Significance
We observed the presence of extrachromosomal circular DNA (eccDNA) in the plasma of pregnant women. We found that the plasma eccDNA molecules were longer than their linear counterparts. Among such eccDNA molecules, those of fetal origin were shorter than those of maternal origin. Characteristic dual-repeat patterns of eccDNA junctions might shed light on their possible generation mechanisms and provide them with distinctive signatures over linear cell-free DNA. Furthermore, the closed circular structure of eccDNA might allow resistance to exonucleases and thus higher stability of these molecules over their linear counterparts. These features of eccDNA provide opportunities for research and biomarker development. This work represents an example in the nascent field of plasma DNA topologics.
Keywords: eccDNA, cell-free DNA, noninvasive prenatal testing, plasma DNA topologics
Abstract
We explored the presence of extrachromosomal circular DNA (eccDNA) in the plasma of pregnant women. Through sequencing following either restriction enzyme or Tn5 transposase treatment, we identified eccDNA molecules in the plasma of pregnant women. These eccDNA molecules showed bimodal size distributions peaking at ∼202 and ∼338 bp with distinct 10-bp periodicity observed throughout the size ranges within both peaks, suggestive of their nucleosomal origin. Also, the predominance of the 338-bp peak of eccDNA indicated that eccDNA had a larger size distribution than linear DNA in human plasma. Moreover, eccDNA of fetal origin were shorter than the maternal eccDNA. Genomic annotation of the overall population of eccDNA molecules revealed a preference of these molecules to be generated from 5′-untranslated regions (5′-UTRs), exonic regions, and CpG island regions. Two sets of trinucleotide repeat motifs flanking the junctional sites of eccDNA supported multiple possible models for eccDNA generation. This work highlights the topologic analysis of plasma DNA, which is an emerging direction for circulating nucleic acid research and applications.
The fragmentation patterns of cell-free DNA (cfDNA) in human plasma is an area of intense research interest (1–4). Recent studies on the size distributions (1), end locations (5), and end motifs (3) revealed that these fragmentation patterns in cfDNA bore relationships with their tissues of origin. In pregnancy, fetal-derived plasma DNA (mainly of placental origin) were observed to be linear fragments of DNA that were shorter than the maternal-derived (mainly of hematopoietic origin) DNA (1, 5, 6). In cancer, tumor-derived cfDNA were detected with smaller sizes and preferred end coordinates from those derived from nonmalignant cells (7–9). Diagnostic applications had been demonstrated for using the fragmentation patterns of plasma DNA in noninvasive prenatal testing and cancer testing (5, 6, 10–12). However, the above-mentioned studies predominantly focused on linear DNA fragments in plasma. We have recently demonstrated that there were different topologic forms (i.e., linear as well as circular) of plasma mitochondrial DNA (mtDNA) (13). Of interest, we observed that circular mtDNA molecules were predominantly of hematopoietic origin, whereas the liver-derived ones were predominantly linear.
In this work, we explored plasma DNA molecules originated from the genome that were of other topological forms. In particular, we focused on extrachromosomal circular DNA (eccDNA) molecules in the plasma of pregnant women. This special form of DNA molecules had previously been observed across different species of organisms from yeast to mouse (14, 15). The sizes of eccDNA varied widely, ranging from dozens of bases to hundreds of thousands of bases, with the majority of them being smaller than 1,000 bp (15, 16). These eccDNA molecules were found to be enriched from genomic regions with high gene densities and GC contents (15, 17). The presence of eccDNA in human and murine plasma had also been reported (18, 19). However, there are no published data on eccDNA in the plasma of pregnant women. Our first goal in this work was therefore to investigate whether a fetus might release eccDNA into the plasma of its pregnant mother, analogous to the presence of linear fetal DNA in the plasma of pregnant women (1, 20). Second, we compared the size profiles between maternal and fetal eccDNA. Last, we explored nucleotide motif signatures flanking the eccDNA junctional sites in the hope of gaining insights into eccDNA generation mechanisms.
Results
Identification of eccDNA in Plasma by MspI Digestion.
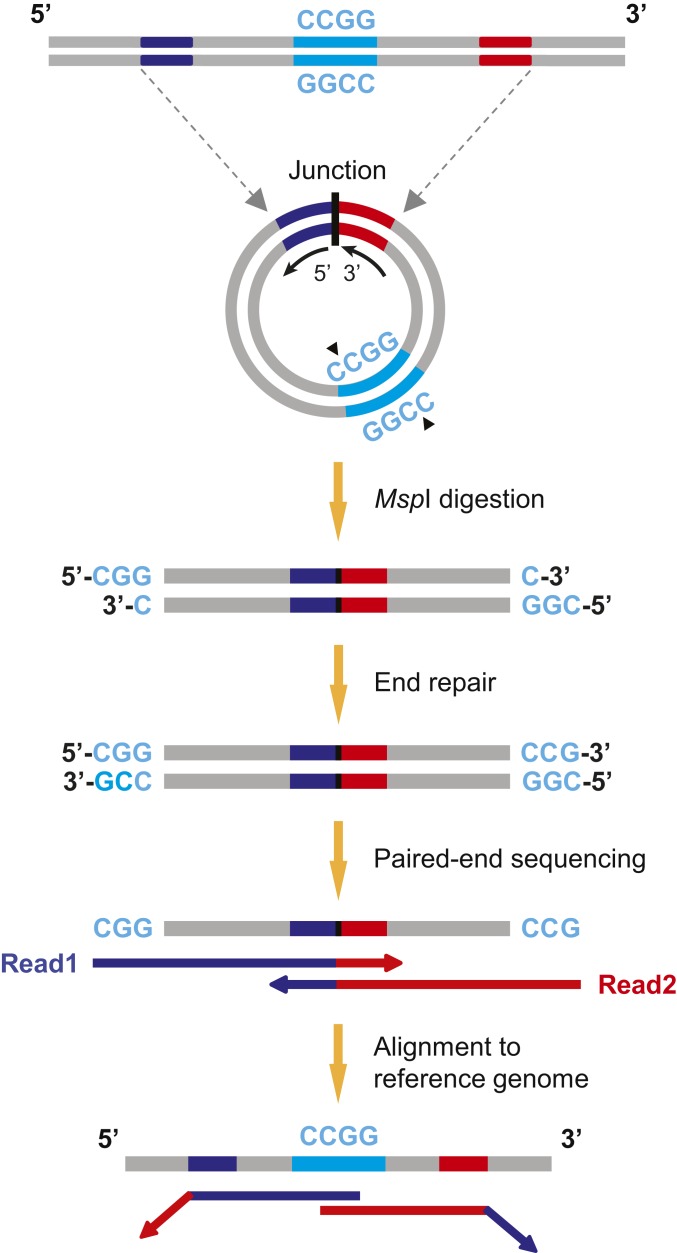
We analyzed plasma DNA samples from five cases of third-trimester pregnancy by MspI digestion. Circular DNA molecules in plasma were first enriched by exonuclease V (exo V) digestion of the background linear DNA. MspI restriction enzyme was then used to linearize the remaining circular DNA, followed by library construction and next-generation sequencing. The workflow of eccDNA detection from plasma DNA is described in Fig. 1, from which we developed bioinformatics algorithms for eccDNA identification and downstream analyses (see details in Materials and Methods). The “junction” indicated the position where two ends of a genomic sequence were ligated, forming a DNA circle.
Fig. 1.
Workflow of eccDNA identification. eccDNA generated from the genome would possess a start (blue) and an end (red) position, which were ligated to form a junctional site. eccDNA molecules in the plasma were cleaved by MspI restriction enzyme, followed by library construction procedures. Paired-end sequencing of the DNA libraries were performed on the Illumina HiSeq 1500/2500 platforms. Sequencing reads were aligned to the reference genome, and algorithms were developed to identify eccDNA. Sequencing reads meeting the four criteria of eccDNA identification (see Materials and Methods for details) were assigned as eccDNA fragment reads.
The plasma eccDNA counts of each sample was normalized as eccDNA per million mappable reads (EPM). The number of mappable reads of each sample used in this calculation was the total number of reads mapped to both chromosomal and mtDNA in that sample. We first confirmed the efficiency of exo V in enriching eccDNA molecules from plasma samples. We compared the EPM values of case 13007 with and without exo V treatment, followed by MspI digestion. We observed a 10,014-fold increase in EPM value after exo V treatment (EPM [exo V + MspI]: 6,409; EPM [MspI only]: 0.64). Thus, exo V treatment could significantly enrich eccDNA molecules. For the five pregnancy cases examined by the MspI approach (exo V + MspI), the median EPM value was 1,462 (range, 844 to 6,409). We further plotted the plasma eccDNA size distributions and compared them with their linear counterparts from the same subjects. Size profiles of these five cases showed that the linear DNA and eccDNA molecules in plasma had distinct size distributions (Fig. 2A). The linear plasma DNA showed a predominant size peak at ∼166 bp with a 10-bp periodic pattern in molecules smaller than 166 bp. Such a size distribution is in concordance with previous reports on linear plasma DNA (1, 21, 22). On the other hand, eccDNA molecules detected from MspI-treated samples showed two major peaks at 202 and 338 bp. The 338-bp peak was around 10 to 30 times more pronounced than the 202-bp peak as indicated by their areas under the curves. The predominant dinucleosomal size signature of plasma eccDNA observed here was in concordance with previous reports of plasma samples from nonpregnant subjects (15, 18, 23). Moreover, there was a distinct 10-bp periodicity throughout the size ranges within each of the peaks. Notably, such small peaks at 10-bp intervals were of almost identical sizes among different cases. The eccDNA size profiles of individual cases and the sizes of each small peak are plotted in SI Appendix, Fig. S1.
Fig. 2.
eccDNA identification by the restriction enzyme (MspI) approach. Plasma samples of five pregnancy cases were analyzed. (A) Size distributions of linear (blue) and eccDNA (red) in the plasma. (B and C) Plots of size distributions of maternal- and fetal-derived plasma eccDNA, respectively. (D) Cumulative frequency plots of maternal- (blue) and fetal-derived (red) eccDNA in plasma.
Detection and Analysis of Fetal eccDNA in Maternal Plasma.
It had previously been shown that fetal-derived linear plasma DNA molecules were generally shorter than the maternal-derived molecules (1, 24). To compare the maternal- and fetal-specific eccDNA in plasma, we classified eccDNA fragments carrying fetal-specific single-nucleotide polymorphism (SNP) alleles as fetal-derived eccDNA, and the ones that carried maternal-specific SNP alleles as maternal-derived eccDNA. Fig. 2 B and C show the size distributions of maternal- and fetal-derived eccDNA of the five pregnancy cases (MspI-treated), respectively. Both maternal and fetal eccDNA exhibited two major peaks at ∼202 and ∼338 bp, with both peaks being sharper for the fetal population. Furthermore, both maternal and fetal eccDNA plots showed a 10-bp periodicity in proximity to both peaks. Fig. 2D shows that the cumulative frequency curve of fetal-specific eccDNA was located on the left of the maternal-specific curve. Hence, fetal-derived eccDNA molecules were generally shorter than the maternal-derived ones. Such size differences between maternal- and fetal-derived eccDNA were thus consistent with those of their linear counterparts (1, 24). Also, the fetal DNA fractions deduced from eccDNA showed a positive correlation with those deduced from linear DNA from the same samples (SI Appendix, Fig. S2).
Verification of eccDNA Characteristics by Tagmentation.
The prerequisite of the need for a CCGG recognition site for MspI digestion had limited the proportion of eccDNA molecules that could be analyzed using this approach. Furthermore, this approach might theoretically be biased toward larger eccDNA molecules for their higher chances to harbor such a recognition site. To further generalize our analysis of plasma eccDNA and to remove this bias, we developed an alternative assay for eccDNA detection in plasma, which made use of Tn5 transposase-based tagmentation. The Tn5 transposase cleaves DNA sequences in a random manner (25), hence removing the limitation imposed by the previously described restriction enzyme-based approach.
We analyzed plasma DNA samples from five cases of third-trimester pregnancy with the tagmentation approach. The median EPM value detected by this approach was 9,638 (range, 5,804 to 20,812), in contrast to that detected by the MspI-based approach of 1,462 (range, 844 to 6,409). Hence, the tagmentation method detected significantly larger numbers of eccDNA molecules in maternal plasma when compared to MspI digestion (P < 0.05, Wilcoxon rank-sum test) (Fig. 3A). We hypothesize that such difference in eccDNA detection sensitivity could be attributed to the distinct DNA cleaving mechanisms between Tn5 and MspI: Tn5 cuts DNA strands in a random manner, while MspI only cuts at CCGG motifs. To test this hypothesis, we calculated the percentages of eccDNA identified by the tagmentation approach harboring CCGG motifs in the five pregnancy cases examined (SI Appendix, Table S1). On average, 19.76% of the eccDNA molecules identified by the tagmentation approach harbored at least one CCGG motif. In other words, approximately one-fifth of eccDNA identified by the tagmentation approach are potentially detectable using the MspI method.
Fig. 3.
Tagmentation-based approach for eccDNA analysis as a verification of the restriction enzyme method. Plasma DNA samples from five pregnancy cases were analyzed by tagmentation. (A) Plasma eccDNA counts detected in pregnancy cases by MspI digestion and tagmentation. P < 0.05, Wilcoxon rank-sum test. EPM, eccDNA per million mappable reads. (B) Size distributions of eccDNA detected by MspI digestion (blue) and tagmentation (red). (C and D) Plots of the size distributions of maternal- and fetal-derived eccDNA, respectively. (E) Cumulative frequency plots of maternal- (blue) and fetal-derived (red) plasma eccDNA.
Fig. 3B compares the size profiles of plasma eccDNA detected by the MspI and tagmentation approaches. Similar to the results following MspI treatment, eccDNA processed by tagmentation exhibited peaks at ∼202 and ∼338 bp with a 10-bp periodicity pattern. Furthermore, eccDNA detected by tagmentation exhibited a higher abundance of small eccDNA (202 bp) than those detected by MspI digestion, which might be explained by the removal of the possible size bias in favor of longer fragments for the restriction enzyme-based approach. However, the 338-bp peak detected by the tagmentation method was still more pronounced, with its area under the curve being approximately twice of that for the 202-bp peak. Moreover, around 95% of the eccDNA molecules identified in the five maternal plasma samples were smaller than 1,000 bp. The eccDNA size profiles for individual cases and the sizes of each peaks with 10-bp intervals are plotted in SI Appendix, Fig. S3. Again, these peak sizes were observed to be identical among all five cases examined.
Fig. 3 C and D show the size profiling of maternal- and fetal-derived eccDNA detected by the tagmentation approach. Both populations exhibited two predominant peaks with the 10-bp periodicity pattern similar to the overall eccDNA population shown in Fig. 3B. The cumulative frequencies plotted in Fig. 3E demonstrated the relatively smaller sizes of fetal eccDNA than the maternal ones. Thus, data generated by tagmentation confirmed a number of findings from the restriction enzyme method: The two major peaks with one at ∼202 bp and another more pronounced at ∼338 bp, a 10-bp periodicity in eccDNA size, and the smaller sizes of fetal eccDNA molecules compared to their maternal counterparts.
Genomic Annotation of eccDNA in Maternal Plasma.
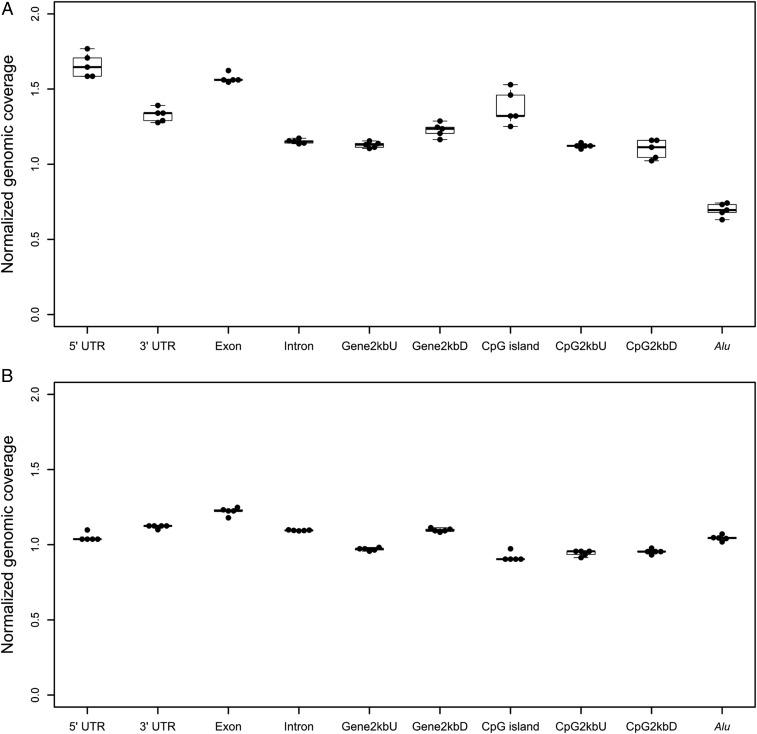
We mapped the overall population of plasma eccDNA detected in the five cases by tagmentation to different classes of genomic elements (Fig. 4A). The normalized genomic distribution of eccDNA in each class of genomic element was termed “normalized genomic coverage,” which was calculated as the percentage of eccDNA mapped to that class of genomic element divided by the percentage of the genome covered by that class of element (see details in Materials and Methods). The eccDNA molecules in maternal plasma were enriched in 5′-untranslated regions (5′-UTRs), exonic regions, and CpG island regions, with relatively low distributions in the Alu repeat regions. Such distribution patterns were in line with those of eccDNA observed from mouse tissues and plasma of nonpregnant human subjects (15, 19). On the other hand, the genomic distributions of linear plasma DNA in different regions were fairly even (Fig. 4B). Thus, the generation of plasma eccDNA from the genome was not entirely random and exhibited certain preferences. We did not observe statistically significant difference in genomic distributions between maternal and fetal eccDNA (SI Appendix, Fig. S4). We also compared the eccDNA junctional sequences among samples to search for identical eccDNA molecules. We found that for any two cases, the median percentage of identical junctional sequences among all junctions was as low as 0.22% (range, 0.17 to 0.26%). There were 322 junctional sequences (0.018%) that could be found in all five cases examined.
Fig. 4.
Genomic distributions of (A) eccDNA and (B) linear DNA in maternal plasma. Plasma DNA samples from five pregnant subjects were analyzed for each groups. Data of eccDNA were obtained from the tagmentation approach. The “normalized genomic coverage” of linear/eccDNA for each class of genomic elements is the percentage of DNA molecules mapped to that class of genomic elements divided by the percentage of the genome covered by that class of elements. CpG2kbD, 2 kb downstream of CpG islands; CpG2kbU, 2 kb upstream of CpG islands; Gene2kbD, 2 kb downstream of genes; Gene2kbU, 2 kb upstream of genes; UTR, untranslated region.
Trinucleotide Motifs Flanking eccDNA Junctions.
We explored whether there were recurrent nucleotide motif patterns flanking the eccDNA junctional sites, which might shed light on the generation mechanisms of these molecules. In the identification of eccDNA junctional sites, we pinpointed the start (the upstream edge in the genome) and end (the downstream edge in the genome) positions of each DNA segment that gave rise to an eccDNA molecule. One could reason that these start and end positions might be where the excision of these fragments from the genome occurred prior to eccDNA formation. Hence, we searched the DNA sequences from 50 bp upstream to 50 bp downstream of the start and end positions of each identified eccDNA molecules for recurrent nucleotide motif signatures flanking these positions. In this approach, the 50-bp sequences within the eccDNA molecules were obtained from sequencing results, while the sequences beyond the range of eccDNA were inferred from the reference genome. We conducted this analysis using data generated by the tagmentation approach due to the larger number of eccDNA molecules identified.
As shown in Fig. 5A, both the start and end positions of eccDNA molecules were flanked by a pair of trinucleotide segments with 4-bp “spacers” in between. This pattern was also observed in eccDNA molecules of sizes at peaks and troughs with 10-bp intervals (SI Appendix, Figs. S5 and S6). Thus, this motif signature was shared by the general eccDNA population in maternal plasma. Upon anchoring the positions of possible motifs and the awareness of their 3-bp length, we further scrutinized the sequences of these motifs. As illustrated in Fig. 5B, we termed these trinucleotide segments as I, II, III, and IV following their genomic orientations, and the spacers in between as S1 and S2 for the start and end positions, respectively. In total, we observed ∼1.2 million different sequence combinations of I, II, III, and IV in our dataset. The sequences of these four trinucleotide segments with the top 20 frequencies were listed. A recurrent pattern was observed from these sequences: motifs I and III were identical (direct repeat 1 or DR1), whereas motifs II and IV were identical (direct repeat 2 or DR2). The percentages of motifs of the top 2,000 frequencies having such trinucleotide repeat signature are listed in SI Appendix, Table S2. To explore whether such dual-direct-repeat pattern is overrepresented in eccDNA, we compared the actual frequency and expected frequency of this pattern. To obtain the expected frequency (assuming random occurrence of eccDNA from the genome) of this pattern, we performed a computer simulation by assigning random positions to the reference genome for each eccDNA lengths obtained from our sequencing data (details in Materials and Methods). Such computer simulation was performed 10 times. The average frequency of this dual-repeat pattern in such simulations was 0.056% (expected frequency). The observed frequency in our sequencing libraries was 3.484%. Thus, the odds ratio of this dual-repeat pattern observed in the actual experiment was 63 times compared to the simulation results. This high degree of overrepresentation suggests that the dual-direct-repeat pattern might be a key factor in the generation of eccDNA.
Fig. 5.
Identification of junctional nucleotide motif patterns. (A) Trinucleotide motif sequences flanking the start and end positions of eccDNA molecules with 4-bp spacers in between. (B) The four trinucleotide segments flanking the start and end positions of eccDNA were labeled as I, II, III, and IV. The 4-bp spacers were termed S1 and S2. Trinucleotide motif sequences of I, II, III, and IV flanking eccDNA junctions with the top 20 frequencies were listed.
Our observation of the dual-direct-repeat pattern is reminiscent of the previously reported 2- to 15-bp direct repeats near eccDNA junctions (15, 18, 26). Such microhomology of eccDNA junctions might favor homologous recombination (SI Appendix, Fig. S7A) and microhomology-mediated end joining (SI Appendix, Fig. S7B) as mechanisms facilitating the circularization of double-stranded DNA (17, 26, 27). Indeed, such models have previously been proposed as possible models for eccDNA generation. However, given the unexpectedly high representation of the dual-repeat signature among eccDNA particularly observed in this study, we pondered whether yet other mechanisms might exist, namely through ssDNA looping and circularization (SI Appendix, Fig. S7C). In a double-strand genomic DNA molecule, the nicking and fraying of one of the strands might create a single-stranded DNA loop if DR1 of the frayed strand is reannealed with the DR1 sequence at the other position of the complementary strand. Once this looped DNA segment is cleaved from the genome and self-ligated, a single-stranded eccDNA molecule would be formed. The intracellular DNA replication machineries might further complete this single-stranded molecule into a double-stranded eccDNA. However, since restriction enzyme digestion or tagmentation operates selectively on double-stranded DNA, we may be underestimating the single-stranded eccDNA in these samples.
Discussion
We have demonstrated the presence of fetal eccDNA in the plasma of pregnant women. Both maternal and fetal eccDNA showed two predominant size peaks at 202 and 338 bp, with both major peaks and small peaks with 10-bp intervals sharper for the fetal population. Such a phenomenon is reminiscent of our previous observation in linear plasma DNA that the fetal-specific DNA demonstrated a reduction in sizes and sharper 10-bp periodic peaks from the overall population of plasma DNA (1). Previous reports demonstrated that cfDNA fragmentation patterns were correlated with nucleosome positioning and cell type-specific gene expression profiles (28–30). We thus proposed that such difference in overall sizes and the 10-bp periodicity sharpness are closely linked to cell type origins (1). For linear cfDNA, the fetal population are predominantly of placental origin. The maternal DNA, on the other hand, are of more heterogenetic origins (hematopoietic, epithelial, hepatic, etc.) (31). Such heterogeneity in cell type origins might shift the overall sizes and blunt the 10-bp periodic peaks of maternal cfDNA. Also, Kumar et al. (18) reported that for lung cancer and ovarian cancer patients, eccDNA detected from presurgery plasma were of larger sizes than those from postsurgery plasma samples. Thus, we hypothesize that the differences in sizes and 10-bp periodicity sharpness between maternal and fetal eccDNA might be explained in a similar fashion as their linear counterparts assuming that the placenta is also the main source of fetal eccDNA.
The signature 166-bp peak of linear plasma DNA reflects a nucleosomal origin of plasma DNA with a core region (∼146 bp) and a linker (∼20 bp) generated from nuclease cleavage during apoptosis (1, 7, 22). On the other hand, the modal sizes of 202 and 338 bp of eccDNA may also be related to the nucleosomal origins of these molecules, with the predominance of the dinucleosomal feature and the sustaining 10-bp periodicity among different sizes of plasma eccDNA molecules (Figs. 2B and 4B) being in contrast to those of linear cfDNA (1, 21). It had been speculated that eccDNA could be formed from DNA sequences wrapped around nucleosomes (15). The modal peaks of 202 bp might represent DNA lengths of approximately one nucleosome core plus two linkers, while that of 338 bp might be of approximately two nucleosome cores plus two linkers. In this light, the shorter the fragment, the higher torsional stiffness it might need to overcome to bring the two ends close enough for ligation (32–34). It has been reported that the torsional stiffness of DNA renders the bending of molecules smaller than 300 bp inefficient (33). On the other hand, the longer the fragment, the lower is the chance for the two fragment ends to be in proximity close enough for ligation. Hence the higher proportion of eccDNA molecules with di-nucleosomal structures (the 338-bp peak) over the mononucleosomal eccDNA (the 202-bp peak), which might represent a “Goldilocks” position between the two above-mentioned factors (Fig. 3B).
Several mechanistic models for eccDNA generation had been proposed previously (17, 27). Some of these propositions were inspired by the observation of microhomology of eccDNA molecules with 2- to 15-bp direct repeats flanking the junctional region (15, 18, 26), pointing to possible homologous recombination-mediated circularizations (SI Appendix, Fig. S7A) or microhomology-mediated end joining of double-stranded DNA (SI Appendix, Fig. S7B) during eccDNA formation (23, 26, 27). DNA replication and transcription processes were also proposed as potential single-stranded DNA-based eccDNA generation mechanisms that might possibly involve replication slippage or R-loop formation (15, 17, 35). Thus, both single- and double-stranded DNA circularization could potentially promote eccDNA generation. These possible models might occur intracellularly as DNA replication, transcription, and repair would require intracellular machineries. In this study, we observed dual direct repeats (DR1 and DR2) flanking the start and end positions of eccDNA molecules (Fig. 5), whose features add to the ones reported previously of single direct-repeat motifs (15, 18, 26). This dual repeat signature might reflect a possible single-strand origin of eccDNA generation mediated by a “lost-and-found” event between two DNA strands, where the nicked single strand dissociated from the original double helix might form a loop by binding to the other direct repeat nearby (SI Appendix, Fig. S7C). Interestingly, in a previous study looking into lymphoblastoid cells treated with chemotherapeutics, cell apoptosis significantly promoted eccDNA production. These data suggested that DNA fragmentation during caspase-driven cell apoptosis might be another important contributor to eccDNA generation (23). Thus, various mechanisms might contribute to the production of eccDNA.
In conclusion, we demonstrated the presence of fetal eccDNA in the plasma of pregnant women and investigated the sizes and explored possible production mechanisms of these molecules. With the closed circular structure, eccDNA molecules might exhibit resistance to exonuclease digestions. Hence, eccDNA might demonstrate higher stability than their linear counterparts. The dual-direct-repeat patterns of eccDNA might also provide distinct signatures of these molecules over linear DNA. It would be interesting for future studies to explore the potential aberrations of maternal plasma eccDNA profiles in different pregnancy-associated disorders, such as preeclampsia and preterm birth. eccDNA in maternal plasma with their increased biostability and distinct molecular signatures might add to the toolbox of the rapidly developing field of noninvasive prenatal testing.
Materials and Methods
Case Recruitment and Sample Processing.
This study was approved by the Joint Chinese University of Hong Kong–Hospital Authority New Territories East Cluster Clinical Research Ethics Committee. Pregnant women attending the antenatal clinic at the Department of Obstetrics and Gynecology, Prince of Wales Hospital, Hong Kong, China, as well as nonpregnant female subjects, were recruited with written informed consent. Peripheral blood samples were collected and centrifuged at 1,600 × g for 10 min at 4 °C. The plasma portion was further centrifuged at 16,000 × g for 10 min at 4 °C to remove residual cells and debris. The buffy coat portion was centrifuged at 5,000 × g for 5 min at room temperature to remove residual plasma. Placental tissues were collected immediately after delivery. Plasma DNA extractions were performed using QIAamp Circulating Nucleic Acid Kit (Qiagen). Buffy coat and placental tissue DNA extractions were performed using QIAamp DNA Mini Kit (Qiagen).
Plasma DNA Library Preparation and Sequencing.
For elimination of linear DNA and enrichment of eccDNA, 25 ng of plasma DNA were treated with 5 units of exonuclease V (New England Biolabs) in a 50-µL reaction system at 37 °C for 5 min, followed by column purification using MinElute Reaction Cleanup Kit (Qiagen). For restriction enzyme-based approach, the resultant circular DNA were digested with MspI (New England Biolabs) according to manufacturer’s instructions. The sequencing libraries of the MspI-digested DNA and linear DNA (30 ng of plasma DNA from each case) were prepared using TruSeq Nano DNA LT Library Prep Kit (Illumina). For tagmentation-based approach, circular DNA enriched from 25 ng of plasma DNA were processed using Nextera XT DNA Library Preparation Kit (Illumina). DNA libraries were sequenced on HiSeq 1500/2500 platforms in Rapid Run mode. All libraries were sequenced as 2 × 250-bp paired-end reads.
Identification and Size Profiling of Plasma eccDNA.
The bioinformatically truncated read1 and read2 consisting of the first 50 bp were used for alignment to a human reference genome using Bowtie 2 (36) in a paired-end mode. For those reads that can be aligned to human genome, if read1 and read2 from the same fragment align to the reference genome in circular DNA-specific mapping directions (as illustrated in Fig. 1), the corresponding reads before truncation were realigned to the reference genome. The paired-end reads with at least one unmappable read in its full length would be used for downstream processing because the unmappability of a read might suggest a junctional site in that fragment. We developed four bioinformatics criteria as listed below for identifying eccDNA fragments:
-
1)
Two reads in a pair showing outward orientations when mapped to the reference genome;
-
2)
CGG sequences at both ends as the result of restriction enzyme cleavage;
-
3)
A 2-base overlap between the two ends;
-
4)
A junctional site detected in one or both of the paired-end reads ligating two sequences of distance from the genome.
To locate an eccDNA in the reference genome, we fine-tuned the realignment for potential eccDNA reads. The first and the last 20 bp from such reads were used as probes (termed probe A and probe B, respectively) to screen for candidate genomic regions forming the possible junctional sequences. In this step, each probe was allowed multiple (≤10) hits in order to maximize the sensitivity for junction detection. If the probe B sequence was not mapped to the downstream of probe A in the reference genome, it would suggest a junctional sequence covered by this read. Next, we established a searching approach to probe the junctional sequences in a single-base resolution. This searching approach was conducted in a “splitting and matching” manner. We used “splitting sites” to divide the original read1 sequence into two parts (part A and part B) and slid these sites along the whole read except for the seed regions to exhaust all combinations of part A and part B, with minimum length of part A and part B no shorter than 18 bp. When the splitting site did not match the actual junction, part A or part B would exhibit certain degrees of unmappability. Once the splitting site exactly matches the actual junction, part A and part B would theoretically be perfectly alignable. Therefore, the splitting site giving rise to the highest mappability of both part A and part B was identified as a junctional site (maximum 2-bp mismatches were allowed). As a result of identifying the junctional site, the exact start and end positions in the genome that were ligated into a junction of the eccDNA would be revealed. The eccDNA sizes were then obtained from the genomic coordinates of the start and end positions. For those fragments with eccDNA-specific outward mapping directions but no junction detected, one cannot exclude the possibility that these are eccDNA fragments as the paired-end 250-bp sequencing length might not be long enough to go through the junctional sequences of all eccDNA. However, in this study, we only focused on sequencing reads with junctional sequences detected in order to avoid false-positive identification of eccDNA and to precisely determine the eccDNA lengths. Having established eccDNA identification algorithms for the MspI approach, we further categorized fragments detected with eccDNA-specific outward mapping directions and junctional sequences as eccDNA reads in the tagmentation approach.
Analysis of Maternal- and Fetal-Derived eccDNA in Maternal Plasma.
Genotyping of genomic DNA from buffy coat (maternal genome) and placenta (fetal genome) were performed with Illumina iScan system using Infinium Omni2.5Exome-8 v1.4 BeadChip. SNPs that were heterozygous in fetal genome and homozygous in maternal genome, or homozygous in fetal genome and heterozygous in maternal genome were defined as informative SNPs. For an allele containing an informative SNP, if the SNP is fetal specific, this allele would be defined as fetal-specific allele. The other allele would be defined as a shared allele. The fetal portions of eccDNA were calculated using the following formula:
where q was the frequency of the fetal-specific alleles, and p was the frequency of shared alleles.
On the other hand, if an allele is carrying a maternal-specific SNP, that allele would be defined as a maternal-specific allele. The frequencies of maternal- and fetal-specific eccDNA of difference sizes were thus determined.
Genomic Annotation of Plasma DNA.
Upon locating the overall populations of linear and eccDNA from plasma in the human genome, the amounts of these DNA molecules with the start positions mapped to the 10 classes of genomic elements were obtained. The theoretical distribution of plasma DNA (linear and eccDNA) in each class of element was predicted as the percentage of genome covered by that class of genomic elements. The normalized genomic coverage of plasma DNA in each class of genomic element was then calculated using the following formula:
Junctional Motifs of eccDNA.
To explore the motif patterns flanking the eccDNA junctions, we scanned the base compositions from 50 bp upstream to 50 bp downstream of the start and end positions for each eccDNA locus. The trinucleotide motifs within the eccDNA molecules were obtained from sequencing results, while the trinucleotide motifs outside of the eccDNA molecules were inferred from the reference genome. The expected frequency of each trinucleotide motif was generated by computer simulation: random new positions in the genome were assigned for the lengths of each eccDNA molecules using BedTools (37). The trinucleotide motif sequences (I, II, III, and IV) flanking the junctions for each randomly assigned eccDNA positions were subsequently inferred from the reference genome. Such simulation for all of the 1.7 million eccDNA molecules identified from the five pregnancy cases had been performed and repeated 10 times. The average frequencies of the dual-direct-repeat pattern observed in the 10 times of simulations was applied as expected frequencies.
Statistical Analysis.
Wilcoxon rank-sum test was applied to compare data from two groups using GraphPad Prism 8.0 (GraphPad Software). Statistical significance was defined as P < 0.05. All probabilities were two-tailed.
Data Availability.
Sequence data for the subjects studied in this work who had consented to data archiving have been deposited at the European Genome–Phenome Archive (EGA) (https://ega-archive.org/datasets/EGAD00001005286) hosted by the European Bioinformatics Institute (EBI) (accession no. EGAS00001003827).
Supplementary Material
Acknowledgments
This work was supported by the Hong Kong Research Grants Council Theme-Based Research Scheme T12-403/15-N. Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation.
Footnotes
Competing interest statement: S.T.K.S., P.J., J.D., L.J., K.C.A.C., R.W.K.C. and Y.M.D.L. have filed a patent application based on the data generated from this work.
Data deposition: Sequence data for the subjects studied in this work who had consented to data archiving have been deposited at the European Genome–Phenome Archive (EGA) (https://ega-archive.org/datasets/EGAD00001005286) hosted by the European Bioinformatics Institute (EBI) (accession no. EGAS00001003827).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914949117/-/DCSupplemental.
References
- 1.Lo Y. M. D., et al. , Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Snyder M. W., Kircher M., Hill A. J., Daza R. M., Shendure J., Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serpas L., et al. , Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc. Natl. Acad. Sci. U.S.A. 116, 641–649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristiano S., et al. , Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun K., et al. , Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing. Proc. Natl. Acad. Sci. U.S.A. 115, E5106–E5114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu S. C. Y., et al. , Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc. Natl. Acad. Sci. U.S.A. 111, 8583–8588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang P., Lo Y. M. D., The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 32, 360–371 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Jiang P., et al. , Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. U.S.A. 112, E1317–E1325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang P., et al. , Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 115, E10925–E10933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straver R., Oudejans C. B. M., Sistermans E. A., Reinders M. J. T., Calculating the fetal fraction for noninvasive prenatal testing based on genome-wide nucleosome profiles. Prenat. Diagn. 36, 614–621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan K. C. A., Leung S.-F., Yeung S.-W., Chan A. T. C., Lo Y. M. D., Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clin. Cancer Res. 14, 4141–4145 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Arko-Boham B., et al. , Circulating cell-free DNA integrity as a diagnostic and prognostic marker for breast and prostate cancers. Cancer Genet. 235-236, 65–71 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Ma M.-J. L., et al. , Topologic analysis of plasma mitochondrial DNA reveals the coexistence of both linear and circular molecules. Clin. Chem. 65, 1161–1170 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Gaubatz J. W., Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res. 237, 271–292 (1990). [DOI] [PubMed] [Google Scholar]
- 15.Shibata Y., et al. , Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336, 82–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Møller H. D., et al. , Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 9, 1069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dillon L. W., et al. , Production of extrachromosomal microDNAs is linked to mismatch repair pathways and transcriptional activity. Cell Rep. 11, 1749–1759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P., et al. , Normal and cancerous tissues release extrachromosomal circular DNA (eccDNA) into the circulation. Mol. Cancer Res. 15, 1197–1205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J., et al. , Molecular characterization of cell-free eccDNAs in human plasma. Sci. Rep. 7, 10968 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo Y. M. D., et al. , Presence of fetal DNA in maternal plasma and serum. Lancet 350, 485–487 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Zheng Y. W. L., et al. , Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: A transplantation model. Clin. Chem. 58, 549–558 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Yu S. C. Y., et al. , High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin. Chem. 59, 1228–1237 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Mehanna P., et al. , Characterization of the microDNA through the response to chemotherapeutics in lymphoblastoid cell lines. PLoS One 12, e0184365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan H. C., Blumenfeld Y. J., Chitkara U., Hudgins L., Quake S. R., Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. U.S.A. 105, 16266–16271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picelli S., et al. , Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Møller H. D., Parsons L., Jørgensen T. S., Botstein D., Regenberg B., Extrachromosomal circular DNA is common in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, E3114–E3122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen T., Kumar P., Koseoglu M. M., Dutta A., Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 34, 270–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun K., et al. , Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 29, 418–427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struhl K., Segal E., Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20, 267–273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov M., Baranova A., Butler T., Spellman P., Mileyko V., Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics 16 (suppl. 13), S1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss J., et al. , Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills J. B., Hagerman P. J., Origin of the intrinsic rigidity of DNA. Nucleic Acids Res. 32, 4055–4059 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagerman P. J., Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 17, 265–286 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Thibault T., et al. , Production of DNA minicircles less than 250 base pairs through a novel concentrated DNA circularization assay enabling minicircle design with NF-κB inhibition activity. Nucleic Acids Res. 45, e26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skourti-Stathaki K., Proudfoot N. J., A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384–1396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data for the subjects studied in this work who had consented to data archiving have been deposited at the European Genome–Phenome Archive (EGA) (https://ega-archive.org/datasets/EGAD00001005286) hosted by the European Bioinformatics Institute (EBI) (accession no. EGAS00001003827).