Significance
Infectious agents, including viruses, activate cellular inflammasomes. Viruses have evolved strategies to parry such host defense mechanisms, and cancer-causing gammaherpesviruses such as Epstein−Barr virus (EBV) are no exception. In distinct contrast, we show that this ubiquitous virus exploits assembly of the NLRP3 inflammasome to kick-start its lytic cascade; this allows EBV to reactivate from quiescence, propagate, and leave the cell when the cell perceives intrinsic or extrinsic threats. This discovery also provides a predictive signature that marks cells in which EBV is poised to enter the lytic phase. Importantly also, with hyperglycemia known to activate the NLRP3 inflammasome, our findings provide a mechanistic basis for recent clinical observations of diabetes as a significant risk factor for EBV-lymphomas in transplant recipients.
Keywords: inflammasome, diabetes, TXNIP, Epstein−Barr virus, KAP1
Abstract
Viruses activate inflammasomes but then subvert resulting inflammatory responses to avoid elimination. We asked whether viruses could instead use such activated or primed inflammasomes to directly aid their propagation and spread. Since herpesviruses are experts at coopting cellular functions, we investigated whether Epstein−Barr virus (EBV), an oncoherpesvirus, exploits inflammasomes to activate its replicative or lytic phase. Indeed, our experiments reveal that EBV exploits several inflammasome sensors to actually activate its replicative phase from quiescence/latency. In particular, TXNIP, a key inflammasome intermediary, causes assembly of the NLRP3 inflammasome, resulting in caspase-1−mediated depletion of the heterochromatin-inducing epigenetic repressor KAP1/TRIM28 in a subpopulation of cells. As a result, only TXNIPhiKAP1lo cells, that is, in a primed/prolytic state, turn expression of the replication/lytic/reactivation switch protein on to enter the replicative phase. Our findings 1) demonstrate that EBV dovetails its escape strategy to a key cellular danger-sensing mechanism, 2) indicate that transcription may be regulated by KAP1 abundance aside from canonical regulation through its posttranslational modification, 3) mechanistically link diabetes, which frequently activates the NLRP3 inflammasome, to deregulation of a tumor virus, and 4) demonstrate that B lymphocytes from NOMID (neonatal onset multisystem inflammatory disease) patients who have NLRP3 mutations and suffer from hyperactive innate responses are defective in controlling a herpesvirus.
Inflammasomes are intracellular multiprotein oligomers that form in response to cell-intrinsic and cell-extrinsic threats. Activation of the inflammasome results in secretion of proinflammatory cytokines IL-1β and IL-18 and can ultimately result in pyroptosis. The inflammasome may be activated by cell-intrinsic damage signals such as reactive oxygen species (ROS), lysosomal instability, and changes in intracellular electrolyte levels. Importantly, as a component of the innate immune system, inflammasome sensors detect foreign molecules, thus forming the first line of defense against infections (1). As a result, infectious agents may be silenced, as in the case of herpesviruses HSV1 and HCMV, or eliminated (2). However, most pathogens successfully avoid elimination by subverting the inflammasome (3, 4). That said, there is also an evolving appreciation of links between inflammation and cancer. Given the constant engagement of inflammasomes with threats to the cell, we looked to a virus to investigate this link—a virus that is highly prevalent, persists for the lifetime of the host, and is able to cause cancer.
As a herpesvirus that not only persists in its host but also causes cancer, Epstein−Barr virus (EBV) is a master manipulator of its host (5). A World Health Organization Group I carcinogen, EBV infects nearly all humans. Although most infections are asymptomatic, EBV is causal to several cancers, including immunocompromise-associated (e.g., posttransplant) lymphomas (PTLD), Burkitt lymphoma (BL), and nasopharyngeal cell carcinoma (6, 7). While EBV expresses several oncoproteins during its lifelong quiescence or latency, transition to its replicative/productive/lytic phase is essential for its persistence in the population as well as causing cancer (8–11). Enhanced transition into the productive phase has also been linked to autoimmune proinflammatory conditions like lupus and multiple sclerosis (12, 13). Since the inflammasome constantly surveys its environment and is frequently activated by a variety of infectious and noninfectious danger triggers, we asked whether inflammasome components or primed/activated inflammasomes provoke EBV to transition into its productive or replicative phase.
We find that EBV actively uses several inflammasome sensors to activate its propagation. Specifically, it uses NLRP3 to turn the EBV replication switch on in response to lytic triggers. Cellular stressors such as hyperglycemia and ROS are known to activate the NLRP3 inflammasome through interactions between NLRP3 and the cellular arrestin protein TXNIP (14). In earlier work, we have demonstrated that the master epigenome regulator KAP1/TRIM28 serves as a barrier to EBV lytic cycle entry by enforcing heterochromatin at EBV lytic genes including BZLF1 (15–17). Our experiments now show that lytic triggers cause assembly of the TXNIP−NLRP3 inflammasome to activate procaspase-1; active caspase-1 then causes partial loss of KAP1/TRIM28 to turn the expression of the viral replication switch protein on within a subpopulation of cells, thereby disrupting quiescence in those cells. Furthermore, this lytic cascade is spontaneously turned on in B cells from NOMID (neonatal onset multisystem inflammatory disease) patients whose myeloid-derived cells are known to exhibit hyperactive inflammatory responses due to a mutation in their NLRP3 gene; our findings therefore also uncover a B lymphocyte-related deficiency in controlling EBV in these patients. Importantly, we find that high glucose functions through the NLRP3 inflammasome to disrupt EBV quiescence, implicating diabetes, also known to activate the same inflammasome, in reactivation of the most prevalent cancer-causing virus.
Results
Inflammasome Components Regulate the Viral Replication Switch.
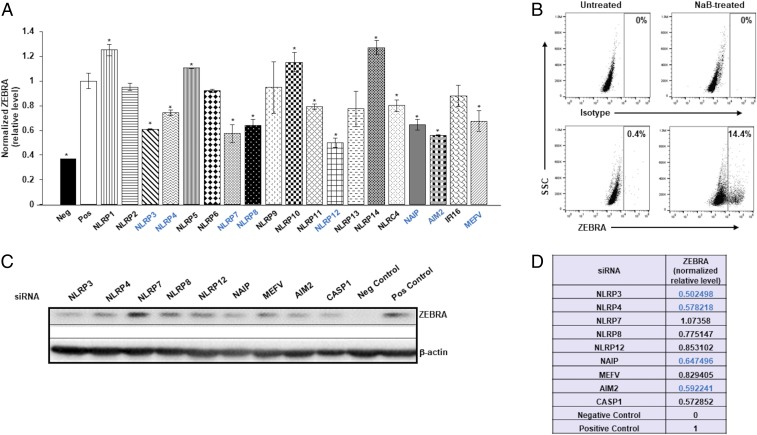
To test the contribution of inflammasome components on the EBV replication switch, we knocked down inflammasome sensor proteins while exposing quiescent/latent EBV+ BL cells (HH514-16) to histone deacetylase inhibitor (HDACi) sodium butyrate (NaB), a chemical inducer of the EBV replicative cycle. On an in-cell Western screen, knockdown of inflammasome sensors resulted in significant changes (gain or loss) in the EBV replication switch protein ZEBRA (Fig. 1A). Since we detected somewhat higher than expected signal in the negative control samples, we verified the level of spontaneous lytic activation in untreated samples; as shown in Fig. 1B, only 0.4% of cells were spontaneously lytic, compared to 14.4% of lytic cells after treatment with NaB. For eight sensors whose knockdown demonstrated at least 25% reduction in ZEBRA in Fig. 1A, we followed up with confirmatory ZEBRA immunoblotting. Of the eight, we validated four sensors, with knockdown of NLRP3 demonstrating the greatest effect on ZEBRA (Fig. 1 C and D). Based on the low numbers of ZEBRA+ cells (Fig. 1B) and lack of detectable ZEBRA protein by immunoblotting (Fig. 1C) in untreated cells, we believe that autofluorescence may have contributed to the signal in negative control samples in in-cell Western experiments (Fig. 1A). Given these results and knowing that a wide variety of danger and pathogen-associated signals can activate NLRP3, we proceeded to investigate its contribution toward activation of the EBV replication switch.
Fig. 1.
Inflammasome sensors regulate the EBV replication switch. (A) HH514-16 BL cells were exposed to control siRNA or inflammasome sensor-directed siRNAs, treated with the replicative/lytic cycle-inducing agent NaB 24 h later, and harvested after another 24 h to assay intracellular levels of the EBV replication switch protein ZEBRA and GAPDH by in-cell Western. (B) HH514-16 BL cells were treated with NaB (versus untreated control), harvested 24 h later, stained with anti-ZEBRA antibody, and subjected to flow cytometry. ZEBRA+ gate was set based on staining with isotype control antibody; numbers represent percent ZEBRA+ cells. (C) Cells treated and harvested as in A were immunoblotted for ZEBRA and β-actin. Negative (Neg) control, untreated cells exposed to control siRNA; positive (Pos) control, NaB-treated cells exposed to control siRNA in A and C. Data in A represent means of three independent experiments; error bars, SEM; *P ≤ 0.05. Sensors whose knockdown resulted in at least 25% reduction in ZEBRA levels compared to control (Pos) in A were tested in C. (D) ZEBRA levels (normalized to β-actin) in siRNA-knocked down cells relative to control siRNA but NaB-treated cells (Pos; shown in C). Inflammasome components whose knockdown resulted in at least 25% loss of ZEBRA in A and D (via in-cell Western and immunoblot) are indicated in blue.
NLRP3 Functions via TXNIP to Activate the Viral Replication Switch.
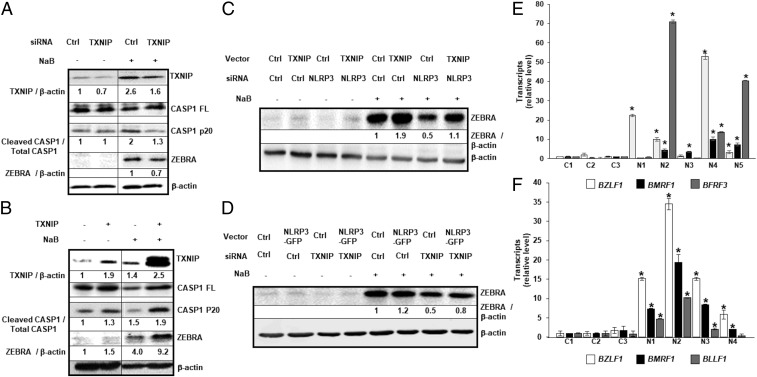
The cellular arrestin protein TXNIP, involved in redox stress pathways and metabolism, can interact with and activate the NLRP3 inflammasome (14). Knockdown of TXNIP resulted in less ZEBRA as well as less cleaved/active caspase-1 relative to control (Fig. 2A); activation of the inflammasome is expected to result in cleavage of caspase-1. In contrast, overexpression of TXNIP in the presence of the lytic trigger resulted in higher levels of both ZEBRA and cleaved caspase-1 (Fig. 2B). We saw similar impacts of TXNIP knockdown on the ability of EBV to activate from latency in an independent EBV+ BL cell line (Akata) (SI Appendix, Fig. S1); EBV in Akata cells is activated through cross-linking of cell surface IgG, a lytic trigger considered more physiologic by some. Importantly, overexpression of TXNIP was also able to partially rescue blunted levels of ZEBRA resulting from NLRP3 knockdown (Fig. 2C and SI Appendix, Fig. S2A). Similarly, in a parallel experiment, we observed that NLRP3 overexpression partially rescued diminished levels of ZEBRA that resulted from knockdown of TXNIP (Fig. 2D and SI Appendix, Fig. S2B). Moreover, knockdown of TXNIP or NLRP3 also resulted in release of fewer encapsidated EBV particles (SI Appendix, Fig. S3). Of note is that activation of the EBV replicative phase did not enhance the ratio of cleaved to full-length IL-1β (SI Appendix, Fig. S4). Thus, TXNIP and NLRP3 cooperate to activate the replication switch protein ZEBRA in the presence of a known lytic trigger, turning the EBV replicative cycle on.
Fig. 2.
NLRP3 activates the EBV replication switch via TXNIP. (A–D) HH514-16 cells were transfected with (A) TXNIP-directed (or control) siRNA, (B) TXNIP (or control) plasmid, and (C and D) combinations of plasmids and siRNAs, exposed to NaB 18 h later, and harvested after another (A and B) 12 h or (C and D) 24 h for immunoblotting with indicated antibodies. Numbers represent ratios between indicated proteins. (E) PBMC from healthy subjects (controls [C]) and NOMID patients (N) were infected with EBV in the presence of FK506 and analyzed 24 h later for transcripts from EBV lytic cycle genes BZLF1, BMRF1, and BFRF3 by qRT-PCR. Transcript levels in each sample were compared to those in control sample 1. (F) Three-week-old EBV-transformed LCL derived from healthy subjects (C) and NOMID patients (N) were harvested for analysis of EBV lytic transcripts BZLF1, BMRF1, and BLLF1 by qRT-PCR. Data represent averages of three independent experiments; error bars, SEM; *P ≤ 0.05.
To address whether EBV residing in cells with preactivated NLRP3 inflammasomes spontaneously switches into the lytic phase, we infected B lymphocytes from patients with NOMID syndrome, a disease characterized by constitutively active NLRP3 inflammasomes (18). Shown in SI Appendix, Table S1 are the NLRP3 mutations in five NOMID patients; all mutations were of germline origin. Following EBV infection, newly infected cells as well as established lymphoblastoid cell lines (LCL) from NOMID B cells demonstrated significantly higher levels of EBV lytic transcripts compared to healthy subject-derived newly infected cells and LCL (Fig. 2 E and F). In addition, NOMID LCL were more likely to undergo spontaneous lytic activation, as shown by ZEBRA+ single cells using flow cytometry (SI Appendix, Fig. S5A). Furthermore, NOMID LCL expressed more lytic proteins, higher levels of cleaved caspase 1, and increased release of encapsidated EBV (SI Appendix, Fig. S5 B and C). Therefore, not only does NLRP3 activate the viral replication switch via TXNIP, but a hyperactive NLRP3 inflammasome (resulting from constitutively active NLRP3) is sufficient, even in the absence of a lytic inducing agent, to activate the EBV replication switch and drive the EBV lytic cycle.
NLRP3 Inflammasome Is Activated Upstream of the Viral Replication Switch.
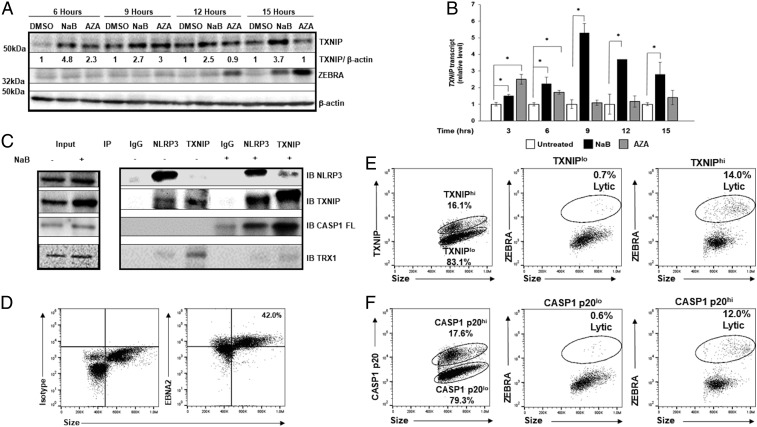
Signals such as increased glucose and ROS result in increased levels of available TXNIP protein, which then triggers the assembly of the inflammasome (14, 19, 20). We therefore examined the temporal relationship of TXNIP levels to expression of the EBV replication switch protein following exposure of cells to two different lytic inducing agents, NaB (an HDAC inhibitor) and AZA (a DNA methyltransferase inhibitor). Compared to control cells, TXNIP transcript and protein levels rose by 3 and 6 h, respectively, whereas ZEBRA protein expression lagged until 12 to 15 h (Fig. 3 A and B). We also observed similar early accumulation of TXNIP transcripts in Mutu I cells (another EBV+ BL cell line) known to respond to both NaB and AZA (SI Appendix, Fig. S6). In contrast, TXNIP levels did not rise in response to Valproic acid (VPA), an HDACi known not to activate EBV from quiescence (SI Appendix, Fig. S7) (21). Remarkably, following cross-linking of IgG on Akata cells, we also observed rapid increases in TXNIP and cleaved caspase-1; these preceded the rise in ZEBRA levels (SI Appendix, Fig. S8). Thus, TXNIP levels rise early in response to distinct lytic triggers in three types of latently infected cells.
Fig. 3.
Early increase in TXNIP and activation of the NLRP3 inflammasome precedes expression of the replication switch. (A and B) HH514-16 cells were exposed to lytic triggers NaB and 5-AZA-2′-deoxycytidine (versus dimethyl sulfoxide [DMSO] control) and harvested at indicated time points for immunoblotting with (A) indicated antibodies or (B) qRT-PCR. Numbers in A represent ratios between indicated proteins. Experiments were performed at least three times, and data in B represent averages of three independent experiments; error bars, SEM; *P ≤ 0.05. (C) HH514-16 cells were treated with NaB (versus untreated control) for 6 h, cell lysates were immunoprecipitated using indicated antibodies (IP), and bound proteins were detected by immunoblotting with indicated antibodies (IB). Input lysates are shown on the left. (D–F) CD19+ B cells from the blood of a patient with PTLD were analyzed by flow cytometry for EBNA2 and size to demarcate EBNA2+ “blast-like” large cells for further analysis; number indicates percent EBNA2+ cells (D). EBV-infected B blast-like cells were analyzed by flow cytometry for TXNIP (E) or cleaved caspase-1 (CASP1 p20; F) expression (Left dot plots), and percent TXNIPhi versus percent TXNIPlo cells (E) or percent CASP1 p20hi versus percent CASP1 p20lo cells (F) expressing the lytic switch protein ZEBRA are shown (Right dot plots). Data from a representative of 3 patients is shown.
We then assessed the effect of TXNIP rise on inflammasome assembly. Indeed, as early as 6 h after exposure to NaB, TXNIP was found in complex with NLRP3 and procaspase-1; this inflammasome assembly was not observed in the absence of NaB (Fig. 3C). Furthermore, thioredoxin-1 (TRX1)−TXNIP complexes, which are typically disrupted in response to ROS (14), were also disrupted concurrent with the assembly of inflammasome in response to NaB (Fig. 3C). These results indicate that increased TXNIP expression as well as dissociation of TXNIP from TRX1 cause activation of NLRP3 inflammasome before ZEBRA expression begins.
To determine the relationship between TXNIP accumulation, caspase-1 cleavage, and ZEBRA expression at the single-cell level and to confirm the relationship between inflammasome activation and activation of the viral replication switch in vivo, we examined proliferating EBV-infected B lymphocytes in the blood of patients with EBV PTLD (Fig. 3D). Fig. 3 E and F shows that cells expressing high levels of TXNIP and cleaved/activated caspase-1 were much more likely to express ZEBRA compared to infected cells expressing low levels of these inflammasome components; moreover, 83% and 72% of lytic cells expressed high levels of activated caspase-1 and TXNIP, respectively. Taken together, these results indicate that NLRP3−TXNIP inflammasome assembly precedes activation of the EBV replication switch and that molecular indicators of active inflammasomes (including cleaved caspase-1) mark B lymphocytes that support the EBV replicative cycle in human blood.
Inflammasome-Activated Caspase-1 Activates the Viral Replication Switch via KAP1.
We have shown that the epigenetic silencer KAP1 transcriptionally restricts ZEBRA (15). Also, KAP1 has two predicted caspase-1 cleavage sites. We therefore tested the effects of modulating caspase-1 levels and activity on KAP1 levels and ZEBRA expression. Exposure to the pan-caspase inhibitor ZVAD, the caspase-1−specific inhibitor YVAD, and small interfering RNA (siRNA) to caspase-1 all resulted in lower levels of ZEBRA accompanied by recovery of KAP1 levels (SI Appendix, Fig. S9 A–C). In contrast, compared to control, overexpression of caspase-1 resulted in increase in the level of ZEBRA and a greater loss of KAP1 (SI Appendix, Fig. S9D). Therefore, caspase-1 activates ZEBRA via depletion of KAP1.
TXNIPhiKAP1lo Cells Represent Cells in a Prolytic State.
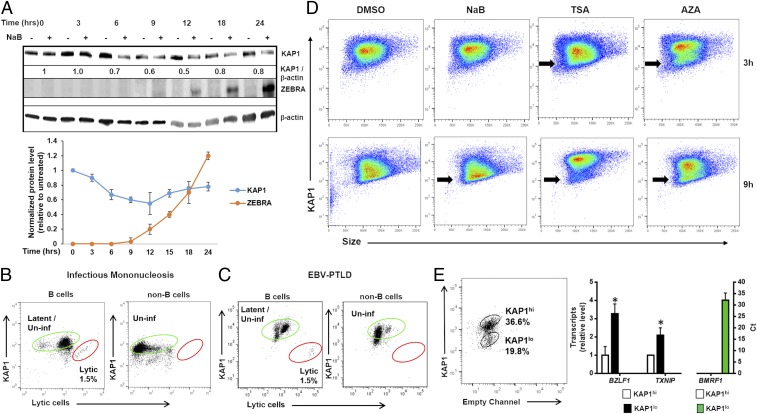
Functional relationships between caspase-1, KAP1, and ZEBRA from experiments above placed KAP1 upstream of ZEBRA. Accordingly, Fig. 4A showed 30% and 50% reduction in KAP1 level at 6 and 12 h, respectively, after exposure to NaB. Similarly, Akata cells exposed to IgG cross-linking demonstrated loss of KAP1 protein prior to lytic activation (SI Appendix, Fig. S10). Consistent with our findings using cultured cells, at the single-cell level, practically all lytic cells in the blood of patients with infectious mononucleosis and PTLD, that is, lytic cells naturally infected with EBV, demonstrated low levels of KAP1 (Fig. 4 B and C); this was in contrast to lytic cells in blood expressing high levels of TXNIP and cleaved caspase-1 (Fig. 3 E and F). In culture, we observed that subpopulations of KAP1lo cells emerge as early as 3 h and are clearly visible by 9 h in response to three different lytic cycle-inducing agents (Fig. 4D). We sorted KAP1lo from KAP1hi cells by flow cytometry and found that KAP1lo cells expressed significantly higher levels of BZLF1 (EBV gene encoding ZEBRA) and TXNIP transcripts compared to KAP1hi cells; furthermore, BMRF1 (transcriptional target of ZEBRA) transcripts were detected only in KAP1lo cells (Fig. 4E). Moreover, tying increased levels of TXNIP from earlier experiments to more-downstream events, we found that knockdown of TXNIP resulted in recovery of KAP1, while overexpression of TXNIP caused loss of KAP1 (SI Appendix, Fig. S11). These results indicate that cells with high levels of TXNIP but low levels of KAP1 (TXNIPhiKAP1lo cells) initiate the viral replicative phase more efficiently, thereby representing a prolytic state, and that changes in TXNIP and KAP1 levels serve as cellular determinants of the viral replication switch.
Fig. 4.
KAP1 loss precedes activation of the EBV replication switch in lytic cells. (A) (Upper) HH514-16 cells were treated with NaB and harvested at different time points for immunoblotting with indicated antibodies. Numbers represent ratios between indicated proteins. (Lower) Mean KAP1 and ZEBRA levels from three independent time course experiments are graphically displayed; error bars, SEM. (B and C) PBMC from the blood of patients with (B) infectious mononucleosis and (C) PTLD were analyzed by flow cytometry for expression of KAP1 and ZEBRA (lytic cells). Percent latent/uninfected and lytic cells are depicted in green and red gates, respectively. (D) HH514-16 cells were exposed to lytic triggers NaB, TSA, or AZA (versus DMSO control) and examined 3 or 9 h later for KAP1 expression by flow cytometry; black arrows, KAP1lo subpopulations. (E) HH514-16 cells were exposed to TSA, FACS-sorted 12 h later into KAP1hi and KAP1lo subpopulations, and followed by qRT-PCR for relative levels of TXNIP transcript and EBV lytic transcript BZLF1, as well as Ct values for lytic transcript BMRF1 in each subpopulation. Data represent averages of three independent experiments; error bars, SEM; *P ≤ 0.05.
High Glucose, a Physiologic Activator of the NLRP3 Inflammasome, Functions via TXNIP to Also Activate the EBV Replication Switch.
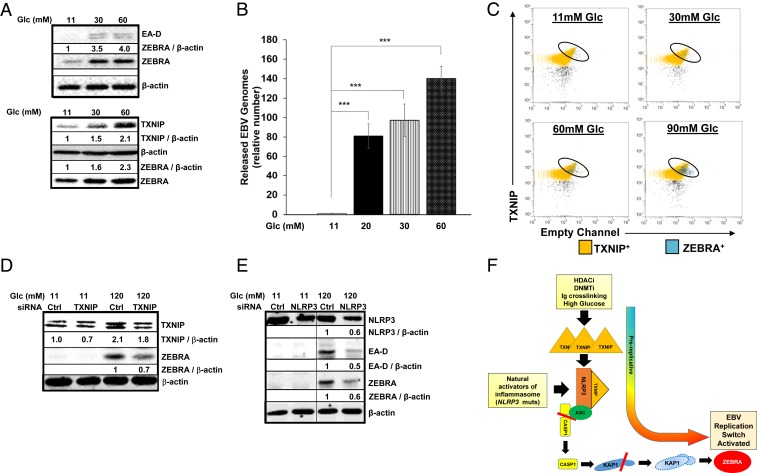
Since high glucose is known to activate the TXNIP−NLRP3 inflammasome (14), and perturbations in glucose levels frequently occur in patients with diabetes, we investigated the effects of high glucose on the EBV replicative phase. Progressive increases in ZEBRA expression and release of encapsidated viral genomes were observed following exposure to high glucose concentrations in both BL lines tested; notably, glucose concentrations of 20 to 30 mM (corresponding to blood glucose levels of 360 to 540 mg/dL) that are often observed in diabetic patients resulted in nearly 4- to 100-fold increase in the numbers of released viral genomes (Fig. 5 A and B and SI Appendix, Fig. S12 A and B). Simultaneously, TXNIP transcript and protein levels increased by 6 h in response to increasing concentrations of glucose (Fig. 5 A, Lower and SI Appendix, Fig. S12 C and D), and, at the single-cell level, ZEBRA-expressing cells demonstrated high levels of TXNIP (Fig. 5C). Mannose, an epimer of glucose known to have no effect on TXNIP or the NLRP3−TXNIP inflammasome (14), was unable to activate the EBV replication switch (SI Appendix, Fig. S12 A, Lower); this control demonstrates that the effect of glucose on the lytic switch was likely not due to changes in osmolarity. Finally, supporting the TXNIP−NLRP3 inflammasome’s involvement in the response to glucose, knockdown of TXNIP or NLRP3 resulted in blunting of ZEBRA levels (Fig. 5 D and E and SI Appendix, Fig. S12E).
Fig. 5.
High glucose triggers the EBV replication switch via TXNIP. (A and B) Mutu I BL cells were seeded in medium containing standard glucose (11 mM) or in the presence of increasing concentrations of glucose. (A) Cells were harvested after 6 h for immunoblotting for TXNIP and ZEBRA levels (Lower), and 24 h for immunoblotting for ZEBRA and EA-D (early lytic gene product) (Upper), while culture media were harvested after 72 h, treated with DNase, and (B) examined for released virus particles by qPCR of the EBV BALF5 gene. Data represent averages of three independent experiments; error bars, SEM; ***P ≤ 0.0005. (C) HH514-16 cells were seeded in media containing indicated concentrations of glucose and harvested 6 h later for staining for TXNIP and ZEBRA followed by flow cytometry. (D) HH514-16 cells were transfected with control or TXNIP-directed siRNA, placed in standard or high-glucose medium for 24 h, and subjected to immunoblotting. Numbers represent ratios between indicated proteins. (E) HH514-16 cells were transfected with control or NLRP3-directed siRNA, placed in standard or high-glucose medium for 24 h, and subjected to immunoblotting. Numbers represent ratios between indicated proteins. (F) Model depicting NLRP3 inflammasome-mediated activation of the EBV replication switch during the “prolytic state.”
Taken together, our results support a model in which several triggers (HDACi, DNMTi, and Ig cross-linking), including natural activators of the NLRP3 inflammasome (glucose, NLRP3-activating mutations, and physiologic activators in patient blood) trigger the loss of the universal corepressor KAP1 in a select subpopulation of cells, thereby sparking expression of the EBV replication switch protein in those cells (Fig. 5E). This TXNIPhiKAP1lo subpopulation marks a prolytic state allowing prediction of cells in which the virus is likely to switch into the lytic phase.
Discussion
While herpesviruses and other viruses have been found to parry host defense mechanisms during different stages of infection, the present findings distinctly demonstrate a virus utilizing an inflammasome to directly trigger its replication. Indeed, to persist, the most prevalent human cancer-causing virus has tied its exit strategy to the most promiscuous danger-sensing mechanism in the cell, capable of sensing both foreign and endogenous dangers. Specifically, the TXNIP−NLRP3 inflammasome depletes KAP1, a barrier to EBV lytic activation. While we have previously shown that KAP1 restricts expression of multiple lytic genes including BZLF1, and is deactivated via phosphorylation after successful entry into the lytic phase leading to amplification of the lytic cascade (15, 17), our present findings demonstrate that depletion instead of phosphorylation of KAP1 triggers entry into the lytic phase through derepression of ZEBRA, the EBV replication switch. This drop in KAP1 level, together with elevation in TXNIP, serves as a prelytic marker to identify cells in which EBV is poised to enter the lytic cycle. Once the virus has entered the lytic phase, however, progress of the lytic cascade and the amount of virus released are dependent on an amplification loop. This amplification relies on the activities of the viral protein kinase which phosphorylates the cellular ATM kinase at S2996; activated ATM then phosphorylates KAP1 at S824 to derepress multiple lytic genes (15–17). Of note is that, although KAP1 depletion was linked to caspase-1 in another study, depletion was observed late in the lytic cascade coincident with activation of apoptotic pathways, and the mechanism of caspase-1 activation was unexplored (22). Our work now links activation of TXNIP−NLRP3 inflammasome to caspase-1 activation resulting in depletion of unphosphorylated KAP1, all upstream of the EBV replication switch.
Others have shown that infection by gammaherpesviruses activates inflammasomes, including IFI16 and AIM2 (23–26). Importantly, they have also shown that these viruses, through unclear mechanisms, block harmful consequences of inflammasome activation to ensure long-term latency. In another study, Orf63, a KSHV (Kaposi’s sarcoma-associated herpesvirus)-encoded protein, was shown to inhibit NLRP1 inflammasome (27). In that context, NLRP1 was found to repress KSHV lytic activation. In contrast, our findings indicate that the NLRP3 inflammasome promotes EBV lytic switch without significant activation of IL-1β. Whether EBV encodes factors to prevent proinflammatory cytokine processing/release or pyroptosis remains to be established.
Metabolic changes such as altered levels of extracellular glucose are sensed via inflammasome components, resulting in assembly of the inflammasome and summary reduction of KAP1 levels in a fraction of cells, thereby triggering the viral lytic switch in only those cells within a population; we find this to be true both in culture systems and in infected B lymphocytes from the blood of patients with EBV-related diseases. Thus, metabolic heterogeneity is at least partly responsible for the asynchronous lytic activation that is routinely observed in latently infected cells. In this way, EBV has tied its replication to a cellular sensing system that is regularly activated by alterations in metabolic states and danger signals.
In type 2 diabetes and obesity, the innate immune response mediated by the NLRP3 inflammasome within myeloid-derived cells contributes to a chronic proinflammatory state resulting in insulin resistance (28–32). Importantly, activation of the NLRP3 inflammasome is also linked to the development of type 1 diabetes (33). While the replicative phase of another herpesvirus (KSHV) was activated by elevated glucose through non-inflammasome-mediated mechanisms, an earlier study failed to demonstrate an effect on the EBV replicative cycle, for unclear reasons (34, 35). However, our findings indicate that, compared to healthy individuals, episodes of hyperglycemia in the context of chronic NLRP3 activation during diabetes may cause greater dysregulation of EBV in B lymphocytes, suggesting connections between diabetes, hyperglycemia, and EBV-related pathology. Indeed, with the replicative phase of EBV necessary for development of EBV-related cancers (8–11, 36, 37), it is noteworthy that a recent study found that diabetes carried the greatest risk for development of EBV PTLD after renal transplantation, a risk similar to that posed by being EBV-naïve before transplantation (38). These links between diabetes, activation of the EBV replication switch via a diabetes-related inflammasome, and EBV PTLD are of relevance given the increasing worldwide rates of both transplantation and diabetes (39, 40). Moreover, these links may extend to other types of EBV cancers such as BL and nasopharyngeal cell carcinoma, also dependent on the viral replicative phase. These cancers are observed in developing countries with rapidly increasing rates of diabetes (41). The relationship between EBV, hyperglycemia, and EBV cancer development warrants further investigation.
Materials and Methods
Cells Lines and Chemical Treatment.
EBV-positive endemic BL cell lines HH514-16 (a gift from George Miller, Yale University, New Haven, CT), Akata (a gift from Benjamin Gewurz, Harvard Medical School, Cambridge, MA), and Mutu I (a gift from Erik Flemington, Tulane University, New Orleans, LA) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). LCL were generated and maintained as described before (42). Sodium butyrate (NaB; 3 mM, 303410; Sigma-Aldrich), Trichostatin A (TSA; 5 µM, T8552-5MG; Sigma-Aldrich), 5-AZA-2′-deoxycytidine (AZA; 5 µM, A3656; Sigma-Aldrich), VPA (5 μM, P4543; Sigma-Aldrich), Rabbit anti-human IgG (1:00, A042301-2; Dako/Agilent), ZVAD-FMK (ZVAD; 20 µM, S7023; Selleckchem), YVAD-FMK (YVAD; 20 µM, 1141-5; Biovision), d-dextrose (Glc; 30 to 150 mM, D16-500; Fisher Scientific), and d-mannose (Man; 30 to 150 mM, AC150600250; ACROS Organics) were used to treat BL cells.
Patients and Ethics Statement.
Peripheral blood mononuclear cells (PBMC) were isolated from the blood of patients with NOMID syndrome who had heterozygous germline mutations in their NLRP3 gene at the National Institute of Allergy and Infectious Diseases (NIAID), infectious mononucleosis at Stony Brook University, and EBV PTLD at Shands Children’s Hospital at the University of Florida. Blood was drawn after obtaining written informed consent from patients, or parents in the case of patients who were minors. The study of human subjects was approved by institutional review boards at NIAID, Stony Brook University, and University of Florida.
Infection of PBMC.
PBMCs from healthy subjects or NOMID donors were infected with EBV (B95-8 strain) in the presence of FK506. FK506 (AG Scientific) was added to the cell suspension at a final concentration of 20 nM for 1 h at 37 °C before infection. B95-8 strain EBV was added at a multiplicity of infection of 1. The flask/plate was swirled to mix and placed in an incubator at 37 °C. Cells were harvested 24 h after infection for analysis of EBV lytic gene expression via qRT-PCR or left in culture to establish LCL.
Plasmids, siRNAs, and Transfection.
Plasmid pCDNA3.1-TXNIP was a gift from Richard Lee at Harvard Medical School, Harvard, MA (43). Plasmid pCDH-Myc-Caspase-1 was a gift from Christian Stehlik at Northwestern University, Evanston, IL (44). Plasmid pEGFP-N2-NLRP3 (45) (Addgene: 73955) was purchased from Addgene, Inc. BACmid p2089 was a gift from Henri-Jacques Delecluse at the German Cancer Research Center, Heidelberg, Germany (46).
All siRNAs targeting human transcripts were reconstituted with nuclease-free water at a concentration of 10 µM. Experiments performed using siRNA targeting TXNIP, CASP1, and NLRP3 were performed using two siRNAs with representative data shown. Specific siRNAs are listed in Table 1.
Table 1.
Specific siRNAs
| siRNA | Supplier | Catalog no. |
| TXNIP-1 | Dharmacon | J-010814–05-0002 |
| TXNIP-2 | Dharmacon | J-010814–06-0002 |
| CASP1-1 | Ambion | s2407 |
| CASP1-2 | Dharmacon | J004401-07–002 |
| NLRP3-1 | Ambion | s41554 |
| NLRP3-2 | Ambion | s41555 |
| NLRP1 | Ambion | s22520 |
| NLRP2 | Ambion | s31176 |
| NLRP4 | Ambion | s45086 |
| NLRP5 | Ambion | s42974 |
| NLRP6 | Ambion | s46905 |
| NLRP7 | Ambion | s47127 |
| NLRP8 | Ambion | s42971 |
| NLRP9 | Ambion | s50314 |
| NLRP10 | Ambion | s50317 |
| NLRP11 | Ambion | s47544 |
| NLRP12 | Ambion | s40729 |
| NLRP13 | Ambion | s42968 |
| NLRP14 | Ambion | s50320 |
| NLRC4 | Ambion | s33828 |
| MEFV | Ambion | s8658 |
| IFI16 | Ambion | s7136 |
| Control siRNA (nontargeting) | Dharmacon | D001810-01–20 |
For nucleofection, cells were subcultured at 5 × 105 cells per mL 24 h prior to transfection, washed twice with phosphate-buffered saline (PBS), and then 1 × 106 cells were transfected with 20 µg of plasmid or 200 pmol of siRNA in 100 µL of total Ingenio solution (MIR50117; Mirus) using an Amaxa Nucleofector II (program A-024). Cells were then seeded into prewarmed complete media at a concentration of 5 × 105 cells per mL and harvested or further treated as indicated.
Antibodies.
Antibodies used include rabbit anti-TXNIP Ab (14715; Cell Signaling Technology), rabbit anti-TRX1 Ab (2429s; Cell Signaling Technology), rabbit anti-cleaved Caspase-1 Ab (4199s; Cell Signaling Technology), rabbit anti-IL1β (12703s; Cell Signaling Technology), rabbit anti-NLRP3 Ab (PA5-21745; Invitrogen), rabbit anti-Caspase-1 Ab (full length, PA5-29342; Thermo Scientific), rabbit anti-KAP1 Ab (A300-274A; Bethyl Laboratories), mouse anti-β-actin Ab (AC-15; Sigma), mouse anti-EA-D Ab (MAB8186; EMD), mouse IgG1 negative isotype Ab (CBL610; EMD Millipore), normal rabbit IgG (sc-2027; Santa Cruz), mouse anti-ZEBRA Ab (a gift from Paul Farrell at Imperial College London, London, UK), mouse anti-CD19-APC mAb (MABF197; Sigma-Aldrich), mouse IgG1-APC isotype Ab (550824; BD Pharmogen), rat anti-EBNA2 mAb (MABE8; EMD Millipore), rat IgG2a isotype Ab (MABF1077Z; EMD Millipore), goat anti-rat IgG-F(ab')2-PE (sc3829; Santa Cruz Biotechnology), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H+L) (AP308P; EMD Millipore), rabbit anti-GFP Ab (G1544; Sigma-Aldrich), HRP-conjugated goat anti-rabbit IgG (H+L)(AP307P; EMD Millipore), fluorescein isothiocyanate-conjugated goat anti-mouse IgG (F0257; Sigma), and Alexa Fluor 647-conjugated goat anti-rabbit IgG (A-21245; Thermo Fisher). All antibodies were used at concentrations and conditions recommended by manufacturers.
Immunoprecipitation and Immunoblotting.
NaB-treated or untreated HH514-16 cells were harvested after 24 h in RIPA buffer (50 mM Tris⋅HCl [pH 7.4], 150 mM NaCl, 1% [vol/vol] Nonidet P-40, 1% [wt/vol] deoxycholate, 1 mM (ethylenedinitrilo)tetraacetic acid, 1× protease and phosphatase inhibitor mixture [catalog no. 5872; Cell Signaling Technology]); 10% of cell lysate was set aside as input, and the remainder was incubated with indicated antibodies and protein A/G agarose (sc-2003; Santa Cruz Biotechnology) for 16 h at 4 °C. Immunoprecipitates were washed with RIPA buffer and subjected to immunoblotting for analysis.
Total cell extracts were electrophoresed in 10% sodium dodecyl sulfate-polyacrylamide gels, transferred onto nitrocellulose membranes, and blocked by using 5% milk. Immunoblots were performed using indicated antibodies and company-recommended concentrations and conditions.
Flow Cytometry and In-Cell Western.
As described previously, cells were fixed with BD Cytofix/Cytoperm solution (554722; BD Bioscience) at room temperature for 15 min protected from light, washed twice with 1× BD Perm/Wash buffer (554723; BD Bioscience), and incubated with indicated primary antibodies for 45 min at room temperature protected from light. After washing, cells were further incubated with corresponding secondary antibodies for 45 min at room temperature and then subjected to flow cytometry or FACS sorting (using fluorochrome-conjugated secondary antibodies secondary antibodies) or in-cell Western (using infrared-dye-based secondary antibodies). Flow cytometry data were acquired on a ThermoFisher Attune NxT Flow cytometer and analyzed using FlowJo software (Tree Star) with placement of gates based on parallel staining with isotype-matched control antibodies. For separation into subpopulations, cells were sorted using a BD Aria II FACS (fluorescent activated cell sorting) instrument; again, cells were gated as indicated in Fig. 4E after comparison to cells stained in parallel with isotype-matched control antibodies. Analysis of in-cell Western was performed using LI-COR Odyssey software in triplicate; here, relative ZEBRA protein levels were compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control.
For flow cytometric analysis of blood cells from PTLD patients in Fig. 3 D–F, we identified infected B lymphoblasts by gating on large B cells that expressed EBV proteins EBNA2. Cells were then analyzed for correlation between expression of ZEBRA and levels of expression of TXNIP or CASP1 p20 (cleaved caspase-1) by staining with appropriate antibodies and gating based on staining with isotype-matched control antibodies. B cells from infectious mononucleosis and PTLD patients in Fig. 4 B and C were similarly stained and analyzed using antibodies to ZEBRA, KAP1, and their respective isotype-matched control antibodies.
qRT-PCR.
Total RNA was isolated from BL cells by using an RNeasy kit (Qiagen) followed by DNase digestion (Promega). RNA was quantitated by using a NanoDrop instrument (Thermo Scientific). RNA (1 µg) was converted to complementary DNA by using MuLV Reverse Transcriptase (New England Biolabs). Relative transcript levels of selected cellular genes were determined with gene-specific primers by using Fast SYBR green Master Mix on a Quant Studio 3 thermocycler (Applied Biosystems) and analyzed using the ΔΔCT method. Primers sequences included the following: forward primer GTAACCCGTTGAACCCCATT and reverse primer CCATCCAATCGGTAGTAGCG for 18S rRNA; forward primer TTCCACAGCCTGCACCAGTG and reverse primer GGCAGAAGCCACCTCACGGT for BZLF1; forward primer ACCTGCCGTTGGATCTTAGTG and reverse primer GGCGTTGTTGGAGTCCTGTG for BMRF1; forward primer AACCAGAATAATCTCCCCAATG and reverse primer CGAGGCACCCCAAAAGTC for BFRF3; forward primer CATGCCGACAAACACCACAG and reverse primer TTGCGTCCTCAGAAGTGACC for BLLF1; forward primer AGGAAGCTCAAAGCCGAACT and reverse primer ACGCTTCTTCTGGAAGACCA for TXNIP; forward primer GCCTCTGTGTGAGACCTGTG and reverse primer AGTACGTTCACCATCCCGAG for KAP1.
Activation of EBV Replicative Cycle via High Glucose.
EBV-positive BL cell lines were split into 3 × 105 cells per m in complete RPMI medium. After 2 d, cells were harvested, counted, and reseeded in high-glucose−containing media at 3 × 105 cells per mL. Treated cells were harvested at indicated time points to assay for EBV replicative cycle activation.
Assay for EBV Load.
Viral DNA in supernatant was quantitated using qPCR by amplifying EBV BALF5 gene with forward primer CGTCTCATTCCCAAGTGTTTC and reverse primer GCCCTTTCCATCCTCGTC or by amplifying EBV BamW using forward primer AGGCTTAGTATACATGCTTCTTGCTTT and reverse primer CCCTGGCTGATGCAACTTG. Released EBV particles were pelleted from supernatant, washed with 1× PBS, and treated with DNase. Absolute EBV genome copy number was determined with a standard curve obtained through qPCR using serially diluted BACmid p2089 as template and BamW primers in SI Appendix, Figs. S3B and S5C. Primers targeting the EBV BALF5 gene region were used to calculate relative released virus compared to normal glucose control (Fig. 5B and SI Appendix, Fig. S12B).
Statistical Analysis.
P values were calculated by comparing the means of two groups of interest using unpaired Student t test.
Supplementary Material
Acknowledgments
E.M.B. was supported by NIH Grant T32 T32AI007539, R.G.-M. was supported by intramural funds from NIH, and S.B.-M. was supported by NIH Grants R01 AI113134 and R41 AI115834, and the University of Florida.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919133117/-/DCSupplemental.
References
- 1.Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Johnson K. E., et al. , IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 10, e1004503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao X., Qi J., Lei X., Wang J., Interactions between enteroviruses and the inflammasome: New insights into viral pathogenesis. Front. Microbiol. 10, 321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodsky I. E., et al. , A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7, 376–387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott R. S., Epstein-Barr virus: A master epigenetic manipulator. Curr. Opin. Virol. 26, 74–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochford R., Moormann A. M., Burkitt’s Lymphoma. Curr. Top. Microbiol. Immunol. 390, 267–285 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Tsao S. W., Tsang C. M., Lo K. W., Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R Soc. Lond. B Biol. Sci. 372, 20160270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong G. K., et al. , Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 79, 13993–14003 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller N., et al. , Hodgkin’s disease and Epstein-Barr virus. Altered antibody pattern before diagnosis. N. Engl. J. Med. 320, 689–695 (1989). [DOI] [PubMed] [Google Scholar]
- 10.Pallesen G., Sandvej K., Hamilton-Dutoit S. J., Rowe M., Young L. S., Activation of Epstein-Barr virus replication in Hodgkin and Reed-Sternberg cells. Blood 78, 1162–1165 (1991). [PubMed] [Google Scholar]
- 11.van Esser J. W., et al. , Epstein-Barr virus (EBV) reactivation is a frequent event after allogeneic stem cell transplantation (SCT) and quantitatively predicts EBV-lymphoproliferative disease following T-cell–depleted SCT. Blood 98, 972–978 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Draborg A. H., Duus K., Houen G., Epstein-Barr virus and systemic lupus erythematosus. Clin. Dev. Immunol. 2012, 370516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pender M. P., Csurhes P. A., Burrows J. M., Burrows S. R., Defective T-cell control of Epstein-Barr virus infection in multiple sclerosis. Clin. Transl. Immunology 6, e126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R., Yazdi A. S., Menu P., Tschopp J., A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Li X., Burton E. M., Bhaduri-McIntosh S., Chloroquine triggers Epstein-Barr virus replication through phosphorylation of KAP1/TRIM28 in Burkitt lymphoma cells. PLoS Pathog. 13, e1006249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., et al. , KRAB-ZFP repressors enforce quiescence of oncogenic human herpesviruses. J. Virol. 92, e00298-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Kozlov S. V., El-Guindy A., Bhaduri-McIntosh S., Retrograde regulation by the viral protein kinase epigenetically sustains the Epstein-Barr Virus latency-to-lytic switch to augment virus production. J. Virol. e00572-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordero M. D., Alcocer-Gómez E., Ryffel B., Gain of function mutation and inflammasome driven diseases in human and mouse models. J. Autoimmun. 91, 13–22 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Fang S., et al. , High glucose condition upregulated Txnip expression level in rat mesangial cells through ROS/MEK/MAPK pathway. Mol. Cell. Biochem. 347, 175–182 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Schulze P. C., et al. , Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. J. Biol. Chem. 279, 30369–30374 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Daigle D., et al. , Valproic acid antagonizes the capacity of other histone deacetylase inhibitors to activate the Epstein-Barr virus lytic cycle. J. Virol. 85, 5628–5643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv D. W., Zhang K., Li R., Interferon regulatory factor 8 regulates caspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction. PLoS Pathog. 14, e1006868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torii Y., et al. , Epstein-Barr virus infection-induced inflammasome activation in human monocytes. PLoS One 12, e0175053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansari M. A., et al. , Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J. Virol. 87, 8606–8623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerur N., et al. , IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9, 363–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh V. V., et al. , Kaposi’s sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J. Virol. 87, 4417–4431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory S. M., et al. , Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331, 330–334 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixit V. D., Nlrp3 inflammasome activation in type 2 diabetes: Is it clinically relevant? Diabetes 62, 22–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant R. W., Dixit V. D., Mechanisms of disease: Inflammasome activation and the development of type 2 diabetes. Front. Immunol. 4, 50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W., et al. , Activation of the TXNIP/NLRP3 inflammasome pathway contributes to inflammation in diabetic retinopathy: A novel inhibitory effect of minocycline. Inflamm. Res. 66, 157–166 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Zhang X., et al. , Reactive oxygen species-induced TXNIP drives fructose-mediated hepatic inflammation and lipid accumulation through NLRP3 inflammasome activation. Antioxid. Redox Signal. 22, 848–870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alhawiti N. M., Al Mahri S., Aziz M. A., Malik S. S., Mohammad S., TXNIP in metabolic regulation: Physiological role and therapeutic outlook. Curr. Drug Targets 18, 1095–1103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu C., et al. , NLRP3 deficiency protects from type 1 diabetes through the regulation of chemotaxis into the pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 112, 11318–11323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye F., et al. , High glucose induces reactivation of latent Kaposi’s sarcoma-associated herpesvirus. J. Virol. 90, 9654–9663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang P. J., et al. , Diabetes and risk of Kaposi’s sarcoma: Effects of high glucose on reactivation and infection of Kaposi’s sarcoma-associated herpesvirus. Oncotarget 8, 80595–80611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montone K. T., et al. , Identification of Epstein-Barr virus lytic activity in post-transplantation lymphoproliferative disease. Mod. Pathol. 9, 621–630 (1996). [PubMed] [Google Scholar]
- 37.Ma S. D., et al. , A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 85, 165–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francis A., Johnson D. W., Teixeira-Pinto A., Craig J. C., Wong G., Incidence and predictors of post-transplant lymphoproliferative disease after kidney transplantation during adulthood and childhood: A registry study. Nephrol. Dial. Transplant. 33, 881–889 (2018). [DOI] [PubMed] [Google Scholar]
- 39.The Global Database on Donation and Transplantation , WHO-ONT statistics on global donation and transplantation. http://www.transplant-observatory.org/. Accessed 7 March 2019.
- 40.Health Resources and Services Administration , Organ donation and transplantation statistics 1991-2018. https://www.organdonor.gov/statistics-stories/statistics/data.html. Accessed 7 March 2019.
- 41.Animaw W., Seyoum Y., Increasing prevalence of diabetes mellitus in a developing country and its related factors. PLoS One 12, e0187670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hui-Yuen J., Koganti S., Bhaduri-McIntosh S., Human B cell immortalization for monoclonal antibody production. Methods Mol. Biol. 1131, 183–189 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Patwari P., Higgins L. J., Chutkow W. A., Yoshioka J., Lee R. T., The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J. Biol. Chem. 281, 21884–21891 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bryan N. B., Dorfleutner A., Rojanasakul Y., Stehlik C., Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 182, 3173–3182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khare S., et al. , An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feederle R., et al. , The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19, 3080–3089 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.