion and Hepatocyte Isolation.
Hepatocytes were isolated from chimeric livers as described (11, 29). Briefly, mice were anesthetized with ketamine/xylazine, and a 24-gauge angiocath was inserted into the inferior vena cava. Then, the portal vein was cut, and the mouse liver was perfused sequentially with PBS−/− supplemented with heparin, Hanks buffered saline solution (HBSS) supplemented with 5 mM ethylenediaminetetraacetic acid (EDTA) and 50 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (Hepes), and finally, HBSS supplemented with 0.05% Type IV collagenase (Sigma Aldrich) and 1 U/mL DNase (Thermo Fisher). After digestion, the liver was disrupted over a 70-μm cell strainer, and the cell suspension (50 mL) was spun at 50 × g for 5 min at 4 °C using an Allegra X-14R Centrifuge (Beckman Coulter). The supernatant was gently aspirated, and the cells were washed once with PBS−/−. The cell pellet was resuspended in 10 mL PBS−/− and gently mixed with equal volume of Percoll working solution (catalog no. 17–0891-01; GE Healthcare). Percoll working solution consisted of 5.4% 10× PBS−/−, 48.6% Percoll, and 46% William’s E medium (catalog no. 12551–032; Life Technologies). The cell suspension was spun at 100 × g for 5 min at 4 °C, and the pellet washed once with PBS. After centrifugation at 50 × g, the cells were resuspended in PBS and left on ice for 45 min. Then, a second round of Percoll was used to further purify the cell suspension. The final pellet was resuspended in W10 plating medium (William’s E medium [WEM] supplemented with 10% FBS, 1% penicillin/streptomycin [catalog no. 15140–12; Life Technologies], 1% 200 mM l-glutamine [catalog no. 25030–081; Life Technologies], 0.1% 50 mg/mL Gentamicin reagent solution [catalog no. 15750–060; Life Technologies], and 0.1% Corning ITS premix [catalog no. 354350; Corning]), and viable cells were counted using trypan blue. Viability was usually above 95% without clumps. If clumps were present, the cell suspension was passed through a 40-μm filter. For adoptive transplant into new FNRG recipient mice, cells were spun at 50 × g for 5 min at 4 °C and resuspended in cold PBS. For each mouse, 65 μL of cell suspension of 0.5 to 1 million cells was injected. For in vitro applications, the cells were kept on ice in W10 plating medium until seeded in various formats.
Hepatocyte Plating and Maintenance.
Once the cells (mpPHH) were isolated and purified with two rounds of Percoll, they were freshly seeded on collagen-coated plates (BD Biosciences) in W10 plating medium. The cells were equally distributed by shaking on a flat surface and then, left on the bench at room temperature for 45 min. Once the cells settled and evenly distributed, they were transferred to a humidified 37 °C incubator. Cryopreserved PHHs were first thawed at 37 °C and then, transferred to 50 mL W10. After centrifugation at 50 × g, the cells were resuspended in W10, counted, and seeded on plates as described for mpPHH. For each plating format, we optimized the seeding density to achieve confluent cultures. To avoid concentrating cells in the middle of the well, we seeded the cells with excess medium. The next day, the cells were washed once with WEM to remove any cell debris and serum. For maintenance medium, we used hepatocyte-defined medium (HDM; catalog no. 05449; Corning) supplemented with 1% penicillin/streptomycin, 1% 200 mM l-glutamine, 0.1% 50 mg/mL Gentamicin, and 2% DMSO (catalog no. 4-x-5; ATCC). Unless otherwise stated, medium was changed every other day. Table 1 summarizes the different formats with seeding densities and medium volumes.
Table 1.
mpPHH seeding density in various plating formats
| Plate format | No. of cells per well | Volume (plating) | Volume (maintenance) |
| 384 well | 10,000 | 50 μL | 50 μL |
| 96 well | 40,000 | 100 μL | 100 μL |
| 24 well | 350,000 | 1.5 mL | 0.5 mL |
| 12 well | 800,000 | 2 mL | 1 mL |
| 6 well | 1,300,000 | 4 mL | 2 mL |
Lentivirus Transduction.
Confluent mpPHH cultures were transduced with RFP-expressing lentiviral pseudoparticles (lentiviral vector SCRPSY expressing red fluorescent protein [SCRPSY-RFP]) in the presence of 4 μg/mL polybrene by spinoculation for 1 h at 1,000 × g at 37 °C. Three days posttransduction, RFP expression reached maximum levels and remained stable over time.
Mobilization of mpPHH for Replating and Retransplantation.
Untransduced or RFP-transduced mpPHH cultures were maintained from days to weeks before a detachment protocol was applied. Specifically, after multiple washes with WEM, the cultures were incubated for 30 min with HBSS (catalog no. 14175–095; Life Technologies) with 10-min interval washes to loosen hepatocyte tight junctions. Then, the cells were incubated with TrypLE Select (10×; catalog no. A12177-01; Life Technologies) for 4 to 5 min, and the cells were gently resuspended in W10 medium. After a 5-min spin at 50 × g, the cells were resuspended in W10, counted, and replated or retransplanted into FNRG mice.
Spheroid 3D Culture Formation.
Confluent mpPHH cultures were transduced with RFP-expressing lentiviral VSV-G pseudoparticles (SCRPSY-RFP) as described above. Ten days later, the cells were detached by brief treatment with TrypLE, washed, and plated in 100 μL W10 plating medium in Ultra-Low Attachment U-bottom 96-well plates (catalog no. 7007; Costar). Plates were centrifuged at 40 × g for 5 min to promote aggregate formation. Spheroid aggregates formed between 1 and 3 d after plating, at which time the cultures were switched to serum-free medium. Spheroids of different size were formed depending on the indicated number of plated cells after 10 d in culture. Spheroid images were acquired on an inverted Zeiss Axiovert 200 microscope fitted with a Perkin-Elmer UltraView spinning disk and Andor iXon 512 × 512 electron multiplying CCD camera; 100 image slices were acquired with 3-μm steps in both the bright-field and fluorescent channels using either transmitted light or 561-nm laser excitation (Spectral Applied) and a 10× Plan-ApoChromat 0.45 numerical aperture objective. Image analysis and editing were performed using ImageJ.
siRNA Transfection and CYP3A4 Activity.
ON-TARGETplus SMARTpool siRNA oligonucleotides (CYP3A4, catalog no. L-008169010005; nontargeting control pool, catalog no. D-001810–10-05; Dharmacon) were delivered to hepatocytes using RNAiMAX Transfection Reagent (Thermo Fisher Scientific) per the manufacturer’s protocols at final concentration of 100 nM in antibiotic-free media (final volume 100 μL). Triplicate wells containing mpPHH were exposed to siRNA duplexes overnight for 24 h and subsequently cultured in supplemented hepatocyte media. Knockdown efficiency was determined by qRT-PCR using specific CYP3A4 primers and using a cell-based P450-Glo CYP3A4 Assay with luciferin-IPA (catalog no. V9002; Promega) according to the manufacturer’s instructions.
Drug Treatments.
Acetaminophen (catalog no. A7085; Sigma), diclofenac (catalog no. D6899; Sigma), clozapine (catalog no. C6305; Sigma), and atorvastatin (catalog no. 10493; Cayman chemical) were dissolved in DMSO and added to mpPHH cultures at the final concentrations indicated.
Plasmodium Infection and Detection.
Pf sporozoites were isolated from salivary glands of Anopheles stephensi bred at John Hopkins University Malaria Research Institute or Harvard School of Public Health. For the entry assay, mpPHHs were exposed to 10,000 sporozoites, washed 3 h later with PBS, and fixed with 4% paraformaldehyde for 15 min at room temperature. After blocking for 30 min in PBS with 2% bovine serum albumin, parasites were detected using a sequential double-staining assay. Cells were first incubated for 1 h with anti-Pf circumsporozoite protein (CSP) antibodies followed by incubation with secondary antibodies and AlexaFluor 488 for another hour. After washing, cells were permeabilized with ice-cold methanol for 10 min at 4 °C and again incubated with anti-PfCSP antibodies and AlexaFluor 594 for 1 h each, with washing in between. Finally, cells were counterstained with Hoechst 33258 (1:5,000), mounted in Aquamount (Lerner Laboratories), and analyzed on a Nikon Eclipse Ti fluorescence microscope. For parasite development assessment, mpPHHs were infected with 40,000 sporozoites, washed 3 h later, and maintained in hepatocyte media supplemented with penicillin/streptomycin (300 μg/mL). Cultures were fixed in ice-cold methanol for 10 min at 4 °C at days 2 and 4 postinfection. Parasites were detected using Pf heat shock protein 70 (HSP70) antibodies followed by AlexaFluor 594 secondary antibodies for 1 h each. Nuclei and cells were mounted as described above. Images were captured, and the parasite area was measured using NIS-Elements Advanced Research imaging software (Nikon).
HBV Infection and Detection.
HBV stock was prepared as previously described (30). Briefly, supernatants from confluent HepDE19 cells (31) (HBV-producing cells) were harvested every 2 d for 3 wk and concentrated using Centricon Plus-70 centrifugal filter devices (Millipore-Sigma). The concentrated stock was aliquoted and kept at −80 °C. The stock concentration was determined as HBV genome equivalents (GEQ) per milliliter using a TaqMan-based qPCR assay. For HBV infections, 4 to 5 d after mpPHH seeding the cells were infected with the indicated concentration of HBV in hepatocyte maintenance medium (HDM with 2% DMSO) supplemented with 4% polyethylene glycol 8000 (catalog no. 81268–250G; Sigma). The plates were spinoculated for 1 h at 1,000 × g at 37 °C. After 24 h, the cells were washed 5× with WEM, and maintenance medium was added. For the 384-well plate format, 500 nM Myrcludex B (32) (HBV entry inhibitor) was incubated with the inoculum and washed out the next day. Secreted HBeAg was measured from culture supernatants every 4 d by loading 50 μL into 96-well plates of a chemiluminescence immunoassay kit according to the manufacturer’s instructions (DiaSino Laboratories Co.). A FLUOstar Omega luminometer was used to read the plates. HBeAg concentrations are expressed in Paul Ehrlich Institute units per milliliter. Hepatocyte cultures were fixed in 4% paraformaldehyde for 20 min at room temperature, washed with PBS, and permeabilized with 0.1% Triton X-100 for 10 min. After extensive washing, cells were incubated for 1 h at room temperature with blocking solution 5% goat serum in PBS (catalog no. 005–000-121; Jackson ImmunoResearch). A rabbit polyclonal anti-HBV core antibody (catalog no. HBP-023–9; Austral Biologicals) was added to the cells at 1:500 dilution in blocking solution and incubated at 4 °C overnight. A goat anti-rabbit AlexaFluor 594 (catalog no. A-11012; Life Technologies) at a dilution of 1:1,000 was used as a secondary antibody. Nuclei were stained with DAPI. Cells were imaged using a Nikon Eclipse TE300 fluorescent microscope and processed with ImageJ.
Immunohistochemistry.
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded murine liver tissues using mouse anti-human FAH (clone 2; Abcam) (13) and rabbit NuMA (Abcam) staining performed as described previously (18). In brief, sections were deparaffinized in xylene (three changes), rehydrated through graded alcohols (three changes 100% ethanol, three changes 95% ethanol), and rinsed in distilled water. Antibody incubation and detection were carried out on a NEXes instrument (Ventana Medical Systems) using Ventana’s reagent buffer and i-View detection kit. Endogenous peroxidase activity was blocked with hydrogen peroxide. Heat-induced epitope retrieval was performed in a 1,200-W microwave oven at 100% power in 10 mM sodium citrate buffer, pH 6.0, for 20 min. Sections were allowed to cool for 30 min and then, rinsed in distilled water. Mouse anti-human FAH or rabbit anti-human NuMA was diluted in PBS and incubated overnight at room temperature. Primary antibody was detected with biotinylated goat anti-mouse followed by application of streptavidin–horseradish peroxidase conjugate. The complex was visualized with 3,3 diaminobenzidine and enhanced with copper sulfate. Matched immunoglobulin isotype, at equivalent concentration and diluted in PBS, was used as negative control. On completion of staining, all slides were washed in distilled water, counterstained with hematoxylin, dehydrated, and mounted with permanent media. Stained slides were scanned at 40× magnification using the Leica Microsystems SCN 400F Whole Slide Scanner. Images were viewed and captured using SlidePath’s Digital Image Hub (Leica Microsystems).
Enzyme-Linked Immunosorbent Assay (ELISA) for Albumin, Transferrin, and Alpha-Antitrypsin.
Serum was obtained from tail veins or retroorbital plexus. Culture supernatants were obtained at indicated time points. hAlb, human transferrin, and human alpha-antitrypsin were quantified using sandwich ELISA methods (Bethyl Labs).
Urea Assay.
Urea concentration in culture supernatants was measured using a colorimetric assay based on the diacetyl-monoxime method (catalog no. 0580–250; Standbio Labs).
Flow Cytometry.
Cells were fixed and stained for human and mouse CD81; human leukocyte antigen (HLA)-A, B, C (BD Biosciences) and RFP expression were measured on an LSRII flow cytometer (BD Biosciences) as described previously (29). Data were analyzed using FlowJo software.
Fluorescence Staining and Imaging.
For immunofluorescence staining, mpPHH cultures were fixed and stained using as primary antibodies a rabbit polyclonal anti-NuMa antibody (catalog no. ab97585; Abcam) and a mouse monoclonal anti-CK18 antibody (catalog no. ab82254; Abcam). AlexaFluor 488 and 594 (Thermo Fisher) of the corresponding species were used as secondary antibodies. Nuclei were stained with DAPI. Cells were imaged using a Nikon Eclipse TE300 fluorescent microscope and processed using ImageJ.
RNA Extraction and Library Preparation.
Total RNA was isolated using TRIzol (catalog no. 15596026; Thermo Fisher) followed by cleaning with RNeasy kit (catalog no. 74104; Qiagen) coupled with on-column DNase I treatment (catalog no. 79254; Qiagen); 250 ng of the DNase-treated RNA was used to generate strand-specific sequencing libraries using TruSeq Stranded mRNA Library Prep (catalog no. 20020594; Illumina). Multiplexed libraries were sequenced as 50-nt single-end reads on a HiSeq2500 system (Illumina) at the Rockefeller University Genomics core facility.
RNA-sequencing Bioinformatics Pipeline.
Transcript alignment and quantification were done using Kallisto (33). Transcript indices for Kallisto were generated from Ensembl release hg38 and included all annotated complementary DNA and noncoding RNA transcripts. Differential expression analysis was then done using Sleuth (34). Gene names and attributes were queried from Ensembl using biomaRt. For genes containing multiple transcripts, we chose the transcript with the highest mean expression across all samples as the representative example for that gene. Heat maps were generated in R Studio.
Statistical Analyses.
Groups were compared by the Student t test. Correlations were calculated using the Spearman correlation coefficient. All statistical analyses were done using Prism 8 software (Graphpad).
Data Availability.
Datasets supporting the findings of this study have been deposited in a publicly available database (Gene Expression Omnibus accession no. GSE130219).
Supplementary Material
Acknowledgments
This work was supported by NIH Fellowship F32DK107164 (to E.M.); fellowships from the German National Academic Foundation and the German Center for Infection Research (to P.P.); and NIH Grants R01HL131093 (to R.W.H. and Y.P.d.J.), R01AI091707 (to C.M.R.), R01DK085713 (to C.M.R.), and K08DK090576 and R01AA027327 (to Y.P.d.J.). K.V. was supported by a Fellowship of the Belgian American Educational Foundation. Further support for this project was provided by grants from the Robertson Foundation (to E.M.). The project was cosponsored by the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable trust (E.M.). This work was supported in part by Bill & Melinda Gates Foundation Grant OPP1023607 (to S.N.B.), Koch Institute Support Grant P30-CA14051 (to S.N.B.) from the National Cancer Institute, and Bloomberg Philanthropies (S.N.B.), which supports the insectary and parasitology core facilities at the Johns Hopkins Bloomberg School of Public Health. S.N.B. is a Howard Hughes Medical Institute Investigator. We thank Dr. Stephan Urban (Heidelberg University) for providing Myrcludex B and members of our laboratories, including Heather Fleming, for helpful suggestions on the work and editing of the manuscript. We also thank the Rockefeller University High-Throughput, Bioimaging, Flow Cytometry and Genomics Resource Centers. The following reagent was obtained through BEI Resources, National Institute of Allergy and Infectious Diseases, NIH: Monoclonal Antibody 2A10 Anti-Plasmodium falciparum Circumsporozoite Protein (produced in vitro), MRA-183A, contributed by Elizabeth Nardin. We thank Julie Vercauteren for graphical assistance.
Footnotes
The authors declare no competing interest.
Data deposition: Datasets supporting the findings of this study are deposited in the publicly available database Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo (accession no. GSE130219).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919035117/-/DCSupplemental.
References
- 1.Kaplowitz N., Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 4, 489–499 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Kidambi S., et al. , Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc. Natl. Acad. Sci. U.S.A. 106, 15714–15719 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gural N., Mancio-Silva L., He J., Bhatia S. N., Engineered livers for infectious diseases. Cell. Mol. Gastroenterol. Hepatol. 5, 131–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heslop J. A., Duncan S. A., The use of human pluripotent stem cells for modeling liver development and disease. Hepatology 69, 1306–1316 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Chen C., Soto-Gutierrez A., Baptista P. M., Spee B., Biotechnology challenges to in vitro maturation of hepatic stem cells. Gastroenterology 154, 1258–1272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu H., et al. , Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606 e19 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Fu G. B., et al. , Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 29, 8–22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy G., et al. , Long-term culture and expansion of primary human hepatocytes. Nat. Biotechnol. 33, 1264–1271 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Azuma H., et al. , Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat. Biotechnol. 25, 903–910 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bissig K. D., et al. , Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J. Clin. Invest. 120, 924–930 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jong Y. P., et al. , Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci. Transl. Med. 6, 254ra129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grompe M., et al. , Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 10, 453–460 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Billerbeck E., et al. , Humanized mice efficiently engrafted with fetal hepatoblasts and syngeneic immune cells develop human monocytes and NK cells. J. Hepatol. 65, 334–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laconi E., et al. , Long-term, near-total liver replacement by transplantation of isolated hepatocytes in rats treated with retrorsine. Am. J. Pathol. 153, 319–329 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlke M. H., et al. , Liver regeneration in a retrorsine/CCl4-induced acute liver failure model: Do bone marrow-derived cells contribute? J. Hepatol. 39, 365–373 (2003). [DOI] [PubMed] [Google Scholar]
- 16.White I. N., Mattocks A. R., Butler W. H., The conversion of the pyrrolizidine alkaloid retrorsine to pyrrolic derivatives in vivo and in vitro and its acute toxicity to various animal species. Chem. Biol. Interact. 6, 207–218 (1973). [DOI] [PubMed] [Google Scholar]
- 17.Samuel A., Jago M. V., Localization in the cell cycle of the antimitotic action of the pyrrolizidine alkaloid, lasiocarpine and of its metabolite, dehydroheliotridine. Chem. Biol. Interact. 10, 185–197 (1975). [DOI] [PubMed] [Google Scholar]
- 18.Raven A., et al. , Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa M., et al. , The reconstituted ‘humanized liver’ in TK-NOG mice is mature and functional. Biochem. Biophys. Res. Commun. 405, 405–410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer D. F., et al. , Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7, 927–933 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Ishida Y., et al. , Novel robust in vitro hepatitis B virus infection model using fresh human hepatocytes isolated from humanized mice. Am. J. Pathol. 185, 1275–1285 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Nishimura M., et al. , Induction of human CYP1A2 and CYP3A4 in primary culture of hepatocytes from chimeric mice with humanized liver. Drug Metab. Pharmacokinet. 20, 121–126 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Zhou X. F., Wang Q., Chu J. X., Liu A. L., Effects of retrorsine on mouse hepatocyte proliferation after liver injury. World J. Gastroenterol. 12, 1439–1442 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bissig K. D., Le T. T., Woods N. B., Verma I. M., Repopulation of adult and neonatal mice with human hepatocytes: A chimeric animal model. Proc. Natl. Acad. Sci. U.S.A. 104, 20507–20511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bierwolf J., et al. , Primary human hepatocytes repopulate livers of mice after in vitro culturing and lentiviral-mediated gene transfer. Tissue Eng. Part A 22, 742–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrus L., et al. , Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology 54, 1901–1912 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grompe M., et al. , Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 7, 2298–2307 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Meuleman P., et al. , Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 41, 847–856 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Vercauteren K., et al. , Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol. Ther. 24, 1042–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michailidis E., et al. , A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci. Rep. 7, 16616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai D., et al. , Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob. Agents Chemother. 56, 4277–4288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni Y., et al. , Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146, 1070–1083 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Pimentel H., Bray N. L., Puente S., Melsted P., Pachter L., Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets supporting the findings of this study have been deposited in a publicly available database (Gene Expression Omnibus accession no. GSE130219).