Significance
Many viral pathogens replicate in the human small intestine where they must invade the epithelial barrier that has evolved to protect the host against microbial assaults. Using a human norovirus strain that requires bile to replicate in stem cell-derived human small intestinal enteroid cultures, we found that conjugated hydrophobic bile acids and ceramide are critical to allow virus entry and subsequent replication in jejunal enteroids. Glycochenodeoxycholic acid treatment of enteroids below the critical micellar concentration leads to multiple cellular responses, including rapid changes in endocytosis and exocytosis dynamics that the bile-requiring human norovirus deftly exploits to overcome the epithelial barrier. Our findings shed light on the role of bile acids and ceramide in human jejunal enterocytes that stimulate viral infection.
Keywords: norovirus, enteroid/organoid, bile acid, acid sphingomyelinase, ceramide
Abstract
Human noroviruses (HuNoVs) cause sporadic and epidemic outbreaks of gastroenteritis in all age groups worldwide. We previously reported that stem cell-derived human intestinal enteroid (HIE) cultures support replication of multiple HuNoV strains and that some strains (e.g., GII.3) replicate only in the presence of bile. Heat- and trypsin-treatment of bile did not reduce GII.3 replication, indicating a nonproteinaceous component in bile functions as an active factor. Here we show that bile acids (BAs) are critical for GII.3 replication and replication correlates with BA hydrophobicity. Using the highly effective BA, glycochenodeoxycholic acid (GCDCA), we show BAs act during the early stage of infection, BA-dependent replication in HIEs is not mediated by detergent effects or classic farnesoid X receptor or Takeda G protein-coupled receptor 5 signaling but involves another G protein-coupled receptor, sphingosine-1-phosphate receptor 2, and BA treatment of HIEs increases particle uptake. We also demonstrate that GCDCA induces multiple cellular responses that promote GII.3 replication in HIEs, including enhancement of 1) endosomal uptake, 2) endosomal acidification and subsequent activity of endosomal/lysosomal enzyme acid sphingomyelinase (ASM), and 3) ceramide levels on the apical membrane. Inhibitors of endosomal acidification or ASM reduce GII.3 infection and exogenous addition of ceramide alone permits infection. Furthermore, inhibition of lysosomal exocytosis of ASM, which is required for ceramide production at the apical surface, decreases GII.3 infection. Together, our results support a model where GII.3 exploits rapid BA-mediated cellular endolysosomal dynamic changes and cellular ceramide to enter and replicate in jejunal HIEs.
Human noroviruses (HuNoVs) are the leading cause of sporadic and epidemic outbreaks of gastrointestinal illness in all age groups worldwide (1–4). These highly contagious pathogens are the leading cause of foodborne disease, and globally the economic burden due to direct health care costs and loss of productivity is estimated to be over $60 billion annually (3, 5–7). Despite the health impact of HuNoV infections, our knowledge of the mechanisms of HuNoV infection and their life cycle of replication is relatively limited, partially due to the longstanding lack of reproducible in vitro cultivation systems despite considerable effort to establish them (8–11).
Noroviruses are phylogenetically classified into 10 genogroups, of which GI, GII, GIV, GVIII, and GIX contain viruses that infect humans. Genogroups are further subdivided into genotypes (e.g., GI.1–9 and GII.1–27) (12, 13). For example, the prototype HuNoV, Norwalk virus is classified as genogroup I genotype 1 (GI.1). Some knowledge about HuNoV tropism and initial interactions with host cells is available from early volunteer studies and studies using virus-like particles (VLPs), which are morphologically and antigenically similar to native virions and are generated from recombinant expression of viral capsid proteins (14, 15). These studies revealed that susceptibility to infection is regulated by the expression of a functional fucosyltransferase 2 gene, with secretor-positive individuals being susceptible to infection with many HuNoV strains and secretor-negative individuals being resistant (16–18). Subsequent VLP studies demonstrated that histo-blood group antigens (HBGAs) are important for initial virus–host interactions; they are considered initial binding factors, and infection with specific HuNoV strains is defined by the host HBGA profile (19–21). In addition, in a variety of cell lines, studies of expression of 1) transfected viral RNA isolated from stools of infected individuals (22, 23), 2) individual HuNoV proteins (24–28), and 3) reverse genetics systems of HuNoV (29, 30) have provided some information about protein localization and function for stages of HuNoV replication after virus entry. Demonstration that HuNoV RNA transfected into cultured cells can replicate and produce particles but not spread throughout transfected cultures that express the proper HBGAs indicated that the block to infection was at an early stage of replication, such as virus-receptor binding and subsequent virus entry (22, 23).
Recently, we established a HuNoV cultivation system using stem cell-derived human small intestinal enteroid (HIE) (31) cultures, which support the replication of multiple HuNoV strains with strain-specific requirements being discovered (32–34). For example, the expression of specific HBGAs required for infection and host strain-specific susceptibility to infection mimic epidemiological host–virus infection patterns. Furthermore, bile is involved in HuNoV replication in HIEs in a strain-dependent manner; bile is essential for the replication of GI.1, GII.3, and GII.17 HuNoVs, while the replication of GII.4 strains is enhanced by bile (32). Bile is required at an early stage of infection, with its effects being on the cells and not the virus. Recent studies evaluating bile acid (BA) binding to HuNoV VLPs confirmed no binding to VLPs from cultivatable viruses, such as GI.1, GII.3, GII.4, and GII.17, while they did bind to other strains (35).
Previous experiments to identify an active factor in bile indicated that heat- and trypsin-treated human bile and bile from several mammalian species support GII.3 and enhance GII.4 replication (32). These results suggested that a nonproteinaceous molecule(s) conserved among mammals is an active factor in bile required for GII.3 replication (32). Bile, which is produced in the liver and stored in the gallbladder prior to secretion into the duodenum, is a complex aqueous secretion composed of many components, including BAs, bile pigments, inorganic salts, fatty acids, and phospholipids (36). BAs are one of the major components of bile synthesized from cholesterol in hepatocytes as cholic acid (CA) and chenodeoxycholic acid (CDCA), or primary unconjugated BAs. These BAs are then conjugated with glycine or taurine to increase water-solubility and to reduce cell toxicity caused by the high hydrophobicity of the primary unconjugated BAs (37). After secretion into the duodenum, secondary BAs are produced from the primary BAs by bacterial metabolism in the colon (36).
To understand the mechanism of replication in small intestinal enterocytes of GII.3 HuNoV, which strictly requires bile, we sought to determine what components of bile support virus replication and how they act in jejunal HIEs. We found that hydrophobic BAs induce cellular changes critical for GII.3 replication, including enhanced endocytosis and altered dynamics of the endo-lysosomal system, which promote rapid acid sphingomyelinase (ASM)-mediated ceramide increases. Ceramide also is sufficient for GII.3 replication. Our results demonstrate unexpected activities of BAs in human jejunal enterocytes.
Results
Hydrophobic BAs Are One Component in Bile That Support GII.3 Replication and Function during the Early Stage of Infection.
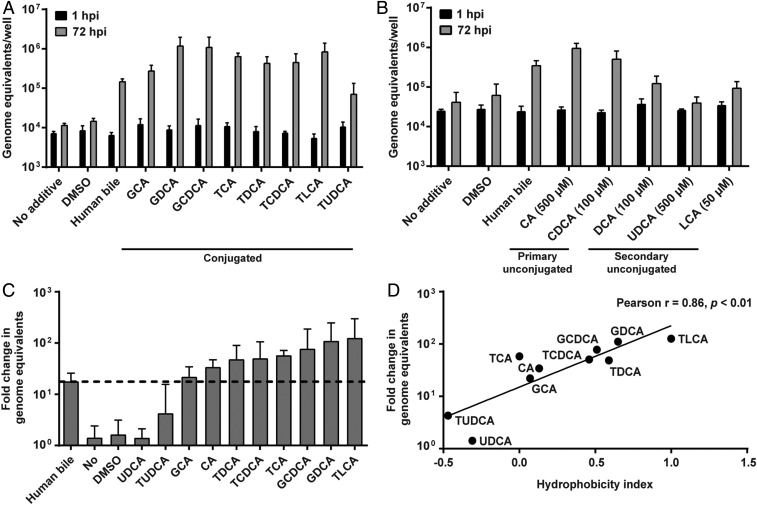
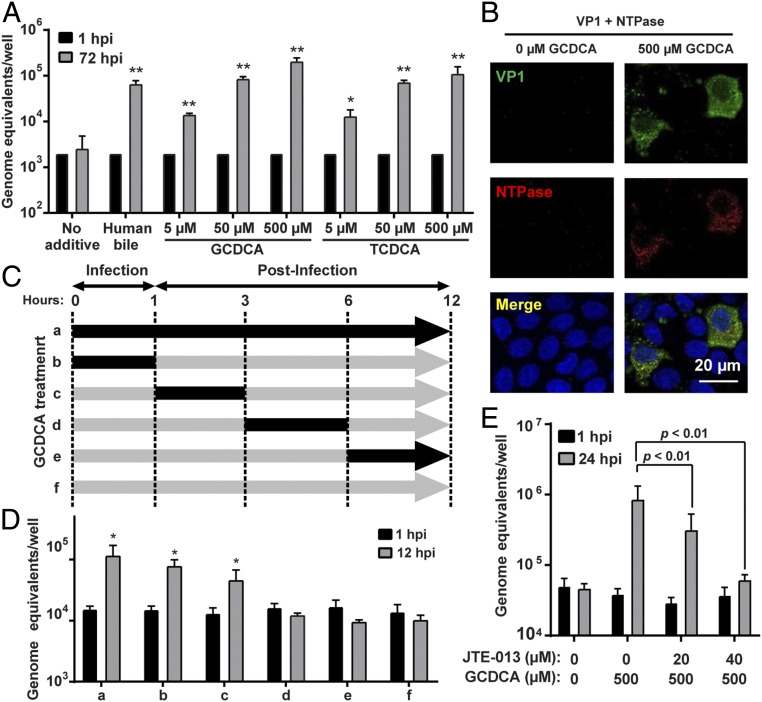
To identify the nonproteinaceous components in bile required for GII.3 replication, we first evaluated whether BAs, major components of bile, might be required for GII.3 infection in a jejunal secretor-positive (J2) HIE line. Tests of human bile treated with the Food and Drug Administration-approved BA sequestrant (cholestyramine) demonstrated a dose-dependent reduction of total BA concentration (SI Appendix, Fig. S1A), and a significant reduction in GII.3 replication in HIEs was associated with decreased BA concentrations (SI Appendix, Fig. S1B). We next evaluated the efficacy of individual BAs to support GII.3 replication; testing the addition of eight conjugated, two primary unconjugated, and three secondary unconjugated BAs to the culture medium revealed most conjugated- and primary unconjugated-BAs enhanced GII.3 replication compared to human bile, while secondary unconjugated-BAs were less effective (Fig. 1 A and B). The BA ursodeoxycholic acid (UDCA) never supported replication. Most BAs were tested at 500 µM; however, because of cytotoxicity at this higher dose, CDCA and deoxycholic acid (DCA) were only tested up to 100 µM and lithocholic acid (LCA) up to 50 µM. Comparison of the fold-change in viral genome equivalents between 1 and 72 h postinfection (hpi) in the presence of individual BAs (used at 500 µM) to the BA hydrophobic indices (38) found that replication is significantly correlated with BA hydrophobicity (Fig. 1 C and D). Because the most hydrophobic BA caused some cell death, we further evaluated a range of concentrations of glycochenodeoxycholic acid (GCDCA) and taurochenodeoxycholic acid (TCDCA), which lacked cytotoxicity at 500 µM, on GII.3 replication. Both GCDCA and TCDCA significantly enhanced GII.3 replication at 5 to 500 µM in a concentration-dependent manner (Fig. 2A). Due to the lack of cytotoxicity at 500 µM, ease of solubility and previous use in an animal calicivirus system (39), we chose to use GCDCA (500 µM) for more detailed studies on how BA enhances GII.3 replication. To ensure the effect of GCDCA was not limited to only J2 HIEs, we first evaluated the effect of this BA on additional HIE lines. GCDCA treatment of HIEs supported GII.3 replication in a second secretor-positive jejunal (J3) line (SI Appendix, Fig. S2A), as well as secretor positive duodenal and ileal HIEs from a single individual (SI Appendix, Fig. S2B). Subsequent mechanistic studies were carried out on J2 HIEs. The expression of GII.3 structural (VP1) and nonstructural (NTPase) proteins was detected by immunofluorescence (IF) microscopy in J2 HIEs only in the presence of GCDCA (Fig. 2B). A time course of BA addition experiment determined that GCDCA addition during the first 3 h of infection is critical for successful GII.3 replication (Fig. 2 C and D), mimicking the early requirement for bile described previously (32). These results indicate that GCDCA facilitates virus entry into cells followed by genome uncoating, nonstructural protein expression, and genome replication, which lead to generation of subgenomic RNA required for production of VP1 to assemble progeny viruses.
Fig. 1.
Hydrophobic BAs are required for GII.3 HuNoV replication. (A and B) HIE monolayers were inoculated with 4.3 × 105 GEs of GII.3 for 1 h at 37 °C, washed twice with CMGF(−) and cultured for 72 h at 37 °C. Viral GEs at 1 and 72 hpi were quantified by RT-qPCR. Data represent the mean of three wells for each treatment and time point. Error bars denote SD. Each experiment was performed two or more times. All subsequent infections were performed and analyzed similarly unless otherwise indicated. During and postinoculation, individual conjugated (A) and unconjugated (B) BAs were added at a final concentration of 500 µM unless otherwise indicated (B). Full chemical names for each BA are defined in the text as they appear. Human bile at the final concentration of 5% (vol/vol) in the medium was used as a positive control and untreated medium as a negative control. (C) Fold-change of GII.3 HuNoV GEs between 1 and 72 hpi in the presence of each BA used at the concentration of 500 µM. (D) A scatter plot of hydrophobicity indices (38) and means of fold-changes of GII.3 GEs. The solid line depicts the best-fit linear regression (R2 = 0.73, P < 0.01). The Pearson correlation coefficient (r) and P value (p) are noted.
Fig. 2.
BAs are required early in GII.3 infection. (A) HIE monolayers were infected as in Fig. 1 with GII.3 in the presence or absence of GCDCA and TCDCA for 72 h. *P < 0.05 and **P < 0.01 comparing GEs at 72 hpi to 1 hpi. (B) HIE monolayers infected with 4.3 × 106 GEs in the presence or absence of 500 µM GCDCA. VP1 and NTPase were detected by confocal laser-scanning microscopy using guinea pig anti-GII.3 VLP (green) and rabbit anti-GII.3 NTPase (red) antisera. Nuclei were stained with DAPI (blue). (Scale bar, 20 µm.) (C) Schematic showing with black arrows when 500 µM GCDCA was added to the medium during GII.3 infection of HIEs. (D) HIE monolayers were infected with GII.3 for 12 h. GCDCA was added to the medium as in C. HIEs were washed three times with CMGF(−) at the end of each period. *P < 0.05 comparing GEs at 12 hpi to 1 hpi. (E) S1PR2 antagonist, JTE-013, and GCDCA were added to the medium at the indicated concentrations and infected with GII.3 as in Fig. 1 for 24 h. P values between conditions are indicated.
BA-Dependent GII.3 Replication in HIEs Is Not Mediated by Detergent Effects, Classic FXR or TGR5 Receptor Signaling, but Involves S1PR2.
BAs act as steroid hormones controlling lipid, glucose, and energy metabolism. Their actions can be implemented through detergent effects or activation of nuclear farnesoid X receptor (FXR) and membrane G protein-coupled receptors (GPCRs), Takeda G protein-coupled receptor 5 (TGR5), and sphingosine-1-phosphate receptor 2 (S1PR2) (40, 41). To begin to understand how the BAs function in jejunal HIEs, we tested whether their natural detergent effects are important for GII.3 infection. Testing of a variety of well-characterized detergents (SDS, Triton X-100, Nonidet P-40, and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate [CHAPS]) showed these treatments did not lead to GII.3 replication (SI Appendix, Fig. S3). BAs are ligands for FXR and TGR5, which are involved in metabolic feedback pathways between the liver and the intestine. High-throughput RNA sequencing (RNA-seq) detected expression of FXR in our jejunal HIE cultures and transcriptional up-regulation of downstream targets of FXR-signaling (FABP6 and FGF19) in HIEs treated with GCDCA for 3 h (SI Appendix, Table S1). Only low expression of TGR5 was observed in untreated differentiated cultures; however, TGR5 was up-regulated with GCDCA treatment. The GCDCA-driven GII.3 replication was not altered by HIE treatment with several agonists (INT-747, GW4064, and fexaramine for FXR; INT-777 and CCDC for TGR5) and an antagonist (guggulsterone for FXR) (SI Appendix, Fig. S4). In contrast, inhibition of S1PR2 using the specific antagonist JTE-013 showed a dose-dependent reduction in GII.3 replication at noncytotoxic concentrations (Fig. 2E and SI Appendix, Fig. S5A). Although expression of S1PR2 in jejunal HIEs is low, protein was detected by Western blot (SI Appendix, Fig. S5B). Recently, BAs were shown to stimulate S1PR2 in liver and intestinal cells which activates downstream signaling pathways (42, 43). These results provide one explanation for how GCDCA may interact with or be taken up into an enterocyte and in turn stimulate GII.3 replication despite its inability to passively enter jejunal cells (44, 45).
BA Enhances Endosomal Uptake and GII.3 Entry to HIEs.
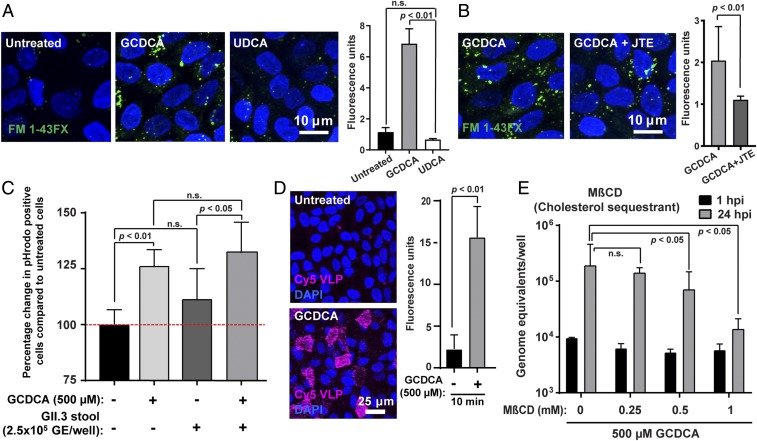
Because GCDCA is required early in GII.3 infection, we sought to determine whether BAs influence viral entry. First, because HBGA expression is important for initial HuNoV binding, we tested whether HBGA expression on HIEs was altered with GCDCA treatment. HBGA expression was evaluated using Ulex Europaeus Agglutinin 1 lectin, which detects the α1,2-linked fucose, and the level of expression was not changed with GCDCA treatment (SI Appendix, Fig. S6A). An ELISA for VLP binding to porcine gastric mucin HBGAs showed no altered binding of GII.3 VLPs when GCDCA was present (SI Appendix, Fig. S6B). Next, we evaluated whether GCDCA alters endocytosis in HIEs using two fluorescent markers that address different aspects of endocytosis: FM1-43FX, a lipophilic dye that is fluorescent only when incorporated into the plasma membrane and labels all endocytic vesicles originating from the apical brush border (46), and pHrodo-dextran, a 10-kDa dextran linked to a pH-sensitive dye that is taken up via endocytosis and is used to mark cellular compartments with low pH (47). In the presence of GCDCA, we observed a rapid and significant up-regulation of FM1-43FX–labeled endocytic vesicles and a corresponding increase in fluorescence intensity at 10 min posttreatment (Fig. 3A). This effect was not observed in HIEs treated with UDCA, the BA that does not enhance GII.3 replication in HIEs. Treatment with JTE-013 significantly decreased the up-regulation of FM1-43FX–labeled endocytic vesicles by GCDCA treatment (Fig. 3B). Elevated endocytosis due to GCDCA treatment was transient, and levels normalized by 60 min posttreatment (SI Appendix, Fig. S7). Using pHrodo dextran, a significant increase in fluorescence was observed by 30 min in the presence of GCDCA (Fig. 3C). Additionally, we found that GII.3 stool inoculum did not significantly elevate internalization of dextran or its transit to acidic compartments (Fig. 3C). We also used fluorescently labeled GII.3 VLPs to analyze the role of GCDCA on particle internalization. Treatment of HIEs with GCDCA significantly increased VLP uptake into the HIEs (Fig. 3D). Addition of methyl-β-cyclodextrin (MβCD) during infection, which sequesters cholesterol from lipid rafts in the plasma membrane and in turn inhibits plasma membrane invagination and subsequent endocytosis, led to the suppression of GII.3 replication (Fig. 3E). Together, these results indicate GCDCA treatment results in a S1PR2-dependent rapid and transient increase in endocytosis coupled with acidification and increased particle (VLP) uptake.
Fig. 3.
GCDCA-mediated endocytosis enhances GII.3 infection. (A) Endocytosis in HIE monolayers treated with either medium alone, 500 µM GCDCA, or 500 µM UDCA was analyzed by confocal laser-scanning microscopy. The endocytic vesicles (green) and nuclei (blue) were visualized by FM1-43FX and DAPI, respectively. (Scale bar, 10 µm.) (Right) Quantitation of the FM1-43FX fluorescence. (B) Endocytosis was monitored as in A in HIE monolayers treated with 500 µM GCDCA in the presence or absence of 40 µM JTE-013 (JTE). Quantitation is to the right. (C) Endocytosis in HIE monolayers in the presence of stool virus was analyzed using pHrodo-labeled dextran (10 kDa) by flow cytometry. Percentage change in the number of pHrodo+ cells treated with additives compared to untreated cells is shown. Untreated pHrodo+ cells were set to 100% (red dashed line). (D) HIE monolayers incubated with Cy5-labeled GII.3 VLPs for 1 h in the presence or absence of GCDCA were observed by confocal laser-scanning microscopy. (E) Cells were pretreated with the cholesterol sequestrant MβCD for 1 h, and then MβCD and GCDCA were added to the medium at the indicated concentrations during infection with GII.3 as in Fig. 1 for 24 h. (A–E) P values between conditions are indicated. n.s., not significant. Error bars denote SD.
BA Induces Endosomal Acidification That Is Required for GII.3 Replication.
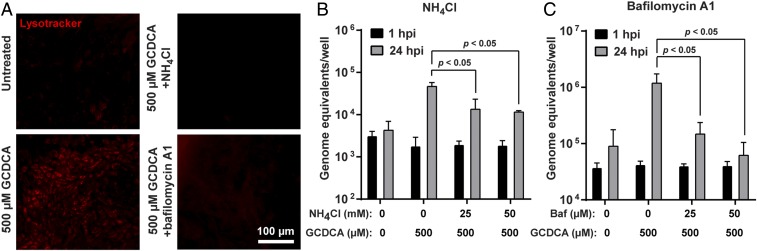
Many viruses, including other caliciviruses, require entry through acidified endosomes (48–50). Therefore, we tested the effect of GCDCA on endosome acidification in GII.3 infection. The pHrodo-dextran results suggested GCDCA treatment targets dextran to acidic compartments in HIEs and hydrophobic BAs are reported to lead to endosomal acidification in a hepatocyte model (51). To determine whether GCDCA caused a significant increase in endo-lysosomal compartments with acidic pH, we used LysoTracker that labels acidic compartments. GCDCA treatment clearly showed enhanced levels of endocytic compartments; this effect of GCDCA was negated by the presence of endosomal acidification inhibitors, such as NH4Cl (neutralizes pH in acidic components) and bafilomycin A1 (inhibits vacuolar-type H+ ATPase) (Fig. 4A). Furthermore, addition of either inhibitor to the medium during GII.3 replication resulted in a significant and dose-dependent reduction of GII.3 replication (Fig. 4 B and C). Together, these results show that BA-induced endosomal acidification occurs in HIEs and that the endosomal acidification is required for GII.3 replication.
Fig. 4.
Endosomal acidification induced by BA is critical for GII.3 replication. (A) HIE monolayers were untreated or treated with differentiation medium containing the indicated additives for 1 h. After two washes with CMGF(−), the cells were incubated with LysoTracker for 10 min and observed by epifluorescence microscopy. Red fluorescence indicates acidic compartments. (Scale bar, 100 µm.) (B and C) HIE monolayers were pretreated with NH4Cl (B) and bafilomycin A1 (C) at the indicated concentrations. Monolayers were infected with GII.3 as in Fig. 1 and harvested at 24 hpi. NH4Cl and bafilomycin A1 were present during and postinoculation. P values between conditions are indicated.
Activity of ASM Is Critical for GII.3 Replication.
To delineate the mechanism by which BA-induced endosomal acidification supports GII.3 replication, we tested the importance of endosomal enzymes activated by acidification. Cathepsins are proteases in acidic endosomes/lysosomes that can alter viral structure by proteolytic cleavage. Ebola virus, reovirus, and other caliciviruses use these structural changes to escape from the endosomal/lysosomal pathway during entry (48, 52, 53). In contrast, the addition of protease inhibitors during GII.3 infection to block cathepsin activity (pepstatin A against cathepsin B and L; E64 against cathepsins D and E) failed to decrease GII.3 replication (SI Appendix, Fig. S8).
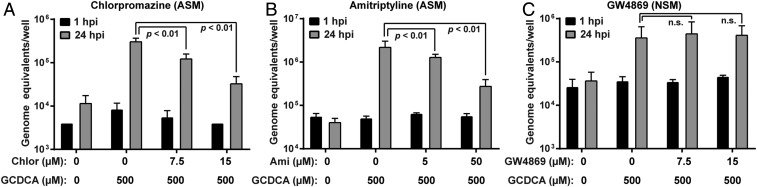
We next evaluated the involvement of another endosomal/lysosomal enzyme, ASM. Two inhibitors of ASM (chlorpromazine and amitriptyline) resulted in a significant decrease in GII.3 replication (Fig. 5 A and B). Inhibition by GW4869 of neutral sphingomyelinase (NSM), another sphingomyelinase that functions in neutral pH conditions, did not change GII.3 replication (Fig. 5C). Taken together, these data indicate the importance of acidic pH conditions for sphingomyelinase activity and endosomes for GII.3 infection.
Fig. 5.
BA-induced ASM activity is required for GII.3 replication. (A–C) HIE monolayers were infected with GII.3 as in Fig. 1 for 24 h. Inhibitors for ASM (A and B) and NSM (C) were present 1 h prior to, during, and postinoculation. P values between conditions are indicated.
Ceramide Plays an Important Role in GII.3 Replication.
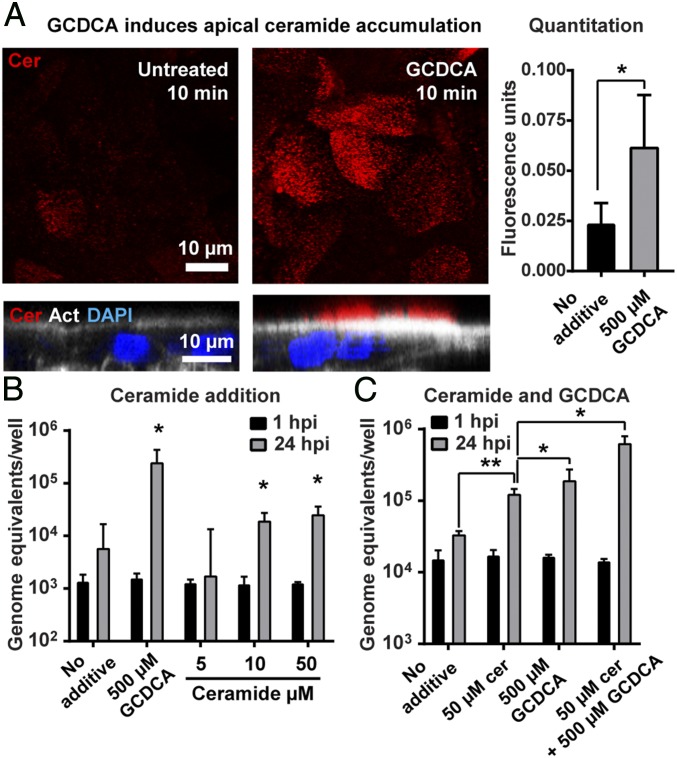
ASM converts sphingomyelin to ceramide in plasma membranes and endocytic membranes. Therefore, we investigated the role of ceramide in GII.3 replication. Strikingly, HIEs treated with GCDCA for 10 min and stained with an anti-ceramide antibody (54) showed significant rapid increases in ceramide at the apical surface (Fig. 6A); this increase was transient as, by 30 min after GCDCA treatment, ceramide levels were similar in untreated and GCDCA-treated cells (SI Appendix, Fig. S9). We next evaluated the exogenous addition of the cell permeant C2-ceramide during GII.3 infection, which is expected to bypass the requirement for BA-activation of ASM at the plasma membrane, and found that ceramide alone supported modest GII.3 replication (Fig. 6B). Furthermore, addition of exogenous C2-ceramide in combination with GCDCA resulted in a significant and synergistic enhancement of GII.3 replication (Fig. 6C). In contrast, the exogenous addition of sphingomyelin, which is a precursor of ceramide, failed to support GII.3 replication (SI Appendix, Fig. S10). To determine whether ceramide acts as a cellular receptor for GII.3, direct binding of GII.3 VLP to a phospholipid panel including ceramide was assayed (SI Appendix, Fig. S11). There was no specific binding of GII.3 to any phospholipid or sphingolipid, including ceramide. As a positive control, cholera toxin B subunit (CTxB) did bind to GM1 ganglioside, its known receptor (SI Appendix, Fig. S11). These results indicate that a specific plasma membrane lipid, ceramide, is sufficient for GII.3 replication. However, ceramide itself is not the cellular receptor for GII.3.
Fig. 6.
Ceramide enhances GII.3 replication. (A) Ceramide levels in HIE monolayers treated with GCDCA were analyzed by IF microscopy (Left). The Upper panels show ceramide staining (red) in optical slices at the apical surface of monolayers in the XY plane, while the Lower panels show orthogonal views. The orthogonal view shows the actin network at the base of the microvilli on the brush border (white, phalloidin) and nuclei (blue, DAPI). (Scale bars, 10 μm.) (Right) Quantification of the fluorescence intensity as described in Materials and Methods. (B and C) The HIE monolayers were infected with GII.3 as in Fig. 1 for 24 h with the indicated additives. Viral GEs at 1 and 24 hpi were quantified by RT-qPCR. (B) *P < 0.05 comparing GEs at 24 hpi to 1 hpi. (C) *P < 0.05 and **P < 0.01 comparing GEs of each sample at 24 hpi.
BA Treatment of HIEs Alters Endosomal–Lysosomal Dynamics.
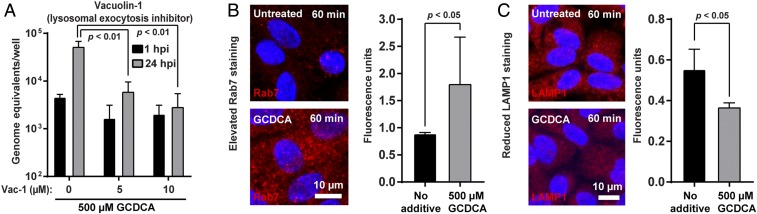
To determine if the increased ceramide on the apical surface occurs as a result of ASM release from lysosomes, vacuolin-1, a lysosomal exocytosis inhibitor, was added to the medium during infection. GII.3 replication was significantly reduced in the presence of vacuolin-1 (Fig. 7A). A time course of vacuolin-1 addition showed that vacuolin-1 acts early in GII.3 replication, indicating inhibition of lysosomal release affects early stages of infection (SI Appendix, Fig. S12). IF microscopy to observe the change of endosomal/lysosomal trafficking induced by BA demonstrated a significant increase of Rab7, a marker of late endosomes, and a decrease in LAMP1, a marker of lysosomes, in HIEs 60 min after treatment with GCDCA compared to nontreated HIEs (Fig. 7 B and C). Taken together, these data indicate GCDCA treatment of HIEs increases the number of late endosomes and decreases the number of lysosomal compartments due to lysosomal exocytosis, which is critical for GII.3 infection. The decrease in LAMP1 signal could also be explained by diffusion of LAMP1 in membranes to below the detection limit of our IF assay through lysosomal fusion with endosomes.
Fig. 7.
Lysosomal exocytosis is critical for GII.3 infection and GCDCA alters the endo-lysosomal dynamics. (A) HIE monolayers were infected as in Fig. 1 with GII.3 in the presence of vacuolin-1 and GCDCA at the indicated concentration for 24 h. Vacuolin-1 was added to the medium 1 h prior to infection and vacuolin-1 and GCDCA were maintained in the medium postinoculation. P values between conditions are indicated. (B and C) HIE monolayers incubated with or without GCDCA for 60 min. Images were acquired by confocal laser-scanning microscope to detect Rab7 (B) and LAMP1 (C) in red, respectively, and nuclei in blue. (Scale bar, 10 μm.) (Right) Quantitation of Rab7 and LAMP1 signals. P values between conditions are indicated.
Discussion
The human small intestinal epithelium, the site of replication for many gastrointestinal viral pathogens, presents a protective environment and barrier against microbial invasion. The luminal milieu is composed of a complex intestinal fluid including pancreatic enzymes, phospholipids, cholesterol, and bile salts deposited into the duodenum as part of the cyclic process of enterohepatic circulation. We previously found bile is required for in vitro replication of a subset of HuNoVs and the effect is on the cells and not on the virus (32). A recent report evaluated BA binding in vitro to human norovirus VLPs and found that BA does not bind several cultivatable HuNoV strains, including GI.1, GII.3, GII.4, and GII.17 (35), consistent with our previous results. Our current study sought to identify active factors in bile and understand the mechanism by which these factors mediate successful replication of GII.3 HuNoV in physiologically active small intestinal HIEs. Our results demonstrate that GII.3 HuNoV exploits hydrophobic, primarily conjugated BAs, a major component of the small intestinal milieu, for entry and infection of jejunal HIEs. We found that GCDCA, used as a model BA, has multiple effects on nontransformed jejunal cultures to promote virus entry and subsequent replication. GCDCA increases endocytosis, endosomal acidification, and stimulates rapid ceramide appearance at the apical plasma membrane, likely through an ASM-dependent pathway. Ceramide alone did not increase endocytosis, indicating that BA is the primary component driving virus uptake. Specificity for GCDCA-driven endocytosis was shown by a lack of endocytosis with UDCA, which also failed to support virus replication. Finally, when ceramide and GCDCA were added together, infection was dramatically increased over GCDCA alone, indicating that the two molecules act synergistically to enhance virus replication.
Most conjugated and primary unconjugated BAs that support replication are present in the small intestine, the primary site of HuNoV replication, whereas secondary unconjugated BAs generated by bacterial modification were less effective or not effective at all (UDCA). The highest concentration of any active BA tested (500 µM) is below the critical micellar concentration. Although the glycine-conjugated BA, GCDCA, can passively be absorbed and enter cells in small amounts in its protonated form, passive absorption seems unlikely in our system because this conjugated BA should be ionized when it is delivered to cells in medium near neutral pH (44, 45). This led to the question whether active hydrophobic BAs identified to be critical for GII.3 replication function as detergents or through a specific receptor.
We tested two hypotheses to understand how GCDCA acts on the human jejunal enteroid cultures to allow virus replication. First, BAs are amphipathic molecules that have detergent-like properties that at low concentration permit BAs to interact with lipids at sublytic levels (55). We found that BA-enhanced virus replication was not simply due to the amphiphilic detergent-like properties of BAs based on testing a set of ionic (SDS), nonionic (Triton X-100, Nonidet P-40), and zwitterionic (CHAPS) detergents. Second, BAs could interact with the cell through specific BA receptors. This hypothesis was prompted by knowing that protein receptors are sensitive to the stereochemistry of their ligand. In our GII.3 infection system, we observe differential activity of CDCA that supports virus replication compared to the inactive UDCA, with these two BAs having similar structures except that UDCA is a 7β-epimer of CDCA. Several receptors associated with BAs were tested. The nuclear BA receptor FXR is traditionally studied in the context of the ileum; however, we found that FXR is well expressed in the jejunal HIEs (fragments per kilobase of transcript per million mapped reads > 20), and GCDCA treatment of the jejunal HIEs resulted in up-regulation of the downstream molecules FGF19 (∼ninefold) and FABP6 (∼fivefold) when compared to cells treated with medium alone. However, this FXR classic BA-signaling pathway was not involved in GII.3 infection because treatment of cells with agonists and an antagonist of FXR resulted in no effect on GII.3 replication in the absence or presence of GCDCA, respectively. These results differ from a BA-FXR activation requirement for hepatitis C virus replication using a replicon system in Huh-7 cells (56, 57). We also found that HuNoV replication was not dependent on BA activation of the GPCR TGR5 that is present in both jejunal and ileal HIEs (ref. 41 and our RNA-seq data). These results are similar to the lack of the FXR or TGR5 pathway involvement in the BA requirement for porcine sapovirus (PoSaV) replication in porcine LLC-PK1 cells (58).
We tested another GPCR, S1PR2, which can bind to hydrophobic BAs (43, 59). Inhibition of S1PR2 also significantly decreased endocytosis caused by GCDCA treatment and resulted in a dose-dependent reduction of GII.3 replication in the presence of GCDCA in jejunal HIEs in conjunction with chemical receptor inhibition. Overall, these data indicate that GCDCA specifically interacts with S1PR2 to promote GII.3 infection and we predict this likely stimulates downstream signaling that supports virus replication. In hepatocytes, BA-activation of S1PR2 stimulates kinase signaling pathways and can modulate lipid and glucose metabolism. The S1PR2 receptor is understudied in the intestine, but some evidence suggests that S1PR2 activation by BAs promotes proliferation (60, 61). In LLC-PK1 cells, BAs directly activate signaling pathways (e.g., MEK/ERK cascades) required for PoSaV replication, although whether this requires a specific receptor remains unclear (62). Future studies will determine which signaling cascades are stimulated by BA-activation of S1PR2 in jejunal HIEs and if the activated signaling is required for HuNoV replication and possibly other caliciviruses in their individual model systems.
Our data indicating that primarily hydrophobic conjugated BAs support GII.3 HuNoV replication are consistent with a requirement for BAs in the replication of other viruses in the Caliciviridae family. This virus family includes Norovirus and four other genera (Lagovirus, Nebovirus, Sapovirus, and Vesivirus). Recently, six new genera, tentatively named Recovirus, Valovirus, Bavovirus, Nacovirus, Salovirus, and Sanovirus, have been added as unclassified caliciviruses (63). The most notable genera in the family are Norovirus and Sapovirus, which cause severe acute gastroenteritis in humans and animals, with noroviruses alone causing over 200,000 deaths annually in children <5 y of age (64). The Cowden strain of PoSaV was the first virus strain in the Sapovirus genus to be cultivated and the presence of porcine intestinal content (IC) fluid filtrate was required for in vitro cultivation in primary porcine kidney cells (65, 66). The IC effects on the growth of PoSaV in cell culture were initially associated with the induction of a protein kinase A signaling pathway by IC, which suggested a novel mechanism for this virus–host relationship dependent on a specific cellular environment (67). Subsequent studies revealed that bile and BAs, specifically GCDCA, function as active factors essential for growth of PoSaV in the continuous cell line LLC-PK1 (39). BAs were reported to induce an increase in cAMP concentration in LLC-PK1 cells that was associated with down-regulation of IFN-mediated signal transducer and activator of transcription 1 (STAT1) activation, a key element in innate immunity (39). This down-regulation was proposed to allow virus replication but this hypothesis has since been questioned based on studies showing that PoSaV replication remains restricted by IFN-mediated signaling pathways, such as IFN, STAT1 and 2′-5′-oligoadenylate synthetases in cells in the presence of GCDCA (68).
Further studies with PoSaV indicated that GCDCA induces endosomal acidification and subsequent ASM activation and ceramide production, resulting in the instability of endosome/lysosome membranes that allow viral escape from the endosomal compartment in LLC-PK1 cells (39, 58, 69). PoSaV, feline calicivirus (FCV), and murine norovirus (MNV-1) particles were shown to require ASM activation, an unexpected result because these latter two viruses were already easily cultivatable in Crandell-Rees feline kidney cells and mouse macrophages (RAW264.7), respectively, and neither virus had previously been thought to require BA for replication (70, 71). Our results indicate that GII.3 replication in HIEs shares some features with these other caliciviruses related to BAs and viral entry steps. In addition to endosomal acidification, we surprisingly found GCDCA likely changes endosomal/lysosomal trafficking and rapidly induces ceramide production on the apical membrane in HIEs. Our observations of increased Rab7, decreased LAMP1 and reduced GII.3 replication when the lysosomal exocytosis inhibitor, vacuolin-1, is added early after infection indicate that GCDCA-induced exocytosis of lysosomes may be involved in the rapid release of lysosomal contents including ASM to the apical surface. It is tempting to speculate that these effects with enhanced endocytosis are mediated through BA-activated S1PR2 GPCR signaling cascades. One limitation of the present study is that our findings relied on chemical inhibitors of ASM to determine its importance. Despite multiple attempts, we were unable to generate an ASM knockout HIE line and we currently lack a genetic model to directly evaluate ASM and infection. Future studies are needed to determine the exact mechanism of action.
Although we demonstrate that exogenous C2-ceramide enhances GII.3 replication (Fig. 6 B and C), the actual sites where the exogenous ceramide functions remain unclear. The C2-ceramide used in this study is permeable to cells and could move to endosomes/lysosomes where it could assist virus release into the cytoplasm. Interestingly, ceramide production to repair endosomal membrane lesions caused by cold treatment is thought to drive replication of PoSaV in LLC-PK1 cells at 4 °C even in the absence of BA (69). Besides ceramide production in endosomes, the emergence of lesions on the cell surface may lead to ceramide production and enhanced endocytosis, which allows PoSaV to enter cells in the absence of BAs. Exogenous C2-ceramide addition has been recently reported for MNV replication in immortalized mouse microglia (BV2) cells (72). In that system, serine palmitoyltransferase was found to be required for sphingomyelin biosynthesis and for MNV replication. Serine palmitoyltransferase was required for the proteinaceous receptor (CD300lf) to obtain a proper functional conformation on cells, and it was proposed that ceramide might bind to the receptor in a lipid pocket (72). CD300lf has also been shown to have two binding sites for BAs using GCDCA and LCA [which is not active for HuNoV at testable concentrations (73)]. An interesting question to address is whether all caliciviruses might require BAs, ceramide, and ASM for their optimal replication. The precise BAs needed for supporting calicivirus replication may also depend on the culture system and the specific pathogenesis of each virus. For example, GCDCA and LCA bind to the MNV P-domain and GCDCA increases cell binding and infection (73). However, no enhancement of infection was observed with taurocholic acid (TCA), glycocholic acid (GCA), CA, or tauroursodeoxycholic acid (TUDCA) (73). In contrast, HuNoV replication is supported in HIEs treated with 500 µM GCDCA, TCA, GCA, and CA. LCA showed no effect at the highest testable concentration with no cytotoxicity, 50 µM. The difference in mechanism between MNV (BA P-domain binding) and HuNoV (BA cellular effects) as well as the difference in specific functional BAs may reflect HuNoV tropism for the small intestine, while some strains of MNV can also replicate in the colon (74–76).
To further define and fully understand the mechanism of GCDCA and ceramide as cofactors for GII.3 HuNoV entry into HIEs, it remains important to discover the cellular receptor for HuNoV. Several known receptors for other caliciviruses are tight junctional proteins, such as species-specific junctional adhesion molecule receptors for FCV, Tulane, and Hom-1 calicivirus (70, 77–79), and occludin for PoSaV (80). These are sufficiently distinct from the CD300lf MNV receptor present on macrophages, dendritic, and tuft cells (81), making it challenging to develop a unifying hypothesis for how GCDCA, ceramide and ASM function to enhance virus entry into cells. These previously identified receptors (except CD300lf) for other caliciviruses are present in many human intestinal cancer cell lines that do not support HuNoV replication, even in the presence of added bile (8, 32), suggesting that the HuNoV receptor is distinct.
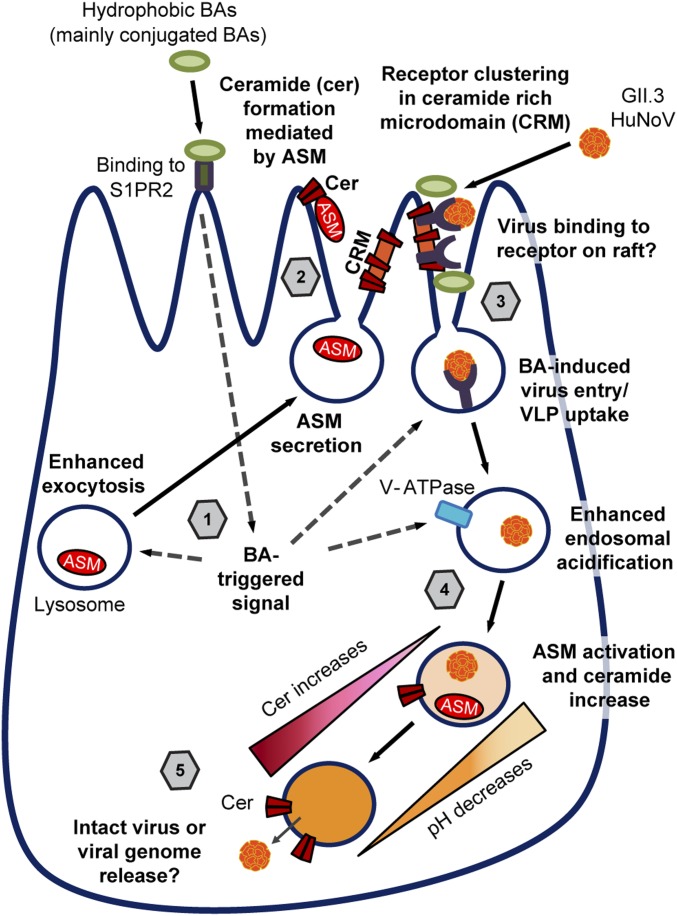
Based on our current results, we propose a model for HuNoV replication (Fig. 8). We predict ceramide is rapidly induced by BA and produced on the cell surface by ASM that mediates the formation of ceramide-rich microdomains, where membrane proteins cluster, and with BA present, rapidly stimulate endocytosis. Epidermal growth factor receptor (EGFR) has been well studied as a model to understand the mechanism of clustering of raft proteins and subsequent endocytosis (82). In a study on EGFR in HIEs, DCA induces EGFR-associated endocytosis mediated by proteolipid assembly and subsequent EGFR clustering (83). However, it is unlikely that the endocytosis induced by EGFR activation is directly involved in GII.3-related endocytosis, because treatment with differentiation medium alone, which contains EGF and is used to grow HIEs, had no effect on GII.3 replication in the absence of BAs. In addition, the observation of ceramide accumulation on the apical membrane and enhanced GII.3 VLP uptake in response to GCDCA treatment leads us to speculate that the formation of ceramide-rich microdomains may be important for clustering of the still unknown HuNoV receptor that subsequently leads to GII.3 entry. Chron1, a GII.3 VLP, derived from a HuNoV strain that caused chronic infection, can bind to sialyl Lewis x (84). This glycan can be present on glycosphingolipid (GSL) structures, including ceramide-containing GSLs. In fact, GII.4 and GI.1 VLPs have been shown to bind GSLs in an HBGA-dependent manner (85–88). The ceramide-rich regions may condense GSLs together to act as an initial binding factor or receptor for GII.3 and other norovirus strains. Confirmation of this hypothesis will require identification of the cellular receptor for HuNoV in human intestinal epithelial cells, and it will be interesting to learn if there are regional-specific receptors or mechanisms of virus entry in the proximal or distal intestinal cells. Furthermore, there are strain- and variant-specific differences in how HuNoVs behave and whether the GII.3 used in our present study binds to GSLs remains to be tested.
Fig. 8.
Proposed model for GII.3 infection in HIEs supported by BA and ceramide. 1) Hydrophobic BAs, mainly conjugated BAs, may bind to BA receptor on the cell surface such as S1PR2 or interact with the cell in an undetermined manner and trigger rapid signals. The BA-mediated signals induce lysosomal exocytosis that releases lysosomal contents including ASM to apical surface. 2) The released ASM produces ceramide in the apical membrane that induces ceramide-rich microdomain formation. 3) The ceramide-rich microdomain likely cluster receptors, by which GII.3 can bind and subsequently enter into cells through BA-signaled uptake. 4) Concurrently, BAs increase endosomal acidification. 5) We hypothesize that virus or viral genome escape from the endosome is mediated by endosomal ASM activation and subsequent local ceramide formation, which increases the permeability of the endosomal membrane permitting release.
Based on our data, after BA treatment, GII.3 entry into cells involves uptake into acidic endosomes. Endosomal conditions and interaction with a yet unknown receptor likely aid in the uncoating of the viral particle, and future studies are required to determine the mechanism. In the PoSaV model, BAs required for endosomal escape and ASM inhibition, which would prevent ceramide generation, led to PoSaV retention in endosomes (58). FCV when bound to its receptor, feline junctional adhesion molecule A, undergoes structural changes that lead the minor capsid protein VP2 to form a portal-like structure in the capsid (89). One hypothesis is that VP2 in HuNoV could form a similar pore structure that would allow the genome to pass through the endosomal membrane. Ceramide can alter membrane curvature, increase lipid flip-flop, and locally destabilize bilayers (90). Based on these studies, we postulate a similar model where GII.3 or the viral genome escapes from endosomes rich in ceramide followed by subsequent translation of the polyprotein, RNA replication, and assembly of new virus particles. Our studies focused on GII.3 HuNoV, but future studies should confirm if the same BA-mediated mechanism is used by other bile-dependent strains. Recently it was shown that HuNoV can be released in vesicles in stool (91). It will be interesting to determine if HuNoV are also released in vesicles from HIE cells and if this is altered by the presence of BA or ceramide.
In summary, we show that hydrophobic BAs mediate multiple effects on HIE cultures early in infection to permit GII.3 HuNoV replication. BAs signal for lysosomal exocytosis to release ASM and generate ceramide-rich microdomains, increase cellular uptake, and increase low pH compartments. Ceramide formation alone did not increase particle uptake but is sufficient for low levels of GII.3 replication and in combination with GCDCA synergistically enhances replication. This study provides unexpected findings of the effect of BAs on the small intestinal jejunal epithelium and provides evidence that, similar to other enteric caliciviruses, GII.3 HuNoV requires BA and ASM-mediated generation of ceramide for successful entry and subsequent replication.
Materials and Methods
Virus, Cells, and Infections.
Infection of GII.3 HuNoV strain TCH04-577 with secretor-positive HIE cultures (see SI Appendix, Table S2 for HBGA phenotypes of HIE lines) was performed as previously described (32–34). In brief, the HIE cultures established previously from adult surgical tissue were grown as multilobular 3D HIEs in Matrigel. For HuNoV infection, the 3D HIEs were dissociated by TrypLE Express (ThermoFisher) and seeded onto collagen IV coated 96-well plates. Following culture in complete medium with growth factors [CMGF(+)] for 24 h, monolayer HIEs were cultured for 5 d in differentiation medium and were treated as described separately or inoculated with HuNoV. The monolayer HIEs were inoculated with stool filtrate containing 4.3 × 105 genome equivalents (GEs; unless otherwise indicated) of GII.3 HuNoV per well for 1 h at 37 °C in 5% CO2 and washed twice with complete medium without growth factors [CMGF(−)] to remove unattached viruses. The inoculated cells were cultured in differentiation medium under different experimental conditions for up to 72 h. Additional, individual additives were supplemented into the medium during and postvirus inoculation, unless otherwise noted. Each experiment was performed at least twice with three technical replicates in each condition. Viral GEs were analyzed by reverse-transcriptase quantitative PCR (RT-qPCR) at 1 h and at indicated later time points for viral replication.
VLPs.
GII.3 TCH04-577 VLPs were produced and purified, as previously described (14, 92).
Bile Acids.
All BAs were purchased from Sigma Aldrich. GCA, glycodeoxycholic acid (GDCA), GCDCA, TCA, taurodeoxycholic acid (TDCA), TCDCA, and TUDCA were dissolved in distilled water. CA, CDCA, DCA, UDCA, LCA, and taurolithocholic acid (TLCA) were dissolved in dimethyl sulfoxide (DMSO).
RT-qPCR.
RT-qPCR was performed as described previously (32). RNA analyzed by RT-qPCR was extracted from the cells and medium in this study. In some experiments, RNA was extracted using MagMAX-96 viral RNA isolation kit (Applied Biosystems) with a Kingfisher Flex Purification System (ThermoFisher) or Direct-zol RNA MiniPrep kit (Zymo) as described by the manufacturer. A standard curve based on a recombinant HuNoV RNA transcript was used to quantitate viral GEs in RNA samples.
IF Staining.
HIE monolayers were grown in glass-bottom 10-well culture slides (Cat. no. 543979) or 96-well plates (Cat. no. 655892) from Greiner Bio-One for all imaging. The HIEs were fixed with fresh 4% paraformaldehyde (PFA) for 20 min at room temperature. For detecting HuNoV proteins, the primary antibodies were incubated overnight at 4 °C. Expression of the capsid protein (VP1) was detected using antisera to guinea pig anti–TCH04-577 VLPs (1:250), or to nonstructural proteins (rabbit anti-U201 NTPase [1:100]) (30, 93). For Rab7 and LAMP1 detection, the HIE monolayers were permeabilized with 0.1% Triton X-100 in PBS for 30 min and blocked with 5% BSA in PBS at room temperature. All of the subsequent steps were performed in PBS + 0.1% Triton X-100. The rabbit anti-Rab7 and anti-LAMP1 antibodies (Cell Signaling Technologies, Cat. Nos. 9367S and 9091S, respectively) were incubated at 4 °C overnight followed by three washes for 5 min each at room temperature. For ceramide detection, HIE monolayers treated with differentiation medium alone or medium with 500 µM GCDCA for either 10 or 30 min at 37 °C. Monolayers were then fixed as described above, permeabilized with 0.05% Triton X-100 in PBS for 5 min at room temperature, and blocked with 0.2% BSA. All subsequent steps were done in PBS without any detergent. Ceramide was detected with rabbit anti-ceramide polyclonal antibody (54). Donkey anti-rabbit 594 Dylight (1:1,000, Rockland) and Alexa Fluor 594 or 488 (1:1,500; Invitrogen) secondary antibodies were used to visualize the viral and cellular proteins. For measuring endocytosis using FM1-43FX, HIE monolayers were treated with warmed medium at 37 °C containing 10 µg/mL of FM1-43FX (ThermoFisher, Cat. no. F35355) alone or with 500 µM GCDCA for either 10 or 60 min at 37 °C. After a quick wash with warmed differentiation medium, the monolayers were fixed in 4% PFA and stained with 300 nM DAPI in PBS for 5 min at room temperature.
Fluorescently Labeled VLPs.
Fluorescently labeled GII.3 VLPs were generated using Cy5 mono reactive dye pack (PA25001, Amersham) as per the manufacturer’s instructions. The labeled VLPs were purified by passing through a G-25 Sephadex column followed by ultracentrifugation at 174,000 × g for 90 min. The labeling was confirmed by colocalizing Cy5 fluorescence and capsid antigen by IF using guinea pig anti-GII.3 VP1 serum and goat anti-guinea pig Alexa Fluor 488. After incubating HIE monolayers on ice for 15 min, 107 Cy5-labeled VLPs were added to each well in cold differentiation medium for 1 h. The cells were washed three times with cold differentiation medium. Then, warmed differentiation medium was added and the cells were incubated for 10 min or 60 min at 37 °C. For imaging, the HIEs were fixed with 4% PFA in PBS, washed and stained for 5 min at room temperature with 300 nM DAPI.
Confocal Microscopy and Quantitation.
HIE monolayers were imaged using a Nikon A1 confocal microscope, as described (32). Quantitation of the fluorescence was done using Fiji software. Briefly, a threshold was set for each confocal stack using the default preset, 12 identical elliptical regions-of-interests (ROIs) were drawn per field and the fluorescence intensity in ROIs was counted. In total, 180 ROIs from three wells were analyzed for each time point and treatment.
Agonists, Antagonists, and Inhibitors.
FXR, TGR5, and S1PR2 were evaluated pharmacologically by using commercially available compounds: FXR agonists (INT747 and GW4064 [Sigma Aldrich]) and antagonist (Z-guggulsterone [Santa Cruz]); TGR5 agonists (INT-777 and CCDC [Cayman Chemical]); S1PR2 antagonist (JTE-013 [Cayman Chemical]). Inhibitors against cathepsins (pepstatin A and E64 [Sigma Aldrich]), ASM (amitriptyline [Sigma Aldrich] and chlorpromazine [Santa Cruz]), and NSM (GW4869 [Santa Cruz]) were used in the current study. Cellular behaviors in the presence of BAs were studied by using NH4Cl (Sigma Aldrich), bafilomycin A1 (Fisher Scientific), MβCD (Sigma), and vacuolin-1 (Santa Cruz).
Sphingolipids.
C2-ceramide was purchased from Enzo Life Sciences and sphingomyelin was purchased from Santa Cruz. Both were dissolved in ethanol and exogenously added to the medium.
Statistical Analysis.
Each experiment was performed two or more times, with three technical replicates of each culture, condition, and time point in each experiment. Data from one representative experiment is presented. All statistical analyses were performed on GraphPad Prism (GraphPad Software). Samples with RNA levels below the limit of detection of the RT-qPCR assay were assigned a value that is one-half the limit of detection of the assay. Comparison between 1-h groups and either 12-, 24-, or 72-h groups was performed using the Student’s t test, with statistical significance determined using the Holm–Sidak method. Where comparisons were made at 24 h or 72 h between treatment groups a two-way ANOVA was performed using Dunnett’s test for post hoc analyses. P < 0.05 was considered statistically significant. Error bars in graphs of infection experiments denote SD.
Data Availability.
All data, including RNA-seq data, used to support the findings of this study are present in the main text and SI Appendix or available from the authors upon request.
Supplementary Material
Acknowledgments
This research was supported by in part by NIH Grant P01 AI57788; the Texas Medical Center Digestive Diseases Center supported by PHS Grants P01AI 057788 (to M.K.E.) and P30 DK 56338 from the NIH; by Agriculture and Food Research Initiative Competitive Grant 2011-68003-30395 from the US Department of Agriculture, National Institute of Food and Agriculture (to M.K.E.); by Japan Agency for Medical Research and Development Grants JP18fk0108034 and JP19fk0108102 (to K.M.); Japan Society for the Promotion of Science KAKENHI Grant JP18K07153 (to K.M.); NIH grants R01AG034389 and R01NS095215 (to E.B.); National Science Foundation Grant 1615874 (to E.B.); Advanced Technology Core Laboratories (Baylor College of Medicine), specifically the Integrated Microscopy Core with funding from CPRIT (RP150578, RP170719); the Genomic and RNA Profiling Core with funding from NIH S10 Grant 1S10OD023469; and the Protein and Monoclonal Antibody Production Core, the Flow Cytometry Core at Baylor College of Medicine with funding from the NIH (CA125123, The Dan L Duncan Comprehensive Cancer Center, and Baylor College of Medicine Office of Research). We thank Xiaomin Yu and Yoko Yamaoka for technical assistance and Alan Hofmann for helpful discussions.
Footnotes
Competing interest statement: M.K.E. is named as an inventor on patents related to cloning and cultivation of the Norwalk virus genome and is a consultant to and received research funding from Takeda Vaccines, Inc. R.L.A. has received research funding from Takeda Vaccines, Inc.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910138117/-/DCSupplemental.
References
- 1.Ahmed S. M., et al. , Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 14, 725–730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall A. J., et al. , Norovirus disease in the United States. Emerg. Infect. Dis. 19, 1198–1205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pires S. M., et al. , Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 10, e0142927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramani S., Atmar R. L., Estes M. K., Epidemiology of human noroviruses and updates on vaccine development. Curr. Opin. Gastroenterol. 30, 25–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartsch S. M., Lopman B. A., Ozawa S., Hall A. J., Lee B. Y., Global economic burden of norovirus gastroenteritis. PLoS One 11, e0151219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belliot G., Lopman B. A., Ambert-Balay K., Pothier P., The burden of norovirus gastroenteritis: An important foodborne and healthcare-related infection. Clin. Microbiol. Infect. 20, 724–730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havelaar A. H., et al. ; World Health Organization Foodborne Disease Burden Epidemiology Reference Group , World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12, e1001923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duizer E., et al. , Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85, 79–87 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Herbst-Kralovetz M. M., et al. , Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg. Infect. Dis. 19, 431–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lay M. K., et al. , Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans. Virology 406, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papafragkou E., Hewitt J., Park G. W., Greening G., Vinjé J., Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PLoS One 8, e63485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroneman A., et al. , Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 158, 2059–2068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chhabra P., et al. , Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 100, 1393–1406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X., Wang M., Graham D. Y., Estes M. K., Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66, 6527–6532 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green K. Y., Lew J. F., Jiang X., Kapikian A. Z., Estes M. K., Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J. Clin. Microbiol. 31, 2185–2191 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marionneau S., et al. , Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122, 1967–1977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindesmith L., et al. , Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9, 548–553 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Hutson A. M., Airaud F., LePendu J., Estes M. K., Atmar R. L., Norwalk virus infection associates with secretor status genotyped from sera. J. Med. Virol. 77, 116–120 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Huang P., et al. , Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: Identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188, 19–31 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Le Pendu J., Ruvoën-Clouet N., Kindberg E., Svensson L., Mendelian resistance to human norovirus infections. Semin. Immunol. 18, 375–386 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan M., Jiang X., Norovirus and its histo-blood group antigen receptors: An answer to a historical puzzle. Trends Microbiol. 13, 285–293 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Guix S., et al. , Norwalk virus RNA is infectious in mammalian cells. J. Virol. 81, 12238–12248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu L., et al. , Replication of human norovirus RNA in mammalian cells reveals lack of interferon response. J. Virol. 90, 8906–8923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotton B. T., et al. , The norovirus NS3 protein is a dynamic lipid- and microtubule-associated protein involved in viral RNA replication. J. Virol. 91, e02138-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettayebi K., Hardy M. E., Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 77, 11790–11797 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Vega V., et al. , Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 78, 4827–4837 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillenbrand B., et al. , Norovirus non-structural protein p20 leads to impaired restitution of epithelial defects by inhibition of actin cytoskeleton remodelling. Scand. J. Gastroenterol. 45, 1307–1319 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Sharp T. M., Guix S., Katayama K., Crawford S. E., Estes M. K., Inhibition of cellular protein secretion by norwalk virus nonstructural protein p22 requires a mimic of an endoplasmic reticulum export signal. PLoS One 5, e13130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asanaka M., et al. , Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 102, 10327–10332 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama K., et al. , Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc. Natl. Acad. Sci. U.S.A. 111, E4043–E4052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato T., et al. , Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Ettayebi K., et al. , Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou W. Y., et al. , Human intestinal enteroids: New models to study gastrointestinal virus infections. Methods Mol. Biol. 1576, 229–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantini V., et al. , Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis. 24, 1453–1464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilic T., Koromyslova A., Hansman G. S., Structural basis for human norovirus capsid binding to bile acids. J. Virol. 93, e01581-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett K. E., “Gastrointestinal physiology” in Ganong’s Review of Medical Physiology (McGraw-Hill Medical, ed. 23, 2009), pp. 429–487. [Google Scholar]
- 37.Kuipers F., Bloks V. W., Groen A. K., Beyond intestinal soap—Bile acids in metabolic control. Nat. Rev. Endocrinol. 10, 488–498 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Heuman D. M., Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J. Lipid Res. 30, 719–730 (1989). [PubMed] [Google Scholar]
- 39.Chang K. O., et al. , Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. U.S.A. 101, 8733–8738 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang J. Y., Bile acid metabolism and signaling. Compr. Physiol. 3, 1191–1212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T., Chiang J. Y., Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66, 948–983 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Studer E., et al. , Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 55, 267–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagahashi M., et al. , The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J. Lipid Res. 57, 1636–1643 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson P. A., Karpen S. J., Intestinal transport and metabolism of bile acids. J. Lipid Res. 56, 1085–1099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietschy J. M., Mechanisms for the intestinal absorption of bile acids. J. Lipid Res. 9, 297–309 (1968). [PubMed] [Google Scholar]
- 46.Hansen G. H., Rasmussen K., Niels-Christiansen L. L., Danielsen E. M., Endocytic trafficking from the small intestinal brush border probed with FM dye. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G708–G715 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Serfass J. M., et al. , Endophilin B2 facilitates endosome maturation in response to growth factor stimulation, autophagy induction, and influenza A virus infection. J. Biol. Chem. 292, 10097–10111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shivanna V., Kim Y., Chang K. O., Endosomal acidification and cathepsin L activity is required for calicivirus replication. Virology 464–465, 287–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart A. D., Brown T. D., Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J. Virol. 80, 7500–7509 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamauchi Y., Helenius A., Virus entry at a glance. J. Cell Sci. 126, 1289–1295 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Becker S., Reinehr R., Grether-Beck S., Eberle A., Häussinger D., Hydrophobic bile salts trigger ceramide formation through endosomal acidification. Biol. Chem. 388, 185–196 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Chandran K., Sullivan N. J., Felbor U., Whelan S. P., Cunningham J. M., Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baer G. S., Ebert D. H., Chung C. J., Erickson A. H., Dermody T. S., Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 73, 9532–9543 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnamurthy K., Dasgupta S., Bieberich E., Development and characterization of a novel anti-ceramide antibody. J. Lipid Res. 48, 968–975 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y., et al. , Bile acids modulate signaling by functional perturbation of plasma membrane domains. J. Biol. Chem. 288, 35660–35670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang K. O., George D. W., Bile acids promote the expression of hepatitis C virus in replicon-harboring cells. J. Virol. 81, 9633–9640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scholtes C., et al. , Enhancement of genotype 1 hepatitis C virus replication by bile acids through FXR. J. Hepatol. 48, 192–199 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Shivanna V., Kim Y., Chang K. O., The crucial role of bile acids in the entry of porcine enteric calicivirus. Virology 456–457, 268–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webster C. R., Anwer M. S., Hydrophobic bile acid apoptosis is regulated by sphingosine-1-phosphate receptor 2 in rat hepatocytes and human hepatocellular carcinoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G865–G873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen T., et al. , Sphingosine-1 phosphate promotes intestinal epithelial cell proliferation via S1PR2. Front. Biosci. 22, 596–608 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Ticho A. L., Malhotra P., Dudeja P. K., Gill R. K., Alrefai W. A., Bile acid receptors and gastrointestinal functions. Liver Res. 3, 31–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soliman M., et al. , Phosphatidylinositol 3-kinase/Akt and MEK/ERK signaling pathways facilitate sapovirus trafficking and late endosomal acidification for viral uncoating in LLC-PK cells. J. Virol. 92, e01674-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desselberger U., Caliciviridae other than noroviruses. Viruses 11, E286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel M. M., et al. , Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14, 1224–1231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flynn W. T., Saif L. J., Serial propagation of porcine enteric calicivirus-like virus in primary porcine kidney cell cultures. J. Clin. Microbiol. 26, 206–212 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parwani A. V., Flynn W. T., Gadfield K. L., Saif L. J., Serial propagation of porcine enteric calicivirus in a continuous cell line. Effect of medium supplementation with intestinal contents or enzymes. Arch. Virol. 120, 115–122 (1991). [DOI] [PubMed] [Google Scholar]
- 67.Chang K.-O., Kim Y., Green K. Y., Saif L. J., Cell-culture propagation of porcine enteric calicivirus mediated by intestinal contents is dependent on the cyclic AMP signaling pathway. Virology 304, 302–310 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Hosmillo M., et al. , Porcine sapovirus replication is restricted by the type I interferon response in cell culture. J. Gen. Virol. 96, 74–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shivanna V., Kim Y., Chang K. O., Ceramide formation mediated by acid sphingomyelinase facilitates endosomal escape of caliciviruses. Virology 483, 218–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ossiboff R. J., Parker J. S., Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J. Virol. 81, 13608–13621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wobus C. E., et al. , Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2, e432 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orchard R. C., Wilen C. B., Virgin H. W., Sphingolipid biosynthesis induces a conformational change in the murine norovirus receptor and facilitates viral infection. Nat. Microbiol. 3, 1109–1114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nelson C. A., et al. , Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc. Natl. Acad. Sci. U.S.A. 115, E9201–E9210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonzalez-Hernandez M. B., et al. , Efficient norovirus and reovirus replication in the mouse intestine requires microfold (M) cells. J. Virol. 88, 6934–6943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nice T. J., Strong D. W., McCune B. T., Pohl C. S., Virgin H. W., A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol. 87, 327–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taube S., et al. , Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J. Virol. 86, 5584–5593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makino A., et al. , Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 80, 4482–4490 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farkas T., Yang K., Le Pendu J., Baines J. D., Cardin R. D., The coxsackievirus and adenovirus receptor, a required host factor for recovirus infection, is a putative enteric calicivirus receptor. J. Virol. 93, e00869-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sosnovtsev S. V., et al. , Identification of human junctional adhesion molecule 1 as a functional receptor for the Hom-1 calicivirus on human cells. MBio 8, e00031-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alfajaro M. M., et al. , Early porcine sapovirus infection disrupts tight junctions and uses occludin as a coreceptor. J. Virol. 93, e01773-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilen C. B., et al. , Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360, 204–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Madshus I. H., Stang E., Internalization and intracellular sorting of the EGF receptor: A model for understanding the mechanisms of receptor trafficking. J. Cell Sci. 122, 3433–3439 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Liang H., Estes M. K., Zhang H., Du G., Zhou Y., Bile acids target proteolipid nano-assemblies of EGFR and phosphatidic acid in the plasma membrane for stimulation of MAPK signaling. PLoS One 13, e0198983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rydell G. E., et al. , Human noroviruses recognize sialyl Lewis x neoglycoprotein. Glycobiology 19, 309–320 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Rydell G. E., Dahlin A. B., Höök F., Larson G., QCM-D studies of human norovirus VLPs binding to glycosphingolipids in supported lipid bilayers reveal strain-specific characteristics. Glycobiology 19, 1176–1184 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Bally M., et al. , Norovirus GII.4 virus-like particles recognize galactosylceramides in domains of planar supported lipid bilayers. Angew. Chem. Int. Ed. Engl. 51, 12020–12024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bally M., et al. , Interaction of single viruslike particles with vesicles containing glycosphingolipids. Phys. Rev. Lett. 107, 188103 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Nasir W., et al. , Histo-blood group antigen presentation is critical for binding of norovirus VLP to glycosphingolipids in model membranes. ACS Chem. Biol. 12, 1288–1296 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Conley M. J., et al. , Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 565, 377–381 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Contreras F. X., Sánchez-Magraner L., Alonso A., Goñi F. M., Transbilayer (flip-flop) lipid motion and lipid scrambling in membranes. FEBS Lett. 584, 1779–1786 (2010). [DOI] [PubMed] [Google Scholar]
- 91.Santiana M., et al. , Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 24, 208–220.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato J., et al. , Effects of disinfectants against norovirus virus-like particles predict norovirus inactivation. Microbiol. Immunol. 60, 609–616 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Karandikar U. C., et al. , Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J. Gen. Virol. 97, 2291–2300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, including RNA-seq data, used to support the findings of this study are present in the main text and SI Appendix or available from the authors upon request.