Abstract
Selenoprotein P (encoded by SELENOP) contains the essential trace element selenium in the form of selenocysteine, which is an analog of cysteine that contains selenium instead of sulfur. Selenoprotein P is a major selenium-containing protein in human plasma and is mainly synthesized in the liver. It functions as a selenium-transporter to maintain antioxidative selenoenzymes in several tissues, such as the brain and testis, and plays a pivotal role in selenium-metabolism and antioxidative defense. A decrease of selenoprotein P and selenoproteins causes various dysfunctions related to oxidative stress. On the other hand, recent studies indicate that excess selenoprotein P exacerbates glucose metabolism and promotes type 2 diabetes. This review focuses on the biological functions of selenoprotein P, particularly its role in selenium-metabolism and antioxidative defense. Furthermore, the effects of excess selenoprotein P on glucose metabolism, and resulting diseases are described. The development of a therapeutic agent that targets excess selenoprotein P is discussed.
Keywords: selenoprotein P, oxidative stress, lipid peroxidation, insulin resistance, insulin secretion, neutralizing antibody
Introduction
Selenoprotein P (SeP, encoded by SELENOP) is a major selenium (Se)-containing protein in human plasma, and the “P” denotes its presence in plasma.(1–3) SeP is synthesized mainly in the liver and secreted to extracellular fluid. SeP contains the essential trace element Se in the form of selenocysteine (Sec), which is an analog of cysteine that contains Se instead of sulfur.(4–6) Twenty-five kinds of selenoproteins have been discovered in humans, including five types of glutathione peroxidases (GPxs) and three types of thioredoxin reductases (TrxRs), which play a pivotal role in antioxidative defense.(7–9) SeP functions as a Se-transporter to maintain antioxidative selenoenzymes in several tissues, such as the brain and testis, and plays a crucial role in the metabolism of Se and antioxidative defense.(10–12) A decrease of SeP and selenoproteins causes various dysfunction related to oxidative stress;(13–16) however, recent studies indicate that excess SeP exacerbates glucose metabolism and promotes type 2 diabetes.(17–19) This review focuses on the biological functions of SeP, particularly its role in Se-metabolism and antioxidative defense. Furthermore, disorders induced by excess SeP that affect the glucose metabolism are summarized, and the development of a therapeutic agent that targets excess SeP is discussed here.
Structure and Function of SeP
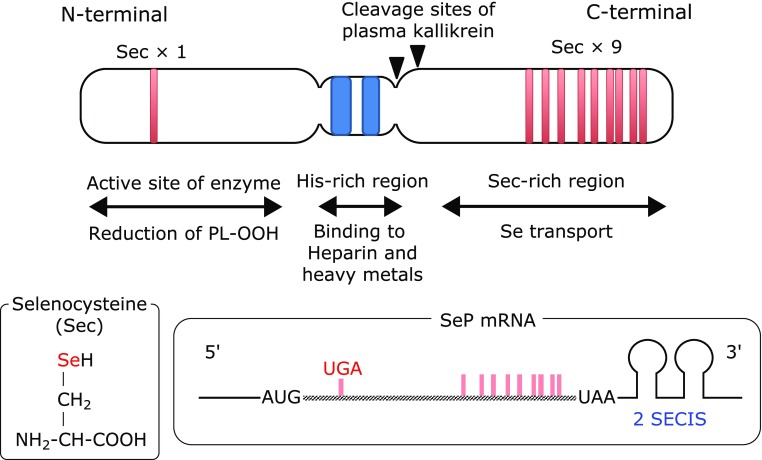
SeP contains ten Sec residues per polypeptide, which are encoded by the UGA stop codon (Fig. 1). SeP is a unique selenoprotein with multiple Sec residues, while other selenoproteins only have one or two Sec residues.(20,21) The distribution of Sec residues in SeP is biased; one Sec is located in the N-terminal region, and the other nine are located in the C-terminal region (Fig. 1). SeP is a multifunctional protein, possessing GPx-like enzyme activity to reduce phospholipid hydroperoxide in the presence of glutathione and Se-transport activity to effectively supply Se to cells.(22–25) Human SeP is a substrate of the serine protease plasma kallikrein, which generates N- and C-terminal fragments.(11) Based on the biological function of these fragments, domain structure of SeP is proposed;(11) Sec in the N-terminal region is the active site of the enzyme that reduces phospholipid hydroperoxide, while the Sec-rich C-terminal region functions as a Se-transporter (Fig. 1). A His-rich region that contains consecutive His residues is located in the middle of SeP. SeP possesses heparin-binding properties, and a typical heparin-binding motif XBBXB (B: a basic amino acid), is found in the His-rich region.(26–28) SeP binds heavy metals such as copper and cadmium, and its consecutive His residues function as a natural, high-affinity His-tag binding site for nickel-nitrilotriacetic acid (Ni-NTA) agarose.(29–31) SeP is also a major methylmercury-binding protein in plasma, suggesting that it plays a role in the detoxication of heavy metals.(32–34)
Fig. 1.
Domain structure of human selenoprotein P. SECIS, Sec insertion sequence.
Significant Role of SeP in Se-metabolism in vivo
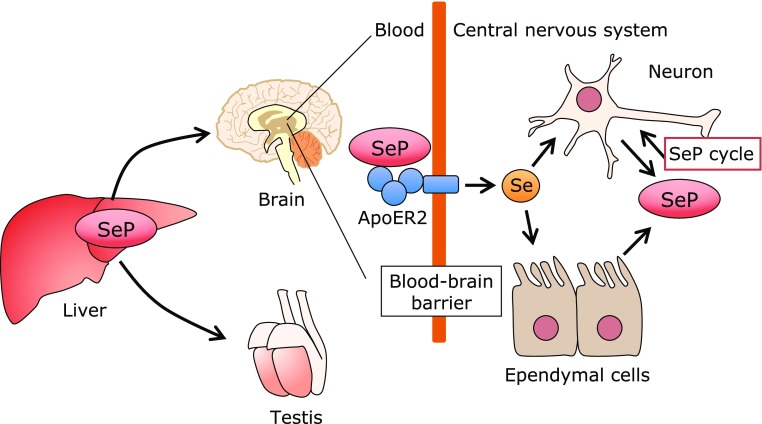
Data regarding the significant role of SeP in Se-metabolism in vivo have been obtained from SeP knockout (KO) mice.(35–39) SeP KO mice can survive on a normal chow diet containing 0.4 mg Se/kg, which contains enough Se to maintain maximum selenoprotein levels in wild type (WT) mice.(40) However, SeP KO mice cannot survive on a Se-deficient diet, indicating the significant role of SeP as a Se-transporter when Se supply is limited. On the other hand, when SeP KO mice are on a normal chow diet with enough Se, Se levels are noticeably low in their brain and testis, which demonstrates the essential role of SeP in delivering Se to these tissues (Fig. 2). One obvious phenotype of SeP KO mice is the failure of spermatogenesis, which cannot be reversed by Se supplementation. Another phenotype is neurological dysfunction when the supply of Se is limited (<0.1 mg Se/kg diet, which satisfies the Se requirement of WT mice); SeP KO mice develop progressive spasticity that requires euthanasia.(41–43) This neurological dysfunction in SeP KO mice is prevented by a chow diet with more than 0.25 mg Se/kg. Three kinds of SeP receptors have been identified, namely apolipoprotein E receptor 2 (ApoER2)/low-density lipoprotein receptor-related protein 8 (LRP8), megalin, and LRP1, which belong to the low-density lipoprotein receptor family.(44–46) Similar phenotypes have been reported in ApoER2 KO mice, suggesting the importance of receptor-mediated uptake of Se in the brain and testis.(47) Similar phenotypes, but with a later onset or less severe implications, in the brain and testis have been reported in Sepp1Δ240–361 mice, in which the Sec-rich C-terminal domain of SeP had been deleted.(41) The interaction between the C-terminal domain of SeP and the YWTD β-propeller domain of ApoER2 has been reported, and the importance of this interaction, particularly in maintaining Se levels in the brain and testis, has been manifested in the phenotypes of these KO mice (Fig. 2).(48) On the other hand, on the normal diet that contains enough Se, Se levels in other tissues of SeP KO mice, expect for the brain and testis, do not differ greatly from those in WT mice, and, in line with these Se levels, other obvious phenotypes do not appear in these KO mice. These findings suggest that SeP-dependent and SeP-independent Se-transport systems exist in vivo. Approximately half of the Se in human plasma is derived from SeP, while extracellular GPx (GPx3) contains 20% of Se.(10) The remaining Se might be present as low molecular weight Se and selenomethionine, which are mainly associated with and/or contained by albumin. In fact, GPx3, selenomethionine, and albumin function as a source of Se for the synthesis of cellular selenoproteins.(10)
Fig. 2.
Selenium transport mechanism via selenoprotein P. Selenoprotein P (SeP) synthesized in the liver is preferentially incorporated by the brain and testis via SeP-receptor ApoER2/LRP8. In the brain, several cells such as neurons and ependymal cells synthesize SeP, which contributes to maintain Se and selenoprotein levels in the brain.
The organ in which SeP is mostly synthesized is the liver, as described above, but SeP-expressing cells are found in several other tissues. Based on blood SeP concentrations of liver-specific SeP KO mice, it is estimated that 60% of SeP is derived from the liver and the remaining 40% from other tissues.(49,50) SeP expression in the brain is important for maintaining Se and selenoprotein levels, and Se concentrations in the brain are preserved in liver specific-SeP KO mice (Fig. 2). SeP expression has been reported in several cell types of the brain, including neurons, and ependymal cells that are responsible for cerebrospinal fluid production.(51,52) Presumably, SeP synthesized in the brain is incorporated to other brain cells and used to synthesize selenoproteins, which help to maintain Se concentrations in the brain (Fig. 2). This system is called the SeP cycle, and it retains selenoproteins in several tissues and cells.
Increase of SeP Levels under High Glucose and High Fat Conditions
Se is an essential trace element, and its deficiency causes numerous dysfunctions due to the decrease of selenoproteins.(53–55) However, recent evidence shows an increase of SeP in type 2 diabetes patients, and excess SeP levels have undesirable effects on the glucose metabolism.(17–19,46,56–58) This indicates that increased SeP levels are a significant therapeutic target for this lifestyle-related disease. The increase of SeP in diabetic patients has been discovered by comprehensive gene expression analysis of mRNA samples obtained from liver biopsies of diabetic patients.(17) A significantly positive correlation between SeP expression levels and biomarkers of diabetes, such as fasting glucose and blood glucose levels after administrations of a glucose tolerance test, has been shown.(17,57–60) To increase SeP levels, enough Se intake is necessary; however, this might not be sufficient. Normal mice chow contains 0.4 mg Se/kg, which causes neither an increase in SeP levels nor high blood glucose levels in normal mice. In contrast, high fat, high sucrose chows containing 0.2 mg Se/kg induce diabetes in mice and increase SeP levels.(17,18) Therefore, it is assumed that SeP levels increase when both high energy food sources and enough Se are available, which allows SeP mRNA levels to increase and to synthesize SeP proteins (Fig. 3). Accordingly, limiting the Se intake may be effective in diabetic patients with high SeP levels; however, this might be difficult, because numerous foods contain trace amounts of Se. In a large-scale clinical trial in humans, it has been reported that the prolonged intake of Se supplements (200 µg/day) increases the risk of type 2 diabetes; however, it is noteworthy that this observation is limited to a population that has high Se baseline blood levels.(56)
Fig. 3.
Selenium metabolism in the liver. Diet-derived Se is used for Sec synthesis or excreted following methylation/glycosylation. Se in the Sec synthesis pathway is used for the synthesis of liver selenoproteins or SeP, which enter the systemic circulation. In addition, small Se-containing compounds bound to albumin and/or selenomethionine also enter the systemic circulation. High glucose and high lipid diets increase SeP mRNA and enhance the synthesis of SeP when enough Sec-tRNA[ser]sec is available.
Metformin is a widely used drug to treat diabetes and lowers SeP expression in the liver.(61–63) Metformin activates AMP-activated kinase (AMPK), and activated AMPK subsequently inactivates the transcription factor FoxO3a and decreases SeP mRNA levels. In the promotor region of SeP, FoxO-binding sites have been identified, and the AMPK-FoxO3 axis is an important pathway that links energy metabolism and SeP expression. In addition, eicosapentaenoic acid (EPA), a major component of ω-3 polyunsaturated fatty acids (PUFAs) contained in fish oil, has been reported to decrease SeP expression in H4IIEC3 hepatocytes.(64–66) EPA is used to improve lipid metabolism and has beneficial effects in the treatment of type 2 diabetes. EPA has been known to activate AMPK; however, EPA decreases the binding of sterol regulatory element-binding protein-1c (SREBP-1c) to the SeP promoter region and lowers SeP levels. These results suggest that SeP levels are transcriptionally regulated by FoxO3a and SREBP-1 activation, which are closely related to glucose and lipid metabolism.
Negative Effects of Increased SeP on Insulin Resistance and Insulin Secretion
Recent studies show that increased SeP in type 2 diabetic patients worsens glucose metabolism via the impairment of insulin resistance and insulin secretion.(17,18) Injection of human SeP protein at a concentration that reflects the increment of SeP concentrations in diabetic patients inhibited insulin signal transduction in normal mice and induced hyperglycemia during the glucose tolerance test.(17,18) SeP KO mice exhibited the resistance against high fat, high sucrose diet-induced hyperglycemia or insulin resistance. Excess SeP levels induced insulin resistance in primary hepatocytes and C2C12 myocytes, a model of skeletal muscles, via the inhibition of AMPK activity.(17) AMPK is activated not only by AMP but also by several factors including reactive oxygen species (ROS).(67–69) It is known that ROS generated during exercise activate AMPK, increase peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α), and exert health-promoting effects.(70–72) These positive effects of ROS generated by exercise are reduced by excess SeP, resulting in a decrease of AMPK phosphorylation and health-promoting effects. This new clinical concept is called “exercise resistance.”(46) SeP is incorporated in the skeletal muscle by LRP1, and the SeP-LRP1 axis is an important regulator of exercise resistance. Furthermore, it has been reported that increased amounts of circulating SeP levels predicted the ineffectiveness of training on the endurance capacity in humans, suggesting that the SeP-LRP1 axis is a significant target for the treatment of diseases associated with a sedentary lifestyle.(46)
It has been shown that excess SeP impairs the function of pancreatic β-cells and decreases insulin secretion.(18) When excess amounts of SeP are added to MIN6 cells, a model for pancreatic β-cells, cellular insulin levels and high glucose-induced insulin secretion decrease significantly. Excess SeP also inhibits the insulin secretion of rat primary pancreatic islets; equally excessive amounts of selenocystine showed similar inhibitory effects on insulin secretion, suggesting a relation with the Se-transport activity of SeP.(18) The impairing effects of excess SeP on pancreatic function have been observed in in vivo experiments, and the injection of purified human SeP protein resulted in the decrease of pancreatic insulin levels, area of islets, and glucose-induced insulin secretion (Fig. 4).(18) SeP injection resulted in a decrease of not only β-cells but also α-cells in the pancreas, which might be accompanied by a rearrangement of the position of these cells in the pancreatic island (Fig. 4). The rearrangement and decrease of both α- and β-cells has been observed in the animal diabetes model and in humans.(73–75) Recently, it has been shown that SeP levels are negatively correlated with the insulinogenic index, an indicator of insulin secretion,(57) suggesting that excess SeP is a considerable therapeutic target to protect pancreas function in patients with type 2 diabetes.
Fig. 4.
Pancreatic β-cell dysfunction induced by excess selenoprotein P. Increased SeP is incorporated by the pancreas, decreases the insulin levels in β cells, and reduces insulin secretion triggered by a high glucose stimulus. Immunohistochemical analysis of the pancreas of SeP- and neutralizing antibody AE2-administered mice indicated that excess SeP alters the cellular distribution of islets. The histochemical analysis is shown in the lower panel: anti-insulin Ab (green, indicative of β-cells) and anti-glucagon Ab (red, indicative of α-cells). Scale bars = 100 µm.
It is also notable that excess SeP impairs angiogenesis by inhibiting vascular endothelial growth factor (VEGF) signaling in vascular endothelial cells.(76) This is a hallmark of vascular complications in type 2 diabetes, and ROS generated by VEGF stimuli are important for the phosphorylation of VEGF receptor 2 (VEGFR2) and extracellular signal-regulated kinase 1/2 (ERK1/2) in human umbilical vein endothelial cells (HUVECs). Treatment with excess SeP inhibited VEGF-stimulated proliferation and the phosphorylation of VEGFR2 and ERK2 in HUVECs, which was significantly improved by the addition of buthionine sulphoximine (BSO), an inhibitor of glutathione synthesis.(76) Therefore, the adverse effects of increased SeP in type 2 diabetes are diverse and might be related to vascular complications.
Significance of SeP Expression in Pulmonary Arterial Hypertension
Recently, it has been described that the increased expression of SeP in pulmonary artery smooth muscle cells (PASMCs) forming lesions of pulmonary arterial hypertension (PAH).(77) PAH-PASMCs are proliferative compared with normal PASMCs, and the pulmonary artery is constricted/occluded by abnormal proliferation of PAH-PASMCs, which induces PAH with right heart failure. High expression of SeP in PAH-PASMCs has been discovered by comprehensive gene and protein expression analysis of PAH-PASMCs and control PASMCs. PASMC-specific SeP KO mice and mice treated with SeP-lowering drugs showed improvement of PAH symptoms; therefore, it is suggested that increased expression of SeP in PAH-PASMCs is a significant mediator of lesion formation in PAH.(77) The decrease of SeP expression by SeP-siRNA treatment inhibits the proliferation of PAH-PASMCs, and these effects are mediated by the SeP-receptor ApoER2.(77) This observation reminds the SeP cycle described above; however, interestingly, the proliferative effects of SeP were not explained by Se-transport activity, namely the addition of selenocystine did not reproduced this effect of SeP, and the proliferation-promoting effects were observed by the overexpression of the mutant in which all Secs were substituted with Cys.(77) Thus, the proliferative effects of increased SeP in PAH-PASMCs are considered to be mediated by autocrine and/or paracrine stimuli from the SeP-receptor ApoER2. Se-independent biological effects of SeP have been recently described in a study on PAH, and it is interesting to speculate about the possibilities to relate not only to PAH, but also other physiological and/or pathological events.
Therapeutic Strategies to Treat Increased SeP
These lines of evidence indicate that increased SeP is a significant therapeutic target for type 2 diabetes and its vascular complications. The strategy to treat excess SeP is shown in Fig. 5. Metformin and EPA decrease SeP mRNA expression.(61,64) However, these diabetes drugs are not effective against PAH-PASMC proliferation, which suggests other molecular mechanisms that increase SeP expression in PAH.(77) To inhibit the SeP expression in PAH-PASMCs, sanguinarine, a plant alkaloid, has been identified by high-throughput screening of 3,336 compounds. Sanguinarine reduced SeP expression and proliferation in PAH-PASMCs and ameliorated PAH in animal models.(77)
Fig. 5.
Therapeutic strategies that target increased SeP in related diseases.
SeP-neutralizing antibodies have been developed to inhibit the Se-transport activity of SeP in vitro and in vivo, and two SeP-neutralizing monoclonal antibodies, namely clones AE2 and AA3, have been identified.(18) The epitope of AE2 is the N-terminal side of the His-rich region, while that of AA3 is the C-terminal region, which is Sec-rich.(78) The C-terminal region of SeP has been reported to bind to the β-propeller domain of ApoER2, while detail binding sites are not fully elucidated in other receptors. Several variants of ApoER2 are reported, and their efficiency of Se-transport activity varies depending on the tissues and type of cells. The significant inhibitory effects of AE2 and heparin sulfate on SeP binding and Se-transport activity suggests the importance of interaction between His-rich region and carbohydrate moiety of the cells.(18) SeP-neutralizing antibodies suppressed the incorporation of injected SeP in the skeletal muscle and pancreas, and improved insulin resistance and insulin secretion that had been impeded by the injection of excess SeP (Fig. 5). SeP-neutralizing antibodies are effective against diabetes in model mice, such as KKAy mice and mice fed with a high fat and high sucrose diet, in which SeP levels are increased.(18) In addition to improved insulin resistance and insulin secretion, SeP-neutralizing antibodies significantly decreased total cholesterol and triglycerides in the liver of KKAy mice, indicating the diverse beneficial effects of SeP-targeting reagents. Selenoprotein I is identified as ethanolamine phosphotransferase 1 (EPT1), playing a significant role in phospholipid synthesis.(79,80) How Se and/or SeP levels regulate lipid metabolism is not fully elucidated, which makes them a potential new target for therapeutic agents.
Conclusions
This brief review focuses on the biological functions of SeP and its implications in several diseases. The change of SeP levels significantly affects the homeostasis of the whole body, and its dysregulation is related to several diseases. These findings strongly support the necessity of SeP determination at clinical sites. Furthermore, they suggest the usefulness of establishing tailor-made treatments based on SeP levels.
Abbreviations
- AMPK
AMP-activated kinase
- ApoER2
apolipoprotein E receptor 2
- BSO
buthionine sulphoximine
- EPA
eicosapentaenoic acid
- ERK
extracellular signal-regulated kinase
- GPx
glutathione peroxidase
- HUVEC
human umbilical vein endothelial cell
- LRP
low-density lipoprotein receptor-related protein
- PAH
pulmonary arterial hypertension
- PASMC
pulmonary artery smooth muscle cell
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-α
- ROS
reactive oxygen species
- Se
selenium
- Sec
selenocysteine
- SeP
selenoprotein P
- SREBP
sterol regulatory element-binding protein
- TrxR
thioredoxin reductase
- VEGF
vascular endothelial growth factor
- VEGFR2
VEGF receptor 2
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta 2009; 1790: 1441–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito Y, Takahashi K. Selenoprotein P. In: Junqiu L, Guimin L, Yang M, eds. Selenoproteins Mimics, Vol. 5. Berlin: Springer-Verlag GmBH, 2012; 77–88. [Google Scholar]
- 3.Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Ann Rev Nutr 2005; 25: 215–235. [DOI] [PubMed] [Google Scholar]
- 4.Hill KE, Lloyd RS, Yang JG, Read R, Burk RF. The cDNA for rat selenoprotein P contains 10 TGA codons in the open reading frame. J Biol Chem 1991; 266: 10050–10053. [PubMed] [Google Scholar]
- 5.Hill KE, Lloyd RS, Burk RF. Conserved nucleotide sequences in the open reading frame and 3' untranslated region of selenoprotein P mRNA. Proc Nat Acad Sci U S A 1993; 90: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S, Hill KE, Caprioli RM, Burk RF. Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that three UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J Biol Chem 2002; 277: 12749–12754. [DOI] [PubMed] [Google Scholar]
- 7.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev 2014; 94: 739–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Holmgren A. Selenoproteins. J Biol Chem 2009; 284: 723–727. [DOI] [PubMed] [Google Scholar]
- 9.Flohé L. The labour pains of biochemical selenology: the history of selenoprotein biosynthesis. Biochim Biophys Acta 2009; 1790: 1389–1403. [DOI] [PubMed] [Google Scholar]
- 10.Saito Y, Takahashi K. Characterization of selenoprotein P as a selenium supply protein. Eur J Biochem 2002; 269: 5746–5751. [DOI] [PubMed] [Google Scholar]
- 11.Saito Y, Sato N, Hirashima M, Takebe G, Nagasawa S, Takahashi K. Domain structure of bi-functional selenoprotein P. Biochem J 2004; 381 (Pt 3): 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burk RF, Hill KE. Regulation of selenium metabolism and transport. Ann Rev Nutr 2015; 35: 109–134. [DOI] [PubMed] [Google Scholar]
- 13.Saito Y, Yoshida Y, Akazawa T, Takahashi K, Niki E. Cell death caused by selenium deficiency and protective effect of antioxidants. J Biol Chem 2003; 278: 39428–39434. [DOI] [PubMed] [Google Scholar]
- 14.Hill KE, Zhou J, McMahan WJ, et al. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem 2003; 278: 13640–13646. [DOI] [PubMed] [Google Scholar]
- 15.Saito Y, Shichiri M, Hamajima T, et al. Enhancement of lipid peroxidation and its amelioration by vitamin E in a subject with mutations in the SBP2 gene. J Lipid Res 2015; 56: 2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T, Kelly VP, Motohashi H, et al. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem 2008; 283: 2021–2030. [DOI] [PubMed] [Google Scholar]
- 17.Misu H, Takamura T, Takayama H, et al. A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab 2010; 12: 483–495. [DOI] [PubMed] [Google Scholar]
- 18.Mita Y, Nakayama K, Inari S, et al. Selenoprotein P-neutralizing antibodies improve insulin secretion and glucose sensitivity in type 2 diabetes mouse models. Nat Commun 2017; 8: 1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellwege JN, Palmer ND, Ziegler JT, et al. Genetic variants in selenoprotein P plasma 1 gene (SEPP1) are associated with fasting insulin and first phase insulin response in Hispanics. Gene 2014; 534: 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito Y, Takahashi K. Selenoprotein P: its structure and functions. J Health Sci 2000; 46: 409–413. [Google Scholar]
- 21.Shetty SP, Shah R, Copeland PR. Regulation of selenocysteine incorporation into the selenium transport protein, selenoprotein P. J Biol Chem 2014; 289: 25317–25326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito Y, Hayashi T, Tanaka A, et al. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein P. J Biol Chem 1999; 274: 2866–2871. [DOI] [PubMed] [Google Scholar]
- 23.Takebe G, Yarimizu J, Saito Y, et al. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem 2002; 277: 41254–41258. [DOI] [PubMed] [Google Scholar]
- 24.Hirashima M, Naruse T, Maeda H, Nozaki C, Saito Y, Takahashi K. Identification of selenoprotein P fragments as a cell-death inhibitory factor. Biol Pharm Bull 2003; 26: 794–798. [DOI] [PubMed] [Google Scholar]
- 25.Rock C, Moos PJ. Selenoprotein P protects cells from lipid hydroperoxides generated by 15-LOX-1. Prostaglandins Leukot Essent Fatty Acids 2010; 83: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hondal RJ, Ma S, Caprioli RM, Hill KE, Burk RF. Heparin-binding histidine and lysine residues of rat selenoprotein P. J Biol Chem 2001; 276: 15823–15831. [DOI] [PubMed] [Google Scholar]
- 27.Arteel GE, Franken S, Kappler J, Sies H. Binding of selenoprotein P to heparin: characterization with surface plasmon resonance. Biol Chem 2000; 381: 265–268. [DOI] [PubMed] [Google Scholar]
- 28.Herrman JL. The properties of a rat serum protein labelled by the injection of sodium selenite. Biochim Biophys Acta 1977; 500: 61–70. [DOI] [PubMed] [Google Scholar]
- 29.Sasakura C, Suzuki KT. Biological interaction between transition metals (Ag, Cd and Hg), selenide/sulfide and selenoprotein P. J Inorg Biochem 1998; 71: 159–162. [DOI] [PubMed] [Google Scholar]
- 30.Du XB, Zheng YB, Wang Z, et al. Inhibitory act of selenoprotein P on Cu+/Cu2+-induced tau aggregation and neurotoxicity. Inorg Chem 2014; 53: 11221–11230. [DOI] [PubMed] [Google Scholar]
- 31.Tujebajeva RM, Harney JW, Berry MJ. Selenoprotein P expression, purification, and immunochemical characterization. J Biol Chem 2000; 275: 6288–6294. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Zhang W, Zhao JT, et al. Selenoprotein P as the major transporter for mercury in serum from methylmercury-poisoned rats. J Trace Elem Med Bio 2018; 50: 589–595. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki KT, Sasakura C, Yoneda S. Binding sites for the (Hg-Se) complex on selenoprotein P. Biochim Biophys Acta 1998; 1429: 102–112. [DOI] [PubMed] [Google Scholar]
- 34.Yoneda S, Suzuki KT. Equimolar Hg-Se complex binds to selenoprotein P. Biochem Biophys Res Commun 1997; 231: 7–11. [DOI] [PubMed] [Google Scholar]
- 35.Hill KE, Motley AK, Winfrey VP, Burk RF. Selenoprotein P is the major selenium transport protein in mouse milk. PloS One 2014; 9: e103486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill KE, Wu S, Motley AK, et al. Production of selenoprotein P (Sepp1) by hepatocytes is central to selenium homeostasis. J Biol Chem 2012; 287: 40414–40424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitts MW, Raman AV, Hashimoto AC, Todorovic C, Nichols RA, Berry MJ. Deletion of selenoprotein P results in impaired function of parvalbumin interneurons and alterations in fear learning and sensorimotor gating. Neuroscience 2012; 208: 58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burk RF, Hill KE, Motley AK, Austin LM, Norsworthy BK. Deletion of selenoprotein P upregulates urinary selenium excretion and depresses whole-body selenium content. Biochim Biophys Acta 2006; 1760: 1789–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schomburg L, Schweizer U, Holtmann B, Flohé L, Sendtner M, Köhrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J 2003; 370 (Pt 2): 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunde RA, Raines AM. Selenium regulation of the selenoprotein and nonselenoprotein transcriptomes in rodents. Adv Nutr 2011; 2: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill KE, Zhou J, Austin LM, et al. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J Biol Chem 2007; 282: 10972–10980. [DOI] [PubMed] [Google Scholar]
- 42.Hill KE, Zhou J, McMahan WJ, Motley AK, Burk RF. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J Nutr 2004; 134: 157–161. [DOI] [PubMed] [Google Scholar]
- 43.Byrns CN, Pitts MW, Gilman CA, Hashimoto AC, Berry MJ. Mice lacking selenoprotein P and selenocysteine lyase exhibit severe neurological dysfunction, neurodegeneration, and audiogenic seizures. J Biol Chem 2014; 289: 9662–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem 2007; 282: 12290–12297. [DOI] [PubMed] [Google Scholar]
- 45.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem 2008; 283: 6854–6860. [DOI] [PubMed] [Google Scholar]
- 46.Misu H, Takayama H, Saito Y, et al. Deficiency of the hepatokine selenoprotein P increases responsiveness to exercise in mice through upregulation of reactive oxygen species and AMP-activated protein kinase in muscle. Nat Med 2017; 23: 508–516. [DOI] [PubMed] [Google Scholar]
- 47.Burk RF, Hill KE, Olson GE, et al. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J Neurosci 2007; 27: 6207–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurokawa S, Bellinger FP, Hill KE, Burk RF, Berry MJ. Isoform-specific binding of selenoprotein P to the β-propeller domain of apolipoprotein E receptor 2 mediates selenium supply. J Biol Chem 2014; 289: 9195–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schweizer U, Streckfuss F, Pelt P, et al. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem J 2005; 386 (Pt 2): 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson DR. More roles for selenoprotein P: local selenium storage and recycling protein in the brain. Biochem J 2005; 386 (Pt 2): e5–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scharpf M, Schweizer U, Arzberger T, Roggendorf W, Schomburg L, Köhrle J. Neuronal and ependymal expression of selenoprotein P in the human brain. J Neural Transm 2007; 114: 877–884. [DOI] [PubMed] [Google Scholar]
- 52.Renko K, Werner M, Renner-Müller I, et al. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J 2008; 409: 741–749. [DOI] [PubMed] [Google Scholar]
- 53.Baum MK, Shor-Posner G, Lai S, et al. High risk of HIV-related mortality is associated with selenium deficiency. J Acquir Immune Defic Syndr Hum Retrovirol 1997; 15: 370–374. [DOI] [PubMed] [Google Scholar]
- 54.Hamajima T, Mushimoto Y, Kobayashi H, Saito Y, Onigata K. Novel compound heterozygous mutations in the SBP2 gene: characteristic clinical manifestations and the implications of GH and triiodothyronine in longitudinal bone growth and maturation. Eur J Endocrinol 2012; 166: 757–764. [DOI] [PubMed] [Google Scholar]
- 55.Saito Y, Noguchi N. 7-Hydroxycholestrol as a possible biomarker of cellular lipid peroxidation: difference between cellular and plasma lipid peroxidation. Biochem Biophys Res Commun 2014; 446: 741–744. [DOI] [PubMed] [Google Scholar]
- 56.Rayman MP. Selenium and human health. Lancet 2012; 379: 1256–1268. [DOI] [PubMed] [Google Scholar]
- 57.Oo SM, Misu H, Saito Y, et al. Serum selenoprotein P, but not selenium, predicts future hyperglycemia in a general Japanese population. Sci Rep 2018; 8: 16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohri K, Misu H, Takayama H, et al. Circulating concentrations of insulin resistance-associated hepatokines, selenoprotein P and leukocyte cell-derived chemotaxin 2, during an oral glucose tolerance test in humans. Biol Pharm Bull 2019; 42: 373–378. [DOI] [PubMed] [Google Scholar]
- 59.Saito Y, Misu H, Takayama H, et al. Comparison of human selenoprotein P determinants in serum between our original methods and commercially available kits. Biol Pharm Bull 2018; 41: 828–832. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka M, Saito Y, Misu H, et al. Development of a sol particle homogeneous immunoassay for measuring full-length selenoprotein P in human serum. J Clin Lab Anal 2016; 30: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takayama H, Misu H, Iwama H, et al. Metformin suppresses expression of the selenoprotein P gene via an AMP-activated kinase (AMPK)/FoxO3a pathway in H4IIEC3 hepatocytes. J Biol Chem 2014; 289: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang DS, Jonker JW, Kato Y, Kusuhara H, Schinkel AH, Sugiyama Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J Pharmacol Exp Ther 2002; 302: 510–515. [DOI] [PubMed] [Google Scholar]
- 63.Shu Y, Sheardown SA, Brown C, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest 2007; 117: 1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tajima-Shirasaki N, Ishii KA, Takayama H, et al. Eicosapentaenoic acid down-regulates expression of the selenoprotein P gene by inhibiting SREBP-1c protein independently of the AMP-activated protein kinase pathway in H4IIEC3 hepatocytes. J Biol Chem 2017; 292: 10791–10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carpentier YA, Portois L, Malaisse WJ. n-3 fatty acids and the metabolic syndrome. Am J Clin Nutr 2006; 83 (6 Suupl): 1499S–1504S. [DOI] [PubMed] [Google Scholar]
- 66.Arita M, Yoshida M, Hong S, et al. Resolvin E1, an endogenous lipid mediator derived from omega-3 eicosapentaenoic acid, protects against 2,4,6-trinitrobenzene sulfonic acid-induced colitis. Proc Nat Acad Sci U S A 2005; 102: 7671–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cantó C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009; 458: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cardaci S, Filomeni G, Ciriolo MR. Redox implications of AMPK-mediated signal transduction beyond energetic clues. J Cell Sci 2012; 125 (Pt 9): 2115–2125. [DOI] [PubMed] [Google Scholar]
- 69.Zadra G, Batista JL, Loda M. Dissecting the dual role of AMPK in cancer: from experimental to human studies. Mol Cancer Res 2015; 13: 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narkar VA, Downes M, Yu RT, et al. AMPK and PPARδ agonists are exercise mimetics. Cell 2008; 134: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 2008; 454: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu J, Ruas JL, Estall JL, et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1α/ATF6α complex. Cell Metabolism 2011; 13: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 2012; 150: 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kharouta M, Miller K, Kim A, et al. No mantle formation in rodent islets -- the prototype of islet revisited. Diabetes Res Clin Pract 2009; 85: 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsuchiyama N, Takamura T, Ando H, et al. Possible role of alpha-cell insulin resistance in exaggerated glucagon responses to arginine in type 2 diabetes. Diabetes Care 2007; 30: 2583–2587. [DOI] [PubMed] [Google Scholar]
- 76.Ishikura K, Misu H, Kumazaki M, et al. Selenoprotein P as a diabetes-associated hepatokine that impairs angiogenesis by inducing VEGF resistance in vascular endothelial cells. Diabetologia 2014; 57: 1968–1976. [DOI] [PubMed] [Google Scholar]
- 77.Kikuchi N, Satoh K, Kurosawa R, et al. Selenoprotein P promotes the development of pulmonary arterial hypertension. Circulation 2018; 138: 600–623. [DOI] [PubMed] [Google Scholar]
- 78.Saito Y, Watanabe Y, Saito E, Honjoh T, Takahashi K. Production and application of monoclonal antibodies to human selenoprotein P. J Health Sci 2001; 47: 346–352. [Google Scholar]
- 79.Horibata Y, Elpeleg O, Eran A, et al. EPT1 (selenoprotein I) is critical for the neural development and maintenance of plasmalogen in humans. J Lipid Res 2018; 59: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horibata Y, Hirabayashi Y. Identification and characterization of human ethanolaminephosphotransferase1. J Lipid Res 2007; 48: 503–508. [DOI] [PubMed] [Google Scholar]